Design, Synthesis, Biological Evaluation and Molecular Docking Studies of New N-Heterocyclic Compounds as Aromatase Inhibitors
Abstract
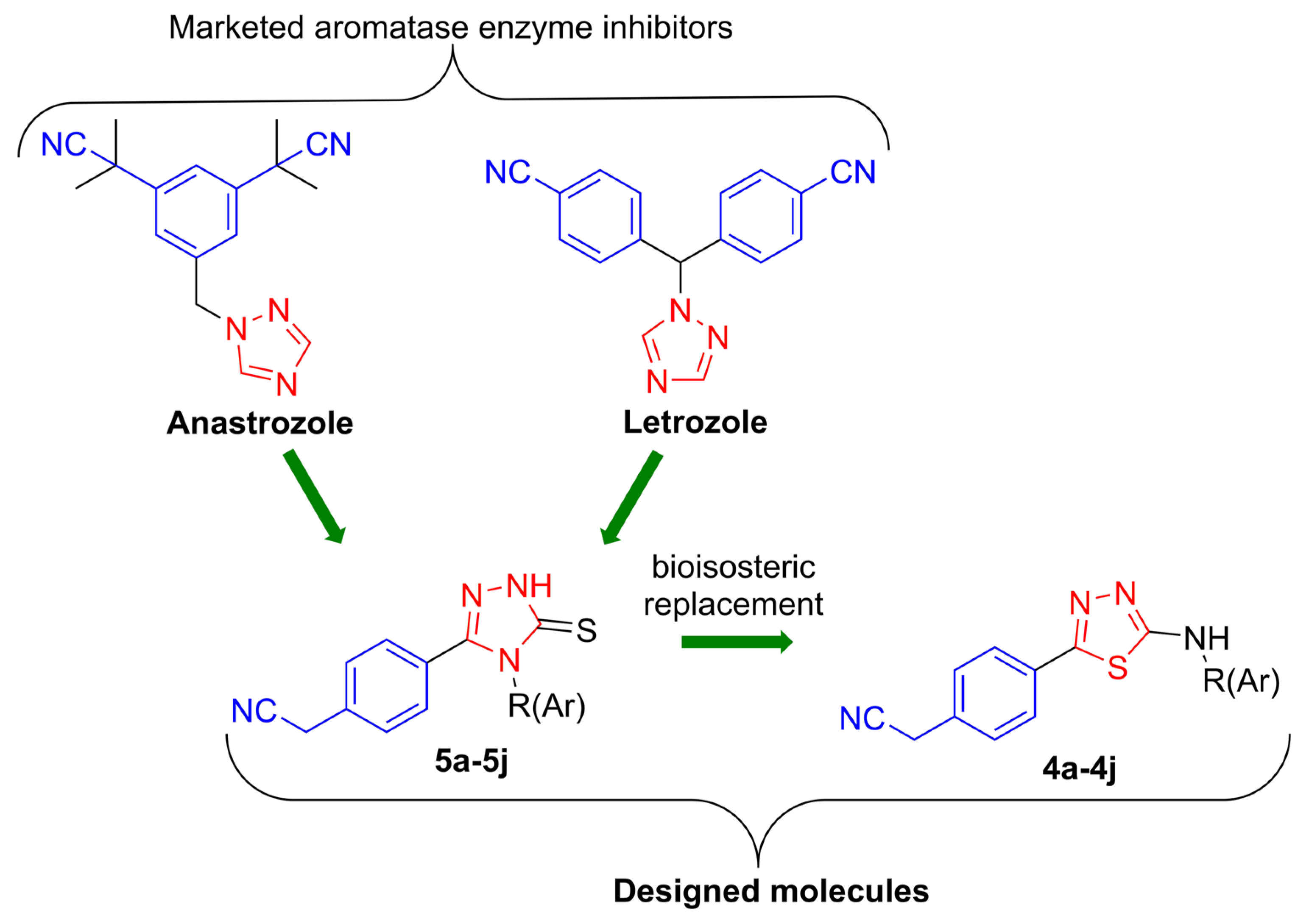
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Activity
Structure–Activity Relationship Studies
- ✓
- Based on both cytotoxic and aromatase activity results, compounds bearing a substituted phenyl ring as the R(Ar) group were found to exhibit higher activity (except for the 4-bromophenyl compound). The highest antiproliferative activity among both thiadiazole (4a–4j) and triazole (5a–5j) compounds was observed in derivatives bearing 4-fluorophenyl, 4-chlorophenyl, 4-methylphenyl, 4-methoxyphenyl, and 4-nitrophenyl groups, respectively.
