Abstract
SMADs are the canonical intracellular effector proteins of the TGF-β (transforming growth factor-β). SMADs translocate from plasma membrane receptors to the nucleus regulated by many SMAD-interacting proteins through phosphorylation and other post-translational modifications that govern their nucleocytoplasmic shuttling and subsequent transcriptional activity. The signaling pathway of TGF-β/SMAD exhibits both tumor-suppressing and tumor-promoting phenotypes in epithelial-derived solid tumors. Collectively, the pleiotropic nature of TGF-β/SMAD signaling presents significant challenges for the development of effective cancer therapies. Here, we review preclinical studies that evaluate the efficacy of inhibitors targeting major SMAD-regulating and/or -interacting proteins, particularly enzymes that may play important roles in epithelial or mesenchymal compartments within solid tumors.
1. Introduction
Transforming growth factor-β (TGF-β) superfamily and resulting canonical SMAD signaling have gained significant attention in cancer research since the mid-1980s because of their significance in regulating central functions of the cell, such as proliferation, apoptosis, adhesion, and differentiation. SMADs specifically function to transmit information from extracellular signals received by TGF-β receptors, registering at the plasma membrane to the nucleus downstream of TGF-β. In the nucleus, SMADs cooperate with transcription factors, co-activators, and co-repressor regulators to control gene expression in a context-dependent manner [1,2]. Importantly, many non-SMAD factors are involved in directly controlling how SMAD proteins function in the TGF-β pathway to support, attenuate, or modulate downstream cellular responses.
TGF-β/SMAD plays a biphasic function during tumor progression, where it can suppress or potentiate tumorigenesis in normal and pre-malignant epithelial cells [3]. Signals downstream of TGF-β produced by tumors in the TME (tumor microenvironment) can also activate tumors to undergo an EMT (epithelial–mesenchymal transition) and/or hybrid/partial EMT [4,5,6,7]. EMT is well known to confer invasive, therapy-resistant, and metastatic properties to cancer cells [8,9,10]. Since TGF-β signaling plays a critical role in tumor progression, targeting the downstream SMAD signals presents an enormous challenge in the effort to develop target therapies to eradicate solid tumors effectively. Previous reviews have highlighted novel approaches to targeting signals downstream of TGF-β that focus on microRNAs (miRNAs), long non-coding RNAs (lnRNAs), deubiquitinating enzymes (DUBs), and protein–protein interactions (PPIs) [11,12,13,14,15]. However, the pleiotropic nature of TGF-β signaling contributes to alternate pathway engagement and tumor escape and drug resistance, creating challenges for clinicians and patients. Preclinical findings demonstrate that the TGF-β pathway can be modulated and/or targeted potentially using different alternatives. Although mouse/human chimeric monoclonal antibodies, receptors ligand traps, synthetic DNA antisense oligomers, therapeutic vaccines, and small-molecule inhibitors [16,17,18,19] are worth mentioning, these therapies commonly disrupt normal physiological functions and provide unsatisfactory results in clinical trials [20,21].
Here, we present a consideration of preclinical studies testing the anti-tumor efficacy of pharmacological targeting SMAD-regulating and/or -interacting proteins. We summarize inhibitors targeting the major SMAD-interacting enzymes involved in each pathway in addition to non-SMAD non-canonical pathways downstream of the TGF-β signaling. Finally, we propose how these inhibitors may contribute to abrogating cancer progression induced by TGF-β/SMAD signaling in the epithelial and/or mesenchymal cell types that are commonly found within the solid TME.
2. Canonical TGF-β/SMAD Signaling
The basic principles underlying TGF-β signaling are well-founded. Ligands within the TGF-β superfamily are known as activins/NODALs, bone morphogenetic proteins (BMPs), and growth differentiation factors (GDFs). Ligand-driven signaling activity depends on the formation of pairs with type I and type II receptors. The receptors (serine/threonine kinase) take part in the formation of dimers with disulfide-linked ligand dimers [22,23]. Upon their activation, the type I receptors are trans-phosphorylated by the type II receptors in the juxta membrane region (TTSGSGSG), which drives activation of the type I receptor kinase domain [24,25,26]. These activated receptor complexes subsequently activate a series of downstream signal transduction proteins known as SMAD proteins via phosphorylation that are transported to the nucleus, regulating various transcriptional programs.
Among the eight SMADs in vertebrates, only five are recognized as R-SMADs (Receptor-SMADs). TGF-β- and activins/NODAL-mediated signals activate SMAD2 and SMAD3 predominantly, whereas GDFs and BMPs initiate the activation of SMAD1, SMAD5, and SMAD8 [27]. Two serines are involved to initiate the phosphorylation of receptor and R-SMADs. Primarily, the phosphorylation event of R-SMADs initiates them to form homomeric complexes and subsequently primes them to form complexes with R-SMADs and SMAD4 (Co-SMAD). After activation, the heteromeric complexes (R-SMADs-SMAD4) finally accumulate in the nucleus and bind to DNA (high-affinity sequences) cooperatively with transcription factors and/or co-factors and co-repressors to regulate transcription (Figure 1) [1,2].
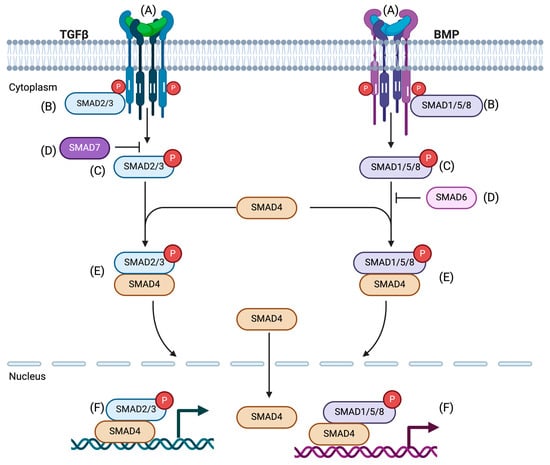
Figure 1.
The TGF-β/SMAD signaling pathway. (A) Ligand binding: A heteromeric complex is formed with an interaction of type II, type I receptors, and TGF-β ligands. (B) Activation of receptor: Ligand binding stimulates the type II receptor to phosphorylate and activate the type I receptor. (C) Activation of R-SMADs: Once the type I receptor is activated, it initiates phosphorylation and activation of the receptor-activated SMADs (R-SMADs, SMAD2, and SMAD3 for TGF-β signal). (D) Inhibition of R-SMAD activation: Inhibitory SMADs, SMAD7, and SMAD6 compete with R-SMADs to intervene with the type I receptor. As a result, R-SMAD activation and transmission of the SMAD signaling is prevented. (E) The complex of R-SMADs and CO-SMAD formation: Activated R-SMADs dissociate from type I receptors to form a complex with the common mediator, CO-SMAD, SMAD4. (F) The complex of R-SMAD and SMAD4 can translocate from cytoplasm to nucleus and bind to transcription factors, resulting in the transcription of target genes. Here, SMAD1, SMAD5, and SMAD8 are shown for BMP signal. Adapted from Molecular Biology of Human Cancers (Springer 2005) [28].
3. Activity and Nucleocytoplasmic Trafficking of SMADS
Mechanisms of SMAD Protein Nucleocytoplasmic Trafficking
SMAD proteins exist in dynamic states in the cytoplasm or nucleus with or without the stimulation of TGF-β, respectively. A study by Nicolás et al. [29] showed that both uninduced and induced cells with TGF-β could exhibit variation in SMAD distribution. Accumulating evidence supports that a few proteins are involved in the activation, retention, and trafficking of SMADs from the cytoplasm to the nucleus and vice versa (Figure 2). Activation begins with phosphorylation of the GS region on the type I receptor TGF-β (TGF-βRI) by the type II receptor (TGF-βRII), which leads to the affinity of receptor I for sxsmotif of R-SMADs at the C-terminal [30]. Thereby, the two C-terminal serines of R-SMADs are phosphorylated with substantial interaction between the R-SMADs and the type I receptor. This event is critical to changing the conformation of R-SMADs and allowing them to dissociate consequently from the receptor complex [31]. In this context, the association of the endocytic protein SARA (SMAD Anchor of Receptor Activation) [32,33] and a few SARA-associated proteins (e.g., cPML [34,35], endofin [32], and Dab2 [36] promote R-SMAD activation by recruiting them to the TGF-β receptor. Phosphorylated R-SMADs can interact with SMAD4 and form heteromeric complexes after dissociation from the TGF-β receptor complexes and subsequently transport to the nucleus [35,36,37,38]. It was also found that hepatocyte growth factor-regulated tyrosine kinase substrate (Hgs) is associated with SARA and promotes phosphorylation of SMADs 1, 5, and 8 induced by BMP signals [39,40]. The conformational change driven by the phosphorylation of R-SMAD monomers supports the dissociation from the receptor complex as well as promotes the interaction between the R-SMAD, which is phosphorylated at the C-terminal in addition to SMAD4 and another R-SMAD. Thus, different compositions of SMAD trimeric complexes can exist as a complex of R-SMAD monomers (activated homo or heterotrimer) and/or a complex of phosphorylated one or two R-SMADs with SMAD4 in the cytoplasm [41,42,43,44].
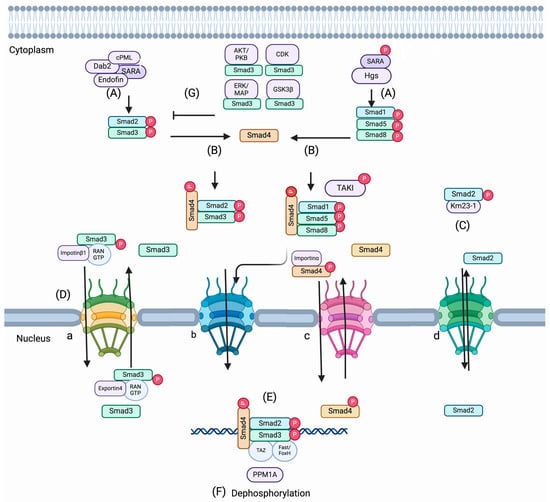
Figure 2.
Activation and nucleocytoplasmic trafficking of SMADs. (A) The association of endocytic protein SARA (SMAD Anchor of Receptor Activation) and a few SARA-associated proteins, cPML, endofin, and Dab2, promote R-SMAD activation R-SMADs to the TGF-β receptor. Hgs (hepatocyte growth factor-regulated tyrosine kinase substrate) is associated with SARA and promotes phosphorylation of SMADs 1, 5, and 8 and TAKI induced by BMP signal (B). Once R-SMADs are phosphorylated and dissociated from the receptor complex, SMAD4 and phosphorylated R-SMADs can interact and make a trimeric complex. (C) A dynein light chain km23-1 interacts with SMAD2 and SMAD3 and assists in nuclear translocation. (D) Nuclear translocation of SMAD complexes is maintained by nuclear pore proteins. Importin/exportin family members and Ran proteins (small Ran GTPase) are also involved in the trafficking of R-SMADS and SMAD4. (a) Importin/exportin-mediated Ran-dependent method; (b) Phospho-dependent method; (c) Importin-mediated Ran-independent method; (d) Importin/exportin-independent and phospho-independent method. (E) SMADs can be reserved in the nucleus in connection with DNA-binding transcription factors, YAP (Yes-associated protein), and co-factors like Fast1/FoxH1 and TAZ. (F) PPM1A functions as a phosphatase and terminates TGF-β signaling. (G) AKT/PKB, ERK/MAP, CDKs, and GSKB3b can inhibit the nuclear accumulation of R-SMADS by phosphorylation with the linker region of R-SMADs.
Nuclear translocation of SMAD complexes is sustained by some nuclear pore proteins. Although MH1 domains of SMAD proteins contain NLS-motifs, the regulation of trafficking of SMADs is governed by divergent methods including importin/exportin-mediated, Ran-dependent (Figure 2(Da)); phospho-dependent (Figure 2(Db,Dc)); importin-mediated Ran-independent (Figure 2(Dc)); and importin-exportin-independent and phospho-independent method (Figure 2(Dd)). Many importin/exportin family members and Ran proteins (small Ran GTPase) are involved in the trafficking of R-SMADs. Importin-β1 interacts at the MH1 domain of SMAD3 and is frequently imported into the nucleus through Ran-depended proteins [45], whereas exportin 4, which has a conserved sequence on the MH2 domain of SMAD3, allows the exportation of SMAD3 from the nucleus in a GTPase manner [46]. Importantly, RanBP3 acts as a selective nuclear protein of SMAD2/SMAD3 in the TGF-β pathway. In mammalian cells (human keratinocyte cells, HaCaT, human HEK293T, human HepG2, mouse myoblast cells C2C12) and Xenopus embryos, RanBP3 can directly identify dephosphorylated SMAD2/SMAD3 produced by the phosphatase activity of nuclear SMAD phosphatase. Thereby, the nuclear export of SMAD2/SMAD3 is augmented in a Ran-dependent manner to terminate the TGF-β signal [47]. On the other hand, importin-α mediates the nuclear translocation of SMAD4 [48]. Moreover, nuclear pore proteins (CAN/Nup214 and Nup15) can regulate the cytoplasmic shuttling of SMAD2, SMAD3, and SMAD4 proteins [49,50]. Further, dynein light chain km23-1 and Km23-2 can interact with SMAD2 and SMAD3, which facilitates the nuclear trafficking of the R-SMADS from cytoplasm to the nucleus for target gene transcription [51,52].
Other proteins, like AKT/PKB [53,54], can directly interact with SMAD3 and inhibit its phosphorylation and nuclear accumulation, further inhibiting SMAD3-mediated transcription and apoptosis. On the other hand, ERK/MAP [55,56] CDKs (2/4) [57,58] and GSKB3b [59,60] can inhibit the nuclear accumulation of R-SMADS by phosphorylating the linker region of R-SMADs in both epithelial and fibroblast cells. Nucleocytoplasmic shuttling is mediated by dephosphorylation, which promotes R-SMAD traveling from the nucleus to the cytoplasm. It was found that PPM1A functions as a phosphatase and terminates TGF-β signaling [61]. A study by Dai et al. [62] shows that PPM1A targets the nuclear exporter RanBp3 and thereby controls the transport of SMAD2/SMAD3 from the nucleus to the cytoplasm. RanBP3 is dephosphorylated at Ser 58, which promotes SMAD2/SMAD3 exportation from the nucleus. Thus, PPM1A provides the maximum RanBP3 exporter function for the effective termination of canonical TGF-β signaling.
Intriguingly, it was also found in epithelial, fibroblast, cancer (Hela cells, small lung cancer cell lines, breast cancer cell lines), and embryonic stem cells where SMADs can be reserved in the nucleus through the interaction with high-affinity DNA-binding transcription factors. YAP (Yes-associated protein), co-factors like Fast1/FoxH1, and TAZ (transcriptional co-activator with PDZ-binding motif) can associate with SMADs [63,64,65,66,67]. The nuclear retention inhibits the export of SMADs from the nucleus to the cytoplasm, and it is speculated that the association of YAP and SMAD3 in the nucleus may prevent the interaction of SMAD3 with exportin 4 or RanBP3. Further, it was found that phospho-SMAD-3 in epithelial cells can block the interaction of SMAD-4 with exportin 1 and thereby promote nuclear accumulation of SMAD4 [68].
