Gene Therapy in the Light of Lifestyle Diseases: Budesonide, Acetaminophen and Simvastatin Modulates rAAV Transduction Efficiency
Abstract
1. Introduction
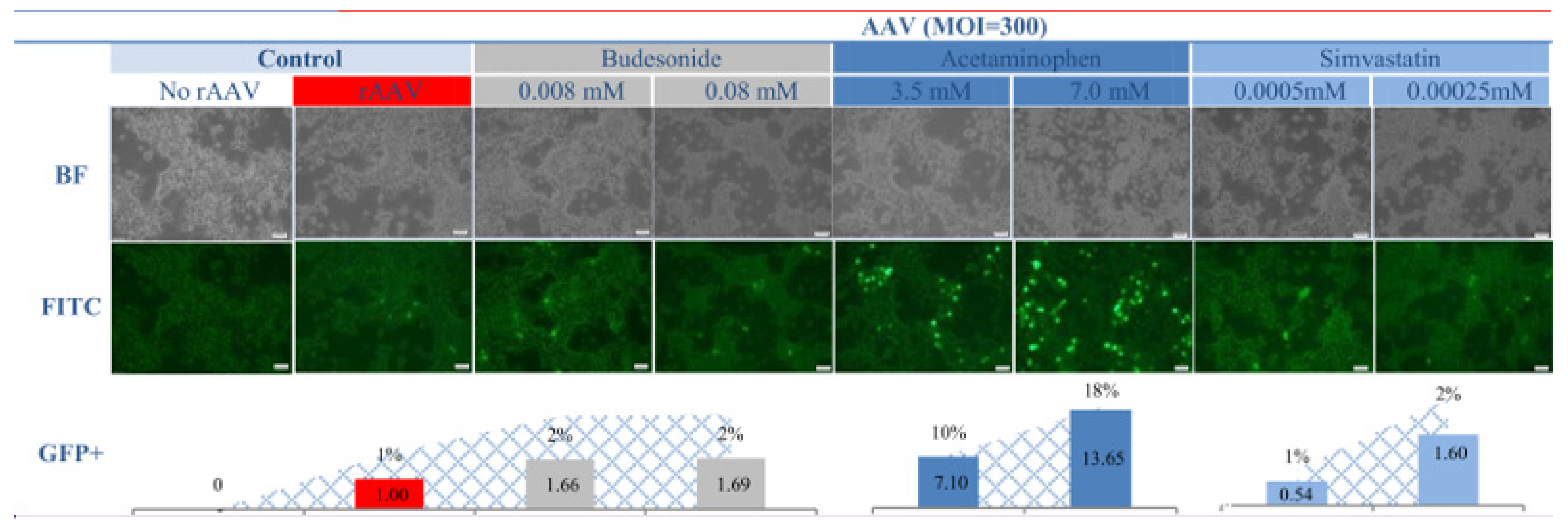
2. Results and Discussion
3. Conclusions
4. Materials and Methods
4.1. Reagents and Materials
Preparation of Drug Solutions
4.2. rAAV Transduction
4.3. Measurement of Transduction Efficiency
- A.
- Real-time PCR for examination of rAAV genome copy number
- B.
