A Hybrid In Silico Approach for Identifying Dual VEGFR/RAS Inhibitors as Potential Anticancer and Anti-Angiogenic Agents
Abstract
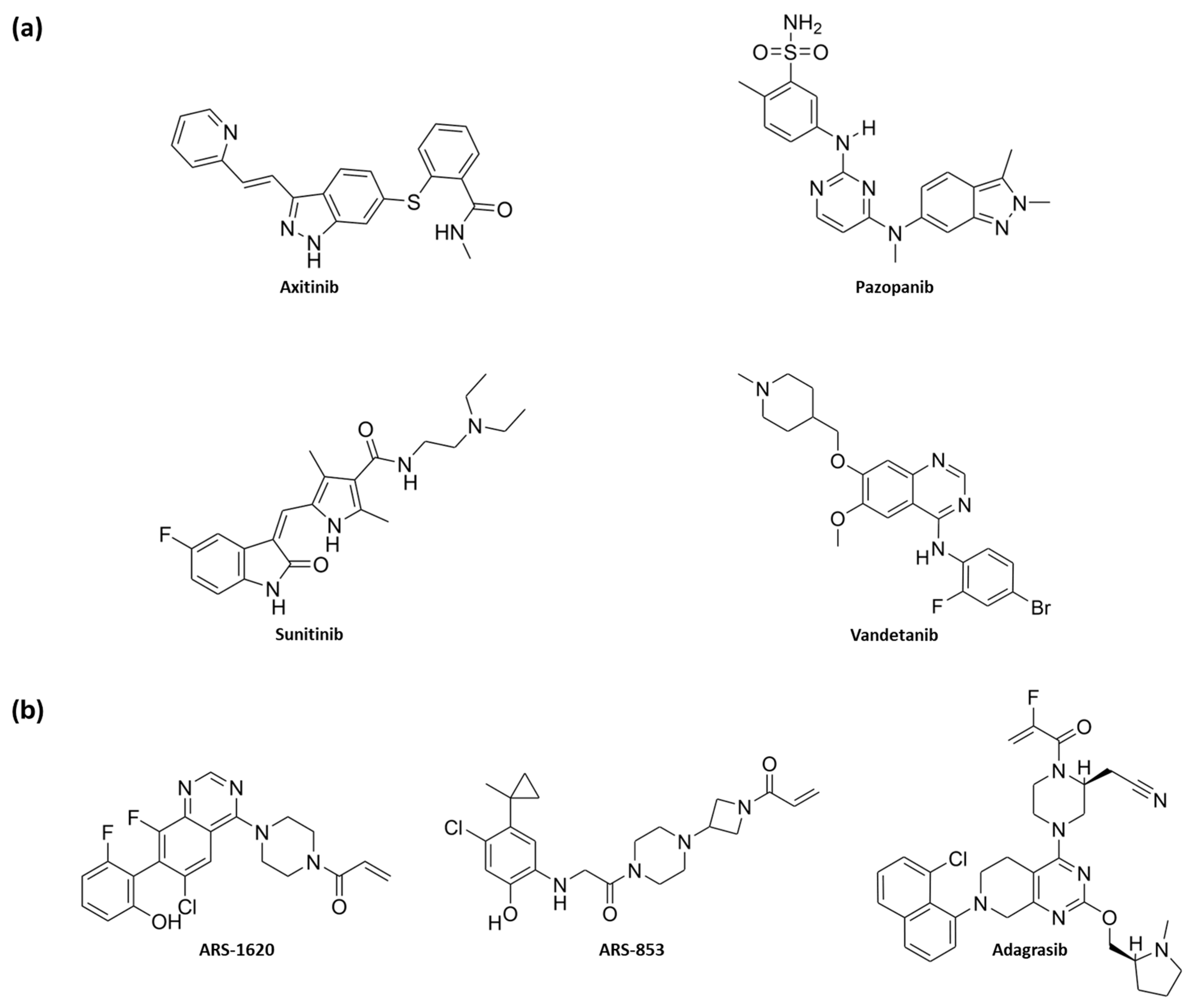
1. Introduction
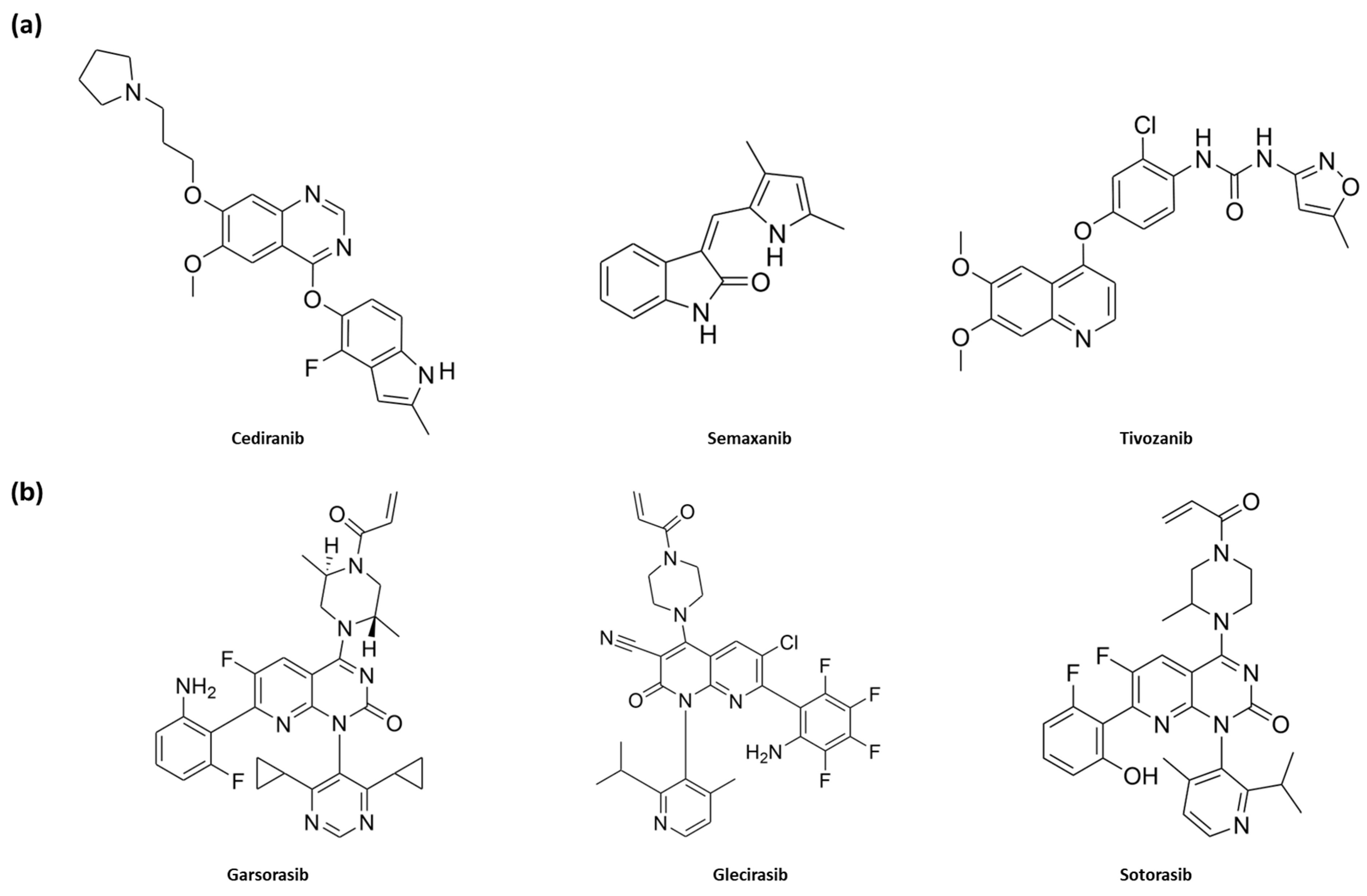
2. Results and Discussion
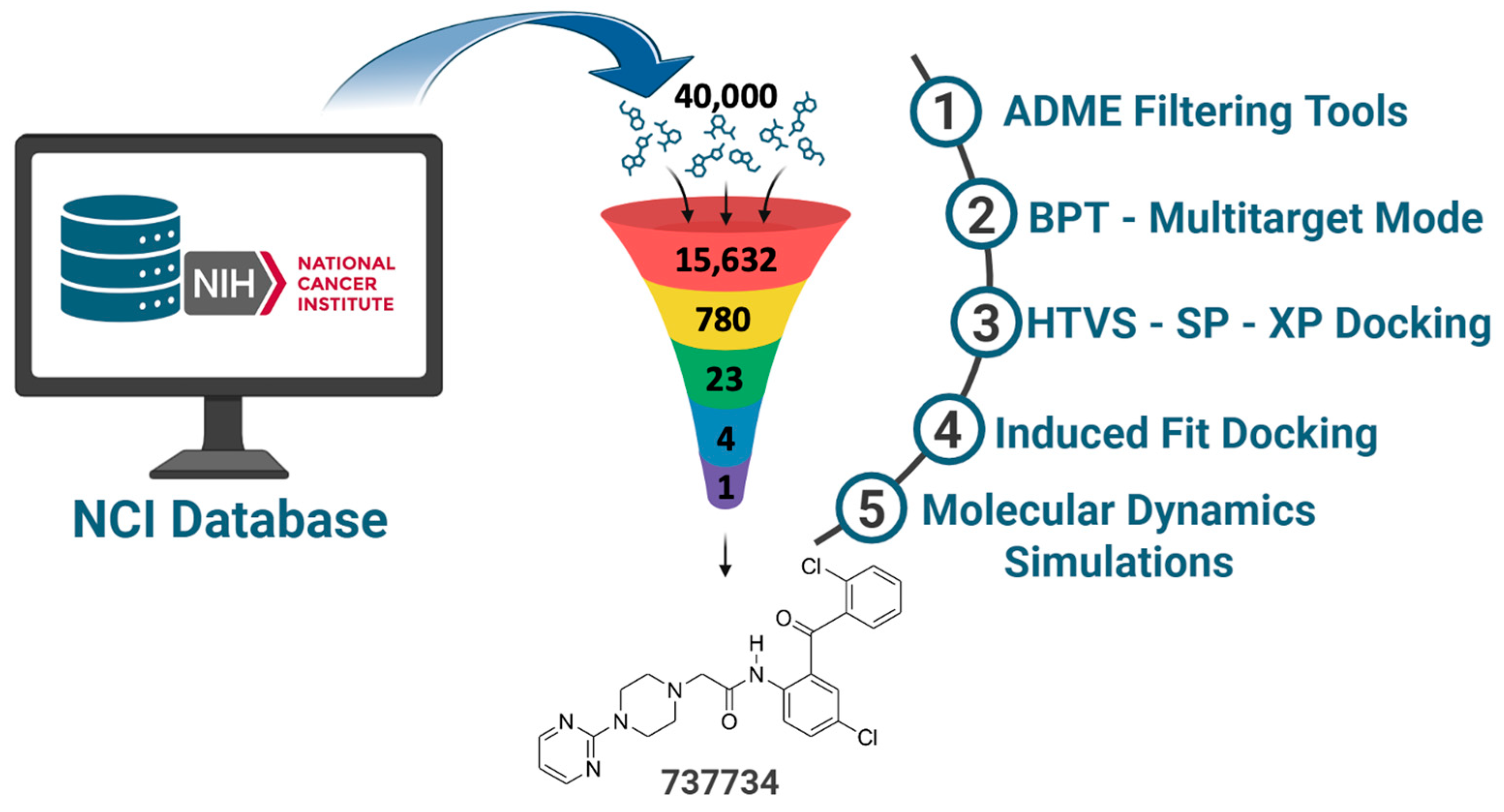
2.1. NCI Database Cleaning Phase
2.2. Ligand-Based Studies
2.2.1. Target Template Building
2.2.2. Biotarget Predictor Tool—Multitarget Mode
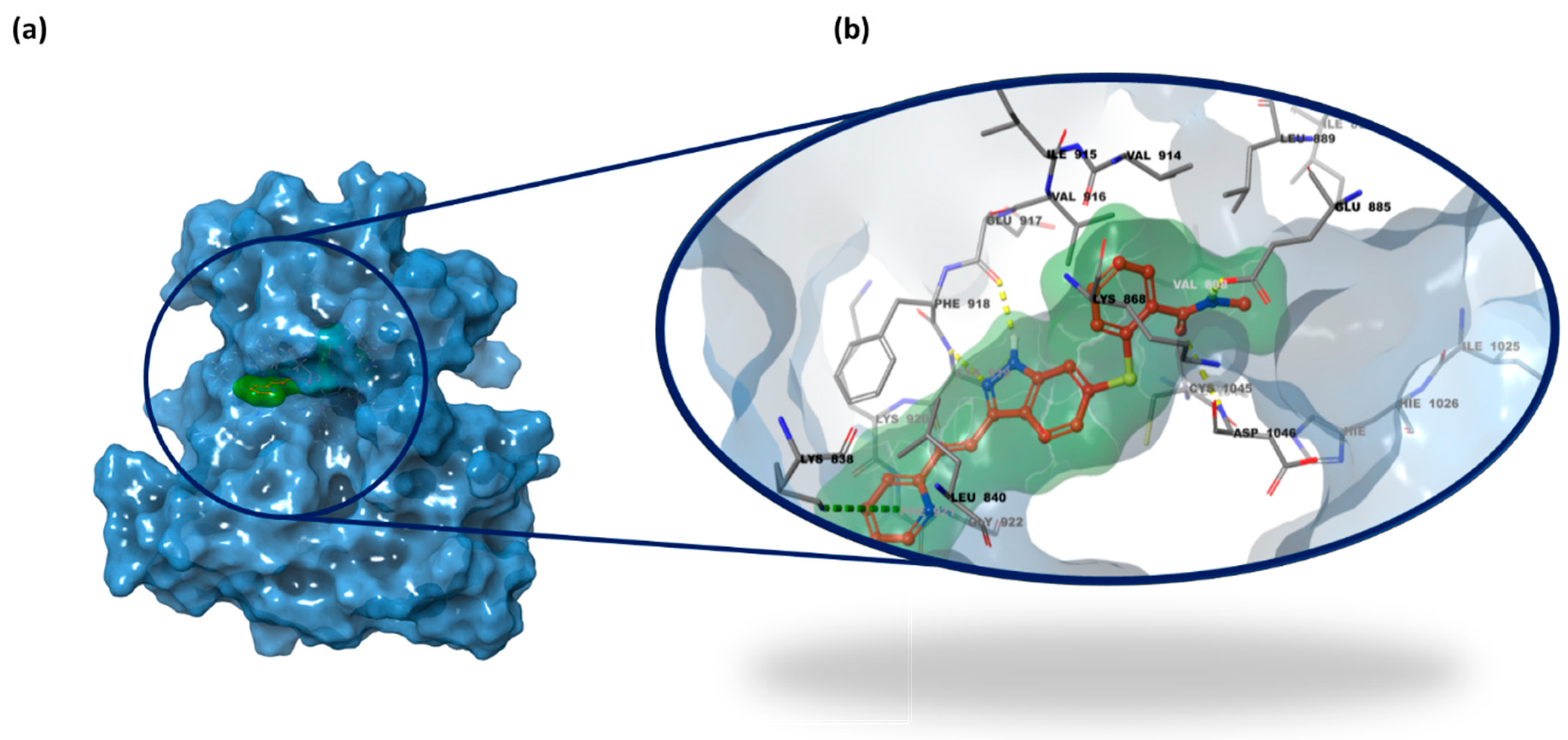
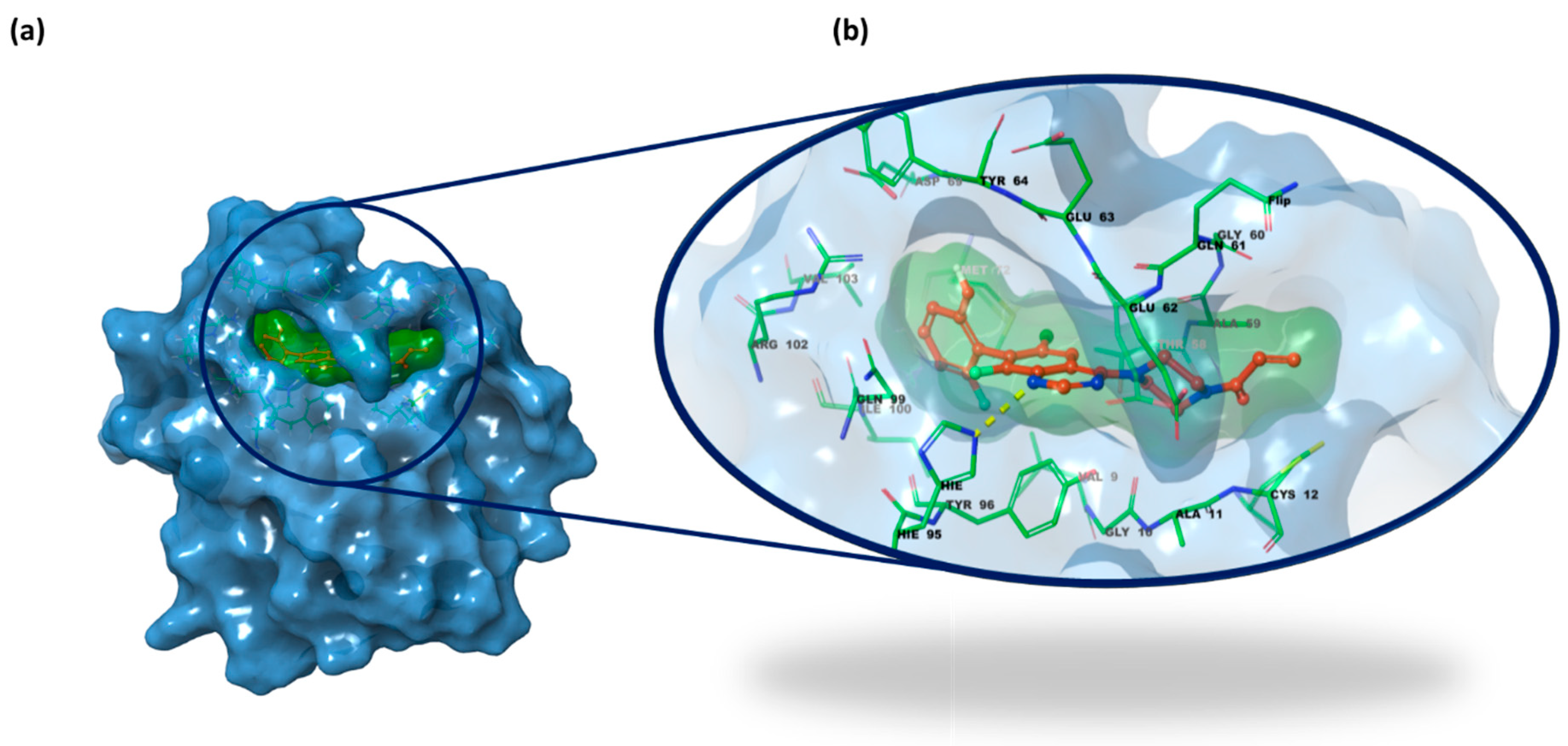
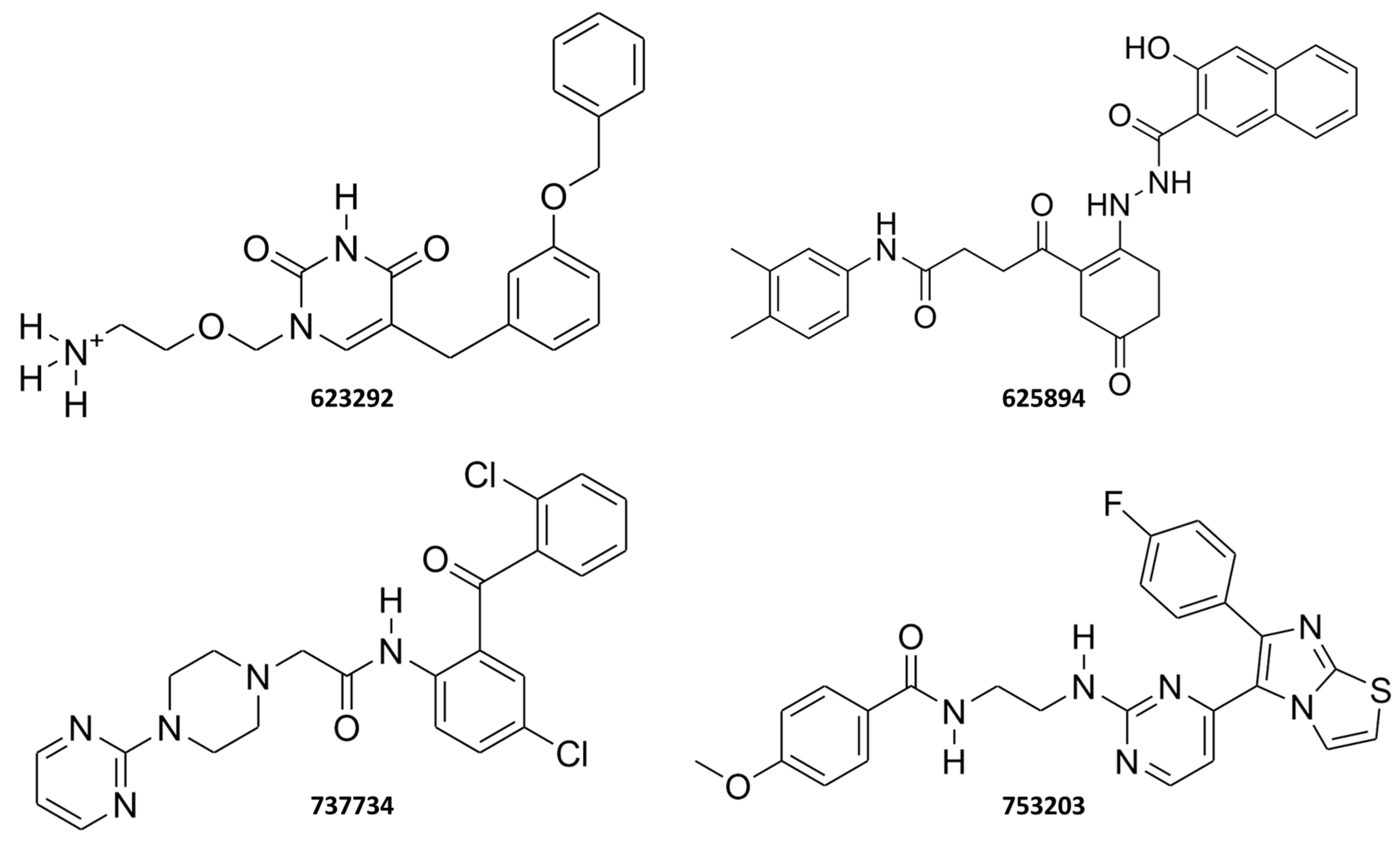
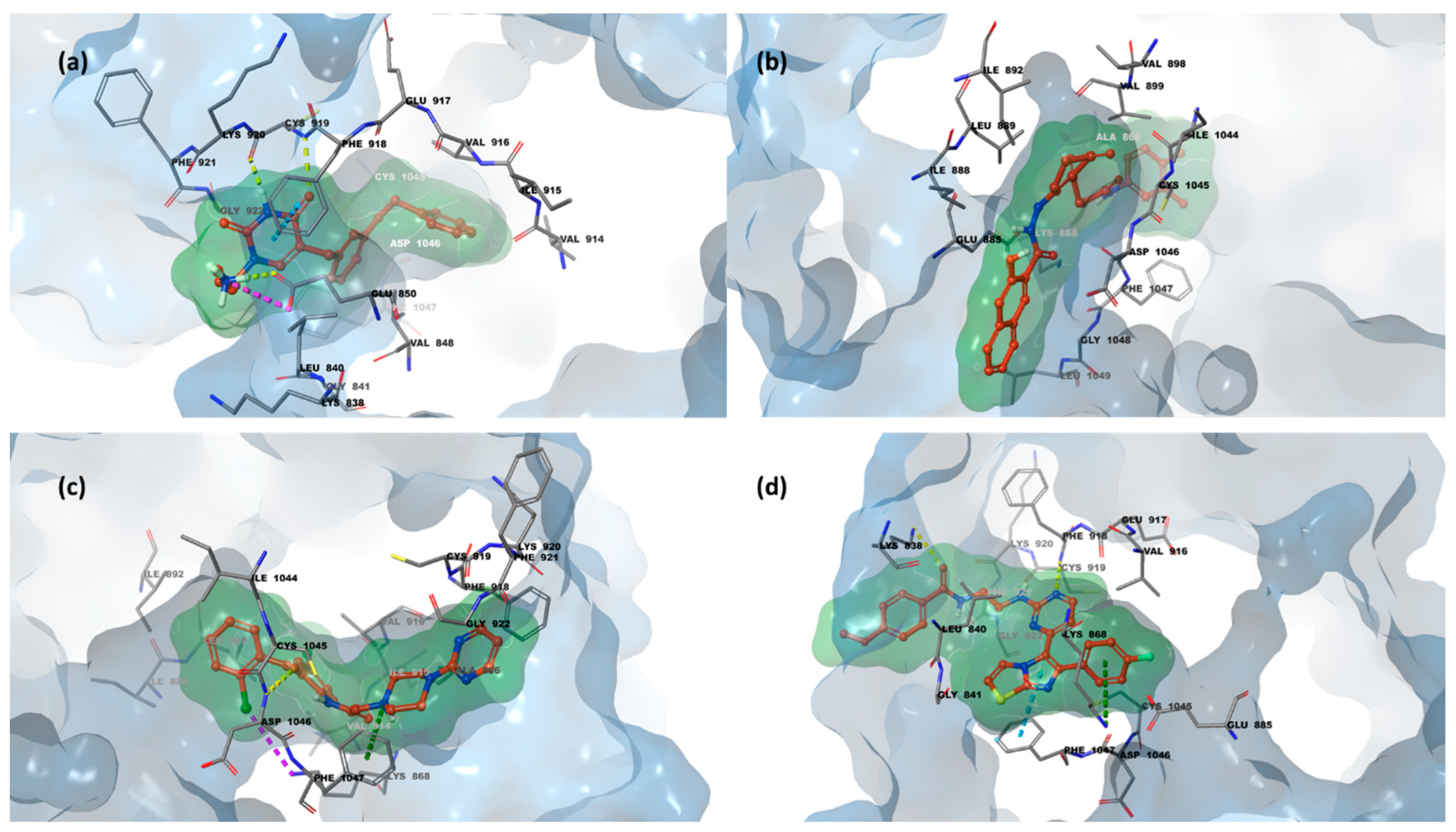
2.3. Structure-Based Studies: Molecular Docking Analysis
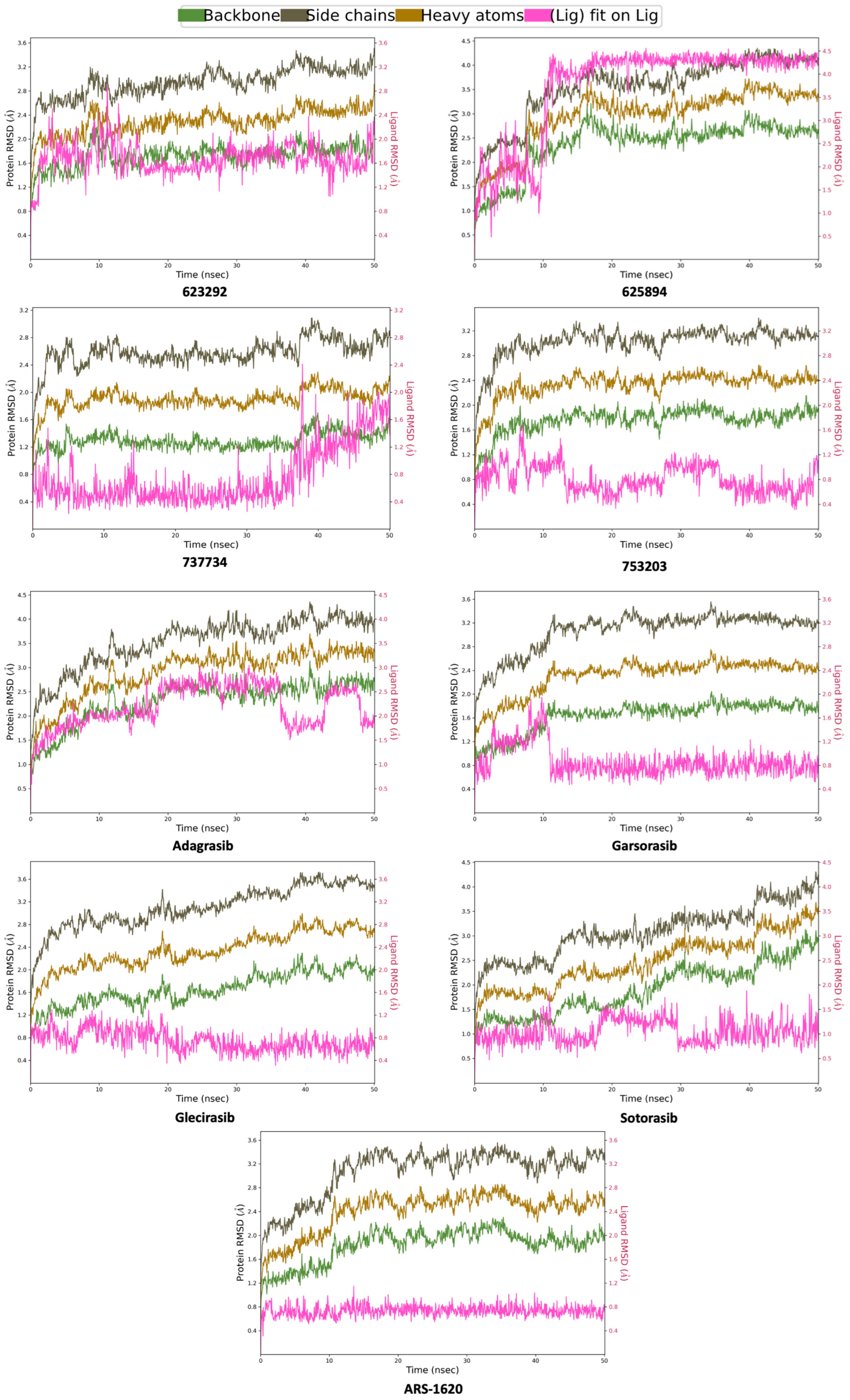
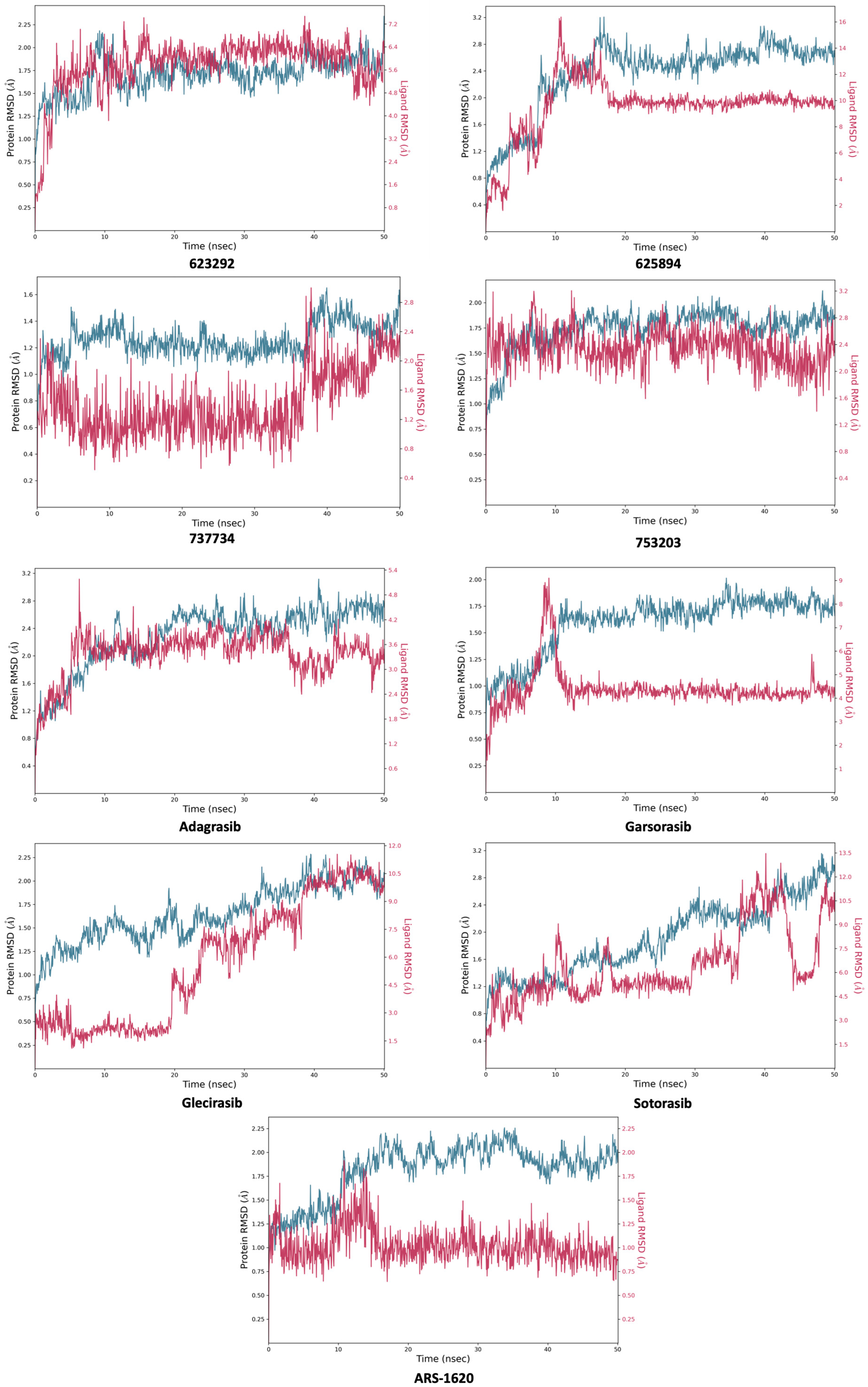
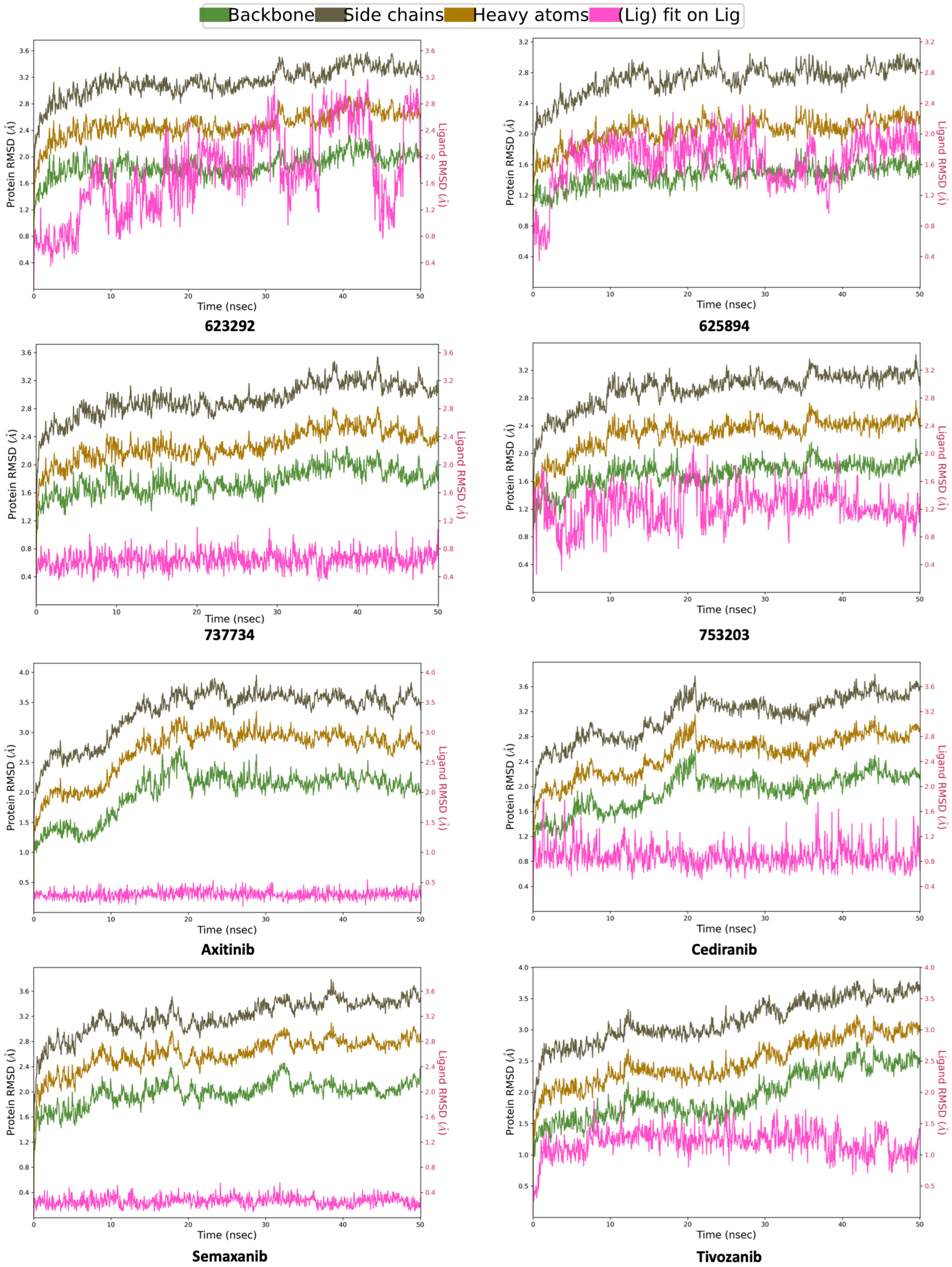
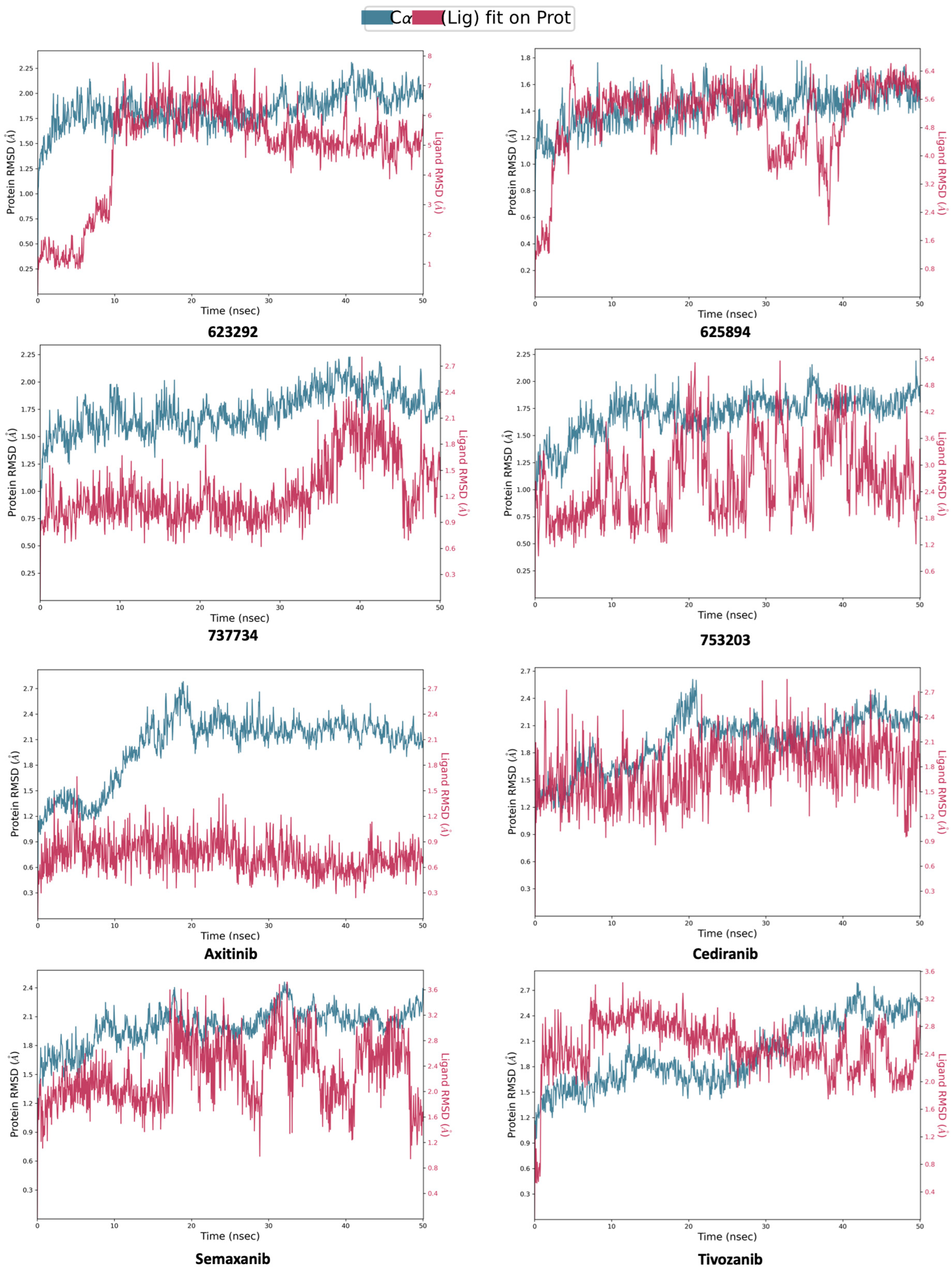
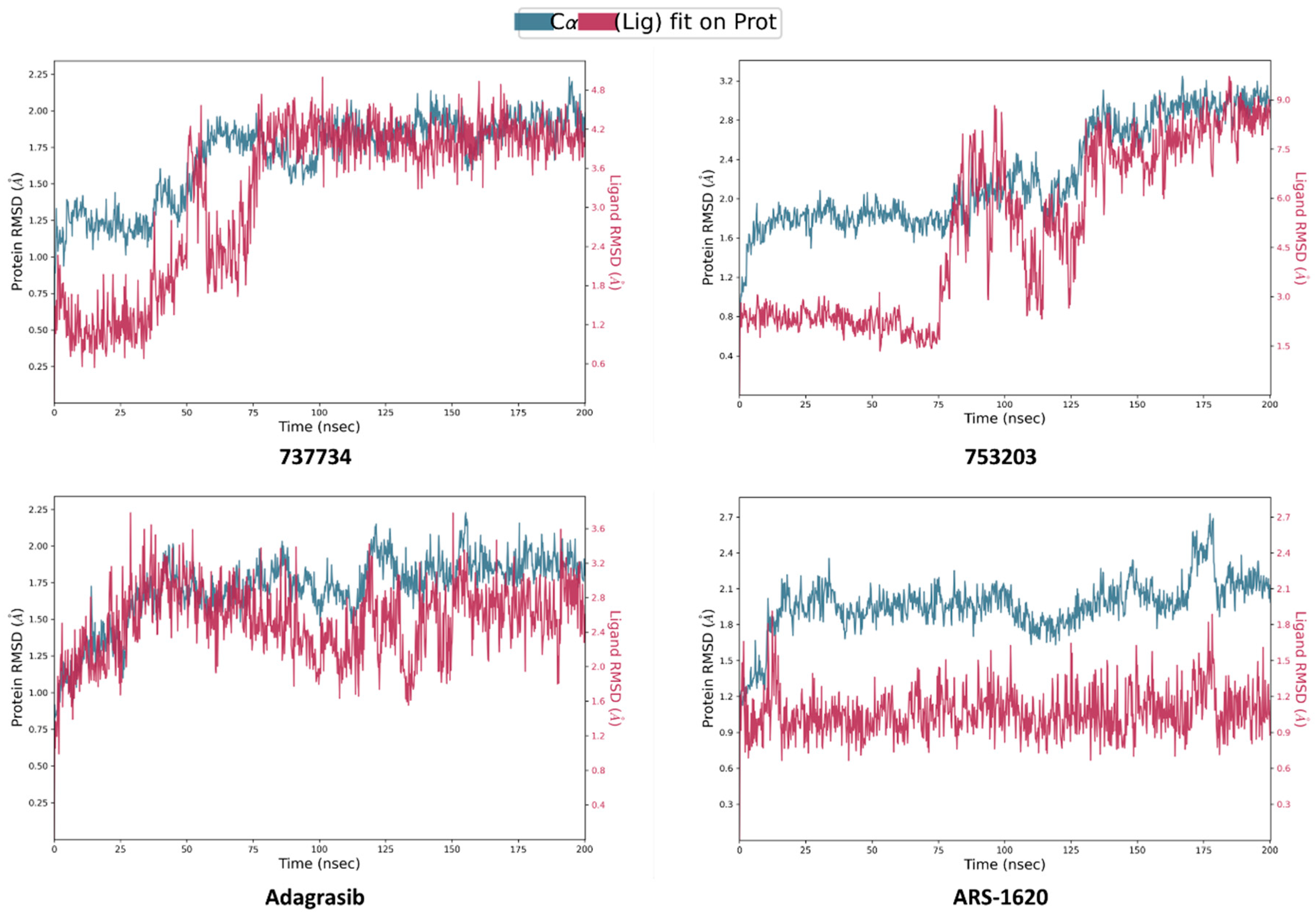
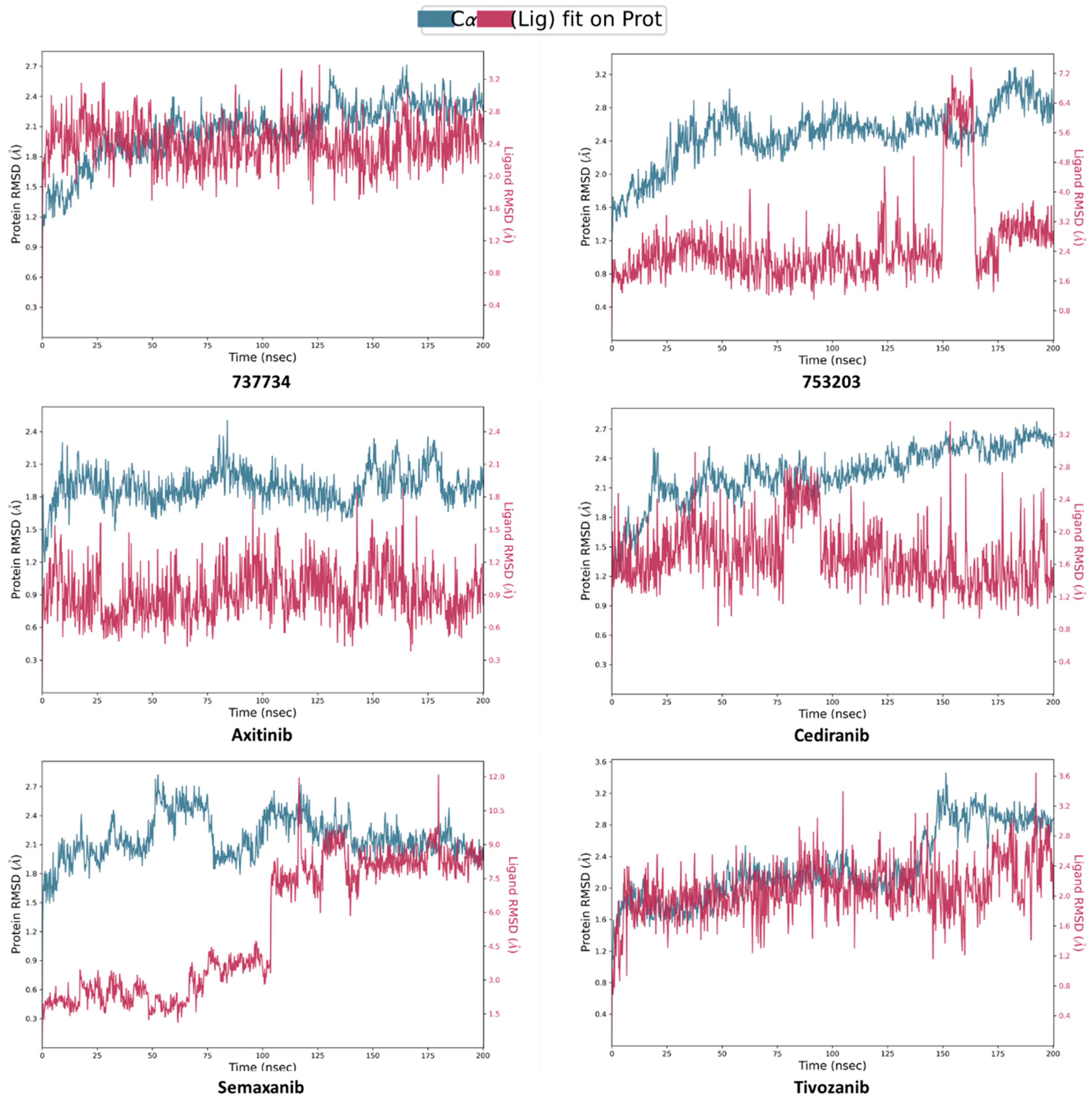
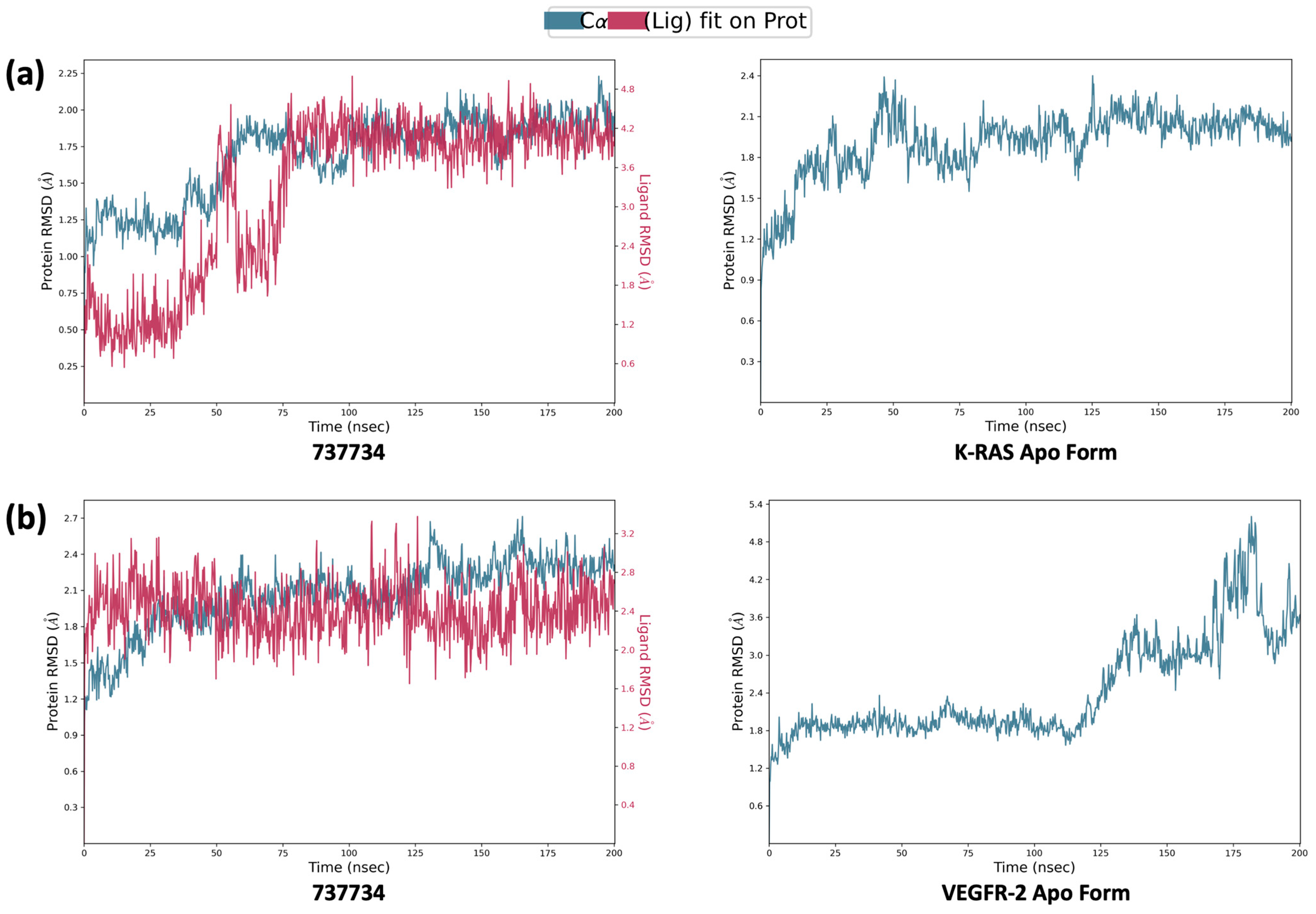
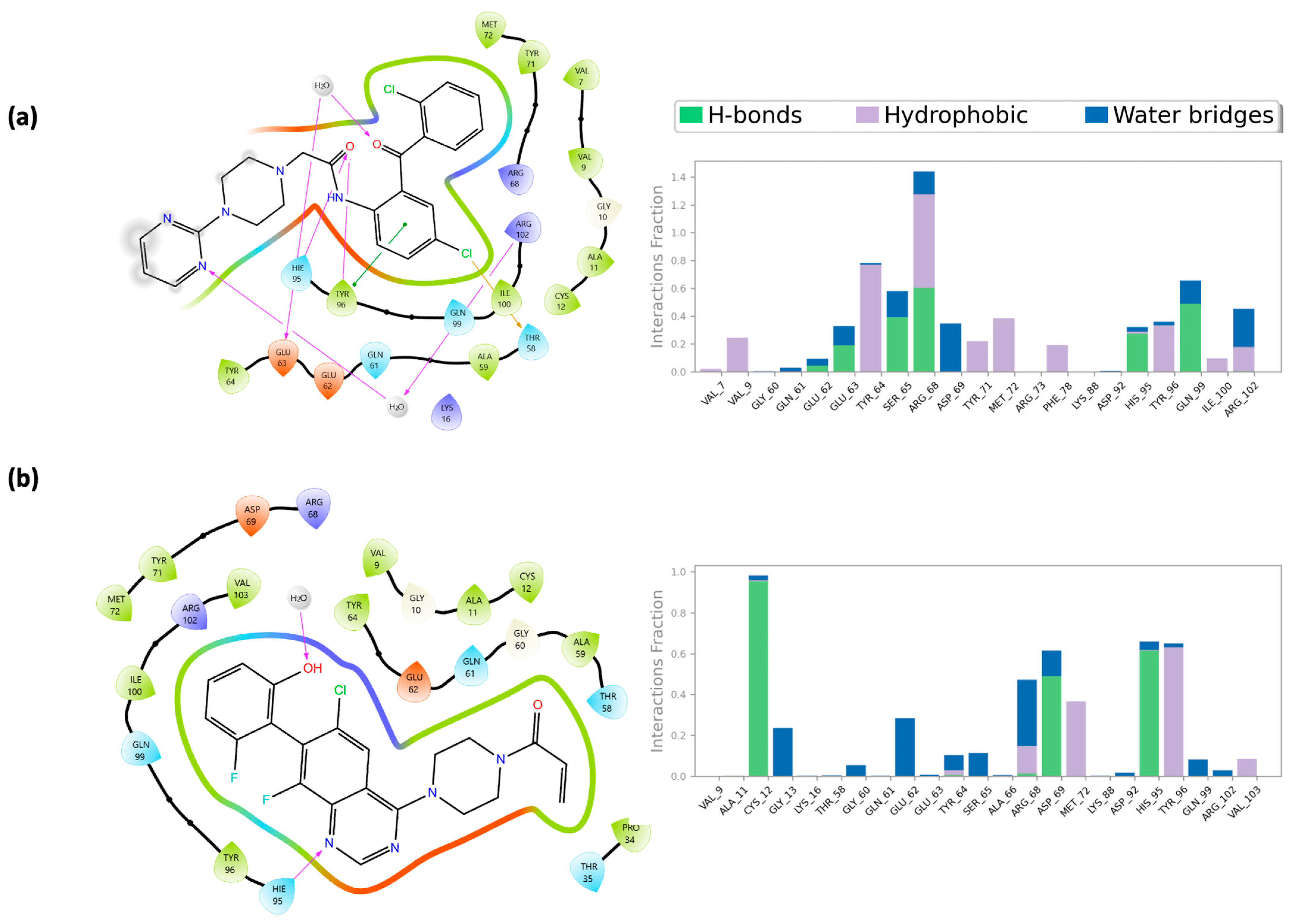
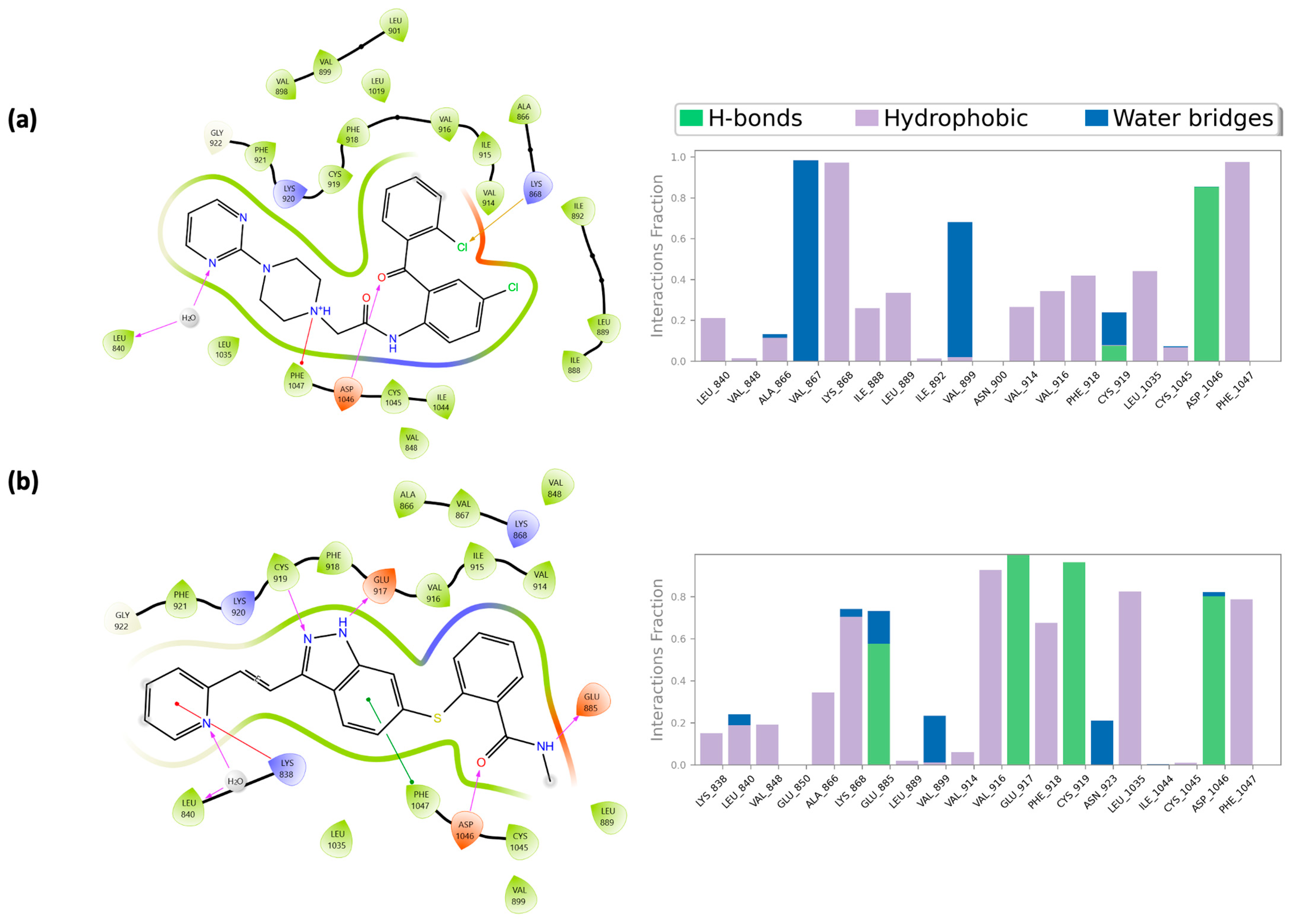
2.4. Molecular Dynamics Simulations
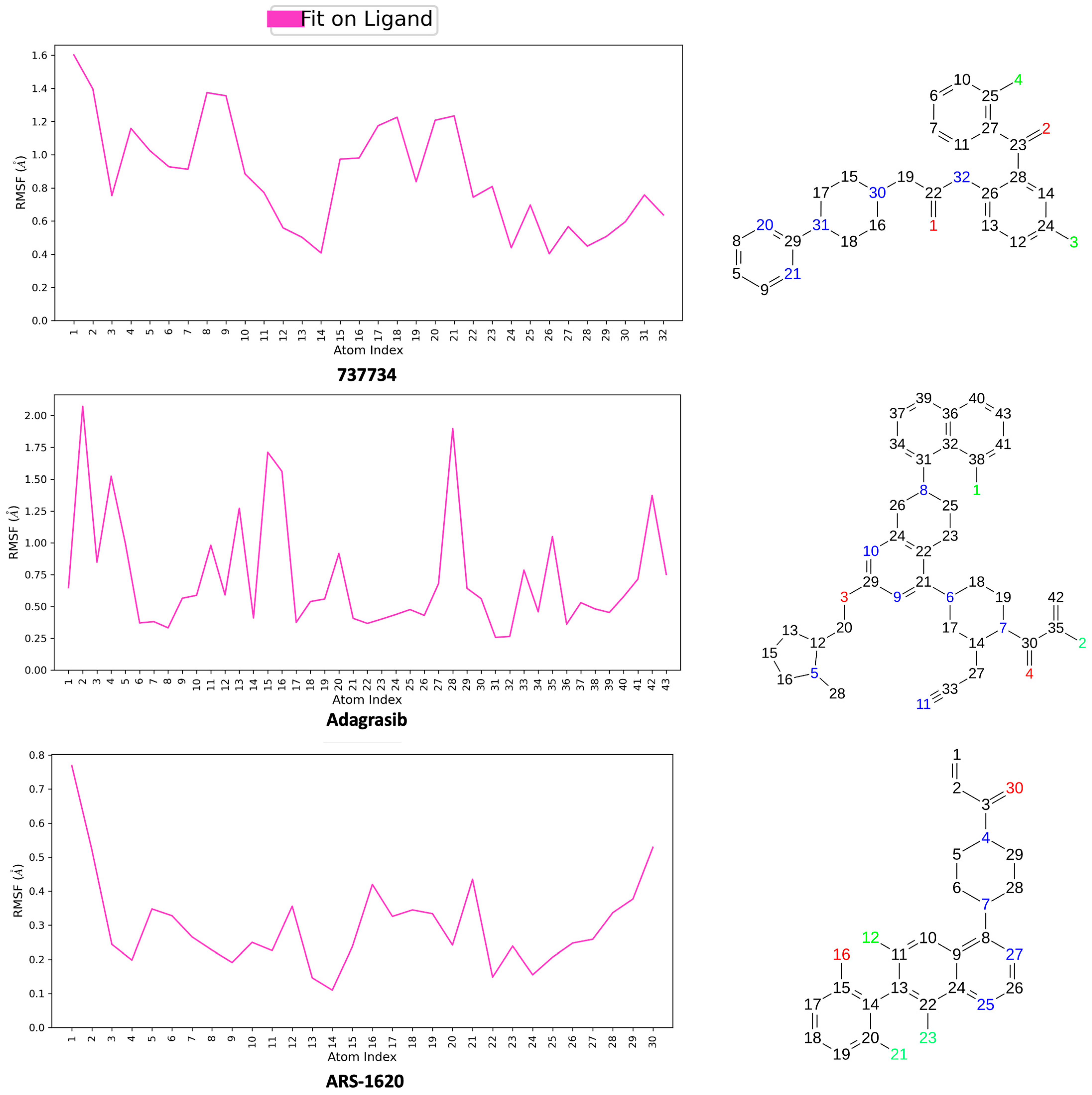
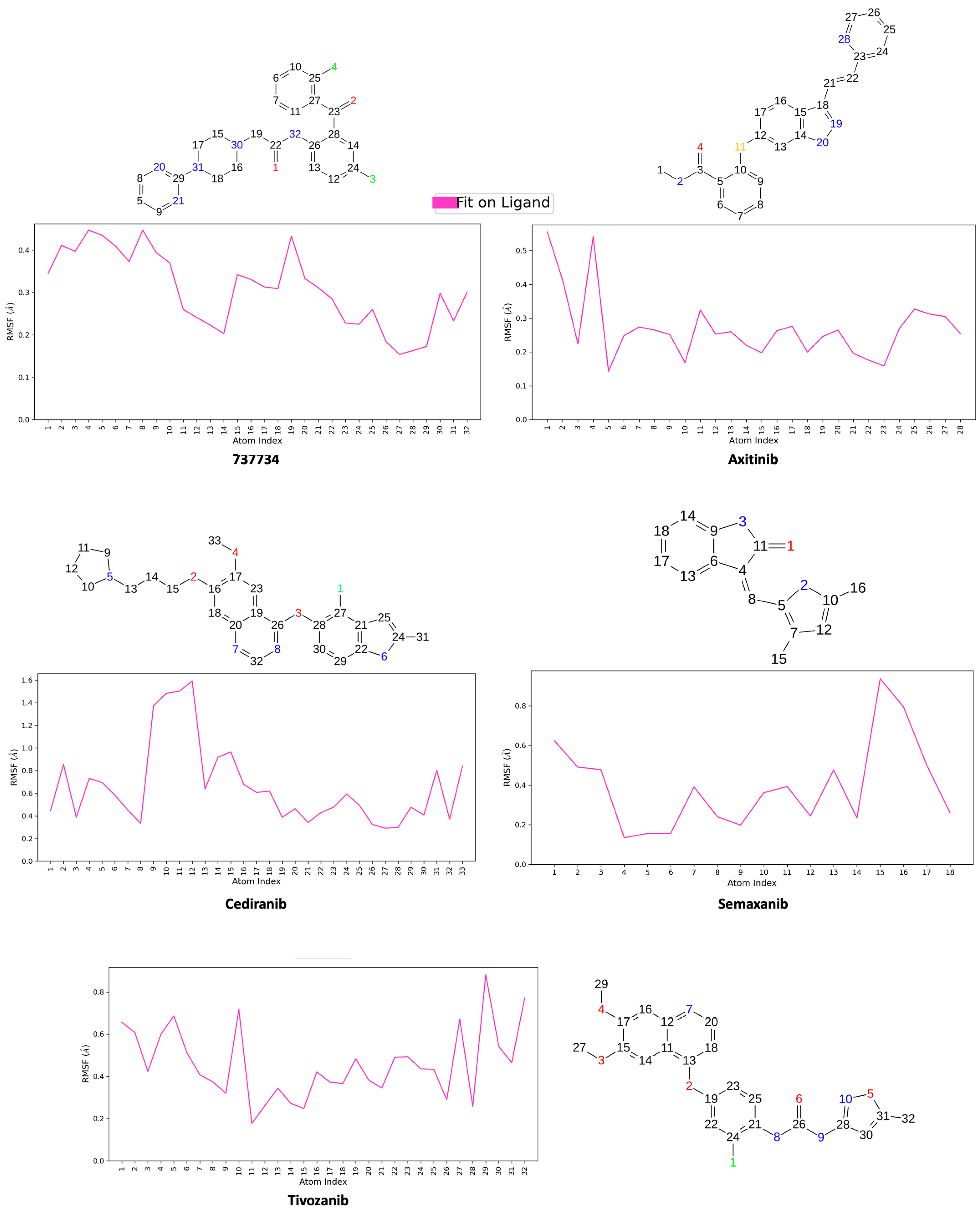
2.5. Integrated Analysis of Amino Acid Interactions Between Compound 737734 and Protein Binding Sites
3. Materials and Methods
3.1. Database Cleaning Phase
3.2. Ligand-Based Studies
3.3. Structure-Based Studies
3.3.1. Ligand Preparation
3.3.2. Protein Preparation
3.3.3. Docking Validation
3.3.4. Molecular Dynamics Simulation
3.3.5. MM-GBSA Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elmadani, M.; Mokaya, P.O.; Omer, A.A.A.; Kiptulon, E.K.; Klara, S.; Orsolya, M. Cancer burden in Europe: A systematic analysis of the GLOBOCAN database (2022). BMC Cancer 2025, 25, 447. [Google Scholar] [CrossRef]
- Li, X.; Deng, Y.; Li, Z.; Zhao, H. A novel angiogenesis-associated risk score predicts prognosis and characterizes the tumor microenvironment in colon cancer. Transl. Cancer Res. 2024, 13, 2094–2107. [Google Scholar] [CrossRef]