Inhibitors targeting the SMAD-interacting enzymes such as AKT/PKB (Ser/Thr kinase/protein kinase B), CDK (cyclin-dependent kinase), GSK GSK3b (axin/glycogen synthase kinase 3 beta), and ERK (extracellular signal-related kinase) are cataloged in Table 1 [69,70,71,72,73,74,75,76,77,78,79,80,81,82,83]. These inhibitors act by modulating the phosphorylation activity of the kinases and thereby regulate their interaction, the R-SMADs, and SMAD-mediated signaling in epithelial and mesenchymal cells. Significant advancements have been observed in the area of small-molecule inhibitors of AKT functions through different mechanisms. In addition, pleckstrin homology domain, ATP-competitive, and allosteric inhibitors of AKT are noteworthy in developing clinical trials, and therapeutic benefits in treating different solid tumors have been observed [83]. A preclinical study of MK-2206 in epithelial morphology of pancreatic cancer cells by Wang et al. [72] showed that AKT phosphorylation is inhibited, and cell proliferation is attenuated by the application of the MK-2206 inhibitor. Further, a combination of MK-2206 with gemcitabine enhanced the inhibition of cancer cell proliferation. It suggests that gemcitabine-mediated AKT activation induced increased cell proliferation [72]. In epithelial cells, AKT can act as one of the major mediators of cell survival and apoptosis and regulates the activity of SMAD3 (Figure 2). In epithelioid pancreatic cancer cells, AKT phosphorylation might enhance the activity of SMAD3 and stimulate SMAD-mediated cell proliferation. It is considered that the inhibition of AKT-phosphorylation by MK-2206 may promote the interaction of AKT with SMAD3 and thereby support the inhibition of SMAD3 activation, nuclear accumulation, SMAD3-mediated transcription, and cell proliferation.

Table 1.
Inhibitors of SMAD-interacting enzymes controlling SMAD nucleocytoplasmic trafficking.
Another potential inhibitor is nitazoxanide (NTZ), which targets CDK1 by inhibiting phosphorylation of CDK1 at Thr161, as studied by Huang et al. [74]. It is thought that CDK1 phosphorylation might enhance the activity of SMAD3 and nucleocytoplasmic trafficking and stimulate SMAD-mediated cell proliferation in epithelioid glioblastoma cells. However, NTZ application might modulate SMAD-3 and CDK1 interaction, resulting in inhibition of SMAD3 activation, nucleocytoplasmic trafficking, transcription, and SMAD3-induced cell proliferation. Thus, modulation of the SMAD signaling by inhibiting the phosphorylation of kinase enzymes suggests that NTZ can be a promising CDK1 inhibitor for the development of clinical trials.
4. Regulation of SMAD Activity by Post-Translational Modifications (PTMs)
4.1. Phosphorylation and Dephosphorylation
Activation of R-SMADs is originated by phosphorylation induced by type I receptors of two serines in their carboxy SSXS motif induced by ligands. R-SMAD2 and R-SMAD3 are phosphorylated by the TβRI/ALK-5 (TGF-β-specific type I receptor); R-SMAD 1, 5, and 8 are phosphorylated by ALK-3/BMPRIA (BMP-specific type I receptor, BMP receptor IA). In addition, the structure of R-SMADs with the MHI and MH2 domains and linker region contains substrate sites for other kinases such as Map kinases, cyclin-dependent kinases (CDKs), extracellular signal-regulated kinase 1 (ERK1), and monopolar spindle kinase (MPS1), which regulate the stability of R-SMADs and subsequent transcriptional responses [84,85]. For example, ERK (extracellular signal-regulated kinase 1) phosphorylates SMAD2 on their MH1 domain and enhances SMAD-mediated transcription in epithelial cells [86]. Not all kinases that target R-SMADs potentiate SMAD-mediated transcription. In epithelium and mesenchymal cells, for example, PKC (protein kinase C) directly phosphorylates SMAD3, but this action inhibits SMAD3-dependent transcription [87]. Still, other R-SMAD phosphorylation events generate a platform for R-SMAD degradation via ubiquitin-dependent processes. A study by Saura et al. [88] shows that PKG1 (cGMP-mediated protein kinase 1)-mediated phosphorylation of SMAD3 at the MH1 domain fosters proteasomal degradation of SMAD3 in endothelial cells. Other small GTPase proteins like Ras promote ubiquitin-dependent SMAD2/3 degradation and stabilization of TβRII via the degradation of the SPSB1 protein [89]. Ras interacts and colocalizes with the SPSB (TβRII negative regulator) on the cell membrane, resulting in SPSB1 degradation [89]. Both ubiquitylation and degradation can also occur through phosphorylation of SMAD3 and SMAD2 by casein kinase 1 gamma 2 (CKIg2) and PAK4 [90,91]. And phosphorylation of SMAD2 by PAK2 confers the inactivation of SMAD2 by interfering with TGFβRI-SMAD2 interaction [92] in epithelial cells. SMAD4 is constitutively phosphorylated in epithelial cells (mink lung epithelial cells, Mv1Lu) and cancer cells (human squamous carcinoma cell lines, HSC-4), as shown by a study by Nakao et al. [93]. Although constitutive phosphorylation of SMAD4 has been observed in epithelial and cancer cells, the site of phosphorylation of SMAD4 remains to be identified. ERK promotes SMAD4 nuclear accumulation and enhances SMAD4-mediated transcriptional activity [94]. Other kinases, such as LKB1 (liver kinase B1) and MAP38, phosphorylate SMAD4 on MH1 and MH2 domains, respectively. LBK1 promotes SMAD4 stability by associating with a scaffolding protein LIPI and enhances the SMAD-dependent transcription as a negative regulator in controlling TGFβ gene responses and EMT [95,96]. On the other hand, MAP38-mediated phosphorylation at Ser343 of SMAD4 drives positive regulation of transcription in response to TGF-β-mediated apoptosis and cell proliferation in a kinase-dependent manner [96].
The dephosphorylation of C-terminal serine plays a significant role in deactivating the R-SMADs and mitigating the SMAD-mediated signaling. PPM1A (PP2Cα), designated as Mg+/Mn+-dependent 1A protein phosphatase, was the first phosphatase identified by Lin et al. [61] that dephosphorylates the SMAD2/3 at the C-terminus SXS motif. The phosphate MTMR4 dephosphorylates the activated SMAD2 and SMAD3 in the endosome [97]. Phosphates such as SCP1, SCP2, and SCP3 can remove linker phosphorylation at specific levels without interfering with phosphorylation at the C-terminal of SMAD2/SMAD3 to enhance the TGF-β signal [98]. On the other hand, in mammal keratinocyte cells and Xenopus embryos, dephosphorylation of SMAD1 by SCP1, SCP2, and SCP3 can attenuate the BMP signal [99].
4.2. Ubiquitylation and Deubiquitylation
Ubiquitylation involves the sequential covalent modification of protein substrates with ubiquitin molecules via the action of E1, E2, and E3 ubiquitin ligases, which mark the substrate for further activity or degradation. Regulation of R-SMADs is coordinated by ubiquitylation and ubiquitin-like modifications, which supports SMADs’ stability and subsequent activity and subcellular localization. Several ubiquitin ligases have been involved in the degradation of R-SMADs. HECT (the homologous E6-AP carboxy terminal) family E3 ligases SMURF1 and SMURF2 [100,101,102], NEED4-2/NEDD4L [103], Hsc70-interacting protein (CHIP) [104], and the RING-finger family E3 ligase SCF complexes such as Rbx1, Skp1, Cullins, F-box proteins [105,106] and Arkadia (also known as RNF111) [107,108] are mentioned here. In brief, SMURF1 promotes ubiquitylation of the R-SMADs SMAD1 and SMAD5 and indicates them for degradation [102]. SMURF2 polyubiquitylates SMAD2 and promotes degradation, whereas SMURF2 monoubiquitylates SMAD3, inhibiting the formation of SMAD3 complex [101,102]. NEDD4-2/NEDD4L ubiquitylates both SMAD2 and SMAD3, while CHIP, SCF (Skp1), and GSK3β only trigger SMAD3 ubiquitylation and degradation after phosphorylation [107]. In addition, Akadia triggers both phospho-SMAD2 and SMAD3 ubiquitylation and proteasomal degradation [108].
Monoubiquitylation also regulates the SMAD4-mediated signaling, and it is worth mentioning that SCFskp2 E3 ligase ubiquitylates SMAD4 at the MH2 domain [109], enabling SMAD4 mutations in cancer to be degraded. The tripartite motif-containing 33 E3 ligase (TRIM33/TIF1γ/Ectodermin) promotes the disruption of the SMAD complex by SMAD4 monoubiquitylation [110,111]. A deubiquitylating enzyme (DUB) can oppose the action of ubiquitylation on SMADs. For example, USP15 (ubiquitin-specific peptidase) removes the monoubiquitylation of SMAD3 and thereby enhances the access of SMAD complexes to its target promoter domains/complexes [112]. Further, DUB, ubiquitin-specific protease 9x (USP9X/FAM), can reverse the function of monoubiquitylation of SMAD4 at Lys519 and inhibit the association of SMAD4 with R-SMADs [113].
4.3. Acetylation, ADP-Ribosylation, and Sumoylation
In addition to phosphorylation and ubiquitylation, acetylation of R-SMADs marks the TGF-β-induced interaction of the transcription co-activators cAMP-response element-binding (CREB) protein (CBP) and p300 by acetyltransferase. SMAD2 is acetylated at the MH1 domain, whereas the MH2 domain of SMAD3 is primarily acetylated, and each acetylation event enhances SMAD-mediated transcription [114,115,116]. ADP-ribosylation is another type of PTM; thereby, one or more ADP-ribose moieties are added to arginine by ADP-ribosyl transferase to control cell signaling and various cellular processes. PRAP1 (ADP-ribose polymerase 1) induces ribosylation of SMAD3 in its MH1 domain and thereby dissociates SMAD3 from the SMAD3/SMAD4 complex. SMAD-mediated TGF-β responses are attenuated by the action PARP-1. Further, overexpression of PARP-1 promotes impaired SMAD3-mediated gene expression and EMT [117]. By SUMOylating, a small ubiquitin-like modifier (SUMO) polypeptide is covalently attached to the target protein. SUMO does not lead to the degradation of the proteins. It was found that SMAD3 was associated with a protein inhibitor of activated STATy (PIAS4, PIASy); SUMO E3 ligase associated with the nuclear matrix could suppress the activity of SMAD3 [118]. Thereby, PIAS4 regulates SMAD-mediated signals by a negative feedback loop. On the other hand, another SUMO E3 ligase, PIAS1 (an inhibitor of activated STAT1), has been identified to promote SMAD4 sumoylation and SMAD-mediated transcription [119].
Table 2 shows the list of inhibitors targeting ERK, PKC (protein kinase C), PKG (protein kinase G), CK1 (casein kinase 1), protein kinase PAK2, PAK4, and LKB1 (liver kinase B1) [120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139]. Others enzyme inhibitors against protein phosphatase, such as (PPM1A/PP2CA, SCP), ubiquitin ligase (NEDD4-2/NEDD4L, HSC70-interacting protein, CHIP, SCFskp1 SCFskp2,) histone acetyltransferase (p300/CREB), and methyltransferase (SET 7/9, SETDB1/ESET, m6 methyltransferase, PRAP1), are listed in Table 2 [140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163]. A study by Vena et al. [131] showed that CK1δ is overexpressed in human pancreatic and bladder epithelial cell lines, and the inhibition of CK1δ by application of SR-309 strongly promotes the antiproliferative effect and sensitizes them to gemcitabine treatments. Casein kinase I (CKI) family consists of different isoforms of alpha (α, β, γ1, γ2, γ3, δ, and ϵ), which have been implicated in critical regulatory roles by interacting with R-SMADs. Figure 3 shows that CK1g2 can promote PTMs of SMAD3 activity by deactivation and degradation of the SMAD3 mediated by ubiquitination and phosphorylation [90]. Considering the negative impact of SMAD-mediated signaling, overexpression of CK1δ might dysregulate R-SMAD activity in epithelial cancer cells and activate further SMAD activity to induce SMAD-mediated protumorigenic signals. Upregulation of deoxycytidine kinase (dCK) using the CK1δ inhibitor, SR-3029, may activate the R-SMADs degradation by phosphorylation and promote an antitumorigenic effect. Another potential inhibitor is FRAX597, a small-molecule ATP-competitive Group I PAK inhibitor. It reduces NF2-deficient schwannoma cell proliferation in culture. In vivo, the inhibitor shows potent anti-tumor activity [133]. In NF2-deficient Schwann cells, originating from mesenchymal stem cells, it might inhibit R-SMAD degradation and promote SMAD-mediated protumorigenic signals. It is considered that the application of FRAX597 further deactivates R-SMADs by inducing PTMs, resulting in anti-proliferative and anti-tumorigenic effects in NF2-deficient Schwann cells. Thus, the FRAX597 inhibitor shows significant potential for the treatment of neurofibromatosis and other cancers.

Table 2.
Inhibitors of SMAD-interacting enzymes controlling SMAD post-translational modification.
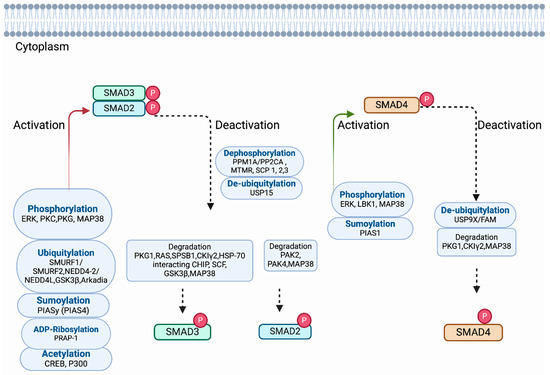
Figure 3.
Regulation of SMADs by post-translational modifications (PTMs). The complexes of R-SMADs and SMAD4 translocate from the cytoplasm to the nucleus and control the expression of target genes. Here, all the proteins are shown in the cytoplasm. The name of each post-translational modification and the associated proteins involved in each mechanism are designated in the box. A solid red arrow and green arrow indicate that R-SMADs and SMAD4 are modified by activation through post-translational modification. The black dashed arrow indicates the degradation of R-SMADs and SMAD4 and whether R-SMADs and SMAD4 are modified by deactivation and degradation.
5. Regulation of SMAD-Mediated Transcription
5.1. Histone Modification
Although SMAD complexes have a low binding affinity, they are guided by many DNA-binding transcription factors, co-factors, repressors, and chromatin modifiers to proficiently control the expression of target genes in a context-dependent manner [164,165,166,167,168,169,170,171]. Here, we discuss and illustrate some SMAD-interacting proteins that regulate SMAD-mediated transcription (Figure 4). It is reported that histone acetyltransferases (HATs) regulate the SMAD-dependent transcription by modifying histones and/or controlling the SMAD activity with chromatin modifiers. Evidence from different studies shows that histone acetyltransferase p300/CBP-associated factor (P/CAF) [172], general control of non-repressed protein 5 (GNC5) [173], switch/sucrose non-fermentable (SWI/SNF) remodeling complex [174], and histone lysine methyltransferase 1 (SETDB1/ESET) [175] are important in executing the function of HATS for SMAD-mediated transcription. The histone acetyltransferases CREB-binding protein (CBP) and p300 govern the SMAD3 transcriptional activity, whereas SMAD4 plays a role as a transcriptional co-activator to stabilize the SMAD complex [176]. P/CAF enhances SMAD3-mediated transactivation driven by the interaction of SMAD3 independently or in association with p300 and [172]. Another HAT, GCN5, plays a role as a co-activator of both TGF-β and BMP-induced SMADs mediated transcription induced by both TGF-β and BMP [173]. SMAD2-mediated transcription is regulated by histone 3 (H3) acetylation with the recruitment of p300 and SWI/SNF and, thereby, confers that chromatin remodeling is necessary for SMAD-mediated transcription [174]. On the other hand, a study by Du et al. [175] shows SMAD-3 mediated recruitment of a histone methaytransferase1, SETDB1/ESET, regulates snail gene expression, EMT, and cancer dissemination.
