In Silico Development of a Chimeric Multi-Epitope Vaccine Targeting Helcococcus kunzii: Coupling Subtractive Proteomics and Reverse Vaccinology for Vaccine Target Discovery
Abstract
1. Introduction
2. Results
2.1. Proteome Subtraction and Candidate Protein Selection
2.2. Antigenicity Prediction
2.3. Epitope Selection Phase
2.4. Multi-Epitope Vaccine Construction: Integration of Adjuvants and Linkers for Synergistic Immune Activation
2.5. Population Coverage Analysis of Selected CTL and HTL Epitopes
2.6. Post-Translational and Physicochemical Analysis of the Vaccine Construct
2.7. Structural Analysis and Validation of the Multi-Epitope Vaccine Construct
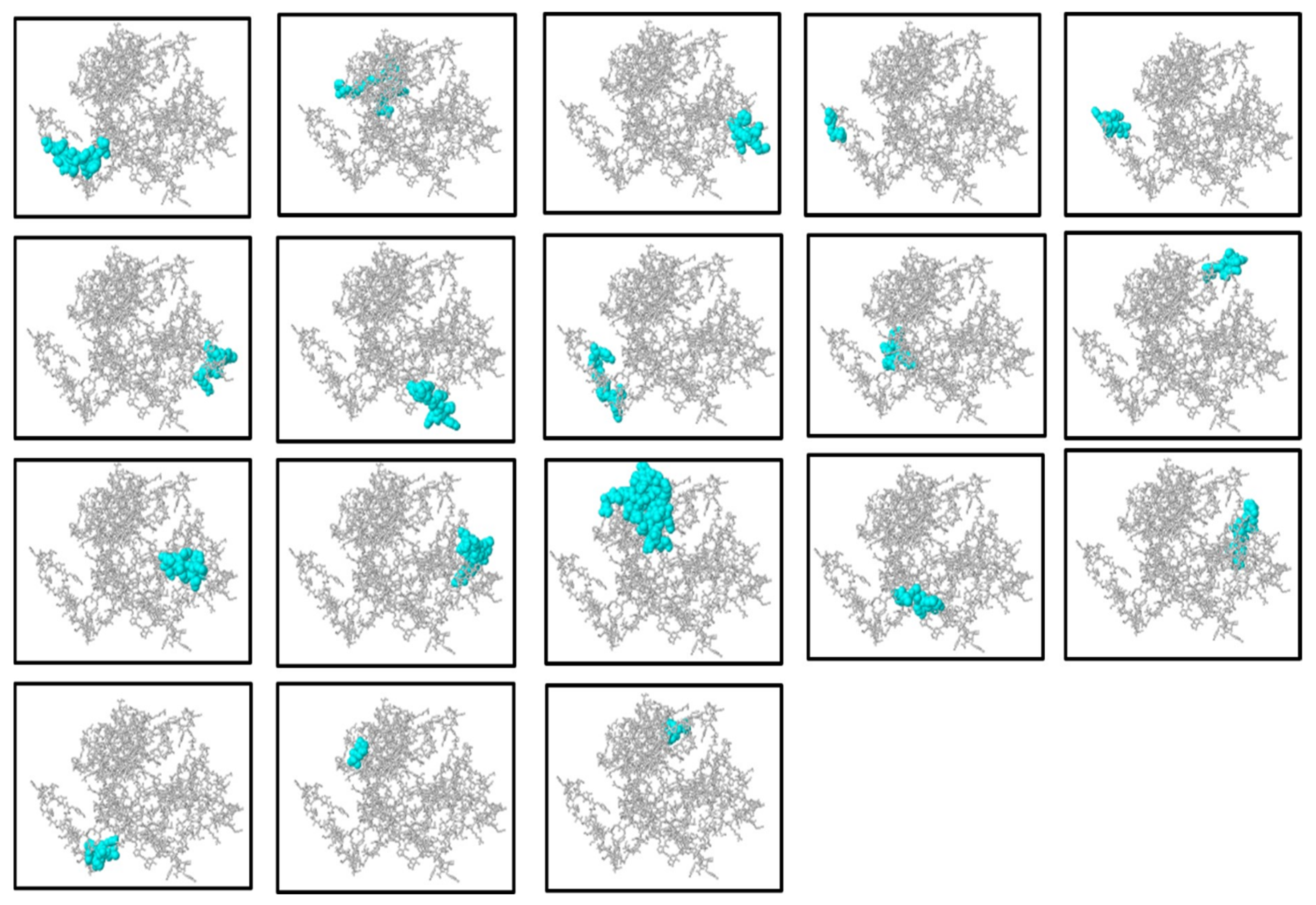
2.8. Prediction and Selection of B-Cell Epitopes
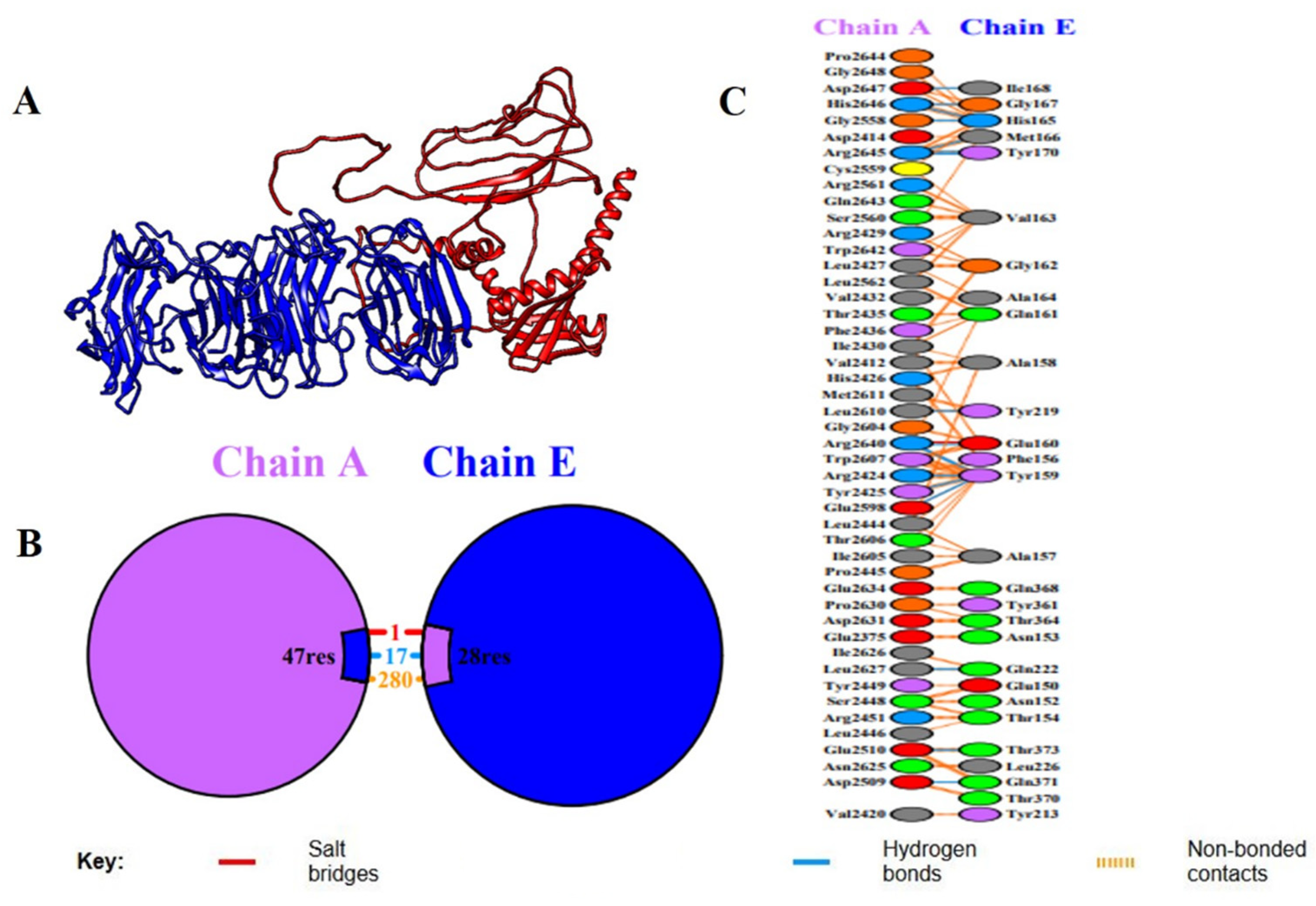
2.9. Molecular Docking Analysis with Host Immune Receptor
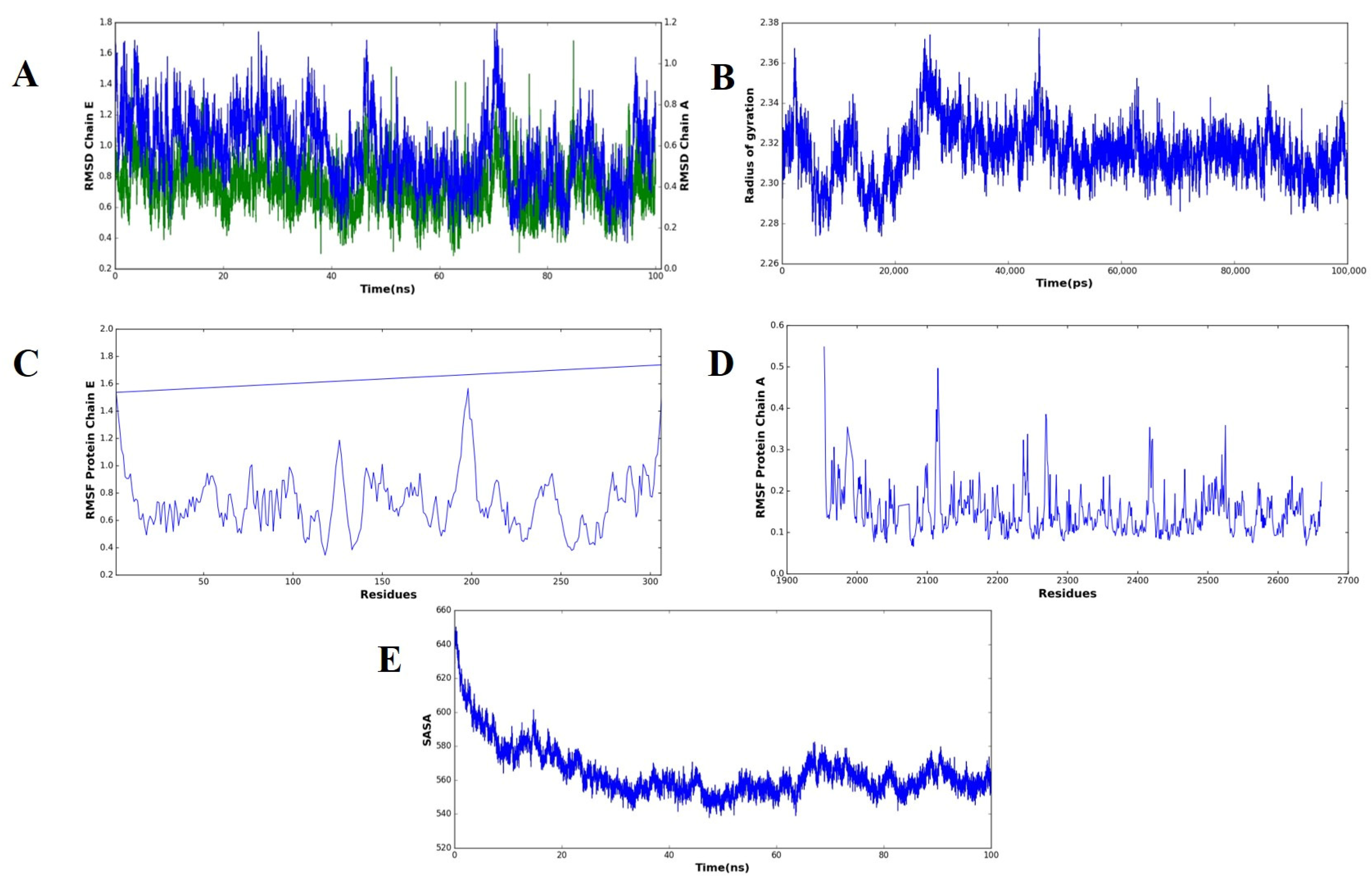
2.10. Molecular Dynamics Simulation of the MEV–TLR4 Complex
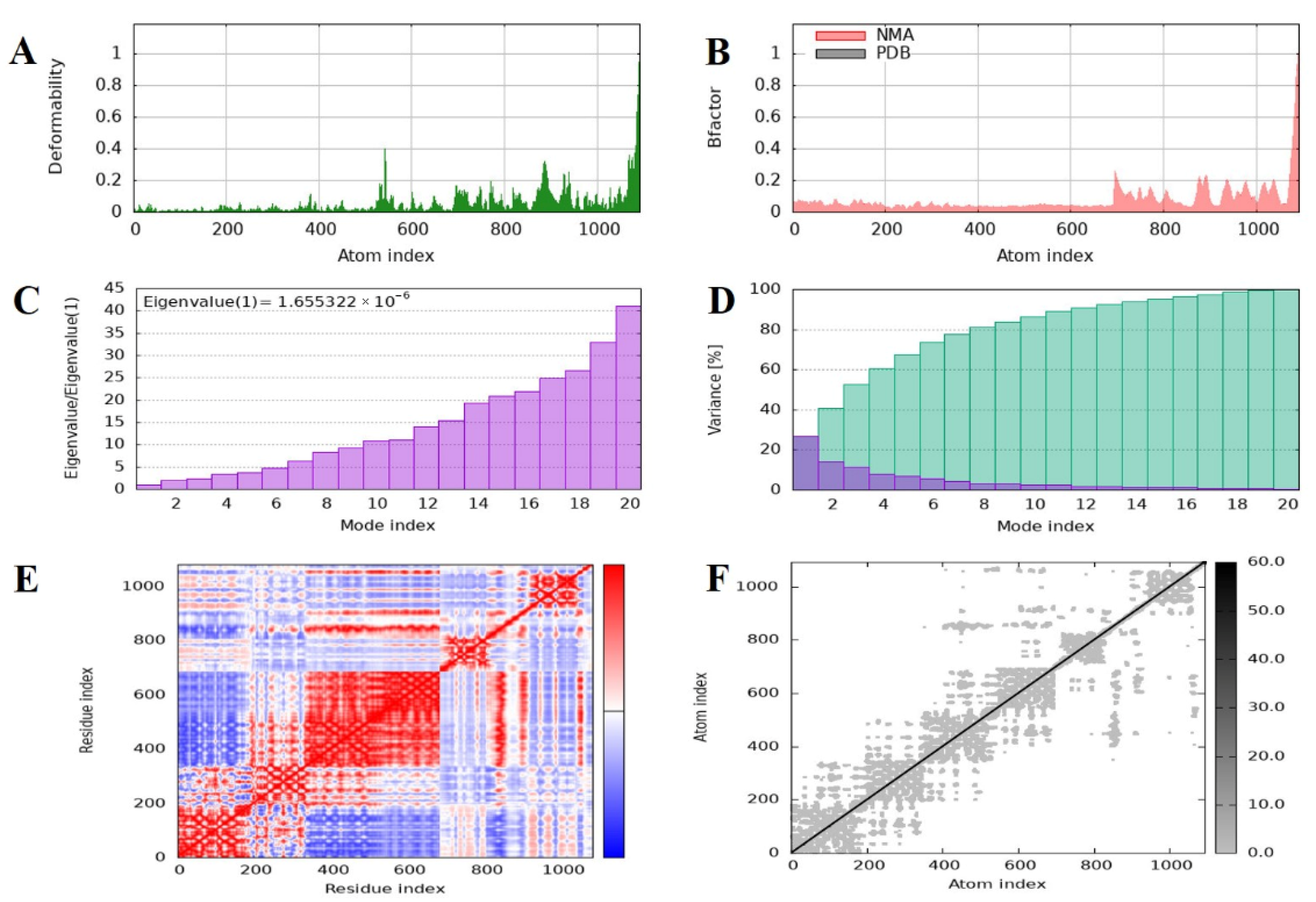
2.11. Normal Mode Analysis of the MEV–TLR4 Complex
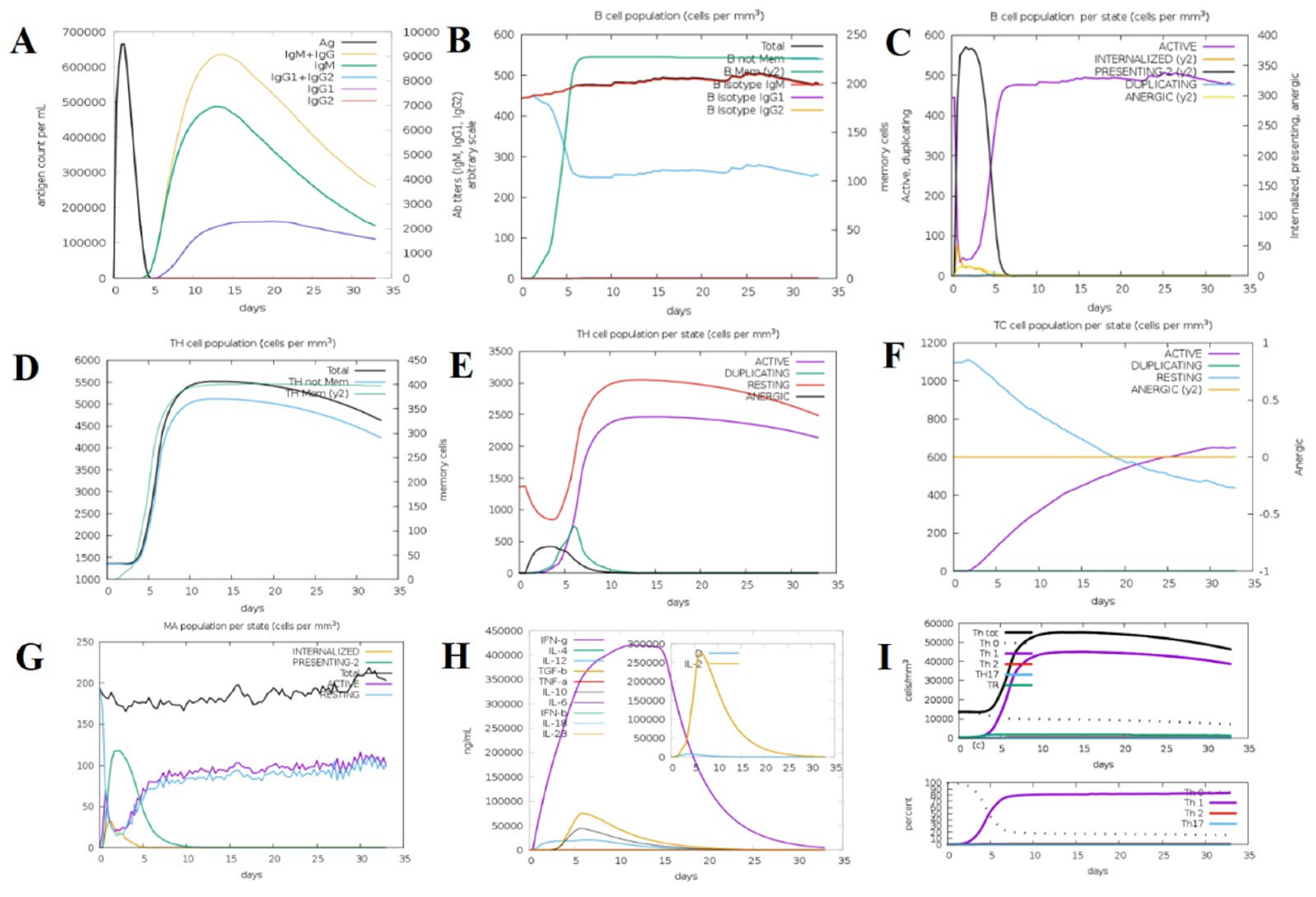
2.12. Immune Simulation Analysis
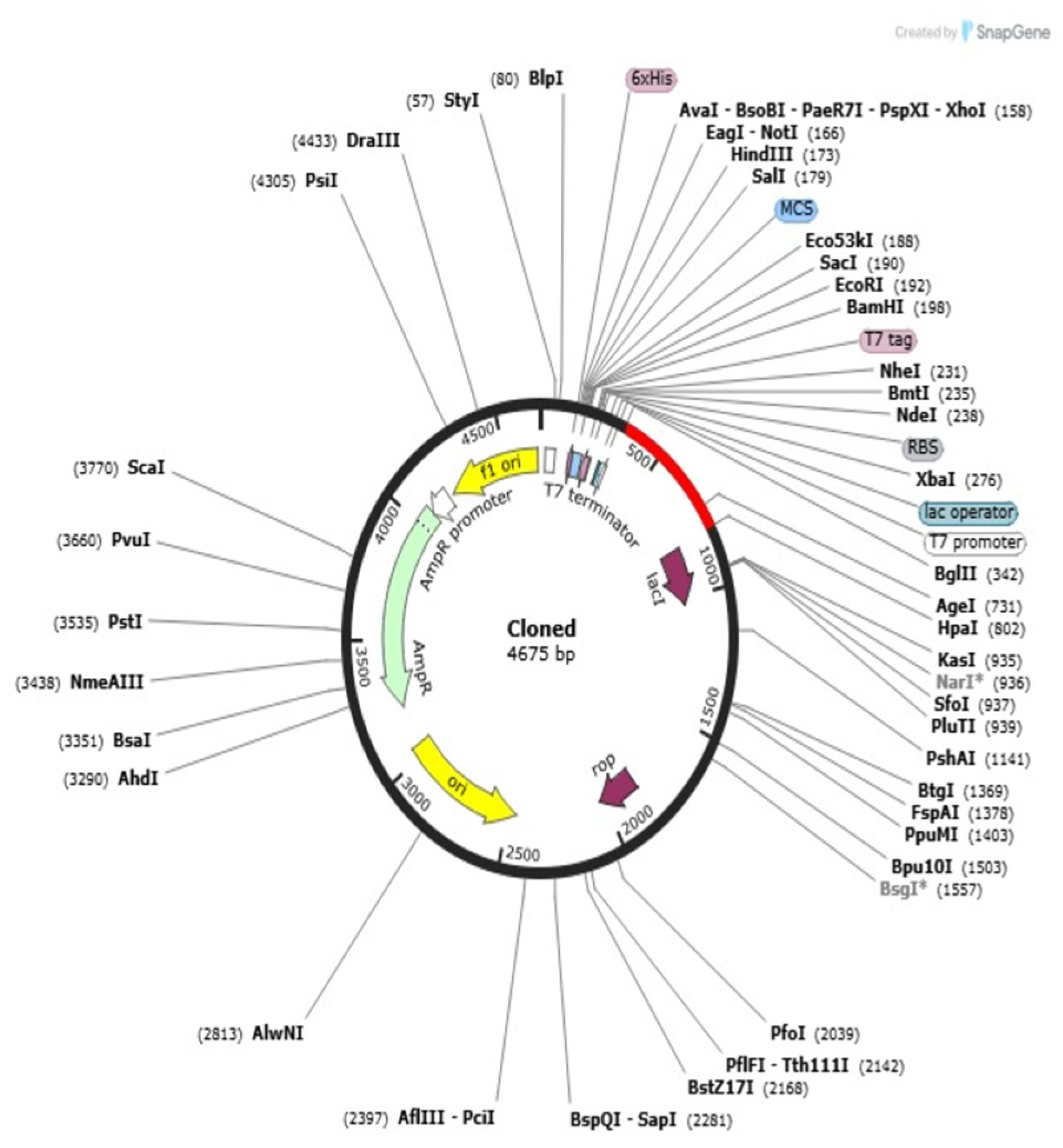
2.13. Codon Adaptation and In Silico Cloning
3. Discussion
4. Materials and Methods
4.1. Protein Sequence Retrieval and Selection
4.2. Prediction and Selection of Cytotoxic T-Lymphocyte (CTL) Epitopes
4.3. Prediction of Helper T-Lymphocyte (HTL) Epitopes
4.4. Prediction and Assessment of B-Cell Epitopes
4.5. Construction of the Multi-Epitope Vaccine (MEV) Sequence
4.6. Secondary Structure Prediction of the Multi-Epitope Vaccine (MEV)
4.7. Tertiary Structure Prediction, Refinement, and Validation of the Multi-Epitope Vaccine (MEV)
4.8. Prediction of Discontinuous B-Cell Epitopes
4.9. Molecular Docking of the MEV Construct with TLR4 Receptor
4.10. Molecular Dynamics Simulation
4.11. Normal Mode Analysis of Docked Complex
4.12. Immune Response Simulation
4.13. Reverse Translation and Codon Optimization
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MEV | Multi-Epitope Vaccine |
| CTL | Cytotoxic T Lymphocyte |
| HTL | Helper T Lymphocyte |
| LBL | Linear B Lymphocyte |
| CTB | Cholera Toxin B Subunit |
| NCBI | National Center for Biotechnology Information |
References
- Mouro, M.; Frade, J.; Chaves, C.; Velez, J. Helcococcus kunzii, a Skin-Colonising Microorganism with Pathogenic Capacity: A Case of Polymicrobial Intracranial Empyema. BMJ Case Rep. 2021, 14, e244600. [Google Scholar] [CrossRef]
- Vergne, A.; Guérin, F.; Lienhard, R.; Le Coustumier, A.; Daurel, C.; Isnard, C.; Marty, N.; Poyart, C.; Cattoir, V. Identification and Clinical Significance of Helcococcus kunzii in Human Samples. J. Clin. Microbiol. 2015, 53, 2703–2705. [Google Scholar] [CrossRef]
- Alaifan, T.; Al-Thaqafi, A.; Munshi, E.; Kaaki, M.; Khojah, A.; Sakhakhni, A. Chest Wall-Necrotizing Soft Tissue Infection Caused by Finegoldia Magna and Helcococcus Kunzii in an Immunocompromised Patient: A Case Report. ASM Case Rep. 2025, 1, e00046-24. [Google Scholar] [CrossRef]
- Pérez-Jorge, C.; Cordero, J.; Marin, M.; Esteban, J. Prosthetic Joint Infection Caused by Helcococcus kunzii. J. Clin. Microbiol. 2012, 50, 528–530. [Google Scholar] [CrossRef]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef]
- Lade, H.; Joo, H.-S.; Kim, J.-S. Molecular Basis of Non-β-Lactam Antibiotics Resistance in Staphylococcus aureus. Antibiotics 2022, 11, 1378. [Google Scholar] [CrossRef]
- Rajput, P.; Nahar, K.S.; Rahman, K.M. Evaluation of Antibiotic Resistance Mechanisms in Gram-Positive Bacteria. Antibiotics 2024, 13, 1197. [Google Scholar] [CrossRef] [PubMed]
- Anju, V.T.; Busi, S.; Imchen, M.; Kumavath, R.; Mohan, M.S.; Salim, S.A.; Subhaswaraj, P.; Dyavaiah, M. Polymicrobial Infections and Biofilms: Clinical Significance and Eradication Strategies. Antibiotics 2022, 11, 1731. [Google Scholar] [CrossRef] [PubMed]
- Micoli, F.; Bagnoli, F.; Rappuoli, R.; Serruto, D. The Role of Vaccines in Combatting Antimicrobial Resistance. Nat. Rev. Microbiol. 2021, 19, 287–302. [Google Scholar] [CrossRef]
- Ghattas, M.; Dwivedi, G.; Lavertu, M.; Alameh, M.-G. Vaccine Technologies and Platforms for Infectious Diseases: Current Progress, Challenges, and Opportunities. Vaccines 2021, 9, 1490. [Google Scholar] [CrossRef] [PubMed]