- Li, X.Y.; Ma, W.N.; Su, L.X.; Shen, Y.; Zhang, L.; Shao, Y.; Wang, D.; Wang, Z.; Wen, M.Z.; Yang, X.T. Association of Angiogenesis Gene Expression With Cancer Prognosis and Immunotherapy Efficacy. Front. Cell Dev. Biol. 2022, 10, 805507. [Google Scholar] [CrossRef] [PubMed]
- Maiborodin, I.; Mansurova, A.; Chernyavskiy, A.; Romanov, A.; Voitcitctkii, V.; Kedrova, A.; Tarkhov, A.; Chernyshova, A.; Krasil’nikov, S. Cancer Angiogenesis and Opportunity of Influence on Tumor by Changing Vascularization. J. Pers. Med. 2022, 12, 327. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Cai, E.; Huang, J.; Yu, P.; Li, K. Prognostic value of vascular endothelial growth factor expression in patients with esophageal cancer: A systematic review and meta-analysis. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1126–1134. [Google Scholar] [CrossRef]
- Dudley, A.C.; Griffioen, A.W. Pathological angiogenesis: Mechanisms and therapeutic strategies. Angiogenesis 2023, 26, 313–347. [Google Scholar] [CrossRef]
- Chung, A.S.; Ferrara, N. Developmental and pathological angiogenesis. Annu. Rev. Cell Dev. Biol. 2011, 27, 563–584. [Google Scholar] [CrossRef]
- Yadav, L.; Puri, N.; Rastogi, V.; Satpute, P.; Sharma, V. Tumour Angiogenesis and Angiogenic Inhibitors: A Review. J. Clin. Diagn. Res. 2015, 9, XE01–XE05. [Google Scholar] [CrossRef]
- Nishida, N.; Yano, H.; Nishida, T.; Kamura, T.; Kojiro, M. Angiogenesis in cancer. Vasc. Health Risk Manag. 2006, 2, 213–219. [Google Scholar] [CrossRef]
- Bielenberg, D.R.; Zetter, B.R. The Contribution of Angiogenesis to the Process of Metastasis. Cancer J. 2015, 21, 267–273. [Google Scholar] [CrossRef]
- Carmeliet, P. VEGF as a key mediator of angiogenesis in cancer. Oncology 2005, 69 (Suppl. S3), 4–10. [Google Scholar] [CrossRef] [PubMed]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef] [PubMed]
