Safety Implications of Modulating Nuclear Receptors: A Comprehensive Analysis from Non-Clinical and Clinical Perspectives
Abstract
1. Introduction
2. Results
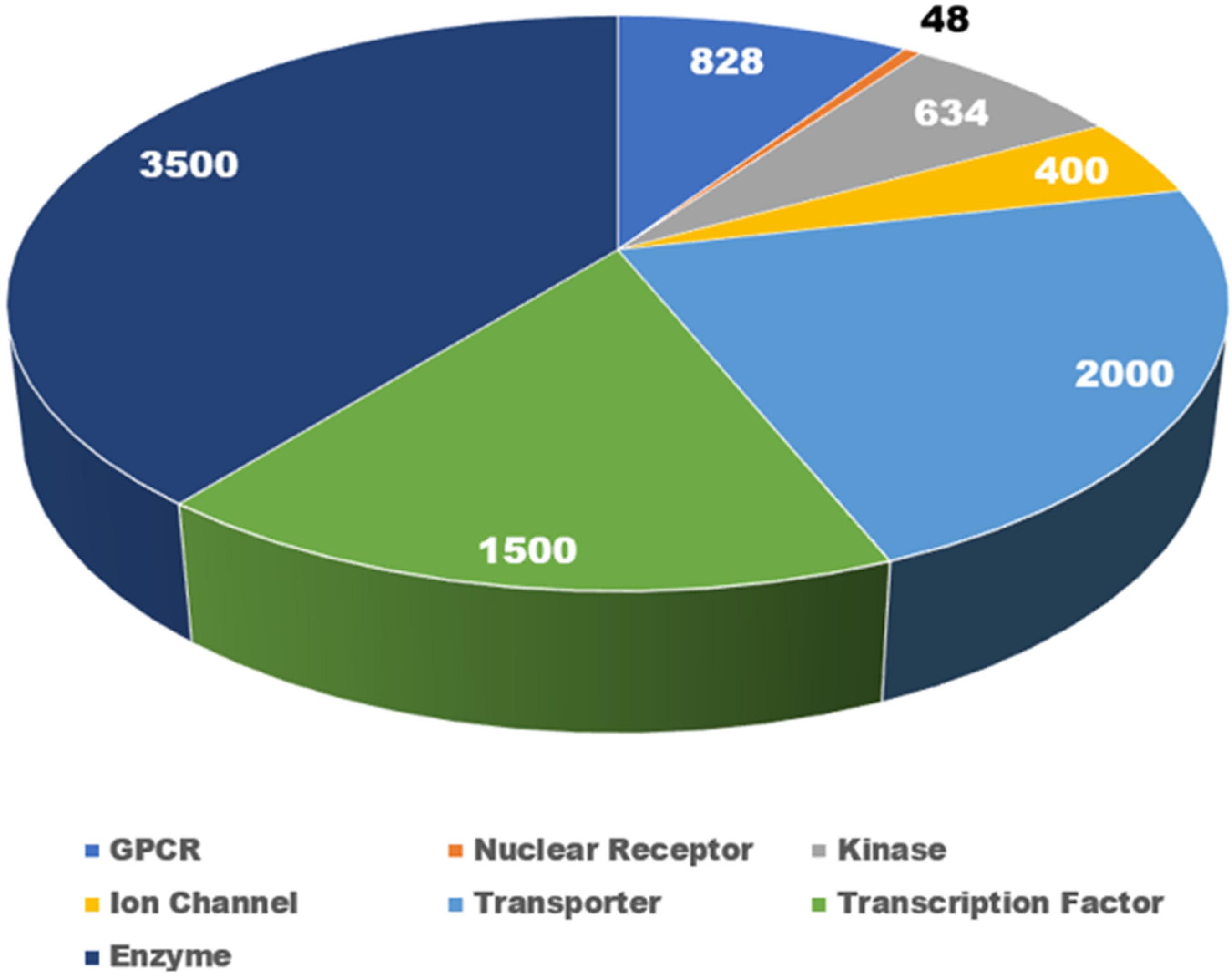
2.1. NR Families and Their Toxicological Implications
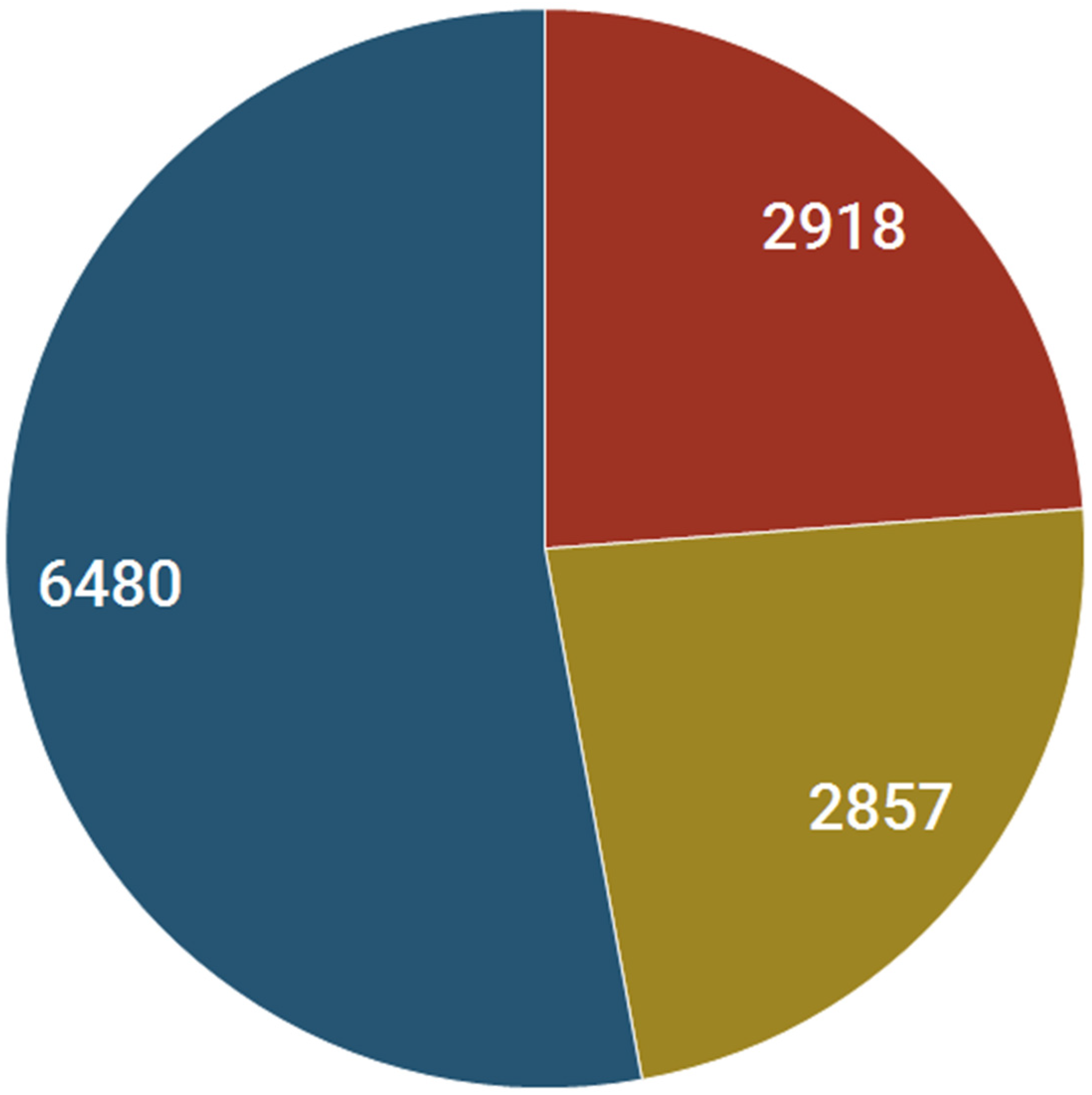
2.2. Small Molecules and Nuclear Receptor Interactions: Safety Alert Analysis
2.3. Detailed Analysis of Select 44 NRs
2.3.1. AR
2.3.2. GR
2.3.3. ER (α and β)
2.3.4. RAR (α, β, and γ)
2.3.5. PXR
2.3.6. PPAR (α, γ, and δ)
2.3.7. NR3C2 (Also Known as the Mineralocorticoid Receptor)
2.3.8. LXR (α and β)
2.3.9. NR5A1 (Steroidogenic Factor 1)
2.3.10. NR1D1 (Rev-Erb)
2.3.11. NR4A1 (Nerve Growth Factor IB)
2.3.12. VDR (Vitamin D Receptor)
2.3.13. THRA and THRB (Thyroid Hormone Receptors)
2.3.14. NR1I3 (Constitutive Androstane Receptor)
2.3.15. NR5A2 (Liver Receptor Homolog 1)
2.3.16. NR2F1 and NR2F2 (COUP-TF1 and COUP-TF2)
2.3.17. NR0B2 (SHP1)
2.3.18. RXR (α, β, and γ)
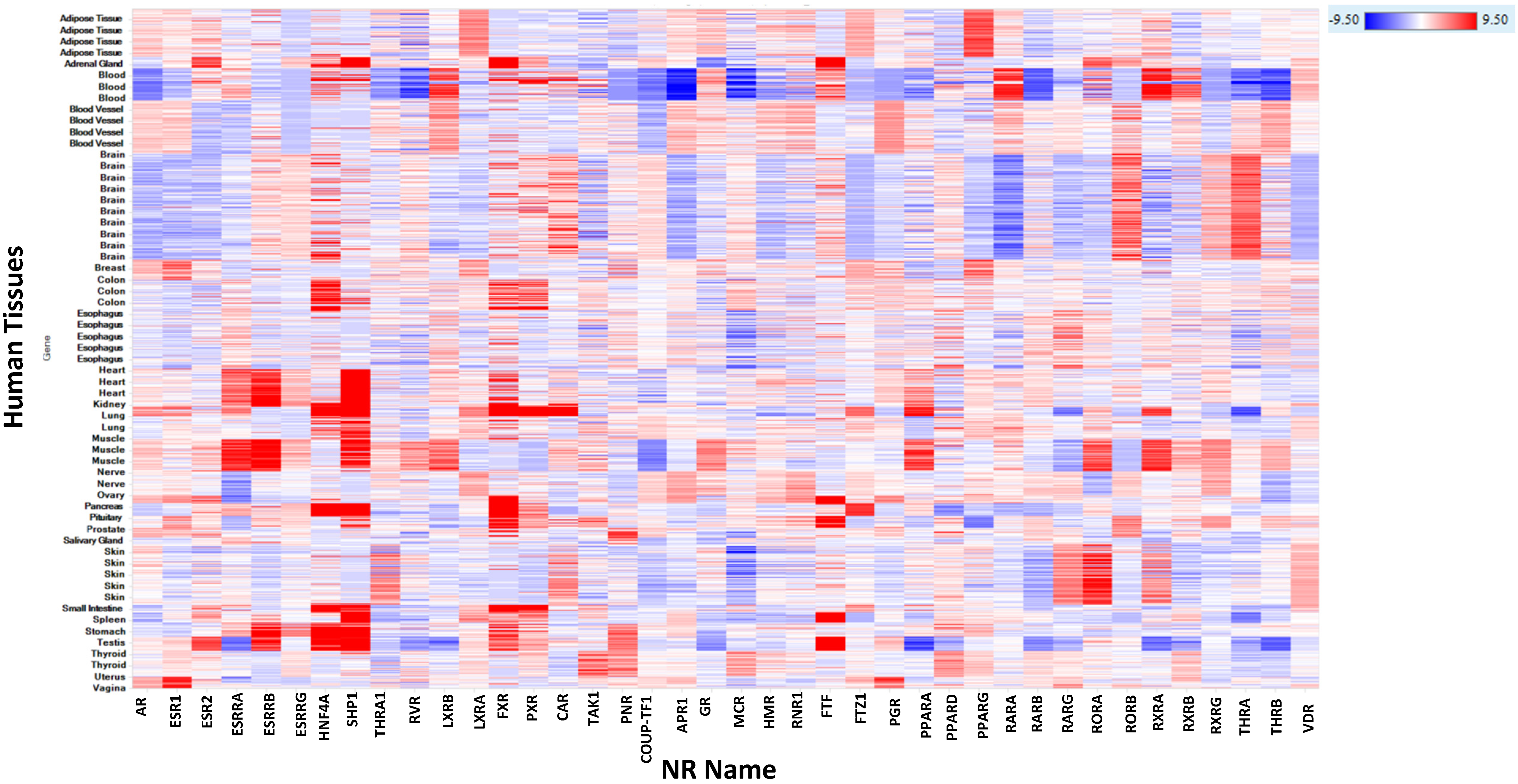
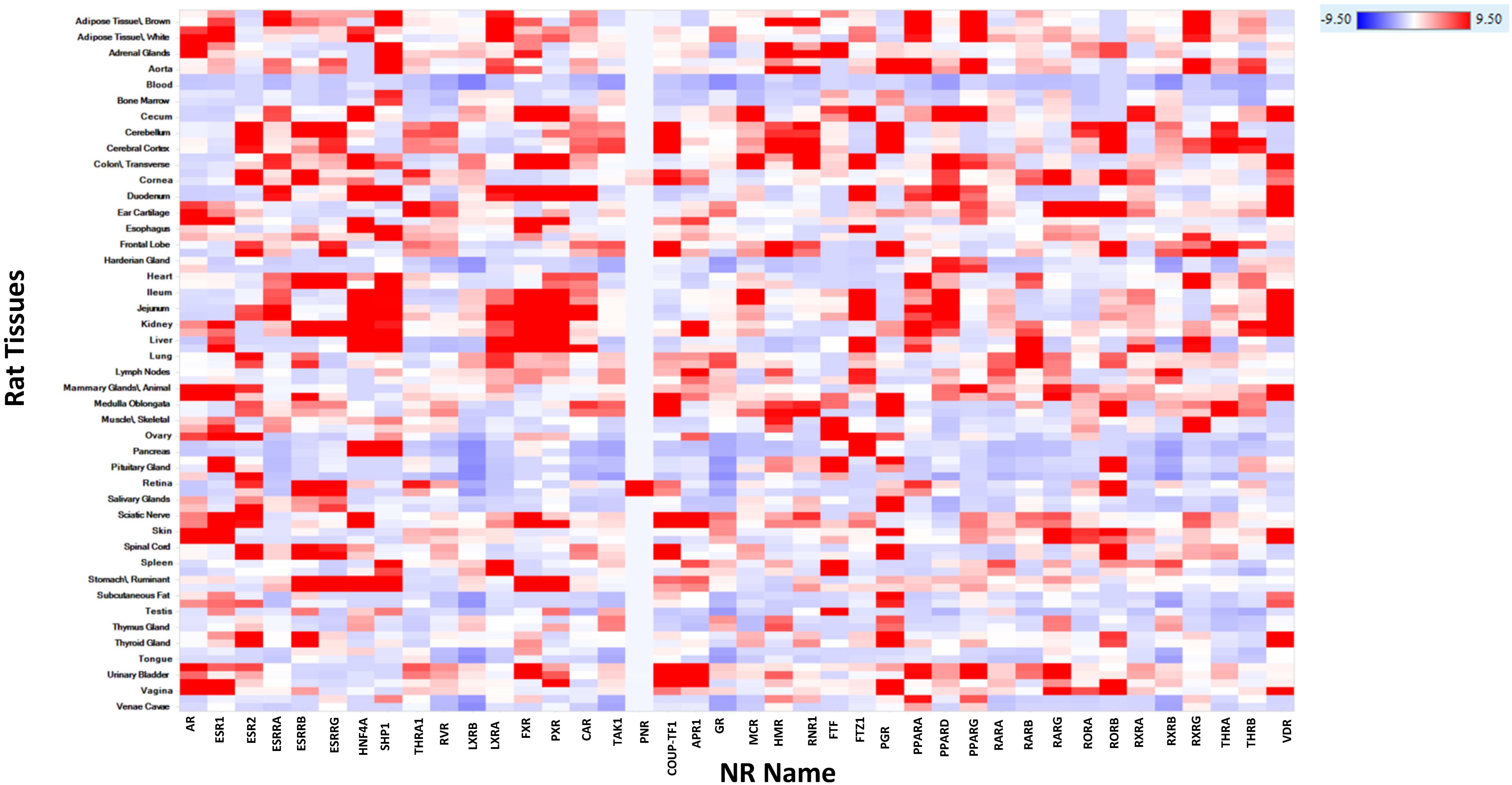
2.3.19. Human, Rat, Mouse, Dog, and Monkey mRNA Expression of NR
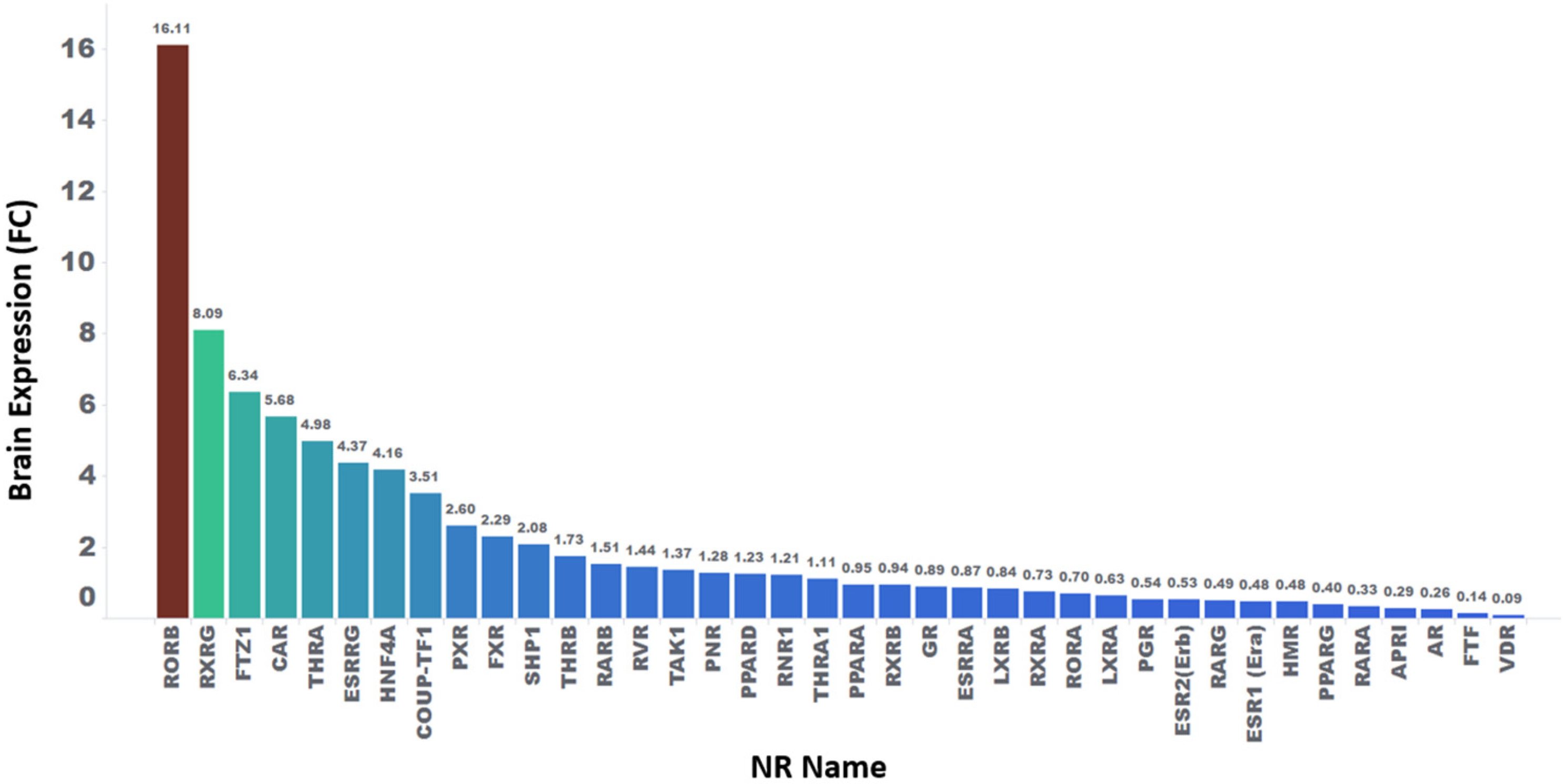
2.4. Statistical Analysis of Relative Fold Change Expression in Various Tissues for NR
3. Discussion
3.1. Improving Drug Safety through Early Target Panel Screening
3.2. NRs: Roles, Safety Risks, and Therapeutic Potential
3.3. Integrative Insights into Small Molecule Interactions with NRs
3.4. Comprehensive Analysis of Hepatic, CNS, and CV Toxicities
3.4.1. Hepatic Toxicity
3.4.2. CNS Toxicity
3.4.3. CV Toxicity
3.4.4. PPARa and g, AR, and GR
3.4.5. Limitations in NR Modulation Analysis
4. Materials and Methods
4.1. Source for NR Gene Names and Safety Information
4.2. Safety Alerts and Gene Expression Data for 44 NRs
4.3. NR Gene Enrichment Analysis across Tissues
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, D.; Gao, W.; Hu, H.; Zhou, S. Why 90% of Clinical Drug Development Fails and How to Improve It? Acta Pharm. Sin. B 2022, 12, 3049–3062. [Google Scholar] [CrossRef]
- Ponder, J.; Rajagopal, R.; Singal, M.; Baker, N.; Patlewicz, G.; Roggen, E.; Cochrane, S.; Sullivan, K. “In Litero” Screening: Retrospective Evaluation of Clinical Evidence to Establish a Reference List of Human Chemical Respiratory Sensitizers. Front. Toxicol. 2022, 4, 916370. [Google Scholar] [CrossRef]
- Li, A.P. A Comprehensive Approach for Drug Safety Assessment. Chem.-Biol. Interact. 2004, 150, 27–33. [Google Scholar] [CrossRef]
- Reese, M.; Sakatis, M.; Ambroso, J.; Harrell, A.; Yang, E.; Chen, L.; Taylor, M.; Baines, I.; Zhu, L.; Ayrton, A.; et al. An Integrated Reactive Metabolite Evaluation Approach to Assess and Reduce Safety Risk during Drug Discovery and Development. Chem.-Biol. Interact. 2011, 192, 60–64. [Google Scholar] [CrossRef]
- Berdigaliyev, N.; Aljofan, M. An Overview of Drug Discovery and Development. Futur. Med. Chem. 2020, 12, 939–947. [Google Scholar] [CrossRef]
- Vo, A.H.; Vleet, T.R.V.; Gupta, R.R.; Liguori, M.J.; Rao, M.S. An Overview of Machine Learning and Big Data for Drug Toxicity Evaluation. Chem. Res. Toxicol. 2020, 33, 20–37. [Google Scholar] [CrossRef]
