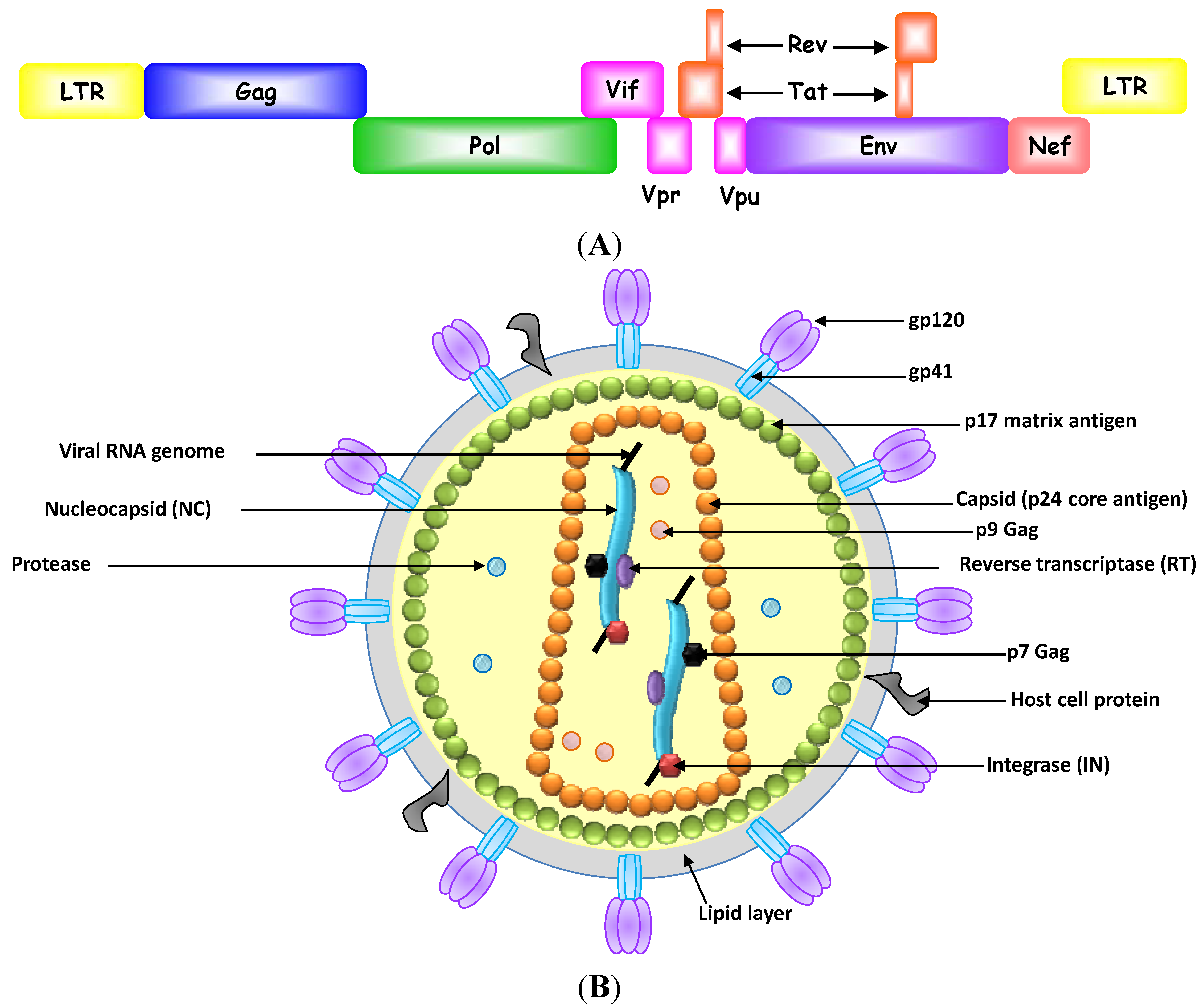
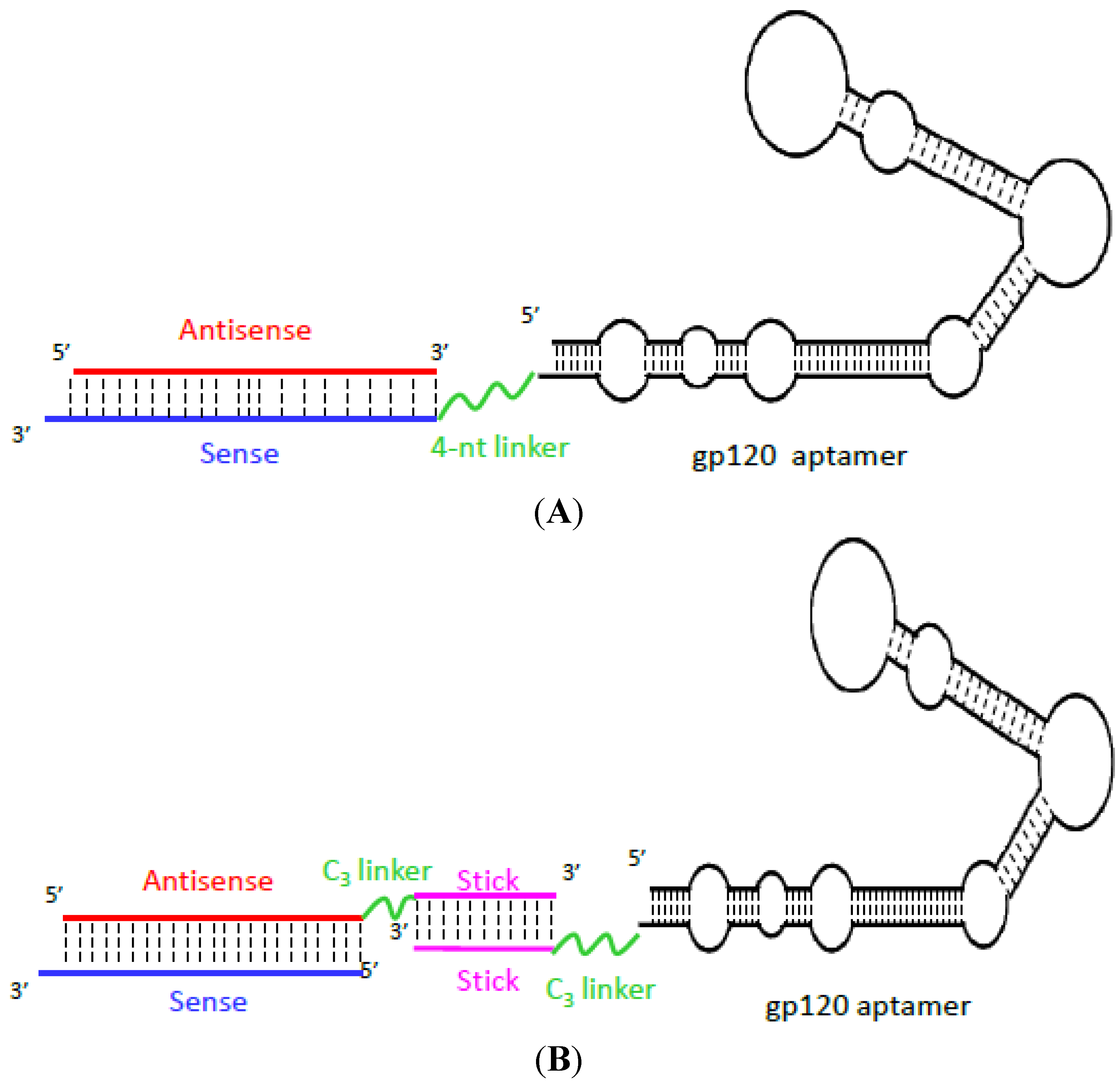
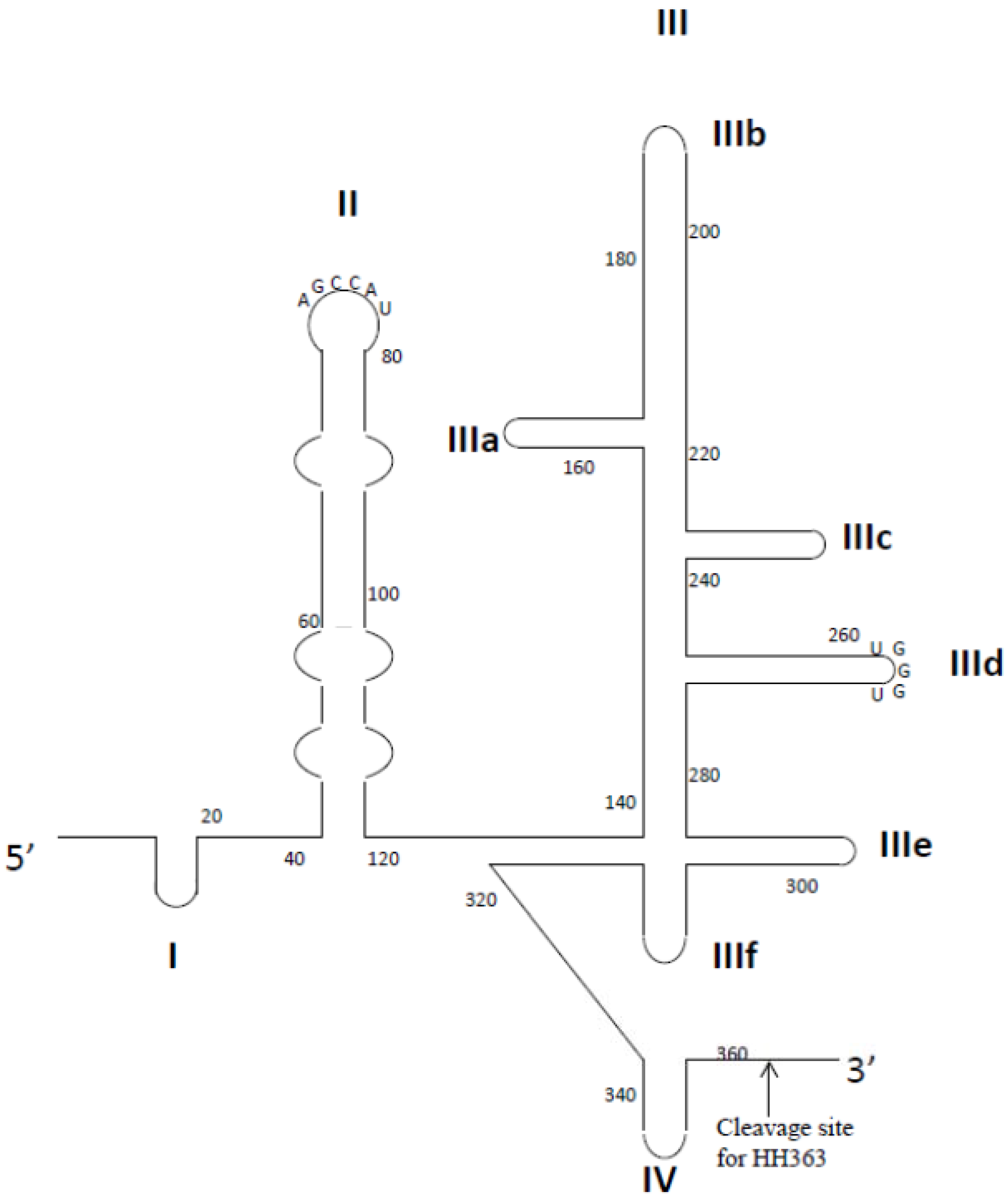
Aptamer-Based Therapeutics: New Approaches to Combat Human Viral Diseases
Abstract
:1. Introduction
3. Conclusions
Acknowledgments
Conflicts of Interest
References
- Haasnoot, J.; Berkhout, B. Nucleic acids-based therapeutics in the battle against pathogenic viruses. Handb. Exp. Pharmacol. 2009, 189, 243–263. [Google Scholar] [CrossRef]
- Mescalchin, A.; Restle, T. Oligomeric nucleic acids as antivirals. Molecules 2011, 16, 1271–1296. [Google Scholar] [CrossRef]
- Alter, G.; Heckerman, D.; Schneidewind, A.; Fadda, L.; Kadie, C.M.; Carlson, J.M.; Oniangue-Ndza, C.; Martin, M.; Li, B.; Khakoo, S.I.; et al. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature 2011, 476, 96–100. [Google Scholar] [CrossRef]
- Ding, Q.; Cao, X.; Lu, J.; Huang, B.; Liu, Y.J.; Kato, N.; Shu, H.B.; Zhong, J. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J. Hepatol. 2013, 59, 52–58. [Google Scholar] [CrossRef]
- Arts, E.J.; Hazuda, D.J. HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med. 2012, 2, a007161. [Google Scholar]
- Gopinath, S.C. Antiviral aptamers. Arch. Virol. 2007, 152, 2137–2157. [Google Scholar] [CrossRef]
- Jordheim, L.P.; Durantel, D.; Zoulim, F.; Dumontet, C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013, 12, 447–464. [Google Scholar] [CrossRef]
- Jones, M.; Nunez, M. Liver toxicity of antiretroviral drugs. Semin. Liver Dis. 2012, 32, 167–176. [Google Scholar] [CrossRef]
- Preston, B.D.; Poiesz, B.J.; Loeb, L.A. Fidelity of HIV-1 reverse transcriptase. Science 1988, 242, 1168–1171. [Google Scholar]
- Brown, A.J.; Cleland, A. Independent evolution of the env and pol genes of HIV-1 during zidovudine therapy. AIDS 1996, 10, 1067–1073. [Google Scholar]
- Kwong, A.D.; Rao, B.G.; Jeang, K.T. Viral and cellular RNA helicases as antiviral targets. Nat. Rev. Drug Discov. 2005, 4, 845–853. [Google Scholar] [CrossRef]
- Kuritzkes, D.R. Drug resistance in HIV-1. Curr. Opin. Virol. 2011, 1, 582–589. [Google Scholar] [CrossRef]
- Garcia-Lerma, J.G.; Nidtha, S.; Blumoff, K.; Weinstock, H.; Heneine, W. Increased ability for selection of zidovudine resistance in a distinct class of wild-type HIV-1 from drug-naive persons. Proc. Natl. Acad. Sci. USA 2001, 98, 13907–13912. [Google Scholar] [CrossRef]
- Schmitz, S.H.; Scheding, S.; Voliotis, D.; Rasokat, H.; Diehl, V.; Schrappe, M. Side effects of AZT prophylaxis after occupational exposure to HIV-infected blood. Ann. Hematol. 1994, 69, 135–138. [Google Scholar] [CrossRef]
- Kuehl, A.K.; Noormohamed, S.E. Recombinant erythropoietin for zidovudine-induced anemia in AIDS. Ann. Pharmacother. 1995, 29, 778–779. [Google Scholar]
- Rachlis, A.; Fanning, M.M. Zidovudine toxicity. Clinical features and management. Drug Saf. 1993, 8, 312–320. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Tuerk, C.; MacDougal-Waugh, S. In vitro evolution of functional nucleic acids: High-affinity RNA ligands of HIV-1 proteins. Gene 1993, 137, 33–39. [Google Scholar] [CrossRef]
- Hernandez, L.I.; Flenker, K.S.; Hernandez, F.J.; Klingelhutz, A.J.; McNamara, J.O.; Giangrande, P.H. Methods for evaluating cell-specific, cell-internalizing RNA aptamers. Pharmaceuticals 2013, 6, 295–319. [Google Scholar] [CrossRef]
- Zhou, J.; Li, H.; Zhang, J.; Piotr, S.; Rossi, J. Development of cell-type specific anti-HIV gp120 aptamers for siRNA delivery. J. Vis. Exp. 2011, 52, 357–368. [Google Scholar]
- Liu, J.; You, M.; Pu, Y.; Liu, H.; Ye, M.; Tan, W. Recent developments in protein and cell-targeted aptamer selection and applications. Curr. Med. Chem. 2011, 18, 4117–4125. [Google Scholar] [CrossRef]
- McNamara, J.O., 2nd; Andrechek, E.R.; Wang, Y.; Viles, K.D.; Rempel, R.E.; Gilboa, E.; Sullenger, B.A.; Giangrande, P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006, 24, 1005–1015. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J.J. Aptamer-targeted RNAi for HIV-1 therapy. Methods Mol. Biol. 2011, 721, 355–371. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J.J. Aptamer-targeted cell-specific RNA interference. Silence 2010, 1, 4. [Google Scholar] [CrossRef]
- Tucker, W.O.; Shum, K.T.; Tanner, J.A. G-quadruplex DNA aptamers and their ligands: Structure, function and application. Curr. Pharm. Des. 2012, 18, 2014–2026. [Google Scholar] [CrossRef]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef]
- Doggrell, S.A. Pegaptanib: The first antiangiogenic agent approved for neovascular macular degeneration. Expert Opin. Pharmacother. 2005, 6, 1421–1423. [Google Scholar] [CrossRef]
- Sundaram, P.; Kurniawan, H.; Byrne, M.E.; Wower, J. Therapeutic RNA aptamers in clinical trials. Eur. J. Pharm. Sci. 2013, 48, 259–271. [Google Scholar] [CrossRef]
- Burnett, J.C.; Rossi, J.J. RNA-based therapeutics: Current progress and future prospects. Chem. Biol. 2012, 19, 60–71. [Google Scholar] [CrossRef]
- Mongelard, F.; Bouvet, P. AS-1411, a guanosine-rich oligonucleotide aptamer targeting nucleolin for the potential treatment of cancer, including acute myeloid leukemia. Curr. Opin. Mol. Ther. 2010, 12, 107–114. [Google Scholar]
- Gilbert, J.C.; DeFeo-Fraulini, T.; Hutabarat, R.M.; Horvath, C.J.; Merlino, P.G.; Marsh, H.N.; Healy, J.M.; Boufakhreddine, S.; Holohan, T.V.; Schaub, R.G. First-in-human evaluation of anti von Willebrand factor therapeutic aptamer ARC1779 in healthy volunteers. Circulation 2007, 116, 2678–2686. [Google Scholar] [CrossRef]
