Trimethyltin(IV) Bearing 3-(4-Methyl-2-oxoquinolin-1(2H)-yl)propanoate Causes Lipid Peroxidation-Mediated Autophagic Cell Death in Human Melanoma A375 Cells
Abstract
1. Introduction
2. Results and Discussion
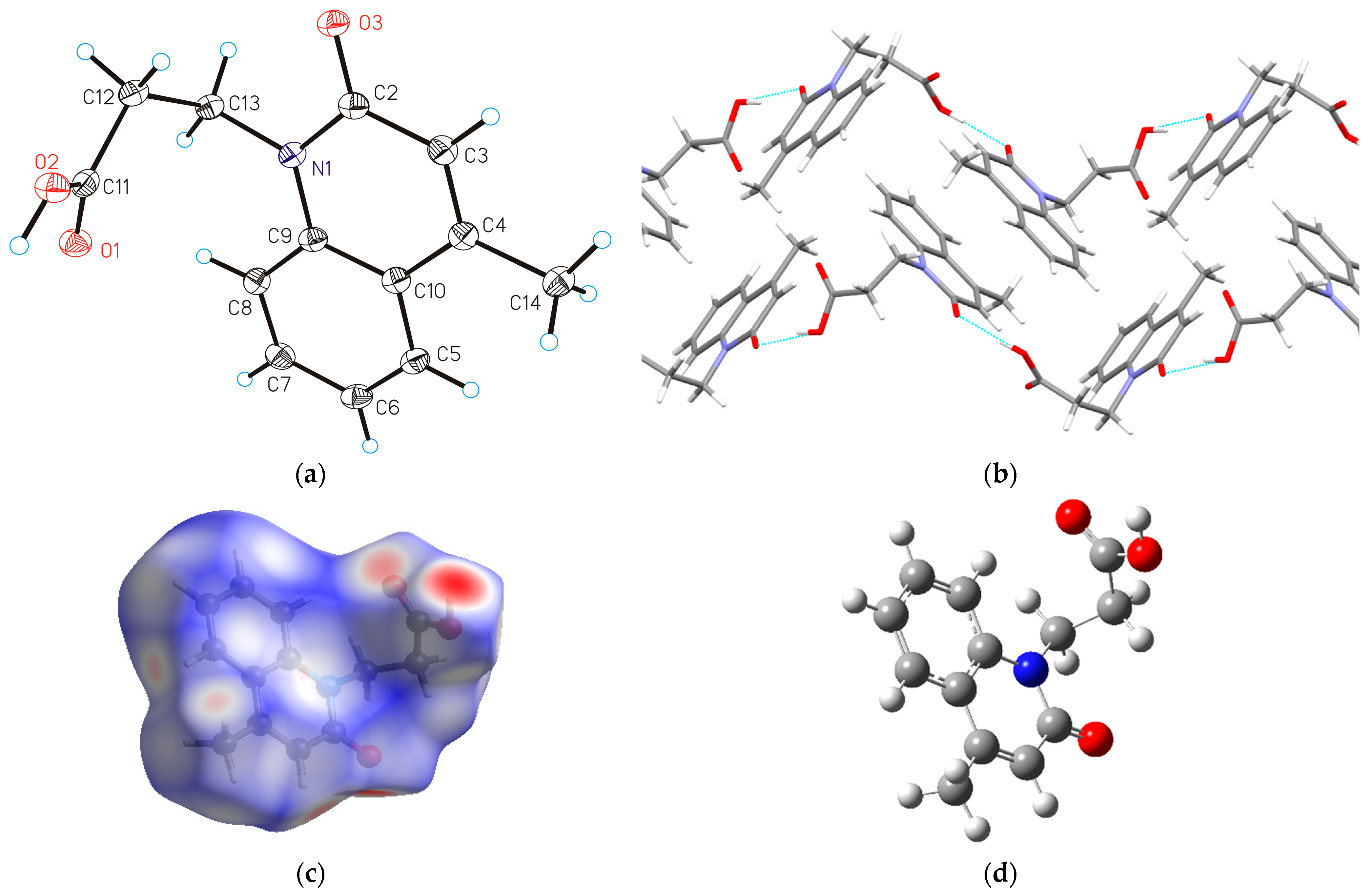
2.1. SC-XRD Analysis, Structure Optimization and Hirshfeld Surface Analysis of HL
2.1.1. Molecular Structure of HL
2.1.2. Geometry Optimization of HL
2.1.3. Hirshfeld Surface Analysis
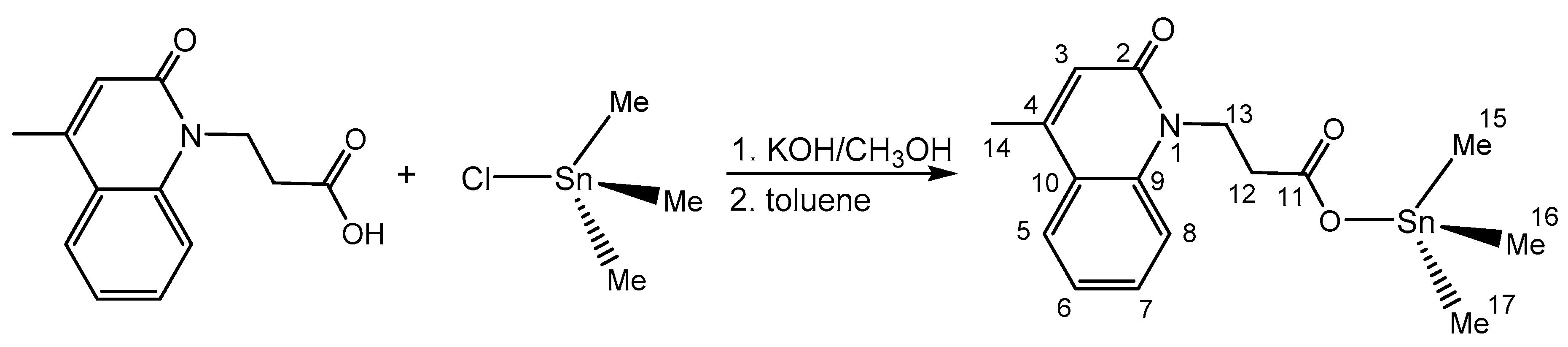