- Heidary, M.; Kaviar, V.H.; Shirani, M.; Ghanavati, R.; Motahar, M.; Sholeh, M.; Ghahramanpour, H.; Khoshnood, S. A Comprehensive Review of the Protein Subunit Vaccines Against COVID-19. Front. Microbiol. 2022, 13, 927306. [Google Scholar] [CrossRef] [PubMed]
- Hajissa, K.; Zakaria, R.; Suppian, R.; Mohamed, Z. Epitope-Based Vaccine as a Universal Vaccination Strategy against Toxoplasma Gondii Infection: A Mini-Review. J. Adv. Vet. Anim. Res. 2019, 6, 174. [Google Scholar] [CrossRef]
- Yuan, L.; Zhang, S.; Bi, R.; Liu, X.; Han, Z.; Li, M.; Liao, X.; Xie, T.; Bai, S.; Xie, Q.; et al. A Broad-Spectrum Multiepitope Vaccine against Seasonal Influenza A and B Viruses in Mice. eBioMedicine 2024, 106, 105269. [Google Scholar] [CrossRef]
- Dingding, H.; Muhammad, S.; Manzoor, I.; Ghaffar, S.A.; Alodaini, H.A.; Moubayed, N.M.; Hatamleh, A.A.; Songxiao, X. Subtractive Proteomics and Reverse-Vaccinology Approaches for Novel Drug Targets and Designing a Chimeric Vaccine against Ruminococcus Gnavus Strain RJX1120. Front. Immunol. 2025, 16, 1555741. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Chiou, C.-C.; Ahmad, S.; Islam, Z.U.; Tanaka, T.; Alouffi, A.; Chen, C.-C.; Almutairi, M.M.; Ali, A. Subtractive Proteomics and Reverse-Vaccinology Approaches for Novel Drug Target Identification and Chimeric Vaccine Development against Bartonella Henselae Strain Houston-1. Bioengineering 2024, 11, 505. [Google Scholar] [CrossRef] [PubMed]
- Bidmos, F.A.; Gladstone, C.A.; Langford, P.R. Reverse Vaccinology. In Molecular Medical Microbiology; Elsevier: Amsterdam, The Netherlands, 2024; pp. 2023–2038. ISBN 978-0-12-818619-0. [Google Scholar]
- Oli, A.N.; Obialor, W.O.; Ifeanyichukwu, M.O.; Odimegwu, D.C.; Okoyeh, J.N.; Emechebe, G.O.; Adejumo, S.A.; Ibeanu, G.C. Immunoinformatics and Vaccine Development: An Overview. ITT 2020, 9, 13–30. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Bhattacharya, M.; Sharma, G.; Lee, S.-S. Immunoinformatics Approach for the Identification and Characterization of T Cell and B Cell Epitopes towards the Peptide-Based Vaccine against SARS-CoV-2. Arch. Med. Res. 2021, 52, 362–370. [Google Scholar] [CrossRef]
- Jespersen, M.C.; Mahajan, S.; Peters, B.; Nielsen, M.; Marcatili, P. Antibody Specific B-Cell Epitope Predictions: Leveraging Information From Antibody-Antigen Protein Complexes. Front. Immunol. 2019, 10, 298. [Google Scholar] [CrossRef]
- Shaw, S.; Bagchi, A.; Ruj, D.; Bhattacharya, S.P.; Biswas, A.; Bhattacharya, A. A Systemic Immunoinformatics Approach to Design Combinatorial Multiepitope Vaccine Candidates against Vector-Borne Bacterial Infections Exploiting the Proteomes of the Causative Agent and Vector for Scrub Typhus. Microbe 2025, 7, 100324. [Google Scholar] [CrossRef]
- Dar, H.A.; Zaheer, T.; Shehroz, M.; Ullah, N.; Naz, K.; Muhammad, S.A.; Zhang, T.; Ali, A. Immunoinformatics-Aided Design and Evaluation of a Potential Multi-Epitope Vaccine against Klebsiella pneumoniae. Vaccines 2019, 7, 88. [Google Scholar] [CrossRef]
- Ghandadi, M. An Immunoinformatic Strategy to Develop New Mycobacterium tuberculosis Multi-Epitope Vaccine. Int. J. Pept. Res. Ther. 2022, 28, 99. [Google Scholar] [CrossRef]
- Margaroni, M.; Agallou, M.; Tsanaktsidou, E.; Kammona, O.; Kiparissides, C.; Karagouni, E. Immunoinformatics Approach to Design a Multi-Epitope Nanovaccine against Leishmania Parasite: Elicitation of Cellular Immune Responses. Vaccines 2023, 11, 304. [Google Scholar] [CrossRef]
- Maharaj, L.; Adeleke, V.T.; Fatoba, A.J.; Adeniyi, A.A.; Tshilwane, S.I.; Adeleke, M.A.; Maharaj, R.; Okpeku, M. Immunoinformatics Approach for Multi-Epitope Vaccine Design against P. Falciparum Malaria. Infect. Genet. Evol. 2021, 92, 104875. [Google Scholar] [CrossRef]
- Peele, K.A.; Srihansa, T.; Krupanidhi, S.; Ayyagari, V.S.; Venkateswarulu, T.C. Design of Multi-Epitope Vaccine Candidate against SARS-CoV-2: A in-Silico Study. J. Biomol. Struct. Dyn. 2021, 39, 3793–3801. [Google Scholar] [CrossRef]
