Jaceosidin Attenuates Sepsis-Induced Myocardial Dysfunction by Promoting SIRT2-Mediated Inhibition of Histone H3K18 Lactylation
Abstract
1. Introduction
2. Results
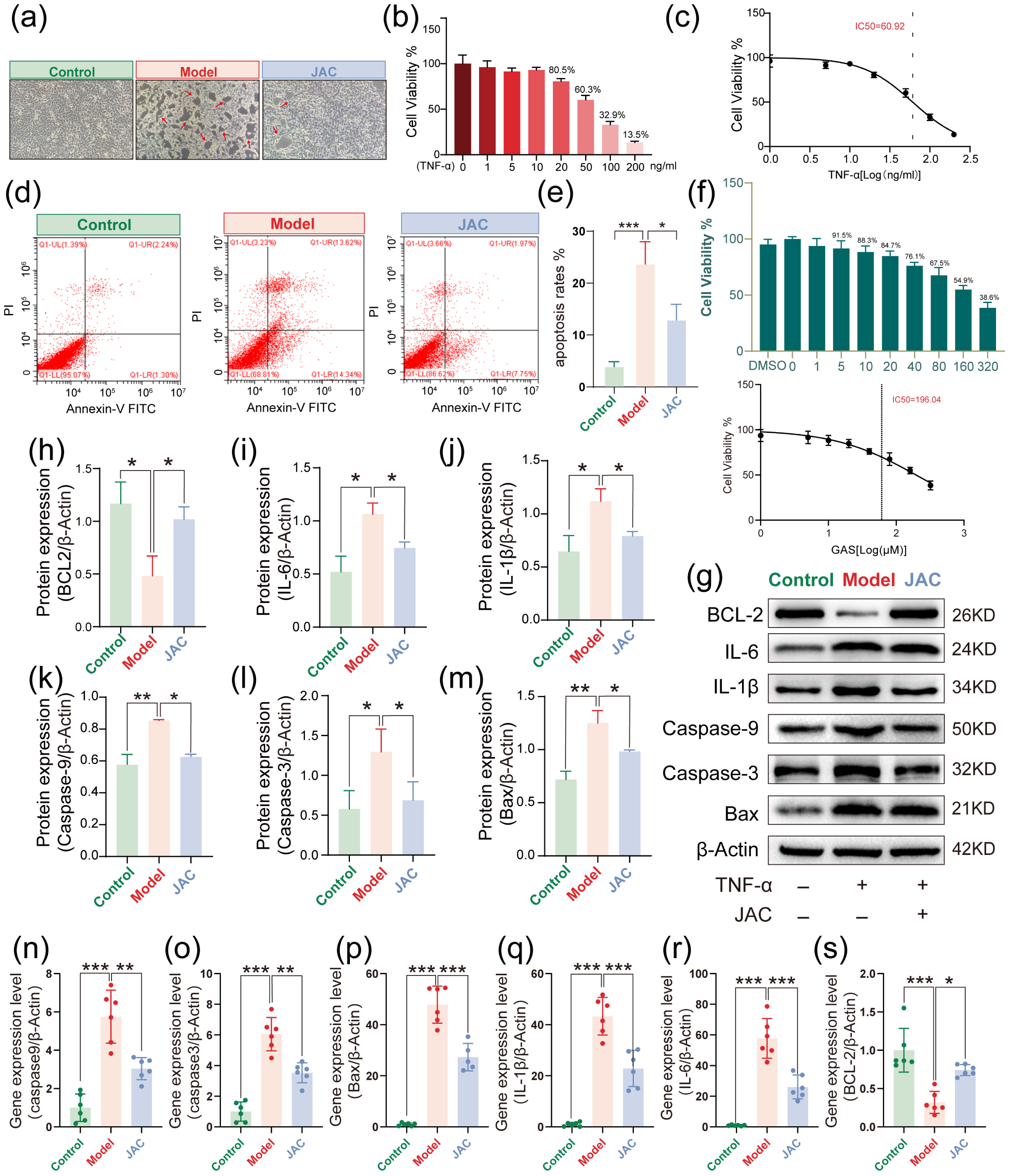
2.1. In Vitro Protective Effects by JAC Against TNF-α-Induced Injury in AC16 Cells
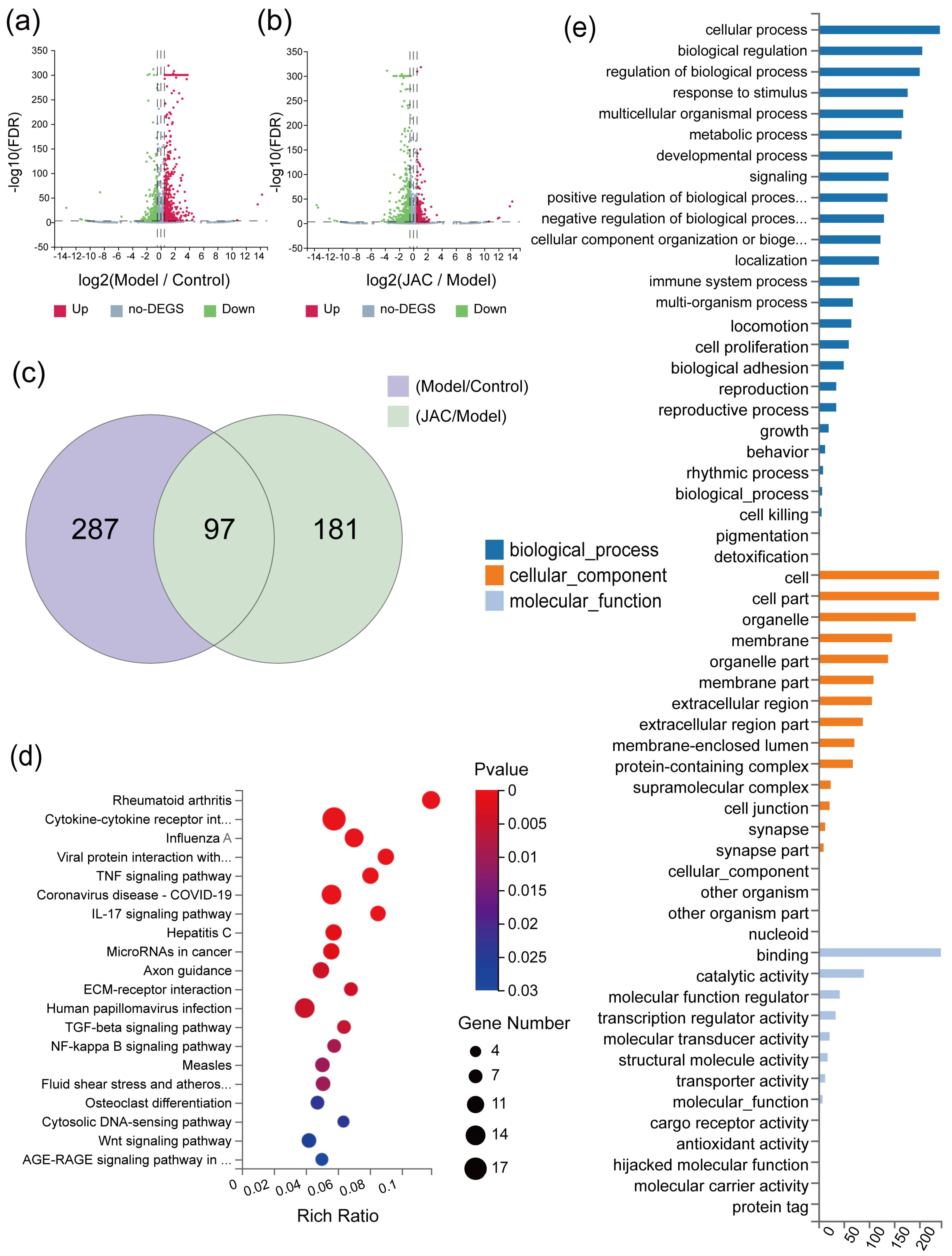
2.2. Gene Expression Profiles of mRNA Sequences in JAC-Treated Cells Under TNF-α Induction
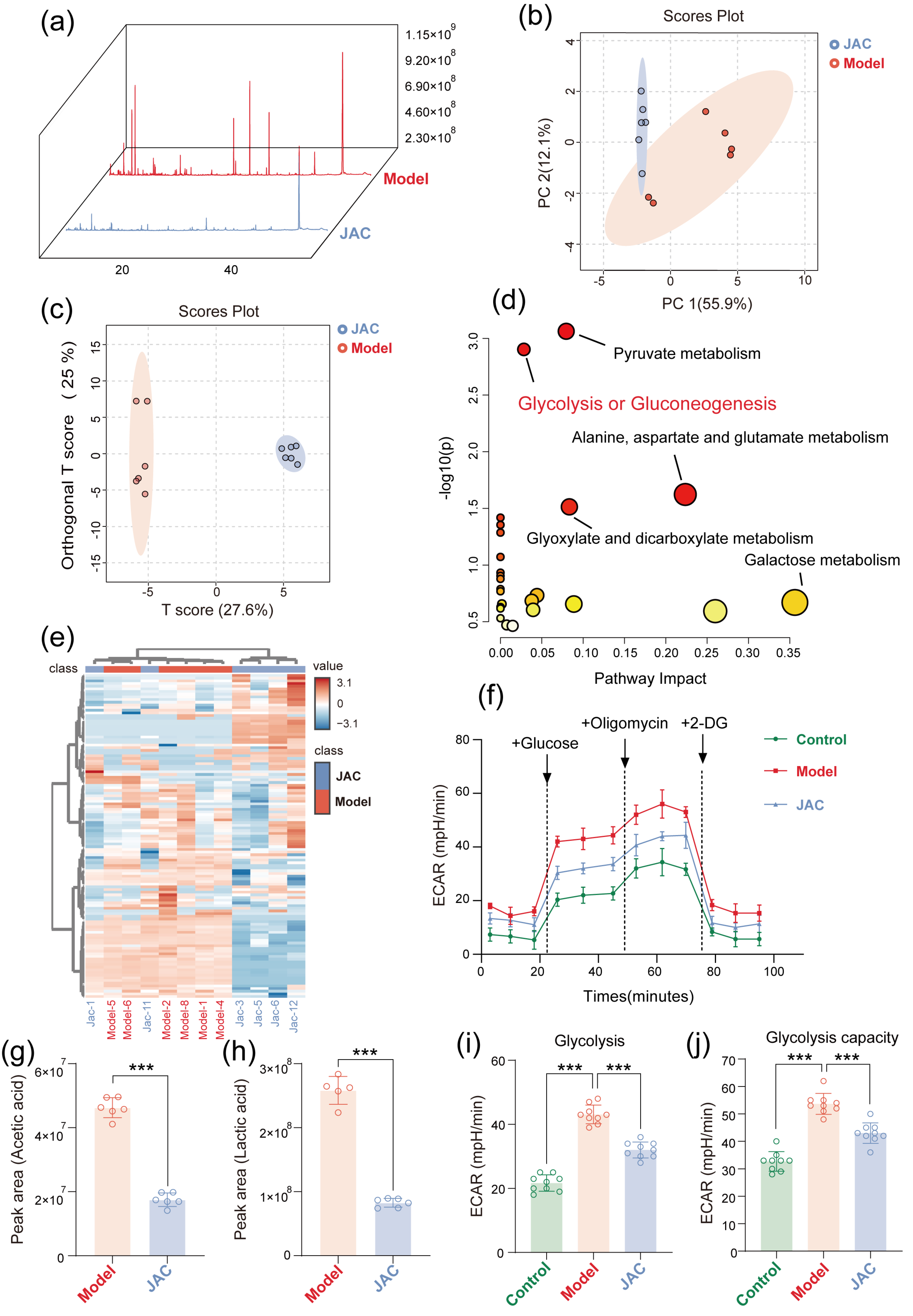
2.3. JAC Remodels the Cellular Metabolic Profile and Alleviates Metabolic Dysfunction Through Gly-Colysis Regulation
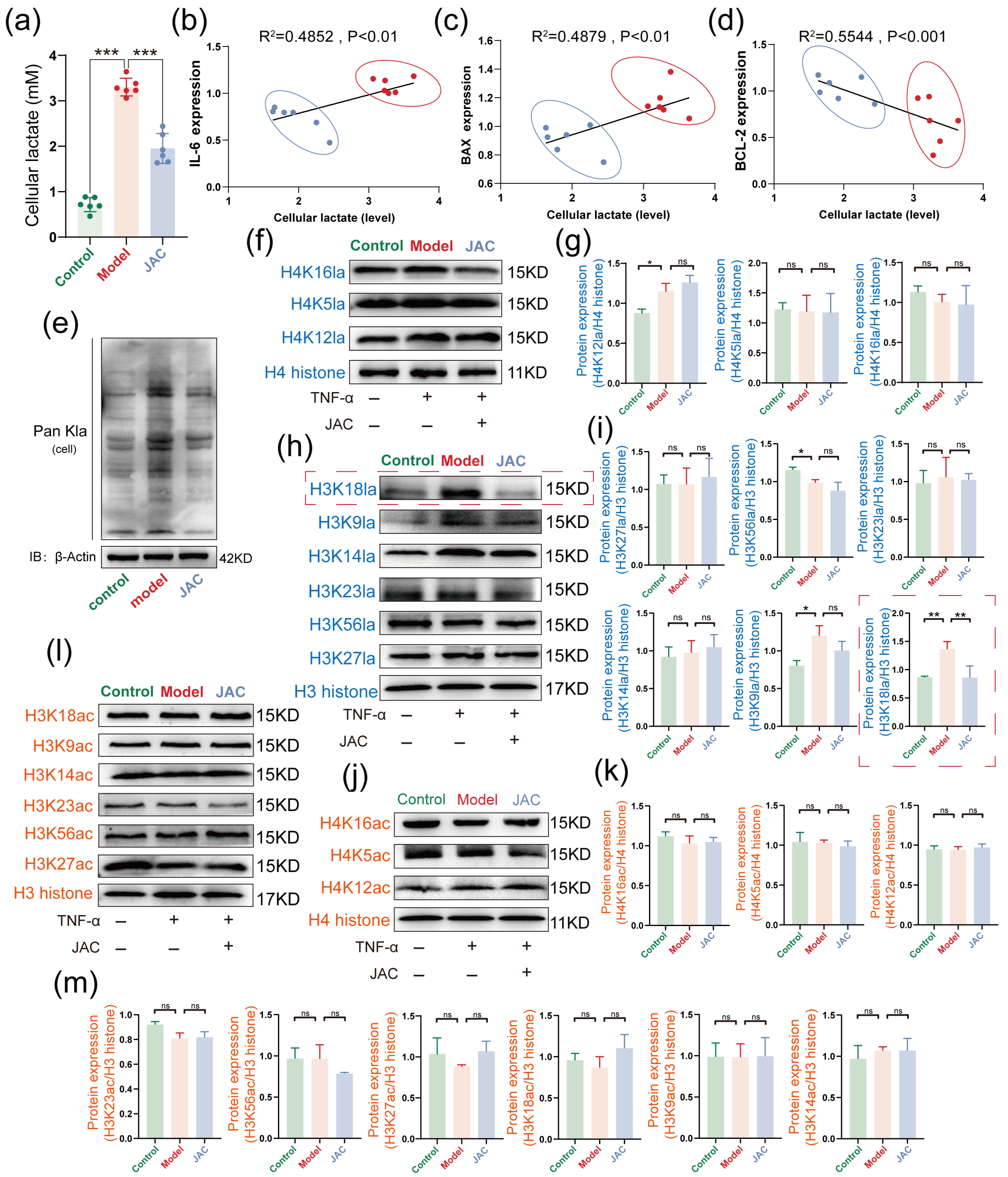
2.4. JAC’s Specific Inhibition of H3K18la and Its Impact on Metabolic-Epigenetic Coupling
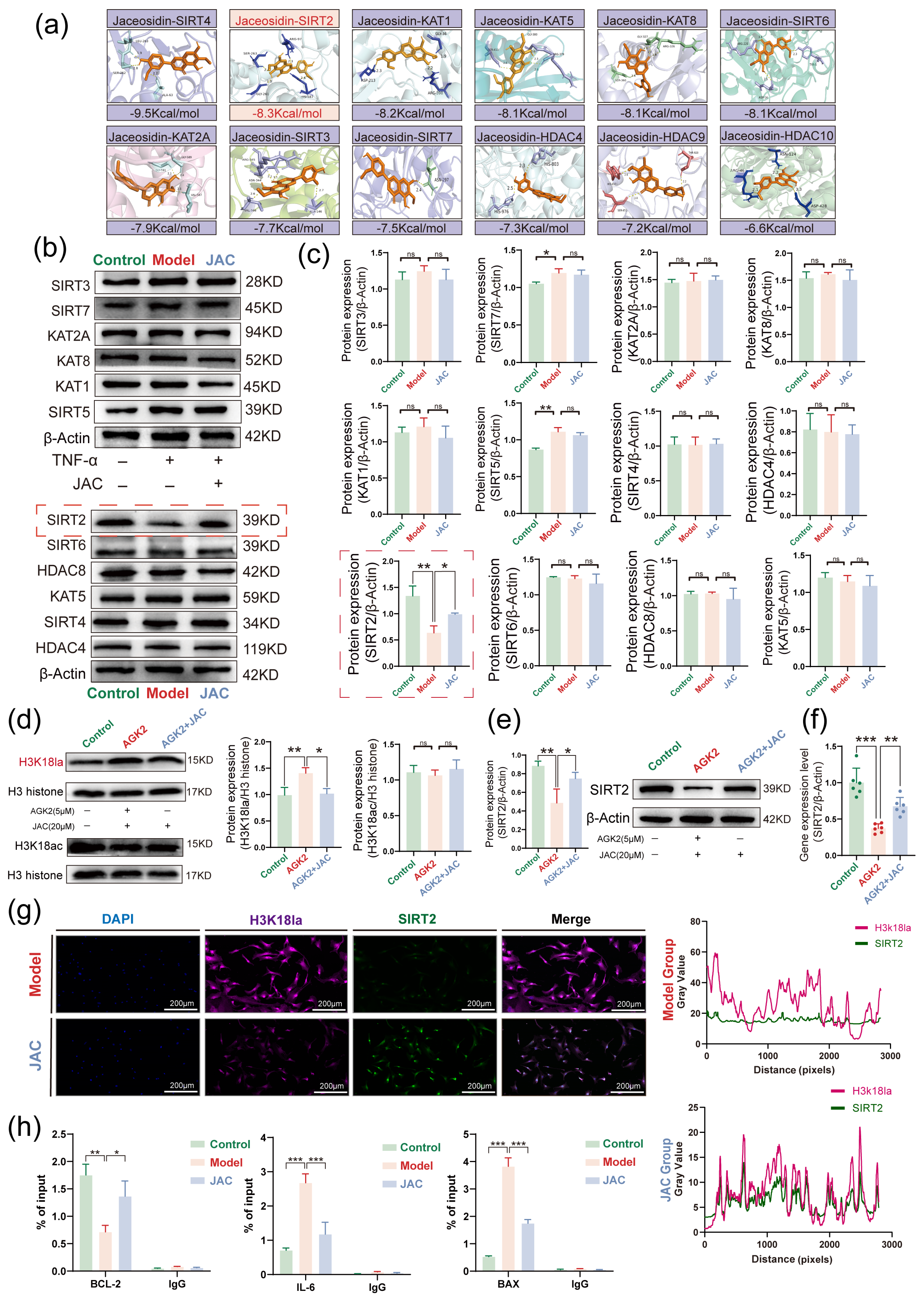
2.5. JAC Regulates SIMD Through SIRT2 Activation and H3K18la Inhibition
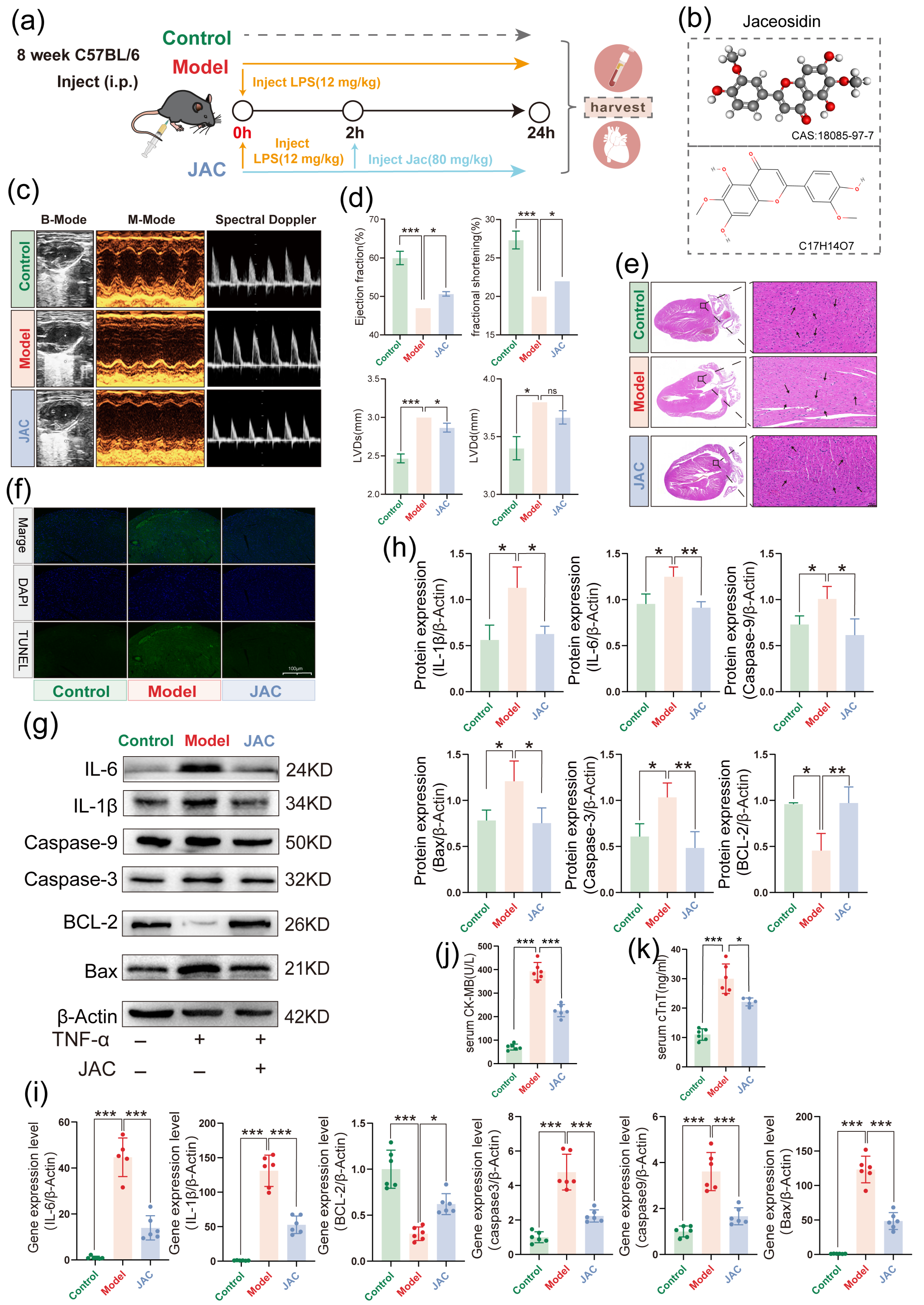
2.6. JAC Rescues Cardiac Function and Mitigates Myocardial Injury in LPS-Induced In Vivo SIMD in Mice
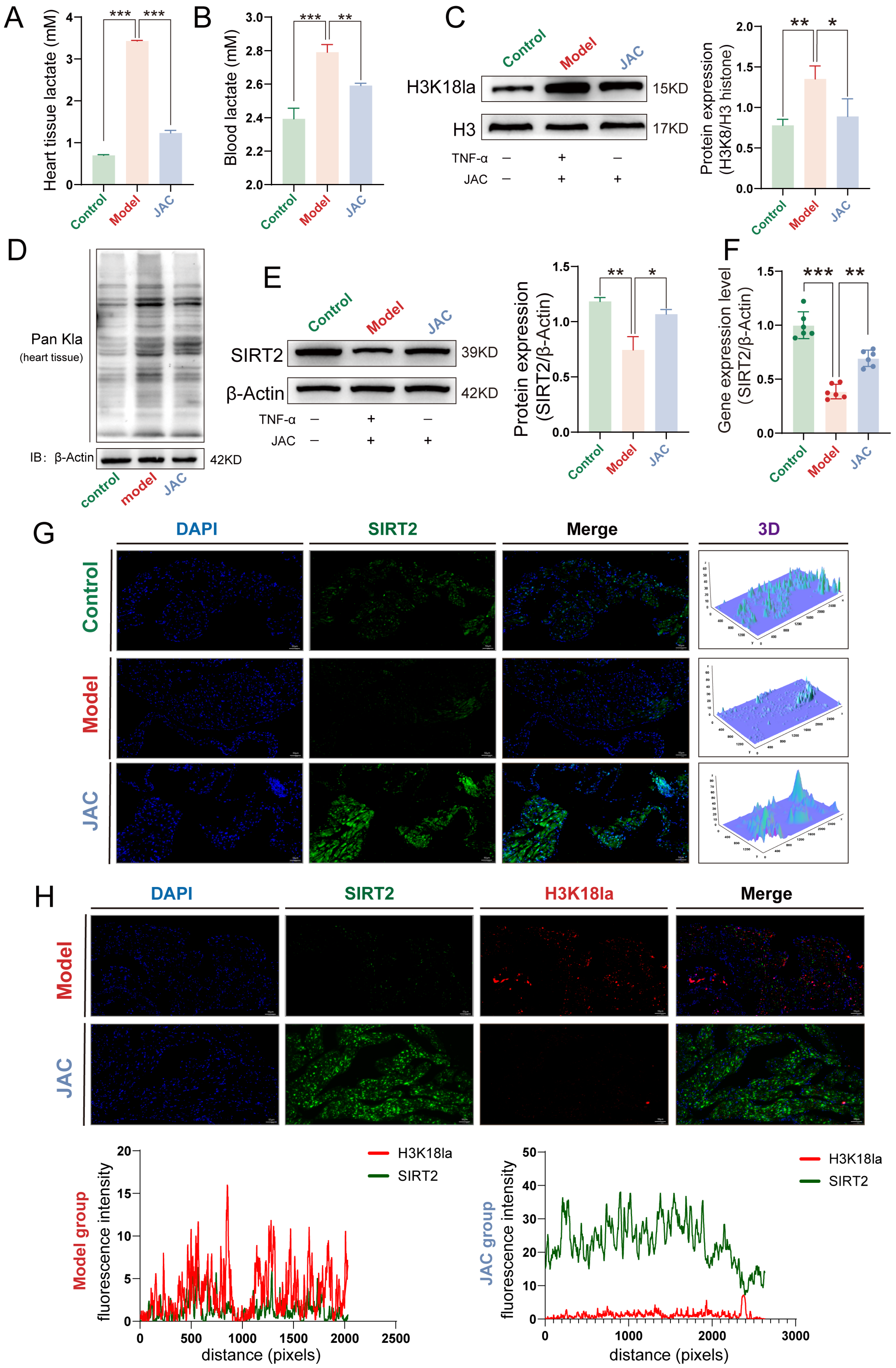
2.7. JAC Alleviates SIMD In Vivo by Modulating Lactate Metabolism and SIRT2-H3K18la Axis
3. Discussion
4. Materials and Methods
4.1. Cell Culture Protocols and Experimental Treatments
4.2. Cell Viability Assay
4.3. Flow Cytometric Analysis of Apoptosis
4.4. RNA Sequencing (RNA-Seq)
4.5. Gas Chromatography-Mass Spectrometer (GC–MS) Metabolome Analysis
4.6. Real-Time Metabolic Flux Analysis Using Seahorse XF Technology
4.7. Lactate Acid Detection
4.8. Molecular Docking
4.9. Immunofluorescence (IF) Assay
4.10. Chromatin Immunoprecipitation Quantitative Polymerase Chain Reaction (ChIP-qPCR) Assay
4.11. Animal Model and Treatment
4.12. Mouse Echocardiography
4.13. Histopathological Analysis
4.14. Enzyme Linked Immunosorbent Assay (ELISA)
4.15. Western Blotting
4.16. Quantitative Real-Time PCR (qRT–PCR)
4.17. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| JAC | Jaceosidin |
| SIMD | Sepsis-induced myocardial dysfunction |
| H3K18la | Histone H3K18 lactylation |
| PAMPs | Pathogen-associated molecular patterns |
| LPS | Lipopolysaccharide |
| TCM | Traditional Chinese medicine |
| DEGs | Differentially expressed genes |