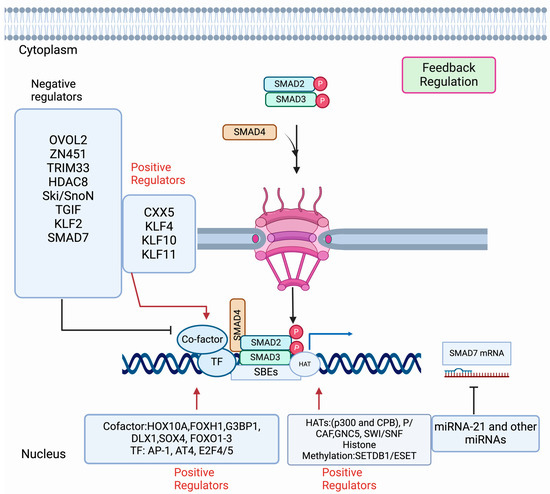
Figure 4.
Regulation of SMAD-mediated transcription. Activation of R-SMADs (SMAD2, SMAD3) by TGF-β receptor type I promotes the complex formation with R-SMADs-SMAD4. Then the trimeric complexes are imported in the nucleus to turn on or off the target genes. The name of the mechanism and the associated proteins are designated in each box. A solid blue arrow indicates SMAD-mediated transcription. The solid red arrow indicates the regulators that act positively on the SMAD-mediated transcription. The solid black inhibitory line shows the inhibition function mediated by the regulators. Diverse regulators [co-factors, transcription factors (TF), Histone acetyltransferase, methyltransferase, etc.] act on the activated SMAD complexes inside the nucleus and control SMAD-mediated transcription in a context-dependent manner.
SMAD-driven transcription acts as positive and negative regulators, thereby maintaining the feedback loop of TGF-β/SMAD signaling. The proteins that act as positive regulators are predominantly FOXA10 (Homeobox A10), FOXH1 (Forkhead pioneer factor), G3BO1 (Ras GTPase-activating protein SH3 domain-binding protein 1), AT4 (activating transcription factor 4), SOX4, DLX1, FOXO1-3, E2F4/5, AP-1, CXXC5, and KLF family member proteins (Figure 4).
HOXA10 binds to the SMAD complex and modulates the expression of snail and slug through m6A modification of mRNA and METTL3 expression by enhancing the SMAD signaling [177]. FOX1 can recruit SMAD2 to SMAD4 and assist in binding the SMAD complex with SMAD3:FOXH1 [178]. G3BP1 is another positive co-factor that acts as a novel binding partner to the SMAD complex by activating SMAD signaling and recruiting the SMAD2/SMAD4 complex [179]. The co-factor SOX4 can occupy many genomic loci with SMAD3 in a cell type-specific manner [180]. Forkhead-binding element (FHBE) resides within the SMAD binding region of the p21Cip1 promoter and associates with SMAD2/SMAD3/SMAD4 complex coprecipitated with FOXO1, FOXO2, and FOXO3 [181]. AP-1 and AT4 are downstream regulators of the SMAD complex and mTORC2 and control TGF-β/SMAD and mTOR/RAC1-RHOA pathways independently [182]. DLX1 can positively modulate the TGF-β/SMAD signal by interaction with SMAD4 localized in the nucleus upon TGF-β1 induction [183]. The transcription factor E2F4/5 controls SMAD-mediated transcription in a promoter-specific manner [184,185]. The complex of SMAD3 and the transcription factors E2F4/5, DP1, and corepressor p107 translocate to the nucleus and interact with SMAD4, distinguishing a combined SMAD-E2F site for arresting the cell cycle [185]. Another novel regulator and coordinator of TGF-β, BMP, and Wint signaling is CXXC5, which associates HDAC1 and competes for binding with SMAD complex in hepatocarcinoma cells. CXXC5 reduces the inhibitory effect of HDAC1, resulting in cycle arrest and apoptosis [186,187]. In this context, some Küppel-like factors (KFLs) are well known and can associate with SMADs and act as DNA-binding transcriptional regulators. There are 17 well-known KLFs that have highly conserved C-terminal DNA-binding domains with three C2H2 zinc finger motifs. Some of them play significant roles in feedback regulation in SMAD-mediated transcription. Among them, KLF4, which is up-regulated in vascular smooth muscle cells, recruits p300 and forms an active complex with SMAD2. Thus, KL4 induces the expression of SM22α and α-SMA [188,189]. KLF10 can facilitate multiple TGF-β-induced functions through the expression of SMAD2, inhibition of SMAD7 expression to control the proliferation of epithelial cells, and development of immune and bone cells [190]. Further, KLF11 associates with SMAD3 and enhances TGF-β-induced growth inhibition by repressing SMAD7 and Myc expression in epithelial and pancreatic cancer cells [191].
Among the negative regulators, zinc finger protein OVOL2, 451, histone-binding protein TRIM33 (also E3 ubiquitin ligase activity), HDAC8 (histone deacetylate 8), Ski/SnoN (Sloan Kettering Institute) and SnoN (Ski novel), TGIF, Sp1/KLF-like zinc-finger protein KLF1, and PAX2 are significant to control SMAD-mediated transcription in a cell and context-dependent manner (Figure 4). The zinc finger protein 451 inhibits the recruitment of p300 to the SMAD complex and represses SMAD-mediated transcription, resulting in the reduced H3 Lys9 acetylation of the promoters of target genes [192]. Further, the recruitment of TRIM33 to chromatin is mediated by SMAD4, which promotes histone modification upon binding of TRIM33 and SMAD2/SMAD3 complexes on the regulatory sequences of the target genes. This modification by histone allows the switching of the chromatin state from the poised to the active state and supports the negative feedback mechanism [193].
HDAC8 is class I histone deacetylase, a novel co-factor of the SMAD3/4 complex. A study by Tang et al. [194] shows that chromatin remodeling represses SIRT7 transcription by making a complex with HDAC8 and SMAD2/SMAD3. In contrast, the reduction of SIRT7 activates TGF-β by a feedback loop, which regulates the TGF-β/SMAD signal. The negative regulator OVOL2 induces the expression of SMAD7, thereby reducing the expression of SMAD4 and interrupting the complex formation by interfering with the complex formation between SMAD2/SMAD3 and SMAD4 [195]. Also, the negative regulators Ski and SnoN interact simultaneously with the R-SMADs and SMAD4 and disrupt the ability of the SMAD complexes to turn on the target genes [196,197]. Ski/SnoN actively recruits a transcriptional co-repressor complex containing N-CoR/HDAC to the targeted promoter by preventing the recruitment and binding of R-SMADs to p300/CBP. It has been shown that 5′TG3′-interacting factor (TGIF) represses TGF-β signaling [198] by binding with R-SMAD/SMAD4 complexes. Further, Guca et al. [199] showed that TGIFI-HD (homeodomain) binds to SMADs in a mutually exclusive manner. Thereby, it provides a transcriptional repression system in a context-dependent manner. Among the KLF family, KLF2 employs a negative feedback loop on TGF-β/SMAD signaling by inducing SMAD7 transcription [200]. A negative feedback loop is maintained to regulate TGF-β/SMAD signaling by the action of I-SMADs. It is found that in the absence of a ligand, TGF-β promotes nuclear accumulation of I-SMADs from the nucleus to cytoplasm and promotes SMAD7 mRNA and, thereby, creates a negative feedback loop that firmly controls TGF-β SMAD signaling [201,202]. Enhanced SMAD7 methylation by SET9-mediated TGF-β/SMAD signaling promotes its association with the E3 ubiquitin ligase Arkadia and enhances its ubiquitylation and degradation in lung fibroblast. Pharmacological inhibition or depletion of Set9 showed that high SMAD7 protein levels inhibited the expression of TGF-β/SMAD-mediated genes encoding extracellular matrix components [203].
5.2. Regulation of SMAD-Mediated Transcriptional Activity Post-Transcriptionally
At the post-transcriptional level, SMAD-mediated transcription is regulated by RNA-binding proteins (RBPs), microRNA (miRNA), and non-coding RNAs (ncRNAs). A study by Tripathi et al. [204] shows that the interaction of phosphorylated T179 of SMAD3a with PCBP1 (RNA-binding protein) promotes alternative cancer stem cell marker CD44 splicing. SMAD3 and PCBP1 can cooperate in the variable regions of exons for CD44 pre-mRNA and modulate spliceosome assembly. As a result, mesenchymal isoform CD44s is expressed over epithelial isoform CD44s. Thus, alternative splicing by SMAD3 plays an important role in tumorigenesis. Interestingly, R-SMADs are found to influence miRNA expression, processing, and maturation of 44 types of miRNAs. R-SMADs can bind to the stem region of miRNA at a consensus sequence and recruit p68 (DDX5), an RNA helicase component, DROSHA (DROSHA/DGCR8/RNA) in a ligand-dependent manner. Thus, the post-transcriptional modification induced by TGF-β and BMP signaling can drive increased expression of mature miRNA-21 after processing of primary transcripts of miRNA-21 (pri-miRNA-21) into precursor miRNA-21 (pre-miRNA-21) [205,206]. In addition to miRNA-21, other miRNAs are directly involved in the feedback regulation of SMAD-mediated signaling, as reported by Yan et al. [207]. Additionally, long-coding RNAs (lncRNAs) are recognized as intermediaries of the TGF-β response. A comprehensive analysis by Adylova et al. [208] shows that the different lncRNAs can play a role in positive and negative regulation of TGF-β/SMAD signaling in different cancer cells. Another post-transcriptional negative feedback has been described by Bertero et al. [209] in human pluripotent stem cells in which R-SMADs can associate with the m6A methyltransferase complex METTL3-METTL4-WTAP. In the nascent transcripts, N6-adenosine methylation on the RNA occurs by the action of the m6A methyltransferase, resulting in destabilizing and degrading the transcripts [209].
Table 3 shows the list of inhibitors targeting histone acetyltransferases (GNC5/PCAF, SWI/SNF, histone lysine methyltransferase (SETDB1/ESET), small GTPases (RAS), histone deacetylases (HDAC1, HDAC8), and methyltransferase (m6A methyltransferase, SET (7/9) methyltransferase) [123,160,210,211,212,213,214,215,216,217,218,219,220,221,222,223,224,225,226,227,228,229,230,231,232,233,234]. Some of the potential inhibitors are highlighted here. The application of microRNAs (miRNAs) for cancer therapy and radiosensitivity in tumors has achieved significant consideration. A study by Shao et al. [217] showed that miRNA-621 could enhance the radiosensitivity of hepatocellular carcinoma (HCC) through direct inhibition of SETDB1 and targeting the 3′UTR of SETDB1. It was reported that miRNA-621 and/or the SETDB1 axis activated the p53-signaling pathway and advanced the radiosensitivity of HCC cells. SETDB1/ESET regulates SMAD-mediated transcription to control snail gene expression in epithelial or mesenchymal cells in a context-dependent manner. It is supported that snail may bind directly to the DNA-binding domain of p53 and reduce the p53 tumor-suppressive function. Since the expression of miRNA-621 and SETDB1 are negatively correlated in HCC tissues, miRNA-621 might enhance the radiosensitivity and active p53 signaling pathway in HCC cells by inhibiting SETDB1 expression. Thus, mi-RNA-621 and/or SETDB1 in epithelial cells of HCC can be potentially used as a novel therapeutic target [217].

Table 3.
Inhibitors of SMAD-interacting enzyme in transcription and post-transcription.
STM2457 is the first bioavailable small-molecule inhibitor of METTL3 (a regulator of m6A methyltransferase) that affects the inhibition of catalytic activity and upregulation of METTL3 increasing PD-L1 and reduction of tumor progression in NSCLC (non-small-cell lung cancer) [233]. Upregulation of METTL3 induced by STM2457 enhances the interaction of METTL3 with the translation initiation machinery to make more circularized mRNA of PDL-1. In this case, STM2457 may inhibit R-SMADs from associating with the m6A methyltransferase complex METTL3 to destabilize and degrade the nascent transcripts of PDL-1. Thus, STM2457 is considered a potential suppressor that provides an inhibitory effect in epithelial cells of NSCL.
The SET domain containing lysine methyltransferase (SETD7/9) is involved in various disease-related signaling pathways with a broad group of substrates. Methyltransferase activity of human SETD7 is inhibited by a selective inhibitor, (R)-PFI-2. It was found that the Hippo pathway was modulated by (R)-PFI-2 with increasing nuclear YAP and YAP-mediated gene expression in epithelial cancer cells (MCF7) and murine embryonic fibroblasts [234]. How (R)-PFI-2 works is not well defined, but a crosstalk between TGF-β/SMAD and Hippo signaling suggests that the binding of YAP to SMAD7 may modulate TGF-β signaling. SMAD7 methylation by SET (7/9)-mediated TGF-β/SMAD signaling might promote its association with E3 ubiquitin ligase, thereby enhancing its ubiquitylation and degradation. YAP can associate with the complexes of R-SMADs-SMAD4 and drive their sub-cellular localization and transcription in a context-dependent manner. Thus, (R)-PFI-2 and related compounds can be valuable inhibitors to target methyltransferases.
6. Non-SMAD, Non-Canonical TGF-β Pathway Control
6.1. ERK/MAP Kinase Signaling
The Ras/Raf/MAPK (MEK)/ERK pathway is one of the important signaling cascades and contributes an important role in tumor cell survival and progression. TGF-β activates the ERK-MAP kinase pathway by direct phosphorylation of ShcA, which subsequently activates downstream signals (Figure 5A). A study in both Mv1Lu mink lung epithelial cells and mouse fibroblasts by Lee et al. [235] showed that phosphorylation of ShcA on tyrosine results in the initiation of a docking site for the downstream mediators Grb2 and Sos, which further activate Ras GTPase, Raf, MEK (MAP kinase/ERK kinase), and EK1/2 kinase. In conjugation with SMADs, Erk regulates the transcription of target genes through the downstream transcription factors and regulates EMT. ERK activation in human keratinocytes (HaCaT) is initiated by the TGF-β receptor complexes, which are localized in lipid rafts. Clathrin-dependent endocytosis of TGF-β receptor complexes initiates SMAD activation, SMAD-mediated transcription, and TGF-β/SMAD-directed epithelial cell plasticity [236].
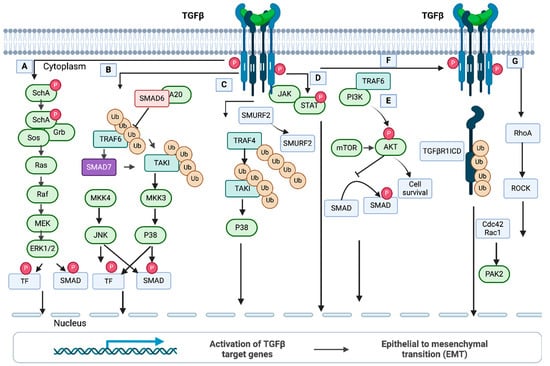
Figure 5.