- Rao, M.S.; Gupta, R.; Liguori, M.J.; Hu, M.; Huang, X.; Mantena, S.R.; Mittelstadt, S.W.; Blomme, E.A.G.; Vleet, T.R.V. Novel Computational Approach to Predict Off-Target Interactions for Small Molecules. Front. Big Data 2019, 2, 25. [Google Scholar] [CrossRef]
- Vleet, T.R.V.; Liguori, M.J.; Lynch, J.J.; Rao, M.; Warder, S. Screening Strategies and Methods for Better Off-Target Liability Prediction and Identification of Small-Molecule Pharmaceuticals. Slas Discov. Adv. Life Sci. R. D 2018, 24, 1–24. [Google Scholar] [CrossRef]
- Lynch, J.J.; Rossignol, E.; Moehrle, J.J.; Vleet, T.R.V.; Marsh, K.C.; Parman, T.; Mirsalis, J.; Ottinger, S.E.; Segreti, J.A.; Rao, M.; et al. Increased Stress Associated with Head-out Plethysmography Testing Can Exacerbate Respiratory Effects and Lead to Mortality in Rats. J. Pharmacol. Toxicol. 2019, 99, 106580. [Google Scholar] [CrossRef]
- Bass, A.S.; Cartwright, M.E.; Mahon, C.; Morrison, R.; Snyder, R.; McNamara, P.; Bradley, P.; Zhou, Y.-Y.; Hunter, J. Exploratory Drug Safety: A Discovery Strategy to Reduce Attrition in Development. J. Pharmacol. Toxicol. Methods 2009, 60, 69–78. [Google Scholar] [CrossRef]
- Lin, A.; Giuliano, C.J.; Palladino, A.; John, K.M.; Abramowicz, C.; Yuan, M.L.; Sausville, E.L.; Lukow, D.A.; Liu, L.; Chait, A.R.; et al. Off-Target Toxicity Is a Common Mechanism of Action of Cancer Drugs Undergoing Clinical Trials. Sci. Transl. Med. 2019, 11, eaaw8412. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, G.; Yan, Z.; Tang, W.; Dasgupta, M.; Hasting, B. ADME Optimization and Toxicity Assessment in Early- and Late-Phase Drug Discovery. Curr. Top. Med. Chem. 2009, 9, 965–980. [Google Scholar] [CrossRef] [PubMed]
- Eddershaw, P.J.; Beresford, A.P.; Bayliss, M.K. ADME/PK as Part of a Rational Approach to Drug Discovery. Drug Discov. Today 2000, 5, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Herman, J.L. Generic Approach to High Throughput ADME Screening for Lead Candidate Optimization. Int. J. Mass. Spectrom. 2004, 238, 107–117. [Google Scholar] [CrossRef]
- Smith, D.A.; Obach, R.S. Metabolites and Safety: What Are the Concerns, and How Should We Address Them? Chem. Res. Toxicol. 2006, 19, 1570–1579. [Google Scholar] [CrossRef] [PubMed]
- Grillo, M.P. Detecting Reactive Drug Metabolites for Reducing the Potential for Drug Toxicity. Expert. Opin. Drug Metab. Toxicol. 2015, 11, 1281–1302. [Google Scholar] [CrossRef]
- Cavero, I.; Guillon, J.-M. Safety Pharmacology Assessment of Drugs with Biased 5-HT2B Receptor Agonism Mediating Cardiac Valvulopathy. J. Pharmacol. Toxicol. Methods 2014, 69, 150–161. [Google Scholar] [CrossRef]
- Ralston, S. Pre-Development Attrition of Pharmaceuticals: How to Identify the Bad Actors Early. Toxico. Sci. 2017, 150, 2323. [Google Scholar]
- Leads, B. New Clinical Development Success Rates and Contributing Factors 2011–2020; BIO: Washington, DC, USA, 2021. [Google Scholar]
- Smith, D.A. Postmarketing Attrition. In Attrition in the Pharmaceutical Industry: Reasons, Implications, and Pathways Forward; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 128–157. [Google Scholar] [CrossRef]