- ✓
- 1,2,4-Triazole derivatives (5a–5j) generally exhibited higher antiproliferative and anti-aromatase activities than 1,3,4-thiadiazole compounds (4a–4j).
- ✓
- Only certain 1,2,4-triazole compounds (5b, 5c, 5e, 5f and 5g) have demonstrated significant inhibitory activity against the aromatase enzyme.
- ✓
- No antiproliferative or anti-aromatase activity has been observed in thiadiazole and triazole compounds bearing alkyl and alicyclic hydrocarbon (4h, 4i, 4j, 5h, 5i and 5j).
- ✓
- Compounds 5c, 5e and 5f in the series were identified as the most potent aromatase inhibitors with IC50 values lower than 0.1 μM.
- ✓
- The anti-aromatase activity was highest with p-Cl substitution as a halogen on the aromatic phenyl ring in the triazole compounds (5a–5j). Anti-aromatase activity also decreased with p-F substitution and disappeared with p-Br substitution. A similar situation was observed in the ranking of the potency of antiproliferative compounds.
2.3. In Silico Studies
2.3.1. Molecular Docking Studies
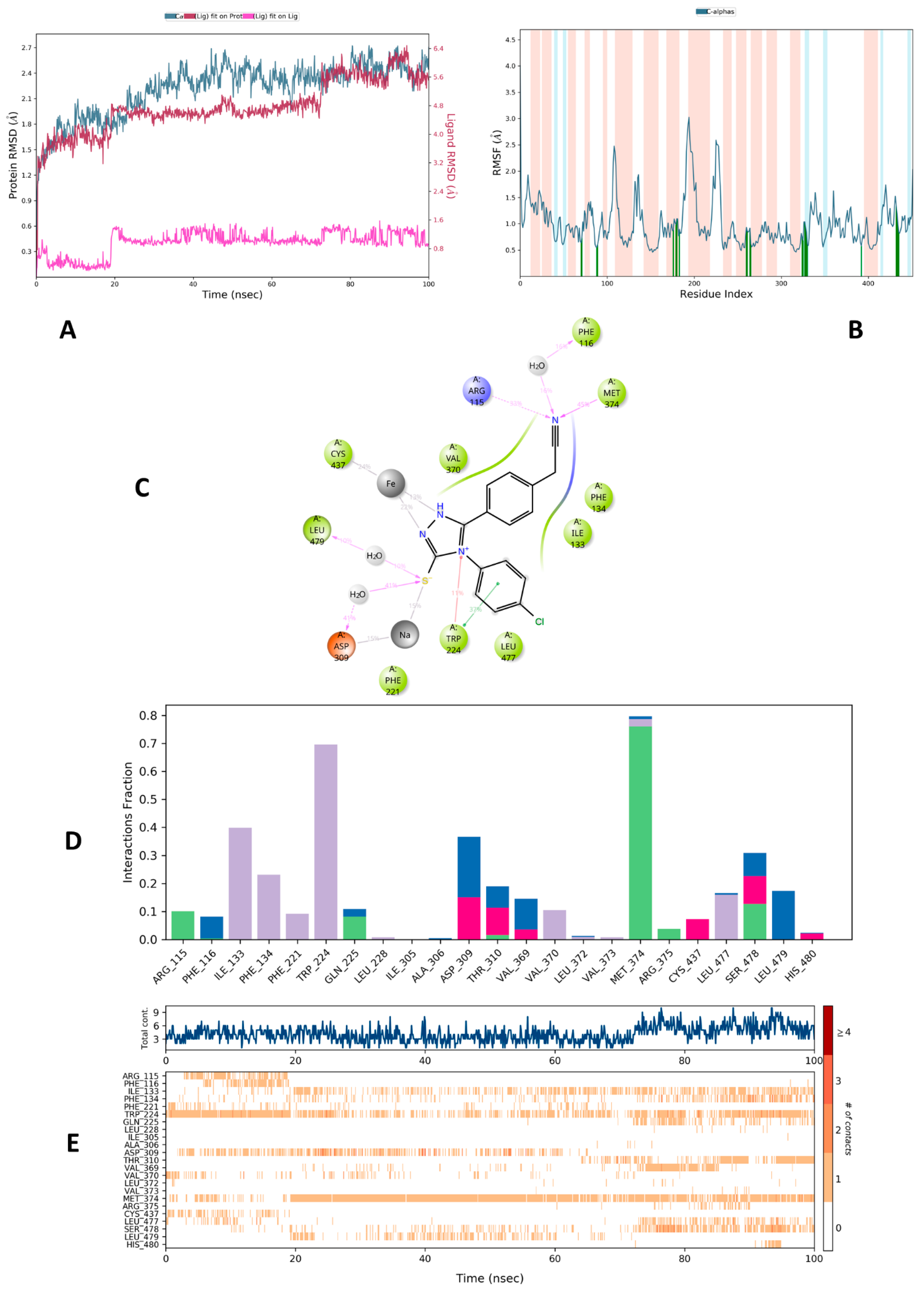
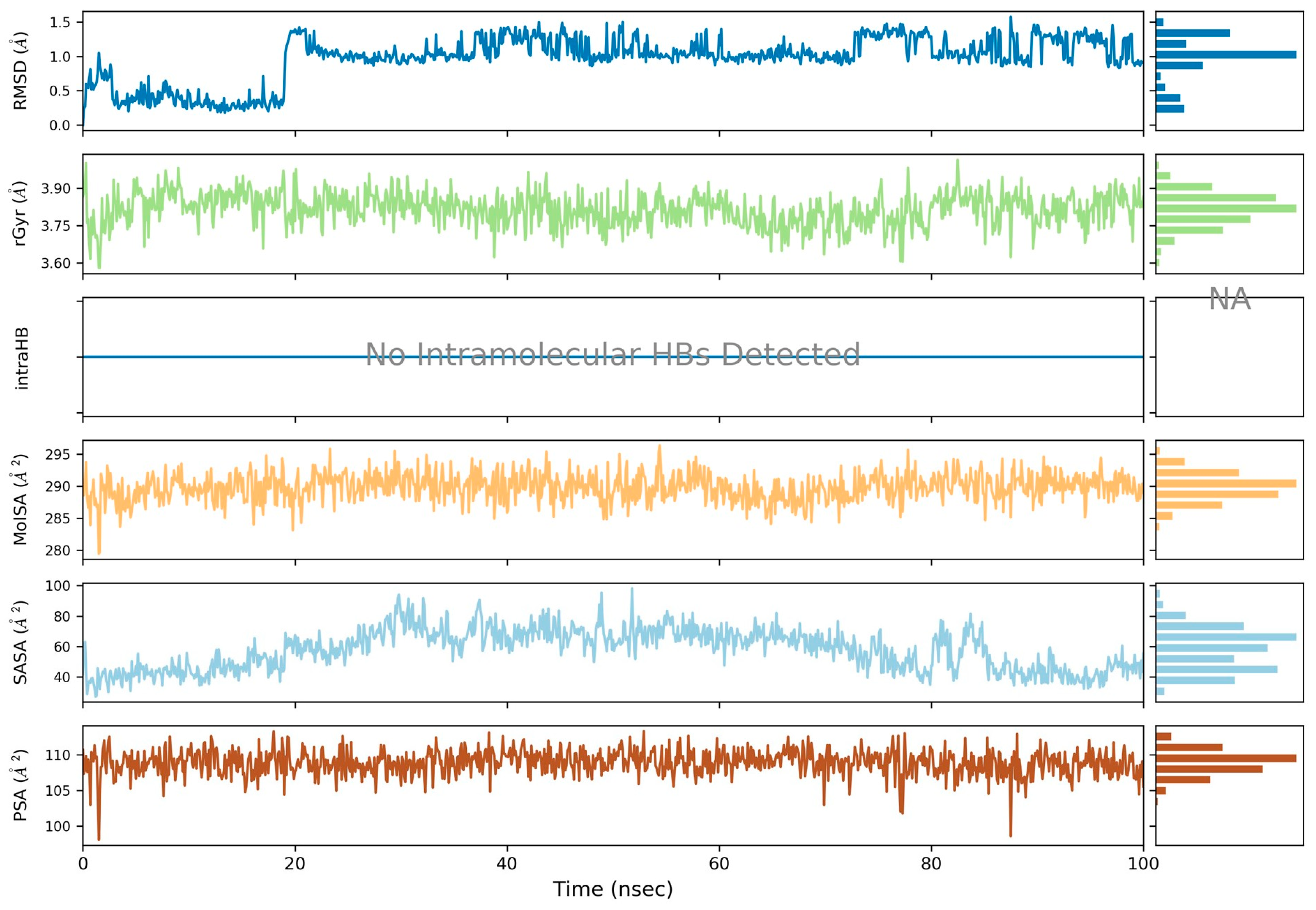
2.3.2. Molecular Dynamic Studies on Aromatase Enzyme
2.3.3. ADME Prediction
3. Materials and Methods
3.1. Chemistry
3.2. Biological Activity
3.2.1. Cytotoxicity Assay
3.2.2. Aromatase Inhibition Assay
3.3. In Silico Studies
3.3.1. Molecular Docking Studies
3.3.2. Molecular Dynamics Simulation
3.3.3. ADME Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MCF-7 | Human breast adenocarcinoma cell |
| NIH3T3 | Mouse embryonic fibroblast cell |
| MD | Molecular dynamics |
| ADME | Absorption, distribution, metabolism and excretion |
| RMSD | Root mean square deviation |
| RMSF | Root mean square fluctuation |
| SASA | Solvent accessible surface area |
| PSA | Polar surface area |
| TLC | Thin-layer chromatography |
References
- Wilkinson, L.; Gathani, T. Understanding breast cancer as a global health concern. Br. J. Radiol. 2022, 95, 20211033. [Google Scholar] [CrossRef] [PubMed]
