Designing Novel Antimicrobial Agents from the Synthetic Antimicrobial Peptide (Pep-38) to Combat Antibiotic Resistance
Abstract
1. Introduction
2. Results
2.1. Peptide Design, Molecular Modelling, and In Silico Analysis
2.1.1. Result of CAMPR3 Analysis
2.1.2. Results of NPS Secondary Structure Analysis
2.1.3. Results Using the Innovagen Calculator
2.1.4. Results of ProtParam/ExPASY and APD3 Analysis
2.1.5. Results of PeptideRanker
2.2. Peptide Synthesis and Purification
2.3. Bacterial Susceptibility Assay
2.4. Antibiofilm Activity Assay
2.5. Hemolytic Assay
2.6. Mammalian Cell Cytotoxicity Assay
2.7. Time–Kill Assay
3. Discussion
4. Materials and Methods
4.1. Materials and Bacterial Strains
4.2. Methods
4.2.1. Peptide Design and In Silico Analysis
CAMPR3
NPS Secondary Structure Analysis
Innovagen Calculator
ProtParam/ExPASy and APD3 Analysis
PeptideRanker
4.2.2. Peptide Synthesis and Purification
4.2.3. Bacterial Susceptibility Assay
Determination of MIC and MBC of the Parent and the Modified Peptides
4.2.4. Hemolytic Assay
4.2.5. Mammalian Cell Cytotoxicity Assay
4.2.6. Antibiofilm Activity Assay
Biofilm Formation
Challenge Plate
Recovery Plate
4.2.7. Time–Kill Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Λ | Lambda |
| µL | Micro-liter |
| µM | Micro-molar |
| AI | Aliphatic index |
| AMR | Antimicrobial resistance |
| AMPs | Antimicrobial peptides |
| ATCC | American Type Culture Collection |
| CAMPR3 | Collection of Antimicrobial Peptides |
| CFU | Colony-forming unit |
| CLSI | Clinical and Laboratory Standards Institute |
| DMSO | Dimethyl sulfoxide |
| EDTA | Ethylenediaminetetraacetic acid |
| ELISA | Enzyme-linked immunosorbent assay |
| EPS | Extracellular polymeric substance |
| EU | European Union |
| GRAVY | Grand average of hydropathy |
| HDP | Host defense peptide |
| HEL | Helical |
| HPLC | High-performance liquid chromatography |
| LA | Lipoteichoic acid |
| LSTM | Long short-term memory |
| MBC | Minimum bactericidal concentration |
| MBEC | Minimum biofilm eradication concentration |
| MDRB | Multi-drug-resistant bacteria |
| MHB | Mueller Hinton broth |
| MIC | Minimum inhibitory concentration |
| MRSA | Multidrug-resistant Staphylococcus aureus. |
| MS | Mass spectrometry |
| MTT | Dimethyl thiazolyl diphenyl tetrazolium |
| NPS | Network protein sequence |
| OD | Optical density |
| PBS | Phosphate buffer saline |
| RBC | Red blood cell |
| RF | Random forest |
| RNN | Recurrent neural network |
| Rpm | Rounds per minute |
| SVM | Support vector machine |
| UK | United Kingdom |
| US | United States |
| WHO | World Health Organization |
References
- Hou, J.; Long, X.; Wang, X.; Li, L.; Mao, D.; Luo, Y.; Ren, H. Global Trend of Antimicrobial Resistance in Common Bacterial Pathogens in Response to Antibiotic Consumption. J. Hazard. Mater. 2023, 442, 130042. [Google Scholar] [CrossRef]
- Dissanayake, S.; He, J.; Yang, S.H.; Brimble, M.A.; Harris, P.W.R.; Cameron, A.J. Flow-Based Fmoc-SPPS Preparation and SAR Study of Cathelicidin-PY Reveals Selective Antimicrobial Activity. Molecules 2023, 28, 1993. [Google Scholar] [CrossRef] [PubMed]
- Almutairy, B. Extensively and Multidrug-Resistant Bacterial Strains: Case Studies of Antibiotics Resistance. Front. Microbiol. 2024, 15, 1381511. [Google Scholar] [CrossRef]
- Sulis, G.; Sayood, S.; Gandra, S. Antimicrobial Resistance in Low- and Middle-Income Countries: Current Status and Future Directions. Expert Rev. Anti Infect. Ther. 2022, 20, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Arzanlou, M.; Chai, W.C.; Venter, H. Intrinsic, Adaptive and Acquired Antimicrobial Resistance in Gram-Negative Bacteria. Essays Biochem. 2017, 61, 49–59. [Google Scholar]
- Bobde, S. Computationally Designed Novel Synthetic Antimicrobial Peptides Against Gram-Negative Bacteria. Ph.D. Thesis, George Mason University, Fairfax, VA, USA, 2021. [Google Scholar]
- Ramezanzadeh, M.; Saeedi, N.; Mesbahfar, E.; Farrokh, P.; Salimi, F.; Rezaei, A. Design and Characterization of New Antimicrobial Peptides Derived from Aurein 1.2 with Enhanced Antibacterial Activity. Biochimie 2021, 181, 42–51. [Google Scholar] [CrossRef]
- Abdel-Qader, D.H.; Albassam, A.; Ismael, N.S.; El-Shara’, A.A.; Shehri, A.; Almutairi, F.S.; Al-Harbi, D.M.; Al Zahrani, M.M.; Chen, L.-C.; Al Mazrouei, N.; et al. Awareness of Antibiotic Use and Resistance in Jordanian Community. J. Prim. Care Community Health 2020, 11, 2150132720961255. [Google Scholar] [CrossRef]