Non-SMAD, non-canonical TGF-β pathways. (A) ERK/MAP kinase pathway. TGF-β activates the ERK/MAP kinase pathway by direct phosphorylation of ShcA, which subsequently activates downstream signals. In conjugation with SMADs, Erk regulates target gene expression and EMT via the downstream TFs (transcription factors). (B) JNK and p38 MAP kinase pathway. The Tumor Necrosis Factor Receptor-Associated Factor 6 (TRAF6) and TGF-β complexes interact after activation of the receptors. TRAF6 is then induced by an autoubiquitylation and subsequently stimulates TAKI through polyubiquitylation. This event can lead to activation of the p38 MAP kinase pathway. (C) The MAP kinase pathway via TRAF4 (Tumor Necrosis Factor Receptor-Associated Factor 4). TRAF4 is auto-ubiquitylated upon ligand binding and is recruited to the TGF-β receptor complex. (D) JAK/STAT (the Janus Kinase/Signal Transducer and Activator of Transcription) pathway. Phosphorylation and activation of STAT is initiated by JAK, which interacts with TGF-β receptor type I. (E) PI3/AKT/mTOR pathway. TRAF6 polyubiquitylates the regulatory subunits of PI3K, p85α. Then, a complex is formed between the TGF-β receptor type I and p85α, resulting in activation of PI3K and AKT. Phosphorylation of AKT prevents SMAD3-mediated signaling. (F) TGF-β type I receptor intracellular domain signaling. The interaction of TRF6 and TGF-β receptor I results in polyubiquitination of TGF-β receptor I at Lys63 and subsequent degradation by TNF-alpha converting enzyme (TACE). TACE is not shown here. The newly formed intracellular domain (ICD) of TGFβ receptor I, named TGFβRI ICD, associates with the transcriptional factor to activate genes. (G) Rho-like GTPase pathway. Activation of Rho GTPase is promoted by both SMAD and non-SMAD-mediated signaling. After stimulation, Rho-like proteins Cdc2 and Rac1 are activated, which drive actin reorganization through signals from PAK2. This figure has been adapted from Tzavlaki and Moustakas, 2020 [43].
6.2. JNK and p38 MAP Kinase Signaling
Jun N-terminal kinases (JNKs) and p38 mitogen-activated protein kinases (MAPKs) significantly coordinate with many signaling mechanisms associated with stresses. In addition, different cellular functions are regulated by the JNK and p38 pathways. In brief, TRAF6 (Tumor Necrosis Factor Associated Receptor-Associated Factor 6) interacts with the TGF-β receptor complex once receptors are activated by ligand binding. An autoubiquitylation modification induces TRAF6, which activates TAKI by polyubiquitylation at Lys63TAK1 and initiates the p38 MAPK pathway (Figure 5B) [237,238]. Importantly, TAK1 is a negative regulator of canonical TGF-β signaling and promotes R-SMAD phosphorylation at the linker region in neural crest-derived mesenchymal cells [239]. In this pathway, I-SMADs play significant roles in regulating p38 MAP kinase signaling. It was found that SMAD7 in prostate cancer cells can act as a scaffolding protein for p38 and its upstream kinases [240]. Also, in AML-12 mouse liver cells and primary hepatocytes, SMAD6 acts as a negative regulator and eliminates K63-linked polyubiquitylation of TRAF6 by recruiting the A20 DUB enzyme induced by TGF-β1 [241].
Another ligase, E3, and TRAF4 activate the MAP kinase pathway (Figure 5C). In this pathway, TRAF4 is auto-ubiquitylated upon ligand binding and is recruited to the TGF-β receptor complex. In breast cancer cells, TRAF4 activates TAKI via polyubiquitylation, which results in the activation of the p38 pathway. Subsequently, SMURF2 is degraded through polyubiquitylation by TRAF4, thereby maintaining the stability of the TGF-β1 receptor [242].
6.3. JAK-STAT Signaling
JAK/STAT (the Janus Kinase Signal Transducer and Activator of Transcription) pathway is a prime regulator of cell function. (Figure 5D). Jak/STAT-mediated downstream effects may vary and drive hematopoiesis, tissue repair, inflammation, immune surveillance, apoptosis, and adipogenesis [243]. A study by Tang et al. [244] shows that JAK1 is a constitutive TGF-βRI that is essentially involved in STAT phosphorylation within a short period after TGF-β stimulation. Once SMADs are activated, another phosphorylation of STAT is started for de novo protein synthesis. Thus, the non-SMAD JAK1/STAT pathway in hepatic stellate cells is essential for the expression of TGF-β subset genes.
6.4. PI3/AKT/mTOR Signaling
The two pathways, PI3K (Phosphatidylinositol-3-kinsae)/Akt and Mammalian Target of Rapamycin (mTOR), play roles in many different cellular functions (Figure 5E). In epithelial cells, the TGF-β type I receptor (TGF-βRI) can bind PI3K and modulate the kinase activity [245]. Further, a study by Hamidi et al. [246] demonstrated that in prostate cancer cells, TRAF6 can polyubiquitylate p85α, a regulatory subunit of PI3K, and make a complex between p85α and TGF-βRI to activate PI3K and AKT. Moreover, Lamouille et al. [247,248] showed that TGF-β can induce mTORC2 (Mammalian Target of Rapamycin Complex 2), which further phosphorylates and activates AKT, resulting in cell size changes and epithelial cell progression through EMT.
6.5. TGF-β Type I Receptor (TGF-βRI) Intracellular Domain Signaling
TGF-βRI intracellular domain signaling (Figure 5F) promotes TGF-β-mediated tumor invasion. It is reported by Mu et al. [249] that in prostate cancer cells, TGF-β uses TRAF6, resulting in Lys63-linked polyubiquitination of TGF-βRI and promoting cleavage of the extracellular domain of TGF-βRI by TACE (TNF-alpha converting enzyme). The newly formed intracellular domain (ICD) of TGF-βRI can bind with p300 for transcription of genes, thereby driving tumor invasion by induction of SNAIL and MM2 [250]. TRAF6 can polyubiquitylate a membrane-bound protein, PS-1 (membrane-embedded protease presenilin-1), to initiate a proteolytic cleavage on TGF-βRI. Thereby, the ICD (the complete intracellular domain) of the receptor is formed to assist the translocation of TGF-βRI-ICD to the nucleus [251]. In the nucleus, TGF-βRI-ICD binds to the promoter and turns on the transcription of the genes encoding TGF-βRI. In prostate cancer, TRAF-6-mediated Lys6-linked ubiquitination of the TGF-βRI intracellular domain is important for the modulation of TGF-β and regulation of other genes controlling the cell cycle, proliferation, differentiation, and migration [251].
6.6. Rho-(like) GTPase Signaling
Rho GTPases encompass a branch of the Ras superfamily with 22 genes in humans, and among them, RhoA, Rac1, and Cdc42 are the best exemplified (Figure 5G). They have a significant role in many cellular processes, mainly in cell morphology regulation, cell adhesion, and cytokinesis production. In many studies of mammalian cells, the constitutive and dominant negative mutants were used to examine the function of Rho GTPases. Bhowmik et al. [252] showed that in epithelial cells, the RhoA-dependent signaling pathway stimulates the formation of stress fibers induced by TGF-β. As a result, epithelial cells can transform into cells with mesenchymal characteristics such as increased N-cadherin expression and motility with the loss of E-cadherin (markers of TGF-β-induced EMT). TGF-β induction can be reduced by blocking RhoA or its downstream target, p160Rock, expressed by the dominant-negative mutants. Another study by Edlund et al. [253] showed that TGF-β1-treated human prostate carcinoma cells (PC-3U) can promptly form lamellipodia by rearranging the actin filament system. This response was independent of SMAD for a short time, but it requires Rho GTPases Cdc42 signaling for the long term.
A regulator of epithelial cell polarity, Par-6, can negatively control Rho-GTPase signaling by cooperating with TGF-β [254]. Par6 is associated with the TGF-βRI, phosphorylated by TGF-βRII, and thereby it recruits SMURF1 ubiquitin ligase (Figure 5G). Rapid degradation of RhoA GTPase is initiated through polyubiquitylation by SMURF1. This introduces the loss of a tight junction of epithelial cells. Further, Wilkes et al. [255] showed that signals from the TGF-β receptor activate PAK2 (STE20 homolog) in mammalian cells. PAK2 activation was observed in fibroblasts (not in epithelial cell cultures) mediated by signals from SMAD2 and/or SMAD3. In fibroblasts, Rac1 and Cdc42 might regulate PAK2 activity induced by TGF-β. However, targeting PAK2 by morpholino antisense oligonucleotides and dominant negative PAK2 can prevent the morphological features. Thus, PAK2 is considered a novel SMAD-independent pathway that distinguishes TGF-β signaling in fibroblasts and epithelial cells.
6.7. Crosstalk between SMAD and Other Signaling Pathway Molecules
The intracellular signaling network, the SMAD pathway, and the other pathways are connected to develop crosstalk. The crosstalk within the pathways plays an important role in the regulation of biological processes. The crosstalk can occur at different levels by altering signaling components, transcriptional modification, and chromatin modification or by direct interactions between intracellular signaling components with SMADs. Here, we briefly mention the crosstalk of SMADS with Yes-associated proteins with a PDZ-binding motif (YAP/TAZ) and interactions with other proteins, such as TRAP-1, km23-1, and PKA. The binding of YAP to SMAD7 by TGF-β and Hippo signaling was the first reported crosstalk that resulted in the increased inhibition of TGF-β signaling [256]. YAP can bind to the PpxY motif and phosphorylate the SMAD1 linker region by CDK9 in mammalian cells [257]. Neural cell (mESC) differentiation by BMP signal is suppressed by YAP-SMAD1 binding and further SMAD-1-dependent transcription. TAZ and YAP can associate with a heteromeric complex of R-SMAD-SMAD4 mediated by TGF-β signaling [258]. In contrast, another study showed that SMAD nuclear localization in response to TGF-β is not dependent on the levels of YAP or TAZ [259]. Also, it was found that the localization of YAP/TAZ can overcome cell cycle arrest stimulated by TGF-β and promote a pro-tumorigenic transcriptional program [260]. It is reported that TGF-β/SMAD/YAP/TAZ crosstalk also plays an important role in cell differentiation, proliferation, and fibrogenesis in a context-dependent manner [259,260].
The protein kinase A (PKA) signaling pathway is described by Wang et al. [261] in mesangial cells. PKA activates TGF-β stimulated cAMP response element-binding protein phosphorylation and expression of fibronectin. A study by Yang et al. [262] reported that PKA is independent of cAMP by aiding an interaction of PKA holoenzyme subunits with activated SMAD2/SMAD3. The interaction domains of SMAD4 and PKA-R and their functional roles are defined by the study. It was shown that amino acids 290–300 of the SMAD4 linker region are critical for the specific interaction of SMAD4 and PKA-R for the regulation of TGF-β-mediated cellular functions (e.g., PKA activity, CREB phosphorylation, induction of p21, and inhibition of growth). In pancreatic cancer cells, SMAD-PKA plays a role in TGF-β-induced EMT and tumor growth.
TLP is a TRAP-1-like protein that acts as an adaptor protein that mediates the interaction with the TGF-β type II receptor and the downstream effector SMADs. Felici et al. [263] proposed through a study that SMAD2 and SMAD3 might be balanced by TLP signaling through the localization of SMAD4 intracellularly. Thus, the specificity of TGF-β transcriptional responses can occur in a context-dependent manner.
Table 4 shows the inhibitors of SMAD-interacting enzymes (Jun N-terminal kinase (JNK), p38 MAP kinase, AKT, TRAF6, RhoA, PKA) that control non-SMAD noncanonical TGF-β pathways [264,265,266,267,268,269,270,271,272,273,274,275,276,277,278,279,280,281,282,283,284,285,286,287,288]. A potential inhibitor, SP0016125, targets c-Jun N-terminal kinase (JNK) and stimulates TGF-β-induced apoptosis of the RBE human cholangiocarcinoma cell line [264]. The result showed that SP0016125 increased the TGF-β-induced SMAD2 and SMAD3 phosphorylation, which promoted TGF-β1-induced transcriptional response and apoptosis in RBE cells. Depletion of SMAD4 reduced the effect of SP600125 on the transcriptional response and apoptosis. Further, TGF-β-induced apoptosis was abolished using the pan-caspase inhibitor Z-VAD-fmk. These findings indicate that SP600125 enhances TGF-β-induced apoptosis of RBE cells by promoting SMAD-dependent caspase activation through a SMAD-dependent pathway. Further, a study by Lu et al. [265] showed a novel pro-apoptotic role in combination with dihydroartemisinin (DHA). In human lung adenocarcinoma cells (ASTC-a-1) induced by DHA, SP600125 synergistically enhances apoptosis through Bax translocation and other intrinsic apoptotic pathways like caspase activation and the mitochondrial pathway. These findings suggest that SP600125 may enhance TGF-β-induced apoptosis in lung adenocarcinoma through a SMAD-dependent caspase pathway. Therefore, SP0016125 is considered a novel therapy for the treatment of cholangiocarcinoma and lung adenocarcinoma epithelial or mesenchymal cells that express JNK.

Table 4.
Inhibitors of SMAD-interacting enzymes that control noncanonical TGF-β pathways.
A potential inhibitor named LY2228820 (an imidazole derivative) is a selective ATP-competitive inhibitor of the α-and β-isoforms of P38α mitogen-activated protein kinase (p38 MAPK) [274]. In vivo cancer models of breast, ovarian, lung, glioma, and myeloma showed that tumor growth was potentially delayed with LY2228820 treatment. Since p38α regulates cytokine (TNF, IL-Iβ, IL-6, IL-8, etc.) production in TME, it is considered that TRAF6 expression in multiple cancer cell lines (MDA-MB-468, OPM-2, A549, A2780) may also control cytokine production. Further, it is hypothesized that auto-ubiquitylation of TRAF6 upon ligand binding might result in recruiting the TGF-β receptor complex, which in turn actives TGF-β/SMAD-mediated target genes to stimulate EMT. Therefore, LY2228820, a p38 MAPK inhibitor, has been optimized for various purposes such as potency, selectivity, bioavailability, and efficacy in animal models of human cancer.
Another important inhibitor of the non-SMAD, noncanonical TGF-β pathway is PKI protein targeting PKA, a well-known regulator of physiological and oncogenic functions. A study by Hoy et al. [289] found that PKI can modulate tumor growth by a molecular switch to drive GPCR-Gαs-CAMP signaling toward EPAC-RAP1 and MAPK. Amplification of PKIA is common in prostate cancer cells, and depletion of PIKA showed reduced migration and tumor growth. To understand the mechanism of PKIA, it is hypothesized that the expression of PIKA and EPAC may modulate MAPK or ERK/MAPK signaling. In conjugation of SMADs, Erk may regulate downstream gene expression for EMT in prostate cancer cells.