| ECAR | Extracellular acidification rate |
| IF | Immunofluorescence |
| CHIP-qPCR | Chromatin immunoprecipitatin coupled with qPCR |
| LVEF | Left Ventricular Eiection Fraction |
| LVFS | Left Ventricular Fractional Shortening |
| LVDd | Left Ventricular end-diastolic internal diameter |
| LVDs | Left ventricular end-systolic internal diameters |
| H&E | Hematoxylin and eosin staining |
| TUNEL | TdT-mediated dUTP nick end labeling staining |
| cTnT | Cardiac troponin T |
| CK-MB | Creatine kinase isoenzymes |
| Kla | Lysine Lactylation |
| H3 | Histone H3 |
| H4 | Histone H4 |
| TNF-α | Tumour necrosis factor alpha |
| IL-6 | Interleukin-6 |
| IL-1β | Interleukin-1β |
| qPCR | Quantitative Real-time polymerase chain reaction |
References
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef]
- Beesley, S.J.; Weber, G.; Sarge, T.; Nikravan, S.; Grissom, C.K.; Lanspa, M.J.; Shahul, S.; Brown, S.M. Septic Cardiomyopathy. Crit. Care Med. 2018, 46, 625–634. [Google Scholar] [CrossRef]
- Winterbottom, F. Treating Sepsis in Patients with Heart Failure. Crit. Care Nurs. Clin. N. Am. 2022, 34, 165–172. [Google Scholar] [CrossRef]
- Hollenberg, S.M.; Singer, M. Pathophysiology of sepsis-induced cardiomyopathy. Nat. Rev. Cardiol. 2021, 18, 424–434. [Google Scholar] [CrossRef]
- Lin, Y.; Xu, Y.; Zhang, Z. Sepsis-Induced Myocardial Dysfunction (SIMD): The Pathophysiological Mechanisms and Therapeutic Strategies Targeting Mitochondria. Inflammation 2020, 43, 1184–1200. [Google Scholar] [CrossRef]
- Zindel, J.; Kubes, P. DAMPs, PAMPs, and LAMPs in Immunity and Sterile Inflammation. Annu. Rev. Pathol. 2020, 15, 493–518. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, R.F.; Sa-Correia, I.; Valvano, M.A. Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol. Rev. 2016, 40, 480–493. [Google Scholar] [CrossRef]
- Li, N.; Zhou, H.; Wu, H.; Wu, Q.; Duan, M.; Deng, W.; Tang, Q. STING-IRF3 contributes to lipopolysaccharide-induced cardiac dysfunction, inflammation, apoptosis and pyroptosis by activating NLRP3. Redox Biol. 2019, 24, 101215. [Google Scholar] [CrossRef] [PubMed]