- Holmes, K.; Roberts, O.L.; Thomas, A.M.; Cross, M.J. Vascular endothelial growth factor receptor-2: Structure, function, intracellular signalling and therapeutic inhibition. Cell. Signal. 2007, 19, 2003–2012. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.; Gordon, E.; Claesson-Welsh, L. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 2016, 17, 611–625. [Google Scholar] [CrossRef]
- Ferrara, N.; Gerber, H.P.; LeCouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef]
- Marques, C.S.; Brandão, P.; Burke, A.J. Targeting Vascular Endothelial Growth Factor Receptor 2 (VEGFR-2): Latest Insights on Synthetic Strategies. Molecules 2024, 29, 5341. [Google Scholar] [CrossRef]
- Kranenburg, O. The KRAS oncogene: Past, present, and future. Biochim. Biophys. Acta 2005, 1756, 81–82. [Google Scholar] [CrossRef]
- Damani Shah, H.; Saranath, D.; Das, S.; Kharkar, P.; Karande, A. In-silico identification of small molecules targeting H-Ras and in-vitro cytotoxicity with caspase-mediated apoptosis in carcinoma cells. J. Cell. Biochem. 2019, 120, 5519–5530. [Google Scholar] [CrossRef]
- Chen, H.; Smaill, J.B.; Liu, T.; Ding, K.; Lu, X. Small-Molecule Inhibitors Directly Targeting KRAS as Anticancer Therapeutics. J. Med. Chem. 2020, 63, 14404–14424. [Google Scholar] [CrossRef]
- Ros, J.; Vaghi, C.; Baraibar, I.; Saoudi González, N.; Rodríguez-Castells, M.; García, A.; Alcaraz, A.; Salva, F.; Tabernero, J.; Elez, E. Targeting KRAS G12C Mutation in Colorectal Cancer, A Review: New Arrows in the Quiver. Int. J. Mol. Sci. 2024, 25, 3304. [Google Scholar] [CrossRef]
- Rak, J.; Kerbel, R.S. Ras regulation of vascular endothelial growth factor and angiogenesis. Methods Enzymol. 2001, 333, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Kranenburg, O.; Gebbink, M.F.; Voest, E.E. Stimulation of angiogenesis by Ras proteins. Biochim. Biophys. Acta 2004, 1654, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Lin, Y.; Wu, Y.; Deng, L.; Zhang, H.; Liao, M.; Peng, Y. MvMRL: A multi-view molecular representation learning method for molecular property prediction. Brief. Bioinform. 