7. TGF-β/SMAD Mediated Progression in Solid Tumors
The TGF-β/SMAD signaling pathway plays a dual role in cancer progression. In addition to promoting apoptosis and cell cycle arrest in epithelial cells, TGF-β/SMAD and non-SMAD signaling enhances EMT, cell migration, invasion, angiogenesis, and stemness. TGF-β/SMAD exhibits a tumor suppressor phenotype in epithelial cells at the early stages of tumorigenesis. In contrast, at the later stages, TGF-β/SMAD signals drive oncogenesis and metastasis in mesenchymal properties of cancer cells (Figure 6). Here, we show SMAD-dependent canonical and non-canonical signals mediated by cancer cells with epithelial, EMT, hybrid/partial EMT, and mesenchymal properties associated with cancer progression and metastasis.
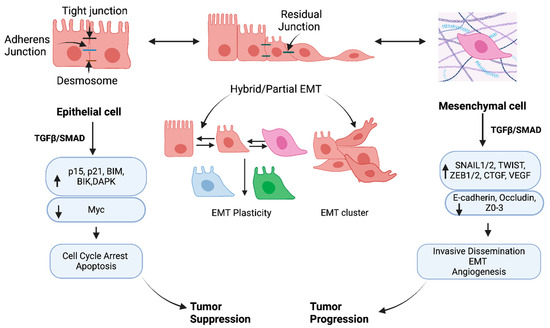
Figure 6.
TGF-β/SMAD mediated tumor suppression and tumor progression in epithelial and mesenchymal cancer cells. In normal epithelium and pre-malignant cells, TGF-β/SMAD promotes cell cycle arrest and apoptosis via induction of CDK inhibitors (G1 Arrest activating Cyclin-Dependent Kinase) p21 and p15, B-cell lymphoma-2 (BCL-2) family members, BIM (BCL2L11) death-associated kinase (DAPK), and BH3-protein BIK and suppression of c-Myc. At the later stage, TGF-β/SMAD-mediated induction of SNAIL1/2, TWIST, and ZEB1/2 is associated with EMT and tumor progression in mesenchymal cells. The intermediate stage is hybrid/partial EMT, having intermediate polarity and loose intercellular junctions. The partial EMT shows plasticity, which stimulates the cell differentiation and clusters of cells for collective migration. This figure has been adapted from Huang et al., 2022 [289].
7.1. Epithelial Cells
The typical characteristics of epithelial cells are described and summarized by Huang et al. [289]. Briefly, epithelial cells comprise strong apical and basal polarity with plasma membranes placed toward and away from the lumen, respectively. Different proteins are found in each membrane. Proteins in the membrane assist in the transportation and localization of targeted molecules to specific cellular regions with various activities (Figure 6). Both SMAD-dependent canonical and non-canonical pathways predominantly regulate non-cancerous epithelial cells at the pre-malignant stage by cell cycle progression through CDK (G1-Arrest Activating Cyclin-Dependent Kinase) inhibitors p15 and p21. Further, the downregulation of an important oncogene, c-Myc, drives the proliferation and inhibits the transcriptional activity of p15 and p21 [3]. Importantly, apoptosis is initiated by the signals modulating the expression of B-cell lymphoma-2 (BCL-2) family members, BIM (BCL2L11), FAS (death receptor fibroblast-associated antigen (FAS)), DAPK (death-associated kinase), BH3-protein BIK, and caspases, which induce both intrinsic and extrinsic apoptosis [290,291,292]. Further, tumor suppression mediated by TGF-β/SMAD signaling has been demonstrated in some solid tumors like breast cancer, prostate cancer, melanoma, and colon cancer [293]. The non-canonical TGF-β/SMAD is linked to p38 MAPK and caspase-8-dependent programmed cell death, which exhibits a tumor suppressor role by inducing apoptosis [294]. Further, to drive tumor cell death by activating programmed cell death, TGF-β/SMAD promotes the regulation of immune cell function [295,296].
7.2. Cancer Cells with EMT, and Hybrid/Partial EMT
The transition of cancer cells is dynamic, allowing cells to go from epithelial to mesenchymal states and vice versa. A change from epithelium to the mesenchymal state is referred to as EMT (epithelial–mesenchymal transition), which allows cells to gain migratory properties by modifying the adhesion molecules expressed by the cells. The reverse process is MET (mesenchymal–epithelial transition), which is associated with loss of migratory properties rather than adopting an apicobasal polarization [4]. Thus, beyond epithelial phenotype, various EMTs can impact cell behaviors, physiology, and ecology, allowing the transition of different phenotypes (Figure 6). The EMT is implemented for pleiotropic signaling through canonical and non-canonical SMAD pathways. Thereby, specific transcription factors (TFs) named EMT-TFs (e.g., SNAIL, ZEB, TWIST) are expressed to regulate EMT and MET.
Along with transcription, miRNA, post-translational and epigenetic regulators mediate EMT in cancer progression [289]. EMT has been established as a spectrum rather than a linear process supported by recent studies [5,6,7] (Figure 6). A study by Pastushenko et al. [6] used different markers, CD106, CD51, and CD61, showing that the different states of the EMT spectrum can form a “hybrid” (Figure 6). A single-cell analysis found that partial/hybrid EMT expresses SNAIL1/2, ZEB1/2, and TWIST1/2 during mouse organogenesis in a manner similar to epithelial gene expression [297]. Further, using the single-RNA sequencing technique with primary and metastatic head and neck squamous cells, it was shown that that cell contained a gene signature of partial EMT [298]. Partial EMT is also highly aggressive and invasive since the cells present several classical features of EMT. Expression of vimentin (VIM), TGF-β1, and extracellular matrix genes are worth mentioning. Although the EMT-TF expression was low, transcription of epithelial genes was still maintained within the partial EMT. The EMT phenotype can be variable, such as pancreatic ductal adenocarcinoma resulting from adjusting epithelial junction proteins that showed no change in expression, contrasting the common repression of epithelial characteristics of cells [299]. Moreover, many carcinoma cells, such as breast and colorectal cancer cells, can utilize this partial/hybrid EMT program to make a cluster of cells contrasting the single-cell pattern, which is connected to traditionally defined EMT mechanisms [299].
In addition, EndMT (endothelial–mesenchymal transition) shows similarity to EMT in the context of molecular processes and phenotypes. The loss of endothelial junctions, EMT-TF activation, and upregulation of mesenchymal markers push EndMT formation. Endothelial cells downregulate the VE (vascular endothelial cadherin), where cell-type-specific changes distinguish EndMT from EMT [300].
7.3. Cancer Cells with Mesenchymal Characteristics
Upon EMT, cells repress their epithelial phenotypes and gain mesenchymal and invasive properties during cancer progression. The hallmarks of EMT are increased N-cadherin and VIM expression with decreased expression of E-cadherin, ZO1, and desmoplakin [301,302,303]. In cancer cells with mesenchyme morphology, the major EMT-TF are zinc finger binding transcription factors, including SNAIL1/2, E-box binding homology frame factors (ZEB1/2), and BHLH (basic Helix–Loop–Helix) factors (TWIST1/2) [304]. Both SNAIL1 and SNAIL2 drive tumorigenesis induced by EMT. During the upregulation of mesenchymal phenotypic markers, including VIM and fibronectin, SNAIL1 directly inhibits TJ formation by decreasing epithelial markers like E-cadherin and claudin expression. E-cadherin positively correlates to patient survival, whereas the overexpression of MMPs provides aggressiveness of tumors [305]. On the other hand, SNAIL2 promotes the loss of cell adhesion and polarity by decreasing e-calmodulin levels, thereby influencing migration and metastasis in breast and ovarian cancer [306,307,308].
In tumor stem cells, ZEB1/2 expression in epithelial cells causes EMT and mesenchymal phenotypes with invasiveness, metastatic, and dedifferentiation potential [309]. In vivo study of pancreatic cancer supports that ZEB1 is essential for tumor invasion and metastasis [310]. Collective data show that ZEB1/2 expression in breast, colorectal, and pancreatic cancers affects poor patient outcomes [311,312,313,314].
Like SNAIL, TWIST1 can suppress the expression of E-cadherin and promote N-cadherin expression, which affects cell adhesion and cell motility [315,316]. Tumor invasion and metastasis are driven by the overexpression of TWIST [317]. TWIST can be activated by various signals, such as HIF-1α (hypoxia-inducible factor-1α), during the progression of EMT. In a hypoxic condition, activation of TWIST by HIF-1α allows the cell to disseminate with metastatic potential [315]. Further, in a mouse model of squamous cell carcinoma, it was found that TWIST can facilitate the cancer cells to undergo EMT and dissemination in the circulation [318].
7.4. Cancer Cells with Stemness Characteristics
Cancer cells have a group of tumor-initiating cells known as cancer stem cells (CSCs), which can differentiate, renew, and generate tumor heterogeneity within the populations of cells [319]. CSCs studied in breast cancer models revealed that EMT correlates with CSC generation by repressing epithelial and mesenchymal properties [320]. It was reported that the expression of CSC and EMT markers were observed in mammary epithelial and carcinoma cells by the direct induction of TGF-β/SMAD signaling, which promotes mammosphere, soft agar colony, and tumor formation [321]. Also, it was shown that autocrine TGF-β/SMAD signaling is crucial within the subpopulation of immortalized breast epithelial cells to maintain its mesenchymal phenotype and tumorigenicity [321]. It is suggested that in this system, BMP signaling may or may not antagonize TGF-β-induced EMT and CSC generation [322]. Thus, enhanced TGF-β/SMAD signaling can promote EMT and CSC properties in cancer cells, further allowing CSC in cancer cell invasion and dissemination.
7.5. Cancer Dissemination and Metastasis
Tumor cells interact with ECM components (collagen, fibronectin, laminin) and cells in the tumor stroma in vivo. In TME, TGF-β/SMAD plays significant roles in cancer cells driven by autonomous tumor cell signaling. Cancer metastasis is promoted in stromal fibroblast and mesenchymal stem cells stimulated by TGF-β/SMAD signaling [8,9,10,323]. Therefore, ECM and stroma are the primary sites for cancer dissemination and metastasis, which are initiated through invasion. Further, intravasation into the blood circulation, extravasation at distant sites, and adaptation at a secondary site within a new microenvironment proceed [324].
Epithelial plasticity plays a vital role in invasion by carcinoma cells and further promotes their dissemination and metastasis [325,326]. Mesenchymal gene expression is often increased in circulating tumor cells, which disseminate through individual as well as collective cell migration [327]. However, many carcinoma cells, such as breast and colorectal cancer cells, can utilize partial/hybrid EMT programs that favor migration as clusters over signal cell migration defined by traditional EMT migration [301]. Interestingly, leading cells in a cluster with mesenchymal characteristics boost invasion, whereas most stalking cells are hybrid/partial EMT or remain epithelial [328]. At the invasive edges, cells with EMT plasticity respond to TGF-β/SMAD signaling to facilitate dissemination through blood vessels or lymphatics [326]. In squamous cell carcinoma, SNAIL induces the expression of claudin-11, a tight junction protein, and advocates cell migration as a tumor cell cluster [278].
Reversible EMT is supported by a model in which transient expression of EMT transcription factors affects the reversion of EMT and metastatic colonization [11,12,13]. TGF-β/SMAD signaling, which stimulates EMT and dissemination, needs to be repressed for metastatic reversal to epithelial morphology [323]. Breast cancer cells can switch from cohesive to single-cell motility mediated by localized and reversible TGF-β/SMAD. To examine SMAD localization in breast cancer progression, Giampieri et al. [323] demonstrated that TGF-β is activated locally and transiently in a motile cell. A transcriptional program involving several factors (SMAD4, NEED9, EGFR, RhoC) can actively modulate cell motility from cohesive to a single cell. Inhibition of signaling can prevent single-cell motility, but collective cell migration was not inhibited. However, TGF-β/SMAD signals can stabilize mesenchymal phenotypes within the tumor cells and are not shown as supportive for leading MET.
A study by Katsuno et al. [329] showed that the prolonged TGF-β/SMAD signals could promote EMT in epithelial cancer cells. Short exposure to TGF-β/SMAD induces the opposite effect and drives MET for colonization in the lung. Moreover, it was shown that the prolonged TGF-β/SMAD signals further modulate mTOR signaling, which contributes to cancer cell stemness and drug resistance. Therefore, it is important to either short exposure of TGF-β/SMAD signals or other signals to balance between TGF-β signaling and switching of EMT to MET. Another study showed that EMT reversed during migration, not in circulation. This study observed MET that was formed after enormous cycles of cell division to reach a secondary site during colonization [11]. Furthermore, it was found that the hybrid EMT has high tumor-initiating and self-renewal capacity in primary cancer cells, especially in breast and prostate cancer [330,331]. Therefore, it is suggested that EMT plasticity and stemness can play a role in the metastatic colonization of tumor cells. Future studies are required to understand the mechanism of EMT plasticity in cancer cell progression, dissemination, and metastasis.
Dormancy is another stage of the tumor cells that allows the cell to be dormant in an arrest phase at primary or secondary sites [332]. A recent review by Fares et al. [331] described that a delayed adaption of disseminating cancer cells (either single invading or cluster cells in circulation) to their secondary at a new microenvironment causes dormancy. TGF-β/SMAD signals induce the tumor suppressor gene DEC2 (chondrocyte 2), which assists cells in entering a quiescence state by inhibiting CDK4 and activating p27.
Tumor dormancy is maintained by the intracellular signals from the two important SMAD-independent pathways, RAS-MEK-ERK/MAK and PI3K-AKT [333].
Some studies [334,335] show that EMT and cancer cell dissemination can occur at the pre-malignant or epithelial stage of tumorigenesis, contrasting with the concept of whether mesenchymal cells aid metastasis in the later stage of tumorigenesis. Before primary tumors are detected, invasive cells with EMT characteristics are detected in models of various solid tumors, such as in pancreas, lung, and breast cancer cells. The evidence supports that such cells can disseminate and then undergo dormancy. Further reactivation of dormancy allows them to form metastatic tumors
8. Conclusions
This review highlights the comprehensive understanding of the functions of SMADs and SMAD-interacting and/or SMAD-signal-modulating proteins in tumorigenesis. The phenotypes of solid tumors are flexible and can switch through epithelial, EMT, partial/hybrid EMT, mesenchymal, and MET states. This phenotypic plasticity creates challenges to developing new therapies for the treatment of cancer. Therefore, we summarize the important enzymes (involving both TGF-β/SMAD and non-SMAD-mediated TGF-β signaling pathways) and the inhibitors targeting the enzymes (Table 1, Table 2, Table 3 and Table 4). The preclinical findings of the inhibitors on solid tumor models have the potential to understand their inhibitory mechanisms and future perspective. The inhibitors can act by different mechanisms that induce cell cycle arrest, apoptosis, modulation of EMT, and dissemination, thereby inhibiting cancer cell proliferation and tumor growth. Although the findings are preliminary, some considerations are required for prospects. Detection of novel therapies is essential to increase efficacy. Further well-designed clinical trials are important to validate safety, effectiveness, and tolerability with the patients.
Author Contributions
Conceptualization, F.R. and J.A.K.; Formal Analysis, F.R. and J.A.K.; Investigation, J.A.K. and N.R.d.B.; Resources, J.A.K.; Writing—Original Draft Preparation, Runa; Review and Editing, J.A.K., N.R.d.B., and G.O.-S.; Visualization, J.A.K. and N.R.d.B.; Supervision, J.A.K. Project Administration, J.A.K.; Funding Acquisition, J.A.K. All authors have read and agreed to the published version of the manuscript.