- Al-Tamimi, M.; Albalawi, H.; Isied, W.; Musallam, A.; Qazzaz, F.; Azab, M.; Abu-raideh, J. Gram-Positive Bacterial Infections and Antibiotics Resistance in Jordan: Current Status and Future Perspective. Jordan Med. J. 2022, 56, 17–44. [Google Scholar] [CrossRef]
- Otsuka, Y. Potent Antibiotics Active against Multidrug-Resistant Gram-Negative Bacteria. Chem. Pharm. Bull. 2020, 68, 182–190. [Google Scholar] [CrossRef]
- Ji, S.; An, F.; Zhang, T.; Lou, M.; Guo, J.; Liu, K.; Zhu, Y.; Wu, J.; Wu, R. Antimicrobial Peptides: An Alternative to Traditional Antibiotics. Eur. J. Med. Chem. 2024, 265, 116072. [Google Scholar] [CrossRef] [PubMed]
- Hetta, H.F.; Sirag, N.; Alsharif, S.M.; Alharbi, A.A.; Alkindy, T.T.; Alkhamali, A.; Albalawi, A.S.; Ramadan, Y.N.; Rashed, Z.I.; Alanazi, F.E. Antimicrobial Peptides: The Game-Changer in the Epic Battle Against Multidrug-Resistant Bacteria. Pharmaceuticals 2024, 17, 1555. [Google Scholar] [CrossRef] [PubMed]
- Min, K.H.; Kim, K.H.; Ki, M.-R.; Pack, S.P. Antimicrobial Peptides and Their Biomedical Applications: A Review. Antibiotics 2024, 13, 794. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Tamanna, N.T.; Sagor, M.S.; Zaki, R.M.; Rabbee, M.F.; Lackner, M. Antimicrobial Peptides: A Promising Solution to the Rising Threat of Antibiotic Resistance. Pharmaceutics 2024, 16, 1542. [Google Scholar] [CrossRef]
- Roque-Borda, C.A.; Primo, L.M.D.G.; Medina-Alarcón, K.P.; Campos, I.C.; Nascimento, C.D.F.; Saraiva, M.M.S.; Berchieri Junior, A.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S.; Perdigão, J.; et al. Antimicrobial Peptides: A Promising Alternative to Conventional Antimicrobials for Combating Polymicrobial Biofilms. Adv. Sci. 2025, 12, 2410893. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The Antimicrobial Peptides and Their Potential Clinical Applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Chen, E.H.-L.; Wang, C.-H.; Liao, Y.-T.; Chan, F.-Y.; Kanaoka, Y.; Uchihashi, T.; Kato, K.; Lai, L.; Chang, Y.-W.; Ho, M.-C.; et al. Visualizing the Membrane Disruption Action of Antimicrobial Peptides by Cryo-Electron Tomography. Nat. Commun. 2023, 14, 5464. [Google Scholar] [CrossRef]
- Duarte-Mata, D.I.; Salinas-Carmona, M.C. Antimicrobial Peptides’ Immune Modulation Role in Intracellular Bacterial Infection. Front. Immunol. 2023, 14, 1119574. [Google Scholar] [CrossRef]
- K R, G.; Balenahalli Narasingappa, R.; Vishnu Vyas, G. Unveiling Mechanisms of Antimicrobial Peptide: Actions beyond the Membranes Disruption. Heliyon 2024, 10, e38079. [Google Scholar] [CrossRef]
- Mwangi, J.; Kamau, P.; Thuku, R.; Lai, R. Design Methods for Antimicrobial Peptides with Improved Performance. Zool. Res. 2023, 44, 1095–1114. [Google Scholar] [CrossRef]
- Wang, Y.; Song, M.; Chang, W. Antimicrobial Peptides and Proteins against Drug-Resistant Pathogens. Cell Surf. 2024, 12, 100135. [Google Scholar] [CrossRef] [PubMed]
- Bucataru, C.; Ciobanasu, C. Antimicrobial Peptides: Opportunities and Challenges in Overcoming Resistance. Microbiol. Res. 2024, 286, 127822. [Google Scholar] [CrossRef] [PubMed]
- Shriwastav, S.; Kaur, N.; Hassan, M.; Ahmed Mohammed, S.; Chauhan, S.; Mittal, D.; Aman, S.; Bibi, A. Antimicrobial Peptides: A Promising Frontier to Combat Antibiotic Resistant Pathogens. Ann. Med. Surg. 2025, 87, 2118–2132. [Google Scholar] [CrossRef]
- Ali, M.; Garg, A.; Srivastava, A.; Arora, P.K. The Role of Antimicrobial Peptides in Overcoming Antibiotic Resistance. Microbe 2025, 7, 100337. [Google Scholar] [CrossRef]
- Zhang, Q. Antimicrobial Peptides: From Discovery to Developmental Applications. Appl. Environ. Microbiol. 2025, 91, e02115-24. [Google Scholar] [CrossRef]
- Zheng, S.; Tu, Y.; Li, B.; Qu, G.; Li, A.; Peng, X.; Li, S.; Shao, C. Antimicrobial Peptide Biological Activity, Delivery Systems and Clinical Translation Status and Challenges. J. Transl. Med. 2025, 23, 292. [Google Scholar] [CrossRef]
- Zhang, H.; Lv, J.; Ma, Z.; Ma, J.; Chen, J. Advances in Antimicrobial Peptides: Mechanisms, Design Innovations, and Biomedical Potential. Molecules 2025, 30, 1529. [Google Scholar] [CrossRef]
- Zhang, T.; Jin, Q.; Ji, J. Antimicrobial Peptides and Their Mimetics: Promising Candidates of Next-Generation Therapeutic Agents Combating Multidrug-Resistant Bacteria. Adv. Biol. 2025, 9, 2400461. [Google Scholar] [CrossRef]
- Bolatchiev, A.; Baturin, V.; Shchetinin, E.; Bolatchieva, E. Novel Antimicrobial Peptides Designed Using a Recurrent Neural Network Reduce Mortality in Experimental Sepsis. Antibiotics 2022, 11, 411. [Google Scholar] [CrossRef]