- Liao, J.; Zhang, X.; Zeng, X.; Zhao, Z.; Sun, T.; Xia, Z.; Jing, H.; Yuan, Y.; Chen, Z.; Gou, Q.; et al. A Rational Designed Multi-Epitope Vaccine Elicited Robust Protective Efficacy against Klebsiella pneumoniae Lung Infection. Biomed. Pharmacother. 2024, 174, 116611. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, N.N.; Barzi, S.M.; Ajdary, S.; Chiani, M.; Yekaninejad, M.S.; Badmasti, F.; Pourmand, M.R. Immunogenic Evaluation of LptD + LtgC as a Bivalent Vaccine Candidate against Neisseria Gonorrhoeae. J. Transl. Med. 2025, 23, 261. [Google Scholar] [CrossRef]
- Mortazavi, B.; Molaei, A.; Fard, N.A. Multi-Epitope Vaccines, from Design to Expression; an in Silico Approach. Hum. Immunol. 2024, 85, 110804. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Xu, L.; Wang, X.; Wang, M.; Cao, Y.; Li, R.; Zheng, K.; Wu, X. A Comprehensive Strategy for the Development of a Multi-Epitope Vaccine Targeting Treponema Pallidum, Utilizing Heat Shock Proteins, Encompassing the Entire Process from Vaccine Design to in Vitro Evaluation of Immunogenicity. Front. Microbiol. 2025, 16, 1551437. [Google Scholar] [CrossRef]
- Garba, B.; Bahaman, A.R.; Zakaria, Z.; Bejo, S.K.; Mutalib, A.R.; Bande, F.; Suleiman, N. Antigenic Potential of a Recombinant Polyvalent DNA Vaccine against Pathogenic Leptospiral Infection. Microb. Pathog. 2018, 124, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.S.; Sethi, G.; Ramadas, K. Design of Multi-Epitope Based Vaccine against Mycobacterium tuberculosis: A Subtractive Proteomics and Reverse Vaccinology Based Immunoinformatics Approach. J. Biomol. Struct. Dyn. 2023, 41, 14116–14134. [Google Scholar] [CrossRef]
- Den Blaauwen, T.; Andreu, J.M.; Monasterio, O. Bacterial Cell Division Proteins as Antibiotic Targets. Bioorg. Chem. 2014, 55, 27–38. [Google Scholar] [CrossRef]
- Kumar, K.; Awasthi, D.; Berger, W.T.; Tonge, P.J.; Slayden, R.A.; Ojima, I. Discovery of Anti-TB Agents That Target the Cell-Division Protein FtsZ. Future Med. Chem. 2010, 2, 1305–1323. [Google Scholar] [CrossRef]
- Kar, T.; Narsaria, U.; Basak, S.; Deb, D.; Castiglione, F.; Mueller, D.M.; Srivastava, A.P. A Candidate Multi-Epitope Vaccine against SARS-CoV-2. Sci. Rep. 2020, 10, 10895. [Google Scholar] [CrossRef] [PubMed]
- Ghaffar, S.A.; Tahir, H.; Muhammad, S.; Shahid, M.; Naqqash, T.; Faisal, M.; Albekairi, T.H.; Alshammari, A.; Albekairi, N.A.; Manzoor, I. Designing of a Multi-Epitopes Based Vaccine against Haemophilius parainfluenzae and Its Validation through Integrated Computational Approaches. Front. Immunol. 2024, 15, 1380732. [Google Scholar] [CrossRef] [PubMed]
- Soltan, M.A.; Behairy, M.Y.; Abdelkader, M.S.; Albogami, S.; Fayad, E.; Eid, R.A.; Darwish, K.M.; Elhady, S.S.; Lotfy, A.M.; Alaa Eldeen, M. In Silico Designing of an Epitope-Based Vaccine Against Common E. Coli Pathotypes. Front. Med. 2022, 9, 829467. [Google Scholar] [CrossRef]
- Stratmann, T. Cholera Toxin Subunit B as Adjuvant––An Accelerator in Protective Immunity and a Break in Autoimmunity. Vaccines 2015, 3, 579–596. [Google Scholar] [CrossRef]
- Zaheer, T.; Waseem, M.; Waqar, W.; Dar, H.A.; Shehroz, M.; Naz, K.; Ishaq, Z.; Ahmad, T.; Ullah, N.; Bakhtiar, S.M.; et al. Anti-COVID-19 Multi-Epitope Vaccine Designs Employing Global Viral Genome Sequences. PeerJ 2020, 8, e9541. [Google Scholar] [CrossRef]
- Zheng, W.; Ruan, J.; Hu, G.; Wang, K.; Hanlon, M.; Gao, J. Analysis of Conformational B-Cell Epitopes in the Antibody-Antigen Complex Using the Depth Function and the Convex Hull. PLoS ONE 2015, 10, e0134835. [Google Scholar] [CrossRef]
- Tahir Ul Qamar, M.; Rehman, A.; Tusleem, K.; Ashfaq, U.A.; Qasim, M.; Zhu, X.; Fatima, I.; Shahid, F.; Chen, L.-L. Designing of a next Generation Multiepitope Based Vaccine (MEV) against SARS-CoV-2: Immunoinformatics and in Silico Approaches. PLoS ONE 2020, 15, e0244176. [Google Scholar] [CrossRef]
- Samad, A.; Ahammad, F.; Nain, Z.; Alam, R.; Imon, R.R.; Hasan, M.; Rahman, M.S. Designing a Multi-Epitope Vaccine against SARS-CoV-2: An Immunoinformatics Approach. J. Biomol. Struct. Dyn. 2022, 40, 14–30. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Arora, N.; Rawat, S.S.; Keshri, A.K.; Singh, N.; Show, S.K.; Kumar, P.; Mishra, A.; Prasad, A. Immunoinformatics Driven Construction of Multi-Epitope Vaccine Candidate against Ascaris lumbricoides Using Its Entire Immunogenic Epitopes. Expert Rev. Vaccines 2021, 20, 1637–1649. [Google Scholar] [CrossRef]