2.2. Me3SnL: Synthesis, Experimental and In Silico Characterization
2.2.1. Chemistry and DFT
2.2.2. UV/Vis Spectrophotometry and IR Spectroscopy
2.2.3. Multinuclear NMR Spectroscopy
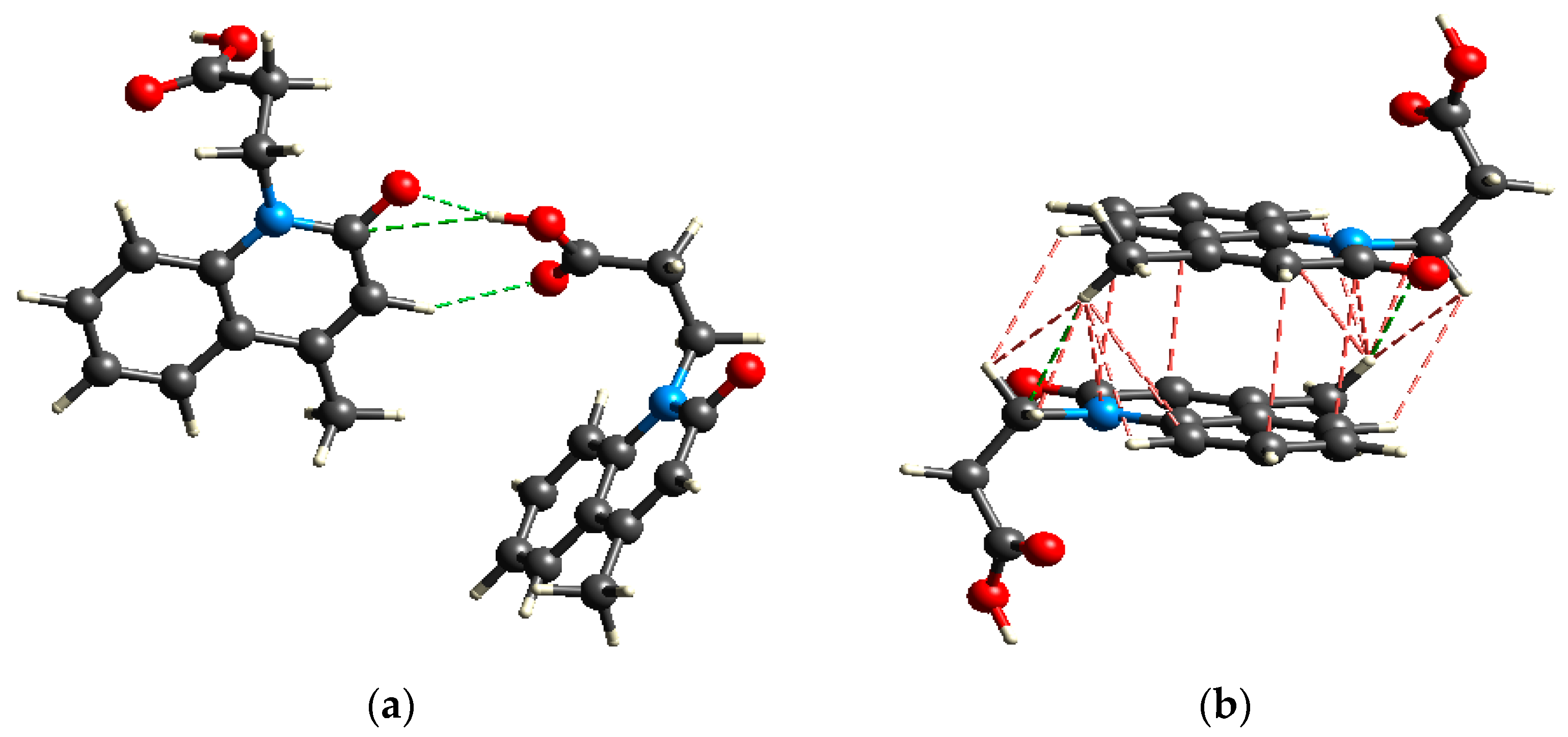
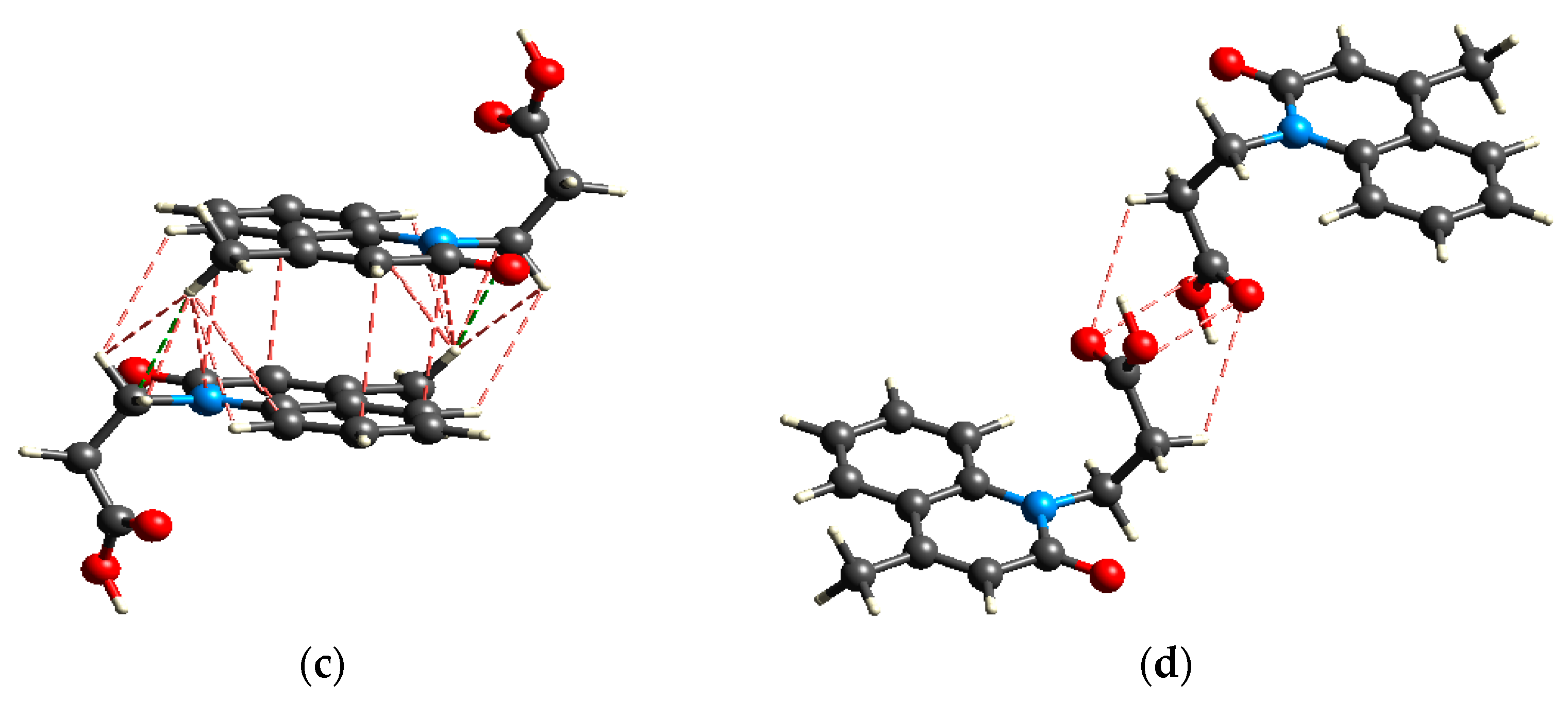
2.2.4. Lipophilicity and Stability
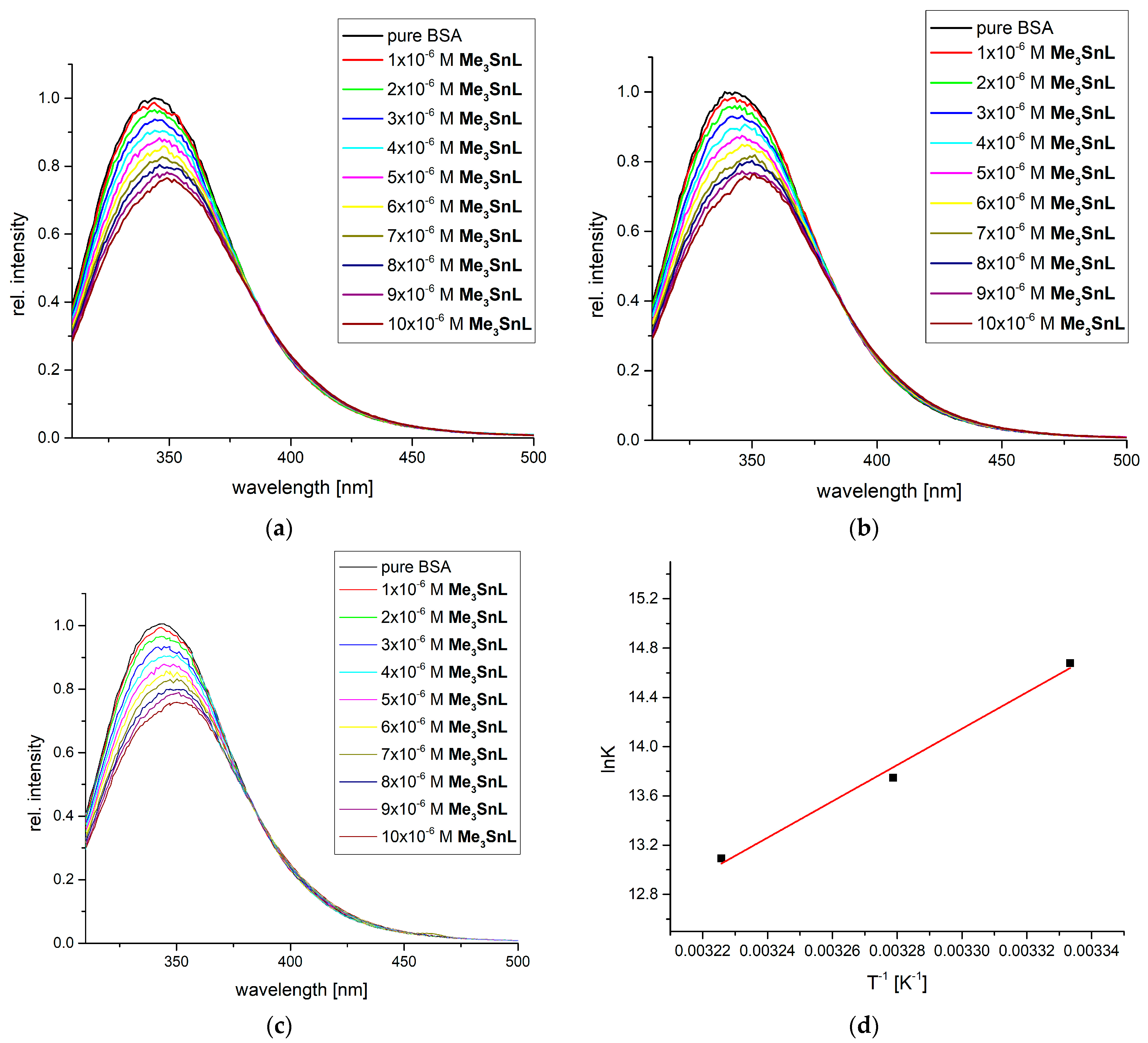
2.3. Protein Binding Affinity of Me3SnL
2.4. In Vitro Screening
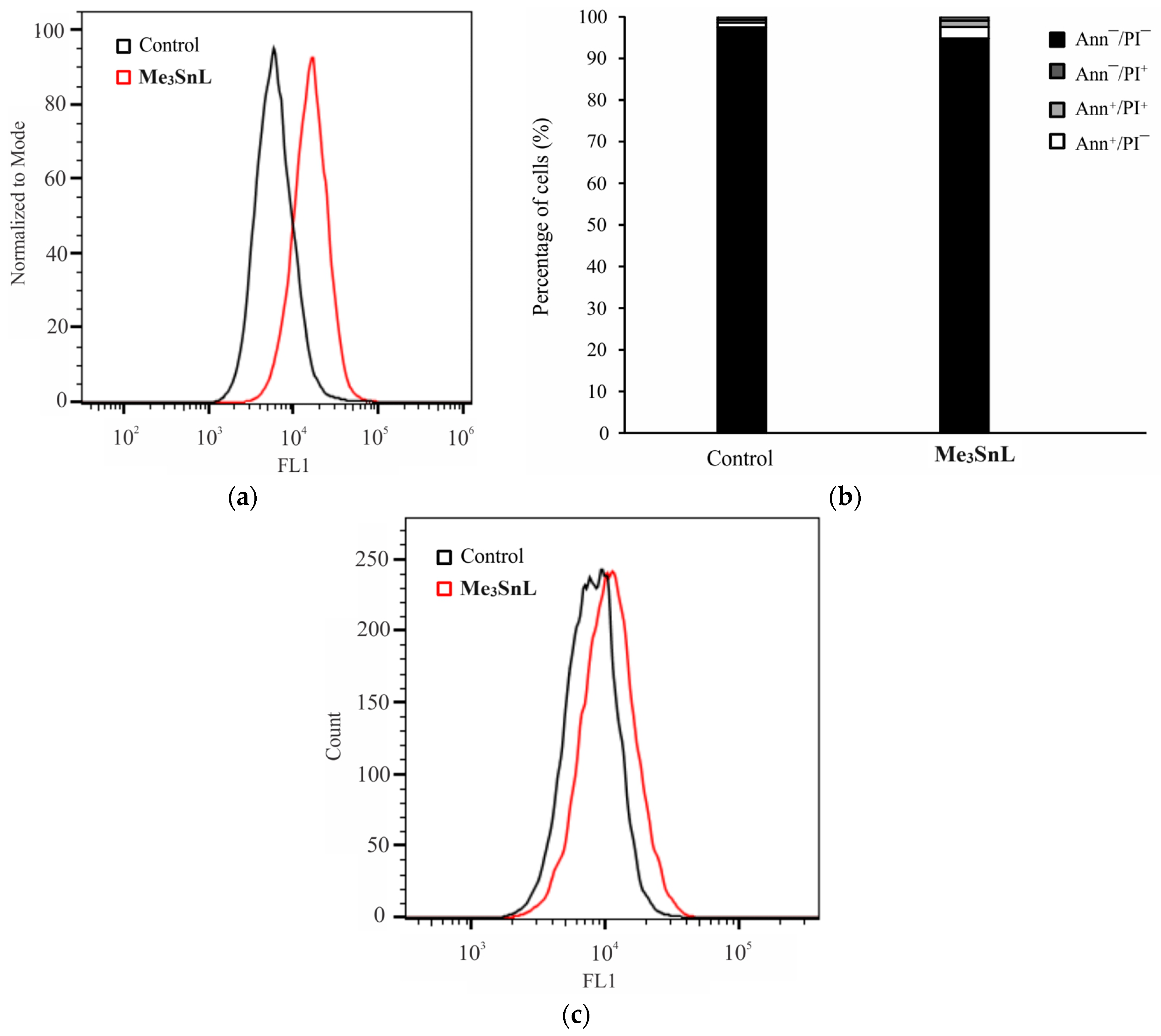
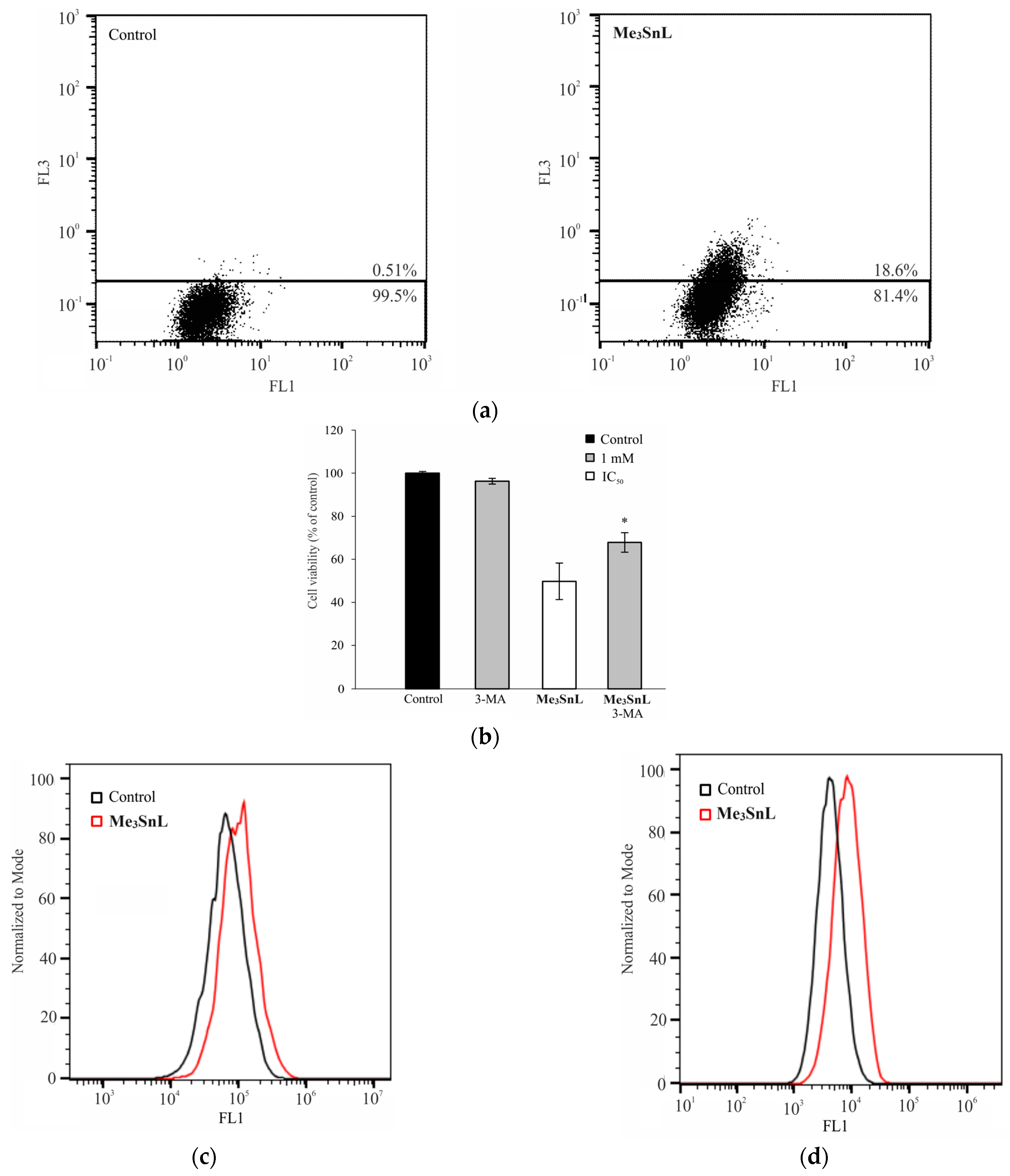