Funding
The work is funded by National Institute of Health (NIH), grunt number (SC1GM121182).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors acknowledge members of the Developmental Oncogene Laboratory at Baylor University and Cal State University Northridge for helpful comments during the drafting of this review. Baylor University College of Arts and Sciences (to J.A.K.) supported this work. The authors acknowledge that BioRender (Scientific Image and Illustration Software, https://www.biorender.com/ (accessed on 28 February 2024)) was used to generate the figures.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Shi, Y.; Massagué, J. Mechanisms of TGF-β Signaling from Cell Membrane to the Nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012, 13, 616–630. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.; Winter, S.L.; Alexandrow, M.G. Cell cycle arrest by transforming growth factor beta1 near G1/S is mediated by acute abrogation of prereplication complex activation involving an Rb-MCM interaction. Mol. Cell Biol. 2010, 30, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.J.; Nieto, M.A. Epithelial-Mesenchymal Transitions in Development and Disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumor transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef]
- Varga, J.; Greten, F.R. Cell plasticity in epithelial homeostasis and tumorigenesis. Nat. Cell Biol. 2017, 19, 1133–1141. [Google Scholar] [CrossRef]
- Massagué, J. TGFbeta in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef]
- Huynh, L.K.; Hipolito, C.J.; Ten Dijke, P. A Perspective on the Development of TGF-β Inhibitors for Cancer Treatment. Biomolecules 2019, 9, 743. [Google Scholar] [CrossRef]
- Suzuki, H.I. MicroRNA Control of TGF-β Signaling. Int. J. Mol. Sci. 2018, 19, 1901. [Google Scholar] [CrossRef]
- Papoutsoglou, P.; Moustakas, A. Long non-coding RNAs and TGF-β signaling in cancer. Cancer Sci. 2020, 111, 2672–2681. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Xie, F.; Zhang, Z.; van Dam, H.; Zhang, L.; Zhou, F. The regulation of TGF-β/SMAD signaling by protein deubiquitination. Protein Cell 2014, 5, 503–517. [Google Scholar] [CrossRef]
- Robertson, I.B.; Rifkin, D.B. Regulation of the Bioavailability of TGF-β and TGF-β-Related Proteins. Cold Spring Harb. Perspect. Biol. 2016, 8, a021907. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, M.; Sang, X.; You, X.; Wang, Y.; Paterson, I.C.; Hong, W.; Yang, X. Development of small molecule inhibitors targeting TGF-β ligand and receptor: Structures, mechanism, preclinical studies and clinical usage. Eur. J. Med. Chem. 2020, 191, 112154. [Google Scholar] [CrossRef]
- Kim, B.-G.; Malek, E.; Choi, S.H.; Ignatz-Hoover, J.J.; Driscoll, J.J. Novel therapies emerging in oncology to target the TGF-β pathway. J. Hematol. Oncol. 2021, 14, 55. [Google Scholar] [CrossRef]
- Huang, C.-Y.; Chung, C.-L.; Hu, T.-H.; Chen, J.-J.; Liu, P.-F.; Chen, C.-L. Recent progress in TGF-β inhibitors for cancer therapy. Biomed. Pharmacother. 2021, 134, 111046. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T.; et al. Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal Transduct. Target. Ther. 2021, 6, 201. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, A.F.; Ten Dijke, P.; Zhu, H.J. On-Target Anti-TGF-β Therapies Are Not Succeeding in Clinical Cancer Treatments: What Are Remaining Challenges? Front. Cell Dev. Biol. 2020, 8, 605. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, D.; Elez, E.; Tabernero, J.; Seoane, J. Clinical development of therapies targeting TGFβ: Current knowledge and future perspectives. Ann. Oncol. 2020, 31, 1336–1349. [Google Scholar] [CrossRef]
- Feng, X.H.; Derynck, R. Specificity and versatility in tgf-beta signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005, 21, 659–693. [Google Scholar] [CrossRef]
- Wakefield, L.M.; Hill, C.S. Beyond TGFβ: Roles of other TGFβ superfamily members in cancer. Nat. Rev. Cancer 2013, 13, 328–341. [Google Scholar] [CrossRef]
- Hata, A.; Chen, Y.G. TGF-β Signaling from Receptors to Smads. Cold Spring Harb. Perspect. Biol. 2016, 8, a022061. [Google Scholar] [CrossRef]
- Hill, C.S. Transcriptional Control by the SMADs. Cold Spring Harb. Perspect. Biol. 2016, 8, a022079. [Google Scholar] [CrossRef]
- Heldin, C.-H.; Moustakas, A. Role of Smads in TGFβ signaling. Cell Tissue Res. 2012, 347, 21–36. [Google Scholar] [CrossRef]
- Miyazawa, K.; Shinozaki, M.; Hara, T.; Furuya, T.; Miyazono, K. Two major Smad pathways in TGF-β superfamily signalling. GenesCells 2002, 7, 1191–1204. [Google Scholar] [CrossRef]
- Locker, J. Book reveiw Review of Molecular Biology of Human Cancers (An Advanced Student’s Text, by Wolfgang Schulz, Department of Urology and Center for Biological and Medical Research, Heinrich Heine University, Düsseldorf) ISBN 1-4020-3185-8. Cancer Causes Control 2005, 16, 191–192. [Google Scholar] [CrossRef]
- Nicolás, F.J.; De Bosscher, K.; Schmierer, B.; Hill, C.S. Analysis of Smad nucleocytoplasmic shuttling in living cells. J. Cell Sci. 2004, 117 Pt 18, 4113–4125. [Google Scholar] [CrossRef] [PubMed]
- Huse, M.; Muir, T.W.; Xu, L.; Chen, Y.-G.; Kuriyan, J.; Massagué, J. The TGFβ Receptor Activation Process: An Inhibitor- to Substrate-Binding Switch. Mol. Cell 2001, 8, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Souchelnytskyi, S.; Tamaki, K.; Engström, U.; Wernstedt, C.; ten Dijke, P.; Heldin, C.-H. Phosphorylation of Ser465 and Ser467 in the C Terminus of Smad2 Mediates Interaction with Smad4 and Is Required for Transforming Growth Factor-β Signaling*. J. Biol. Chem. 1997, 272, 28107–28115. [Google Scholar] [CrossRef]
- Panopoulou, E.; Gillooly, D.J.; Wrana, J.L.; Zerial, M.; Stenmark, H.; Murphy, C.; Fotsis, T. Early endosomal regulation of Smad-dependent signaling in endothelial cells. J. Biol. Chem. 2002, 277, 18046–18052. [Google Scholar] [CrossRef] [PubMed]
- Tsukazaki, T.; Chiang, T.A.; Davison, A.F.; Attisano, L.; Wrana, J.L. SARA, a FYVE Domain Protein that Recruits Smad2 to the TGFβ Receptor. Cell 1998, 95, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.K.; Bergmann, S.; Pandolfi, P.P. Cytoplasmic PML function in TGF-beta signalling. Nature 2004, 431, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Gao, Y.; Lin, H.-K. Cytoplasmic PML: From Molecular Regulation to Biological Functions. J. Cell. Biochem. 2014, 115, 812–818. [Google Scholar] [CrossRef]
- Hocevar, B.A.; Smine, A.; Xu, X.-X.; Howe, P.H. The adaptor molecule Disabled-2 links the transforming growth factor β receptors to the Smad pathway. EMBO J. 2001, 20, 2789–2801. [Google Scholar] [CrossRef]
- Heldin, C.-H.; Miyazono, K.; ten Dijke, P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature 1997, 390, 465–471. [Google Scholar] [CrossRef]
- Wu, G.; Chen, Y.G.; Ozdamar, B.; Gyuricza, C.A.; Chong, P.A.; Wrana, J.L.; Massagué, J.; Shi, Y. Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science 2000, 287, 92–97. [Google Scholar] [CrossRef]
- Miura, S.; Takeshita, T.; Asao, H.; Kimura, Y.; Murata, K.; Sasaki, Y.; Hanai, J.I.; Beppu, H.; Tsukazaki, T.; Wrana, J.L.; et al. Hgs (Hrs), a FYVE domain protein, is involved in Smad signaling through cooperation with SARA. Mol. Cell Biol. 2000, 20, 9346–9355. [Google Scholar] [CrossRef] [PubMed]
- Miura, S.; Mishina, Y. Hepatocyte growth factor–regulated tyrosine kinase substrate (Hgs) is involved in BMP signaling through phosphorylation of smads and TAK1 in early mouse embryo. Dev. Dyn. 2011, 240, 2474–2481. [Google Scholar] [CrossRef] [PubMed]
- Chacko, B.M.; Qin, B.Y.; Tiwari, A.; Shi, G.; Lam, S.; Hayward, L.J.; De Caestecker, M.; Lin, K. Structural basis of heteromeric smad protein assembly in TGF-beta signaling. Mol. Cell 2004, 15, 813–823. [Google Scholar] [CrossRef]
- Kawabata, M.; Inoue, H.; Hanyu, A.; Imamura, T.; Miyazono, K. Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J. 1998, 17, 4056–4065. [Google Scholar] [CrossRef] [PubMed]
- Tzavlaki, K.; Moustakas, A. TGF-β Signaling. Biomolecules 2020, 10, 487. [Google Scholar] [CrossRef] [PubMed]
- Lucarelli, P.; Schilling, M.; Kreutz, C.; Vlasov, A.; Boehm, M.E.; Iwamoto, N.; Steiert, B.; Lattermann, S.; Wäsch, M.; Stepath, M.; et al. Resolving the Combinatorial Complexity of Smad Protein Complex Formation and Its Link to Gene Expression. Cell Syst. 2018, 6, 75–89.e11. [Google Scholar] [CrossRef] [PubMed]
- Kurisaki, A.; Kose, S.; Yoneda, Y.; Heldin, C.H.; Moustakas, A. Transforming growth factor-beta induces nuclear import of Smad3 in an importin-beta1 and Ran-dependent manner. Mol. Biol. Cell 2001, 12, 1079–1091. [Google Scholar] [CrossRef] [PubMed]
- Kurisaki, A.; Kurisaki, K.; Kowanetz, M.; Sugino, H.; Yoneda, Y.; Heldin, C.H.; Moustakas, A. The mechanism of nuclear export of Smad3 involves exportin 4 and Ran. Mol. Cell Biol. 2006, 26, 1318–1332. [Google Scholar] [CrossRef]
- Dai, F.; Lin, X.; Chang, C.; Feng, X.H. Nuclear export of Smad2 and Smad3 by RanBP3 facilitates termination of TGF-beta signaling. Dev. Cell 2009, 16, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Latek, R.; Lodish, H.F. An extended bipartite nuclear localization signal in Smad4 is required for its nuclear import and transcriptional activity. Oncogene 2003, 22, 1057–1069. [Google Scholar] [CrossRef]
- Xu, L.; Kang, Y.; Çöl, S.; Massagué, J. Smad2 Nucleocytoplasmic Shuttling by Nucleoporins CAN/Nup214 and Nup153 Feeds TGFβ Signaling Complexes in the Cytoplasm and Nucleus. Mol. Cell 2002, 10, 271–282. [Google Scholar] [CrossRef]
- Xu, L.; Alarcón, C.; Çöl, S.; Massaguè, J. Distinct Domain Utilization by Smad3 and Smad4 for Nucleoporin Interaction and Nuclear Import*. J. Biol. Chem. 2003, 278, 42569–42577. [Google Scholar] [CrossRef]
- Jin, Q.; Ding, W.; Mulder, K.M. Requirement for the Dynein Light Chain km23-1 in a Smad2-dependent Transforming Growth Factor-β Signaling Pathway*. J. Biol. Chem. 2007, 282, 19122–19132. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Ding, W.; Mulder, K.M. The TGFβ Receptor-interacting Protein km23-1/DYNLRB1 Plays an Adaptor Role in TGFβ1 Autoinduction via Its Association with Ras*. J. Biol. Chem. 2012, 287, 26453–26463. [Google Scholar] [CrossRef]
- Conery, A.R.; Cao, Y.; Thompson, E.A.; Townsend, C.M., Jr.; Ko, T.C.; Luo, K. Akt interacts directly with Smad3 to regulate the sensitivity to TGF-beta induced apoptosis. Nat. Cell Biol. 2004, 6, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Remy, I.; Montmarquette, A.; Michnick, S.W. PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nat. Cell Biol. 2004, 6, 358–365. [Google Scholar] [CrossRef]
- Kretzschmar, M.; Liu, F.; Hata, A.; Doody, J.; Massagué, J. The TGF-beta family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 1997, 11, 984–995. [Google Scholar] [CrossRef]
- Matsuura, I.; Wang, G.; He, D.; Liu, F. Identification and Characterization of ERK MAP Kinase Phosphorylation Sites in Smad3. Biochemistry 2005, 44, 12546–12553. [Google Scholar] [CrossRef]
- Wang, G.; Matsuura, I.; He, D.; Liu, F. Transforming growth factor-{beta}-inducible phosphorylation of Smad3. J. Biol. Chem. 2009, 284, 9663–9673. [Google Scholar] [CrossRef]
- Matsuura, I.; Denissova, N.G.; Wang, G.; He, D.; Long, J.; Liu, F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature 2004, 430, 226–231. [Google Scholar] [CrossRef]
- Fuentealba, L.C.; Eivers, E.; Ikeda, A.; Hurtado, C.; Kuroda, H.; Pera, E.M.; De Robertis, E.M. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell 2007, 131, 980–993. [Google Scholar] [CrossRef] [PubMed]
- Millet, C.; Yamashita, M.; Heller, M.; Yu, L.R.; Veenstra, T.D.; Zhang, Y.E. A negative feedback control of transforming growth factor-beta signaling by glycogen synthase kinase 3-mediated Smad3 linker phosphorylation at Ser-204. J. Biol. Chem. 2009, 284, 19808–19816. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Duan, X.; Liang, Y.-Y.; Su, Y.; Wrighton, K.H.; Long, J.; Hu, M.; Davis, C.M.; Wang, J.; Brunicardi, F.C.; et al. PPM1A Functions as a Smad Phosphatase to Terminate TGFβ Signaling. Cell 2006, 125, 915–928. [Google Scholar] [CrossRef] [PubMed]
- Dai, F.; Shen, T.; Li, Z.; Lin, X.; Feng, X.H. PPM1A dephosphorylates RanBP3 to enable efficient nuclear export of Smad2 and Smad3. EMBO Rep. 2011, 12, 1175–1181. [Google Scholar] [CrossRef]