- Houghton, S.C.; Hankinson, S.E. Cancer Progress and Priorities: Breast Cancer. Cancer Epidemiol. Biomark. Prev. 2021, 30, 822–844. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Breast Cancer Facts & Figures 2024–2025; American Cancer Society: Atlanta, GA, USA, 2024. [Google Scholar]
- PDQ Adult Treatment Editorial Board. Breast Cancer Treatment (PDQ®): Patient Version 2024 Dec 11. In PDQ Cancer Information Summaries [Internet]; National Cancer Institute (US): Bethesda, MD, USA, 2002. Available online: https://www.ncbi.nlm.nih.gov/books/NBK65969/ (accessed on 1 November 2025).
- Lumachi, F.; Santeufemia, D.A.; Basso, S.M. Current medical treatment of estrogen receptor-positive breast cancer. World J. Biol. Chem. 2015, 6, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Roleira, F.M.F.; Costa, S.C.; Gomes, A.R.; Varela, C.L.; Amaral, C.; Augusto, T.V.; Correia-da-Silva, G.; Romeo, I.; Costa, G.; Alcaro, S.; et al. Design, synthesis, biological activity evaluation and structure-activity relationships of new steroidal aromatase inhibitors. The case of C-ring and 7β substituted steroids. Bioorg. Chem. 2023, 131, 106286. [Google Scholar] [CrossRef]
- Al-Wahaibi, L.H.; Abou-Zied, H.A.; Abdelrahman, M.H.; Morcoss, M.M.; Trembleau, L.; Youssif, B.G.M.; Bräse, S. Design and synthesis new indole-based aromatase/iNOS inhibitors with apoptotic antiproliferative activity. Front. Chem. 2024, 12, 1432920. [Google Scholar] [CrossRef]
- Caciolla, J.; Bisi, A.; Belluti, F.; Rampa, A.; Gobbi, S. Reconsidering Aromatase for Breast Cancer Treatment: New Roles for an Old Target. Molecules 2020, 25, 5351. [Google Scholar] [CrossRef]
- Bhatia, N.; Thareja, S. Aromatase inhibitors for the treatment of breast cancer: An overview (2019–2023). Bioorg. Chem. 2024, 151, 107607. [Google Scholar] [CrossRef]
- Słopień, R.; Męczekalski, B. Aromatase inhibitors in the treatment of endometriosis. Prz. Menopauzalny 2016, 15, 43–47. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, L.; Liu, Y.; Zhang, J.; Zheng, L.; Zheng, M. Adverse Event Profiles of the Third-Generation Aromatase Inhibitors: Analysis of Spontaneous Reports Submitted to FAERS. Biomedicines 2024, 12, 1708. [Google Scholar] [CrossRef]