- Vinores, S.A. Pegaptanib in the treatment of wet, age-related macular degeneration. Int. J. Nanomed. 2006, 1, 263–268. [Google Scholar]
- Zhou, J.; Rossi, J.J. Therapeutic potential of aptamer-siRNA conjugates for treatment of HIV-1. BioDrugs 2012, 26, 393–400. [Google Scholar]
- Zhou, J.; Rossi, J.J. Progress in RNAi-based antiviral therapeutics. Methods Mol. Biol. 2011, 721, 67–75. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J.J. Cell-specific aptamer-mediated targeted drug delivery. Oligonucleotides 2011, 21, 1–10. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J.J. Bivalent aptamers deliver the punch. Chem. Biol. 2008, 15, 644–645. [Google Scholar] [CrossRef]
- Kanwar, J.R.; Roy, K.; Kanwar, R.K. Chimeric aptamers in cancer cell-targeted drug delivery. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 459–477. [Google Scholar]
- Khati, M. The future of aptamers in medicine. J. Clin. Pathol. 2010, 63, 480–487. [Google Scholar] [CrossRef]
- Cheung, Y.W.; Kwok, J.; Law, A.W.; Watt, R.M.; Kotaka, M.; Tanner, J.A. Structural basis for discriminatory recognition of Plasmodium lactate dehydrogenase by a DNA aptamer. Proc. Natl. Acad. Sci. USA 2013, 110, 15967–15972. [Google Scholar]
- Bunka, D.H.; Stockley, P.G. Aptamers come of age—at last. Nat. Rev. Microbiol. 2006, 4, 588–596. [Google Scholar] [CrossRef]
- Tuerk, C.; MacDougal, S.; Gold, L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc. Natl. Acad. Sci. USA 1992, 89, 6988–6992. [Google Scholar] [CrossRef]
- Jaeger, J.; Restle, T.; Steitz, T.A. The structure of HIV-1 reverse transcriptase complexed with an RNA pseudoknot inhibitor. EMBO J. 1998, 17, 4535–4542. [Google Scholar] [CrossRef]
- Guo, P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010, 5, 833–842. [Google Scholar] [CrossRef]
- Guo, P.; Haque, F.; Hallahan, B.; Reif, R.; Li, H. Uniqueness, advantages, challenges, solutions, and perspectives in therapeutics applying RNA nanotechnology. Nucleic Acid Ther. 2012, 22, 226–245. [Google Scholar]
- Shukla, G.C.; Haque, F.; Tor, Y.; Wilhelmsson, L.M.; Toulme, J.J.; Isambert, H.; Guo, P.; Rossi, J.J.; Tenenbaum, S.A.; Shapiro, B.A. A boost for the emerging field of RNA nanotechnology. ACS Nano 2011, 5, 3405–3418. [Google Scholar] [CrossRef]
- Tarapore, P.; Shu, Y.; Guo, P.; Ho, S.M. Application of phi29 motor pRNA for targeted therapeutic delivery of siRNA silencing metallothionein-IIA and survivin in ovarian cancers. Mol. Ther. 2011, 19, 386–394. [Google Scholar] [CrossRef]
- Shu, Y.; Cinier, M.; Shu, D.; Guo, P. Assembly of multifunctional phi29 pRNA nanoparticles for specific delivery of siRNA and other therapeutics to targeted cells. Methods 2011, 54, 204–214. [Google Scholar] [CrossRef]
- Shu, Y.; Haque, F.; Shu, D.; Li, W.; Zhu, Z.; Kotb, M.; Lyubchenko, Y.; Guo, P. Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. RNA 2013, 19, 767–777. [Google Scholar] [CrossRef]
- Neff, C.P.; Zhou, J.; Remling, L.; Kuruvilla, J.; Zhang, J.; Li, H.; Smith, D.D.; Swiderski, P.; Rossi, J.J.; Akkina, R. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci. Transl. Med. 2011, 3. [Google Scholar] [CrossRef]
- Bai, J.; Banda, N.; Lee, N.S.; Rossi, J.; Akkina, R. RNA-based anti-HIV-1 gene therapeutic constructs in SCID-hu mouse model. Mol. Ther. 2002, 6, 770–782. [Google Scholar] [CrossRef]
- Ahmad, K.M.; Xiao, Y.; Soh, H.T. Selection is more intelligent than design: Improving the affinity of a bivalent ligand through directed evolution. Nucleic Acids Res. 2012, 40, 11777–11783. [Google Scholar] [CrossRef]
- Ye, M.; Hu, J.; Peng, M.; Liu, J.; Liu, H.; Zhao, X.; Tan, W. Generating aptamers by cell-SELEX for applications in molecular medicine. Int. J. Mol. Sci. 2012, 13, 3341–3353. [Google Scholar] [CrossRef]
- Dua, P.; Kim, S.; Lee, D.K. Nucleic acid aptamers targeting cell-surface proteins. Methods 2011, 54, 215–225. [Google Scholar] [CrossRef]
- Tang, Z.; Parekh, P.; Turner, P.; Moyer, R.W.; Tan, W. Generating aptamers for recognition of virus-infected cells. Clin. Chem. 2009, 55, 813–822. [Google Scholar] [CrossRef]
- Richman, D.D.; Margolis, D.M.; Delaney, M.; Greene, W.C.; Hazuda, D.; Pomerantz, R.J. The challenge of finding a cure for HIV infection. Science 2009, 323, 1304–1307. [Google Scholar] [CrossRef]
- Joshi, P.J.; Fisher, T.S.; Prasad, V.R. Anti-HIV inhibitors based on nucleic acids: Emergence of aptamers as potent antivirals. Curr. Drug Targets Infect. Disord. 2003, 3, 383–400. [Google Scholar] [CrossRef]
- Scherer, L.; Rossi, J.J.; Weinberg, M.S. Progress and prospects: RNA-based therapies for treatment of HIV infection. Gene Ther. 2007, 14, 1057–1064. [Google Scholar] [CrossRef]
- Held, D.M.; Kissel, J.D.; Patterson, J.T.; Nickens, D.G.; Burke, D.H. HIV-1 inactivation by nucleic acid aptamers. Front. Biosci. 2006, 11, 89–112. [Google Scholar] [CrossRef]
- Jager, J.; Pata, J.D. Getting a grip: Polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 1999, 9, 21–28. [Google Scholar] [CrossRef]
- De Clercq, E. Chemotherapeutic approaches to the treatment of the acquired immune deficiency syndrome (AIDS). J. Med. Chem. 1986, 29, 1561–1569. [Google Scholar] [CrossRef]
- De Clercq, E. Perspectives of non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the therapy of HIV-1 infection. Farmaco 1999, 54, 26–45. [Google Scholar] [CrossRef]