- Hale, J.D.; Hancock, R.E. Alternative Mechanisms of Action of Cationic Antimicrobial Peptides on Bacteria. Expert Rev. Anti Infect. Ther. 2007, 5, 951–959. [Google Scholar] [CrossRef]
- Sánchez-Vásquez, L.; Silva-Sanchez, J.; Jiménez-Vargas, J.M.; Rodríguez-Romero, A.; Muñoz-Garay, C.; Rodríguez, M.C.; Gurrola, G.B.; Possani, L.D. Enhanced Antimicrobial Activity of Novel Synthetic Peptides Derived from Vejovine and Hadrurin. Biochim. Biophys. Acta BBA-Gen. Subj. 2013, 1830, 3427–3436. [Google Scholar] [CrossRef] [PubMed]
- Jindal, M.H.; Le, C.F.; Mohd Yusof, M.Y.; Sekaran, S.D. Net Charge, Hydrophobicity and Specific Amino Acids Contribute to the Activity of Antimicrobial Peptides. J. Health Transl. Med. 2014, 17, 1–7. [Google Scholar]
- Saidemberg, D.M.; Baptista-Saidemberg, N.B.; Palma, M.S. Chemometric Analysis of Hymenoptera Toxins and Defensins: A Model for Predicting the Biological Activity of Novel Peptides from Venoms and Hemolymph. Peptides 2011, 32, 1924–1933. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Huang, H.; Aweya, J.J.; Zheng, Z.; Liu, G.; Zhang, Y. PvHS9 is a novel in silico predicted antimicrobial peptide derived from hemocyanin of Penaeus vannamei. Aquaculture 2021, 530, 735926. [Google Scholar] [CrossRef]
- Koosehlar, E.; Mohabatkar, H.; Behbahani, M. In Silico and In Vitro Evaluations of the Antibacterial Activities of HIV-1 Nef Peptides against Pseudomonas aeruginosa. Int. J. Mol. Cell Med. 2024, 13, 46–63. [Google Scholar]
- Deslouches, B.; Phadke, S.M.; Lazarevic, V.; Cascio, M.; Islam, K.; Montelaro, R.C.; Mietzner, T.A. De Novo Generation of Cationic Antimicrobial Peptides: Influence of Length and Tryptophan Substitution on Antimicrobial Activity. Antimicrob. Agents Chemother. 2005, 49, 316–322. [Google Scholar] [CrossRef]
- Sato, H.; Feix, J.B. Lysine-Enriched Cecropin-Mellitin Antimicrobial Peptides with Enhanced Selectivity. Antimicrob. Agents Chemother. 2008, 52, 4463–4465. [Google Scholar] [CrossRef]
- Hilpert, K.; Elliott, M.R.; Volkmer-Engert, R.; Henklein, P.; Donini, O.; Zhou, Q.; Winkler, D.F.H.; Hancock, R.E.W. Sequence Requirements and an Optimization Strategy for Short Antimicrobial Peptides. Chem. Biol. 2006, 13, 1101–1107. [Google Scholar] [CrossRef]
- Zhang, K.; Yang, N.; Mao, R.; Hao, Y.; Teng, D.; Wang, J. An Amphipathic Peptide Combats Multidrug-Resistant Staphylococcus aureus by Disrupting Membrane Integrity. Commun. Biol. 2024, 7, 1582. [Google Scholar] [CrossRef]
- Matthyssen, T.; Li, W.; Holden, J.A.; Lenzo, J.C.; Hadjigol, S.; O’Brien-Simpson, N.M. Dimerization and lysine substitution of melittin have differing effects on bacteria. Front. Pharmacol. 2024, 15, 1443497. [Google Scholar] [CrossRef]
- Lee, P.-C.; Chu, C.-C.; Tsai, Y.-J.; Chuang, Y.-C.; Lung, F.-D. Design, Synthesis, and Antimicrobial Activities of Novel Functional Peptides against Gram-Positive and Gram-Negative Bacteria. Chem. Biol. Drug Des. 2019, 94, 1537–1544. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Hernández, D.; Juárez-González, V.R.; Bustamante, V.H.; Martínez-Martínez, L.L.; Ramírez, V.; Balleza, D.; Quintero-Hernández, V. Conformational Flexibility and Net Charge are Key Determinants for the Antimicrobial Activity of Peptide Uy234 Against Multidrug-resistant Bacteria. Int. J. Pept. Res. Ther. 2024, 30, 79. [Google Scholar] [CrossRef]
- Gagat, P.; Ostrówka, M.; Duda-Madej, A.; Mackiewicz, P. Enhancing Antimicrobial Peptide Activity through Modifications of Charge, Hydrophobicity, and Structure. Int. J. Mol. Sci. 2024, 25, 10821. [Google Scholar] [CrossRef]
- Kang, H.K.; Seo, C.H.; Luchian, T.; Park, Y. Pse-T2, an Antimicrobial Peptide with High-Level, Broad-Spectrum Antimicrobial Potency and Skin Biocompatibility against Multidrug-Resistant Pseudomonas Aeruginosa Infection. Antimicrob. Agents Chemother. 2018, 62, e01493-18. [Google Scholar] [CrossRef]
- Ansari, J.M.; Abraham, N.M.; Massaro, J.; Murphy, K.; Smith-Carpenter, J.; Fikrig, E. Anti-Biofilm Activity of a Self-Aggregating Peptide against Streptococcus Mutans. Front. Microbiol. 2017, 8, 488. [Google Scholar] [CrossRef]
- Pokharel, K.; Dawadi, B.R.; Shrestha, L.B. Role of Biofilm in Bacterial Infection and Antimicrobial Resistance. J. Nepal Med. Assoc. 2022, 60, 836–840. [Google Scholar] [CrossRef]
- Duffey, M.; Jumde, R.P.; Da Costa, R.M.A.; Ropponen, H.-K.; Blasco, B.; Piddock, L.J.V. Extending the Potency and Lifespan of Antibiotics: Inhibitors of Gram-Negative Bacterial Efflux Pumps. ACS Infect. Dis. 2024, 10, 1458–1482. [Google Scholar] [CrossRef]