- Fluorescent imaging
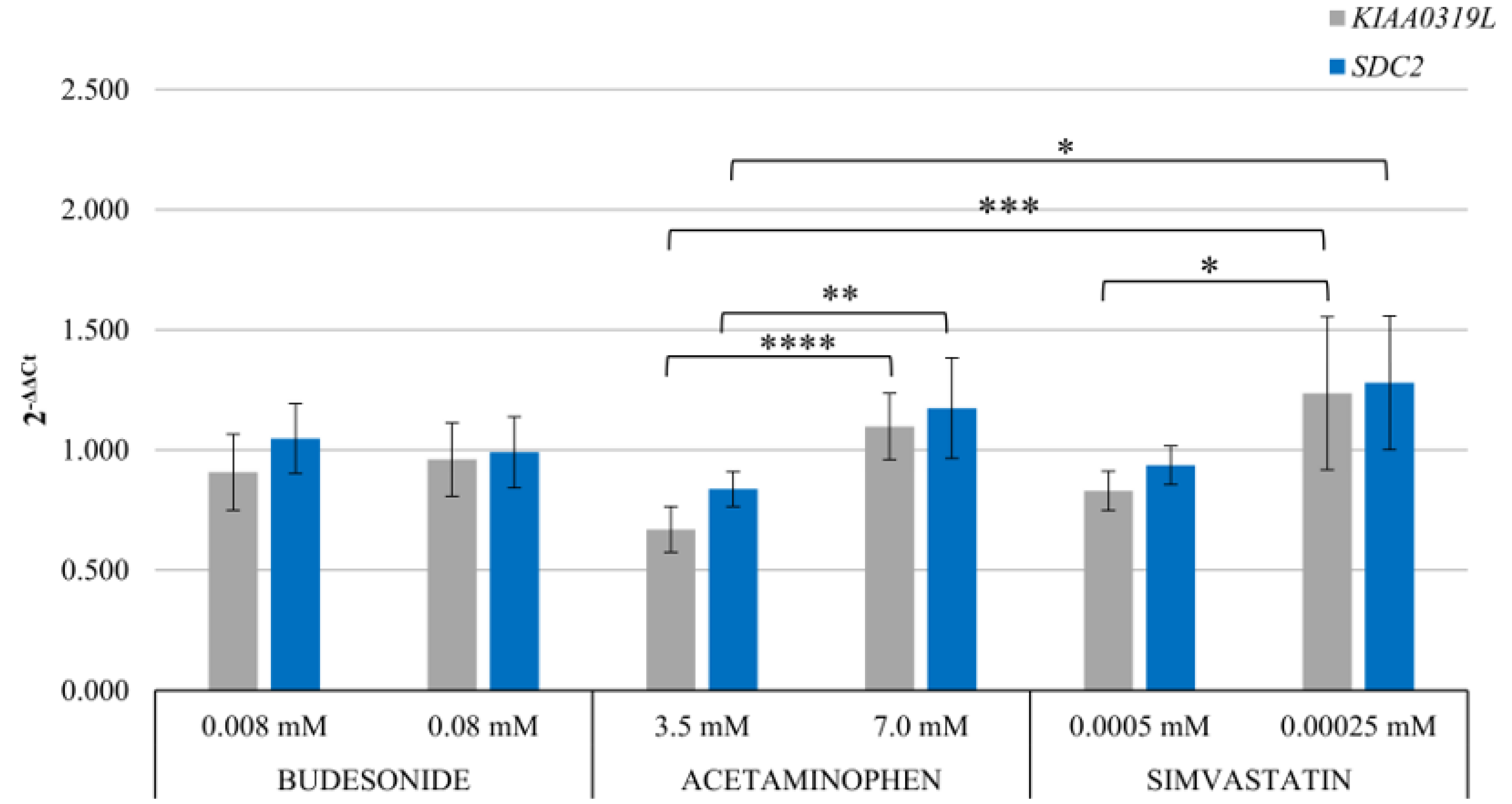
4.4. Assessment of the Expression of rAAV Receptors
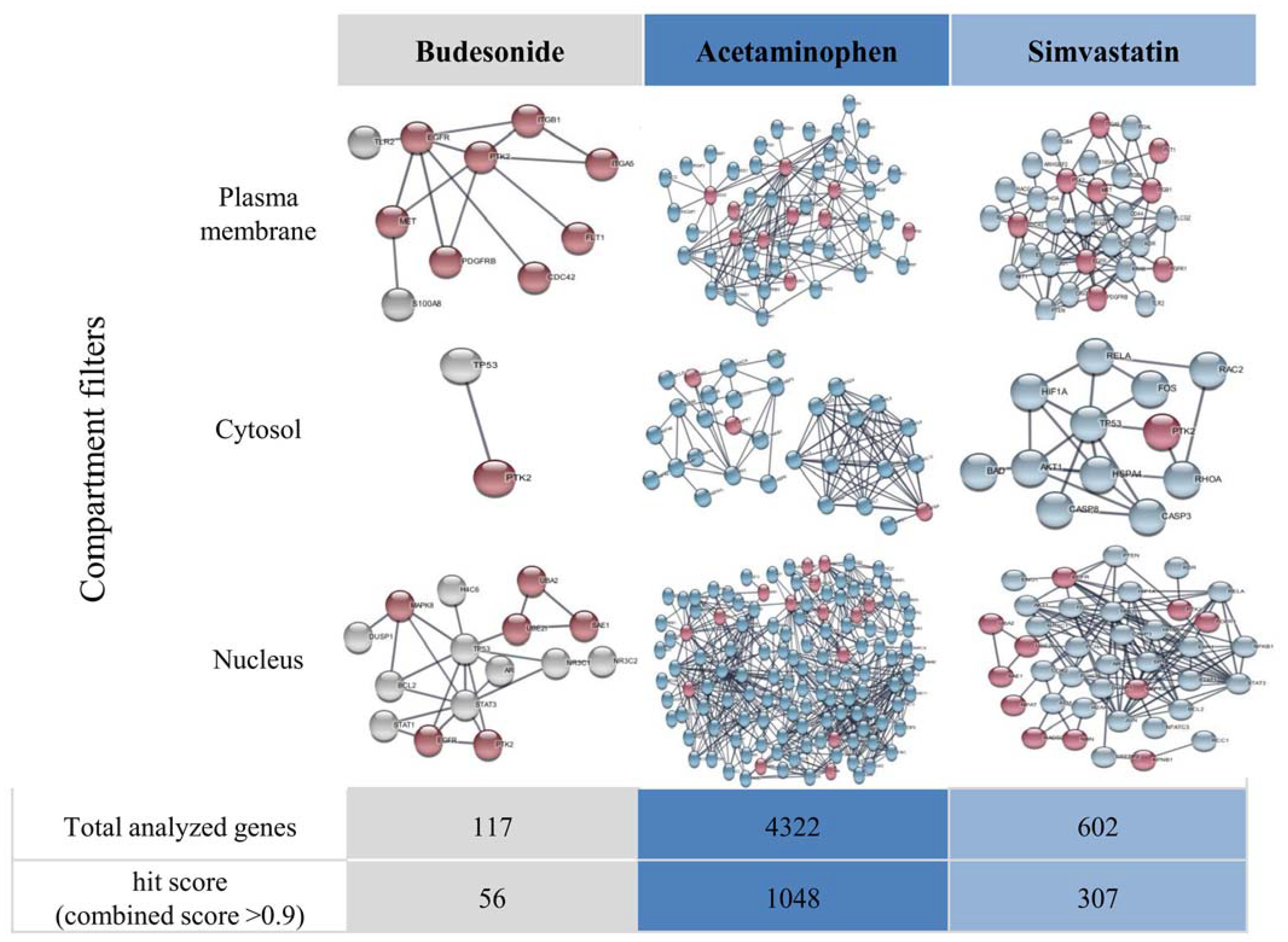
5. Visualization of Functional Gene Interactions in Drug-Exposed Cells
6. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, D.; Tai, P.W.L.; Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019, 18, 358–378. [Google Scholar] [CrossRef] [PubMed]
- Au, H.K.E.; Isalan, M.; Mielcarek, M. Gene Therapy Advances: A Meta-Analysis of AAV Usage in Clinical Settings. Front. Med. 2021, 9, 809118. [Google Scholar] [CrossRef] [PubMed]
- Heo, Y.-A. Etranacogene dezaparvovec: First approval. Drugs 2023, 83, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Keam, S.J. Eladocagene exuparvovec: First approval. Drugs 2022, 82, 1427–1432. [Google Scholar] [CrossRef]
- Philippidis, A. BioMarin’s ROCTAVIAN wins food and drug administration approval as first gene therapy for severe hemophilia A. Hum. Gene Ther. 2023, 34, 665–668. [Google Scholar] [CrossRef]
- Zittersteijn, H.A.; Gonçalves, M.; Hoeben, R.C. A primer to gene therapy: Progress, prospects, and problems. J. Inherit. Metab. Dis. 2021, 44, 54–71. [Google Scholar] [CrossRef]
- Srivastava, A.; Mallela, K.M.G.; Deorkar, N.; Brophy, G. Manufacturing Challenges and Rational Formulation Development for AAV Viral Vectors. J. Pharm. Sci. 2021, 110, 2609–2624. [Google Scholar] [CrossRef]
- Pupo, A.; Fernández, A.; Low, S.H.; François, A.; Suárez-Amarán, L.; Samulski, R.J. AAV vectors: The Rubik’s cube of human gene therapy. Mol. Ther. 2022, 30, 3515–3541. [Google Scholar] [CrossRef]
- Stone, D.; Aubert, M.; Jerome, K.R. Adeno-associated virus vectors and neurotoxicity—Lessons from preclinical and human studies. Gene Ther. 2023, 1–14. [Google Scholar] [CrossRef]
