Regeneration of Hair Cells: Making Sense of All the Noise
Abstract
: Hearing loss affects hundreds of millions of people worldwide by dampening or cutting off their auditory connection to the world. Current treatments for sensorineural hearing loss (SNHL) with cochlear implants are not perfect, leaving regenerative medicine as the logical avenue to a perfect cure. Multiple routes to regeneration of damaged hair cells have been proposed and are actively pursued. Each route not only requires a keen understanding of the molecular basis of ear development but also faces the practical limitations of stem cell regulation in the delicate inner ear where topology of cell distribution is essential. Improvements in our molecular understanding of the minimal essential genes necessary for hair cell formation and recent advances in stem cell manipulation, such as seen with inducible pluripotent stem cells (iPSCs) and epidermal neural crest stem cells (EPI-NCSCs), have opened new possibilities to advance research in translational stem cell therapies for individuals with hearing loss. Despite this, more detailed network maps of gene expression are needed, including an appreciation for the roles of microRNAs (miRs), key regulators of transcriptional gene networks. To harness the true potential of stem cells for hair cell regeneration, basic science and clinical medicine must work together to expedite the transition from bench to bedside by elucidating the full mechanisms of inner ear hair cell development, including a focus on the role of miRs, and adapting this knowledge safely and efficiently to stem cell technologies.1. Introduction
Sensorineural hearing loss (SNHL) afflicts over 278 million individuals worldwide and nearly half of all individuals over the age of 65 [1-3]. This multifactorial disease results from gene mutations, ototoxic drugs, environmental insults, or aging, and is often irreversible. Hearing loss imparts a substantial financial cost to society and bears an emotional and quality of life burden to the affected individual along with his/her family [1,4,5]. Current treatments with cochlear implants require an invasive surgical procedure without faithfully recapitulating normal hearing. Cochlear implants do not represent a perfect cure but rather a crutch to obtain some hearing capability [1, 6-12]. Prevention of hearing loss and regenerative medicine are two alternative pathways toward a possible perfect cure [13-15]. While prevention of hearing loss through protection of hair cells is ideal, it is not always possible [16-19], leaving regenerative medicine the sole ex post facto option for a permanent and restorative treatment of hearing loss. However, regeneration of damaged sensory epithelia in the mammalian inner ear is complicated as differentiated adult inner ear neurosensory tissue cannot re-enter the cell cycle and be used to replace neighboring cells, as is spontaneously accomplished in non-mammalian systems. This absence of proliferative capacity necessitates the use of exogenous stem cells as a medium for hair cell restoration, unless possibilities are discovered to jumpstart adult neurosensory epithelia proliferation.
The organ of Corti is a highly specialized sense organ with polarized and highly ordered inner and outer hair cells positioned in a stereotypic pattern on the basilar membrane surrounded by multiple, specialized supporting cells. Inner hair cells are innervated by approximately 95% of all sensory neurons and play the major role in sound perception while outer hair cells serve to fine-tune the hearing sensitivity. SNHL results primarily from damage to the inner hair cells in the organ of Corti. Regenerative treatment must therefore be aimed at replacing damaged organ of Corti inner hair cells; this can be done by either regenerating an entirely new organ of Corti or by replacing only the damaged hair cells, on a one-by-one basis. As inner hair cells die, morphologic changes occur to the organ of Corti including eventual loss of afferent neurons, making replacement of lost hair cells on a one-by-one basis problematic, in particular, years after loss of hair cells and subsequent organ of Corti dedifferentiation. In the absence of hair cells and key neurotrophins, originating from organ of Corti cells, spiral ganglion neurons are progressively lost [20,21]. The organ of Corti's central projections [20] form a complex tonotopic map that may not be capable of being faithfully recapitulated in a regenerated organ of Corti as brain plasticity declines with age [22-26]. If central projections cannot be retained in patients suffering from SNHL for years, not only must the individual hair cells be reformed, but so too must the neurons. In such cases, regeneration of the entire organ of Corti (with its delaminating sensory neurons) may provide the only feasible therapeutic option. This approach requires an initial jumpstart of the organ of Corti developmental process using stem cells transplanted into the cochlea, after which the context within the cochlea may suffice to activate neurosensory cell developmental programs to form a replacement organ of Corti. However, it is unlikely that an adult cochlea will provide the contextual environment needed as developmental signaling has long been turned off [27]. Therefore, stepwise cell fate restriction of transplanted stem cells using molecular signals required for organ of Corti induction, specification, and neurosensory cell formation is needed. Alternatively, the development program for the organ of Corti must be re-initiated to reform through proliferation and differentiation the lost organ. Additionally, applications of neurotrophic factors may be needed to facilitate a functional connection between the brain and the inner ear.
In patients suffering from short-term hearing loss, regeneration of damaged portions of the organ of Corti on a one-by-one basis may be possible as sensory neurons have not undergone apoptosis and the organ of Corti has not dedifferentiated. This therapeutic option requires a similar molecular understanding of neurosensory cell development but in addition requires exact targeting of prosensory cells to regions of damaged sensory epithelia while retaining proper orientation, structure, and functionality of healthy neighboring tissue. Transdifferentiation of neighboring cells cannot be used in mammals as they would deplete supporting cell populations as supporting cells are currently unable to re-enter the cell cycle [28-38]. In vitro attempts using stem cells have been initiated to reconstitute hair cells in the murine system and have been shown to be capable of differentiating into hair cell like cells [33,39-46]. However, despite great advances, it is apparent that we do not quite understand the molecular basis of hair cell development in vitro or in vivo or have the ability to control the placement of stem cells and/or hair cell precursors to damaged tissue.
Evolutionarily, the inability of mammalian neurosensory cell re-entry into the cell cycle may be due to the increased cellular diversity and complexity of the mammalian cochlea compared to non-mammalian species, where spontaneous proliferation does occur to restore lost hair cells [4,45,46]. For example, avian systems have two cell types (one supporting cell and one hair cell with graded size change) whereas mammalian cochleae have two distinct hair cell types (inner and outer) in a unique topographic distribution and at least six morphologically distinct supporting cell types in topographic restricted distribution in the organ of Corti [2]. Molecularly, a number of changes make cell re-entry difficult. In mice, hair cell precursors exit the cell cycle in an apex to base gradient from embryonic day 12.5 (E12.5) to E14.5 [47,48]. After cell cycle exit, re-entry is permanently prohibited by an increased concentration of cell cycle inhibitors, including p27Kip1 (now CDKN1) [49], an increased phosphorylation burden of the tumor suppressor retinoblastoma (pRB) [50], and a corresponding relative decrease in proto-oncogene (including Myc) levels. This evolutionary loss of proliferation may have evolved to protect the delicate inner ear from additional proliferation which would disrupt the existing stereotyped organization and has been shown to be impossible without causing cell death in adult hair cells [27,51]. In mitotically quiescent hair cell precursors, a bidirectional wave of Atoh1, originating in the middle-base portion of the cochlea [52], specifies hair cell fate [53-57] and stabilizes the cell fate of surrounding cells to differentiate as supporting cells [58-60] (Figure 1).
Non-mammalian transdifferentiation and spontaneous cell cycle re-entry are seemingly incompatible with the mammalian system as far we understand it. Ideally, differentiated mammalian cells can be forced to re-enter the cell cycle. Forced expansion of proliferation through manipulation of both pRB and especially p27Kip1 lasts until P14 in mice but human restoration is needed much later than P14 mouse equivalent. Furthermore, hair cells formed during this time are not viable. While cell specification [47-49,52-57] may render cells incapable of cell cycle entry by changing their molecular signature and enacting epigenetic changes [61-64], inhibiting tumor suppressors and increasing endogenous levels of proto-oncogene may reinitiate the cell cycle for the generation of new hair cells and supporting cells. Important to this is a greater understanding of Myc, implicated in both hair cell precursor proliferation and stem cell maintenance [65-69]. Clearly, a more complete understanding of proliferative control in the mammalian ear, including the regulatory network of miRs, may provide insights into the ability of neurosensory cells to re-enter the cell cycle. If the cell cycle could be reinitiated in adult and senescent ears to form either an asymmetrically dividing supporting cell or a symmetrically dividing inner ear stem cell, this could provide ample precursors available for transdifferentiation and/or cell fate re-specification (Figure 1). Until we possess the ability to restart proliferation in the adult ear, transplantation of stem cells seems to provide the sole avenue to hair cell regeneration and for enhancing our understanding of the molecular steps to turn a naïve cell into a hair cell. Some reports suggest the presence of an inner ear stem cell in mammals but the usefulness or potential for regeneration of these cells have not been studied. While a stem cell population in the ear would provide the ideal medium to manipulate hearing restoration, the same principles that would be applied to an ear specific stem cell population can be used with exogenous stem cells for the same purpose. Indeed, lessons learned through manipulation of stem cells in vitro may prove useful for the manipulation of ear stem cells in vivo. In conclusion, the apparent inability for adult mammalian neurosensory cells to re-enter the cycle makes stem cells the only viable therapeutic option for patients with SNHL.
In summary, both replacement of the entire organ of Corti and the replacement of lost hair cells on a one-by-one basis require the use of stem cells as adult mammalian inner ear neurosensory cells cannot be forced to re-enter the cell cycle as far as we understand it currently. As a result, the potential use of stem cells as therapy for hearing loss has blossomed [1,70-73]. Because of the varied cell types necessary for permanent hearing loss restoration (i.e. inner ear hair cells, sensory neurons), using either multipotent or pluripotent stem cells transplanted to the cochlea followed by incremental inductive cues provides a reasonable therapeutic option. Further progress to a perfect and permanent cure demands first, an understanding of stem cell sources and differentiation potentials; second, a comprehensive knowledge of the minimally essential genes required for ear development, organ of Corti formation, and hair cell maintenance; and last, the technological savvy to safely and non-invasively introduce stem cells, their differentiated derivatives, and/or the appropriate genetic therapy into a damaged cochlea.
2. Stem Cells
Stem cells are self-renewing cells with variable differentiation potentials capable of both symmetric and asymmetric division. Stem cells are categorically divided into four groups depending on their potency: unipotent, multipotent, pluripotent, totipotent (Figure 2). The potency, or ability of a stem cell to differentiate into a specific cell type, is a property of intracellular gene regulation, epigenetics, and contextual interactions (the so-called stem cell niche) [74-80]. Stem cells exist throughout the body, but the skin, with both hair follicles and fibroblasts, are an easily accessible and non-invasive source of both multipotent and pluripotent stem cells. Depending on the potency of the stem cell, which ranges from the ability to make every cell type (totipotent) to the ability to make just a single cell type (unipotent), different stepwise cell fate restricting decisions must be used to direct the differentiation of the stem cell into a specific functional cell. Transplanted stem cells can survive in the cochlea to varying degrees but the cell type it differentiates into depends on the type of stem cell it originated from and the molecular signals it receives [81]. Directing the cell fate of stem cells requires knowledge of regulation of the cell cycle and the genes needed for specification of the desired tissue [1,39]. For example, a pluripotent stem cell (such as an iPSC) must first be stepwise restricted to an ectodermal fate, then directed toward an inner ear neurosensory cell fate, and finally to a hair cell fate. In contrast, an ectodermal multipotent stem cell (such as EPI-NCSC) need only be restricted to a neurosensory cell fate and then to a hair cell fate (Figure 1). Obviously, fewer local regulations (as seems to be the case in postnatal ears) [82] driving the direction of differentiation must be substituted by more specification prior to injection of cells to ensure that hair cells differentiate in the right type at the right place.
2.1. Unipotent Stem Cells: A One-Way Road
Unipotent stem cells have the ability to differentiate into one cell type (or maximally two or three and then are termed bi- or tri-potent). Unipotent stem cells are present in nearly all tissue and primarily function to repair or replace aging or damaged tissue [83]. Unipotent stem cells are often considered progenitor cells [84] and conform to Hayflick's limit which states that after 50-70 population doublings, the progenitor cells undergo apoptosis [84,85]. Unipotent stem cells do not respond to inductive cues along other lineages [84]. While unipotent stem cells are found throughout the body, their inability to differentiate into multiple cell types limits unipotent cells' therapeutic usefulness.
2.2. Multipotent Stem Cells: A Bulge of Hope
Multipotent stem cells have the ability to differentiate into multiple related cell types. In 2006, it was proposed to use the bulge region of hair follicles as a source of multipotent stem cells [60] as these epidermal neural crest stem cells (EPI-NCSC) [86-88] are capable of differentiating to cells of neural crest origin [89-91]. Using EPI-NCSCs, differentiation into a number of cells of ectodermal lineage has been accomplished [86,89,92], specifically neurons, where a proof of principle experiment was shown to repair a spinal cord injury [93]. Importantly to hair cell therapy, the bulge region is Gata3 positive (Figure 1). Gata3 is essential for neurosensory development [94,95], most likely by defining a threshold within which other transcription factors operate to achieve their specific functions. Another multipotent and ectodermally defined stem cell population, neural stem cells (NSCs) have been shown to express hair cell markers when transplanted into the scala tympani [96-100]. NSCs have been isolated from the cochlear nucleus [101], providing a potential source of stem cells to the inner ear. Together, the accessibility and differentiation potential of multipotent stem cells provides a realistic and exciting avenue towards regenerative medicine for patients with hearing loss.
2.3. Pluripotent Stem Cells: ESCs and iPSCs
Pluripotent stem cells retain the ability to differentiate into nearly all tissue, including any of the three germ layers: ectoderm, endoderm, and mesoderm. Recent reprogramming of mouse embryonic fibroblasts (MEFs) into inducible Pluripotent Stem Cells (iPSCs) with the transcription factors Oct-4, Sox-2, Klf-4, and C-Myc [1,102-105] have provided an abundant and ethical alternative to embryonic stem cells (ESCs) [106,107]. Since the reprogramming technique was originally detailed in 2006 [107], a number of changes have made the induction more efficient, safer, and closer to clinical use [102,104,106,108-114]. ESCs are another example of pluripotent stem cells and are the focus of ethical debates due to their tremendous differentiation potential [115-117]. While possessing great differentiation capability, ESCs are susceptible to immune system rejection [118] whereas iPSCs were believed to be accepted [119,120]. However, recent findings show some immunogenicity of iPSCs, possibly due to expression changes due to the in vitro conditions they are kept in [121]. Both iPSCs and ESC transplanted in the cochlea differentiate into native tissue but not as efficiently as multipotent stem cells. ESCs transplanted into the spiral ganglion expressed neural markers [122-127] but are unable to express hair cell markers [81,128,129]. In general, pluripotent stem cells perform worse in the cochlea compared to multipotent stem cells. This decreased survival and efficiency is likely due to context [81,97] as the molecular signature is different between ESCs and iPSCs than their counterpart cells of the developing embryo [130] and the fact that less naïve/more differentiated stem cells (i.e. neural stem cells) have differentiated along the same developmental pathway.