- Son, M.; Lee, Y.; Hwang, H.; Hyun, S.; Yu, J. Disruption of Interactions between Hydrophobic Residues on Nonpolar Faces Is a Key Determinant in Decreasing Hemolysis and Increasing Antimicrobial Activities of α-Helical Amphipathic Peptides. ChemMedChem 2013, 8, 1638–1642. [Google Scholar] [CrossRef]
- Ma, Z.; Yang, J.; Han, J.; Gao, L.; Liu, H.; Lu, Z.; Zhao, H.; Bie, X. Insights into the Antimicrobial Activity and Cytotoxicity of Engineered α-Helical Peptide Amphiphiles. J. Med. Chem. 2016, 59, 10946–10962. [Google Scholar] [CrossRef]
- Clark, S.; Jowitt, T.A.; Harris, L.K.; Knight, C.G.; Dobson, C.B. The lexicon of antimicrobial peptides: A complete set of arginine and tryptophan sequences. Commun. Biol. 2021, 4, 605. [Google Scholar] [CrossRef]
- Rathore, A.S.; Kumar, N.; Choudhury, S.; Mehta, N.K.; Raghava, G.P.S. Prediction of hemolytic peptides and their hemolytic concentration. Commun. Biol. 2025, 8, 176. [Google Scholar] [CrossRef] [PubMed]
- Arenas, I.; Ibarra, M.A.; Santana, F.L.; Villegas, E.; Hancock, R.E.W.; Corzo, G. In Vitro and In Vivo Antibiotic Capacity of Two Host Defense Peptides. Microbiol. Spectr. 2020, 8, e01318-21. [Google Scholar] [CrossRef]
- Böttger, R.; Hoffmann, R.; Knappe, D. Differential stability of therapeutic peptides with different proteolytic cleavage sites in blood, plasma and serum. PLoS ONE 2017, 12, e0176791. [Google Scholar] [CrossRef] [PubMed]
- Zapadka, K.L.; Becher, F.J.; Gomes dos Santos, A.L.; Jackson, S.E. Factors affecting the physical stability (aggregation) of peptide and protein therapeutics. Interface Focus 2017, 7, 20170030. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, D.; Nagarajan, T.; Roy, N.; Kulkarni, O.; Ravichandran, S.; Mishra, M.; Chakravortty, D.; Chandra, N. Computational antimicrobial peptide design and evaluation against multidrug-resistant clinical isolates of bacteria. J. Biol. Chem. 2018, 293, 3492–3509. [Google Scholar] [CrossRef]
- Gawde, U.; Chakraborty, S.; Waghu, F.H.; Barai, R.S.; Khanderkar, A.; Indraguru, R.; Shirsat, T.; Idicula-Thomas, S. CAMPR4: A Database of Natural and Synthetic Antimicrobial Peptides. Nucleic Acids Res. 2023, 51, D377–D383. [Google Scholar] [CrossRef]
- Guermeur, Y. Combinaison de Classifieurs Statistiques: Application à La Prédiction de La Structure Secondaire Des Protéines. Ph.D. Thesis, Universite Paris 6 (Pierre et Marie Curie University), Paris, France, 1997. [Google Scholar]
- Huang, Y.; He, L.; Li, G.; Zhai, N.; Jiang, H.; Chen, Y. Role of Helicity of α-Helical Antimicrobial Peptides to Improve Specificity. Protein Cell 2014, 5, 631–642. [Google Scholar] [CrossRef]
- Osorio, D.; Rondón-Villarreal, P.; Torres Sáez, R. Peptides: A Package for Data Mining of Antimicrobial Peptides. R. J. 2015, 7, 4–14. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Guruprasad, K.; Reddy, B.V.; Pandit, M.W. Correlation between Stability of a Protein and Its Dipeptide Composition: A Novel Approach for Predicting in Vivo Stability of a Protein from Its Primary Sequence. Protein Eng. 1990, 4, 155–161. [Google Scholar] [CrossRef]
- Ikai, A. Thermostability and Aliphatic Index of Globular Proteins. J. Biochem. 1980, 88, 1895–1898. [Google Scholar] [PubMed]
- Terziyski, Z.; Terziyska, M.; Hadzhikoleva, S.; Desseva, I. A Software Tool for Data Mining of Physicochemical Properties of Peptides. BIO Web Conf. 2023, 58, 03007. [Google Scholar] [CrossRef]
- Boman, H.G. Antibacterial Peptides: Basic Facts and Emerging Concepts. J. Intern. Med. 2003, 254, 197–215. [Google Scholar] [CrossRef] [PubMed]
- Pearman, N.A.; Ronander, E.; Smith, A.M.; Morris, G.A. The Identification and Characterisation of Novel Bioactive Peptides Derived from Porcine Liver. Curr. Res. Food Sci. 2020, 3, 314–321. [Google Scholar] [CrossRef]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance. Pathogens 2021, 10, 165. [Google Scholar] [CrossRef]
- Verma, P. Methods for Determining Bactericidal Activity and Antimicrobial Interactions: Synergy Testing, Time-Kill Curves, and Population Analysis. In Antimicrobial Susceptibility Testing Protocols; Schwalbe, R., Steele-Moore, L., Goodwin, A., Eds.; CRC Press: Boca Raton, FL, USA, 2007; pp. 275–298. [Google Scholar]
- CLSI Document M100-S21; Performance Standards For Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute (CLSI): Malvern, PA, USA, 2011; Volume 31.