2.5. Mechanism of Action
3. Materials and Methods
3.1. General Remarks
3.2. Synthesis of Me3SnL
3.3. Crystal Structure Determination and Refinement
3.4. Computational Methods
3.4.1. Hirshfeld Surface Analysis
3.4.2. Structure Optimization and Spectral Characterization
3.5. Lipophilicity Assay
3.6. Spectrofluorimetric Investigation of the BSA Binding Affinity
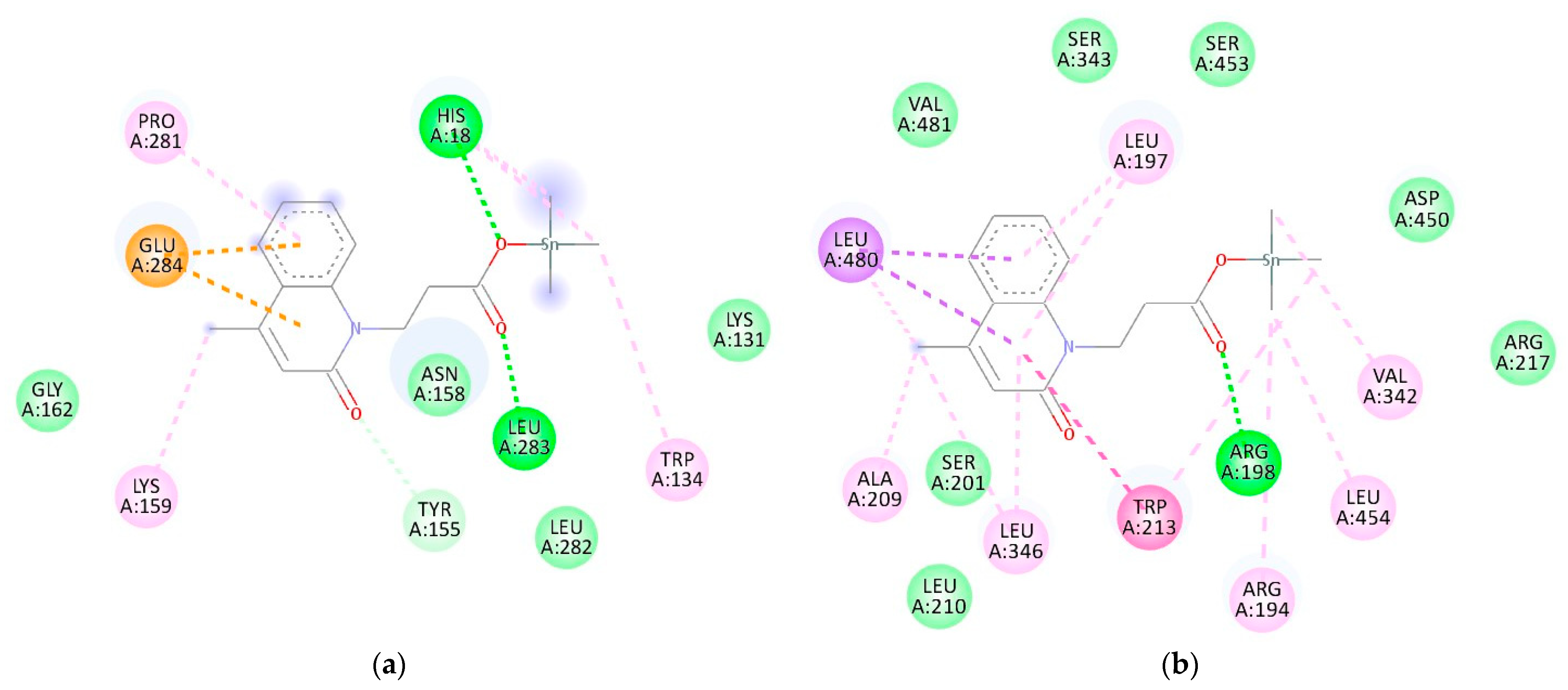
3.7. Molecular Docking
3.8. In Vitro Studies
3.8.1. Reagents and Cells
3.8.2. Determination of Cell Viability (MTT and CV Assays)
3.8.3. Annexin V/Propidium Iodide (PI)
3.8.4. ApoStat Staining
3.8.5. AO Staining
3.8.6. CFSE Staining
3.8.7. Measurement of Reactive Oxygen and Nitrogen Species (ROS/RNS) Generation
3.8.8. Detection of Lipid Peroxidation
3.8.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cancer—Symptoms and Causes. Available online: https://www.mayoclinic.org/diseases-conditions/cancer/symptoms-causes/syc-20370588 (accessed on 26 January 2023).