Both iPSCs and EPI-NCSCs are easily accessible from skin tissue and are promising sources to use for hair cell regeneration (Figure 1 and Figure 3). When stem cells are transplanted in the cochlea, it is necessary to guide the exogenous stem cell towards a hair cell fate through sequential induction of hair cell signals [39,81]. What factors are minimally needed is not yet known, but the current focus in developmental ear biology is on identifying the essential genes involved in ear development: otic placode induction, organ of Corti specification, and neurosensory development. Many reviews exist on this topic but we will attempt to briefly cover the essential genes necessary for stem cell applications in the next section.
3. Otic Placode Induction, Organ of Corti Specification, and Neurosensory Development Requires a Cornucopia of Genes that Play both Integrated and Independent Functions
3.1. Ear Development: Otic Placode Induction and Organ of Corti Specification
A number of genes are essential to each step of ear formation. Fgf3, Fgf8, Fgf10 [131-134] and Wnt1/3a/8a [135-137] are sufficient as diffusible signals from the hindbrain to induce nearby ectoderm to thicken and form the presumptive otic placode. Induction changes the molecular signature of the placodal region [138-140] and includes the upregulation of Dlx5, Dlx6, Eya1, Foxg1, Foxi1, Gata3, Gbx2, Hes2, Hmx2, Hmx3, Pax2, Six1, Sox9 and Spry1 in the preplacode and invaginating placode. Many of these known and other unknown genes are required for axis formation and neurosensory formation as their absence leads to abnormal ear development. Pluripotent stem cells must first be restricted to an otic-ectodermal fate. Among others, signaling of the Fgfs, Dkk1, Sis1 and Igf have been shown to promote ectodermal fate while inhibiting both endodermal and mesodermal fates [39] (Figure 3). Importantly, cochlear duct elongation and patterning of the organ of Corti are dependent on Sox2 [141], Jagged1 [58], Gata3 [95,132,142], Lmx1a [143], Foxg1 [15,135,144] and Shh [145]. Stem cells aimed at forming inner ear hair cells must also be responsive to these signals (Figure 3). After translocation of naïve stem cells to the cochlea, contextual clues may suffice to provide the determination necessary for the differentiation and function of hair cells [104]. However, it is more likely that in the adult, the necessary co-factors are no longer expressed. Therefore, manipulating the above sets of genes is essential to priming stem cells to adopt an organ of Corti prosensory and subsequently a neurosensory cell fate.
3.2. Neurosensory Cell Differentiation and Maintenance
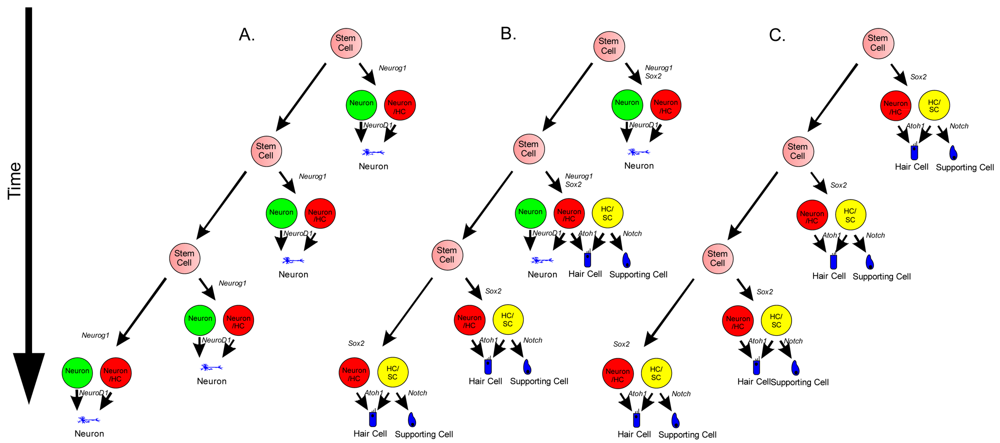
Neurosensory cells consist of hair cells, supporting cells, and sensory neurons. Neurosensory precursor populations follow nearly identical cell fate restriction pathways and appear to have a differentiation hierarchy from sensory neuron, to hair cell, and to supporting cell (Figure 1). Furthermore, it is believed that stem cells defined within the ear possess the capability to produce three distinct subsets of cells with either clonal [only neurons (Figure 4A) or only hair cell/supporting cells (Figure 4C)] or temporal restrictions [initially neurons form and later hair cells/supporting cells form (Figure 4B)] [60]. Terminally differentiated neurosensory cells are the ultimate goal of regenerative medicine.
3.2.1. Sensory Neurons
Sensory neuron formation requires a number of genes that are used to specify the neurosensory domain and differentiate pro-neuronal cells into neurons. Mutations in Eya1 and Six1 illustrate the loss of sensory neurons while mutations in Gata3 and Shh result in mice that have a reduction in sensory neurons [95,146,147]. Minimally, sensory neurons are dependent on Neurog1 [148,149] and NeuroD1 [56,150-153] as undifferentiated neuronal cells differentiate into hair cells in the absence of NeuroD1 [56,151] (Figure 1 and Figure 4). Neurotrophins Bdnf and Ntf3 are required for proper guidance and long-term survival of sensory neurons [154-157].
3.2.2. Hair Cells and Supporting Cells
Hair cell development and maintenance requires a set of genes mostly unique to sensory neurons to facilitate the placement, orientation, and maintenance of hair cells throughout all the sensory epithelia. Formation of hair cells requires proliferation of precursor populations within a specified organ of Corti, differentiation to mature hair cells, and maintenance of hair cells. Precursor proliferation controls the correct number of hair cells and is controlled through regulation of the cell cycle, in part by Myc [27,68,69]. After hair cell precursors exit to the post-mitotic state, upregulation of Atoh1 by E14.5 forces hair cell differentiation [47,158,159]. Lateral inhibition through the DELTA/NOTCH system inhibits hair cell fate for neighboring cells and forces adjacent cells to become supporting cells [15,160-162]. Downstream to Atoh1 are cell maintenance genes Pou4f3, Gfi1, Barhl1 and miR183 family members [15,163-166] (Figure 5). In the absence of these genes, hair cells and other organ of Corti cells undergo progressive cell death [1,165-168].
4. Role of miRs: A Small Name for a Big Player
If transcription factors are the hands on a clock, miRs may well be the gears that really make the clock tick. MiRs are small non-coding 20-25 nucleotide single stranded RNAs that have a two to eight nucleotide “seed” region that interacts with the 3′ end of mature mRNAs. MiRs repress translation of mRNA or degrade mRNA depending on degree of complementarity of the seed region to the target mRNA. MiRs are not fully functional until processed from their immature strands. Abnormalities of processing enzymes, specifically Dicer knockouts, have revealed a large impact of miRs (and other small RNAs) on the proper development of the embryo. Dicer cleaves the hairpin loop of the pre-miRNA to form the mature miRNA and controls regulatory activity [169]. Pax2-Cre Dicer conditional knockouts (CKOs) lose the ability to differentiate into the three germ layers and show defective differentiation [171] and conditional deletion of Dicer with Foxg1-Cre results in loss of almost all cochlear neurosensory elements [174]. However, Dicer also plays other roles making the correlation with miR loss somewhat tenuous. The DICER enzyme is also necessary for siRNA production, an area not well understood in its rapidly growing significance that will not be dealt with here.
4.1. miR Control of Stem Cell Self-Renewal
Both ESCs and iPSCs hold great regenerative potential, but this potential is lost if the stem cell state is lost. A number of genes control the transcriptional regulatory circuit necessary for self-renewal including Nanog, Oct4, C-Myc, Sox2, and Klf4 [175-179] (Figure 6). MiRs are essential to stem cell maintenance and play an important role in stem cell differentiation into hair cells [180]. Throughout the body, miRs are thought to regulate a wide array of genes with a subset of these miRs also expressed in stem cells [180]. Much like the Dicer knockouts fail to develop a proper embryo, stem cells deficient in miR processing enzymes do not have proper proliferation or differentiation [181-183]. Identifying which miRs are important is difficult due to a lack of annotation and sheer numbers (over 400 mature miRs in humans) [184,185]. Nonetheless, the four core transcription factors needed for stem cell identity co-occupy 55 distinct transcription units thought to regulate 81 mature miRs [180]. Among these miRs are miR-9, 124, 135, 148/152, 290/371 cluster, 302 cluster, 363 cluster, 615 and 708, all of which show conserved binding [180]. While this may not be overly surprising as the four core transcription factors are thought to co-occupy over 14,000 sites on the human genome [180], it shows the importance of miRs to stem cell control (Figure 6).
To maintain the stem cell state, the core transcription factors regulate two sets of miRs, the first set that is actively expressed in stem cells, the other set that is silenced in stem cells and that are important in differentiation [180]. Important to this balance is the function of Myc, whose inhibition leads to the loss of self-renewal, while activation of Myc retains the ability of self-renewal [68,192,193]. Myc is both regulated by and controls the regulation of a number of miRs, including miR141, 200, 249 [180,194,195], all important in self-renewal programs. The miRs in the 290-295 and 302 clusters are implicated in cell proliferation and are potential targets of the Mycs [196,197]. This suggests that Myc is an essential gene highly regulated by miRs to control the balance between self-renewal and differentiation (Figure 7). When the balance shifts away from proliferation, miR activity that was silenced in the stem cell state will repress the stem cell fate to favor the differentiated state (Figure 6 and Figure 7).
4.2. miR Control of Stem Cell Differentiation
During differentiation, stem cells must silence their self-renewal program [192]. miRs 9 [198-200], 134, 203, [201-203], 296, and 470 repress Nanog, Oct4, and Sox2 [204, 205]. Oct4 and miR145 create a negative feedback loop that switches stem cells between self-renewal and differentiation [171]. In stem cells, miR145 is low but is highly upregulated upon differentiation and acts to inhibit Klf4, Oct4, Sox2 and Myc [206]. Exogenous increase of cellular miR145 decreases self-renewal capacity and induces stem cells into an ectodermal cell lineage [171]. Once started along the ectodermal fate, miR124 increases (due to its inhibitor REST decreasing) and leads to the differentiation of progenitor cells to specific cell types, including neurons [207]. In the bulge region of hair follicles in the skin, miR125 (ortholog of lin-4) is critical for exit of stem cells from the cell cycle, and for their differentiation [208-210]. Once cells are differentiated, a number of miRs stabilize this cell fate, including the let-7 family [192,211]. Among the nine-membered let-7 miR family functions is repression of the Myc family [211], suggesting one mechanism that maintains the differentiated state and prohibits cell cycle re-entry (Figure 7) [192,212-216]. These examples show the importance of miRs in exiting self-renewal, forced entry into differentiation, and how feedback loops are essential in the control of this process (Figure 6).
4.3. miR Control of Hair Cell Differentiation
There are over one hundred miRs present in the cochlea and vestibular systems [172,217,218]. Dicer CKOs have shown the importance of miRs (and other small RNAs) to both ear development and hair cell maintenance. Foxg1-Cre Dicer CKOs result in loss of almost all cochlear neurosensory elements [174] while Pax2-Cre Dicer CKOs [169] and Pou4f3-Cre Dicer CKOs [172] show less complete loss.
In vertebrate sensory end organs, miR183 is highly conserved [1,219] and is formed from the same primary transcript as miR96 and 182 [220]. During ear formation, the miR183 family is restricted to hair cells and neurons. Levels of miR183 were characterized in the Dicer CKO murine malformed ear and a graded residual expression was seen which correlated to the degree of differentiation seen in the remaining sensory epithelium [169]. Mutations to a miR183 family subunit lead to hearing loss. A mutation in the seed region of miR96 also leads to an autosomal dominant progressive hearing loss [170,220]. These mice have irregular hair cell bundles and have a progressive hair cell loss after birth [221]. In non-mammalian systems, knockdowns of the miR183 family resulted in loss/reduction of hair cells and sensory ganglia [222,223]. Other miRs including miR15, 99, 100, 125, 133 all may play important roles in hair cell development and maintenance [172,173].
“Where there is smoke, there is fire”. This age old idiom holds true for our current knowledge of miRs. While in its infancy, we know miRs play a crucial role in the balance between proliferation and differentiation of the stem cell state and that the miRs are essential for the proper development and maintenance of tissues throughout the body, including the ear. Full understanding of the miRs is required prior to in vivo manipulation as deregulation of miRs is implicated in a number of tumors [192,229-232].
5. Gene Delivery: Penetrating the Problem
Gene therapy has been achieved in principle in other systems [233-236] but the stereotypic order of cells combined with the complexity of the organ of Corti cellular distribution remains a roadblock in the ear. We have previously discussed the importance of understanding stem cell regulation and what genes are necessary for stem cell differentiation to form functional hair cells, but all this falls on deaf ears unless the stem cells can target the appropriate tissue in vivo and the needed genes (i.e. Atoh1) can be regulated within the stem cells. Therefore, technical consideration must be given to both the targeting of stem cells to damaged tissue (and their survival) and the regulated gene expression within the stem cells (or in vivo newly proliferated cells).
The endolymph can safely accept up to 0.5 μL of stem cells. Stem cells are capable of efficient migration from the point of injection through the entire scala media [40,96,237]. A number of attempts have been made to target damaged tissue in the ear using various stem cells including neural stem cells [96], embryonic stem cells [128,238], and bone-marrow derived stem cells [239,240] but many of these cells were immunorejected or otherwise had unremarkable results [40]. To date, targeting and survival of stem cells remains the limiting factor in applying stem cell therapy to the ear.
Once established in damaged tissue, regulation of gene expression in the stem cell is necessary for tissue restoration. Regulation of stem cells in vitro has successfully established hair cell like cells, but manipulation of stem cells in vivo has not been accomplished. However, the ear is an ideal playground for viral transduction due to the encasement by the bony mastoid [241]. Three viral vectors are commonly used to target the inner ear tissue: Adenovirus (AV), Adeno-Associated Virus (AAV) and Lentivirus (LV). Each of these three viruses target cochlear cell types. AV targets hair cells, supporting cells, spiral ganglion neurons, fibrocytes, stria vascularis, and Reissner's membrane [40]. AAV targets hair cells, supporting cells, spiral limbus, spiral ganglion neurons, Reissner's membrane, and the spiral ligament [40]. LV targets spiral ganglion neurons, fibrocytes and Reissner's membrane [40]. Each of these vectors have been used to successfully transfect target cells with different genes: GDNF [242], HGF [243], Catalase [244], BDNF [245-247], NTF-3 [248], Atoh1 [249], EGFP [250, 251], and GJB2 [252] in the inner ear. Unfortunately, each of these vectors have shown off-target effects, notably in vivo injections of Atoh1 in the mammalian cochlea have resulted in both normal hair cell formation [158,249,253,254] and Atoh1 positive hair cells outside the organ of Corti [255]. While viruses do have some tropic specificity, they can target ectopic tissue. Furthermore, viruses themselves may pose a health risk if injecting DNA. The ability to stably and specifically target damaged tissue safely with both exogenous stem cells and the necessary genes is the final step necessary for hearing loss restoration. Multiple laboratories are working on other stem cell targeting and gene regulation expression methods including the use of nanoparticles [256] and other non-viral vectors [257-260] that will be safer, more efficient [261] and will reduce auditory trauma [237,262-264]. For example, regulated expression of target genes could be accomplished through the conditional activation of non-mammalian promoters that drive hair cell specific genes in target tissue, not ectopic sites [265]. Regardless of method, implanted stem cells must be cell fate restricted through regulated gene expression to achieve gene therapy in the ear.