- Dosler, S.; Karaaslan, E. Inhibition and Destruction of Pseudomonas aeruginosa Biofilms by Antibiotics and Antimicrobial Peptides. Peptides 2014, 62, 32–37. [Google Scholar] [CrossRef]
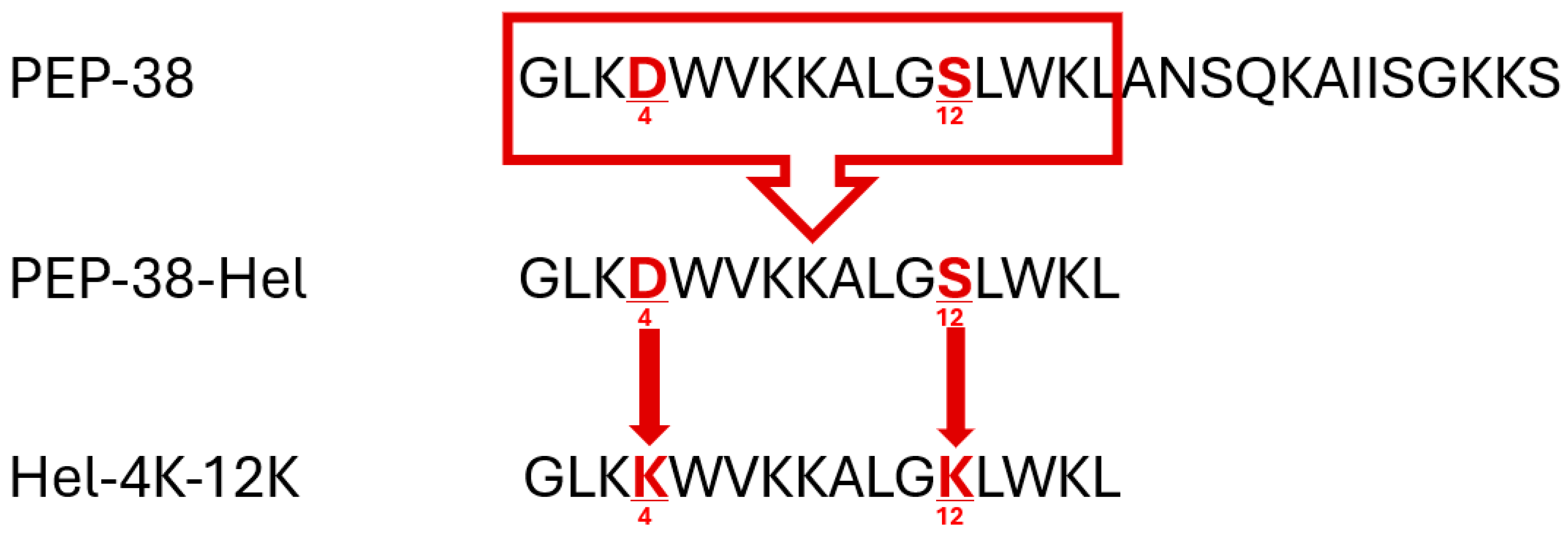
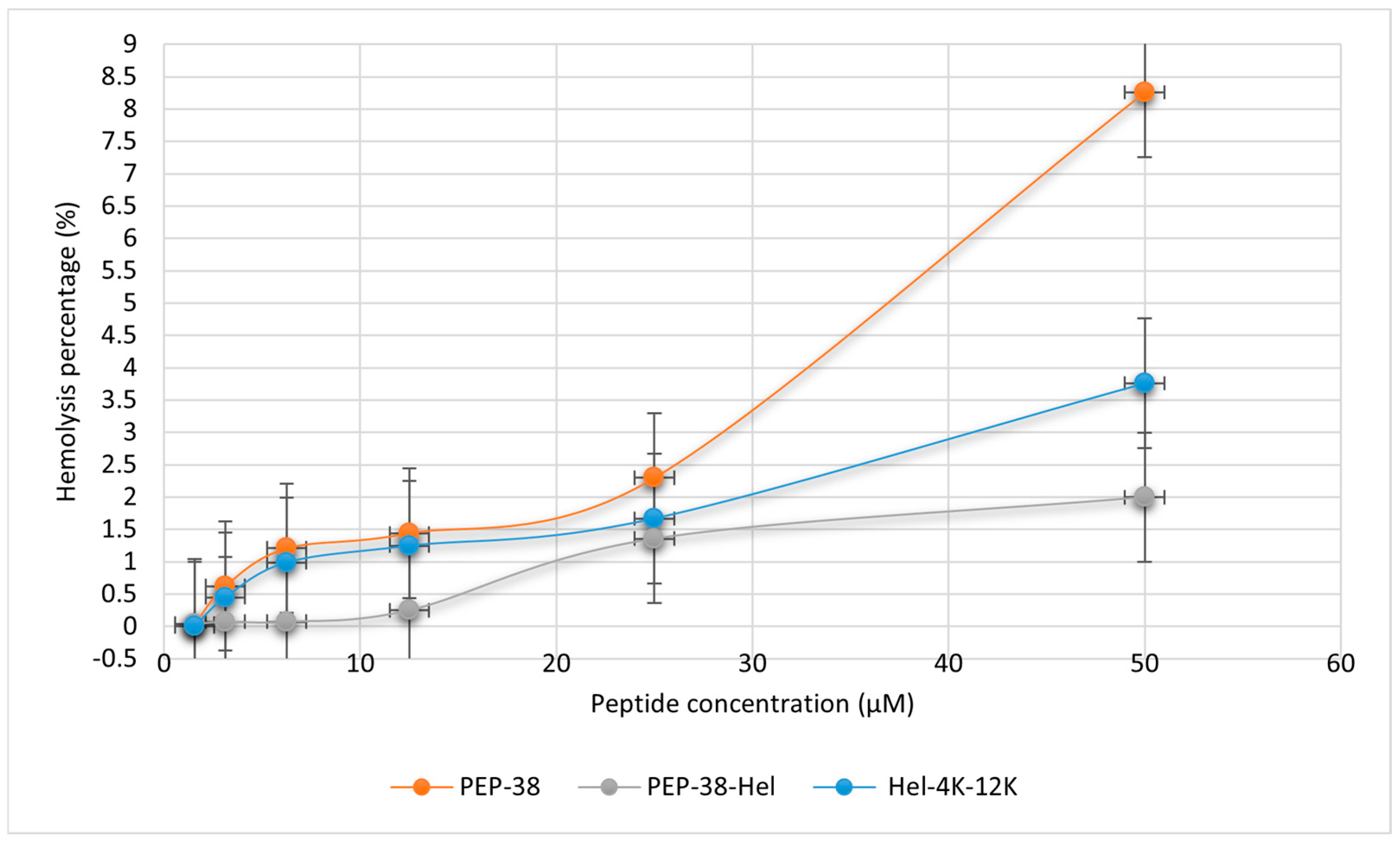
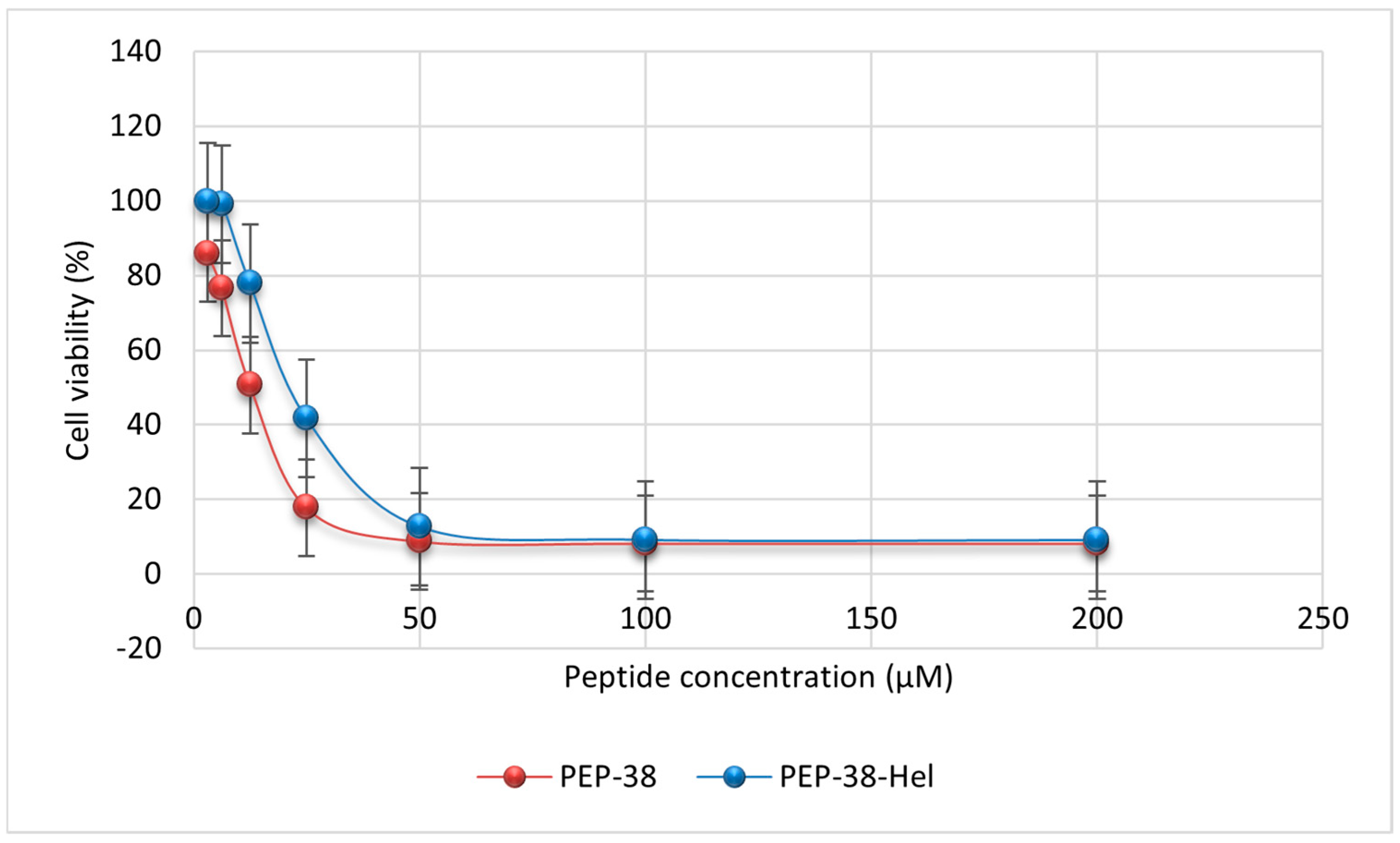
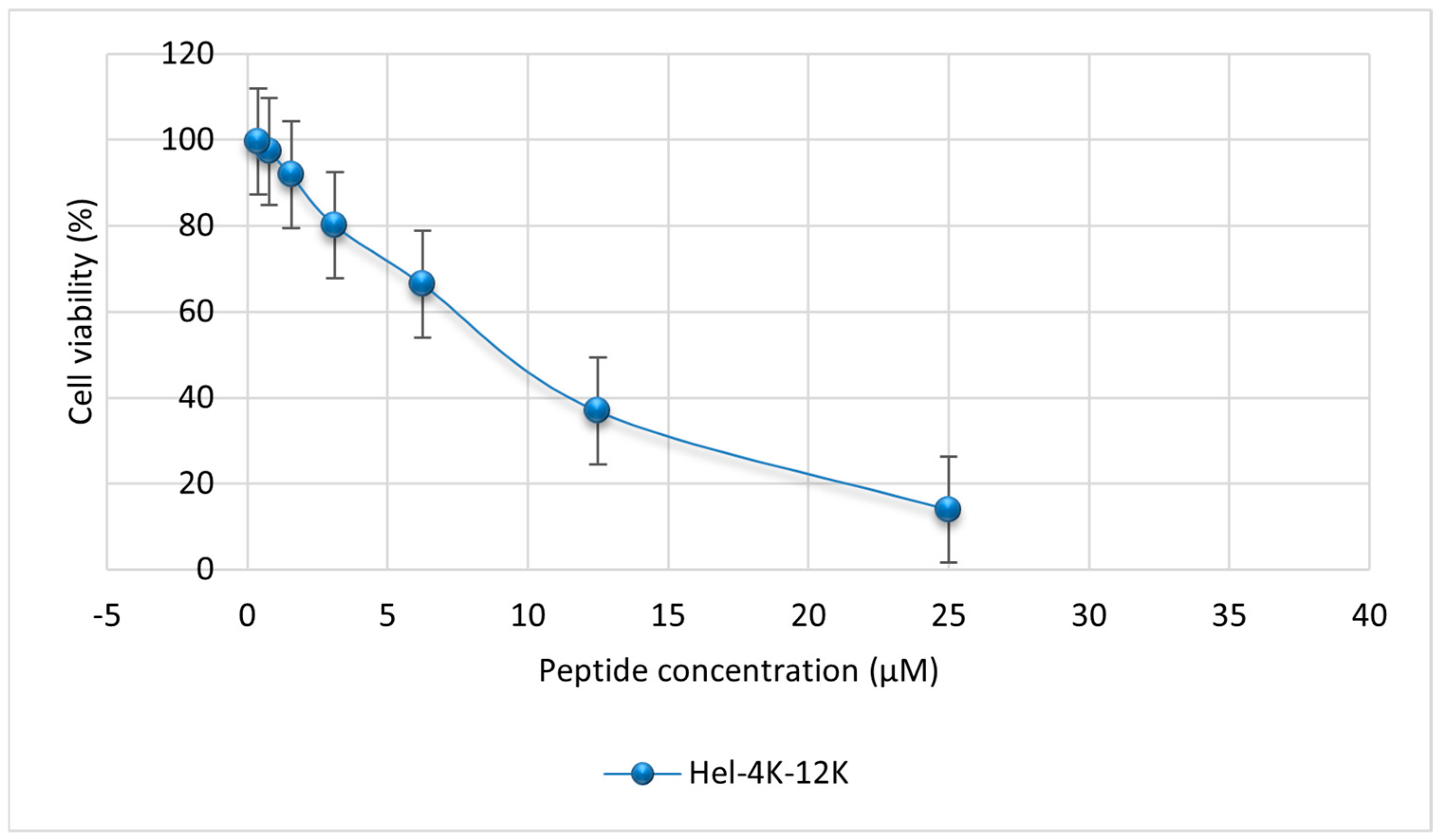

| Peptide ID. | Position | Sequence |
|---|---|---|
| Hel-4R | 4-R | GLKRWVKKALGSLWKL |
| Hel-4K | 4-K | GLKKWVKKALGSLWKL |
| Hel-4W | 4-W | GLKWWVKKALGSLWKL |
| Hel-11K | 11-K | GLKDWVKKALKSLWKL |
| Hel-12K | 12-K | GLKDWVKKALGKLWKL |
| Peptide ID. | Peptide Sequence | a.a. no. | Helicity | Net Charge |
|---|---|---|---|---|
| PEP-38 | GLKDWVKKALGSLWKLANSQKAIISGKKS | 29 | 51.72% | +6 |
| PEP-38-Hel | GLKDWVKKALGSLWKL | 16 | 81.25% | +3 |
| Hel-4R | GLKRWVKKALGSLWKL | 16 | 81.25% | +5 |
| Hel-4W | GLKWWVKKALGSLWKL | 16 | 68.75% | +4 |
| Hel-4K | GLKKWVKKALGSLWKL | 16 | 81.25% | +5 |
| Hel-11K | GLKDWVKKALKSLWKL | 16 | 81.25% | +4 |
| Hel-12K | GLKDWVKKALGKLWKL | 16 | 81.25% | +4 |
| Peptide ID. | Peptide Sequence | Instability Index | Aliphatic Index | GRAVY | Boman Index (Kcal/mol) |
|---|---|---|---|---|---|
| PEP-38 | GLKDWVKKALGSLWKLANSQKAIISGKKS | 11.76 | 101.3 | −0.352 | 0.92 |
| PEP-38-Hel | GLKDWVKKALGSLWKL | 6.46 | 121.88 | −0.081 | 0.14 |