- Kensch, O.; Connolly, B.A.; Steinhoff, H.J.; McGregor, A.; Goody, R.S.; Restle, T. HIV-1 reverse transcriptase-pseudoknot RNA aptamer interaction has a binding affinity in the low picomolar range coupled with high specificity. J. Biol. Chem. 2000, 275, 18271–18278. [Google Scholar]
- Chaloin, L.; Lehmann, M.J.; Sczakiel, G.; Restle, T. Endogenous expression of a high-affinity pseudoknot RNA aptamer suppresses replication of HIV-1. Nucleic Acids Res. 2002, 30, 4001–4008. [Google Scholar] [CrossRef]
- Li, N.; Wang, Y.; Pothukuchy, A.; Syrett, A.; Husain, N.; Gopalakrisha, S.; Kosaraju, P.; Ellington, A.D. Aptamers that recognize drug-resistant HIV-1 reverse transcriptase. Nucleic Acids Res. 2008, 36, 6739–6751. [Google Scholar] [CrossRef]
- Andreola, M.L.; Pileur, F.; Calmels, C.; Ventura, M.; Tarrago-Litvak, L.; Toulme, J.J.; Litvak, S. DNA aptamers selected against the HIV-1 RNase H display in vitro antiviral activity. Biochemistry 2001, 40, 10087–10094. [Google Scholar] [CrossRef]
- Somasunderam, A.; Ferguson, M.R.; Rojo, D.R.; Thiviyanathan, V.; Li, X.; O’Brien, W.A.; Gorenstein, D.G. Combinatorial selection, inhibition, and antiviral activity of DNA thioaptamers targeting the RNase H domain of HIV-1 reverse transcriptase. Biochemistry 2005, 44, 10388–10395. [Google Scholar] [CrossRef]
- DeStefano, J.J.; Nair, G.R. Novel aptamer inhibitors of human immunodeficiency virus reverse transcriptase. Oligonucleotides 2008, 18, 133–144. [Google Scholar] [CrossRef]
- Schneider, D.J.; Feigon, J.; Hostomsky, Z.; Gold, L. High-affinity ssDNA inhibitors of the reverse transcriptase of type 1 human immunodeficiency virus. Biochemistry 1995, 34, 9599–9610. [Google Scholar] [CrossRef]
- Kissel, J.D.; Held, D.M.; Hardy, R.W.; Burke, D.H. Single-stranded DNA aptamer RT1t49 inhibits RT polymerase and RNase H functions of HIV type 1, HIV type 2, and SIVCPZ RTs. AIDS Res. Hum. Retroviruses 2007, 23, 699–708. [Google Scholar] [CrossRef]
- Lai, Y.T.; DeStefano, J.J. DNA aptamers to human immunodeficiency virus reverse transcriptase selected by a primer-free SELEX method: Characterization and comparison with other aptamers. Nucleic Acid Ther. 2012, 22, 162–176. [Google Scholar]
- Whatley, A.S.; Ditzler, M.A.; Lange, M.J.; Biondi, E.; Sawyer, A.W.; Chang, J.L.; Franken, J.D.; Burke, D.H. Potent Inhibition of HIV-1 Reverse Transcriptase and Replication by Nonpseudoknot, “UCAA-motif” RNA Aptamers. Mol. Ther. Nucleic Acids 2013, 2, e71. [Google Scholar] [CrossRef]
- Ditzler, M.A.; Lange, M.J.; Bose, D.; Bottoms, C.A.; Virkler, K.F.; Sawyer, A.W.; Whatley, A.S.; Spollen, W.; Givan, S.A.; Burke, D.H. High-throughput sequence analysis reveals structural diversity and improved potency among RNA inhibitors of HIV reverse transcriptase. Nucleic Acids Res. 2013, 41, 1873–1884. [Google Scholar] [CrossRef]
- De Soultrait, V.R.; Lozach, P.Y.; Altmeyer, R.; Tarrago-Litvak, L.; Litvak, S.; Andreola, M.L. DNA aptamers derived from HIV-1 RNase H inhibitors are strong anti-integrase agents. J. Mol. Biol. 2002, 324, 195–203. [Google Scholar] [CrossRef]
- Faure-Perraud, A.; Metifiot, M.; Reigadas, S.; Recordon-Pinson, P.; Parissi, V.; Ventura, M.; Andreola, M.L. The guanine-quadruplex aptamer 93del inhibits HIV-1 replication ex vivo by interfering with viral entry, reverse transcription and integration. Antivir. Ther. 2011, 16, 383–394. [Google Scholar] [CrossRef]
- Jing, N.; Hogan, M.E. Structure-activity of tetrad-forming oligonucleotides as a potent anti-HIV therapeutic drug. J. Biol. Chem. 1998, 273, 34992–34999. [Google Scholar] [CrossRef]
- Ramalingam, D.; Duclair, S.; Datta, S.A.; Ellington, A.; Rein, A.; Prasad, V.R. RNA aptamers directed to human immunodeficiency virus type 1 Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J. Virol. 2011, 85, 305–314. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, M.Y.; Lee, J.H.; You, J.C.; Jeong, S. Selection and stabilization of the RNA aptamers against the human immunodeficiency virus type-1 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2002, 291, 925–931. [Google Scholar] [CrossRef]
- Kim, M.Y.; Jeong, S. RNA aptamers that bind the nucleocapsid protein contain pseudoknots. Mol. Cells 2003, 16, 413–417. [Google Scholar]
- Sayer, N.; Ibrahim, J.; Turner, K.; Tahiri-Alaoui, A.; James, W. Structural characterization of a 2'F-RNA aptamer that binds a HIV-1 SU glycoprotein, gp120. Biochem. Biophys. Res. Commun. 2002, 293, 924–931. [Google Scholar] [CrossRef]
- Mufhandu, H.T.; Gray, E.S.; Madiga, M.C.; Tumba, N.; Alexandre, K.B.; Khoza, T.; Wibmer, C.K.; Moore, P.L.; Morris, L.; Khati, M. UCLA1, a synthetic derivative of a gp120 RNA aptamer, inhibits entry of human immunodeficiency virus type 1 subtype C. J. Virol. 2012, 86, 4989–4999. [Google Scholar] [CrossRef]
- Zhou, J.; Neff, C.P.; Liu, X.; Zhang, J.; Li, H.; Smith, D.D.; Swiderski, P.; Aboellail, T.; Huang, Y.; Du, Q.; et al. Systemic administration of combinatorial dsiRNAs via nanoparticles efficiently suppresses HIV-1 infection in humanized mice. Mol. Ther. 2011, 19, 2228–2238. [Google Scholar] [CrossRef]
- Zhou, J.; Swiderski, P.; Li, H.; Zhang, J.; Neff, C.P.; Akkina, R.; Rossi, J.J. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009, 37, 3094–3109. [Google Scholar] [CrossRef]