- Blomme, E.A.G.; Will, Y. Toxicology Strategies for Drug Discovery: Present and Future. Chem. Res. Toxicol. 2016, 29, 473–504. [Google Scholar] [CrossRef]
- Leeson, P.D. Molecular Inflation, Attrition and the Rule of Five. Adv. Drug Deliv. Rev. 2016, 101, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Price, D.A.; Blagg, J.; Jones, L.; Greene, N.; Wager, T. Physicochemical Drug Properties Associated with in Vivo Toxicological Outcomes: A Review. Expert. Opin. Drug Metab. Toxicol. 2009, 5, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Bowes, J.; Brown, A.J.; Hamon, J.; Jarolimek, W.; Sridhar, A.; Waldron, G.; Whitebread, S. Reducing Safety-Related Drug Attrition: The Use of in Vitro Pharmacological Profiling. Nat. Rev. Drug Discov. 2012, 11, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Whitebread, S.; Dumotier, B.; Armstrong, D.; Fekete, A.; Chen, S.; Hartmann, A.; Muller, P.Y.; Urban, L. Secondary Pharmacology: Screening and Interpretation of off-Target Activities—Focus on Translation. Drug Discov. Today 2016, 21, 1232–1242. [Google Scholar] [CrossRef] [PubMed]
- Alexander, S.P.H.; Cidlowski, J.A.; Kelly, E.; Mathie, A.; Peters5, J.A.; Veale4, E.L.; Armstrong6, J.F.; Faccenda6, E.; Harding6, S.D.; Pawson6, A.J.; et al. The Concise Guide to Pharmacology 2021/22: Nuclear Hormone Receptors. Br. J. Pharmacol. 2021, 178, S246–S263. [Google Scholar] [CrossRef]
- Rao, M.; McDuffie, E.; Sachs, C. Artificial Intelligence/Machine Learning-Driven Small Molecule Repurposing via Off-Target Prediction and Transcriptomics. Toxics 2023, 11, 875. [Google Scholar] [CrossRef] [PubMed]
- Woods, C.G.; Heuvel, J.P.V.; Rusyn, I. Genomic Profiling in Nuclear Receptor-Mediated Toxicity. Toxicol. Pathol. 2007, 35, 474–494. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.J.; Vleet, T.R.V.; Mittelstadt, S.W.; Blomme, E.A.G. Potential Functional and Pathological Side Effects Related to Off-Target Pharmacological Activity. J. Pharmacol. Toxicol. Methods 2017, 87, 108–126. [Google Scholar] [CrossRef] [PubMed]
- Bendels, S.; Bissantz, C.; Fasching, B.; Gerebtzoff, G.; Guba, W.; Kansy, M.; Migeon, J.; Mohr, S.; Peters, J.-U.; Tillier, F.; et al. Safety Screening in Early Drug Discovery: An Optimized Assay Panel. J. Pharmacol. Toxicol. Methods 2019, 99, 106609. [Google Scholar] [CrossRef]
- Robinson-Rechavi, M.; Carpentier, A.-S.; Duffraisse, M.; Laudet, V. How Many Nuclear Hormone Receptors Are There in the Human Genome? Trends Genet. 2001, 17, 554–556. [Google Scholar] [CrossRef]
- Carithers, L.J.; Moore, H.M. The Genotype-Tissue Expression (GTEx) Project. Biopreserv. Biobank. 2015, 13, 307–308. [Google Scholar] [CrossRef] [PubMed]
- Amaral, P.; Carbonell-Sala, S.; Vega, F.M.D.L.; Faial, T.; Frankish, A.; Gingeras, T.; Guigo, R.; Harrow, J.L.; Hatzigeorgiou, A.G.; Johnson, R.; et al. The Status of the Human Gene Catalogue. arXiv 2023, arXiv:2303.13996. [Google Scholar] [CrossRef] [PubMed]