6. Conclusion: There is Hope!
With the dawn of gene therapy and technological booms in stem cell technology, regenerative medicine appears to be the wave of the future, promising cures to a wide array of human diseases. While regenerative medicine is still in its infancy and years away from clinical adaptation, it represents new interactions between basic scientists and clinicians. Unprecedented in applicability, regeneration of any tissue requires a solid foundational knowledge of the molecular basis of developmental processes and stem cell regulation. As basic scientists dredge through thousands of genes in the hopes of elucidating the minimal essential gene networks required for induction of directed differentiation and clinicians have increasing numbers of patients asking about permanent cures, the race from bench to bedside has never been more important as well as challenging. Progress in network analysis highlights the controllability of a cell network and the minimal nodes needed to control that network [266].
Millions of patients suffer from hearing loss resulting from lost hair cells in the organ of Corti. Gene therapy and stem cells may offer a permanent cure for afflicted individuals, but the complexity of the ear compared to other systems, makes regeneration of hair cells a daunting task. Yet, the past ten years have shown an incredible advancement in the molecular understanding of ear development [15,267]. A continued improvement in understanding of ear development amalgamated with the nonstop advancements of gene therapy and stem cell biology may someday be fine-tuned music to the ears of many.
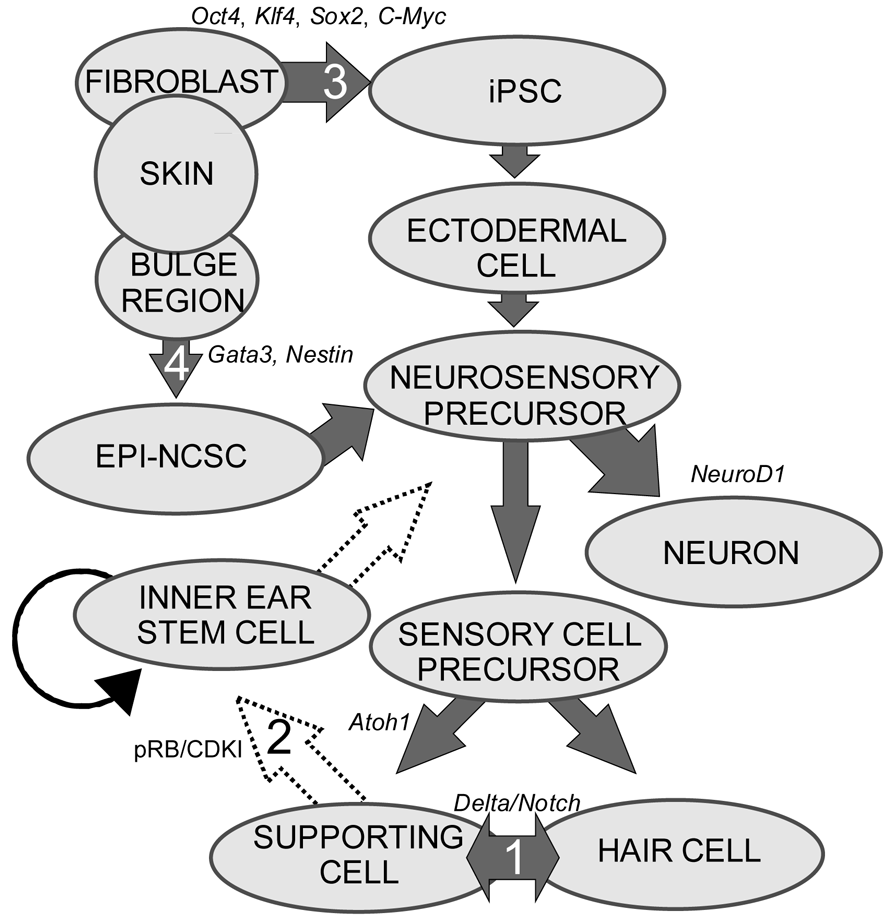
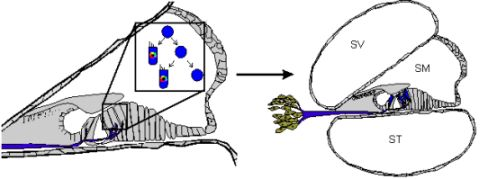
1. Transdifferentiation. As hair cells differentiate, they express Atoh1, and drive the DELTA/NOTCH signaling cascade to force neighboring cells to adopt a supporting cell fate. By inhibiting the NOTCH pathway, it may be possible to change the cell fate from a supporting cell to a hair cell. In mammals, this method is not advisable, unless more cells proliferate. 2. Inner ear stem cells. A small subpopulation of cycling cells (either endogenous stem cells or made from differentiated supporting cells) will provide a source of cells for directed differentiation and replacement of nearby damaged tissue. These populations have not been well characterized and there is no current technique to restart controlled proliferation giving rise to long-term viable hair cells in the adult in vivo. However, it is hypothesized that by manipulating the balance between proto-oncogenes (Myc) and tumor suppressors (pRB and CDKIs), in addition to higher level regulation of tissue specific factors through localization of surface receptors and ligands, re-entry in the cell cycle may be possible. 3. Neural crest stem cells. Gata3 and Nestin positive multipotent stem cells in the bulge region of hair follicles have the capacity to differentiate into various cell types. Gata3 is an important gene for the development of the organ of Corti. Selecting proper subpopulations of EPI-NCSCs may provide the necessary foundation to forming novel hair cells. 4. Inducible Pluripotent Stem Cells (iPSCs). IPSCs derived from mouse embryonic fibroblasts, through the forced expression of C-Myc, Klf4, Sox2, and Oct4, have been important in providing alternative pluripotent cells from embryonic stem cells. From the pluripotent and multipotent state, an ectodermal and neurosensory fate restriction is necessary to form hair cells. Neurosensory cells can form neurons with the expression of both Neurogenin1 (Neurog1) and NeuroD1 while hair cells form in the absence of NeuroD1 combined with Atoh1 upregulation. Both EPI-NCSCs and iPSCs can be derived from the skin of the donor. Adapted from Fritzsch et al [60].
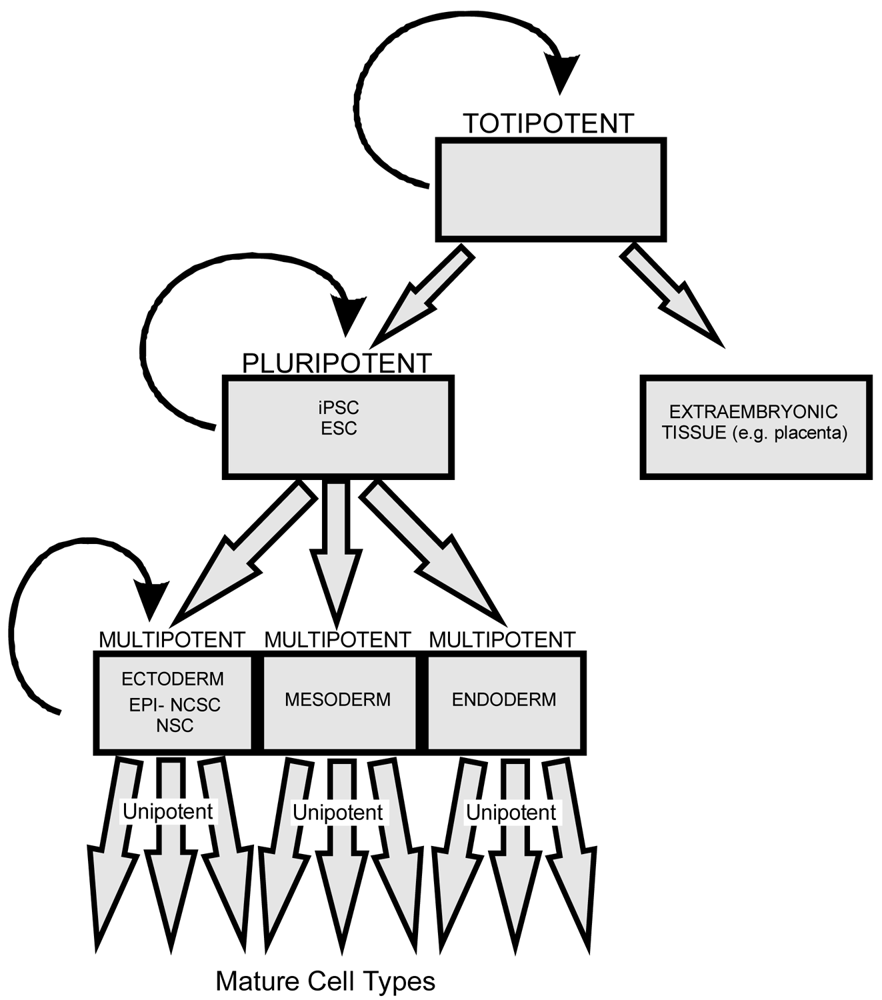
All stem cells have the ability to divide symmetrically and asymmetrically. As a stem cell asymmetrically divides it loses the potential to differentiate into cells of other lineage. Totipotent stem cells have the ability to form any cells derived from both pluripotent stem cells and extraembryonic tissue (including the placenta). Pluripotent stem cells, including both iPSCs and embryonic stem cells (ESCs), have the ability to differentiate into all three germ layers (ectoderm, mesoderm, and endoderm). Multipotent stem cells such as EPI-NCSCs and NSCs have less differentiation potential than pluripotent stem cells but both can form multiple cells of related lineage. Unipotent stem cells possess the least differentiation potential and can generate only a limited array of cell types.
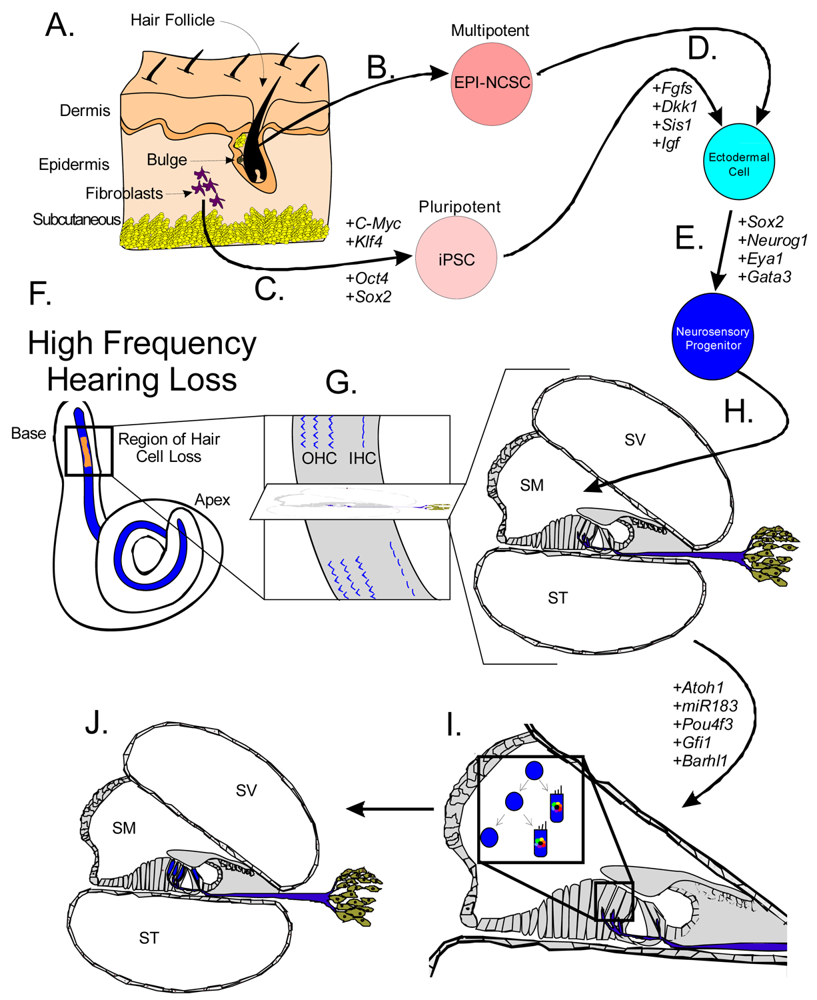
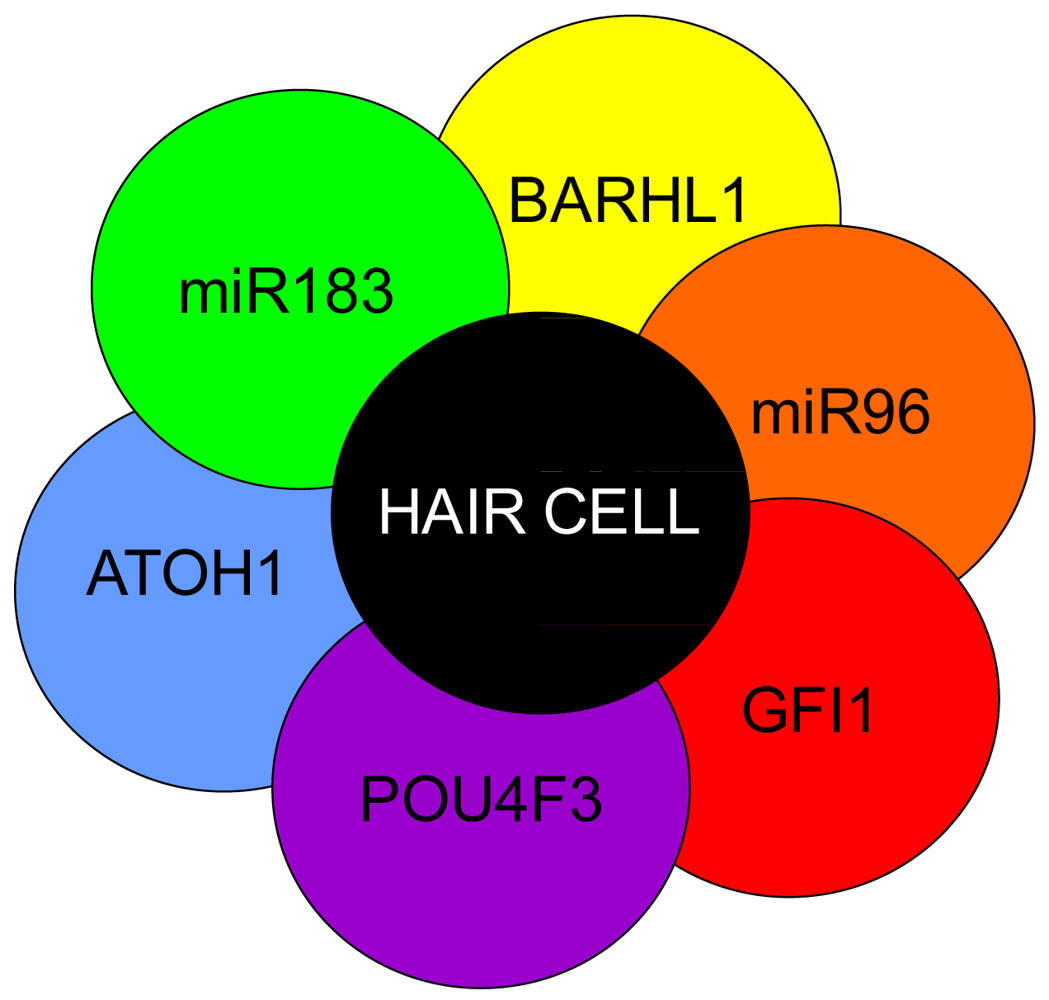
Atoh1 is required for differentiation into hair cells whereas Pou4f3, Gfi1 and Barhl1 are downstream to Atoh1 and are important for the maintenance of the hair cells. A number of miRs have been shown to be essential in the formation of hair cells and include the miR183 family (miR96, 182, and 183) as their losses result in hearing loss [1,169-171]. Additionally, miRs 15, 99, 100, 125, 133 may also be important to hair cell development but have not been well characterized [172,173]. Adapted from Pauley et al [1].