- Balodi, D.C.; Anand, A.; Ramalingam, K.; Yadav, S.; Goyal, N. Dipeptidylcarboxypeptidase of Leishmania Donovani: A Potential Vaccine Molecule against Experimental Visceral Leishmaniasis. Cell. Immunol. 2022, 375, 104529. [Google Scholar] [CrossRef]
- Dikhit, M.R.; Kumar, A.; Das, S.; Dehury, B.; Rout, A.K.; Jamal, F.; Sahoo, G.C.; Topno, R.K.; Pandey, K.; Das, V.N.R.; et al. Identification of Potential MHC Class-II-Restricted Epitopes Derived from Leishmania donovani Antigens by Reverse Vaccinology and Evaluation of Their CD4+ T-Cell Responsiveness against Visceral Leishmaniasis. Front. Immunol. 2017, 8, 1763. [Google Scholar] [CrossRef] [PubMed]
- Makrides, S.C. Strategies for Achieving High-Level Expression of Genes in Escherichia coli. Microbiol. Rev. 1996, 60, 512–538. [Google Scholar] [CrossRef]
- The UniProt Consortium The Universal Protein Resource (UniProt). Nucleic Acids Res. 2007, 36, D190–D195. [CrossRef]
- The UniProt Consortium. UniProt: A Hub for Protein Information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Wei, W.; Ning, L.-W.; Ye, Y.-N.; Guo, F.-B. Geptop: A Gene Essentiality Prediction Tool for Sequenced Bacterial Genomes Based on Orthology and Phylogeny. PLoS ONE 2013, 8, e72343. [Google Scholar] [CrossRef] [PubMed]
- Chao, P.; Zhang, X.; Zhang, L.; Yang, A.; Wang, Y.; Chen, X. Proteomics-Based Vaccine Targets Annotation and Design of Multi-Epitope Vaccine against Antibiotic-Resistant Streptococcus gallolyticus. Sci. Rep. 2024, 14, 4836. [Google Scholar] [CrossRef] [PubMed]
- Wen, Q.-F.; Liu, S.; Dong, C.; Guo, H.-X.; Gao, Y.-Z.; Guo, F.-B. Geptop 2.0: An Updated, More Precise, and Faster Geptop Server for Identification of Prokaryotic Essential Genes. Front. Microbiol. 2019, 10, 1236. [Google Scholar] [CrossRef]
- McGinnis, S.; Madden, T.L. BLAST: At the Core of a Powerful and Diverse Set of Sequence Analysis Tools. Nucleic Acids Res. 2004, 32, W20–W25. [Google Scholar] [CrossRef]
- Pertsemlidis, A.; Fondon, J.W. Having a BLAST with Bioinformatics (and Avoiding BLASTphemy). Genome Biol. 2001, 2, reviews2002.1. [Google Scholar] [CrossRef]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved Protein Subcellular Localization Prediction with Refined Localization Subcategories and Predictive Capabilities for All Prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Acar, M.B.; Ayaz-Güner, Ş.; Güner, H.; Dinç, G.; Ulu Kılıç, A.; Doğanay, M.; Özcan, S. A Subtractive Proteomics Approach for the Identification of Immunodominant Acinetobacter Baumannii Vaccine Candidate Proteins. Front. Immunol. 2022, 13, 1001633. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, F.; Textor, J.; Van Den Bogaart, G. Transmembrane Helices Are an Overlooked Source of Major Histocompatibility Complex Class I Epitopes. Front. Immunol. 2017, 8, 1118. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A Server for Prediction of Protective Antigens, Tumour Antigens and Subunit Vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Ong, E.; Cooke, M.F.; Huffman, A.; Xiang, Z.; Wong, M.U.; Wang, H.; Seetharaman, M.; Valdez, N.; He, Y. Vaxign2: The Second Generation of the First Web-Based Vaccine Design Program Using Reverse Vaccinology and Machine Learning. Nucleic Acids Res. 2021, 49, W671–W678. [Google Scholar] [CrossRef]
- Weigelin, B.; Den Boer, A.T.; Wagena, E.; Broen, K.; Dolstra, H.; De Boer, R.J.; Figdor, C.G.; Textor, J.; Friedl, P. Cytotoxic T Cells Are Able to Efficiently Eliminate Cancer Cells by Additive Cytotoxicity. Nat. Commun. 2021, 12, 5217. [Google Scholar] [CrossRef]
- Hewitt, E.W. The MHC Class I Antigen Presentation Pathway: Strategies for Viral Immune Evasion. Immunology 2003, 110, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Rencilin, C.F.; Rosy, J.C.; Mohan, M.; Coico, R.; Sundar, K. Identification of SARS-CoV-2 CTL Epitopes for Development of a Multivalent Subunit Vaccine for COVID-19. Infect. Genet. Evol. 2021, 89, 104712. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.; Andreatta, M.; Peters, B.; Buus, S. Immunoinformatics: Predicting Peptide–MHC Binding. Annu. Rev. Biomed. Data Sci. 2020, 3, 191–215. [Google Scholar] [CrossRef]
- Hisham, Y.; Ashhab, Y.; Hwang, S.-H.; Kim, D.-E. Identification of Highly Conserved SARS-CoV-2 Antigenic Epitopes with Wide Coverage Using Reverse Vaccinology Approach. Viruses 2021, 13, 787. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2—A Server for in Silico Prediction of Allergens. J. Mol. Model 2014, 20, 2278. [Google Scholar] [CrossRef]