- Yang, X.; Teng, Y.; Hou, N.; Fan, X.; Cheng, X.; Li, J.; Wang, L.; Wang, Y.; Wu, X.; Yang, X. Delayed Re-epithelialization in Ppm1a Gene-deficient Mice Is Mediated by Enhanced Activation of Smad2*. J. Biol. Chem. 2011, 286, 42267–42273. [Google Scholar] [CrossRef]
- Lo Sardo, F.; Strano, S.; Blandino, G. YAP and TAZ in Lung Cancer: Oncogenic Role and Clinical Targeting. Cancers 2018, 10, 137. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zeng, C.; Ye, S.; Dai, X.; He, Q.; Yang, B.; Zhu, H. Yes-associated protein (YAP) and transcriptional coactivator with a PDZ-binding motif (TAZ): A nexus between hypoxia and cancer. Acta Pharm. Sin. B 2020, 10, 947–960. [Google Scholar] [CrossRef]
- Hiemer, S.E.; Szymaniak, A.D.; Varelas, X. The transcriptional regulators TAZ and YAP direct transforming growth factor β-induced tumorigenic phenotypes in breast cancer cells. J. Biol. Chem. 2014, 289, 13461–13474. [Google Scholar] [CrossRef] [PubMed]
- Varelas, X.; Sakuma, R.; Samavarchi-Tehrani, P.; Peerani, R.; Rao, B.M.; Dembowy, J.; Yaffe, M.B.; Zandstra, P.W.; Wrana, J.L. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 2008, 10, 837–848. [Google Scholar] [CrossRef]
- Chen, H.B.; Rud, J.G.; Lin, K.; Xu, L. Nuclear targeting of transforming growth factor-beta-activated Smad complexes. J. Biol. Chem. 2005, 280, 21329–21336. [Google Scholar] [CrossRef]
- Chen, Y.L.; Law, P.Y.; Loh, H.H. Inhibition of Akt/Protein Kinase B Signaling by Naltrindole in Small Cell Lung Cancer Cells. Cancer Res. 2004, 64, 8723–8730. [Google Scholar] [CrossRef]
- DeFeo-Jones, D.; Barnett, S.F.; Fu, S.; Hancock, P.J.; Haskell, K.M.; Leander, K.R.; McAvoy, E.; Robinson, R.G.; Duggan, M.E.; Lindsley, C.W.; et al. Tumor cell sensitization to apoptotic stimuli by selective inhibition of specific Akt/PKB family members. Mol. Cancer Ther. 2005, 4, 271–279. [Google Scholar] [CrossRef]
- Lin, J.; Sampath, D.; Nannini, M.A.; Lee, B.B.; Degtyarev, M.; Oeh, J.; Savage, H.; Guan, Z.; Hong, R.; Kassees, R.; et al. Targeting Activated Akt with GDC-0068, a Novel Selective Akt Inhibitor That Is Efficacious in Multiple Tumor Models. Clin. Cancer Res. 2013, 19, 1760–1772. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Luo, G.; Qiu, Z. Akt inhibitor MK-2206 reduces pancreatic cancer cell viability and increases the efficacy of gemcitabine. Oncol. Lett. 2020, 19, 1999–2004. [Google Scholar] [CrossRef]
- Malkomes, P.; Lunger, I.; Luetticke, A.; Oppermann, E.; Haetscher, N.; Serve, H.; Holzer, K.; Bechstein, W.O.; Rieger, M.A. Selective AKT Inhibition by MK-2206 Represses Colorectal Cancer-Initiating Stem Cells. Ann. Surg. Oncol. 2016, 23, 2849–2857. [Google Scholar] [CrossRef]
- Huang, S.; Xiao, J.; Wu, J.; Liu, J.; Feng, X.; Yang, C.; Xiang, D.; Luo, S. Tizoxanide Promotes Apoptosis in Glioblastoma by Inhibiting CDK1 Activity. Front. Pharmacol. 2022, 13, 895573. [Google Scholar] [CrossRef] [PubMed]
- Nong, H.B.; Zhang, Y.N.; Bai, Y.G.; Zhang, Q.; Liu, M.F.; Zhou, Q.; Shi, Z.H.; Zeng, G.F.; Zong, S.H. Adapalene Inhibits Prostate Cancer Cell Proliferation In Vitro and In Vivo by Inducing DNA Damage, S-phase Cell Cycle Arrest, and Apoptosis. Front. Pharmacol. 2022, 13, 801624. [Google Scholar] [CrossRef] [PubMed]
- Mehraj, U.; Wani, N.A.; Hamid, A.; Alkhanani, M.; Almilaibary, A.; Mir, M.A. Adapalene inhibits the growth of triple-negative breast cancer cells by S-phase arrest and potentiates the antitumor efficacy of GDC-0941. Front. Pharmacol. 2022, 13, 958443. [Google Scholar] [CrossRef]
- Decker, J.T.; Ma, J.A.; Shea, L.D.; Jeruss, J.S. Implications of TGFβ Signaling and CDK Inhibition for the Treatment of Breast Cancer. Cancers 2021, 13, 5343. [Google Scholar] [CrossRef]
- Tarasewicz, E.; Rivas, L.; Hamdan, R.; Dokic, D.; Parimi, V.; Bernabe, B.P.; Thomas, A.; Shea, L.D.; Jeruss, J.S. Inhibition of CDK-mediated phosphorylation of Smad3 results in decreased oncogenesis in triple negative breast cancer cells. Cell Cycle 2014, 13, 3191–3201. [Google Scholar] [CrossRef]
- Zhu, Y.; Ke, K.-B.; Xia, Z.-K.; Li, H.-J.; Su, R.; Dong, C.; Zhou, F.-M.; Wang, L.; Chen, R.; Wu, S.-G.; et al. Discovery of vanoxerine dihydrochloride as a CDK2/4/6 triple-inhibitor for the treatment of human hepatocellular carcinoma. Mol. Med. 2021, 27, 15. [Google Scholar] [CrossRef]
- Duda, P.; Akula, S.M.; Abrams, S.L.; Steelman, L.S.; Martelli, A.M.; Cocco, L.; Ratti, S.; Candido, S.; Libra, M.; Montalto, G.; et al. Targeting GSK3 and Associated Signaling Pathways Involved in Cancer. Cells 2020, 9, 1110. [Google Scholar] [CrossRef]
- Kazi, A.; Xiang, S.; Yang, H.; Delitto, D.; Trevino, J.; Jiang, R.H.Y.; Ayaz, M.; Lawrence, H.R.; Kennedy, P.; Sebti, S.M. GSK3 suppression upregulates β-catenin and c-Myc to abrogate KRas-dependent tumors. Nat. Commun. 2018, 9, 5154. [Google Scholar] [CrossRef] [PubMed]
- Abushahba, W.; Olabisi, O.O.; Jeong, B.-S.; Boregowda, R.K.; Wen, Y.; Liu, F.; Goydos, J.S.; Lasfar, A.; Cohen-Solal, K.A. Non-Canonical Smads Phosphorylation Induced by the Glutamate Release Inhibitor, Riluzole, through GSK3 Activation in Melanoma. PLoS ONE 2012, 7, e47312. [Google Scholar] [CrossRef] [PubMed]
- Otręba, M.; Sjölander, J.J.; Grøtli, M.; Sunnerhagen, P. A Small Molecule Targeting Human MEK1/2 Enhances ERK and p38 Phosphorylation under Oxidative Stress or with Phenothiazines. Life 2021, 11, 297. [Google Scholar] [CrossRef] [PubMed]
- Mattmann, M.E.; Stoops, S.L.; Lindsley, C.W. Inhibition of Akt with small molecules and biologics: Historical perspective and current status of the patent landscape. Expert. Opin. Ther. Pat. 2011, 21, 1309–1338. [Google Scholar] [CrossRef]
- Xu, P.; Lin, X.; Feng, X.H. Posttranslational Regulation of Smads. Cold Spring Harb. Perspect. Biol. 2016, 8, a022087. [Google Scholar] [CrossRef]
- Funaba, M.; Zimmerman, C.M.; Mathews, L.S. Modulation of Smad2-mediated Signaling by Extracellular Signal-regulated Kinase*. J. Biol. Chem. 2002, 277, 41361–41368. [Google Scholar] [CrossRef]
- Yakymovych, I.; Ten Dijke, P.; Heldin, C.H.; Souchelnytskyi, S. Regulation of Smad signaling by protein kinase C. Faseb J. 2001, 15, 553–555. [Google Scholar] [CrossRef]
- Saura, M.; Zaragoza, C.; Herranz, B.; Griera, M.; Diez-Marqués, L.; Rodriguez-Puyol, D.; Rodriguez-Puyol, M. Nitric Oxide Regulates Transforming Growth Factor-β Signaling in Endothelial Cells. Circ. Res. 2005, 97, 1115–1123. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Iaria, J.; Simpson, R.J.; Zhu, H.-J. Ras enhances TGF-β signaling by decreasing cellular protein levels of its type II receptor negative regulator SPSB1. Cell Commun. Signal. 2018, 16, 10. [Google Scholar] [CrossRef]
- Guo, X.; Waddell, D.S.; Wang, W.; Wang, Z.; Liberati, N.T.; Yong, S.; Liu, X.; Wang, X.F. Ligand-dependent ubiquitination of Smad3 is regulated by casein kinase 1 gamma 2, an inhibitor of TGF-beta signaling. Oncogene 2008, 27, 7235–7247. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, Y.; Zhang, H.; Liu, F.; Cheng, Z.; Wang, D.; Wang, G.; Xu, H.; Zhao, Y.; Cao, L.; et al. Oncogenic PAK4 regulates Smad2/3 axis involving gastric tumorigenesis. Oncogene 2014, 33, 3473–3484. [Google Scholar] [CrossRef]
- Yan, X.; Zhang, J.; Sun, Q.; Tuazon, P.T.; Wu, X.; Traugh, J.A.; Chen, Y.-G. p21-activated Kinase 2 (PAK2) Inhibits TGF-β Signaling in Madin-Darby Canine Kidney (MDCK) Epithelial Cells by Interfering with the Receptor-Smad Interaction. J. Biol. Chem. 2012, 287, 13705–13712. [Google Scholar] [CrossRef] [PubMed]
- Nakao, A.; Imamura, T.; Souchelnytskyi, S.; Kawabata, M.; Ishisaki, A.; Oeda, E.; Tamaki, K.; Hanai, J.; Heldin, C.H.; Miyazono, K.; et al. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997, 16, 5353–5362. [Google Scholar] [CrossRef] [PubMed]
- Roelen, B.A.; Cohen, O.S.; Raychowdhury, M.K.; Chadee, D.N.; Zhang, Y.; Kyriakis, J.M.; Alessandrini, A.A.; Lin, H.Y. Phosphorylation of threonine 276 in Smad4 is involved in transforming growth factor-beta-induced nuclear accumulation. Am. J. Physiol. Cell Physiol. 2003, 285, C823–C830. [Google Scholar] [CrossRef] [PubMed]
- Morén, A.; Raja, E.; Heldin, C.H.; Moustakas, A. Negative regulation of TGFβ signaling by the kinase LKB1 and the scaffolding protein LIP1. J. Biol. Chem. 2011, 286, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Seong, H.A.; Jung, H.; Ha, H. Murine protein serine/threonine kinase 38 stimulates TGF-beta signaling in a kinase-dependent manner via direct phosphorylation of Smad proteins. J. Biol. Chem. 2010, 285, 30959–30970. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Pan, L.; Qin, X.; Chen, H.; Xu, Y.; Chen, Y.; Tang, H. MTMR4 Attenuates Transforming Growth Factor β (TGFβ) Signaling by Dephosphorylating R-Smads in Endosomes*. J. Biol. Chem. 2010, 285, 8454–8462. [Google Scholar] [CrossRef] [PubMed]
- Wrighton, K.H.; Willis, D.; Long, J.; Liu, F.; Lin, X.; Feng, X.-H. Small C-terminal Domain Phosphatases Dephosphorylate the Regulatory Linker Regions of Smad2 and Smad3 to Enhance Transforming Growth Factor-β Signaling*. J. Biol. Chem. 2006, 281, 38365–38375. [Google Scholar] [CrossRef] [PubMed]
- Knockaert, M.; Sapkota, G.; Alarcón, C.; Massagué, J.; Brivanlou, A.H. Unique players in the BMP pathway: Small C-terminal domain phosphatases dephosphorylate Smad1 to attenuate BMP signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 11940–11945. [Google Scholar] [CrossRef]
- Lin, X.; Liang, M.; Feng, X.-H. Smurf2 Is a Ubiquitin E3 Ligase Mediating Proteasome-dependent Degradation of Smad2 in Transforming Growth Factor-β Signaling*210. J. Biol. Chem. 2000, 275, 36818–36822. [Google Scholar] [CrossRef]
- Zhang, Y.; Chang, C.; Gehling, D.J.; Hemmati-Brivanlou, A.; Derynck, R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 2001, 98, 974–979. [Google Scholar] [CrossRef]
- Zhu, H.; Kavsak, P.; Abdollah, S.; Wrana, J.L.; Thomsen, G.H. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 1999, 400, 687–693. [Google Scholar] [CrossRef]
- Kuratomi, G.; Komuro, A.; Goto, K.; Shinozaki, M.; Miyazawa, K.; Miyazono, K.; Imamura, T. NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-beta (transforming growth factor-beta) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-beta type I receptor. Biochem. J. 2005, 386 Pt 3, 461–470. [Google Scholar] [CrossRef]
- Xin, H.; Xu, X.; Li, L.; Ning, H.; Rong, Y.; Shang, Y.; Wang, Y.; Fu, X.Y.; Chang, Z. CHIP controls the sensitivity of transforming growth factor-beta signaling by modulating the basal level of Smad3 through ubiquitin-mediated degradation. J. Biol. Chem. 2005, 280, 20842–20850. [Google Scholar] [CrossRef]
- Fukuchi, M.; Imamura, T.; Chiba, T.; Ebisawa, T.; Kawabata, M.; Tanaka, K.; Miyazono, K. Ligand-dependent Degradation of Smad3 by a Ubiquitin Ligase Complex of ROC1 and Associated Proteins. Mol. Biol. Cell 2001, 12, 1431–1443. [Google Scholar] [CrossRef]
- Guo, X.; Ramirez, A.; Waddell, D.S.; Li, Z.; Liu, X.; Wang, X.-F. Axin and GSK3-β control Smad3 protein stability and modulate TGF-β signaling. Genes Dev. 2008, 22, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Levy, L.; Howell, M.; Das, D.; Harkin, S.; Episkopou, V.; Hill, C.S. Arkadia Activates Smad3/Smad4-Dependent Transcription by Triggering Signal-Induced SnoN Degradation. Mol. Cell. Biol. 2007, 27, 6068–6083. [Google Scholar] [CrossRef] [PubMed]
- Mavrakis, K.J.; Andrew, R.L.; Lee, K.L.; Petropoulou, C.; Dixon, J.E.; Navaratnam, N.; Norris, D.P.; Episkopou, V. Arkadia Enhances Nodal/TGF-β Signaling by Coupling Phospho-Smad2/3 Activity and Turnover. PLoS Biol. 2007, 5, e67. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Liang, Y.-Y.; Wrighton, K.; Ungermannova, D.; Wang, X.-P.; Brunicardi, F.C.; Liu, X.; Feng, X.-H.; Lin, X. Ubiquitination and Proteolysis of Cancer-Derived Smad4 Mutants by SCFSkp2. Mol. Cell. Biol. 2004, 24, 7524–7537. [Google Scholar] [CrossRef]