- Cicchinelli, S.; Pignataro, G.; Gemma, S.; Piccioni, A.; Picozzi, D.; Ojetti, V.; Franceschi, F.; Candelli, M. PAMPs and DAMPs in Sepsis: A Review of Their Molecular Features and Potential Clinical Implications. Int. J. Mol. Sci. 2024, 25, 962. [Google Scholar] [CrossRef] [PubMed]
- Kuzmich, N.N.; Sivak, K.V.; Chubarev, V.N.; Porozov, Y.B.; Savateeva-Lyubimova, T.N.; Peri, F. TLR4 Signaling Pathway Modulators as Potential Therapeutics in Inflammation and Sepsis. Vaccines 2017, 5, 34. [Google Scholar] [CrossRef]
- Tang, J.; Tam, E.; Song, E.; Xu, A.; Sweeney, G. Crosstalk between myocardial autophagy and sterile inflammation in the development of heart failure. Autophagy Rep. 2024, 3, 2320605. [Google Scholar] [CrossRef]
- Prescott, H.C.; Angus, D.C. Enhancing Recovery From Sepsis: A Review. JAMA 2018, 319, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, C.; Zhou, T.; Xie, F.; Liu, Z.; Xu, H.; Liu, M.; Wang, S.; Li, L.; Chi, Q.; et al. Lumican promotes calcific aortic valve disease through H3 histone lactylation. Eur. Heart J. 2024, 45, 3871–3885. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xia, Y.; Qu, L.; Liu, Y.; Liu, X.; Xu, K. Cardamonin inhibits osteogenic differentiation of human valve interstitial cells and ameliorates aortic valve calcification via interfering in the NF-kappaB/NLRP3 inflammasome pathway. Food Funct. 2021, 12, 11808–11818. [Google Scholar] [CrossRef]
- Liu, M.; Li, F.; Huang, Y.; Zhou, T.; Chen, S.; Li, G.; Shi, J.; Dong, N.; Xu, K. Caffeic Acid Phenethyl Ester Ameliorates Calcification by Inhibiting Activation of the AKT/NF-kappaB/NLRP3 Inflammasome Pathway in Human Aortic Valve Interstitial Cells. Front. Pharmacol. 2020, 11, 826. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, F.; Li, G. Traditional Chinese medicine and lung cancer—From theory to practice. Biomed. Pharmacother. 2021, 137, 111381. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, L.; Du, B.; Liu, Y.; Xing, J.; Guo, S.; Li, L.; Chen, H. Protection against Doxorubicin-Related Cardiotoxicity by Jaceosidin Involves the Sirt1 Signaling Pathway. Oxid. Med. Cell. Longev. 2021, 2021, 9984330. [Google Scholar] [CrossRef]