- Mintaev, R.R.; Glazkova, D.V.; Bogoslovskaya, E.V.; Shipulin, G.A. Immunogenic Epitope Prediction to Create a Universal Influenza Vaccine. Heliyon 2022, 8, e09364. [Google Scholar] [CrossRef] [PubMed]
- Yaseen, A.R.; Suleman, M.; Qadri, A.S.; Asghar, A.; Arshad, I.; Khan, D.M. Development of Conserved Multi-Epitopes Based Hybrid Vaccine against SARS-CoV-2 Variants: An Immunoinformatic Approach. Silico Pharmacol. 2023, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.; Fikes, J.; Hoffman, S.; Franke, E.; Sacci, J.; Appella, E.; Chisari, F.V.; Guidotti, L.G.; Chesnut, R.W.; Livingston, B.; et al. The Optimization of Helper T Lymphocyte (HTL) Function in Vaccine Development. Immunol. Res. 1998, 18, 79–92. [Google Scholar] [CrossRef]
- Frei, R.; Steinle, J.; Birchler, T.; Loeliger, S.; Roduit, C.; Steinhoff, D.; Seibl, R.; Büchner, K.; Seger, R.; Reith, W.; et al. MHC Class II Molecules Enhance Toll-Like Receptor Mediated Innate Immune Responses. PLoS ONE 2010, 5, e8808. [Google Scholar] [CrossRef] [PubMed]
- Jagadeb, M.; Pattanaik, K.P.; Rath, S.N.; Sonawane, A. Identification and Evaluation of Immunogenic MHC-I and MHC-II Binding Peptides from Mycobacterium tuberculosis. Comput. Biol. Med. 2021, 130, 104203. [Google Scholar] [CrossRef]
- Wang, P.; Sidney, J.; Dow, C.; Mothé, B.; Sette, A.; Peters, B. A Systematic Assessment of MHC Class II Peptide Binding Predictions and Evaluation of a Consensus Approach. PLoS Comput. Biol. 2008, 4, e1000048. [Google Scholar] [CrossRef]
- Ahmad, H.I.; Jabbar, A.; Mushtaq, N.; Javed, Z.; Hayyat, M.U.; Bashir, J.; Naseeb, I.; Abideen, Z.U.; Ahmad, N.; Chen, J. Immune Tolerance vs. Immune Resistance: The Interaction Between Host and Pathogens in Infectious Diseases. Front. Vet. Sci. 2022, 9, 827407. [Google Scholar] [CrossRef]
- Kottarathil, A.; Murugan, G.; Rajkumar, D.S.; Chandran, A.K.; Elumalai, V.; Padmanaban, R. Designing Multi-Epitope-Based Vaccine Targeting Immunogenic Proteins of Streptococcus Mutans Using Immunoinformatics to Prevent Caries. Microbe 2025, 7, 100320. [Google Scholar] [CrossRef]
- Cyster, J.G.; Wilson, P.C. Antibody Modulation of B Cell Responses—Incorporating Positive and Negative Feedback. Immunity 2024, 57, 1466–1481. [Google Scholar] [CrossRef]
- Singh, H.; Ansari, H.R.; Raghava, G.P.S. Improved Method for Linear B-Cell Epitope Prediction Using Antigen’s Primary Sequence. PLoS ONE 2013, 8, e62216. [Google Scholar] [CrossRef]
- Singh, J.; Malik, D.; Raina, A. Immuno-Informatics Approach for B-Cell and T-Cell Epitope Based Peptide Vaccine Design against Novel COVID-19 Virus. Vaccine 2021, 39, 1087–1095. [Google Scholar] [CrossRef]
- Mukherjee, S.; Tworowski, D.; Detroja, R.; Mukherjee, S.B.; Frenkel-Morgenstern, M. Immunoinformatics and Structural Analysis for Identification of Immunodominant Epitopes in SARS-CoV-2 as Potential Vaccine Targets. Vaccines 2020, 8, 290. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Selvaraj, G.; Gopalan, V.; Padmanabhan, P.; Ramesh, K.; Govindan, K.; Chandran, A.; Dhandapani, P.; Krishnasamy, K.; Kitambi, S. Epitope Identification and Designing a Potent Multi-Epitope Vaccine Construct against SARS-CoV-2 Including the Emerging Variants. J. Glob. Infect Dis. 2022, 14, 24. [Google Scholar] [CrossRef]
- Sarkar, I.; Garg, R.; Van Drunen Littel-van Den Hurk, S. Selection of Adjuvants for Vaccines Targeting Specific Pathogens. Expert Rev. Vaccines 2019, 18, 505–521. [Google Scholar] [CrossRef]
- Hou, J.; Liu, Y.; Hsi, J.; Wang, H.; Tao, R.; Shao, Y. Cholera Toxin B Subunit Acts as a Potent Systemic Adjuvant for HIV-1 DNA Vaccination Intramuscularly in Mice. Hum. Vaccines Immunother. 2014, 10, 1274–1283. [Google Scholar] [CrossRef]
- Liljeqvist, S.; Ståhl, S.; Andréoni, C.; Binz, H.; Uhlén, M.; Murby, M. Fusions to the Cholera Toxin B Subunit: Influence on Pentamerization and GM1 Binding. J. Immunol. Methods 1997, 210, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Najeebullah; Haq, I.U.; Rahiyab, M.; Ali, S.S.; Khan, I.; Iqbal, A. Rational In-Silico Design of a Multi-Epitope Vaccine against Human Rhinovirus an Immune Simulation and Molecular Dynamics Simulation Approach. Vacunas 2025, 26, 500427. [Google Scholar] [CrossRef]