2024, 25, bbae298. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, Q.; Chen, X.; Weng, M.; Huang, Y.; Chen, Q.; Liu, X.; Huang, H.; Feng, Y.; Zhou, H.; et al. Targeting SOX13 inhibits assembly of respiratory chain supercomplexes to overcome ferroptosis resistance in gastric cancer. Nat. Commun. 2024, 15, 4296. [Google Scholar] [CrossRef]
- Bao, M.H.; Li, G.Y.; Huang, X.S.; Tang, L.; Dong, L.P.; Li, J.M. Long Noncoding RNA LINC00657 Acting as a miR-590-3p Sponge to Facilitate Low Concentration Oxidized Low-Density Lipoprotein-Induced Angiogenesis. Mol. Pharmacol. 2018, 93, 368–375. [Google Scholar] [CrossRef]
- Yang, H.; He, C.; Bi, Y.; Zhu, X.; Deng, D.; Ran, T.; Ji, X. Synergistic effect of VEGF and SDF-1α in endothelial progenitor cells and vascular smooth muscle cells. Front. Pharmacol. 2022, 13, 914347. [Google Scholar] [CrossRef]
- Liu, P.W.; Liu, Z.Y.; Deng, S.J.; Zhang, X.; Wang, Z.B.; Wu, N.Y.; Liu, C.S.; Hu, M.H.; Wang, J.; Li, H. A Pyroptosis-Related LncRNA Signature for Predicting Prognosis, Immune Features and Drug Sensitivity in Ovarian Cancer. OncoTargets Ther. 2025, 18, 585–601. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Wang, Y.; Lin, C.; Zhang, D.; Chen, J.; Ouyang, L.; Wu, F.; Zhang, J.; Chen, L. Recent progress on vascular endothelial growth factor receptor inhibitors with dual targeting capabilities for tumor therapy. J. Hematol. Oncol. 2022, 15, 89. [Google Scholar] [CrossRef]
- Tarallo, V.; De Falco, S. The vascular endothelial growth factors and receptors family: Up to now the only target for anti-angiogenesis therapy. Int. J. Biochem. Cell Biol. 2015, 64, 185–189. [Google Scholar] [CrossRef]
- Lugano, R.; Ramachandran, M.; Dimberg, A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell. Mol. Life Sci. 2020, 77, 1745–1770. [Google Scholar] [CrossRef]