- Frigo, D.E.; Bondesson, M.; Williams, C. Nuclear Receptors: From Molecular Mechanisms to Therapeutics. Essays Biochem. 2021, 65, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Grünewald, S. Sequence, Structure and Ligand Binding Evolution of Rhodopsin-Like G Protein-Coupled Receptors: A Crystal Structure-Based Phylogenetic Analysis. PLoS ONE 2015, 10, e0123533. [Google Scholar] [CrossRef]
- Essegian, D.; Khurana, R.; Stathias, V.; Schürer, S.C. The Clinical Kinase Index: A Method to Prioritize Understudied Kinases as Drug Targets for the Treatment of Cancer. Cell Rep. Med. 2020, 1, 100128. [Google Scholar] [CrossRef]
- Clare, J.J. Targeting Voltage-Gated Sodium Channels for Pain Therapy. Expert Opin. Investig. Drugs 2010, 19, 45–62. [Google Scholar] [CrossRef]
- Sahoo, S.; Aurich, M.K.; Jonsson, J.J.; Thiele, I. Membrane Transporters in a Human Genome-Scale Metabolic Knowledgebase and Their Implications for Disease. Front. Physiol. 2014, 5, 91. [Google Scholar] [CrossRef]
- Wingender, E.; Schoeps, T.; Haubrock, M.; Dönitz, J. TFClass: A Classification of Human Transcription Factors and Their Rodent Orthologs. Nucleic Acids Res. 2015, 43, D97–D102. [Google Scholar] [CrossRef]
- Babbi, G.; Baldazzi, D.; Savojardo, C.; Luigi, M.P.; Casadio, R. Highlighting Human Enzymes Active in Different Metabolic Pathways and Diseases: The Case Study of EC 1.2.3.1 and EC 2.3.1.9. Biomedicines 2020, 8, 250. [Google Scholar] [CrossRef]
- Inagaki, T.; Choi, M.; Moschetta, A.; Peng, L.; Cummins, C.L.; McDonald, J.G.; Luo, G.; Jones, S.A.; Goodwin, B.; Richardson, J.A.; et al. Fibroblast Growth Factor 15 Functions as an Enterohepatic Signal to Regulate Bile Acid Homeostasis. Cell Metab. 2005, 2, 217–225. [Google Scholar] [CrossRef]
- Pognan, F.; Beilmann, M.; Boonen, H.C.M.; Czich, A.; Dear, G.; Hewitt, P.; Mow, T.; Oinonen, T.; Roth, A.; Steger-Hartmann, T.; et al. The Evolving Role of Investigative Toxicology in the Pharmaceutical Industry. Nat. Rev. Drug Discov. 2023, 22, 317–335. [Google Scholar] [CrossRef] [PubMed]
- Buckley, L.A.; Bebenek, I.; Cornwell, P.D.; Hodowanec, A.; Jensen, E.C.; Murphy, C.; Ghantous, H.N. Drug Development 101: A Primer. Int. J. Toxicol. 2020, 39, 379–396. [Google Scholar] [CrossRef] [PubMed]
- Namdari, R.; Jones, K.; Chuang, S.S.; Cruchten, S.V.; Dincer, Z.; Downes, N.; Mikkelsen, L.F.; Harding, J.; Jäckel, S.; Jacobsen, B.; et al. Species Selection for Nonclinical Safety Assessment of Drug Candidates: Examples of Current Industry Practice. Regul. Toxicol. Pharmacol. 2021, 126, 105029. [Google Scholar] [CrossRef]
- Clark, M.; Steger-Hartmann, T. A Big Data Approach to the Concordance of the Toxicity of Pharmaceuticals in Animals and Humans. Regul. Toxicol. Pharmacol. 2018, 96, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Olson, H.; Betton, G.; Robinson, D.; Thomas, K.; Monro, A.; Kolaja, G.; Lilly, P.; Sanders, J.; Sipes, G.; Bracken, W.; et al. Concordance of the Toxicity of Pharmaceuticals in Humans and in Animals. Regul. Toxicol. Pharmacol. 2000, 32, 56–67. [Google Scholar] [CrossRef]
- Shah, R.R. Can Pharmacogenetics Help Rescue Drugs Withdrawn from the Market? Pharmacogenomics 2006, 7, 889–908. [Google Scholar] [CrossRef]