- Cancer. Available online: https://www.who.int/health-topics/cancer (accessed on 26 January 2023).
- Devi, J.; Kumar, B.; Taxak, B. Recent Advancements of Organotin(IV) Complexes Derived from Hydrazone and Thiosemicarbazone Ligands as Potential Anticancer Agents. Inorg. Chem. Commun. 2022, 139, 109208. [Google Scholar] [CrossRef]
- Cisplatin—NCI. Available online: https://www.cancer.gov/about-cancer/treatment/drugs/cisplatin (accessed on 23 October 2023).
- Weiss, R.B.; Christian, M.C. New Cisplatin Analogues in Development: A Review. Drugs 1993, 46, 360–377. [Google Scholar] [CrossRef]
- Cisplatin Side Effects: Common, Severe, Long Term. Available online: https://www.drugs.com/sfx/cisplatin-side-effects.html (accessed on 12 February 2023).
- Raguz, S.; Yagüe, E. Resistance to Chemotherapy: New Treatments and Novel Insights into an Old Problem. Br. J. Cancer 2008, 99, 387–391. [Google Scholar] [CrossRef]
- Kumar, M.; Abbas, Z.; Tuli, H.S.; Rani, A. Organotin Complexes with Promising Therapeutic Potential. Curr. Pharmacol. Rep. 2020, 6, 167–181. [Google Scholar] [CrossRef]
- Alama, A.; Tasso, B.; Novelli, F.; Sparatore, F. Organometallic Compounds in Oncology: Implications of Novel Organotins as Antitumor Agents. Drug Discov. Today 2009, 14, 500–508. [Google Scholar] [CrossRef]
- García-López, M.C.; Muñoz-Flores, B.M.; Jiménez-Pérez, V.M.; Moggio, I.; Arias, E.; Chan-Navarro, R.; Santillan, R. Synthesis and Photophysical Characterization of Organotin Compounds Derived from Schiff Bases for Organic Light Emitting Diodes. Dyes Pigments 2014, 106, 188–196. [Google Scholar] [CrossRef]
- Vieriu, S.-M.; Someşan, A.-A.; Silvestru, C.; Licarete, E.; Banciu, M.; Varga, R.A. Synthesis, Structural Characterization and in Vitro Antiproliferative Effects of Novel Organotin(iv) Compounds with Nicotinate and Isonicotinate Moieties on Carcinoma Cells. New J. Chem. 2021, 45, 1020–1028. [Google Scholar] [CrossRef]
- Gielen, M. Review: Organotin Compounds and Their Therapeutic Potential: A Report from the Organometallic Chemistry Department of the Free University of Brussels. Appl. Organomet. Chem. 2002, 16, 481–494. [Google Scholar] [CrossRef]
- Banti, C.N.; Hadjikakou, S.K.; Sismanoglu, T.; Hadjiliadis, N. Anti-Proliferative and Antitumor Activity of Organotin(IV) Compounds. An Overview of the Last Decade and Future Perspectives. J. Inorg. Biochem. 2019, 194, 114–152. [Google Scholar] [CrossRef] [PubMed]
- Tariq, M.; Sirajuddin, M.; Ali, S.; Khalid, N.; Tahir, M.N.; Khan, H.; Ansari, T.M. Pharmacological Investigations and Petra/Osiris/Molinspiration (POM) Analyses of Newly Synthesized Potentially Bioactive Organotin(IV) Carboxylates. J. Photochem. Photobiol. B Biol. 2016, 158, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Syed Annuar, S.N.; Kamaludin, N.F.; Awang, N.; Chan, K.M. Cellular Basis of Organotin(IV) Derivatives as Anticancer Metallodrugs: A Review. Front. Chem. 2021, 9, 657599. [Google Scholar] [CrossRef]
- Ullah, H.; Previtali, V.; Mihigo, H.B.; Twamley, B.; Rauf, M.K.; Javed, F.; Waseem, A.; Baker, R.J.; Rozas, I. Structure-Activity Relationships of New Organotin(IV) Anticancer Agents and Their Cytotoxicity Profile on HL-60, MCF-7 and HeLa Human Cancer Cell Lines. Eur. J. Med. Chem. 2019, 181, 111544. [Google Scholar] [CrossRef]
- Saeed, A.; Channar, P.A.; Larik, F.A.; Jabeen, F.; Muqadar, U.; Saeed, S.; Flörke, U.; Ismail, H.; Dilshad, E.; Mirza, B. Design, Synthesis, Molecular Docking Studies of Organotin-Drug Derivatives as Multi-Target Agents against Antibacterial, Antifungal, α-Amylase, α-Glucosidase and Butyrylcholinesterase. Inorganica Chim. Acta 2017, 464, 204–213. [Google Scholar] [CrossRef]
- Pantelić, N.Đ.; Božić, B.; Zmejkovski, B.B.; Banjac, N.R.; Dojčinović, B.; Wessjohann, L.A.; Kaluđerović, G.N. In Vitro Evaluation of Antiproliferative Properties of Novel Organotin(IV) Carboxylate Compounds with Propanoic Acid Derivatives on a Panel of Human Cancer Cell Lines. Molecules 2021, 26, 3199. [Google Scholar] [CrossRef] [PubMed]
- Kamaludin, N.F.; Ismail, N.; Awang, N.; Mohamad, R.; Pim, N.U. Cytotoxicity Evaluation and the Mode of Cell Death of K562 Cells Induced by Organotin (IV) (2-Methoxyethyl) Methyldithiocarbamate Compounds. J. Appl. Pharm. Sci. 2019, 9, 10–15. [Google Scholar] [CrossRef]
- Kamaludin, N.F.; Awang, N.; Baba, I.; Hamid, A.; Meng, C.K. Synthesis, Characterization and Crystal Structure of Organotin(IV) N-Butyl-N-Phenyldithiocarbamate Compounds and Their Cytotoxicity in Human Leukemia Cell Lines. Pak. J. Biol. Sci. 2013, 16, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Pandey, N.; Tilak, R.; Yadav, S.K.; Mishra, H.; Pokharia, S. New Triorganotin(IV) Complexes of Quinolone Antibacterial Drug Sparfloxacin: Synthesis, Structural Characterization, DFT Studies and Biological Activity: Triorganotin(IV) Complexes of Sparfloxacin. Appl. Organomet. Chem. 2018, 32, e4324. [Google Scholar] [CrossRef]