| Hel-4R | GLKRWVKKALGSLWKL | 62.63 | 121.88 | −0.144 | 0.52 |
| Hel-4W | GLKWWVKKALGSLWKL | 6.46 | 121.88 | 0.081 | −0.55 |
| Hel-4K | GLKKWVKKALGSLWKL | 6.46 | 121.88 | −0.106 | −0.05 |
| Hel-11K | GLKDWVKKALKSLWKL | 1.15 | 121.88 | −0.300 | 0.54 |
| Hel-12K | GLKDWVKKALGKLWKL | −4.16 | 121.88 | −0.275 | 0.27 |
| Peptide ID. | Peptide Sequence | a.a no. | Helicity | Net Charge | Instability Index | Aliphatic Index | GRAVY | Boman IndexKcal/mol |
|---|---|---|---|---|---|---|---|---|
| Hel-4R-11K | GLKRWVKKALKSLWKL | 16 | 81.25% | +6 | 57.33 | 121.88 | −0.362 | 0.93 |
| Hel-4R-12K | GLKRWVKKALGKLWKL | 16 | 81.25% | +6 | 52.02 | 121.88 | −0.338 | 0.66 |
| Hel-4R-11K-12K | GLKRWVKKALKKLWKL | 16 | 81.25% | +7 | 52.02 | 121.88 | −0.556 | 1.06 |
| Hel-4W-11K | GLKWWVKKALKSLWKL | 16 | 68.75% | +5 | 1.15 | 121.88 | −0.138 | −0.14 |
| Hel-4W-12K | GLKWWVKKALGKLWKL | 16 | 68.75% | +5 | −4.16 | 121.88 | −0.113 | −0.41 |
| Hel-4W-11K-12K | GLKWWVKKALKKLWKL | 16 | 68.75% | +6 | −4.16 | 121.88 | −0.331 | −0.01 |
| Hel-4K-11K | GLKKWVKKALKSLWKL | 16 | 81.25% | +6 | 1.15 | 121.88 | −0.325 | 0.34 |
| Hel-4K-12K | GLKKWVKKALGKLWKL | 16 | 81.25% | +6 | −4.16 | 121.88 | −0.300 | 0.07 |
| Hel-11K-12K | GLKDWVKKALKKLWKL | 16 | 81.25% | +5 | −4.16 | 121.88 | −0.494 | 0.68 |
| Hel-4K-11K-12K | GLKKWVKKALKKLWKL | 16 | 81.25% | +7 | −4.16 | 121.88 | −0.519 | 0.48 |