- Duan, D. Lethal immunotoxicity in high-dose systemic AAV therapy. Mol. Ther. 2023, 31, 3123–3126. [Google Scholar] [CrossRef]
- Cao, D.; Byrne, B.J.; de Jong, Y.P.; Terhorst, C.; Duan, D.; Herzog, R.W.; Kumar, S.R. Innate Immune Sensing of Adeno-Associated Virus Vectors. Hum. Gene Ther. 2024, 35, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Costa-Verdera, H.; Unzu, C.; Valeri, E.; Adriouch, S.; González Aseguinolaza, G.; Mingozzi, F.; Kajaste-Rudnitski, A. Understanding and tackling immune responses to adeno-associated viral vectors. Hum. Gene Ther. 2023, 34, 836–852. [Google Scholar] [CrossRef] [PubMed]
- Boutin, S.; Monteilhet, V.; Veron, P.; Leborgne, C.; Benveniste, O.; Montus, M.F.; Masurier, C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: Implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010, 21, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Liu, S.; Ou, L. rAAV immunogenicity, toxicity, and durability in 255 clinical trials: A meta-analysis. Front. Immunol. 2022, 13, 1001263. [Google Scholar] [CrossRef]
- Srivastava, A. Rationale and strategies for the development of safe and effective optimized AAV vectors for human gene therapy. Mol. Ther. -Nucleic Acids 2023, 32, 949–959. [Google Scholar] [CrossRef]
- Tickner, Z.J.; Farzan, M. Riboswitches for Controlled Expression of Therapeutic Transgenes Delivered by Adeno-Associated Viral Vectors. Pharmaceuticals 2021, 14, 554. [Google Scholar] [CrossRef]
- Guilbaud, M.; Devaux, M.; Couzinié, C.; Le Duff, J.; Toromanoff, A.; Vandamme, C.; Jaulin, N.; Gernoux, G.; Larcher, T.; Moullier, P.; et al. Five Years of Successful Inducible Transgene Expression Following Locoregional Adeno-Associated Virus Delivery in Nonhuman Primates with No Detectable Immunity. Hum. Gene Ther. 2019, 30, 802–813. [Google Scholar] [CrossRef]