- Aly, A.A.; Ramadan, M.; Abuo-Rahma, G.E.-D.A.; Elshaier, Y.A.M.M.; Elbastawesy, M.A.I.; Brown, A.B.; Bräse, S. Chapter Three—Quinolones as Prospective Drugs: Their Syntheses and Biological Applications. In Advances in Heterocyclic Chemistry; Scriven, E.F.V., Ramsden, C.A., Eds.; Academic Press: Cambridge, MA, USA, 2021; Volume 135, pp. 147–196. [Google Scholar]
- Uivarosi, V. Metal Complexes of Quinolone Antibiotics and Their Applications: An Update. Molecules 2013, 18, 11153–11197. [Google Scholar] [CrossRef] [PubMed]
- Greeff, J.; Joubert, J.; Malan, S.F.; van Dyk, S. Antioxidant Properties of 4-Quinolones and Structurally Related Flavones. Bioorg. Med. Chem. 2012, 20, 809–818. [Google Scholar] [CrossRef]
- Senerovic, L.; Opsenica, D.; Moric, I.; Aleksic, I.; Spasić, M.; Vasiljevic, B. Quinolines and Quinolones as Antibacterial, Antifungal, Anti-Virulence, Antiviral and Anti-Parasitic Agents. Adv. Exp. Med. Biol. 2020, 1282, 37–69. [Google Scholar] [CrossRef]
- Ahmed, S.; Bhatti, M.H.; Ali, S.; Ahmed, F. Organotin (IV) Derivatives of 1-Ethyl-1,4-Dihydro-7-Methyl-4-Oxo- 1,8-Naphthyridine-3-Carboxylic Acid (Nalidixic Acid): Synthesis, Structural Elucidation and Biological Activities. Turk. J. Chem. 2006, 30, 193–202. [Google Scholar]
- Kasalović, M.P.; Jelača, S.; Maksimović-Ivanić, D.; Lađarević, J.; Radovanović, L.; Božić, B.; Mijatović, S.; Pantelić, N.Đ.; Kaluđerović, G.N. Novel Diphenyltin(IV) Complexes with Carboxylato N-Functionalized 2-Quinolone Ligands: Synthesis, Characterization and in Vitro Anticancer Studies. J. Inorg. Biochem. 2024, 250, 112399. [Google Scholar] [CrossRef]
- Dimić, D.S.; Kaluđerović, G.N.; Avdović, E.H.; Milenković, D.A.; Živanović, M.N.; Potočňák, I.; Samoľová, E.; Dimitrijević, M.S.; Saso, L.; Marković, Z.S.; et al. Synthesis, Crystallographic, Quantum Chemical, Antitumor, and Molecular Docking/Dynamic Studies of 4-Hydroxycoumarin-Neurotransmitter Derivatives. Int. J. Mol. Sci. 2022, 23, 1001. [Google Scholar] [CrossRef]
- Su, H.-Q.; Zhang, R.-F.; Guo, Q.; Wang, J.; Li, Q.-L.; Du, X.-M.; Ru, J.; Zhang, Q.-F.; Ma, C.-L. Five Organotin Complexes Derived from Hydroxycinnamic Acid Ligands: Synthesis, Structure, in Vitro Cytostatic Activity and Binding Interaction with BSA. J. Mol. Struct. 2022, 1247, 131290. [Google Scholar] [CrossRef]
- Milenković, D.; Avdović, E.; Dimić, D.; Sudha, S.; Ramarajan, D.; Milanović, Ž.; Trifunović, S.; Marković, Z.S. Vibrational and Hirshfeld Surface Analyses, Quantum Chemical Calculations, and Molecular Docking Studies of Coumarin Derivative 3-(1-m-Toluidinoethylidene)-Chromane-2,4-Dione and Its Corresponding Palladium(II) Complex. J. Mol. Struct. 2020, 1209, 127935. [Google Scholar] [CrossRef]
- Gak Simić, K.; Đorđević, I.; Lazić, A.; Radovanović, L.; Petković-Benazzouz, M.; Rogan, J.; Trišović, N.; Janjić, G. On the Supramolecular Outcomes of Fluorination of Cyclohexane-5-Spirohydantoin Derivatives. CrystEngComm 2021, 23, 2606–2622. [Google Scholar] [CrossRef]
- Janjić, G.V.; Jelić, S.T.; Trišović, N.P.; Popović, D.M.; Dordević, I.S.; Milčić, M.K. New Theoretical Insight into Fluorination and Fluorine-Fluorine Interactions as a Driving Force in Crystal Structures. Cryst. Growth Des. 2020, 20, 2943–2951. [Google Scholar] [CrossRef]
- Kavitha, E.; Ramarajan, D.; Rakić, A.; Dimić, D.; Sudha, S.; Nirmala, P.N. Structural, Spectroscopic, Quantum Chemical, and Molecular Docking Investigation of (E)-N’-(2,5-Dimethoxybenzylidene)Picolinohydrazide. J. Mol. Struct. 2022, 1253, 132259. [Google Scholar] [CrossRef]
- Nikolić, D.; Genčić, M.; Aksić, J.; Radulović, N.; Dimić, D.; Kaluđerović, G.N. Diorganotin(IV) Complexes with Hydroxamic Acids Derivatives of Some Histone Deacetylases Inhibitors. J. Serbian Chem. Soc. 2023, 88, 64. [Google Scholar] [CrossRef]
- Cantón-Diaz, A.; Muñoz-Flores, B.M.; Berrones-Reyes, J.; Moggio, I.; Arias, E.; Turlakov, G.; Santillán, R.; Jiménez-Pérez, V.M. Organotin Compounds Bearing C3-Symmetric Schiff Base: Microwave-Assisted Multicomponent Synthesis and Their Photophysical Properties. J. Organomet. Chem. 2021, 954–955, 122111. [Google Scholar] [CrossRef]
- Novák, M.; Turek, J.; Milasheuskaya, Y.; Růžičková, Z.; Podzimek, Š.; Jambor, R. N-Donor Stabilized Tin(II) Cations as Efficient ROP Catalysts for the Synthesis of Linear and Star-Shaped PLAs via the Activated Monomer Mechanism. Dalton Trans. 2021, 50, 16039–16052. [Google Scholar] [CrossRef] [PubMed]
- Goerigk, F.; Birchall, N.; Feil, C.M.; Nieger, M.; Gudat, D. Reactions of Imidazolio-Phosphides with Organotin Chlorides: Surprisingly Diverse. Eur. J. Inorg. Chem. 2022, 2022, e202101026. [Google Scholar] [CrossRef]