- Fantacuzzi, M.; Gallorini, M.; Gambacorta, N.; Ammazzalorso, A.; Aturki, Z.; Balaha, M.; Carradori, S.; Giampietro, L.; Maccallini, C.; Cataldi, A.; et al. Design, Synthesis and Biological Evaluation of Aromatase Inhibitors Based on Sulfonates and Sulfonamides of Resveratrol. Pharmaceuticals 2021, 14, 984. [Google Scholar] [CrossRef]
- Rashdan, H.R.M.; Shehadi, I.A. Triazoles Synthesis & Applications as Nonsteroidal Aromatase Inhibitors for Hormone-Dependent Breast Cancer Treatment. Heteroat. Chem. 2022, 2022, 5349279. [Google Scholar] [CrossRef]
- Korani, M. Aromatase inhibitors in male: A literature review. Med. Clin. Pract. 2023, 6, 100356. [Google Scholar] [CrossRef]
- Karakuş, S.; Başçıl, E.; Tok, F.; Erdoğan, Ö.; Çevik, Ö.; Başoğlu, F. Synthesis, biological evaluation and molecular docking studies of novel 1,3,4-thiadiazoles as potential anticancer agents and human carbonic anhydrase inhibitors. Mol. Divers. 2024, 28, 3801–3815. [Google Scholar] [CrossRef] [PubMed]
- Kołodziej, P.; Wujec, M.; Doligalska, M.; Makuch-Kocka, A.; Khylyuk, D.; Bogucki, J.; Demkowska-Kutrzepa, M.; Roczeń-Karczmarz, M.; Studzińska, M.; Tomczuk, K.; et al. Synthesis and anthelmintic activity of novel thiosemicarbazide and 1,2,4-triazole derivatives: In vitro, in vivo, and in silico study. J. Adv. Res. 2024, 60, 57–73. [Google Scholar] [CrossRef]
- Sıcak, Y. Synthesis, predictions of drug-likeness, and pharmacokinetic properties of some chiral thioureas as potent enzyme inhibition agents. Turk. J. Chem. 2022, 46, 665–676. [Google Scholar] [CrossRef]
- Sicak, Y. Design and antiproliferative and antioxidant activities of furan-based thiosemicarbazides and 1,2,4-triazoles: Their structure-activity relationship and SwissADME predictions. Med. Chem. Res. 2021, 30, 1557–1568. [Google Scholar] [CrossRef]