- Yamamoto, R.; Katahira, M.; Nishikawa, S.; Baba, T.; Taira, K.; Kumar, P.K. A novel RNA motif that binds efficiently and specifically to the Ttat protein of HIV and inhibits the trans-activation by Tat of transcription in vitro and in vivo. Genes Cells 2000, 5, 371–388. [Google Scholar] [CrossRef]
- Jensen, K.B.; Green, L.; MacDougal-Waugh, S.; Tuerk, C. Characterization of an in vitro-selected RNA ligand to the HIV-1 Rev protein. J. Mol. Biol. 1994, 235, 237–247. [Google Scholar] [CrossRef]
- Boiziau, C.; Dausse, E.; Yurchenko, L.; Toulme, J.J. DNA aptamers selected against the HIV-1 trans-activation-responsive RNA element form RNA-DNA kissing complexes. J. Biol. Chem. 1999, 274, 12730–12737. [Google Scholar] [CrossRef]
- Duconge, F.; Toulme, J.J. In vitro selection identifies key determinants for loop-loop interactions: RNA aptamers selective for the TAR RNA element of HIV-1. RNA 1999, 5, 1605–1614. [Google Scholar] [CrossRef]
- Watrin, M.; von Pelchrzim, F.; Dausse, E.; Schroeder, R.; Toulme, J.J. In vitro selection of RNA aptamers derived from a genomic human library against the TAR RNA element of HIV-1. Biochemistry 2009, 48, 6278–6284. [Google Scholar] [CrossRef]
- Sekkai, D.; Dausse, E.; di Primo, C.; Darfeuille, F.; Boiziau, C.; Toulme, J.J. In vitro selection of DNA aptamers against the HIV-1 TAR RNA hairpin. Antisense Nucleic Acid Drug Dev. 2002, 12, 265–274. [Google Scholar] [CrossRef]
- Kumar, P.K.; Machida, K.; Urvil, P.T.; Kakiuchi, N.; Vishnuvardhan, D.; Shimotohno, K.; Taira, K.; Nishikawa, S. Isolation of RNA aptamers specific to the NS3 protein of hepatitis C virus from a pool of completely random RNA. Virology 1997, 237, 270–282. [Google Scholar] [CrossRef]
- Fukuda, K.; Vishnuvardhan, D.; Sekiya, S.; Hwang, J.; Kakiuchi, N.; Taira, K.; Shimotohno, K.; Kumar, P.K.; Nishikawa, S. Isolation and characterization of RNA aptamers specific for the hepatitis C virus nonstructural protein 3 protease. Eur. J. Biochem. 2000, 267, 3685–3694. [Google Scholar] [CrossRef]
- Nishikawa, F.; Funaji, K.; Fukuda, K.; Nishikawa, S. In vitro selection of RNA aptamers against the HCV NS3 helicase domain. Oligonucleotides 2004, 14, 114–129. [Google Scholar] [CrossRef]
- Biroccio, A.; Hamm, J.; Incitti, I.; de Francesco, R.; Tomei, L. Selection of RNA aptamers that are specific and high-affinity ligands of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 2002, 76, 3688–3696. [Google Scholar] [CrossRef]
- Bellecave, P.; Cazenave, C.; Rumi, J.; Staedel, C.; Cosnefroy, O.; Andreola, M.L.; Ventura, M.; Tarrago-Litvak, L.; Astier-Gin, T. Inhibition of hepatitis C virus (HCV) RNA polymerase by DNA aptamers: Mechanism of inhibition of in vitro RNA synthesis and effect on HCV-infected cells. Antimicrob. Agents Chemother. 2008, 52, 2097–2110. [Google Scholar] [CrossRef]
- Lee, C.H.; Lee, Y.J.; Kim, J.H.; Lim, J.H.; Kim, J.H.; Han, W.; Lee, S.H.; Noh, G.J.; Lee, S.W. Inhibition of hepatitis C virus (HCV) replication by specific RNA aptamers against HCV NS5B RNA replicase. J. Virol. 2013, 87, 7064–7074. [Google Scholar] [CrossRef]
- Marton, S.; Romero-Lopez, C.; Berzal-Herranz, A. RNA aptamer-mediated interference of HCV replication by targeting the CRE-5BSL3.2 domain. J. Viral Hepat. 2013, 20, 103–112. [Google Scholar] [CrossRef]
- Kikuchi, K.; Umehara, T.; Fukuda, K.; Hwang, J.; Kuno, A.; Hasegawa, T.; Nishikawa, S. RNA aptamers targeted to domain II of hepatitis C virus IRES that bind to its apical loop region. J. Biochem. 2003, 133, 263–270. [Google Scholar] [CrossRef]
- Kikuchi, K.; Umehara, T.; Fukuda, K.; Kuno, A.; Hasegawa, T.; Nishikawa, S. A hepatitis C virus (HCV) internal ribosome entry site (IRES) domain III-IV-targeted aptamer inhibits translation by binding to an apical loop of domain IIId. Nucleic Acids Res. 2005, 33, 683–692. [Google Scholar] [CrossRef]
- Romero-Lopez, C.; Barroso-delJesus, A.; Puerta-Fernandez, E.; Berzal-Herranz, A. Interfering with hepatitis C virus IRES activity using RNA molecules identified by a novel in vitro selection method. Biol. Chem. 2005, 386, 183–190. [Google Scholar]
- Romero-Lopez, C.; Diaz-Gonzalez, R.; Barroso-delJesus, A.; Berzal-Herranz, A. Inhibition of hepatitis C virus replication and internal ribosome entry site-dependent translation by an RNA molecule. J. Gen. Virol. 2009, 90, 1659–1669. [Google Scholar] [CrossRef]
- Liu, J.; Yang, Y.; Hu, B.; Ma, Z.Y.; Huang, H.P.; Yu, Y.; Liu, S.P.; Lu, M.J.; Yang, D.L. Development of HBsAg-binding aptamers that bind HepG2.2.15 cells via HBV surface antigen. Virol. Sin. 2010, 25, 27–35. [Google Scholar] [CrossRef]
- Feng, H.; Beck, J.; Nassal, M.; Hu, K.H. A SELEX-screened aptamer of human hepatitis B virus RNA encapsidation signal suppresses viral replication. PLoS One 2011, 6, e27862. [Google Scholar] [CrossRef]
- Shum, K.T.; Tanner, J.A. Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. ChemBioChem 2008, 9, 3037–3045. [Google Scholar] [CrossRef]
- Jang, K.J.; Lee, N.R.; Yeo, W.S.; Jeong, Y.J.; Kim, D.E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem. Biophys. Res. Commun. 2008, 366, 738–744. [Google Scholar] [CrossRef]
- Jeon, S.H.; Kayhan, B.; Ben-Yedidia, T.; Arnon, R. A DNA aptamer prevents influenza infection by blocking the receptor binding region of the viral hemagglutinin. J. Biol. Chem. 2004, 279, 48410–48419. [Google Scholar] [CrossRef]