- Uprety, D.; Adjei, A.A. KRAS: From undruggable to a druggable Cancer Target. Cancer Treat. Rev. 2020, 89, 102070. [Google Scholar] [CrossRef] [PubMed]
- Nussinov, R.; Jang, H. Direct K-Ras Inhibitors to Treat Cancers: Progress, New Insights, and Approaches to Treat Resistance. Annu. Rev. Pharmacol. Toxicol. 2024, 64, 231–253. [Google Scholar] [CrossRef] [PubMed]
- Costa, N.S.; Lima, L.R.; Cruz, J.N.; Santos, I.V.F.; Silva, R.C.; Maciel, A.A.; Barros, E.S.; Andrade, M.L.D.S.; Ramos, R.S.; Kimani, N.M.; et al. Identification of Inhibitors with Potential Anti-Prostate Cancer Activity: A Chemoinformatics Approach. Pharmaceuticals 2025, 18, 888. [Google Scholar] [CrossRef] [PubMed]
- Martorana, A.; Gentile, C.; Lauria, A. In Silico Insights into the SARS CoV-2 Main Protease Suggest NADH Endogenous Defences in the Control of the Pandemic Coronavirus Infection. Viruses 2020, 12, 805. [Google Scholar] [CrossRef]
- Modi, S.J.; Kulkarni, V.M. Exploration of structural requirements for the inhibition of VEGFR-2 tyrosine kinase: Binding site analysis of type II, ‘DFG-out’ inhibitors. J. Biomol. Struct. Dyn. 2022, 40, 5712–5727. [Google Scholar] [CrossRef]
- Modi, S.J.; Kulkarni, V.M. Vascular Endothelial Growth Factor Receptor (VEGFR-2)/KDR Inhibitors: Medicinal Chemistry Perspective. Med. Drug Discov. 2019, 2, 100009. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, M.; Wang, Y.; Fan, Y.; Chen, X.; Yang, Y.; Hua, Y.; Xie, W.; Lu, T.; Tang, W.; et al. Protein-ligand interaction-guided discovery of novel VEGFR-2 inhibitors. J. Biomol. Struct. Dyn. 2020, 38, 2559–2574. [Google Scholar] [CrossRef]
- McTigue, M.; Murray, B.W.; Chen, J.H.; Deng, Y.L.; Solowiej, J.; Kania, R.S. Molecular conformations, interactions, and properties associated with drug efficiency and clinical performance among VEGFR TK inhibitors. Proc. Natl. Acad. Sci. USA 2012, 109, 18281–18289. [Google Scholar] [CrossRef]