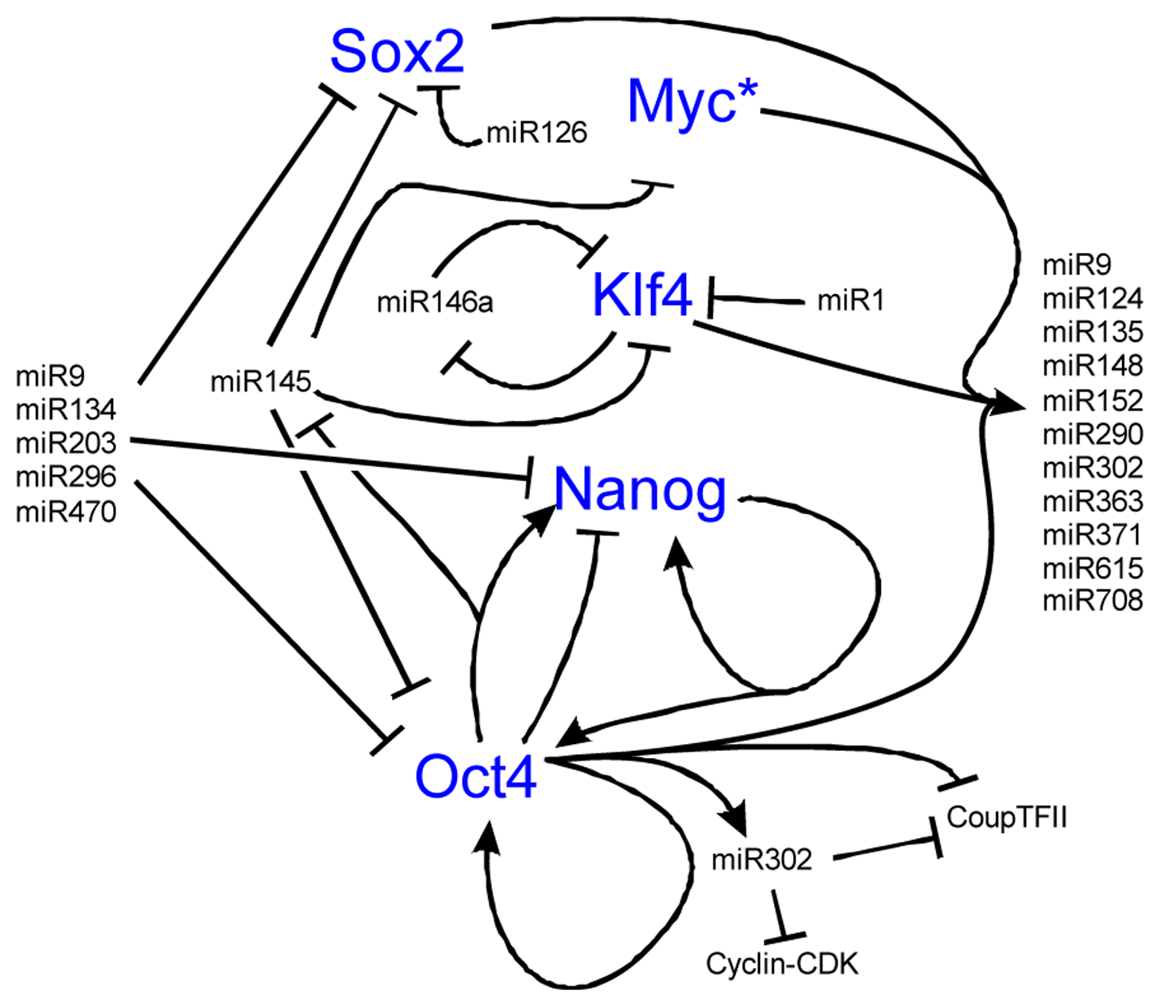
Nanog is known to be important for the maintenance of the stem cell state in a feedback loop with Oct4. While there are a number of transcriptional networks essential to the balance between stem cell self-renewal and differentiation, we omit the transcription factors to highlight the tight regulation and feedback loops that miRs play in stem cell control. Over eighty miRs are activated by the key stem cell transcription factors while other miRs are silenced. Most of these miRs have not been well characterized functionally but those that have display both positive and negative feedback loops as illustrated. Notably, miR145 is repressed by Oct4 to maintain pluripotency but at the onset of differentiation, lower levels of Oct4 allow miR145 to repress the pluripotent state through repression of Oct4, Sox2, Klf4, and Myc. Also note known feedback loops with miR9 and miR146. Among the 81 miRs upregulated by the binding of the four transcription factors is miR9, which after activation represses Sox2, Nanog, and Oct4. MiR146 forms a double negative feedback loop with Klf4. Together, the miRs are important to both maintaining the pluripotent state and when the balance is shifted towards differentiation, maintains the differentiated state by inhibiting stem cell transcription factors. Interactive pathways adapted from [114,186-191]. * See Figure 7 for Myc regulation.
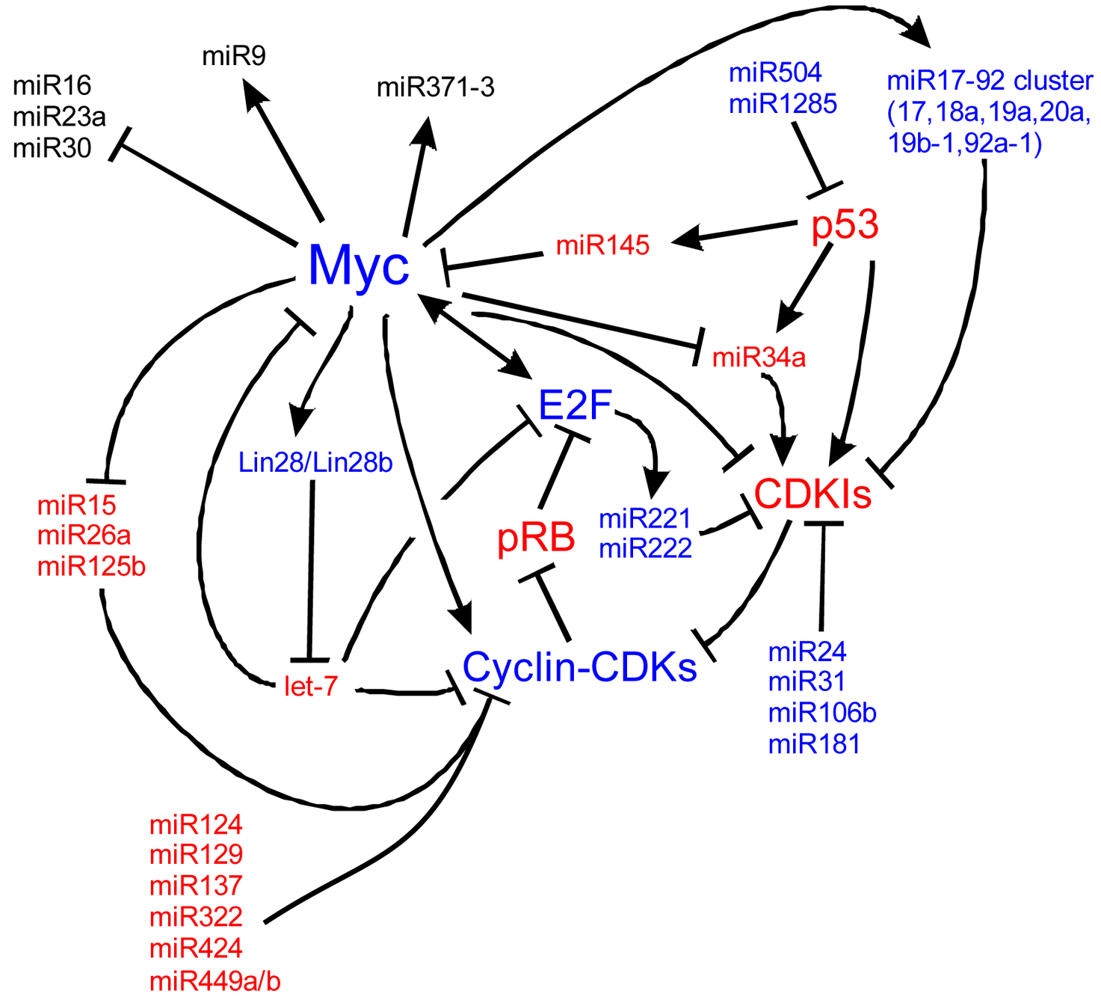
Myc is also implicated in cancers, cell cycle regulation, apoptosis, metabolism, and cellular differentiation. Myc's regulation is complex. Here we detail many of the known upstream and downstream miR regulators of Myc and the cell cycle. In blue are molecules that are important for proliferation and in red, molecules suppressing proliferation to favor cell cycle exit. At the core of the cell cycle is the tightly regulated G1-S checkpoint which is regulated by the phosphorylation state of retinoblastoma (pRB). When unphosphorylated, pRB binds E2F family members which are then unable to bind S-phase promoting genes. Phosphorylation of pRB releases E2F and the cell is able to enter the S-phase of the cell cycle. Myc is a central cog in cell cycle regulation through its signaling of miRs, the cyclins, and its repression of cyclin dependent kinase inhibitors (CDKIs) such as p27Kip1. Positive Myc regulation of a number of miRs, including the miR17-92 cluster, represses cell cycle inhibition. Negative regulation of other miRs, including miR15, 26a and 125b, de-represses negative regulation of the cell cycle. Other miRs, not currently characterized to be responsive to Mycs, are also responsible for cell cycle dynamics. Understanding the regulation of Myc, of the miRs and of the cell cycle is essential to manipulating stem cells and ultimately, manipulating these pathways may be the key to forced re-entry of hair cells into the cell cycle. Myc pathways adapted from [224-228].
Acknowledgements
Grant funding for Bernd Fritzsch was provided through the NIH and NIDCD RO1-DC055095590. We thank the NIH P30 grants (11507900 and DC010362), the University of Iowa Carver College of Medicine, Medical Scientist Training Program, and the Office of the Vice President for Research for support.
Conflict of Interest
The authors declare no conflict of interest.