- Bozkurt, E.; Sıcak, Y.; Oruç-Emre, E.E.; Karaküçük-İyidoğan, A.; Öztürk, M. Design and Bioevaluation of Novel Hydrazide-Hydrazones Derived from 4-Acetyl-N-Substituted Benzenesulfonamide. Russ. J. Bioorg. Chem. 2020, 46, 702–714. [Google Scholar] [CrossRef]
- Tok, F.; Baltaş, N.; Abas, B.İ.; Kozan, B.; Kaya, S.; Tatar-Yılmaz, G.; Çevik, Ö. Synthesis, Biological Evaluation and Molecular Docking Studies of New 4-(Cyanomethyl)-N’-Substituted Benzohydrazide Derivatives as Anti-Alzheimer Agents. Drug Dev. Res. 2025, 86, e70166. [Google Scholar] [CrossRef]
- Başaran, E.; Sıcak, Y.; Sogukomerogullari, H.G.; Karaküçük-İyidoğan, A.; Oruç-Emre, E.E.; Sönmez, M.; Öztürk, M. Synthesis of novel chiral metal complexes derived from chiral thiosemicarbazide ligands as potential antioxidant agents. Chirality 2019, 31, 434–444. [Google Scholar] [CrossRef]
- Karaküçük-Iyidogan, A.; Basaran, E.; Tatar-Yılmaz, G.; Oruç-Emre, E.E. Development of new chiral 1,2,4-triazole-3-thiones and 1,3,4-thiadiazoles with promising in vivo anticonvulsant activity targeting GABAergic system and voltage-gated sodium channels (VGSCs). Bioorg. Chem. 2024, 151, 107662. [Google Scholar] [CrossRef]
- Sağlık, B.N.; Ilgın, S.; Özkay, Y. Synthesis of new donepezil analogues and investigation of their effects on cholinesterase enzymes. Eur. J. Med. Chem. 2016, 124, 1026–1040. [Google Scholar] [CrossRef] [PubMed]
- Osmaniye, D.; Görgülü, Ş.; Sağlık, B.N.; Levent, S.; Özkay, Y.; Kaplancıklı, Z.A. Design, synthesis, in vitro and in silico studies of some novel thiazole-dihydrofuran derivatives as aromatase inhibitors. Bioorg. Chem. 2021, 114, 105123. [Google Scholar] [CrossRef]
- Berridge, M.V.; Herst, P.M.; Tan, A.S. Tetrazolium dyes as tools in cell biology: New insights into their cellular reduction. Biotechnol. Annu. Rev. 2005, 11, 127–152. [Google Scholar]
- BioVision, Aromatase (CYP19A) Inhibitor Screening Kit (Fluorometric) (Catalog No: K984-100) Manual. Available online: https://www.biovision.com/documentation/datasheets/K984.pdf (accessed on 1 August 2025).