- Huang, L.; Guo, Z.; Wang, F.; Fu, L. KRAS mutation: From undruggable to druggable in cancer. Signal Transduct. Target. Ther. 2021, 6, 386. [Google Scholar] [CrossRef]
- Parasrampuria, D.A.; Yu, A.; Haddish-Berhane, N. Chapter 13-KRAS: Structure, function, and development of anticancer drugs. In Cancer-Leading Proteases; Gupta, S.P., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 359–389. [Google Scholar]
- Pandey, D.; Chauhan, S.C.; Kashyap, V.K.; Roy, K.K. Structural insights into small-molecule KRAS inhibitors for targeting KRAS mutant cancers. Eur. J. Med. Chem. 2024, 277, 116771. [Google Scholar] [CrossRef]
- Janes, M.R.; Zhang, J.; Li, L.S.; Hansen, R.; Peters, U.; Guo, X.; Chen, Y.; Babbar, A.; Firdaus, S.J.; Darjania, L.; et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018, 172, 578–589.e517. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Lauria, A.; Mannino, S.; Gentile, C.; Mannino, G.; Martorana, A.; Peri, D. DRUDIT: Web-based DRUgs DIscovery Tools to design small molecules as modulators of biological targets. Bioinformatics 2020, 36, 1562–1569. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2021–2: LigPrep; Schrödinger, LLC: New York, NY, USA, 2021.
- Schrödinger Release 2021-2: Protein Preparation Wizard; Epik, Schrödinger, LLC: New York, NY, USA; Impact, Schrödinger, LLC: New York, NY, USA; Prime, Schrödinger, LLC: New York, NY, USA, 2021.
- Banks, J.L.; Beard, H.S.; Cao, Y.; Cho, A.E.; Damm, W.; Farid, R.; Felts, A.K.; Halgren, T.A.; Mainz, D.T.; Maple, J.R.; et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- RCSB PDB. Available online: www.rcsb.org (accessed on 29 June 2022).
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]

| K-RAS | VEGFR-2 | ||
|---|---|---|---|
| Title | IFDScore | Title | IFDScore |
| ARS-1620 | −466.025 | 753203 | −669.154 |