| Substitution Position | Probability | Peptide Sequence |
|---|---|---|
| PEP-38 | 0.917525 | GLKDWVKKALGSLWKLANSQKAIISGKKS |
| PEP-38-Hel | 0.912204 | GLKDWVKKALGSLWKL |
| Hel-4K-12K | 0.910295 | GLKKWVKKALGKLWKL |
| Hel-11K-12K | 0.904663 | GLKDWVKKALKKLWKL |
| Hel-4K-11K-12K | 0.904145 | GLKKWVKKALKKLWKL |
| Hel-4K-11K | 0.899625 | GLKKWVKKALKSLWKL |
| Peptide Name | Sequence | a.a.no. | M.W. (g/mol) |
|---|---|---|---|
| PEP-38 | GLKDWVKKALGSLWKLANSQKAIISGKKS | 29 | 3155.73 |
| PEP-38-Hel | GLKDWVKKALGSLWKL | 16 | 1842.23 |
| Hel-4K-12K | GLKKWVKKALGKLWKL | 16 | 1896.41 |
| Bacterial Strain | ATCC Number | PEP-38 | PEP-38-Hel | Hel-4K-12K | |||
|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | ||
| E. coli | 25922 | 25 µM | 25 µM | 50 µM | 50 µM | 3.125 µM | 3.125 µM |
| E. coli | BAA-2452 | 25 µM | 25 µM | NA 1 | NA 1 | 6.25 µM | 6.25 µM |
| S. aureus | 29213 | 25 µM | 25 µM | 50 µM | 50 µM | 3.125 µM | 3.125 µM |
| S. aureus | BAA-44 | 12.5 µM | 12.5µM | 12.5 µM | 12.5 µM | 3.125 µM | 3.125 µM |
| Bacterial Strain | Peptide ID. | MBEC (μM) |
|---|---|---|
| Staphylococcus aureus (ATCC BAA-44) | PEP-38 | 12.5 |
| PEP-38-Hel | 25 | |
| Hel-4K-12K | 6.25 |
| Peptide ID. | IC50 (μM) |
|---|---|
| PEP-38 | 13.25 |
| PEP-38-Hel | 24.89 |
| Hel-4K-12K | 7.96 |
| ATCC Number | Bacterial Strain | Characteristics |