- Osmaniye, D.; Levent, S.; Sağlık, B.N.; Karaduman, A.B.; Özkay, Y.; Kaplancıklı, Z.A. Novel imidazole derivatives as potential aromatase and monoamine oxidase-B inhibitors against breast cancer. New J. Chem. 2022, 46, 7442–7451. [Google Scholar] [CrossRef]
- Evren, A.E.; Nuha, D.; Dawbaa, S.; Sağlık, B.N.; Yurttaş, L. Synthesis of novel thiazolyl hydrazone derivatives as potent dual monoamine oxidase-aromatase inhibitors. Eur. J. Med. Chem. 2022, 229, 114097. [Google Scholar] [CrossRef]
- Ghosh, D.; Griswold, J.; Erman, M.; Pangborn, W. Structural basis for androgen specificity and oestrogen synthesis in human aromatase. Nature 2009, 457, 219–223. [Google Scholar] [CrossRef]
- Maestro, Version 10.6; Schrödinger, LLC: New York, NY, USA, 2020.
- Schrödinger, LLC. LigPrep, Version 3.8; Schrödinger, LLC: New York, NY, USA, 2020.
- Schrödinger, LLC. Glide, Version 7.1; Schrödinger, LLC: New York, NY, USA, 2020.
- M.D.I. Tools. Schrödinger Release 2018-3: Prime, 2018; Schrödinger, LLC: New York, NY, USA, 2020. [Google Scholar]
- Osmaniye, D.; Evren, A.E.; Karaca, Ş.; Özkay, Y.; Kaplancıklı, Z.A. Novel thiadiazol derivatives; design, synthesis, biological activity, molecular docking and molecular dynamics. J. Mol. Struct. 2023, 1272, 134171. [Google Scholar] [CrossRef]



| Compounds | MCF-7 | NIH3T3 | Selectivity Index | Aromatase Enzyme Inhibition |
|---|---|---|---|---|
| 4a | 66.325 ± 1.859 | 83.057 ± 3.025 | 1.252 | - |
| 4b | 47.854 ± 2.012 | >100 | >2.090 | - |
| 4c | 20.759 ± 1.021 | >100 | >4.817 | - |
| 4d | >100 | >100 | - | - |
| 4e | 30.250 ± 1.045 | 69.410 ± 2.108 | 2.295 | - |
| 4f | 24.007 ± 1.006 | >100 | >4.165 | - |
| 4g | 52.459 ± 1.367 | 84.306 ± 3.148 | 1.607 | - |
| 4h | >100 | >100 | - | - |
| 4i | >100 | >100 | - | - |
| 4j | >100 | >100 | - | - |
| 5a | 70.145 ± 2.859 | >100 | >1.426 | - |
| 5b | 9.623 ± 0.357 | >100 | >10.392 | 2.224 ± 0.110 |
| 5c | 3.142 ± 0.139 | >100 | >31.827 | 0.064 ± 0.003 |
| 5d | 47.122 ± 1.856 | 84.325 ± 3.067 | 1.790 | - |
| 5e | 7.569 ± 0.248 | 68.332 ± 2.956 | 9.028 | 0.097 ± 0.003 |
| 5f | 4.758 ± 0.196 | >100 | >21.017 | 0.092 ± 0.004 |
| 5g | 10.415 ± 0.337 | 84.126 ± 3.455 | 8.077 | 1.023 ± 0.050 |
| 5h | >100 | >100 | - | - |
| 5i | >100 | >100 | - | - |
| 5j | >100 | >100 | - | - |
| Doxorubicin | 1.940 ± 0.084 | >100 | - | - |
| Letrozole | 0.875 ± 0.038 | >100 | - | 0.031 ± 0.001 |
| Comp. | Moiety | Pi-pi Interactions | Cation-pi Interactions | Salt Bridge | Hydrogen Bonds | Halogen Bonds |