- Gauthier, T.; Yao, C.; Dowdy, T.; Jin, W.; Lim, Y.J.; Patino, L.C.; Liu, N.; Ohlemacher, S.I.; Bynum, A.; Kazmi, R.; et al. TGF-beta uncouples glycolysis and inflammation in macrophages and controls survival during sepsis. Sci. Signal 2023, 16, eade0385. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Song, Y.; Li, J.; Li, Y.; Yu, Y.; Wang, X. Potential biomarker for diagnosis and therapy of sepsis: Lactylation. Immun. Inflamm. Dis. 2023, 11, e1042. [Google Scholar] [CrossRef]
- Wang, C.; Wang, S.; Wang, Z.; Han, J.; Jiang, N.; Qu, L.; Xu, K. Andrographolide regulates H3 histone lactylation by interfering with p300 to alleviate aortic valve calcification. Br. J. Pharmacol. 2024, 181, 1843–1856. [Google Scholar] [CrossRef]
- Wu, J.; Wang, S.; Gao, P.; Wang, S.; Yu, H.; Du, Q.; Liu, M.; Hou, S.; Jiang, S.; Xu, H.; et al. Jatrorrhizine Alleviates Calcific Aortic Valve Disease via Interfering With Glycolysis Targeting ALDOA K42 Lactylation. Phytother. Res. 2025, 39, 3212–3224. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.L.; Wei, X.C.; Guo, L.Q.; Zhao, L.; Chen, X.H.; Cui, Y.D.; Yuan, J.; Chen, D.F.; Zhang, J. The therapeutic effects of Jaceosidin on lipopolysaccharide-induced acute lung injury in mice. J. Pharmacol. Sci. 2019, 140, 228–235. [Google Scholar] [CrossRef]
- Lee, C.C.; Yang, C.Y.; Su, B.A.; Hsieh, C.C.; Hong, M.Y.; Lee, C.H.; Ko, W.C. The Hypotension Period after Initiation of Appropriate Antimicrobial Administration Is Crucial for Survival of Bacteremia Patients Initially Experiencing Severe Sepsis and Septic Shock. J. Clin. Med. 2020, 9, 2617. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Jang, D.; Jeon, J.; Cho, C.; Choi, S.; Han, S.J.; Oh, E.; Nam, J.; Park, C.H.; Shin, Y.S.; et al. Seomae mugwort and jaceosidin attenuate osteoarthritic cartilage damage by blocking IkappaB degradation in mice. J. Cell. Mol. Med. 2020, 24, 8126–8137. [Google Scholar] [CrossRef]
- Ouyang, Z.; Li, W.; Meng, Q.; Zhang, Q.; Wang, X.; Elgehama, A.; Wu, X.; Shen, Y.; Sun, Y.; Wu, X.; et al. A natural compound jaceosidin ameliorates endoplasmic reticulum stress and insulin resistance via upregulation of SERCA2b. Biomed. Pharmacother. 2017, 89, 1286–1296. [Google Scholar] [CrossRef]