- Cheng, C.; Dong, J.; Yao, L.; Chen, A.; Jia, R.; Huan, L.; Guo, J.; Shu, Y.; Zhang, Z. Potent inhibition of human influenza H5N1 virus by oligonucleotides derived by SELEX. Biochem. Biophys. Res. Commun. 2008, 366, 670–674. [Google Scholar] [CrossRef]
- Gopinath, S.C.; Misono, T.S.; Kawasaki, K.; Mizuno, T.; Imai, M.; Odagiri, T.; Kumar, P.K. An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J. Gen. Virol. 2006, 87, 479–487. [Google Scholar] [CrossRef]
- Misono, T.S.; Kumar, P.K. Selection of RNA aptamers against human influenza virus hemagglutinin using surface plasmon resonance. Anal. Biochem. 2005, 342, 312–317. [Google Scholar] [CrossRef]
- Wongphatcharachai, M.; Wang, P.; Enomoto, S.; Webby, R.J.; Gramer, M.R.; Amonsin, A.; Sreevatsan, S. Neutralizing DNA aptamers against swine influenza H3N2 viruses. J. Clin. Microbiol. 2013, 51, 46–54. [Google Scholar] [CrossRef]
- Choi, S.K.; Lee, C.; Lee, K.S.; Choe, S.Y.; Mo, I.P.; Seong, R.H.; Hong, S.; Jeon, S.H. DNA aptamers against the receptor binding region of hemagglutinin prevent avian influenza viral infection. Mol. Cells 2011, 32, 527–533. [Google Scholar] [CrossRef]
- Park, S.Y.; Kim, S.; Yoon, H.; Kim, K.B.; Kalme, S.S.; Oh, S.; Song, C.S.; Kim, D.E. Selection of an antiviral RNA aptamer against hemagglutinin of the subtype H5 avian influenza virus. Nucleic Acid Ther. 2011, 21, 395–402. [Google Scholar] [CrossRef]
- Gopinath, S.C.; Sakamaki, Y.; Kawasaki, K.; Kumar, P.K. An efficient RNA aptamer against human influenza B virus hemagglutinin. J. Biochem. 2006, 139, 837–846. [Google Scholar] [CrossRef]
- Liang, H.R.; Hu, G.Q.; Zhang, T.; Yang, Y.J.; Zhao, L.L.; Qi, Y.L.; Wang, H.L.; Gao, Y.W.; Yang, S.T.; Xia, X.Z. Isolation of ssDNA aptamers that inhibit rabies virus. Int. Immunopharmacol. 2012, 14, 341–347. [Google Scholar] [CrossRef]
- Liang, H.R.; Liu, Q.; Zheng, X.X.; Gai, W.W.; Xue, X.H.; Hu, G.Q.; Wu, H.X.; Wang, H.L.; Yang, S.T.; Xia, X.Z. Aptamers targeting rabies virus-infected cells inhibit viral replication both in vitro and in vivo. Virus Res. 2013, 173, 398–403. [Google Scholar] [CrossRef]
- Toscano-Garibay, J.D.; Benitez-Hess, M.L.; Alvarez-Salas, L.M. Isolation and characterization of an RNA aptamer for the HPV-16 E7 oncoprotein. Arch. Med. Res. 2011, 42, 88–96. [Google Scholar] [CrossRef]
- Graham, J.C.; Zarbl, H. Use of cell-SELEX to generate DNA aptamers as molecular probes of HPV-associated cervical cancer cells. PLoS One 2012, 7, e36103. [Google Scholar] [CrossRef]
- Nicol, C.; Cesur, O.; Forrest, S.; Belyaeva, T.A.; Bunka, D.H.; Blair, G.E.; Stonehouse, N.J. An RNA aptamer provides a novel approach for the induction of apoptosis by targeting the HPV16 E7 oncoprotein. PLoS One 2013, 8, e64781. [Google Scholar]
- Nicol, C.; Bunka, D.H.; Blair, G.E.; Stonehouse, N.J. Effects of single nucleotide changes on the binding and activity of RNA aptamers to human papillomavirus 16 E7 oncoprotein. Biochem. Biophys. Res. Commun. 2011, 405, 417–421. [Google Scholar] [CrossRef]
- Gopinath, S.C.; Hayashi, K.; Kumar, P.K. Aptamer that binds to the gD protein of herpes simplex virus 1 and efficiently inhibits viral entry. J. Virol. 2012, 86, 6732–6744. [Google Scholar] [CrossRef]
- Moore, M.D.; Bunka, D.H.; Forzan, M.; Spear, P.G.; Stockley, P.G.; McGowan, I.; James, W. Generation of neutralizing aptamers against herpes simplex virus type 2: Potential components of multivalent microbicides. J. Gen. Virol. 2011, 92, 1493–1499. [Google Scholar] [CrossRef]
- Nickens, D.G.; Patterson, J.T.; Burke, D.H. Inhibition of HIV-1 reverse transcriptase by RNA aptamers in Escherichia coli. RNA 2003, 9, 1029–1033. [Google Scholar] [CrossRef]
- Joshi, P.; Prasad, V.R. Potent inhibition of human immunodeficiency virus type 1 replication by template analog reverse transcriptase inhibitors derived by SELEX (systematic evolution of ligands by exponential enrichment). J. Virol. 2002, 76, 6545–6557. [Google Scholar] [CrossRef]
- Lange, M.J.; Sharma, T.K.; Whatley, A.S.; Landon, L.A.; Tempesta, M.A.; Johnson, M.C.; Burke, D.H. Robust suppression of HIV replication by intracellularly expressed reverse transcriptase aptamers is independent of ribozyme processing. Mol. Ther. 2012, 20, 2304–2314. [Google Scholar] [CrossRef]
- Asante-Appiah, E.; Skalka, A.M. Molecular mechanisms in retrovirus DNA integration. Antiviral Res. 1997, 36, 139–156. [Google Scholar] [CrossRef]
- Li, T.; Shi, L.; Wang, E.; Dong, S. Multifunctional G-quadruplex aptamers and their application to protein detection. Chemistry 2009, 15, 1036–1042. [Google Scholar] [CrossRef]
- Phan, A.T.; Kuryavyi, V.; Ma, J.B.; Faure, A.; Andreola, M.L.; Patel, D.J. An interlocked dimeric parallel-stranded DNA quadruplex: A potent inhibitor of HIV-1 integrase. Proc. Natl. Acad. Sci. USA 2005, 102, 634–639. [Google Scholar]
- Chou, S.H.; Chin, K.H.; Wang, A.H. DNA aptamers as potential anti-HIV agents. Trends Biochem. Sci. 2005, 30, 231–234. [Google Scholar] [CrossRef]
- Mazumder, A.; Neamati, N.; Ojwang, J.O.; Sunder, S.; Rando, R.F.; Pommier, Y. Inhibition of the human immunodeficiency virus type 1 integrase by guanosine quartet structures. Biochemistry 1996, 35, 13762–13771. [Google Scholar] [CrossRef]
- Magbanua, E.; Zivkovic, T.; Hansen, B.; Beschorner, N.; Meyer, C.; Lorenzen, I.; Grotzinger, J.; Hauber, J.; Torda, A.E.; Mayer, G.; et al. d(GGGT) 4 and r(GGGU) 4 are both HIV-1 inhibitors and interleukin-6 receptor aptamers. RNA Biol. 2013, 10, 216–227. [Google Scholar] [CrossRef]