|---|---|---|---|---|---|---|
| 5c | Phenyl | HEM | - | - | - | - |
| Phenyl ring of Phe134 | ||||||
| Triazole | HEM | HEM | HEM | - | - | |
| Triazole-N | - | - | - | HEM | - | |
| Sulphur | - | - | HEM | - | - | |
| 4-CI-phenyl | Indole ring of Trp224 | - | - | - | Hydroxyl of Ser478 | |
| 5e | Phenyl | HEM | - | - | - | - |
| Phenyl ring of Phe134 | ||||||
| Triazole | HEM | HEM | HEM | - | - | |
| Triazole-N | - | - | - | HEM | - | |
| Sulphur | - | - | HEM | - | - | |
| 5f | Phenyl | HEM | - | - | - | - |
| Phenyl ring of Phe134 | ||||||
| Triazole | HEM | HEM | HEM | - | - | |
| Triazole-N | - | - | - | HEM | - | |
| Sulphur | - | - | HEM | - | - | |
| 4-OCH3-phenyl | Indole ring of Trp224 | - | - | - | - |
| Complex | RMSD | Rg | RMSF |
|---|---|---|---|
| 5c-3EQM | 2.4 Å | 3.7–3.9 Å | Arg115 (0.64 Å), Phe116 (0.75 Å), Ile133 (0.55 Å), Phe134 (0.61 Å), Phe221 (0.89 Å), Trp224 (0.98 Å), Gln225 (1.09 Å), Leu228 (0.90 Å), Ile305 (0.93 Å), Ala306 (0.86 Å), Asp309 (0.92 Å), Thr310 (0.62 Å), Val369 (0.75 Å), Val370 (0.69 Å), Leu372 (1.01 Å), Val373 (0.93 Å), Met374 (0.83 Å), Arg375 (0.61 Å), Cys437 (0.58 Å), Leu477 (1.25 Å), Ser478 (1.09 Å), Leu479 (1.00 Å), His480 (0.89 Å) |
| Complex | Amino Acids Interacting Above 10% | Interaction Fractions |
|---|---|---|
| 5c-3EQM | Arg115 (H-bond: 33%) Phe116 (Water mediated H-bond: 16%) Trp224 (π-π interaction: 37%) Trp224 (Cation-π interaction: 11%) Asp309 (Water mediated H-bond: 41%) Met374 (H-bond: 45%) Leu479 (Water mediated H-bond: 10%) HEM (Metal coordination 13% and 22%) | Water mediated H-bond (blue) Phe116, Gln225, Ala306, Asp309, Thr310, Val369, Leu372, Met374, Leu477, Ser478, Leu479, His480 |
| H-bond (green) Arg115, Phe116, Gln225, Ile305, Thr310, Met374, Arg375, Ser478 | ||
| Ionic interaction (pink) Asp309, Thr310, Val369, Cys437, Ser478, His480 | ||
| Hydrophobic interaction (purple) Ile133, Phe134, Phe221, Trp224, Leu228, Ile305, Val370, Leu372, Val373, Met374, Leu477, His480 |
| Comp. | MW | n-ROTB | HBA | HBD | TPSA | Log P | Log S | Lipinski | Ghose | Veber | Egan | Muegge |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4a | 292.36 | 4 | 3 | 1 | 89.84 | 2.55 | −4.23 | Yes | Yes | Yes | Yes | Yes |
| 4b | 310.35 | 4 | 4 | 1 | 89.84 | 2.94 | −4.38 | Yes | Yes | Yes | Yes | Yes |
| 4c | 326.80 | 4 | 3 | 1 | 89.84 | 3.06 | −4.81 | Yes | Yes | Yes | Yes | Yes |
| 4d | 371.25 | 4 | 3 | 1 | 89.84 | 3.18 | −5.13 | Yes | Yes | Yes | Yes | Yes |