|---|---|---|
| 25922 | Escherichia coli (E. coli) | Control strain |
| BAA-2452 | Escherichia coli (E. coli) | Resistant strain |
| 29213 | Staphylococcus aureus (S. aureus) | Control strain |
| BAA-44 | Staphylococcus aureus (S. aureus) | Resistant strain |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Tall, Y.; Alkurdi, Y.; Alshraiedeh, N.; Sabi, S.H. Designing Novel Antimicrobial Agents from the Synthetic Antimicrobial Peptide (Pep-38) to Combat Antibiotic Resistance. Pharmaceuticals 2025, 18, 862. https://doi.org/10.3390/ph18060862
Al Tall Y, Alkurdi Y, Alshraiedeh N, Sabi SH. Designing Novel Antimicrobial Agents from the Synthetic Antimicrobial Peptide (Pep-38) to Combat Antibiotic Resistance. Pharmaceuticals. 2025; 18(6):862. https://doi.org/10.3390/ph18060862
Chicago/Turabian StyleAl Tall, Yara, Yasmeen Alkurdi, Nid’A Alshraiedeh, and Salsabeel H. Sabi. 2025. "Designing Novel Antimicrobial Agents from the Synthetic Antimicrobial Peptide (Pep-38) to Combat Antibiotic Resistance" Pharmaceuticals 18, no. 6: 862. https://doi.org/10.3390/ph18060862
APA StyleAl Tall, Y., Alkurdi, Y., Alshraiedeh, N., & Sabi, S. H. (2025). Designing Novel Antimicrobial Agents from the Synthetic Antimicrobial Peptide (Pep-38) to Combat Antibiotic Resistance. Pharmaceuticals, 18(6), 862. https://doi.org/10.3390/ph18060862