- Gheysen, D.; Jacobs, E.; de Foresta, F.; Thiriart, C.; Francotte, M.; Thines, D.; de Wilde, M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 1989, 59, 103–112. [Google Scholar]
- Wills, J.W.; Craven, R.C. Form, function, and use of retroviral gag proteins. AIDS 1991, 5, 639–654. [Google Scholar] [CrossRef]
- Demirov, D.G.; Freed, E.O. Retrovirus budding. Virus Res. 2004, 106, 87–102. [Google Scholar] [CrossRef]
- Rein, A.; Henderson, L.E.; Levin, J.G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: Significance for viral replication. Trends Biochem. Sci. 1998, 23, 297–301. [Google Scholar] [CrossRef]
- De Guzman, R.N.; Wu, Z.R.; Stalling, C.C.; Pappalardo, L.; Borer, P.N.; Summers, M.F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 1998, 279, 384–388. [Google Scholar] [CrossRef]
- Kim, M.Y.; Jeong, S. Inhibition of the functions of the nucleocapsid protein of human immunodeficiency virus-1 by an RNA aptamer. Biochem. Biophys. Res. Commun. 2004, 320, 1181–1186. [Google Scholar] [CrossRef]
- Kwong, P.D.; Wyatt, R.; Robinson, J.; Sweet, R.W.; Sodroski, J.; Hendrickson, W.A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 1998, 393, 648–659. [Google Scholar] [CrossRef]
- Sattentau, Q.J.; Moore, J.P. The role of CD4 in HIV binding and entry. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1993, 342, 59–66. [Google Scholar] [CrossRef]
- Ugolini, S.; Mondor, I.; Sattentau, Q.J. HIV-1 attachment: Another look. Trends Microbiol. 1999, 7, 144–149. [Google Scholar] [CrossRef]
- Khati, M.; Schuman, M.; Ibrahim, J.; Sattentau, Q.; Gordon, S.; James, W. Neutralization of infectivity of diverse R5 clinical isolates of human immunodeficiency virus type 1 by gp120-binding 2'F-RNA aptamers. J. Virol. 2003, 77, 12692–12698. [Google Scholar] [CrossRef]
- Dey, A.K.; Griffiths, C.; Lea, S.M.; James, W. Structural characterization of an anti-gp120 RNA aptamer that neutralizes R5 strains of HIV-1. RNA 2005, 11, 873–884. [Google Scholar] [CrossRef]
- Dey, A.K.; Khati, M.; Tang, M.; Wyatt, R.; Lea, S.M.; James, W. An aptamer that neutralizes R5 strains of human immunodeficiency virus type 1 blocks gp120-CCR5 interaction. J. Virol. 2005, 79, 13806–13810. [Google Scholar] [CrossRef]
- Cohen, C.; Forzan, M.; Sproat, B.; Pantophlet, R.; McGowan, I.; Burton, D.; James, W. An aptamer that neutralizes R5 strains of HIV-1 binds to core residues of gp120 in the CCR5 binding site. Virology 2008, 381, 46–54. [Google Scholar] [CrossRef]
- Zhou, J.; Li, H.; Li, S.; Zaia, J.; Rossi, J.J. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol. Ther. 2008, 16, 1481–1489. [Google Scholar] [CrossRef]
- Zhou, J.; Neff, C.P.; Swiderski, P.; Li, H.; Smith, D.D.; Aboellail, T.; Remling-Mulder, L.; Akkina, R.; Rossi, J.J. Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a chemically synthesized aptamer with a sticky bridge. Mol. Ther. 2013, 21, 192–200. [Google Scholar] [CrossRef]
- Arya, S.K.; Guo, C.; Josephs, S.F.; Wong-Staal, F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III). Science 1985, 229, 69–73. [Google Scholar]
- Berkhout, B.; Silverman, R.H.; Jeang, K.T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 1989, 59, 273–282. [Google Scholar] [CrossRef]
- Ensoli, B.; Barillari, G.; Salahuddin, S.Z.; Gallo, R.C.; Wong-Staal, F. Tat protein of HIV-1 stimulates growth of cells derived from Kaposi’s sarcoma lesions of AIDS patients. Nature 1990, 345, 84–86. [Google Scholar] [CrossRef]
- Cullen, B.R.; Greene, W.C. Regulatory pathways governing HIV-1 replication. Cell 1989, 58, 423–426. [Google Scholar] [CrossRef]
- Rosen, C.A.; Pavlakis, G.N. Tat and Rev: Positive regulators of HIV gene expression. AIDS 1990, 4, 499–509. [Google Scholar] [CrossRef]
- Kjems, J.; Frankel, A.D.; Sharp, P.A. Specific regulation of mRNA splicing in vitro by a peptide from HIV-1 Rev. Cell 1991, 67, 169–178. [Google Scholar] [CrossRef]
- Konopka, K.; Lee, N.S.; Rossi, J.; Duzgunes, N. Rev-binding aptamer and CMV promoter act as decoys to inhibit HIV replication. Gene 2000, 255, 235–244. [Google Scholar] [CrossRef]
- Gait, M.J.; Karn, J. RNA recognition by the human immunodeficiency virus Tat and Rev proteins. Trends Biochem. Sci. 1993, 18, 255–259. [Google Scholar] [CrossRef]
- Gatignol, A.; Duarte, M.; Daviet, L.; Chang, Y.N.; Jeang, K.T. Sequential steps in Tat trans-activation of HIV-1 mediated through cellular DNA, RNA, and protein binding factors. Gene Expr. 1996, 5, 217–228. [Google Scholar]
- Zimmermann, B.; Bilusic, I.; Lorenz, C.; Schroeder, R. Genomic SELEX: A discovery tool for genomic aptamers. Methods 2010, 52, 125–132. [Google Scholar] [CrossRef]
- Li, M.; Rossi, J.J. Lentiviral vector delivery of siRNA and shRNA encoding genes into cultured and primary hematopoietic cells. Methods Mol. Biol. 2005, 309, 261–272. [Google Scholar]
- DiGiusto, D.L.; Krishnan, A.; Li, L.; Li, H.; Li, S.; Rao, A.; Mi, S.; Yam, P.; Stinson, S.; Kalos, M.; et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2010, 2, 36–ra43. [Google Scholar]
- Wasley, A.; Alter, M.J. Epidemiology of hepatitis C: Geographic differences and temporal trends. Semin. Liver Dis. 2000, 20, 1–16. [Google Scholar] [CrossRef]
- Hanazaki, K. Antiviral therapy for chronic hepatitis B: A review. Curr. Drug Targets Inflamm. Allergy 2004, 3, 63–70. [Google Scholar] [CrossRef]