- Słyk, Ż.; Stachowiak, N.; Małecki, M. Recombinant Adeno-Associated Virus Vectors for Gene Therapy of the Central Nervous System: Delivery Routes and Clinical Aspects. Biomedicines 2024, 12, 1523. [Google Scholar] [CrossRef]
- Coroadinha, A.S. Host Cell Restriction Factors Blocking Efficient Vector Transduction: Challenges in Lentiviral and Adeno-Associated Vector Based Gene Therapies. Cells 2023, 12, 732. [Google Scholar] [CrossRef]
- Mano, M.; Ippodrino, R.; Zentilin, L.; Zacchigna, S.; Giacca, M. Genome-wide RNAi screening identifies host restriction factors critical for in vivo AAV transduction. Proc. Natl. Acad. Sci. USA 2015, 112, 11276–11281. [Google Scholar] [CrossRef]
- Zajkowska, A.; Czajka, M.; Gulik, K.; Gawrychowski, K.; Małecki, M. Profiling of microRNA as a tool to introduce rAAV vectors in gene therapy of breast cancer: A preliminary report. Adv. Clin. Exp. Med. Off. Organ Wroc. Med. Univ. 2023, 32, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Bieńkowska, A.; Ducher, M.; Orzechowska, M.; Słyk, Ż.; Ciepiela, O.; Jaworowski, J.; Małecki, M. Increased temperature-related adeno-associated virus vectors transduction of ovarian cancer cells—Essential signatures of AAV receptor and heat shock proteins. Exp. Ther. Med. 2019, 18, 4718–4732. [Google Scholar] [CrossRef] [PubMed]
- Bieńkowska, A.; Kuźmicka, W.; Ciepiela, O.; Ochocki, J.; Małecki, M. Increased Temperature Facilitates Adeno-Associated Virus Vector Transduction of Colorectal Cancer Cell Lines in a Manner Dependent on Heat Shock Protein Signature. BioMed Res. Int. 2020, 1, 9107140. [Google Scholar] [CrossRef] [PubMed]