- Umar, A.; Haque, A.; Alghamdi, Y.S.; Mashraqi, M.M.; Rehman, A.; Shahid, F.; Khurshid, M.; Ashfaq, U.A. Development of a Candidate Multi-Epitope Subunit Vaccine against Klebsiella Aerogenes: Subtractive Proteomics and Immuno-Informatics Approach. Vaccines 2021, 9, 1373. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Jiang, H.; Qiu, M.; Liu, L.; Zou, S.; Li, Y.; Guo, Q.; Han, N.; Sun, Y.; Wang, K.; et al. Multi-Epitope Vaccine Design Using an Immunoinformatic Approach for SARS-CoV-2. Pathogens 2021, 10, 737. [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPMA: Significant Improvements in Protein Secondary Structure Prediction by Consensus Prediction from Multiple Alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef]
- McGuffin, L.J.; Bryson, K.; Jones, D.T. The PSIPRED Protein Structure Prediction Server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef]
- Sarvmeili, J.; Baghban Kohnehrouz, B.; Gholizadeh, A.; Shanehbandi, D.; Ofoghi, H. Immunoinformatics Design of a Structural Proteins Driven Multi-Epitope Candidate Vaccine against Different SARS-CoV-2 Variants Based on Fynomer. Sci. Rep. 2024, 14, 10297. [Google Scholar] [CrossRef]
- Saadh, M.J.; Ahmed, H.H.; Kareem, R.A.; Baldaniya, L.; Verma, L.; Prasad, G.V.S.; Chahar, M.; Taher, W.M.; Alwan, M.; Jawad, M.J.; et al. Design of a Novel Multi-Epitope Vaccine Candidate against Yersinia Pestis Using Advanced Immunoinformatics Approaches: An in Silico Study. Biochem. Biophys. Rep. 2024, 40, 101871. [Google Scholar] [CrossRef]
- Bertoline, L.M.F.; Lima, A.N.; Krieger, J.E.; Teixeira, S.K. Before and after AlphaFold2: An Overview of Protein Structure Prediction. Front. Bioinform. 2023, 3, 1120370. [Google Scholar] [CrossRef]
- Skolnick, J.; Gao, M.; Zhou, H.; Singh, S. AlphaFold 2: Why It Works and Its Implications for Understanding the Relationships of Protein Sequence, Structure, and Function. J. Chem. Inf. Model. 2021, 61, 4827–4831. [Google Scholar] [CrossRef] [PubMed]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein Structure Refinement Driven by Side-Chain Repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef]
- Wang, W.; Xia, M.; Chen, J.; Deng, F.; Yuan, R.; Zhang, X.; Shen, F. Data Set for Phylogenetic Tree and RAMPAGE Ramachandran Plot Analysis of SODs in Gossypium raimondii and G. Arboreum. Data Brief 2016, 9, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Sefid, F.; Rasooli, I.; Jahangiri, A. In Silico Determination and Validation of Baumannii Acinetobactin Utilization A Structure and Ligand Binding Site. BioMed Res. Int. 2013, 2013, 172784. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Tang, F.; Wang, J.; Cheng, P.; Wang, L.; Gong, W. Immunoinformatic-Based Multi-Epitope Vaccine Design for Co-Infection of Mycobacterium tuberculosis and SARS-CoV-2. J. Pers. Med. 2023, 13, 116. [Google Scholar] [CrossRef]
- Ponomarenko, J.; Bui, H.-H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. ElliPro: A New Structure-Based Tool for the Prediction of Antibody Epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef] [PubMed]
- Ansari, H.; Raghava, G.P. Identification of Conformational B-Cell Epitopes in an Antigen from Its Primary Sequence. Immunome Res. 2010, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Duquesnoy, R.J.; Marrari, M. Usefulness of the ElliPro Epitope Predictor Program in Defining the Repertoire of HLA-ABC Eplets. Hum. Immunol. 2017, 78, 481–488. [Google Scholar] [CrossRef] [PubMed]
- De Groot, A.S.; Moise, L.; McMurry, J.A.; Martin, W. Epitope-Based Immunome-Derived Vaccines: A Strategy for Improved Design and Safety. In Clinical Applications of Immunomics; Falus, A., Ed.; Springer US: New York, NY, USA, 2009; pp. 39–69. ISBN 978-0-387-79207-1. [Google Scholar]
- Ali, A.; Ali, S.L.; Alamri, A.; Khatrawi, E.M.; Baiduissenova, A.; Suleimenova, F.; Mishra, V.K.; Khan, A.; Dusmagambetov, M.; Askarova, G. Multi-Epitope-Based Vaccine Models Prioritization against Astrovirus MLB1 Using Immunoinformatics and Reverse Vaccinology Approaches. J. Genet. Eng. Biotechnol. 2025, 23, 100451. [Google Scholar] [CrossRef]