References
- Pauley, S.; Kopecky, B.; Beisel, K.; Soukup, G.; Fritzsch, B. Stem cells and molecular strategies to restore hearing. Panminerva Med. 2008, 50, 41–53. [Google Scholar]
- Oshima, K.; Suchert, S.; Blevins, N.H.; Heller, S. Curing hearing loss: Patient expectations, health care practitioners, and basic science. J. Commun. Disord. 2010, 43, 311–318. [Google Scholar]
- Jongkamonwiwat, N.; Zine, A.; Rivolta, M.N. Stem cell based therapy in the inner ear: Appropriate donor cell types and routes for transplantation. Curr. Drug Targets 2010, 11, 888–897. [Google Scholar]
- Brigande, J.V.; Heller, S. Quo vadis, hair cell regeneration? Nat. Neurosci. 2009, 12, 679–685. [Google Scholar]
- Stucky, S.R.; Wolf, K.E.; Kuo, T. The economic effect of age-related hearing loss: National, state, and local estimates, 2002 and 2030. J. Am. Geriatr. Soc. 2010, 58, 618–619. [Google Scholar]
- Peterson, N.R.; Pisoni, D.B.; Miyamoto, R.T. Cochlear implants and spoken language processing abilities: Review and assessment of the literature. Restor. Neurol. Neurosci. 2010, 28, 237–250. [Google Scholar]
- Chang, Y.P.; Fu, Q.J. Effects of talker variability on vowel recognition in cochlear implants. J. Speech Lang. Hear. Res. 2006, 49, 1331–1341. [Google Scholar]
- Colletti, L.; Mandala, M.; Zoccante, L.; Shannon, R.V.; Colletti, V. Infants versus older children fitted with cochlear implants: Performance over 10 years. Int. J. Pediatr. Otorhinolaryngol. 2011, 75, 504–509. [Google Scholar]
- Bent, T.; Buchwald, A.; Pisoni, D.B. Perceptual adaptation and intelligibility of multiple talkers for two types of degraded speech. J. Acoust. Soc. Am. 2009, 126, 2660–2669. [Google Scholar]
- Gifford, R.H.; Revit, L.J. Speech perception for adult cochlear implant recipients in a realistic background noise: Effectiveness of preprocessing strategies and external options for improving speech recognition in noise. J. Am. Acad. Audiol. 2010, 21, 441–451, quiz 487-448. [Google Scholar]
- Renton, J.P.; Xu, N.; Clark, J.J.; Hansen, M.R. Interaction of neurotrophin signaling with Bcl-2 localized to the mitochondria and endoplasmic reticulum on spiral ganglion neuron survival and neurite growth. J. Neurosci. Res. 2010, 88, 2239–2251. [Google Scholar]
- Roehm, P.C.; Hansen, M.R. Strategies to preserve or regenerate spiral ganglion neurons. Curr. Opin. Otolaryngol. Head Neck Surg. 2005, 13, 294–300. [Google Scholar]
- Coleman, J.; Huang, X.; Liu, J.; Kopke, R.; Jackson, R. Dosing study on the effectiveness of salicylate/N-acetylcysteine for prevention of noise-induced hearing loss. Noise Health 2010, 12, 159–165. [Google Scholar]
- Nader, M.E.; Theoret, Y.; Saliba, I. The role of intratympanic lactate injection in the prevention of cisplatin-induced ototoxicity. Laryngoscope 2010, 120, 1208–1213. [Google Scholar]
- Fritzsch, B.; Jahan, I.; Pan, N.; Kersigo, J.; Duncan, J.; Kopecky, B. Dissecting the molecular basis of organ of Corti development: Where are we now? Hear. Res. 2011. [Google Scholar]
- Alford, R.L. Nonsyndromic hereditary hearing loss. Adv. Otorhinolaryngol. 2011, 70, 37–42. [Google Scholar]
- Korver, A.M.; Admiraal, R.J.; Kant, S.G.; Dekker, F.W.; Wever, C.C.; Kunst, H.P.; Frijns, J.H.; Oudesluys-Murphy, A.M. Causes of permanent childhood hearing impairment. Laryngoscope 2011, 121, 409–416. [Google Scholar]
- Shearer, A.E.; DeLuca, A.P.; Hildebrand, M.S.; Taylor, K.R.; Gurrola, J., 2nd; Scherer, S.; Scheetz, T.E.; Smith, R.J. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2010, 107, 21104–21109. [Google Scholar]
- Wormald, R.; Viani, L.; Lynch, S.A.; Green, A.J. Sensorineural hearing loss in children. Ir. Med. J. 2010, 103, 51–54. [Google Scholar]
- Yang, T.; Kersigo, J.; Jahan, I.; Pan, N.; Fritzsch, B. The molecular basis of making spiral ganglion neurons and connecting them to hair cells of the organ of Corti. Hear. Res. 2011. [Google Scholar] [CrossRef]
- Pan, N.; Jahan, I.; Kersigo, J.; Kopecky, B.; Santi, P.; Johnson, S.; Schmitz, H.; Fritzsch, B. Conditional deletion of Atoh1 using Pax2-Cre results in viable mice without differentiated cochlear hair cells that have lost most of the organ of Corti. Hear. Res. 2010, 275, 66–80. [Google Scholar]
- Hu, Z.; Ulfendahl, M.; Prieskorn, D.M.; Olivius, P.; Miller, J.M. Functional evaluation of a cell replacement therapy in the inner ear. Otol. Neurotol. 2009, 30, 551–558. [Google Scholar]
- Leake, P.A.; Hradek, G.T.; Bonham, B.H.; Snyder, R.L. Topography of auditory nerve projections to the cochlear nucleus in cats after neonatal deafness and electrical stimulation by a cochlear implant. J. Assoc. Res. Otolaryngol. 2008, 9, 349–372. [Google Scholar]
- Fallon, J.B.; Irvine, D.R.; Shepherd, R.K. Cochlear implants and brain plasticity. Hear. Res. 2008, 238, 110–117. [Google Scholar]
- Ruttiger, L.; Panford-Walsh, R.; Schimmang, T.; Tan, J.; Zimmermann, U.; Rohbock, K.; Kopschall, I.; Limberger, A.; Muller, M.; Fraenzer, J.T.; Cimerman, J.; Knipper, M. BDNF mRNA expression and protein localization are changed in age-related hearing loss. Neurobiol. Aging 2007, 28, 586–601. [Google Scholar]
- Webber, A.; Raz, Y. Axon guidance cues in auditory development. Anat. Rec. A Discov. Mol. Cell Evol. Biol. 2006, 288, 390–396. [Google Scholar]
- Mantela, J.; Jiang, Z.; Ylikoski, J.; Fritzsch, B.; Zacksenhaus, E.; Pirvola, U. The retinoblastoma gene pathway regulates the postmitotic state of hair cells of the mouse inner ear. Development 2005, 132, 2377–2388. [Google Scholar]
- Frucht, C.S.; Santos-Sacchi, J.; Navaratnam, D.S. MicroRNA181a plays a key role in hair cell regeneration in the avian auditory epithelium. Neurosci. Lett. 2011, 493, 44–48. [Google Scholar]
- Saunders, J.C. The role of hair cell regeneration in an avian model of inner ear injury and repair from acoustic trauma. ILAR J. 2010, 51, 326–337. [Google Scholar]
- Warchol, M.E. Sensory regeneration in the vertebrate inner ear: Differences at the levels of cells and species. Hear. Res. 2010. [Google Scholar]
- Slattery, E.L.; Warchol, M.E. Cisplatin ototoxicity blocks sensory regeneration in the avian inner ear. J. Neurosci. 2010, 30, 3473–3481. [Google Scholar]
- Bird, J.E.; Daudet, N.; Warchol, M.E.; Gale, J.E. Supporting cells eliminate dying sensory hair cells to maintain epithelial integrity in the avian inner ear. J. Neurosci. 2010, 30, 12545–12556. [Google Scholar]
- Kwan, T.; White, P.M.; Segil, N. Development and regeneration of the inner ear. Ann. N.Y. Acad. Sci. 2009, 1170, 28–33. [Google Scholar]
- Brignull, H.R.; Raible, D.W.; Stone, J.S. Feathers and fins: non-mammalian models for hair cell regeneration. Brain Res. 2009, 1277, 12–23. [Google Scholar]
- Daudet, N.; Gibson, R.; Shang, J.; Bernard, A.; Lewis, J.; Stone, J. Notch regulation of progenitor cell behavior in quiescent and regenerating auditory epithelium of mature birds. Dev. Biol. 2009, 326, 86–100. [Google Scholar]
- Liu, Z.; Zuo, J. Cell cycle regulation in hair cell development and regeneration in the mouse cochlea. Cell Cycle 2008, 7, 2129–2133. [Google Scholar]
- Cotanche, D.A. Genetic and pharmacological intervention for treatment/prevention of hearing loss. J. Commun. Disord. 2008, 41, 421–443. [Google Scholar]
- Cotanche, D.A.; Kaiser, C.L. Hair cell fate decisions in cochlear development and regeneration. Hear. Res. 2010, 266, 18–25. [Google Scholar]
- Oshima, K.; Shin, K.; Diensthuber, M.; Peng, A.W.; Ricci, A.J.; Heller, S. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell 2010, 141, 704–716. [Google Scholar]
- Hildebrand, M.S.; Newton, S.S.; Gubbels, S.P.; Sheffield, A.M.; Kochhar, A.; de Silva, M.G.; Dahl, H.H.; Rose, S.D.; Behlke, M.A.; Smith, R.J. Advances in molecular and cellular therapies for hearing loss. Mol. Ther. 2008, 16, 224–236. [Google Scholar]
- Gubbels, S.P.; Woessner, D.W.; Mitchell, J.C.; Ricci, A.J.; Brigande, J.V. Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature 2008, 455, 537–541. [Google Scholar]
- Praetorius, M.; Hsu, C.; Baker, K.; Brough, D.E.; Plinkert, P.; Staecker, H. Adenovector-mediated hair cell regeneration is affected by promoter type. Acta Otolaryngol. 2010, 130, 215–222. [Google Scholar]
- Groves, A.K. The challenge of hair cell regeneration. Exp. Biol. Med. (Maywood) 2010, 235, 434–446. [Google Scholar]
- White, P.M.; Doetzlhofer, A.; Lee, Y.S.; Groves, A.K.; Segil, N. Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature 2006, 441, 984–987. [Google Scholar]
- Batts, S.A.; Shoemaker, C.R.; Raphael, Y. Notch signaling and Hes labeling in the normal and drug-damaged organ of Corti. Hear. Res. 2009, 249, 15–22. [Google Scholar]
- Izumikawa, M.; Batts, S.A.; Miyazawa, T.; Swiderski, D.L.; Raphael, Y. Response of the flat cochlear epithelium to forced expression of Atoh1. Hear. Res. 2008, 240, 52–56. [Google Scholar]
- Matei, V.; Pauley, S.; Kaing, S.; Rowitch, D.; Beisel, K.W.; Morris, K.; Feng, F.; Jones, K.; Lee, J.; Fritzsch, B. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev. Dyn. 2005, 234, 633–650. [Google Scholar]
- Ruben, R.J. Development of the inner ear of the mouse: A radioautographic study of terminal mitoses. Acta Otolaryngol. 1967, 220 Suppl, 1–44. [Google Scholar]
- Lee, Y.S.; Liu, F.; Segil, N. A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development 2006, 133, 2817–2826. [Google Scholar]
- Rocha-Sanchez, S.; Scheetz, L.; Contreras, M.; Weston, M.; Korte, M.; McGee, J.; Walsh, E.J. Mature mice lacking Rbl2/p130 gene have supernumery inner ear hair cells and supporting cells. J. Neurosci. 2011. In press. [Google Scholar]
- Weber, T.; Corbett, M.K.; Chow, L.M.; Valentine, M.B.; Baker, S.J.; Zuo, J. Rapid cell-cycle reentry and cell death after acute inactivation of the retinoblastoma gene product in postnatal cochlear hair cells. Proc. Natl. Acad. Sci. USA 2008, 105, 781–785. [Google Scholar]
- Chen, P.; Johnson, J.E.; Zoghbi, H.Y.; Segil, N. The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development 2002, 129, 2495–2505. [Google Scholar]
- Bermingham, N.A.; Hassan, B.A.; Price, S.D.; Vollrath, M.A.; Ben-Arie, N.; Eatock, R.A.; Bellen, H.J.; Lysakowski, A.; Zoghbi, H.Y. Math1: An essential gene for the generation of inner ear hair cells. Science 1999, 284, 1837–1841. [Google Scholar]
- Dabdoub, A.; Puligilla, C.; Jones, J.M.; Fritzsch, B.; Cheah, K.S.; Pevny, L.H.; Kelley, M.W. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc. Natl. Acad. Sci. USA 2008, 105, 18396–18401. [Google Scholar]
- Fritzsch, B.; Eberl, D.F.; Beisel, K.W. The role of bHLH genes in ear development and evolution: Revisiting a 10-year-old hypothesis. Cell Mol. Life Sci. 2010, 67, 3089–3099. [Google Scholar]
- Jahan, I.; Pan, N.; Kersigo, J.; Fritzsch, B. Neurod1 suppresses hair cell differentiation in ear ganglia and regulates hair cell subtype development in the cochlea. PLoS One 2010, 5, e11661. [Google Scholar]
- Roybon, L.; Hjalt, T.; Stott, S.; Guillemot, F.; Li, J.Y.; Brundin, P. Neurogenin2 directs granule neuroblast production and amplification while NeuroD1 specifies neuronal fate during hippocampal neurogenesis. PLoS One 2009, 4, e4779. [Google Scholar]
- Brooker, R.; Hozumi, K.; Lewis, J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development 2006, 133, 1277–1286. [Google Scholar]
- Kiernan, A.E.; Xu, J.; Gridley, T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2006, 2, e4. [Google Scholar]
- Fritzsch, B.; Beisel, K.W.; Hansen, L.A. The molecular basis of neurosensory cell formation in ear development: A blueprint for hair cell and sensory neuron regeneration? Bioessays 2006, 28, 1181–1193. [Google Scholar]
- Slattery, E.L.; Speck, J.D.; Warchol, M.E. Epigenetic influences on sensory regeneration: Histone deacetylases regulate supporting cell proliferation in the avian utricle. J. Assoc. Res. Otolaryngol. 2009, 10, 341–353. [Google Scholar]
- Batts, S.A.; Raphael, Y. Transdifferentiation and its applicability for inner ear therapy. Hear. Res. 2007, 227, 41–47. [Google Scholar]
- Mutai, H.; Nagashima, R.; Sugitani, Y.; Noda, T.; Fujii, M.; Matsunaga, T. Expression of Pou3f3/Brn-1 and its genomic methylation in developing auditory epithelium. Dev. Neurobiol. 2009, 69, 913–930. [Google Scholar]
- Frye, M.; Fisher, A.G.; Watt, F.M. Epidermal stem cells are defined by global histone modifications that are altered by Myc-induced differentiation. PLoS One 2007, 2, e763. [Google Scholar]
- Brigande, J.V.; Heller, S. Quo vadis, hair cell regeneration? Nat. Neurosci. 2009, 12, 679–685. [Google Scholar]
- Lou, X.; Zhang, Y.; Yuan, C. Multipotent stem cells from the young rat inner ear. Neurosci. Lett. 2007, 416, 28–33. [Google Scholar]
- Kim, Y.H.; Raphael, Y. Cell division and maintenance of epithelial integrity in the deafened auditory epithelium. Cell Cycle 2007, 6, 612–619. [Google Scholar]
- Kopecky, B.; Santi, P.; Johnson, S.; Schmitz, H.; Fritzsch, B. Conditional deletion of N-Myc disrupts neurosensory and non-sensory development of the ear. Dev. Dyn. 2011, 240, 1373–1390. [Google Scholar]
- Dominguez-Frutos, E.; Lopez-Hernandez, I.; Vendrell, V.; Neves, J.; Gallozzi, M.; Gutsche, K.; Quintana, L.; Sharpe, J.; Knoepfler, P.S.; Eisenman, R.; et al. N-myc controls proliferation, morphogenesis and patterning of the inner ear. J. Neurosci. 2011, 31, 7178–7189. [Google Scholar]
- Vlastarakos, P.V.; Nikolopoulos, T.P.; Tavoulari, E.; Papacharalambous, G.; Tzagaroulakis, A.; Dazert, S. Sensory cell regeneration and stem cells: What we have already achieved in the management of deafness. Otol. Neurotol. 2008, 29, 758–768. [Google Scholar]
- Coleman, B.; de Silva, M.G.; Shepherd, R.K. Concise review: The potential of stem cells for auditory neuron generation and replacement. Stem Cells 2007, 25, 2685–2694. [Google Scholar]
- Nakagawa, T.; Ito, J. Cell therapy for inner ear diseases. Curr. Pharm. Des. 2005, 11, 1203–1207. [Google Scholar]
- Li, H.; Corrales, C.E.; Edge, A.; Heller, S. Stem cells as therapy for hearing loss. Trends Mol. Med. 2004, 10, 309–315. [Google Scholar]
- Ridky, T.W.; Khavari, P.A. The hair follicle bulge stem cell niche resists transformation by the hedgehog pathway. Cell Stem Cell 6, 292–294.