- Lee, T.H.; Jung, H.; Park, K.H.; Bang, M.H.; Baek, N.I.; Kim, J. Jaceosidin, a natural flavone, promotes angiogenesis via activation of VEGFR2/FAK/PI3K/AKT/NF-kappaB signaling pathways in endothelial cells. Exp. Biol. Med. 2014, 239, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Mager, C.E.; Mormol, J.M.; Shelton, E.D.; Murphy, P.R.; Bowman, B.A.; Barley, T.J.; Wang, X.; Linn, S.C.; Liu, K.; Nelin, L.D.; et al. p38 MAPK and MKP-1 control the glycolytic program via the bifunctional glycolysis regulator PFKFB3 during sepsis. J. Biol. Chem. 2023, 299, 103043. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, H.; Wang, C.; Wu, J.; Zan, J.; Liu, Y. Epigenetic Modulation by Lactylation in Sepsis: Linking Metabolism to Immune Dysfunction. J. Inflamm. Res. 2025, 18, 7357–7367. [Google Scholar] [CrossRef]
- Wu, D.; Spencer, C.B.; Ortoga, L.; Zhang, H.; Miao, C. Histone lactylation-regulated METTL3 promotes ferroptosis via m6A-modification on ACSL4 in sepsis-associated lung injury. Redox Biol. 2024, 74, 103194. [Google Scholar] [CrossRef]
- Li, F.; Si, W.; Xia, L.; Yin, D.; Wei, T.; Tao, M.; Cui, X.; Yang, J.; Hong, T.; Wei, R. Positive feedback regulation between glycolysis and histone lactylation drives oncogenesis in pancreatic ductal adenocarcinoma. Mol. Cancer 2024, 23, 90. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, J.; Hong, T.; Chen, X.; Cui, L. SIRT2: Controversy and multiple roles in disease and physiology. Ageing Res. Rev. 2019, 55, 100961. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, Y.; Yang, B.; Sun, S.; Zhang, P.; Luo, Z.; Feng, T.; Cui, Z.; Zhu, T.; Li, Y.; et al. Lactylation of METTL16 promotes cuproptosis via m(6)A-modification on FDX1 mRNA in gastric cancer. Nat. Commun. 2023, 14, 6523. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Guo, S.; Sun, J.; Zhao, Y.; Liu, C. Lactate and lactylation in cardiovascular diseases: Current progress and future perspectives. Metabolism 2024, 158, 155957. [Google Scholar] [CrossRef]