- Czajka, M.; Zajkowska, A.; Gawlak, M.; Bujalska-Zadrozny, M.; Malecki, M. Mosaic Recombinant Adeno-associated Virus Vector rAAV/DJ/CAG for Targeted Gene Delivery to Melanoma Cells Metastasized to the Lung. Anticancer. Res. 2020, 40, 4425–4444. [Google Scholar] [CrossRef]
- Nathwani, A.C.; Cochrane, M.; McIntosh, J.; Ng, C.Y.C.; Zhou, J.; Gray, J.T.; Davidoff, A.M. Enhancing transduction of the liver by adeno-associated viral vectors. Gene Ther. 2009, 16, 60–69. [Google Scholar] [CrossRef]
- Hölscher, C.; Sonntag, F.; Henrich, K.; Chen, Q.; Beneke, J.; Matula, P.; Rohr, K.; Kaderali, L.; Beil, N.; Erfle, H.; et al. The SUMOylation Pathway Restricts Gene Transduction by Adeno-Associated Viruses. PLoS Pathog. 2015, 11, e1005281. [Google Scholar] [CrossRef]
- Chai, Z.; Zhang, X.; Dobbins, A.L.; Samulski, R.J.; Merricks, E.P.; Nichols, T.C.; Li, C. Dexamethasone Transiently Enhances Transgene Expression in the Liver When Administered at Late-Phase Post Long-Term Adeno-Associated Virus Transduction. Hum. Gene Ther. 2022, 33, 119–130. [Google Scholar] [CrossRef]
- Business Research Insights. Paracetamol Market Size. 2024. Available online: https://www.businessresearchinsights.com/market-reports/paracetamol-market-102048 (accessed on 11 September 2024).
- Business Research Insights. Budesonide Market Size 2024. Available online: https://www.businessresearchinsights.com/market-reports/budesonide-market-102187 (accessed on 11 September 2024).
- Business Research Insights. Statin Market Size. 2024. Available online: https://www.businessresearchinsights.com/market-reports/statin-market-108449 (accessed on 11 September 2024).