- Appleford, P.J.; Woollard, A. RUNX genes find a niche in stem cell biology. J. Cell Biochem. 2009, 108, 14–21. [Google Scholar]
- Nishikawa, S.I.; Osawa, M.; Yonetani, S.; Torikai-Nishikawa, S.; Freter, R. Niche required for inducing quiescent stem cells. Cold Spring Harb. Symp. Quant. Biol. 2008, 73, 67–71. [Google Scholar]
- Mitsiadis, T.A.; Barrandon, O.; Rochat, A.; Barrandon, Y.; de Bari, C. Stem cell niches in mammals. Exp. Cell. Res. 2007, 313, 3377–3385. [Google Scholar]
- Braun, K.M.; Prowse, D.M. Distinct epidermal stem cell compartments are maintained by independent niche microenvironments. Stem Cell Rev. 2006, 2, 221–231. [Google Scholar]
- Moore, K.A.; Lemischka, I.R. Stem cells and their niches. Science 2006, 311, 1880–1885. [Google Scholar]
- Li, L.; Xie, T. Stem cell niche: Structure and function. Annu. Rev. Cell Dev. Biol. 2005, 21, 605–631. [Google Scholar]
- Parker, M.A. Biotechnology in the treatment of sensorineural hearing loss: Foundations and future of hair cell regeneration. J. Speech Lang. Hear. Res. 2011. [Google Scholar]
- Pan, N.; Jahan, I.; Kersigo, J.; Kopecky, B.; Santi, P.; Johnson, S.; Schmitz, H.; Fritzsch, B. Conditional deletion of Atoh1 using Pax2-Cre results in viable mice without differentiated cochlear hair cells that have lost most of the organ of Corti. Hear. Res. 2011, 275, 66–80. [Google Scholar]
- Hwang, N.S.; Zhang, C.; Hwang, Y.S.; Varghese, S. Mesenchymal stem cell differentiation and roles in regenerative medicine. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009, 1, 97–106. [Google Scholar]
- Young, H.E.; Black, A.C., Jr. Adult stem cells. Anat. Rec. A Discov. Mol. Cell Evol. Biol. 2004, 276, 75–102. [Google Scholar]
- Hayflick, L. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 1965, 37, 614–636. [Google Scholar]
- El Seady, R.; Huisman, M.A.; Lowik, C.W.; Frijns, J.H. Uncomplicated differentiation of stem cells into bipolar neurons and myelinating glia. Biochem. Biophys. Res. Commun. 2008, 376, 358–362. [Google Scholar]
- Krejci, E.; Grim, M. Isolation and characterization of neural crest stem cells from adult human hair follicles. Folia Biol. (Praha) 2010, 56, 149–157. [Google Scholar]
- Sieber-Blum, M. Epidermal neural crest stem cells and their use in mouse models of spinal cord injury. Brain Res. Bull. 2010, 83, 189–193. [Google Scholar]
- Sieber-Blum, M.; Hu, Y. Epidermal neural crest stem cells (EPI-NCSC) and pluripotency. Stem Cell Rev. 2008, 4, 256–260. [Google Scholar]
- Sulewski, R.; Kirsner, R.S. The multipotent nature of hair bulge cells. J. Invest. Dermatol. 2010, 130, 1198. [Google Scholar]
- Zhang, Y.V.; White, B.S.; Shalloway, D.I.; Tumbar, T. Stem cell dynamics in mouse hair follicles: A story from cell division counting and single cell lineage tracing. Cell Cycle 2010, 9, 1504–1510. [Google Scholar]
- Sieber-Blum, M.; Grim, M. The adult hair follicle: cradle for pluripotent neural crest stem cells. Birth Defects Res. C Embryo Today 2004, 72, 162–172. [Google Scholar]
- Sieber-Blum, M.; Schnell, L.; Grim, M.; Hu, Y.F.; Schneider, R.; Schwab, M.E. Characterization of epidermal neural crest stem cell (EPI-NCSC) grafts in the lesioned spinal cord. Mol. Cell Neurosci. 2006, 32, 67–81. [Google Scholar]
- Duncan, J.; Lim, K.L.; Engel, J.D.; Fritzsch, B. Limited vestibular epithelia can develop in the absence of GATA3. Int. J. Dev. Bio. 2011. In Press. [Google Scholar]
- Karis, A.; Pata, I.; van Doorninck, J.H.; Grosveld, F.; de Zeeuw, C.I.; de Caprona, D.; Fritzsch, B. Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morphogenesis of the ear. J. Comp. Neurol. 2001, 429, 615–630. [Google Scholar]
- Ito, J.; Kojima, K.; Kawaguchi, S. Survival of neural stem cells in the cochlea. Acta Otolaryngol. 2001, 121, 140–142. [Google Scholar]
- Parker, M.A.; Corliss, D.A.; Gray, B.; Anderson, J.K.; Bobbin, R.P.; Snyder, E.Y.; Cotanche, D.A. Neural stem cells injected into the sound-damaged cochlea migrate throughout the cochlea and express markers of hair cells, supporting cells, and spiral ganglion cells. Hear. Res. 2007, 232, 29–43. [Google Scholar]
- Wei, D.; Levic, S.; Nie, L.; Gao, W.Q.; Petit, C.; Jones, E.G.; Yamoah, E.N. Cells of adult brain germinal zone have properties akin to hair cells and can be used to replace inner ear sensory cells after damage. Proc. Natl. Acad. Sci. USA 2008, 105, 21000–21005. [Google Scholar]
- Slack, J. Skinny dipping for stem cells. Nat. Cell Biol. 2001, 3, E205–E206. [Google Scholar]
- Hunt, D.P.; Jahoda, C.; Chandran, S. Multipotent skin-derived precursors: From biology to clinical translation. Curr. Opin. Biotechnol. 2009, 20, 522–530. [Google Scholar]
- Rak, K.; Wasielewski, N.V.; Radeloff, A.; Volkers, J.; Scherzed, A.; Jablonka, S.; Hagen, R.; Mlynski, R. Isolation and characterization of neural stem cells from the neonatal rat cochlear nucleus. Cell Tissue Res. 2011, 343, 499–508. [Google Scholar]
- Zhou, H.; Wu, S.; Joo, J.Y.; Zhu, S.; Han, D.W.; Lin, T.; Trauger, S.; Bien, G.; Yao, S.; Zhu, Y.; et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 2009, 4, 381–384. [Google Scholar]
- Yamanaka, S. Elite and stochastic models for induced pluripotent stem cell generation. Nature 2009, 460, 49–52. [Google Scholar]
- Beisel, K.; Hansen, L.; Soukup, G.; Fritzsch, B. Regenerating cochlear hair cells: Quo vadis stem cell. Cell Tissue Res. 2008, 333, 373–379. [Google Scholar]
- Yamanaka, S.; Takahashi, K. Induction of pluripotent stem cells from mouse fibroblast cultures. Tanpakushitsu Kakusan Koso 2006, 51, 2346–2351. [Google Scholar]
- Robbins, R.D.; Prasain, N.; Maier, B.F.; Yoder, M.C.; Mirmira, R.G. Inducible pluripotent stem cells: Not quite ready for prime time? Curr. Opin. Organ Transplant 2010, 15, 61–67. [Google Scholar]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar]
- Niibe, K.; Kawamura, Y.; Araki, D.; Morikawa, S.; Miura, K.; Suzuki, S.; Shimmura, S.; Sunabori, T.; Mabuchi, Y.; Nagai, Y.; et al. Purified mesenchymal stem cells are an efficient source for iPS cell induction. PLoS One 2011, 6, e17610. [Google Scholar]
- Wang, Y.; Adjaye, J. A Cyclic AMP analog, 8-Br-cAMP, enhances the induction of pluripotency in human fibroblast cells. Stem Cell Rev. 2010, 7, 331–341. [Google Scholar]
- Okita, K.; Yamanaka, S. Induction of pluripotency by defined factors. Exp. Cell Res. 2010, 316, 2565–2570. [Google Scholar]
- Masip, M.; Veiga, A.; Izpisua Belmonte, J.C.; Simon, C. Reprogramming with defined factors: From induced pluripotency to induced transdifferentiation. Mol. Hum. Reprod. 2010, 16, 856–868. [Google Scholar]
- Tani, J.; Umbas, R. Recent progressions in stem cell research: breakthroughs achieved and challenges faced. Acta Med. Indones. 2009, 41, 30–35. [Google Scholar]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar]
- Rosa, A.; Brivanlou, A.H. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011, 30, 237–248. [Google Scholar]
- Fadel, H.E. Developments in stem cell research and therapeutic cloning: Islamic ethical positions, a review. Bioethics 2010. [Google Scholar] [CrossRef]
- Chester, R.; Sackstein, R. Embryonic stem cell-based therapeutics: Balancing scientific progress and bioethics. Health Matrix Clevel. 2010, 20, 203–217. [Google Scholar]
- Block, W. A libertarian perspective on the stem cell debate: Compromising the uncompromisible. J. Med. Philos. 2010, 35, 429–448. [Google Scholar]
- Lui, K.O.; Waldmann, H.; Fairchild, P.J. Embryonic stem cells: Overcoming the immunological barriers to cell replacement therapy. Curr. Stem Cell Res. Ther. 2009, 4, 70–80. [Google Scholar]
- Wang, Y.; Mah, N.; Prigione, A.; Wolfrum, K.; Andrade-Navarro, M.A.; Adjaye, J. A transcriptional roadmap to the induction of pluripotency in somatic cells. Stem Cell Rev. 2010, 6, 282–296. [Google Scholar]
- Prigione, A.; Adjaye, J. Modulation of mitochondrial biogenesis and bioenergetic metabolism upon in vitro and in vivo differentiation of human ES and iPS cells. Int. J. Dev. Biol. 2010, 54, 1729–1741. [Google Scholar]
- Zhao, T.; Zhang, Z.N.; Rong, Z.; Xu, Y. Immunogenicity of induced pluripotent stem cells. Nature 2011, 474, 2112–215. [Google Scholar]
- Hu, Z.; Andang, M.; Ni, D.; Ulfendahl, M. Neural cograft stimulates the survival and differentiation of embryonic stem cells in the adult mammalian auditory system. Brain Res. 2005, 1051, 137–144. [Google Scholar]
- Lang, H.; Schulte, B.A.; Goddard, J.C.; Hedrick, M.; Schulte, J.B.; Wei, L.; Schmiedt, R.A. Transplantation of mouse embryonic stem cells into the cochlea of an auditory-neuropathy animal model: Effects of timing after injury. J. Assoc. Res. Otolaryngol. 2008, 9, 225–240. [Google Scholar]
- Corrales, C.E.; Pan, L.; Li, H.; Liberman, M.C.; Heller, S.; Edge, A.S. Engraftment and differentiation of embryonic stem cell-derived neural progenitor cells in the cochlear nerve trunk: Growth of processes into the organ of Corti. J. Neurobiol. 2006, 66, 1489–1500. [Google Scholar]
- Martinez-Monedero, R.; Corrales, C.E.; Cuajungco, M.P.; Heller, S.; Edge, A.S. Reinnervation of hair cells by auditory neurons after selective removal of spiral ganglion neurons. J. Neurobiol. 2006, 66, 319–331. [Google Scholar]
- Okano, T.; Nakagawa, T.; Endo, T.; Kim, T.S.; Kita, T.; Tamura, T.; Matsumoto, M.; Ohno, T.; Sakamoto, T.; Iguchi, F.; et al. Engraftment of embryonic stem cell-derived neurons into the cochlear modiolus. Neuroreport 2005, 16, 1919–1922. [Google Scholar]
- Shi, F.; Corrales, C.E.; Liberman, M.C.; Edge, A.S. BMP4 induction of sensory neurons from human embryonic stem cells and reinnervation of sensory epithelium. Eur. J. Neurosci. 2007, 26, 3016–3023. [Google Scholar]
- Sakamoto, T.; Nakagawa, T.; Endo, T.; Kim, T.S.; Iguchi, F.; Naito, Y.; Sasai, Y.; Ito, J. Fates of mouse embryonic stem cells transplanted into the inner ears of adult mice and embryonic chickens. Acta Otolaryngol. Suppl. 2004, 515, 48–52. [Google Scholar]
- Tan, B.T.; Lee, M.M.; Ruan, R. Bone-marrow-derived cells that home to acoustic deafened cochlea preserved their hematopoietic identity. J. Comp. Neurol. 2008, 509, 167–179. [Google Scholar]
- Tang, F.; Barbacioru, C.; Bao, S.; Lee, C.; Nordman, E.; Wang, X.; Lao, K.; Surani, M.A. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell 2010, 6, 468–478. [Google Scholar]
- Urness, L.D.; Paxton, C.N.; Wang, X.; Schoenwolf, G.C.; Mansour, S.L. FGF signaling regulates otic placode induction and refinement by controlling both ectodermal target genes and hindbrain Wnt8a. Dev. Biol. 2010, 340, 595–604. [Google Scholar]
- Lillevali, K.; Haugas, M.; Matilainen, T.; Pussinen, C.; Karis, A.; Salminen, M. Gata3 is required for early morphogenesis and Fgf10 expression during otic development. Mech. Dev. 2006, 123, 415–429. [Google Scholar]
- Mansour, S.L.; Goddard, J.M.; Capecchi, M.R. Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development 1993, 117, 13–28. [Google Scholar]
- Hatch, E.P.; Noyes, C.A.; Wang, X.; Wright, T.J.; Mansour, S.L. Fgf3 is required for dorsal patterning and morphogenesis of the inner ear epithelium. Development 2007, 134, 3615–3625. [Google Scholar]
- Bok, J.; Chang, W.; Wu, D.K. Patterning and morphogenesis of the vertebrate inner ear. Int. J. Dev. Biol. 2007, 51, 521–533. [Google Scholar]
- Riccomagno, M.M.; Takada, S.; Epstein, D.J. Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes Dev. 2005, 19, 1612–1623. [Google Scholar]
- Choo, D.; Ward, J.; Reece, A.; Dou, H.; Lin, Z.; Greinwald, J. Molecular mechanisms underlying inner ear patterning defects in kreisler mutants. Dev. Biol. 2006, 289, 308–317. [Google Scholar]
- Streit, A. The preplacodal region: An ectodermal domain with multipotential progenitors that contribute to sense organs and cranial sensory ganglia. Int. J. Dev. Biol. 2007, 51, 447–461. [Google Scholar]
- Ohyama, T.; Groves, A.K. Expression of mouse Foxi class genes in early craniofacial development. Dev. Dyn. 2004, 231, 640–646. [Google Scholar]
- Bouchard, M.; de Caprona, D.; Busslinger, M.; Xu, P.; Fritzsch, B. Pax2 and Pax8 cooperate in mouse inner ear morphogenesis and innervation. BMC Dev. Biol. 2010, 10, 89. [Google Scholar]