- Kato, N. Genome of human hepatitis C virus (HCV): Gene organization, sequence diversity, and variation. Microb. Comp. Genomics 2000, 5, 129–151. [Google Scholar]
- Failla, C.; Tomei, L.; de Francesco, R. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol. 1994, 68, 3753–3760. [Google Scholar]
- Kim, D.W.; Gwack, Y.; Han, J.H.; Choe, J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 1995, 215, 160–166. [Google Scholar] [CrossRef]
- Nishikawa, F.; Kakiuchi, N.; Funaji, K.; Fukuda, K.; Sekiya, S.; Nishikawa, S. Inhibition of HCV NS3 protease by RNA aptamers in cells. Nucleic Acids Res. 2003, 31, 1935–1943. [Google Scholar] [CrossRef]
- Fukuda, K.; Umehara, T.; Sekiya, S.; Kunio, K.; Hasegawa, T.; Nishikawa, S. An RNA ligand inhibits hepatitis C virus NS3 protease and helicase activities. Biochem. Biophys. Res. Commun. 2004, 325, 670–675. [Google Scholar] [CrossRef]
- Umehara, T.; Fukuda, K.; Nishikawa, F.; Kohara, M.; Hasegawa, T.; Nishikawa, S. Rational design of dual-functional aptamers that inhibit the protease and helicase activities of HCV NS3. J. Biochem. 2005, 137, 339–347. [Google Scholar] [CrossRef]
- Varshney, J.; Sharma, P.K.; Sharma, A. A review on an update of NS5B polymerase hepatitis C virus inhibitors. Eur. Rev. Med. Pharmacol. Sci. 2012, 16, 667–671. [Google Scholar]
- Tsukiyama-Kohara, K.; Iizuka, N.; Kohara, M.; Nomoto, A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992, 66, 1476–1483. [Google Scholar]
- Gallego, J.; Varani, G. The hepatitis C virus internal ribosome-entry site: A new target for antiviral research. Biochem. Soc. Trans. 2002, 30, 140–145. [Google Scholar] [CrossRef]
- Kikuchi, K.; Umehara, T.; Nishikawa, F.; Fukuda, K.; Hasegawa, T.; Nishikawa, S. Increased inhibitory ability of conjugated RNA aptamers against the HCV IRES. Biochem. Biophys. Res. Commun. 2009, 386, 118–123. [Google Scholar] [CrossRef]
- Chen, Y.; Cheng, G.; Mahato, R.I. RNAi for treating hepatitis B viral infection. Pharm. Res. 2008, 25, 72–86. [Google Scholar] [CrossRef]
- Zoulim, F. Hepatitis B virus resistance to antiviral drugs: Where are we going? Liver Int. 2011, 31, 111–116. [Google Scholar] [CrossRef]
- Seeger, C.; Mason, W.S. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000, 64, 51–68. [Google Scholar] [CrossRef]
- Summers, J.; Mason, W.S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 1982, 29, 403–415. [Google Scholar] [CrossRef]
- Wang, G.H.; Seeger, C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell 1992, 71, 663–670. [Google Scholar] [CrossRef]
- Hirsch, R.C.; Lavine, J.E.; Chang, L.J.; Varmus, H.E.; Ganem, D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as wel as for reverse transcription. Nature 1990, 344, 552–555. [Google Scholar] [CrossRef]
- Flodell, S.; Petersen, M.; Girard, F.; Zdunek, J.; Kidd-Ljunggren, K.; Schleucher, J.; Wijmenga, S. Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal. Nucleic Acids Res. 2006, 34, 4449–4457. [Google Scholar] [CrossRef]
- Peiris, J.S.; Lai, S.T.; Poon, L.L.; Guan, Y.; Yam, L.Y.; Lim, W.; Nicholls, J.; Yee, W.K.; Yan, W.W.; Cheung, M.T.; et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003, 361, 1319–1325. [Google Scholar] [CrossRef]
- Stadler, K.; Masignani, V.; Eickmann, M.; Becker, S.; Abrignani, S.; Klenk, H.D.; Rappuoli, R. SARS—beginning to understand a new virus. Nat. Rev. Microbiol. 2003, 1, 209–218. [Google Scholar] [CrossRef]
- Tanner, J.A.; Watt, R.M.; Chai, Y.B.; Lu, L.Y.; Lin, M.C.; Peiris, J.S.; Poon, L.L.; Kung, H.F.; Huang, J.D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5' to 3' viral helicases. J. Biol. Chem. 2003, 278, 39578–39582. [Google Scholar] [CrossRef]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Van Boheemen, S.; de Graaf, M.; Lauber, C.; Bestebroer, T.M.; Raj, V.S.; Zaki, A.M.; Osterhaus, A.D.; Haagmans, B.L.; Gorbalenya, A.E.; Snijder, E.J.; et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 2012. [Google Scholar] [CrossRef]
- Josset, L.; Menachery, V.D.; Gralinski, L.E.; Agnihothram, S.; Sova, P.; Carter, V.S.; Yount, B.L.; Graham, R.L.; Baric, R.S.; Katze, M.G. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio 2013. [Google Scholar] [CrossRef]
- Elderfield, R.; Barclay, W. Influenza pandemics. Adv. Exp. Med. Biol. 2011, 719, 81–103. [Google Scholar] [CrossRef]
- Webster, R.G. 1918 Spanish influenza: The secrets remain elusive. Proc. Natl. Acad. Sci. USA 1999, 96, 1164–1166. [Google Scholar] [CrossRef]
- Bai, T.; Zhou, J.; Shu, Y. Serologic study for influenza A (H7N9) among high-risk groups in China. N. Engl. J. Med. 2013, 368, 2339–2340. [Google Scholar] [CrossRef]
- Watanabe, Y.; Ibrahim, M.S.; Suzuki, Y.; Ikuta, K. The changing nature of avian influenza A virus (H5N1). Trends Microbiol. 2012, 20, 11–20. [Google Scholar] [CrossRef]
- Gultyaev, A.P.; Fouchier, R.A.; Olsthoorn, R.C. Influenza virus RNA structure: Unique and common features. Int. Rev. Immunol. 2010, 29, 533–556. [Google Scholar] [CrossRef]
- Saladino, R.; Barontini, M.; Crucianelli, M.; Nencioni, L.; Sgarbanti, R.; Palamara, A.T. Current advances in anti-influenza therapy. Curr. Med. Chem. 2010, 17, 2101–2140. [Google Scholar] [CrossRef]
- Stropkovska, A.; Janulikova, J.; Vareckova, E. Trends in development of the influenza vaccine with broader cross-protection. Acta Virol. 2010, 54, 7–19. [Google Scholar] [CrossRef]