| adagrasib | −465.595 | 737734 | −668.884 |
| 753203 | −464.212 | cediranib | −667.339 |
| 625894 | −462.446 | axitinib | −667.088 |
| 737734 | −462.348 | tivozanib | −666.343 |
| 623292 | −462.176 | 623292 | −664.999 |
| 654632 | −460.126 | 641220 | −664.389 |
| 751605 | −459.988 | 625894 | −663.527 |
| 766494 | −459.022 | 720567 | −663.426 |
| 736273 | −458.777 | 697231 | −663.010 |
| 697231 | −458.666 | 751605 | −662.414 |
| 735330 | −458.103 | 678359 | −662.108 |
| 169874 | −457.836 | 735330 | −661.753 |
| 641220 | −457.630 | 707444 | −660.894 |
| 697673 | −457.491 | 747856 | −660.605 |
| 747856 | −457.411 | 697673 | −660.516 |
| 744650 | −456.930 | 744650 | −660.441 |
| 707444 | −456.861 | 766494 | −660.166 |
| 707776 | −456.732 | 169874 | −660.023 |
| 720567 | −456.708 | 736273 | −659.949 |
| 733308 | −456.185 | 697227 | −659.573 |
| 647945 | −456.120 | 654632 | −659.458 |
| sotorasib | −455.351 | 733308 | −659.224 |
| 665325 | −455.236 | 665325 | −658.868 |
| garsorasib | −455.067 | 707776 | −656.454 |
| 697227 | −454.700 | 647945 | −656.049 |
| glecirasib | −453.644 | semaxanib | −655.604 |
| 678359 | −452.451 | ||
| K-RAS | VEGFR-2 | ||||
|---|---|---|---|---|---|
| Title | ARS-1620 | 737734 | Title | Axitinib | 737734 |
| Val7 | X | Lys838 | X | ||
| Val9 | X | X | Leu840 | X | X |
| Gly10 | X | X | Val848 | X | X |
| Ala11 | X | X | Ala866 | X | X |
| Cys12 | X | X | Val867 | X | |
| Lys16 | X | X | Lys868 | X | X |
| Pro34 | X | Glu885 | X | ||
| Thr35 | X | Ile888 | X | ||
| Thr58 | X | X | Leu889 | X | X |
| Ala59 | X | X | Ile892 | X | |
| Gly60 | X | Val898 | X | ||
| Gln61 | X | X | Val899 | X | X |
| Glu62 | X | X | Leu901 | X | |
| Glu63 | X | Val914 | X | X | |
| Tyr64 | X | X | Ile915 | X | X |
| Arg68 | X | X | Val916 | X | X |
| Asp69 | X | Glu917 | X * | ||
| Tyr71 | X | X | Phe918 | X | X |
| Met72 | X | X | Cys919 | X * | X |
| Hie95 | X * | X * | Lys920 | X | X |