- Kiernan, A.E.; Pelling, A.L.; Leung, K.K.; Tang, A.S.; Bell, D.M.; Tease, C.; Lovell-Badge, R.; Steel, K.P.; Cheah, K.S. Sox2 is required for sensory organ development in the mammalian inner ear. Nature 2005, 434, 1031–1035. [Google Scholar]
- Milo, M.; Cacciabue-Rivolta, D.; Kneebone, A.; van Doorninck, H.; Johnson, C.; Lawoko-Kerali, G.; Niranjan, M.; Rivolta, M.; Holley, M. Genomic analysis of the function of the transcription factor gata3 during development of the mammalian inner ear. PLoS One 2009, 4, e7144. [Google Scholar]
- Nichols, D.H.; Pauley, S.; Jahan, I.; Beisel, K.W.; Millen, K.J.; Fritzsch, B. Lmx1a is required for segregation of sensory epithelia and normal ear histogenesis and morphogenesis. Cell Tissue Res. 2008, 334, 339–358. [Google Scholar]
- Pauley, S.; Lai, E.; Fritzsch, B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev. Dyn. 2006, 235, 2470–2482. [Google Scholar]
- Bok, J.; Dolson, D.K.; Hill, P.; Ruther, U.; Epstein, D.J.; Wu, D.K. Opposing gradients of Gli repressor and activators mediate Shh signaling along the dorsoventral axis of the inner ear. Development 2007, 134, 1713–1722. [Google Scholar]
- Riccomagno, M.M.; Martinu, L.; Mulheisen, M.; Wu, D.K.; Epstein, D.J. Specification of the mammalian cochlea is dependent on Sonic hedgehog. Genes Dev. 2002, 16, 2365–2378. [Google Scholar]
- Zheng, W.; Huang, L.; Wei, Z.B.; Silvius, D.; Tang, B.; Xu, P.X. The role of Six1 in mammalian auditory system development. Development 2003, 130, 3989–4000. [Google Scholar]
- Ma, Q.; Chen, Z.; Del Barco Barrantes, I.; de la Pompa, J.L.; Anderson, D.J. Neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron 1998, 20, 469–482. [Google Scholar]
- Ma, Q.; Anderson, D.J.; Fritzsch, B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J. Assoc. Res. Otolaryngol. 2000, 1, 129–143. [Google Scholar]
- Liu, M.; Pereira, F.A.; Price, S.D.; Chu, M.J.; Shope, C.; Himes, D.; Eatock, R.A.; Brownell, W.E.; Lysakowski, A.; Tsai, M.J. Essential role of BETA2/NeuroD1 in development of the vestibular and auditory systems. Genes Dev. 2000, 14, 2839–2854. [Google Scholar]
- Jahan, I.; Kersigo, J.; Pan, N.; Fritzsch, B. Neurod1 regulates survival and formation of connections in mouse ear and brain. Cell Tissue Res. 2010, 341, 95–110. [Google Scholar]
- Fritzsch, B.; Beisel, K.W. Molecular conservation and novelties in vertebrate ear development. Curr. Top Dev. Biol. 2003, 57, 1–44. [Google Scholar]
- Kim, W.Y.; Fritzsch, B.; Serls, A.; Bakel, L.A.; Huang, E.J.; Reichardt, L.F.; Barth, D.S.; Lee, J.E. NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development 2001, 128, 417–426. [Google Scholar]
- Fritzsch, B.; Tessarollo, L.; Coppola, E.; Reichardt, L.F. Neurotrophins in the ear: Their roles in sensory neuron survival and fiber guidance. Prog. Brain Res. 2004, 146, 265–278. [Google Scholar]
- Fekete, D.M.; Campero, A.M. Axon guidance in the Inner Ear; University of the Basque Country Press: Bilbao, ESPAGNE, 2007; pp. 549–556. [Google Scholar]
- Tessarollo, L.; Coppola, V.; Fritzsch, B. NT-3 replacement with brain-derived neurotrophic factor redirects vestibular nerve fibers to the cochlea. J. Neurosci. 2004, 24, 2575–2584. [Google Scholar]
- Sciarretta, C.; Fritzsch, B.; Beisel, K.; Rocha-Sanchez, S.M.; Buniello, A.; Horn, J.M.; Minichiello, L. PLCgamma-activated signalling is essential for TrkB mediated sensory neuron structural plasticity. BMC Dev. Biol. 2010, 10, 103. [Google Scholar]
- Bermingham, N.A.; Hassan, B.A.; Price, S.D.; Vollrath, M.A.; Ben-Arie, N.; Eatock, R.A.; Bellen, H.J.; Lysakowski, A.; Zoghbi, H.Y. Math1: An essential gene for the generation of inner ear hair cells. Science 1999, 284, 1837–1841. [Google Scholar]
- Fritzsch, B.; Matei, V.A.; Nichols, D.H.; Bermingham, N.; Jones, K.; Beisel, K.W.; Wang, V.Y. Atoh1 null mice show directed afferent fiber growth to undifferentiated ear sensory epithelia followed by incomplete fiber retention. Dev. Dyn. 2005, 233, 570–583. [Google Scholar]
- Zine, A.; Aubert, A.; Qiu, J.; Therianos, S.; Guillemot, F.; Kageyama, R.; de Ribaupierre, F. Hes1 and Hes5 activities are required for the normal development of the hair cells in the mammalian inner ear. J. Neurosci. 2001, 21, 4712–4720. [Google Scholar]
- Doetzlhofer, A.; Basch, M.L.; Ohyama, T.; Gessler, M.; Groves, A.K.; Segil, N. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev. Cell 2009, 16, 58–69. [Google Scholar]
- Ohyama, T.; Basch, M.L.; Mishina, Y.; Lyons, K.M.; Segil, N.; Groves, A.K. BMP signaling is necessary for patterning the sensory and nonsensory regions of the developing mammalian cochlea. J. Neurosci. 2010, 30, 15044–15051. [Google Scholar]
- Xiang, M.; Maklad, A.; Pirvola, U.; Fritzsch, B. Brn3c null mutant mice show long-term, incomplete retention of some afferent inner ear innervation. BMC Neurosci. 2003, 4, 2. [Google Scholar]
- Erkman, L.; McEvilly, R.J.; Luo, L.; Ryan, A.K.; Hooshmand, F.; O'Connell, S.M.; Keithley, E.M.; Rapaport, D.H.; Ryan, A.F.; Rosenfeld, M.G. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature 1996, 381, 603–606. [Google Scholar]
- Li, S.; Price, S.M.; Cahill, H.; Ryugo, D.K.; Shen, M.M.; Xiang, M. Hearing loss caused by progressive degeneration of cochlear hair cells in mice deficient for the Barhl1 homeobox gene. Development 2002, 129, 3523–3532. [Google Scholar]
- Wallis, D.; Hamblen, M.; Zhou, Y.; Venken, K.J.; Schumacher, A.; Grimes, H.L.; Zoghbi, H.Y.; Orkin, S.H.; Bellen, H.J. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development 2003, 130, 221–232. [Google Scholar]
- Chellappa, R.; Li, S.; Pauley, S.; Jahan, I.; Jin, K.; Xiang, M. Barhl1 regulatory sequences required for cell-specific gene expression and autoregulation in the inner ear and central nervous system. Mol. Cell. Biol. 2008, 28, 1905–1914. [Google Scholar]
- Hertzano, R.; Montcouquiol, M.; Rashi-Elkeles, S.; Elkon, R.; Yucel, R.; Frankel, W.N.; Rechavi, G.; Moroy, T.; Friedman, T.B.; Kelley, M.W.; Avraham, K.B. Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum. Mol. Genet. 2004, 13, 2143–2153. [Google Scholar]
- Soukup, G.A.; Fritzsch, B.; Pierce, M.L.; Weston, M.D.; Jahan, I.; McManus, M.T.; Harfe, B.D. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev. Biol. 2009, 328, 328–341. [Google Scholar]
- Mencia, A.; Modamio-Hoybjor, S.; Redshaw, N.; Morin, M.; Mayo-Merino, F.; Olavarrieta, L.; Aguirre, L.A.; Del Castillo, I.; Steel, K.P.; Dalmay, T.; Moreno, F.; Moreno-Pelayo, M.A. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009, 41, 609–613. [Google Scholar]
- Xu, N.; Papagiannakopoulos, T.; Pan, G.; Thomson, J.A.; Kosik, K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 2009, 137, 647–658. [Google Scholar]
- Friedman, L.M.; Dror, A.A.; Mor, E.; Tenne, T.; Toren, G.; Satoh, T.; Biesemeier, D.J.; Shomron, N.; Fekete, D.M.; Hornstein, E.; et al. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc. Natl. Acad. Sci. USA 2009, 106, 7915–7920. [Google Scholar]
- Weston, M.D.; Pierce, M.L.; Rocha-Sanchez, S.; Beisel, K.W.; Soukup, G.A. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006, 1111, 95–104. [Google Scholar]
- Kersigo, J.; D'Angelo, A.; Gray, B.D.; Soukup, G.A.; Fritzsch, B. The role of sensory organs and the forebrain for the development of the craniofacial shape as revealed by Foxg1-cre-mediated microRNA loss. Genesis 2011. [Google Scholar]
- Young, R.A. Control of the embryonic stem cell state. Cell 2011, 144, 940–954. [Google Scholar]
- Silva, J.; Barrandon, O.; Nichols, J.; Kawaguchi, J.; Theunissen, T.W.; Smith, A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008, 6, e253. [Google Scholar]
- Jaenisch, R.; Young, R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 2008, 132, 567–582. [Google Scholar]
- Pera, M.F.; Tam, P.P. Extrinsic regulation of pluripotent stem cells. Nature 2010, 465, 713–720. [Google Scholar]
- Yamanaka, S.; Blau, H.M. Nuclear reprogramming to a pluripotent state by three approaches. Nature 2010, 465, 704–712. [Google Scholar]
- Marson, A.; Levine, S.S.; Cole, M.F.; Frampton, G.M.; Brambrink, T.; Johnstone, S.; Guenther, M.G.; Johnston, W.K.; Wernig, M.; Newman, J.; et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 2008, 134, 521–533. [Google Scholar]
- Kanellopoulou, C.; Muljo, S.A.; Kung, A.L.; Ganesan, S.; Drapkin, R.; Jenuwein, T.; Livingston, D.M.; Rajewsky, K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005, 19, 489–501. [Google Scholar]
- Murchison, E.P.; Partridge, J.F.; Tam, O.H.; Cheloufi, S.; Hannon, G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA 2005, 102, 12135–12140. [Google Scholar]
- Wang, Y.; Medvid, R.; Melton, C.; Jaenisch, R.; Blelloch, R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007, 39, 380–385. [Google Scholar]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Dyce, P.W.; Toms, D.; Li, J. Stem cells and germ cells: MicroRNA and gene expression signatures. Histol. Histopathol. 2010, 25, 505–513. [Google Scholar]
- Wu, Y.; Liu, S.; Xin, H.; Jiang, J.; Younglai, E.; Sun, S.; Wang, H. Up-regulation of microRNA-145 promotes differentiation by repressing OCT4 in human endometrial adenocarcinoma cells. Cancer 2011. [Google Scholar] [CrossRef]
- Riggi, N.; Suva, M.L.; de Vito, C.; Provero, P.; Stehle, J.C.; Baumer, K.; Cironi, L.; Janiszewska, M.; Petricevic, T.; Suva, D.; et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010, 24, 916–932. [Google Scholar]
- Otsubo, T.; Akiyama, Y.; Hashimoto, Y.; Shimada, S.; Goto, K.; Yuasa, Y. MicroRNA-126 inhibits SOX2 expression and contributes to gastric carcinogenesis. PLoS One 2011, 6, e16617. [Google Scholar]
- Xie, C.; Huang, H.; Sun, X.; Guo, Y.; Hamblin, M.; Ritchie, R.P.; Garcia-Barrio, M.T.; Zhang, J.; Chen, Y.E. MicroRNA-1 regulates smooth muscle cell differentiation by repressing Kruppel-like factor 4. Stem Cells Dev. 2011, 20, 205–210. [Google Scholar]
- Balazsi, G.; van Oudenaarden, A.; Collins, J.J. Cellular decision making and biological noise: From microbes to mammals. Cell 2011, 144, 910–925. [Google Scholar]
- Melton, C.; Blelloch, R. MicroRNA regulation of embryonic stem cell self-renewal and differentiation. Adv. Exp. Med. Biol. 2010, 695, 105–117. [Google Scholar]
- Cartwright, P.; McLean, C.; Sheppard, A.; Rivett, D.; Jones, K.; Dalton, S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 2005, 132, 885–896. [Google Scholar]
- Lin, C.H.; Jackson, A.L.; Guo, J.; Linsley, P.S.; Eisenman, R.N. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009, 28, 3157–3170. [Google Scholar]
- Judson, R.L.; Babiarz, J.E.; Venere, M.; Blelloch, R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol. 2009, 27, 459–461. [Google Scholar]
- O'Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar]
- Voorhoeve, P.M.; Le Sage, C.; Schrier, M.; Gillis, A.J.; Stoop, H.; Nagel, R.; Liu, Y.P.; van Duijse, J.; Drost, J.; Griekspoor, A.; et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 2006, 124, 1169–1181. [Google Scholar]
- Delaloy, C.; Liu, L.; Lee, J.A.; Su, H.; Shen, F.; Yang, G.Y.; Young, W.L.; Ivey, K.N.; Gao, F.B. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 2010, 6, 323–335. [Google Scholar]
- Krichevsky, A.M.; Sonntag, K.C.; Isacson, O.; Kosik, K.S. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 2006, 24, 857–864. [Google Scholar]
- Zhao, C.; Sun, G.; Li, S.; Shi, Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 2009, 16, 365–371. [Google Scholar]
- Zhang, L.; Stokes, N.; Polak, L.; Fuchs, E. Specific microRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment. Cell Stem Cell 2011, 8, 294–308. [Google Scholar]
- Lena, A.M.; Shalom-Feuerstein, R.; Rivetti di Val Cervo, P.; Aberdam, D.; Knight, R.A.; Melino, G.; Candi, E. MiR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008, 15, 1187–1195. [Google Scholar]
- Yi, R.; Poy, M.N.; Stoffel, M.; Fuchs, E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature 2008, 452, 225–229. [Google Scholar]
- Tay, Y.; Zhang, J.; Thomson, A.M.; Lim, B.; Rigoutsos, I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008, 455, 1124–1128. [Google Scholar]
- Tay, Y.M.; Tam, W.L.; Ang, Y.S.; Gaughwin, P.M.; Yang, H.; Wang, W.; Liu, R.; George, J.; Ng, H.H.; Perera, R.J.; et al. MicroRNA-134 modulates the differentiation of mouse embryonic stem cells, where it causes post-transcriptional attenuation of Nanog and LRH1. Stem Cells 2008, 26, 17–29. [Google Scholar]