- Guengerich, F.P. Mechanisms of Drug Toxicity and Relevance to Pharmaceutical Development. Drug Metab. Pharmacok. 2011, 26, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Wensing, G.; Ighrayeb, I.-A.; Boix, O.; Böttcher, M. The Safety of Healthy Volunteers in First-in-Man Trials—An Analysis of Studies Conducted at the Bayer in-House Ward from 2000 to 2005. Int J. Clin. Pharmacol. Ther. 2010, 48, 563–570. [Google Scholar] [CrossRef]
- Valentin, J.-P.; Hammond, T. Safety and Secondary Pharmacology: Successes, Threats, Challenges and Opportunities. J. Pharmacol. Toxicol. Methods 2008, 58, 77–87. [Google Scholar] [CrossRef]
- Weaver, R.J.; Valentin, J.-P. Today’s Challenges to De-Risk and Predict Drug Safety in Human “Mind-the-Gap”. Toxicol. Sci. 2018, 167, 307–321. [Google Scholar] [CrossRef]
- Waring, M.J.; Arrowsmith, J.; Leach, A.R.; Leeson, P.D.; Mandrell, S.; Owen, R.M.; Pairaudeau, G.; Pennie, W.D.; Pickett, S.D.; Wang, J.; et al. An Analysis of the Attrition of Drug Candidates from Four Major Pharmaceutical Companies. Nat. Rev. Drug Discov. 2015, 14, 475–486. [Google Scholar] [CrossRef]
- Sutherland, J.J.; Yonchev, D.; Fekete, A.; Urban, L. A Preclinical Secondary Pharmacology Resource Illuminates Target-Adverse Drug Reaction Associations of Marketed Drugs. Nat. Commun. 2023, 14, 4323. [Google Scholar] [CrossRef]
- Hamon, J.; Whitebread, S.; Techer-Etienne, V.; Coq, H.L.; Azzaoui, K.; Urban, L. In Vitro Safety Pharmacology Profiling: What Else beyond HERG? Futur. Med. Chem. 2009, 1, 645–665. [Google Scholar] [CrossRef] [PubMed]
- Papoian, T.; Chiu, H.-J.; Elayan, I.; Jagadeesh, G.; Khan, I.; Laniyonu, A.A.; Li, C.X.; Saulnier, M.; Simpson, N.; Yang, B. Secondary Pharmacology Data to Assess Potential Off-Target Activity of New Drugs: A Regulatory Perspective. Nat. Rev. Drug Discov. 2015, 14, 294. [Google Scholar] [CrossRef]
- Urban, L.; Whitebread, S.; Hamon, J.; Mikhailov, D.; Azzaoui, K. Polypharmacology in Drug Discovery; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; pp. 15–46. [Google Scholar] [CrossRef]
- Brennan, R.J.; Jenkinson, S.; Brown, A.; Delaunois, A.; Dumotier, B.; Pannirselvam, M.; Rao, M.; Ribeiro, L.R.; Schmidt, F.; Sibony, A.; et al. The State of the Art in Secondary Pharmacology and Its Impact on the Safety of New Medicines. Nat. Rev. Drug Discov. 2024, 1–21. [Google Scholar] [CrossRef]
- Olefsky, J.M. Nuclear Receptor Minireview Series. J. Biol. Chem. 2001, 276, 36863–36864. [Google Scholar] [CrossRef]
- Sar, P. Nuclear Receptor: Structure and Function. Prog. Mol. Biol. Transl. Sci. 2022, 196, 209–227. [Google Scholar] [CrossRef] [PubMed]
- Bosscher, K.D.; Desmet, S.J.; Clarisse, D.; Estébanez-Perpiña, E.; Brunsveld, L. Nuclear Receptor Crosstalk—Defining the Mechanisms for Therapeutic Innovation. Nat. Rev. Endocrinol. 2020, 16, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Urquhart, B.L.; Tirona, R.G.; Kim, R.B. Nuclear Receptors and the Regulation of Drug-Metabolizing Enzymes and Drug Transporters: Implications for Interindividual Variability in Response to Drugs. J. Clin. Pharmacol. 2007, 47, 566–578. [Google Scholar] [CrossRef]