- Qu, L.; Wang, C.; Xu, H.; Li, L.; Liu, Y.; Wan, Q.; Xu, K. Atractylodin targets GLA to regulate D-mannose metabolism to inhibit osteogenic differentiation of human valve interstitial cells and ameliorate aortic valve calcification. Phytother. Res. 2023, 37, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef]
- Yu, H.; Du, Q.; Wu, J.; Feng, F.; Hou, S.; Liu, M.; Wang, S.; Liu, X.; Wang, C.; Xu, K. Gastrodin regulates H3K14la through the CDT2-KAT2A axis to treat Sepsis-induced myocardial dysfunction. Int. Immunopharmacol. 2025, 161, 115065. [Google Scholar] [CrossRef]
- Zhang, M.; Zhu, C.X.; Luo, Z.Y.; Liu, J.H.; Khan, M.T.; Sun, Y.W.; Wei, D.Q.; Zhang, Y.J. Exploring Key Proteins, Pathways and Oxygen Usage Bias of Proteins and Metabolites in Melanoma. J. Comput. Biophys. Chem. 2023. [Google Scholar] [CrossRef]
- Ndongson-Dongmo, B.; Lang, G.P.; Mece, O.; Hechaichi, N.; Lajqi, T.; Hoyer, D.; Brodhun, M.; Heller, R.; Wetzker, R.; Franz, M.; et al. Reduced ambient temperature exacerbates SIRS-induced cardiac autonomic dysregulation and myocardial dysfunction in mice. Basic Res. Cardiol. 2019, 114, 26. [Google Scholar] [CrossRef]
- Yao, Y.Y.; Bian, L.G.; Yang, P.; Sui, Y.; Li, R.; Chen, Y.L.; Sun, L.; Ai, Q.L.; Zhong, L.M.; Lu, D. Gastrodin attenuates proliferation and inflammatory responses in activated microglia through Wnt/beta-catenin signaling pathway. Brain Res. 2019, 1717, 190–203. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Y.; Wang, F.; Xie, G.; Liu, S.; Li, Z.; Wang, P.; Liu, J.; Lin, L. Gastrodin regulates the TLR4/TRAF6/NF-kappaB pathway to reduce neuroinflammation and microglial activation in an AD model. Phytomedicine 2024, 128, 155518. [Google Scholar] [CrossRef]
| Antibody | Cat No | Producer |
|---|---|---|
| TNF-α | 346654 | ZenBio, Chengdu, China |
| IL-1β | 660092 | ZenBio, China |
| IL-6 | 347023 | ZenBio, China |
| BAX | R380709 | ZenBio, China |
| BCL-2 | 381702 | ZenBio, China |
| Caspase-9 | 680095 | ZenBio, China |
| Caspase-3 | R22842 | ZenBio, China |
| β-Actin | R51031 | ZenBio, China |
| Pan Kla | PTM-1425 | PTM BIO, Hangzhou, China |
| Anti-L-Lactyl-Histone H4 (Lys16) | PTM-1417RM | PTM BIO, China |