| Tyr96 | X | X * | Phe921 | X | X |
| Gln99 | X | X | Gly922 | X | X |
| Ile100 | X | X | Leu1019 | X | |
| Arg102 | X | X | Leu1035 | X | X |
| Val103 | X | Ile1044 | X | ||
| Cys1045 | X | X | |||
| Asp1046 | X * | X * | |||
| Phe1047 | X | X | |||
| Tot. | 23 | 20 | Tot. | 22 | 25 |
| K-RAS | |||||
| Complex | Prime Coulomb | Prime vdW | Prime Solvent | Prime Hbond | Prime Energy |
| 737734 | −6302.17 | −851.78 | −1609.49 | −133.84 | −9040.7 |
| ARS-1620 | −6305.19 | −847.79 | −1626.10 | −133.56 | −9063.7 |
| VEGFR-2 | |||||
| Complex | Prime Coulomb | Prime vdW | Prime Solvent | Prime Hbond | Prime Energy |
| 737734 | −9172.15 | −1549.90 | −1824.98 | −142.24 | −13,112.3 |
| Axitinib | −9151.63 | −1578.12 | −1744.62 | −145.96 | −13,056.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bono, A.; La Monica, G.; Alamia, F.; Tocco, D.; Lauria, A.; Martorana, A. A Hybrid In Silico Approach for Identifying Dual VEGFR/RAS Inhibitors as Potential Anticancer and Anti-Angiogenic Agents. Pharmaceuticals 2025, 18, 1579. https://doi.org/10.3390/ph18101579
Bono A, La Monica G, Alamia F, Tocco D, Lauria A, Martorana A. A Hybrid In Silico Approach for Identifying Dual VEGFR/RAS Inhibitors as Potential Anticancer and Anti-Angiogenic Agents. Pharmaceuticals. 2025; 18(10):1579. https://doi.org/10.3390/ph18101579
Chicago/Turabian StyleBono, Alessia, Gabriele La Monica, Federica Alamia, Dennis Tocco, Antonino Lauria, and Annamaria Martorana. 2025. "A Hybrid In Silico Approach for Identifying Dual VEGFR/RAS Inhibitors as Potential Anticancer and Anti-Angiogenic Agents" Pharmaceuticals 18, no. 10: 1579. https://doi.org/10.3390/ph18101579
APA StyleBono, A., La Monica, G., Alamia, F., Tocco, D., Lauria, A., & Martorana, A. (2025). A Hybrid In Silico Approach for Identifying Dual VEGFR/RAS Inhibitors as Potential Anticancer and Anti-Angiogenic Agents. Pharmaceuticals, 18(10), 1579. https://doi.org/10.3390/ph18101579