- Freo, U.; Ruocco, C.; Valerio, A.; Scagnol, I.; Nisoli, E. Paracetamol: A Review of Guideline Recommendations. J. Clin. Med. 2021, 10, 3420. [Google Scholar] [CrossRef]
- Kalola, U.K.; Ambati, S. StatPearls; StatPearls Publishing: Treasure Island, Finland, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK563201/ (accessed on 11 September 2024).
- Issa, S.S.; Shaimardanova, A.A.; Solovyeva, V.V.; Rizvanov, A.A. Various AAV Serotypes and Their Applications in Gene Therapy: An Overview. Cells 2023, 12, 785. [Google Scholar] [CrossRef]
- Chowdhury, E.A.; Meno-Tetang, G.; Chang, H.Y.; Wu, S.; Huang, H.W.; Jamier, T.; Chandran, J.; Shah, D.K. Current progress and limitations of AAV mediated delivery of protein therapeutic genes and the importance of developing quantitative pharmacokinetic/pharmacodynamic (PK/PD) models. Adv. Drug Deliv. Rev. 2021, 170, 214–237. [Google Scholar] [CrossRef]
- Liu, S.; Chowdhury, E.A.; Xu, V.; Jerez, A.; Mahmood, L.; Ly, B.Q.; Le, H.K.; Nguyen, A.; Rajwade, A.; Meno-Tetang, G.; et al. Whole-Body Disposition and Physiologically Based Pharmacokinetic Modeling of Adeno-Associated Viruses and the Transgene Product. J. Pharm. Sci. 2024, 113, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhu, M.; Zhang, Y.; Diao, Y. Crossing the blood-brain barrier with AAV vectors. Metab. Brain Dis. 2021, 36, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, T.K.; Samulski, R.J. Addressing high dose AAV toxicity—‘one and done’ or ‘slower and lower’? Expert Opin. Biol. Ther. 2022, 22, 1067–1071. [Google Scholar] [CrossRef] [PubMed]
- Touahri, Y.; Dixit, R.; Kofoed, R.H.; Miloska, K.; Park, E.; Raeisossadati, R.; Markham-Coultes, K.; David, L.A.; Rijal, H.; Zhao, J.; et al. Focused ultrasound as a novel strategy for noninvasive gene delivery to retinal Müller glia. Theranostics 2020, 10, 2982–2999. [Google Scholar] [CrossRef]
- Corti, M.; Cleaver, B.; Clément, N.; Conlon, T.J.; Faris, K.J.; Wang, G.; Benson, J.; Tarantal, A.F.; Fuller, D.; Herzog, R.W.; et al. Evaluation of Readministration of a Recombinant Adeno-Associated Virus Vector Expressing Acid Alpha-Glucosidase in Pompe Disease: Preclinical to Clinical Planning. Hum. Gene Therapy. Clin. Dev. 2015, 26, 185–193. [Google Scholar] [CrossRef]
- Musa, A.; Ghoraie, L.S.; Zhang, S.D.; Glazko, G.; Yli-Harja, O.; Dehmer, M.; Haibe-Kains, B.; Emmert-Streib, F. A review of connectivity map and computational approaches in pharmacogenomics. Brief. Bioinform. 2018, 19, 506–523. [Google Scholar] [CrossRef]
- Nicolson, S.C.; Li, C.; Hirsch, M.L.; Setola, V.; Samulski, R.J. Identification and Validation of Small Molecules That Enhance Recombinant Adeno-associated Virus Transduction following High-Throughput Screens. J. Virol. 2016, 90, 7019–7031. [Google Scholar] [CrossRef]
- Grimm, D.; Lee Joyce, S.; Wang, L.; Desai, T.; Akache, B.; Storm Theresa, A.; Kay Mark, A. In Vitro and In Vivo Gene Therapy Vector Evolution via Multispecies Interbreeding and Retargeting of Adeno-Associated Viruses. J. Virol. 2008, 82, 5887–5911. [Google Scholar] [CrossRef]