- Cordes, K.R.; Sheehy, N.T.; White, M.P.; Berry, E.C.; Morton, S.U.; Muth, A.N.; Lee, T.H.; Miano, J.M.; Ivey, K.N.; Srivastava, D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009. [Google Scholar]
- Papagiannakopoulos, T.; Kosik, K.S. MicroRNA-124: Micromanager of neurogenesis. Cell Stem Cell 2009, 4, 375–376. [Google Scholar]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar]
- Wu, L.; Belasco, J.G. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 2005, 25, 9198–9208. [Google Scholar]
- Rybak, A.; Fuchs, H.; Smirnova, L.; Brandt, C.; Pohl, E.E.; Nitsch, R.; Wulczyn, F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell. Biol. 2008, 10, 987–993. [Google Scholar]
- Melton, C.; Judson, R.L.; Blelloch, R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 2010, 463, 621–626. [Google Scholar]
- Johnson, C.D.; Esquela-Kerscher, A.; Stefani, G.; Byrom, M.; Kelnar, K.; Ovcharenko, D.; Wilson, M.; Wang, X.; Shelton, J.; Shingara, J.; et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007, 67, 7713–7722. [Google Scholar]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA 2008, 105, 3903–3908. [Google Scholar]
- Schultz, J.; Lorenz, P.; Gross, G.; Ibrahim, S.; Kunz, M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008, 18, 549–557. [Google Scholar]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Muller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar]
- Weston, M.D.; Pierce, M.L.; Jensen-Smith, H.C.; Fritzsch, B.; Rocha-Sanchez, S.; Beisel, K.W.; Soukup, G.A. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev. Dyn. 2011, 240, 808–819. [Google Scholar]
- Soukup, G.A. Little but loud: Small RNAs have a resounding affect on ear development. Brain Res. 2009, 1277, 104–114. [Google Scholar]
- Pierce, M.L.; Weston, M.D.; Fritzsch, B.; Gabel, H.W.; Ruvkun, G.; Soukup, G.A. MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol. Dev. 2008, 10, 106–113. [Google Scholar]
- Weston, M.D.; Soukup, G.A. MicroRNAs sound off. Genome Med. 2009, 1, 59. [Google Scholar]
- Lewis, M.A.; Quint, E.; Glazier, A.M.; Fuchs, H.; de Angelis, M.H.; Langford, C.; van Dongen, S.; Abreu-Goodger, C.; Piipari, M.; Redshaw, N.; et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat. Genet. 2009, 41, 614–618. [Google Scholar]
- Li, H.; Kloosterman, W.; Fekete, D.M. MicroRNA-183 family members regulate sensorineural fates in the inner ear. J. Neurosci. 2010, 30, 3254–3263. [Google Scholar]
- Li, H.; Fekete, D.M. MicroRNAs in hair cell development and deafness. Curr. Opin. Otolaryngol. Head Neck Surg. 2010, 18, 459–465. [Google Scholar]
- Bueno, M.J.; Malumbres, M. MicroRNAs and the cell cycle. Biochim. Biophys. Acta 2011, 1812, 592–601. [Google Scholar]
- Sotillo, E.; Laver, T.; Mellert, H.; Schelter, J.M.; Cleary, M.A.; McMahon, S.; Thomas-Tikhonenko, A. Myc overexpression brings out unexpected antiapoptotic effects of miR-34a. Oncogene 2011. [Google Scholar]
- Bui, T.V.; Mendell, J.T. Myc: Maestro of MicroRNAs. Genes Cancer 2010, 1, 568–575. [Google Scholar]
- Smith, K.N.; Singh, A.M.; Dalton, S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell 2010, 7, 343–354. [Google Scholar]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010, 12, 247–256. [Google Scholar]
- Ivanovska, I.; Ball, A.S.; Diaz, R.L.; Magnus, J.F.; Kibukawa, M.; Schelter, J.M.; Kobayashi, S.V.; Lim, L.; Burchard, J.; Jackson, A.L.; et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell. Biol. 2008, 28, 2167–2174. [Google Scholar]
- Petrocca, F.; Visone, R.; Onelli, M.R.; Shah, M.H.; Nicoloso, M.S.; de Martino, I.; Iliopoulos, D.; Pilozzi, E.; Liu, C.G.; Negrini, M.; et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 2008, 13, 272–286. [Google Scholar]
- Bussing, I.; Slack, F.J.; Grosshans, H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008, 14, 400–409. [Google Scholar]
- Ivey, K.N.; Srivastava, D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell 2010, 7, 36–41. [Google Scholar]
- Rideout, W.M., 3rd; Hochedlinger, K.; Kyba, M.; Daley, G.Q.; Jaenisch, R. Correction of a genetic defect by nuclear transplantation and combined cell and gene therapy. Cell 2002, 109, 17–27. [Google Scholar]
- Bjorklund, L.M.; Sanchez-Pernaute, R.; Chung, S.; Andersson, T.; Chen, I.Y.; McNaught, K.S.; Brownell, A.L.; Jenkins, B.G.; Wahlestedt, C.; Kim, K.S.; et al. Embryonic stem cells develop into functional dopaminergic neurons after transplantation in a Parkinson rat model. Proc. Natl. Acad. Sci. USA 2002, 99, 2344–2349. [Google Scholar]
- Orlic, D. Adult bone marrow stem cells regenerate myocardium in ischemic heart disease. Ann. N.Y. Acad. Sci. 2003, 996, 152–157. [Google Scholar]
- Jackson, K.A.; Majka, S.M.; Wang, H.; Pocius, J.; Hartley, C.J.; Majesky, M.W.; Entman, M.L.; Michael, L.H.; Hirschi, K.K.; Goodell, M.A. Regeneration of ischemic cardiac muscle and vascular endothelium by adult stem cells. J. Clin. Invest. 2001, 107, 1395–1402. [Google Scholar]
- Kakigi, A.; Takeda, T. Effect of artificial endolymph injection into the cochlear duct on the endocochlear potential. Hear. Res. 1998, 116, 113–118. [Google Scholar]
- Hu, Z.; Ulfendahl, M.; Olivius, N.P. Central migration of neuronal tissue and embryonic stem cells following transplantation along the adult auditory nerve. Brain Res. 2004, 1026, 68–73. [Google Scholar]
- Lang, H.; Ebihara, Y.; Schmiedt, R.A.; Minamiguchi, H.; Zhou, D.; Smythe, N.; Liu, L.; Ogawa, M.; Schulte, B.A. Contribution of bone marrow hematopoietic stem cells to adult mouse inner ear: Mesenchymal cells and fibrocytes. J. Comp. Neurol. 2006, 496, 187–201. [Google Scholar]
- Matsuoka, A.J.; Kondo, T.; Miyamoto, R.T.; Hashino, E. In vivo and in vitro characterization of bone marrow-derived stem cells in the cochlea. Laryngoscope 2006, 116, 1363–1367. [Google Scholar]
- Kesser, B.W.; Lalwani, A.K. Gene therapy and stem cell transplantation: Strategies for hearing restoration. Adv. Otorhinolaryngol. 2009, 66, 64–86. [Google Scholar]
- Yagi, M.; Kanzaki, S.; Kawamoto, K.; Shin, B.; Shah, P.P.; Magal, E.; Sheng, J.; Raphael, Y. Spiral ganglion neurons are protected from degeneration by GDNF gene therapy. J. Assoc. Res. Otolaryngol. 2000, 1, 315–325. [Google Scholar]
- Oshima, K.; Shimamura, M.; Mizuno, S.; Tamai, K.; Doi, K.; Morishita, R.; Nakamura, T.; Kubo, T.; Kaneda, Y. Intrathecal injection of HVJ-E containing HGF gene to cerebrospinal fluid can prevent and ameliorate hearing impairment in rats. FASEB J. 2004, 18, 212–214. [Google Scholar]
- Kawamoto, K.; Sha, S.H.; Minoda, R.; Izumikawa, M.; Kuriyama, H.; Schacht, J.; Raphael, Y. Antioxidant gene therapy can protect hearing and hair cells from ototoxicity. Mol. Ther. 2004, 9, 173–181. [Google Scholar]
- Nakaizumi, T.; Kawamoto, K.; Minoda, R.; Raphael, Y. Adenovirus-mediated expression of brain-derived neurotrophic factor protects spiral ganglion neurons from ototoxic damage. Audiol. Neurootol. 2004, 9, 135–143. [Google Scholar]
- Okano, T.; Nakagawa, T.; Kita, T.; Endo, T.; Ito, J. Cell-gene delivery of brain-derived neurotrophic factor to the mouse inner ear. Mol. Ther. 2006, 14, 866–871. [Google Scholar]
- Rejali, D.; Lee, V.A.; Abrashkin, K.A.; Humayun, N.; Swiderski, D.L.; Raphael, Y. Cochlear implants and ex vivo BDNF gene therapy protect spiral ganglion neurons. Hear. Res. 2007, 228, 180–187. [Google Scholar]
- Deng, Z.; Wang, J.; Qiu, J.; Liu, S.; Wang, C.; Yang, A. Protection of NT3 gene transfection on the guinea pig cochlea treated with gentamicin. Lin Chuang Er Bi Yan Hou Ke Za Zhi 2004, 18, 231–233. [Google Scholar]
- Izumikawa, M.; Minoda, R.; Kawamoto, K.; Abrashkin, K.A.; Swiderski, D.L.; Dolan, D.F.; Brough, D.E.; Raphael, Y. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat. Med. 2005, 11, 271–276. [Google Scholar]
- Bedrosian, J.C.; Gratton, M.A.; Brigande, J.V.; Tang, W.; Landau, J.; Bennett, J. In vivo delivery of recombinant viruses to the fetal murine cochlea: Transduction characteristics and long-term effects on auditory function. Mol. Ther. 2006, 14, 328–335. [Google Scholar]
- Liu, Y.; Okada, T.; Sheykholeslami, K.; Shimazaki, K.; Nomoto, T.; Muramatsu, S.; Kanazawa, T.; Takeuchi, K.; Ajalli, R.; Mizukami, H.; et al. Specific and efficient transduction of Cochlear inner hair cells with recombinant adeno-associated virus type 3 vector. Mol. Ther. 2005, 12, 725–733. [Google Scholar]
- Maeda, Y.; Fukushima, K.; Nishizaki, K.; Smith, R.J. In vitro and in vivo suppression of GJB2 expression by RNA interference. Hum. Mol. Genet. 2005, 14, 1641–1650. [Google Scholar]
- Zheng, J.L.; Gao, W.Q. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat. Neurosci. 2000, 3, 580–586. [Google Scholar]
- Kawamoto, K.; Ishimoto, S.; Minoda, R.; Brough, D.E.; Raphael, Y. Math1 gene transfer generates new cochlear hair cells in mature guinea pigs in vivo. J. Neurosci 2003, 23, 4395–4400. [Google Scholar]
- Sheffield, A.M.; Gubbels, S.P.; Hildebrand, M.S.; Newton, S.S.; Chiorini, J.A.; di Pasquale, G.; Smith, R.J. Viral vector tropism for supporting cells in the developing murine cochlea. Hear. Res. 2011. [Google Scholar]
- Pathak, A.; Patnaik, S.; Gupta, K.C. Polyethylenimine derived nanoparticles for efficient gene delivery. Nucleic. Acids Symp. Ser. (Oxf.) 2009, 53, 57–58. [Google Scholar]
- Patnaik, S.; Arif, M.; Pathak, A.; Singh, N.; Gupta, K.C. PEI-alginate nanocomposites: Efficient non-viral vectors for nucleic acids. Int. J. Pharm. 2010, 385, 194–202. [Google Scholar]
- Patnaik, S.; Arif, M.; Pathak, A.; Kurupati, R.; Singh, Y.; Gupta, K.C. Cross-linked polyethylenimine-hexametaphosphate nanoparticles to deliver nucleic acids therapeutics. Nanomedicine 2010, 6, 344–354. [Google Scholar]
- Pathak, A.; Patnaik, S.; Gupta, K.C. Recent trends in non-viral vector-mediated gene delivery. Biotechnol. J. 2009, 4, 1559–1572. [Google Scholar]
- Pathak, A.; Swami, A.; Patnaik, S.; Jain, S.; Chuttani, K.; Mishra, A.K.; Vyas, S.P.; Kumar, P.; Gupta, K.C. Efficient tumor targeting by polysaccharide decked polyethylenimine based nanocomposites. J. Biomed. Nanotechnol. 2009, 5, 264–277. [Google Scholar]
- Snyder, B.R.; Boulis, N.M.; Federici, T. Viral vector-mediated gene transfer for CNS disease. Expert. Opin. Biol. Ther. 2010, 10, 381–394. [Google Scholar]
- Takeuchi, S.; Takeda, T.; Saito, H. Pressure relationship between perilymph and endolymph associated with endolymphatic infusion. Ann. Otol. Rhinol. Laryngol. 1991, 100, 244–248. [Google Scholar]
- Salt, A.N.; DeMott, J.E. Longitudinal endolymph movements induced by perilymphatic injections. Hear. Res. 1998, 123, 137–147. [Google Scholar]
- Iguchi, F.; Nakagawa, T.; Tateya, I.; Kim, T.S.; Endo, T.; Taniguchi, Z.; Naito, Y.; Ito, J. Trophic support of mouse inner ear by neural stem cell transplantation. Neuroreport 2003, 14, 77–80. [Google Scholar]
- Reyes, D.Y.; Zuber, P. Activation of transcription initiation by Spx: Formation of transcription complex and identification of a Cis-acting element required for transcriptional activation. Mol. Microbiol. 2008, 69, 765–779. [Google Scholar]
- Liu, Y.Y.; Slotine, J.J.; Barabasi, A.L. Controllability of complex networks. Nature 2011, 473, 167–173. [Google Scholar]
- Fritzsch, B.; Eberl, D.F.; Beisel, K.W. The role of bHLH genes in ear development and evolution: Revisiting a 10-year-old hypothesis. Cell Mol. Life Sci. 2010, 67, 3089–3099. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kopecky, B.; Fritzsch, B. Regeneration of Hair Cells: Making Sense of All the Noise. Pharmaceuticals 2011, 4, 848-879. https://doi.org/10.3390/ph4060848
Kopecky B, Fritzsch B. Regeneration of Hair Cells: Making Sense of All the Noise. Pharmaceuticals. 2011; 4(6):848-879. https://doi.org/10.3390/ph4060848
Chicago/Turabian StyleKopecky, Benjamin, and Bernd Fritzsch. 2011. "Regeneration of Hair Cells: Making Sense of All the Noise" Pharmaceuticals 4, no. 6: 848-879. https://doi.org/10.3390/ph4060848
APA StyleKopecky, B., & Fritzsch, B. (2011). Regeneration of Hair Cells: Making Sense of All the Noise. Pharmaceuticals, 4(6), 848-879. https://doi.org/10.3390/ph4060848