- Sudhakar, K.; Dugar, N.; Jupudi, S.; Ashwin, R.; Gowthamarajan, K. Unveiling Molecular Insights: In Silico Exploration of TLR4 Antagonist for Management of Dry Eye Syndrome. BMJ Open Ophth 2024, 9, e001610. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro Web Server for Protein–Protein Docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Zhuang, L.; Ali, A.; Yang, L.; Ye, Z.; Li, L.; Ni, R.; An, Y.; Ali, S.L.; Gong, W. Leveraging Computer-Aided Design and Artificial Intelligence to Develop a next-Generation Multi-Epitope Tuberculosis Vaccine Candidate. Infect. Med. 2024, 3, 100148. [Google Scholar] [CrossRef]
- Giri-Rachman, E.A.; Kurnianti, A.M.F.; Rizarullah; Setyadi, A.H.; Artarini, A.; Tan, M.I.; Riani, C.; Natalia, D.; Aditama, R.; Nugrahapraja, H. An Immunoinformatics Approach in Designing High-Coverage mRNA Multi-Epitope Vaccine against Multivariant SARS-CoV-2. J. Genet. Eng. Biotechnol. 2025, 23, 100524. [Google Scholar] [CrossRef]
- Jyotisha; Singh, S.; Qureshi, I.A. Multi-Epitope Vaccine against SARS-CoV-2 Applying Immunoinformatics and Molecular Dynamics Simulation Approaches. J. Biomol. Struct. Dyn. 2022, 40, 2917–2933. [Google Scholar] [CrossRef]
- Moradkasani, S.; Esmaeili, S.; Asadi Karam, M.R.; Mostafavi, E.; Shahbazi, B.; Salek Farrokhi, A.; Chiani, M.; Badmasti, F. Development of a Multi-Epitope Vaccine from Outer Membrane Proteins and Identification of Novel Drug Targets against Francisella Tularensis: An In Silico Approach. Front. Immunol. 2025, 16, 1479862. [Google Scholar] [CrossRef] [PubMed]
- AlMalki, F. In Silico Subtractive Proteome Analysis to Design Multi-Epitope-Based Subunit Vaccine against Eikenella corrodens. J. Microbiol. Biotechnol. 2024, 35, e2410015. [Google Scholar] [CrossRef]
- Shang, K.; Zhu, Y.; Tian, T.; Shi, H.; Yin, Z.; He, Y.; Shi, J.; Ding, J.; Zhang, F. Development of a Novel Multi-Epitope Vaccine for Brucellosis Prevention. Heliyon 2024, 10, e34721. [Google Scholar] [CrossRef]
- López-Blanco, J.R.; Aliaga, J.I.; Quintana-Ortí, E.S.; Chacón, P. iMODS: Internal Coordinates Normal Mode Analysis Server. Nucleic Acids Res. 2014, 42, W271–W276. [Google Scholar] [CrossRef]
- Bauer, J.A.; Pavlović, J.; Bauerová-Hlinková, V. Normal Mode Analysis as a Routine Part of a Structural Investigation. Molecules 2019, 24, 3293. [Google Scholar] [CrossRef]
- Bahar, I.; Lezon, T.R.; Bakan, A.; Shrivastava, I.H. Normal Mode Analysis of Biomolecular Structures: Functional Mechanisms of Membrane Proteins. Chem. Rev. 2010, 110, 1463–1497. [Google Scholar] [CrossRef]
- Skjaerven, L.; Martinez, A.; Reuter, N. Principal Component and Normal Mode Analysis of Proteins; a Quantitative Comparison Using the GroEL Subunit. Proteins 2011, 79, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Orellana, L. Large-Scale Conformational Changes and Protein Function: Breaking the in Silico Barrier. Front. Mol. Biosci. 2019, 6, 117. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, L.; Zhao, Y.; Yang, L.; Li, L.; Ye, Z.; Ali, A.; An, Y.; Ni, R.; Ali, S.L.; Gong, W. Harnessing Bioinformatics for the Development of a Promising Multi-Epitope Vaccine against Tuberculosis: The ZL9810L Vaccine. Decod. Infect. Transm. 2024, 2, 100026. [Google Scholar] [CrossRef]
- Baldazzi, V.; Castiglione, F.; Bernaschi, M. An Enhanced Agent Based Model of the Immune System Response. Cell. Immunol. 2006, 244, 77–79. [Google Scholar] [CrossRef]
- Beig, M.; Sholeh, M.; Moradkasani, S.; Shahbazi, B.; Badmasti, F. Development of a Multi-Epitope Vaccine against Acinetobacter Baumannii: A Comprehensive Approach to Combating Antimicrobial Resistance. PLoS ONE 2025, 20, e0319191. [Google Scholar] [CrossRef]
- Fatahi, G.; Abdollahi, M.; Nashtahosseini, Z.; Minoo, S.; Mostafavi, M.; Saeidi, K. Designing of an Efficient DC-Inducing Multi-Epitope Vaccine against Epstein Barr Virus Targeting the GP350 Using Immunoinformatics and Molecular Dynamic Simulation. Biochem. Biophys. Rep. 2025, 42, 101966. [Google Scholar] [CrossRef]
- Grote, A.; Hiller, K.; Scheer, M.; Munch, R.; Nortemann, B.; Hempel, D.C.; Jahn, D. JCat: A Novel Tool to Adapt Codon Usage of a Target Gene to Its Potential Expression Host. Nucleic Acids Res. 2005, 33, W526–W531. [Google Scholar] [CrossRef]
- Chaudhary, N.; Singh, N.K.; Tyagi, A.; Kumari, A. A Detailed Analysis of Codon Usages Bias and Influencing Factors in the Nucleocapsid Gene of Nipah Virus. Microbe 2023, 1, 100014. [Google Scholar] [CrossRef]