| Histone H4 | PTM-1015RM | PTM BIO, China |
| Anti-L-Lactyl-Histone H4 (Lys5) | PTM-1409 | PTM BIO, China |
| Anti-L-Lactyl-Histone H4 (Lys12) | PTM-1411 | PTM BIO, China |
| Histone H3 | PTM-6613 | PTM BIO, China |
| Anti-L-Lactyl-Histone H3 (Lys14) | PTM-1414RM | PTM BIO, China |
| Anti-L-Lactyl-Histone H3 (Lys18) | PTM-1406RM | PTM BIO, China |
| Anti-L-Lactyl-Histone H3 (Lys9) | PTM-1419RM | PTM BIO, China |
| Anti-L-Lactyl-Histone H3 (Lys23) | PTM-1413RM | PTM BIO, China |
| Anti-L-Lactyl-Histone H3 (Lys56) | PTM-1421RM | PTM BIO, China |
| Anti-L-Lactyl-Histone H3 (Lys27) | PTM-1428 | PTM BIO, China |
| Anti-Acetyl-Histone H4 (Lys16) | PTM-122 | PTM BIO, China |
| Anti-Acetyl-Histone H4 (Lys5) | PTM-119 | PTM BIO, China |
| Anti-Acetyl-Histone H4 (Lys12) | PTM-121RM | PTM BIO, China |
| Anti-Acetyl-Histone H3 (Lys14) | PTM-113RM | PTM BIO, China |
| Anti-Acetyl-Histone H3 (Lys18) | PTM-114RM | PTM BIO, China |
| Anti-Acetyl-Histone H3 (Lys9) | PTM-112RM | PTM BIO, China |
| Anti-Acetyl-Histone H3 (Lys23) | PTM-115RM | PTM BIO, China |
| Anti-Acetyl-Histone H3 (Lys56) | PTM-162 | PTM BIO, China |
| Anti-Acetyl-Histone H3 (Lys27) | PTM-116RM | PTM BIO, China |
| SIRT7 Polyclonal antibody | 12994-1-AP | Proteintech, Rosemont, IL, USA |
| SIRT4 Rabbit pAb | 862208 | ZenBio, China |
| SIRT2 Rabbit mAb | R25722 | ZenBio, China |
| SIRT6 Rabbit mAb | R381408 | ZenBio, China |
| SIRT3 Rabbit mAb | R25724 | ZenBio, China |
| SIRT5 Rabbit mAb | R381473 | ZenBio, China |
| HDAC8 Rabbit mAb | R381478 | ZenBio, China |
| HDAC4 Rabbit pAb | R381467 | ZenBio, China |
| KAT8 Rabbit mAb | R383056 | ZenBio, China |
| KAT1 Rabbit mAb | R383080 | ZenBio, China |
| KAT5 Mouse mAb | 221268 | ZenBio, China |
| KAT2A Rabbit mAb | R24422 | ZenBio, China |
| Gene | Primer (5′-3′) |
|---|---|
| H-IL-1β | F: AGCACCTTCTTTCCCTTCATCTT R: CACCACTTGTTGCTCCATATCCT |
| M-IL-1β | F: CAGCACATCAACAAGAGCTTCAG R: GAGGATGGGCTCTTCTTCAAAGA |
| H-IL-6 | F: AACATGTGTGAAAGCAGCAAAGA R: CTCTGGCTTGTTCCTCACTACTC |
| M-IL-6 | F: GTATGAACAACGATGATGCACTTG R: CTCTCTGAAGGACTCTGGCTTTG |