- Weatherman, R.V.; Fletterick, R.J.; Scanlan, T.S. Nuclear-Receptor Ligands and Ligand-Binding Domains. Annu. Rev. Biochem. 1999, 68, 559–581. [Google Scholar] [CrossRef]
- Grün, F.; Blumberg, B. Perturbed Nuclear Receptor Signaling by Environmental Obesogens as Emerging Factors in the Obesity Crisis. Rev. Endocr. Metab. Disord. 2007, 8, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Achermann, J.C.; Schwabe, J.; Fairall, L.; Chatterjee, K. Genetic Disorders of Nuclear Receptors. J. Clin. Investig. 2017, 127, 1181–1192. [Google Scholar] [CrossRef] [PubMed]
- Xu, P. Nuclear Receptors in Health and Diseases. Int. J. Mol. Sci. 2023, 24, 9153. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.T.; Collins, J.L.; Pearce, K.H. The Nuclear Receptor Superfamily and Drug Discovery. ChemMedChem 2006, 1, 504–523. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; O’Malley, B.W. Front Matter. In Nuclear Receptor Coregulators and Human Diseases; World Scientific: Singapore, 2008; pp. i–xi. [Google Scholar] [CrossRef]
- Germain, P.; Staels, B.; Dacquet, C.; Spedding, M.; Laudet, V. Overview of Nomenclature of Nuclear Receptors. Pharmacol. Rev. 2006, 58, 685–704. [Google Scholar] [CrossRef]
- Novac, N.; Heinzel, T. Nuclear Receptors: Overview and Classification. Curr. Drug Target.-Inflamm. Allergy 2004, 3, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Sever, R.; Glass, C.K. Signaling by Nuclear Receptors. Cold Spring Harb. Perspect. Biol. 2013, 5, a016709. [Google Scholar] [CrossRef]
- Lehrer, S.; Rheinstein, P.H. RORB, an Alzheimer’s Disease Susceptibility Gene, Is Associated with Viral Encephalitis, an Alzheimer’s Disease Risk Factor. Clin. Neurol. Neurosurg. 2023, 233, 107984. [Google Scholar] [CrossRef]
- Janani, C.; Kumari, B.D.R. PPAR Gamma Gene—A Review. Diabetes Metab. Syndr. Clin. Res. Rev. 2015, 9, 46–50. [Google Scholar] [CrossRef]
- Spiegelman, B.M. PPAR-Gamma: Adipogenic Regulator and Thiazolidinedione Receptor. Diabetes 1998, 47, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Chinetti, G.; Lestavel, S.; Bocher, V.; Remaley, A.T.; Neve, B.; Torra, I.P.; Teissier, E.; Minnich, A.; Jaye, M.; Duverger, N.; et al. PPAR-α and PPAR-γ Activators Induce Cholesterol Removal from Human Macrophage Foam Cells through Stimulation of the ABCA1 Pathway. Nat. Med. 2001, 7, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Green, S. PPAR: A Mediator of Peroxisome Proliferator Action. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1995, 333, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Gilde, A.J.; Bilsen, M.V. Peroxisome Proliferator-activated Receptors (PPARS): Regulators of Gene Expression in Heart and Skeletal Muscle. Acta Physiol. Scand. 2003, 178, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Montaigne, D.; Butruille, L.; Staels, B. PPAR Control of Metabolism and Cardiovascular Functions. Nat. Rev. Cardiol. 2021, 18, 809–823. [Google Scholar] [CrossRef] [PubMed]
- Barger, P.M.; Kelly, D.P. PPAR Signaling in the Control of Cardiac Energy Metabolism. Trends Cardiovasc. Med. 2000, 10, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Liss, K.H.H.; Finck, B.N. PPARs and Nonalcoholic Fatty Liver Disease. Biochimie 2017, 136, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.S.; Reddy, J.K. PPARα in the Pathogenesis of Fatty Liver Disease. Hepatology 2004, 40, 783–786. [Google Scholar] [CrossRef]