- Dimic, D.; Petkovic, M. Control of a Photoswitching Chelator by Metal Ions: DFT, NBO, and QTAIM Analysis. Int. J. Quantum Chem. 2016, 116, 27–34. [Google Scholar] [CrossRef]
- Muhammad, N.; Zia-ur-Rehman; Ali, S.; Meetsma, A.; Shaheen, F. Organotin(IV) 4-Methoxyphenylethanoates: Synthesis, Spectroscopic Characterization, X-Ray Structures and in Vitro Anticancer Activity against Human Prostate Cell Lines (PC-3). Inorganica Chim. Acta 2009, 362, 2842–2848. [Google Scholar] [CrossRef]
- Ahmad, M.S.; Hussain, M.; Hanif, M.; Ali, S.; Qayyum, M.; Mirza, B. Di- and Triorganotin(IV) Esters of 3,4-Methylenedioxyphenylpropenoic Acid: Synthesis, Spectroscopic Characterization and Biological Screening for Antimicrobial, Cytotoxic and Antitumor Activities. Chem. Biol. Drug Des. 2008, 71, 568. [Google Scholar] [CrossRef]
- Davies, A.G.; Smith, P.J. 11—Tin. In Comprehensive Organometallic Chemistry; Wilkinson, G., Stone, F.G.A., Abel, E.W., Eds.; Pergamon: Oxford, UK, 1982; pp. 519–627. ISBN 978-0-08-046518-0. [Google Scholar]
- Adeyemi, J.O.; Onwudiwe, D.C. Organotin(IV) Dithiocarbamate Complexes: Chemistry and Biological Activity. Molecules 2018, 23, 2571. [Google Scholar] [CrossRef]
- Sirajuddin, M.; Ali, S.; Tahir, M.N. Pharmacological Investigation of Mono-, Di- and Tri-Organotin(IV) Derivatives of Carbodithioates: Design, Spectroscopic Characterization, Interaction with SS-DNA and POM Analyses. Inorganica Chim. Acta 2016, 439, 145–158. [Google Scholar] [CrossRef]
- Lockhart, T.P.; Manders, W.F. Structure Determination by NMR Spectroscopy. Dependence of |2J(119Sn,1H)| on the Me-Sn-Me Angle in Methyltin(IV) Compounds. Inorg. Chem. 1986, 25, 892–895. [Google Scholar] [CrossRef]
- Debnath, P.; Singh, K.S.; Devi, T.S.; Singh, S.S.; Butcher, R.J.; Sieroń, L.; Maniukiewicz, W. Synthesis, Characterization, Crystal Structures and Anti-Diabetic Activity of Organotin (IV) Complexes with 2-(4-Hydroxynaphthylazo)-Benzoic Acid. Inorganica Chim. Acta 2020, 510, 119736. [Google Scholar] [CrossRef]
- Aziz-ur-Rehman; Hussain, M.; Zia-ur-Rehman; Rauf, A.; Nasim, F.-H.; Tahir, A.A.; Ali, S. New Tetrahedral, Square-Pyramidal, Trigonal-Bipyramidal and Octahedral Organotin(IV) 4-Ethoxycarbonylpiperazine-1-Carbodithioates: Synthesis, Structural Properties and Biological Applications. J. Organomet. Chem. 2010, 695, 1526–1532. [Google Scholar] [CrossRef]
- Mehmood, M.; Din, I.-U.; Raheel, A.; Haq, I.-U.; Tahir, M.N. Preparation, structural elucidation and biocidal applications of trimethyltin(IV) complexes derived from substituted carboxylic acids. Helyion 2020, 6, e05156. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Stephens, C.; Lucena, M.I.; Andrade, R.J. Idiosyncratic Drug-Induced Liver Injury: Mechanisms and Susceptibility Factors. In Comprehensive Toxicology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 625–650. ISBN 978-0-08-100601-6. [Google Scholar]
- Xu, L.; Hong, M.; Yang, Y.; Cui, J.; Li, C. Synthesis, Structural Characterization, in Vitro Cytotoxicities, and BSA Interaction of Di-Organotin(IV) Complexes Derived from Salicylaldehyde Nicotinoyl Hydrazone. J. Coord. Chem. 2016, 69, 2598–2609. [Google Scholar] [CrossRef]
- Wang, F.; Yin, H.; Cui, J.; Zhang, Y.; Geng, H.; Hong, M. Synthesis, structural characterization, in vitro cytotoxicities, DNA-binding and BSA interaction of diorganotin (IV) complexes derived from hydrazone Schiff base. J. Organomet. Chem. 2014, 759, 83–91. [Google Scholar] [CrossRef]
- Liu, K.; Yan, H.; Chang, G.; Li, Z.; Niu, M.; Hong, M. Organotin(IV) complexes derived from hydrazone Schiff base: Synthesis, crystal structure, in vitro cytotoxicity and DNA/BSA interactions. Inorganica Chim. Acta 2017, 464, 137–146. [Google Scholar] [CrossRef]
- Bujacz, A.; Zielinski, K.; Sekula, B. Structural studies of bovine, equine, and leporine serum albumin complexes with naproxen. Proteins Struct. Funct. Bioinform. 2014, 82, 2199–2208. [Google Scholar] [CrossRef] [PubMed]
- Su, L.-J.; Zhang, J.-H.; Gomez, H.; Murugan, R.; Hong, X.; Xu, D.; Jiang, F.; Peng, Z.-Y. Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis. Oxid. Med. Cell Longev. 2019, 2019, 5080843. [Google Scholar] [CrossRef]
- Ye, F.; Kaneko, H.; Hayashi, Y.; Takayama, K.; Hwang, S.-J.; Nishizawa, Y.; Kimoto, R.; Nagasaka, Y.; Tsunekawa, T.; Matsuura, T.; et al. Malondialdehyde Induces Autophagy Dysfunction and VEGF Secretion in the Retinal Pigment Epithelium in Age-Related Macular Degeneration. Free Radic. Biol. Med. 2016, 94, 121–134. [Google Scholar] [CrossRef]
- Haberzettl, P.; Hill, B.G. Oxidized Lipids Activate Autophagy in a JNK-Dependent Manner by Stimulating the Endoplasmic Reticulum Stress Response. Redox Biol. 2013, 1, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. A short history of SHELX. Acta Cryst. Sect. A 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer, 17; University of Western Australia: Perth, Australia, 2017.