- Katada, Y.; Kobayashi, K.; Tsubota, K.; Kurihara, T. Evaluation of AAV-DJ vector for retinal gene therapy. PeerJ 2019, 7, e6317. [Google Scholar] [CrossRef]
- Yoo, M.; Shin, J.; Kim, J.; Ryall, K.A.; Lee, K.; Lee, S.; Jeon, M.; Kang, J.; Tan, A.C. DSigDB: Drug signatures database for gene set analysis. Bioinformatics 2015, 31, 3069–3071. [Google Scholar] [CrossRef]
- Chen, Y.W.; Diamante, G.; Ding, J.; Nghiem, T.X.; Yang, J.; Ha, S.M.; Cohn, P.; Arneson, D.; Blencowe, M.; Garcia, J.; et al. PharmOmics: A species- and tissue-specific drug signature database and gene-network-based drug repositioning tool. iScience 2022, 25, 104052. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Freshour, S.L.; Kiwala, S.; Cotto, K.C.; Coffman, A.C.; McMichael, J.F.; Song, J.J.; Griffith, M.; Griffith, O.L.; Wagner, A.H. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021, 49, D1144–D1151. [Google Scholar] [CrossRef] [PubMed]
- Pillay, S.; Meyer, N.L.; Puschnik, A.S.; Davulcu, O.; Diep, J.; Ishikawa, Y.; Jae, L.T.; Wosen, J.E.; Nagamine, C.M.; Chapman, M.S.; et al. An essential receptor for adeno-associated virus infection. Nature 2016, 530, 108–112. [Google Scholar] [CrossRef]
- Zhong, L.; Zhao, W.; Wu, J.; Li, B.; Zolotukhin, S.; Govindasamy, L.; Agbandje-McKenna, M.; Srivastava, A. A dual role of EGFR protein tyrosine kinase signaling in ubiquitination of AAV2 capsids and viral second-strand DNA synthesis. Mol. Ther. J. Am. Soc. Gene Ther. 2007, 15, 1323–1330. [Google Scholar] [CrossRef]
- Aurnhammer, C.; Haase, M.; Muether, N.; Hausl, M.; Rauschhuber, C.; Huber, I.; Nitschko, H.; Busch, U.; Sing, A.; Ehrhardt, A.; et al. Universal real-time PCR for the detection and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum. Gene Ther. Methods 2012, 23, 18–28. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Słyk, Ż.; Stachowiak, N.; Małecki, M. Gene Therapy in the Light of Lifestyle Diseases: Budesonide, Acetaminophen and Simvastatin Modulates rAAV Transduction Efficiency. Pharmaceuticals 2024, 17, 1213. https://doi.org/10.3390/ph17091213
Słyk Ż, Stachowiak N, Małecki M. Gene Therapy in the Light of Lifestyle Diseases: Budesonide, Acetaminophen and Simvastatin Modulates rAAV Transduction Efficiency. Pharmaceuticals. 2024; 17(9):1213. https://doi.org/10.3390/ph17091213
Chicago/Turabian StyleSłyk, Żaneta, Natalia Stachowiak, and Maciej Małecki. 2024. "Gene Therapy in the Light of Lifestyle Diseases: Budesonide, Acetaminophen and Simvastatin Modulates rAAV Transduction Efficiency" Pharmaceuticals 17, no. 9: 1213. https://doi.org/10.3390/ph17091213
APA StyleSłyk, Ż., Stachowiak, N., & Małecki, M. (2024). Gene Therapy in the Light of Lifestyle Diseases: Budesonide, Acetaminophen and Simvastatin Modulates rAAV Transduction Efficiency. Pharmaceuticals, 17(9), 1213. https://doi.org/10.3390/ph17091213