- Rao, M.; Nassiri, V.; Alhambra, C.; Snoeys, J.; Goethem, F.V.; Irrechukwu, O.; Aleo, M.D.; Geys, H.; Mitra, K.; Will, Y. AI/ML Models to Predict the Severity of Drug-Induced Liver Injury for Small Molecules. Chem. Res. Toxicol. 2023, 36, 1129–1139. [Google Scholar] [CrossRef]
- Pettinelli, P.; Videla, L.A. Up-Regulation of PPAR-γ MRNA Expression in the Liver of Obese Patients: An Additional Reinforcing Lipogenic Mechanism to SREBP-1c Induction. Mol. Endocrinol. 2011, 25, 547. [Google Scholar] [CrossRef][Green Version]
- Thompson, E.A. PPARγ Physiology and Pathology in Gastrointestinal Epithelial Cells. Mol. Cells 2007, 24, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Bassaganya-Riera, J.; Reynolds, K.; Martino-Catt, S.; Cui, Y.; Hennighausen, L.; Gonzalez, F.; Rohrer, J.; Benninghoff, A.U.; Hontecillas, R. Activation of PPAR γ and δ by Conjugated Linoleic Acid Mediates Protection from Experimental Inflammatory Bowel Disease. Gastroenterology 2004, 127, 777–791. [Google Scholar] [CrossRef] [PubMed]
- Froment, P. PPARs and RXRs in Male and Female Fertility and Reproduction. PPAR Res. 2008, 2008, 637490. [Google Scholar] [CrossRef] [PubMed]
- Bogacka, I.; Kurzynska, A.; Bogacki, M.; Chojnowska, K. Peroxisome Proliferator-Activated Receptors in the Regulation of Female Reproductive Functions. Folia Histochem. Cytobiol. 2015, 53, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Froment, P.; Gizard, F.; Defever, D.; Staels, B.; Dupont, J.; Monget, P. Peroxisome Proliferator-Activated Receptors in Reproductive Tissues: From Gametogenesis to Parturition. J. Endocrinol. 2006, 189, 199–209. [Google Scholar] [CrossRef]
- Cariou, B.; Charbonnel, B.; Staels, B. Thiazolidinediones and PPARγ Agonists: Time for a Reassessment. Trends Endocrinol. Metab. 2012, 23, 205–215. [Google Scholar] [CrossRef]
- Won, J.C. Thiazolidinediones (TZDs). In Stroke Revisited: Diabetes in Stroke; Stroke Revisited; Springer: Singapore, 2021; pp. 131–141. [Google Scholar] [CrossRef]
- Stumvoll, M.; Häring, H.-U. Glitazones: Clinical Effects and Molecular Mechanisms. Ann. Med. 2002, 34, 217–224. [Google Scholar] [CrossRef]
- Gale, E.A. Lessons from the Glitazones: A Story of Drug Development. Lancet 2001, 357, 1870–1875. [Google Scholar] [CrossRef]
- Mudaliar, S.; Henry, R.R. New Oral Therapies for Type 2 Diabetes Mellitus: The Glitazones or Insulin Sensitizers1. Annu. Rev. Med. 2001, 52, 239–257. [Google Scholar] [CrossRef]
- Todd, P.A.; Ward, A. Gemfibrozil. Drugs 1988, 36, 314–339. [Google Scholar] [CrossRef]
- Gemfibrozil-Induced Myopathy. Inpharma Wkly. 1991, 810, 17. [CrossRef]
- Hahn, M.; Sriharan, K.; McFarland, M.S. Gemfibrozil-Induced Myositis in a Patient with Normal Renal Function. Ann. Pharmacother. 2010, 44, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Oda, S.; Tsuneyama, K.; Urano, Y.; Yokoi, T. Establishment of a Mouse Model of Troglitazone-induced Liver Injury and Analysis of Its Hepatotoxic Mechanism. J. Appl. Toxicol. 2019, 39, 1541–1556. [Google Scholar] [CrossRef] [PubMed]