| H-TNF-α | F: CTGTAGCCCATGTTGTAGCAAAC R: TTGAAGAGGACCTGGGAGTAGAT |
| M-TNF-α | F: GACCCTCACACTCAGATCATCTT R: CCTTGAAGAGAACCTGGGAGTAG |
| H-BAX | F: GCTTCAGGGTTTCATCCAGGATC R: ATCCTCTGCAGCTCCATGTTACT |
| M-BAX | F: ATCCTCTGCAGCTCCATGTTACT R: TCATCCTCTGCAGCTCCATATTG |
| H-Bcl-2 | F: GGATTGTGGCCTTCTTTGAGTTC R: CTTCAGAGACAGCCAGGAGAAAT |
| M-Bcl-2 | F: GGATTGTGGCCTTCTTTGAGTTC R: CTTCAGAGACAGCCAGGAGAAAT |
| H-caspase-9 | F: TGTCCTACTCTACTTTCCCAGGT R: CCCTTTCACCGAAACAGCATTAG |
| M-caspase-9 | F: CCCGTGGACATTGGTTCTGG R: GAGGAAGGGCAGAAGTTCACA |
| H-caspase-3 | F: TGAGCCATGGTGAAGAAGGAATAA R: CCCGGGTAAGAATGTGCATAAAT |
| M-caspase-3 | F: CAGCCAACCTCAGAGAGACATT R: TTTCAGTTCAACAGGCCCATTTG |
| H-SIRT2 | F: TGCGGAACTTATTCTCCCAGA R: GGAGAGCGAAAGTCGGGGAT |
| M-SIRT2 | F: GCAGAACATAGACACGCTGG R: CCCTGGGAGTTGCTTCTGAGA |
| H-β-actin | F: CTAGGCGGACTGTTACTGAGC R: ATGTTTGCTCCAACCAACTGC |
| M-β-actin | F: AAATCTGGCACCACACCTTCTAC R: CAGCCTGGATAGCAACGTACAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Yu, H.; Liu, M.; Hou, S.; Wu, J.; Du, Q.; Feng, F.; Wang, S.; Wang, C.; Xu, K. Jaceosidin Attenuates Sepsis-Induced Myocardial Dysfunction by Promoting SIRT2-Mediated Inhibition of Histone H3K18 Lactylation. Pharmaceuticals 2026, 19, 97. https://doi.org/10.3390/ph19010097
Yu H, Liu M, Hou S, Wu J, Du Q, Feng F, Wang S, Wang C, Xu K. Jaceosidin Attenuates Sepsis-Induced Myocardial Dysfunction by Promoting SIRT2-Mediated Inhibition of Histone H3K18 Lactylation. Pharmaceuticals. 2026; 19(1):97. https://doi.org/10.3390/ph19010097
Chicago/Turabian StyleYu, Huiming, Minfu Liu, Shuwan Hou, Jiaqin Wu, Qianqian Du, Fan Feng, Sixiang Wang, Chunli Wang, and Kang Xu. 2026. "Jaceosidin Attenuates Sepsis-Induced Myocardial Dysfunction by Promoting SIRT2-Mediated Inhibition of Histone H3K18 Lactylation" Pharmaceuticals 19, no. 1: 97. https://doi.org/10.3390/ph19010097
APA StyleYu, H., Liu, M., Hou, S., Wu, J., Du, Q., Feng, F., Wang, S., Wang, C., & Xu, K. (2026). Jaceosidin Attenuates Sepsis-Induced Myocardial Dysfunction by Promoting SIRT2-Mediated Inhibition of Histone H3K18 Lactylation. Pharmaceuticals, 19(1), 97. https://doi.org/10.3390/ph19010097