| 4e | 306.38 | 4 | 3 | 1 | 89.84 | 2.79 | −4.52 | Yes | Yes | Yes | Yes | Yes |
| 4f | 322.38 | 5 | 4 | 1 | 99.07 | 2.23 | −4.28 | Yes | Yes | Yes | Yes | Yes |
| 4g | 337.36 | 5 | 5 | 1 | 135.66 | 2.33 | −4.26 | Yes | Yes | Yes | No | Yes |
| 4h | 230.29 | 3 | 3 | 1 | 89.84 | 1.19 | −2.84 | Yes | Yes | Yes | Yes | Yes |
| 4i | 244.32 | 4 | 3 | 1 | 89.84 | 1.47 | −3.06 | Yes | Yes | Yes | Yes | Yes |
| 4j | 298.41 | 4 | 3 | 1 | 89.84 | 2.51 | −4.21 | Yes | Yes | Yes | Yes | Yes |
| 5a | 292.36 | 3 | 2 | 1 | 89.49 | 2.61 | −3.85 | Yes | Yes | Yes | Yes | Yes |
| 5b | 310.35 | 3 | 3 | 1 | 89.49 | 2.60 | −4.00 | Yes | Yes | Yes | Yes | Yes |
| 5c | 326.80 | 3 | 2 | 1 | 89.49 | 2.72 | −4.43 | Yes | Yes | Yes | Yes | Yes |
| 5d | 371.25 | 3 | 2 | 1 | 89.49 | 2.84 | −4.75 | Yes | Yes | Yes | Yes | Yes |
| 5e | 306.38 | 3 | 2 | 1 | 89.49 | 2.45 | −4.14 | Yes | Yes | Yes | Yes | Yes |
| 5f | 322.38 | 4 | 3 | 1 | 98.72 | 1.89 | −3.90 | Yes | Yes | Yes | Yes | Yes |
| 5g | 337.36 | 4 | 4 | 1 | 135.31 | 1.99 | −3.88 | Yes | Yes | Yes | No | Yes |
| 5h | 230.29 | 2 | 2 | 1 | 89.49 | 0.84 | −2.46 | Yes | Yes | Yes | Yes | Yes |
| 5i | 244.32 | 3 | 2 | 1 | 89.49 | 1.12 | −2.68 | Yes | Yes | Yes | Yes | Yes |
| 5j | 298.41 | 3 | 2 | 1 | 89.49 | 2.57 | −3.83 | Yes | Yes | Yes | Yes | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Tok, F.; Sağlık Özkan, B.N.; Özkay, Y.; Kaplancıklı, Z.A. Design, Synthesis, Biological Evaluation and Molecular Docking Studies of New N-Heterocyclic Compounds as Aromatase Inhibitors. Pharmaceuticals 2026, 19, 224. https://doi.org/10.3390/ph19020224
Tok F, Sağlık Özkan BN, Özkay Y, Kaplancıklı ZA. Design, Synthesis, Biological Evaluation and Molecular Docking Studies of New N-Heterocyclic Compounds as Aromatase Inhibitors. Pharmaceuticals. 2026; 19(2):224. https://doi.org/10.3390/ph19020224
Chicago/Turabian StyleTok, Fatih, Begüm Nurpelin Sağlık Özkan, Yusuf Özkay, and Zafer Asım Kaplancıklı. 2026. "Design, Synthesis, Biological Evaluation and Molecular Docking Studies of New N-Heterocyclic Compounds as Aromatase Inhibitors" Pharmaceuticals 19, no. 2: 224. https://doi.org/10.3390/ph19020224
APA StyleTok, F., Sağlık Özkan, B. N., Özkay, Y., & Kaplancıklı, Z. A. (2026). Design, Synthesis, Biological Evaluation and Molecular Docking Studies of New N-Heterocyclic Compounds as Aromatase Inhibitors. Pharmaceuticals, 19(2), 224. https://doi.org/10.3390/ph19020224