- Paremskaia, A.I.; Kogan, A.A.; Murashkina, A.; Naumova, D.A.; Satish, A.; Abramov, I.S.; Feoktistova, S.G.; Mityaeva, O.N.; Deviatkin, A.A.; Volchkov, P.Y. Codon-Optimization in Gene Therapy: Promises, Prospects and Challenges. Front. Bioeng. Biotechnol. 2024, 12, 1371596. [Google Scholar] [CrossRef]
- Xu, L.; Xie, N.; Liu, Y.; Tang, H.; Li, T.; Peng, J.; Li, R. Development of a Novel Multi-Epitope Vaccine Against STREPTOCOCCUS ANGINOSUS Infection via Reverse Vaccinology Approach. Immunology 2025, 175, 339–358. [Google Scholar] [CrossRef]
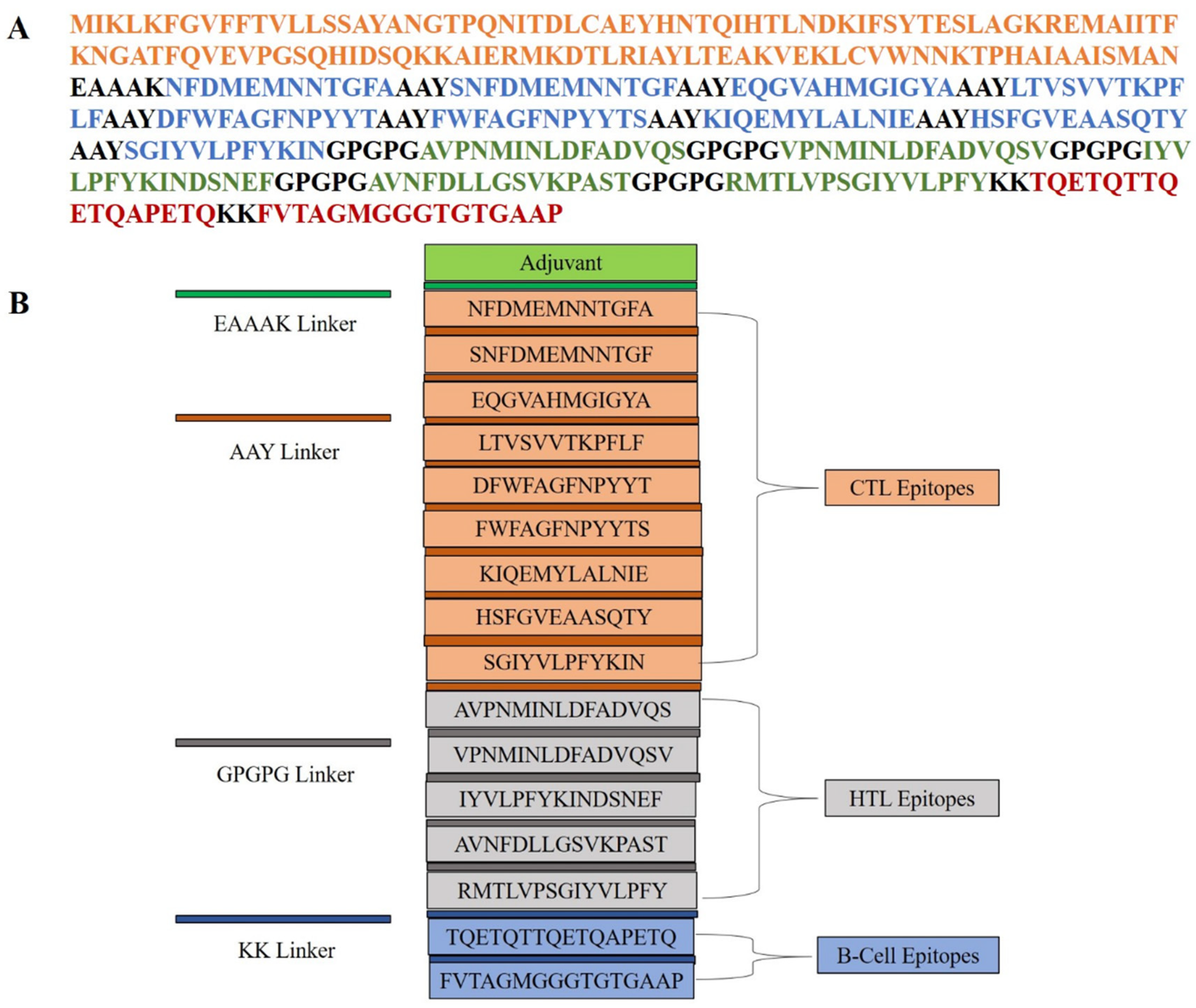
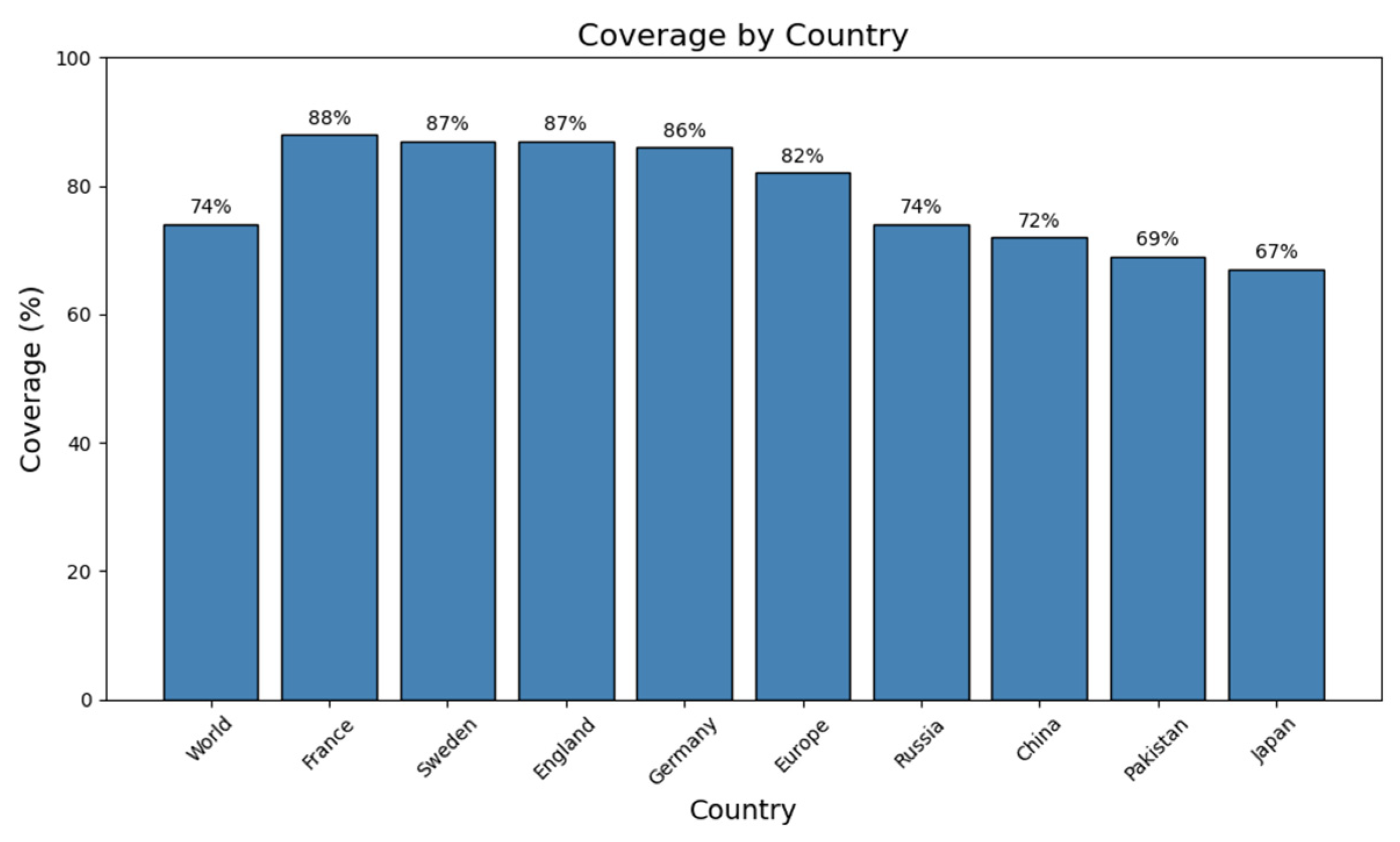
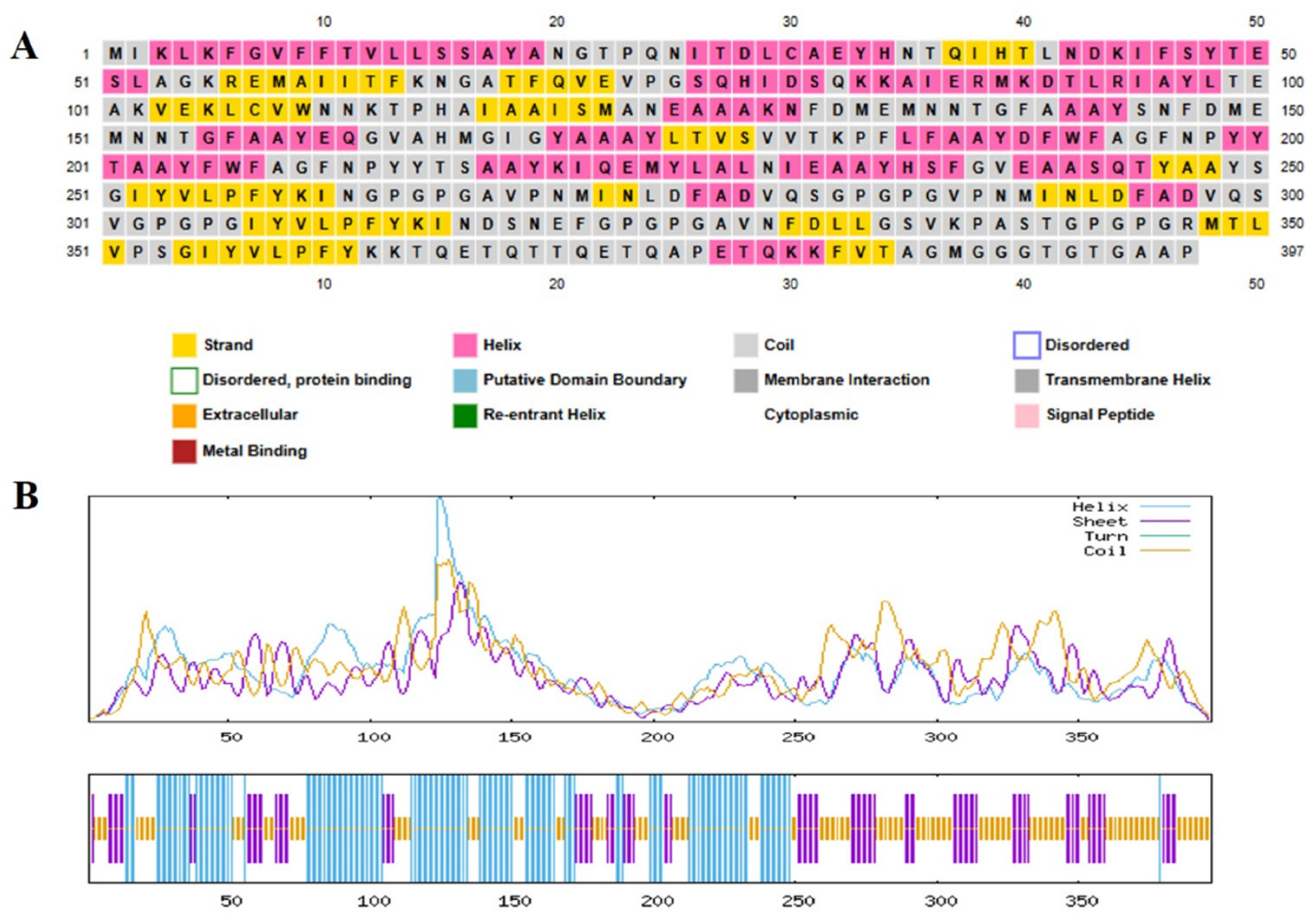
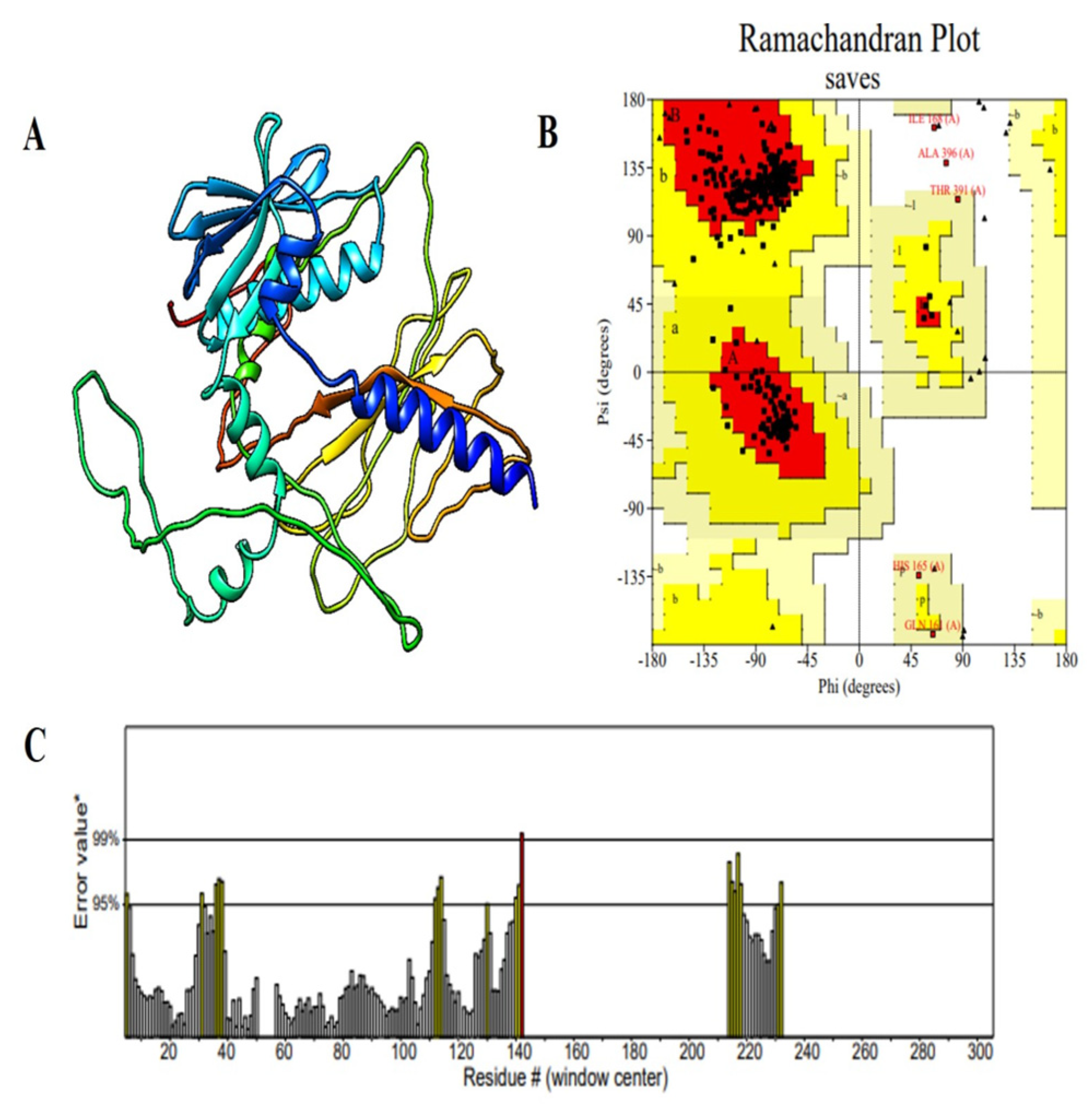
| Accession No. | Protein | Antigenicity | Allergenicity | Toxicity |
|---|---|---|---|---|
| H3NNL9 | Peptidoglycan glycosyltransferase FtsW | 0.5169 | Non-allergenic | Non-toxic |
| H3NNK7 | Cell division protein FtsZ | 0.5439 | Non-allergenic | Non-toxic |
| Epitope | Protein | Allele | Position | Antigenicity | Immunogenicity |
|---|---|---|---|---|---|
| NFDMEMNNTGFA | Cell division protein FtsZ | HLA-B*15:02 | 3–14 | 1.1028 | −0.12206 |
| SNFDMEMNNTGF | Cell division protein FtsZ | HLA-B*15:02 | 2–13 | 0.8996 | −0.14401 |
| EQGVAHMGIGYA | Cell division protein FtsZ | HLA-A*26:01 | 222–233 | 0.6956 | 0.13586 |
| LTVSVVTKPFLF | Cell division protein FtsZ | HLA-B*58:01 HLA-B*57:01 | 129–140 | 0.6866 | −0.16299 |
| DFWFAGFNPYYT | Peptidoglycan glycosyltransferase FtsW | HLA-A*29:02 HLA-B*15:02 | 767–778 | 1.3204 | 0.35278 |
| FWFAGFNPYYTS | Peptidoglycan glycosyltransferase FtsW | HLA-A*29:02 HLA-B*15:02 | 768–779 | 1.2422 | 0.22335 |
| KIQEMYLALNIE | Peptidoglycan glycosyltransferase FtsW | HLA-B*44:03, HLA-B*44:02, HLA-A*32:01, HLA-B*40:02 | 158–169 | 1.2288 | −0.02229 |
| HSFGVEAASQTY | Peptidoglycan glycosyltransferase FtsW | HLA-A*30:02, HLA-B*57:01, HLA-A*01:01, HLA-B*15:01, HLA-B*35:01 | 190–201 | 1.1133 | 0.05519 |
| SGIYVLPFYKIN | Peptidoglycan glycosyltransferase FtsW | HLA-A*11:01, HLA-A*03:01 | 439–450 | 1.0653 | 0.06436 |
| Epitope | Protein | Allele | Position | Antigenicity | Immunogenicity |
|---|---|---|---|---|---|
| AVPNMINLDFADVQS | Cell division protein FtsZ | HLA-DRB1*03:06, HLA-DRB1*03:07, HLA-DRB1*03:08 | 204–218 | 0.9513 | 0.09313 |
| VPNMINLDFADVQSV | Cell division protein FtsZ | HLA-DRB1*03:06, HLA-DRB1*03:07, HLA-DRB1*03:08 | 205–219 | 0.898 | −0.02656 |
| IYVLPFYKINDSNE | Peptidoglycan glycosyltransferase FtsW | HLA-DRB1*04:21, HLA-DRB1*04:26 | 441–455 | 1.0957 | −0.06595 |
| AVNFDLLGSVKPAST | Peptidoglycan glycosyltransferase FtsW | HLA-DRB1*11:07 | 288–302 | 0.8503 | −0.25628 |
| RMTLVPSGIYVLPFY | Peptidoglycan glycosyltransferase FtsW | HLA-DRB1*07:03 | 433–447 | 0.7638 | 0.11754 |
| Epitope | Protein | Score | Position | Antigenicity | Immunogenicity |
|---|---|---|---|---|---|
| TQETQTTQETQAPETQ | Peptidoglycan glycosyltransferase FtsW | 0.87 | 1074 | 1.5134 | 0.0789 |
| FVTAGMGGGTGTGAAP | Cell division protein FtsZ | 0.79 | 80 | 1.9827 | 0.20935 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Allemailem, K.S. In Silico Development of a Chimeric Multi-Epitope Vaccine Targeting Helcococcus kunzii: Coupling Subtractive Proteomics and Reverse Vaccinology for Vaccine Target Discovery. Pharmaceuticals 2025, 18, 1258. https://doi.org/10.3390/ph18091258
Allemailem KS. In Silico Development of a Chimeric Multi-Epitope Vaccine Targeting Helcococcus kunzii: Coupling Subtractive Proteomics and Reverse Vaccinology for Vaccine Target Discovery. Pharmaceuticals. 2025; 18(9):1258. https://doi.org/10.3390/ph18091258
Chicago/Turabian StyleAllemailem, Khaled S. 2025. "In Silico Development of a Chimeric Multi-Epitope Vaccine Targeting Helcococcus kunzii: Coupling Subtractive Proteomics and Reverse Vaccinology for Vaccine Target Discovery" Pharmaceuticals 18, no. 9: 1258. https://doi.org/10.3390/ph18091258
APA StyleAllemailem, K. S. (2025). In Silico Development of a Chimeric Multi-Epitope Vaccine Targeting Helcococcus kunzii: Coupling Subtractive Proteomics and Reverse Vaccinology for Vaccine Target Discovery. Pharmaceuticals, 18(9), 1258. https://doi.org/10.3390/ph18091258