- Burton, D.R.; Poignard, P.; Stanfield, R.L.; Wilson, I.A. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 2012, 337, 183–186. [Google Scholar] [CrossRef]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef]
- Nicholson, K.G.; Wood, J.M.; Zambon, M. Influenza. Lancet 2003, 362, 1733–1745. [Google Scholar] [CrossRef]
- Yousaf, M.Z.; Qasim, M.; Zia, S.; Khan, M.; Ashfaq, U.A.; Khan, S. Rabies molecular virology, diagnosis, prevention and treatment. Virol. J. 2012, 9, 50. [Google Scholar] [CrossRef]
- Dietzschold, B.; Schnell, M.; Koprowski, H. Pathogenesis of rabies. Curr. Top. Microbiol. Immunol. 2005, 292, 45–56. [Google Scholar] [CrossRef]
- Stanley, M. Pathology and epidemiology of HPV infection in females. Gynecol. Oncol. 2010, 117, S5–S10. [Google Scholar] [CrossRef]
- McLaughlin-Drubin, M.E.; Meyers, J.; Munger, K. Cancer associated human papillomaviruses. Curr. Opin. Virol. 2012, 2, 459–466. [Google Scholar] [CrossRef]
- Wilczynski, S.P.; Bergen, S.; Walker, J.; Liao, S.Y.; Pearlman, L.F. Human papillomaviruses and cervical cancer: Analysis of histopathologic features associated with different viral types. Hum. Pathol. 1988, 19, 697–704. [Google Scholar] [CrossRef]
- Thomas, M.; Narayan, N.; Pim, D.; Tomaic, V.; Massimi, P.; Nagasaka, K.; Kranjec, C.; Gammoh, N.; Banks, L. Human papillomaviruses, cervical cancer and cell polarity. Oncogene 2008, 27, 7018–7030. [Google Scholar] [CrossRef]
- DiMaio, D.; Liao, J.B. Human papillomaviruses and cervical cancer. Adv. Virus Res. 2006, 66, 125–159. [Google Scholar] [CrossRef]
- Mineta, H.; Ogino, T.; Amano, H.M.; Ohkawa, Y.; Araki, K.; Takebayashi, S.; Miura, K. Human papilloma virus (HPV) type 16 and 18 detected in head and neck squamous cell carcinoma. Anticancer Res. 1998, 18, 4765–4768. [Google Scholar]
- Schiller, J.T.; Lowy, D.R. Understanding and learning from the success of prophylactic human papillomavirus vaccines. Nat. Rev. Microbiol. 2012, 10, 681–692. [Google Scholar] [CrossRef]
- Ghittoni, R.; Accardi, R.; Hasan, U.; Gheit, T.; Sylla, B.; Tommasino, M. The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes 2010, 40, 1–13. [Google Scholar] [CrossRef]
- Nauenburg, S.; Zwerschke, W.; Jansen-Durr, P. Induction of apoptosis in cervical carcinoma cells by peptide aptamers that bind to the HPV-16 E7 oncoprotein. FASEB J. 2001, 15, 592–594. [Google Scholar]
- Finzer, P.; Aguilar-Lemarroy, A.; Rosl, F. The role of human papillomavirus oncoproteins E6 and E7 in apoptosis. Cancer Lett. 2002, 188, 15–24. [Google Scholar] [CrossRef]
- Maldonado, E.; Cabrejos, M.E.; Banks, L.; Allende, J.E. Human papillomavirus-16 E7 protein inhibits the DNA interaction of the TATA binding transcription factor. J. Cell. Biochem. 2002, 85, 663–669. [Google Scholar] [CrossRef]
- Bernat, A.; Avvakumov, N.; Mymryk, J.S.; Banks, L. Interaction between the HPV E7 oncoprotein and the transcriptional coactivator p300. Oncogene 2003, 22, 7871–7881. [Google Scholar] [CrossRef]
- Draper, K.G.; Frink, R.J.; Devi, G.B.; Swain, M.; Galloway, D.; Wagner, E.K. Herpes simplex virus types 1 and 2 homology in the region between 0.58 and 0.68 map units. J. Virol. 1984, 52, 615–623. [Google Scholar]
- Rozenberg, F.; Deback, C.; Agut, H. Herpes simplex encephalitis: From virus to therapy. Infect. Disord. Drug Targets 2011, 11, 235–250. [Google Scholar] [CrossRef]
- Reske, A.; Pollara, G.; Krummenacher, C.; Chain, B.M.; Katz, D.R. Understanding HSV-1 entry glycoproteins. Rev. Med. Virol. 2007, 17, 205–215. [Google Scholar] [CrossRef]
- Lasky, L.A.; Dowbenko, D.J. DNA sequence analysis of the type-common glycoprotein-D genes of herpes simplex virus types 1 and 2. DNA 1984, 3, 23–29. [Google Scholar] [CrossRef]
- Zhou, J.; Shum, K.T.; Burnett, J.C.; Rossi, J.J. Nanoparticle-based delivery of RNAi therapeutics: Progress and challenges. Pharmaceuticals 2013, 6, 85–107. [Google Scholar] [CrossRef]
- James, W. Aptamers in the virologists’ toolkit. J. Gen. Virol. 2007, 88, 351–364. [Google Scholar] [CrossRef]
- Moore, M.D.; Cookson, J.; Coventry, V.K.; Sproat, B.; Rabe, L.; Cranston, R.D.; McGowan, I.; James, W. Protection of HIV neutralizing aptamers against rectal and vaginal nucleases: Implications for RNA-based therapeutics. J. Biol. Chem. 2011, 286, 2526–2535. [Google Scholar] [CrossRef]
- Wheeler, L.A.; Vrbanac, V.; Trifonova, R.; Brehm, M.A.; Gilboa-Geffen, A.; Tanno, S.; Greiner, D.L.; Luster, A.D.; Tager, A.M.; Lieberman, J. Durable knockdown and protection from HIV transmission in humanized mice treated with gel-formulated CD4 aptamer-siRNA chimeras. Mol. Ther. 2013, 21, 1378–1389. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Shum, K.-T.; Zhou, J.; Rossi, J.J. Aptamer-Based Therapeutics: New Approaches to Combat Human Viral Diseases. Pharmaceuticals 2013, 6, 1507-1542. https://doi.org/10.3390/ph6121507
Shum K-T, Zhou J, Rossi JJ. Aptamer-Based Therapeutics: New Approaches to Combat Human Viral Diseases. Pharmaceuticals. 2013; 6(12):1507-1542. https://doi.org/10.3390/ph6121507
Chicago/Turabian StyleShum, Ka-To, Jiehua Zhou, and John J. Rossi. 2013. "Aptamer-Based Therapeutics: New Approaches to Combat Human Viral Diseases" Pharmaceuticals 6, no. 12: 1507-1542. https://doi.org/10.3390/ph6121507
APA StyleShum, K.-T., Zhou, J., & Rossi, J. J. (2013). Aptamer-Based Therapeutics: New Approaches to Combat Human Viral Diseases. Pharmaceuticals, 6(12), 1507-1542. https://doi.org/10.3390/ph6121507