- Shah, F.; Leung, L.; Barton, H.A.; Will, Y.; Rodrigues, A.D.; Greene, N.; Aleo, M.D. Setting Clinical Exposure Levels of Concern for Drug-Induced Liver Injury (DILI) Using Mechanistic in Vitro Assays. Toxicol. Sci. 2015, 147, 500–514. [Google Scholar] [CrossRef] [PubMed]
- Venkatachalam, G.; Kumar, A.P.; Sakharkar, K.R.; Thangavel, S.; Clement, M.-V.; Sakharkar, M.K. PPARγ Disease Gene Network and Identification of Therapeutic Targets for Prostate Cancer. J. Drug Target. 2011, 19, 781–796. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, A.U.; Bagla, S.; Ravindranath, Y. Identification of a Novel Variant in Phosphoglycerate Kinase-1 (PGK1) in an African-American Child (PGK1 Detroit). Pediatr. Hematol. Oncol. 2019, 36, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.-S.; Huang, P.-S.; Chang, S.-N.; Wu, C.-K.; Hwang, J.-J.; Chuang, E.Y.; Tsai, C.-T. Genome-Wide Copy Number Variation Association Study of Atrial Fibrillation Related Thromboembolic Stroke. J. Clin. Med. 2019, 8, 332. [Google Scholar] [CrossRef]
- Krause, C.; Suwada, K.; Blomme, E.A.G.; Kowalkowski, K.; Liguori, M.J.; Mahalingaiah, P.K.; Mittelstadt, S.; Peterson, R.; Rendino, L.; Vo, A.; et al. Preclinical Species Gene Expression Database: Development and Meta-Analysis. Front. Genet. 2023, 13, 1078050. [Google Scholar] [CrossRef]


| Nuclear Receptor Target | Number of FDA-Approved Drugs |
|---|---|
| GR | 51 |
| AR | 25 |
| PGR | 20 |
| ER | 19 |
| PPARa | 12 |
| VDR | 12 |
| RAR | 9 |
| MCR | 8 |
| PPARg | 8 |
| FXR | 5 |
| THRA | 2 |
| PPARd | 1 |
| SHP | 1 |
| Target—Action | Novartis | AstraZeneca | FDA | AbbVie | Roche | Target Consensus |
|---|---|---|---|---|---|---|
| AR—Agonists | 0 | 1 | 1 | 1 | 1 | 4 |
| AR—Antagonists | 0 | 1 | 1 | 1 | 1 | 4 |
| GR—Agonists | 0 | 1 | 0 | 1 | 1 | 3 |
| PPARg—Agonists | 0 | 0 | 1 | 1 | 1 | 3 |
| ER—Agonists | 0 | 0 | 1 | 0 | 1 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rao, M.; McDuffie, E.; Srivastava, S.; Plaisted, W.; Sachs, C. Safety Implications of Modulating Nuclear Receptors: A Comprehensive Analysis from Non-Clinical and Clinical Perspectives. Pharmaceuticals 2024, 17, 875. https://doi.org/10.3390/ph17070875
Rao M, McDuffie E, Srivastava S, Plaisted W, Sachs C. Safety Implications of Modulating Nuclear Receptors: A Comprehensive Analysis from Non-Clinical and Clinical Perspectives. Pharmaceuticals. 2024; 17(7):875. https://doi.org/10.3390/ph17070875
Chicago/Turabian StyleRao, Mohan, Eric McDuffie, Sanjay Srivastava, Warren Plaisted, and Clifford Sachs. 2024. "Safety Implications of Modulating Nuclear Receptors: A Comprehensive Analysis from Non-Clinical and Clinical Perspectives" Pharmaceuticals 17, no. 7: 875. https://doi.org/10.3390/ph17070875
APA StyleRao, M., McDuffie, E., Srivastava, S., Plaisted, W., & Sachs, C. (2024). Safety Implications of Modulating Nuclear Receptors: A Comprehensive Analysis from Non-Clinical and Clinical Perspectives. Pharmaceuticals, 17(7), 875. https://doi.org/10.3390/ph17070875