- Spackman, M.A.; Byrom, P.G. A Novel Definition of a Molecule in a Crystal. Chem. Phys. Lett. 1997, 267, 215–220. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld Surface Analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Becke, A.D. Density-Functional Thermochemistry. III. The Role of Exact Exchange. J. Chem. Phys. 1993, 98, 5648. [Google Scholar] [CrossRef]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the Damping Function in Dispersion Corrected Density Functional Theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef]
- Dunning, T.H. Gaussian Basis Sets for Use in Correlated Molecular Calculations. I. The Atoms Boron through Neon and Hydrogen. J. Chem. Phys. 1989, 90, 1007. [Google Scholar] [CrossRef]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Reed, A.E.; Curtiss, L.A.; Weinhold, F. Intermolecular Interactions from a Natural Bond Orbital, Donor-Acceptor Viewpoint. Chem. Rev. 1988, 88, 899–926. [Google Scholar] [CrossRef]
- Reed, A.E.; Weinstock, R.B.; Weinhold, F. Natural Population Analysis. J. Chem. Phys. 1985, 83, 735–746. [Google Scholar] [CrossRef]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef]
- Zieliński, R.; Szymusiak, H. Application of Dft B3Lyp/Giao and B3Lyp/Csgt Methods for Interpretation of Nmr Spectra of Flavonoids. Pol. J. Food Nutr. Sci. 2003, 12, 157–162. [Google Scholar]
- Bohmann, J.A.; Weinhold, F.; Farrar, T.C. Natural Chemical Shielding Analysis of Nuclear Magnetic Resonance Shielding Tensors from Gauge-Including Atomic Orbital Calculations. J. Chem. Phys. 1997, 107, 1173. [Google Scholar] [CrossRef]
- Puckett, C.A.; Barton, J.K. Methods to Explore Cellular Uptake of Ruthenium Complexes. J. Am. Chem. Soc. 2007, 129, 46–47. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. AMDock: A versatile graphical tool for assisting molecular docking with Autodock Vina and Autodock4. Biol. Direct 2020, 15, 12. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Milanović, Ž.B.; Antonijević, M.R.; Amić, A.D.; Avdović, E.H.; Dimić, D.S.; Milenković, D.A.; Marković, Z.S. Inhibitory activity of quercetin, its metabolite, and standard antiviral drugs towards enzymes essential for SARS-CoV-2: The role of acid–base equilibria. RSC Adv. 2021, 11, 2838–2847. [Google Scholar] [CrossRef] [PubMed]
- Dassault Systèmes BIOVIA. Discovery Studio Modeling Environment (Release 2017); BIOVIA: San Diego, CA, USA, 2016. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4:Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]

| 1H | 13C | ||||
|---|---|---|---|---|---|
| H atom | Exp. [ppm] | Calc. [ppm] | C atom | Exp. [ppm] | Calc. [ppm] |
| C15,16,17–H | 0.58 | −0.21 | C15,16,17 | −2.75 | −0.39 |
| C14–H | 2.46 | 2.36 | C14 | 18.49 | 22.09 |
| C12–H | 2.73 | 2.55 | C12 | 32.23 | 36.17 |
| C13–H | 4.57 | 4.34 | C13 | 38.16 | 40.91 |
| C3–H | 6.58 | 6.45 | C8 | 113.79 | 112.64 |
| C6–H | 7.26 | 7.31 | C6 | 120.34 | 118.93 |
| C8–H | 7.48 | 7.31 | C3 | 121.13 | 120.47 |
| C5–H | 7.57 | 7.59 | C10 | 121.39 | 124.67 |
| C7–H | 7.72 | 7.64 | C5 | 124.90 | 124.68 |
| R | 0.998 | C7 | 130.05 | 130.54 | |
| MAE [ppm] | 0.19 | C9 | 138.13 | 140.23 | |
| C4 | 146.9 | 148.18 | |||
| C2 | 161.19 | 157.36 | |||
| C11 | 175.52 | 174.34 | |||
| R | 0.999 | ||||
| MAE [ppm] | 2.02 | ||||
| Compound | Assays | MCF-7 | A375 | HCT116 | 4T1 | B16 | CT26 | MRC5 |
|---|---|---|---|---|---|---|---|---|
| HL ** | MTT [µM] | >200 | n.d.*** | |||||
| CV [µM] | >200 | n.d. | ||||||
| Me3SnL | MTT [µM] | 5.7 ± 0.2 | 5.4 ± 0.5 | 6.2 ± 0.4 | 17.2 ± 2.4 | 8.1 ± 0.15 | 11.9 ± 1.5 | / |
| CV [µM] | 11.4 ± 2.7 | 10.9 ± 1.9 | 8.0 ± 0.4 | 18.9 ± 1.8 | 10.6 ± 1.0 | 16.1 ± 2.0 | 93.9 ± 2.2 | |
| Ph2SnL2 ** | MTT [µM] | 0.10 ± 0.00 | 0.30 ± 0.05 | 0.15 ± 0.02 | 0.30 ± 0.03 | 0.36 ± 0.01 | 0.30 ± 0.01 | n.d. |
| CV [µM] | 0.20 ± 0.02 | 0.36 ± 0.05 | 0.26 ± 0.02 | 0.40 ± 0.01 | 0.39 ± 0.07 | 0.40 ± 0.07 | n.d. | |
| Cisplatin ** | MTT [µM] | 5.00 ± 0.70 | 8.10 ± 0.70 | 5.00 ± 2.30 | 1.60 ± 0.20 | 8.30 ± 1.60 | 4.30 ± 0.20 | n.d. |
| CV [µM] | 12.60 ± 2.50 | 10.70 ± 1.60 | 12.30 ± 3.50 | 2.50 ± 0.10 | 18.00 ± 1.80 | 4.70 ± 0.40 | n.d. | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasalović, M.P.; Dimić, D.; Jelača, S.; Maksimović-Ivanić, D.; Mijatović, S.; Zmejkovski, B.B.; Schreiner, S.H.F.; Rüffer, T.; Pantelić, N.Đ.; Kaluđerović, G.N. Trimethyltin(IV) Bearing 3-(4-Methyl-2-oxoquinolin-1(2H)-yl)propanoate Causes Lipid Peroxidation-Mediated Autophagic Cell Death in Human Melanoma A375 Cells. Pharmaceuticals 2024, 17, 372. https://doi.org/10.3390/ph17030372
Kasalović MP, Dimić D, Jelača S, Maksimović-Ivanić D, Mijatović S, Zmejkovski BB, Schreiner SHF, Rüffer T, Pantelić NĐ, Kaluđerović GN. Trimethyltin(IV) Bearing 3-(4-Methyl-2-oxoquinolin-1(2H)-yl)propanoate Causes Lipid Peroxidation-Mediated Autophagic Cell Death in Human Melanoma A375 Cells. Pharmaceuticals. 2024; 17(3):372. https://doi.org/10.3390/ph17030372
Chicago/Turabian StyleKasalović, Marijana P., Dušan Dimić, Sanja Jelača, Danijela Maksimović-Ivanić, Sanja Mijatović, Bojana B. Zmejkovski, Simon H. F. Schreiner, Tobias Rüffer, Nebojša Đ. Pantelić, and Goran N. Kaluđerović. 2024. "Trimethyltin(IV) Bearing 3-(4-Methyl-2-oxoquinolin-1(2H)-yl)propanoate Causes Lipid Peroxidation-Mediated Autophagic Cell Death in Human Melanoma A375 Cells" Pharmaceuticals 17, no. 3: 372. https://doi.org/10.3390/ph17030372
APA StyleKasalović, M. P., Dimić, D., Jelača, S., Maksimović-Ivanić, D., Mijatović, S., Zmejkovski, B. B., Schreiner, S. H. F., Rüffer, T., Pantelić, N. Đ., & Kaluđerović, G. N. (2024). Trimethyltin(IV) Bearing 3-(4-Methyl-2-oxoquinolin-1(2H)-yl)propanoate Causes Lipid Peroxidation-Mediated Autophagic Cell Death in Human Melanoma A375 Cells. Pharmaceuticals, 17(3), 372. https://doi.org/10.3390/ph17030372