- Agricola, E.; Randall, R.A.; Gaarenstroom, T.; Dupont, S.; Hill, C.S. Recruitment of TIF1γ to Chromatin via Its PHD Finger-Bromodomain Activates Its Ubiquitin Ligase and Transcriptional Repressor Activities. Mol. Cell 2011, 43, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Zacchigna, L.; Cordenonsi, M.; Soligo, S.; Adorno, M.; Rugge, M.; Piccolo, S. Germ-Layer Specification and Control of Cell Growth by Ectodermin, a Smad4 Ubiquitin Ligase. Cell 2005, 121, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Inui, M.; Manfrin, A.; Mamidi, A.; Martello, G.; Morsut, L.; Soligo, S.; Enzo, E.; Moro, S.; Polo, S.; Dupont, S.; et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat. Cell Biol. 2011, 13, 1368–1375. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Mamidi, A.; Cordenonsi, M.; Montagner, M.; Zacchigna, L.; Adorno, M.; Martello, G.; Stinchfield, M.J.; Soligo, S.; Morsut, L.; et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 2009, 136, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Simonsson, M.; Kanduri, M.; Grönroos, E.; Heldin, C.-H.; Ericsson, J. The DNA Binding Activities of Smad2 and Smad3 Are Regulated by Coactivator-mediated Acetylation*. J. Biol. Chem. 2006, 281, 39870–39880. [Google Scholar] [CrossRef]
- Inoue, Y.; Itoh, Y.; Abe, K.; Okamoto, T.; Daitoku, H.; Fukamizu, A.; Onozaki, K.; Hayashi, H. Smad3 is acetylated by p300/CBP to regulate its transactivation activity. Oncogene 2007, 26, 500–508. [Google Scholar] [CrossRef]
- Tu, A.W.; Luo, K. Acetylation of Smad2 by the co-activator p300 regulates activin and transforming growth factor beta response. J. Biol. Chem. 2007, 282, 21187–21196. [Google Scholar] [CrossRef] [PubMed]
- Lönn, P.; van der Heide, L.P.; Dahl, M.; Hellman, U.; Heldin, C.H.; Moustakas, A. PARP-1 attenuates Smad-mediated transcription. Mol. Cell 2010, 40, 521–532. [Google Scholar] [CrossRef]
- Imoto, S.; Sugiyama, K.; Muromoto, R.; Sato, N.; Yamamoto, T.; Matsuda, T. Regulation of transforming growth factor-beta signaling by protein inhibitor of activated STAT, PIASy through Smad3. J. Biol. Chem. 2003, 278, 34253–34258. [Google Scholar] [CrossRef]
- Liang, M.; Melchior, F.; Feng, X.H.; Lin, X. Regulation of Smad4 sumoylation and transforming growth factor-beta signaling by protein inhibitor of activated STAT1. J. Biol. Chem. 2004, 279, 22857–22865. [Google Scholar] [CrossRef]
- Hayashida, T.; Decaestecker, M.; Schnaper, H.W. Cross-talk between ERK MAP kinase and Smad signaling pathways enhances TGF-beta-dependent responses in human mesangial cells. Faseb J. 2003, 17, 1576–1578. [Google Scholar] [CrossRef]
- Ragusa, M.; Statello, L.; Maugeri, M.; Majorana, A.; Barbagallo, D.; Salito, L.; Sammito, M.; Santonocito, M.; Angelica, R.; Cavallaro, A.; et al. Specific alterations of the microRNA transcriptome and global network structure in colorectal cancer after treatment with MAPK/ERK inhibitors. J. Mol. Med. 2012, 90, 1421–1438. [Google Scholar] [CrossRef] [PubMed]
- Samadani, R.; Zhang, J.; Brophy, A.; Oashi, T.; Priyakumar, U.D.; Raman, E.P.; St John, F.J.; Jung, K.Y.; Fletcher, S.; Pozharski, E.; et al. Small-molecule inhibitors of ERK-mediated immediate early gene expression and proliferation of melanoma cells expressing mutated BRaf. Biochem. J. 2015, 467, 425–438. [Google Scholar] [CrossRef] [PubMed]
- Kaoud, T.S.; Johnson, W.H.; Ebelt, N.D.; Piserchio, A.; Zamora-Olivares, D.; Van Ravenstein, S.X.; Pridgen, J.R.; Edupuganti, R.; Sammons, R.; Cano, M.; et al. Modulating multi-functional ERK complexes by covalent targeting of a recruitment site in vivo. Nat. Commun. 2019, 10, 5232. [Google Scholar] [CrossRef] [PubMed]
- Herrero, A.; Pinto, A.; Colón-Bolea, P.; Casar, B.; Jones, M.; Agudo-Ibáñez, L.; Vidal, R.; Tenbaum, S.P.; Nuciforo, P.; Valdizán, E.M.; et al. Small Molecule Inhibition of ERK Dimerization Prevents Tumorigenesis by RAS-ERK Pathway Oncogenes. Cancer Cell 2015, 28, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Marques da Fonseca, L.; Jacques da Silva, L.R.; Santos dos Reis, J.; Rodrigues da Costa Santos, M.A.; de Sousa Chaves, V.; Monteiro da Costa, K.; Sa-Diniz, J.d.N.; Freire de Lima, C.G.; Morrot, A.; Nunes Franklim, T.; et al. Piperine Inhibits TGF-β Signaling Pathways and Disrupts EMT-Related Events in Human Lung Adenocarcinoma Cells. Medicines 2020, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Huang, J.; Yuan, Z.; Feng, S.; Xie, Y.; Ma, W. Protein kinase C inhibitor chelerythrine selectively inhibits proliferation of triple-negative breast cancer cells. Sci. Rep. 2017, 7, 2022. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, Y.; Yang, Q.; Zhang, Y.; Liu, H.; Huang, M.H.; Wang, R.; Lu, F. PKC signal amplification suppresses non-small cell lung cancer growth by promoting p21 expression and phosphorylation. Heliyon 2022, 8, e10657. [Google Scholar] [CrossRef]
- Dükel, M.; Tavsan, Z.; Erdogan, D.; Erkan Gök, D.; Ayar Kayali, H. Protein kinase C Inhibitors selectively modulate dynamics of cell adhesion molecules and cell death in human colon cancer cells. Cell Adhes. Migr. 2019, 13, 83–97. [Google Scholar] [CrossRef]
- Whitt, J.D.; Li, N.; Tinsley, H.N.; Chen, X.; Zhang, W.; Li, Y.; Gary, B.D.; Keeton, A.B.; Xi, Y.; Abadi, A.H.; et al. A Novel Sulindac Derivative that Potently Suppresses Colon Tumor Cell Growth by Inhibiting cGMP Phosphodiesterase and β-Catenin Transcriptional Activity. Cancer Prev. Res. 2012, 5, 822–833. [Google Scholar] [CrossRef]
- Tinsley, H.N.; Mathew, B.; Chen, X.; Maxuitenko, Y.Y.; Li, N.; Lowe, W.M.; Whitt, J.D.; Zhang, W.; Gary, B.D.; Keeton, A.B.; et al. Novel Non-Cyclooxygenase Inhibitory Derivative of Sulindac Inhibits Breast Cancer Cell Growth In Vitro and Reduces Mammary Tumorigenesis in Rats. Cancers 2023, 15, 646. [Google Scholar] [CrossRef]
- Vena, F.; Bayle, S.; Nieto, A.; Quereda, V.; Aceti, M.; Frydman, S.M.; Sansil, S.S.; Grant, W.; Monastyrskyi, A.; McDonald, P.; et al. Targeting Casein Kinase 1 Delta Sensitizes Pancreatic and Bladder Cancer Cells to Gemcitabine Treatment by Upregulating Deoxycytidine Kinase. Mol. Cancer Ther. 2020, 19, 1623–1635. [Google Scholar] [CrossRef]
- Liu, M.; Hu, Y.; Lu, S.; Lu, M.; Li, J.; Chang, H.; Jia, H.; Zhou, M.; Ren, F.; Zhong, J. IC261, a specific inhibitor of CK1δ/ε, promotes aerobic glycolysis through p53-dependent mechanisms in colon cancer. Int. J. Biol. Sci. 2020, 16, 882–892. [Google Scholar] [CrossRef]
- Licciulli, S.; Maksimoska, J.; Zhou, C.; Troutman, S.; Kota, S.; Liu, Q.; Duron, S.; Campbell, D.; Chernoff, J.; Field, J.; et al. FRAX597, a small molecule inhibitor of the p21-activated kinases, inhibits tumorigenesis of neurofibromatosis type 2 (NF2)-associated Schwannomas. J. Biol. Chem. 2013, 288, 29105–29114. [Google Scholar] [CrossRef] [PubMed]
- Murray, B.W.; Guo, C.; Piraino, J.; Westwick, J.K.; Zhang, C.; Lamerdin, J.; Dagostino, E.; Knighton, D.; Loi, C.M.; Zager, M.; et al. Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. Proc. Natl. Acad. Sci. USA 2010, 107, 9446–9451. [Google Scholar] [CrossRef]
- Hao, C.; Zhao, F.; Song, H.; Guo, J.; Li, X.; Jiang, X.; Huan, R.; Song, S.; Zhang, Q.; Wang, R.; et al. Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors. J. Med. Chem. 2018, 61, 265–285. [Google Scholar] [CrossRef]
- Song, P.; Zhao, F.; Li, D.; Qu, J.; Yao, M.; Su, Y.; Wang, H.; Zhou, M.; Wang, Y.; Gao, Y.; et al. Synthesis of selective PAK4 inhibitors for lung metastasis of lung cancer and melanoma cells. Acta Pharm. Sin. B 2022, 12, 2905–2922. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, L.; McNamara, K.L.; Li, D.; Borgman, C.L.; McDermott, U.; Brandstetter, K.A.; Padera, R.F.; Chirieac, L.R.; Settleman, J.E.; Wong, K.K. Sunitinib prolongs survival in genetically engineered mouse models of multistep lung carcinogenesis. Cancer Prev. Res. 2009, 2, 330–337. [Google Scholar] [CrossRef]
- Andrade-Vieira, R.; Goguen, D.; Bentley, H.A.; Bowen, C.V.; Marignani, P.A. Pre-clinical study of drug combinations that reduce breast cancer burden due to aberrant mTOR and metabolism promoted by LKB1 loss. Oncotarget 2014, 5, 12738–12752. [Google Scholar] [CrossRef]
- Zheng, B.; Jeong, J.H.; Asara, J.M.; Yuan, Y.Y.; Granter, S.R.; Chin, L.; Cantley, L.C. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol. Cell 2009, 33, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Farrington, C.C.; Yuan, E.; Mazhar, S.; Izadmehr, S.; Hurst, L.; Allen-Petersen, B.L.; Janghorban, M.; Chung, E.; Wolczanski, G.; Galsky, M.; et al. Protein phosphatase 2A activation as a therapeutic strategy for managing MYC-driven cancers. J. Biol. Chem. 2020, 295, 757–770. [Google Scholar] [CrossRef]
- McClinch, K.; Avelar, R.A.; Callejas, D.; Izadmehr, S.; Wiredja, D.; Perl, A.; Sangodkar, J.; Kastrinsky, D.B.; Schlatzer, D.; Cooper, M.; et al. Small-Molecule Activators of Protein Phosphatase 2A for the Treatment of Castration-Resistant Prostate Cancer. Cancer Res. 2018, 78, 2065–2080. [Google Scholar] [CrossRef]
- Wang, L.; Ge, S.; Zhou, F. MicroRNA-487a-3p inhibits the growth and invasiveness of oral squamous cell carcinoma by targeting PPM1A. Bioengineered 2021, 12, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Kauko, O.; O’Connor, C.M.; Kulesskiy, E.; Sangodkar, J.; Aakula, A.; Izadmehr, S.; Yetukuri, L.; Yadav, B.; Padzik, A.; Laajala, T.D.; et al. PP2A inhibition is a druggable MEK inhibitor resistance mechanism in KRAS-mutant lung cancer cells. Sci. Transl. Med. 2018, 10, eaaq1093. [Google Scholar] [CrossRef] [PubMed]
- Sowa, N.; Horie, T.; Kuwabara, Y.; Baba, O.; Watanabe, S.; Nishi, H.; Kinoshita, M.; Takanabe-Mori, R.; Wada, H.; Shimatsu, A.; et al. MicroRNA 26b encoded by the intron of small CTD phosphatase (SCP) 1 has an antagonistic effect on its host gene. J. Cell Biochem. 2012, 113, 3455–3465. [Google Scholar] [CrossRef]
- Matsuoka, H.; Ando, K.; Swayze, E.J.; Unan, E.C.; Mathew, J.; Hu, Q.; Tsuda, Y.; Nakashima, Y.; Saeki, H.; Oki, E.; et al. CTDSP1 inhibitor rabeprazole regulates DNA-PKcs dependent topoisomerase I degradation and irinotecan drug resistance in colorectal cancer. PLoS ONE 2020, 15, e0228002. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Zhang, Y.; Zhou, X.; Ma, H.; Yao, H.; Ji, F. Rabeprazole exhibits antiproliferative effects on human gastric cancer cell lines. Oncol. Lett. 2014, 8, 1739–1744. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Deng, J.; Yuan, J.; Tang, X.; Wang, Y.; Chen, H.; Liu, Y.; Zhou, L. Curcumin exerts its tumor suppressive function via inhibition of NEDD4 oncoprotein in glioma cancer cells. Int. J. Oncol. 2017, 51, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yu, Y.; Wang, W.; Jiang, Y.; Li, Y.; Jiang, X.; Qiao, Y.; Chen, L.; Zhao, X.; Liu, J.; et al. Targeting the E3 ligase NEDD4 as a novel therapeutic strategy for IGF1 signal pathway-driven gastric cancer. Oncogene 2023, 42, 1072–1087. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, J.J.; Zhou, S.J.; Chen, J.; Hu, Q.; Pu, J.X.; Lu, J.L. Diosgenin inhibits the expression of NEDD4 in prostate cancer cells. Am. J. Transl. Res. 2019, 11, 3461–3471. [Google Scholar]
- Leu, J.I.; Pimkina, J.; Frank, A.; Murphy, M.E.; George, D.L. A small molecule inhibitor of inducible heat shock protein 70. Mol. Cell 2009, 36, 15–27. [Google Scholar] [CrossRef]
- Dublang, L.; Underhaug, J.; Flydal, M.I.; Velasco-Carneros, L.; Maréchal, J.D.; Moro, F.; Boyano, M.D.; Martinez, A.; Muga, A. Inhibition of the Human Hsc70 System by Small Ligands as a Potential Anticancer Approach. Cancers 2021, 13, 2936. [Google Scholar] [CrossRef]
- Liu, Y.Q.; Wang, X.L.; Cheng, X.; Lu, Y.Z.; Wang, G.Z.; Li, X.C.; Zhang, J.; Wen, Z.S.; Huang, Z.L.; Gao, Q.L.; et al. Skp1 in lung cancer: Clinical significance and therapeutic efficacy of its small molecule inhibitors. Oncotarget 2015, 6, 34953–34967. [Google Scholar] [CrossRef]
- Hussain, M.; Lu, Y.; Tariq, M.; Jiang, H.; Shu, Y.; Luo, S.; Zhu, Q.; Zhang, J.; Liu, J. A small-molecule Skp1 inhibitor elicits cell death by p53-dependent mechanism. iScience 2022, 25, 104591. [Google Scholar] [CrossRef]
- Wu, J.W.; Krawitz, A.R.; Chai, J.; Li, W.; Zhang, F.; Luo, K.; Shi, Y. Structural mechanism of Smad4 recognition by the nuclear oncoprotein Ski: Insights on Ski-mediated repression of TGF-beta signaling. Cell 2002, 111, 357–367. [Google Scholar] [CrossRef]
- Zhou, L.; Yu, X.; Li, M.; Gong, G.; Liu, W.; Li, T.; Zuo, H.; Li, W.; Gao, F.; Liu, H. Cdh1-mediated Skp2 degradation by dioscin reprogrammes aerobic glycolysis and inhibits colorectal cancer cells growth. EBioMedicine 2020, 51, 102570. [Google Scholar] [CrossRef]
- Chan, C.H.; Morrow, J.K.; Li, C.F.; Gao, Y.; Jin, G.; Moten, A.; Stagg, L.J.; Ladbury, J.E.; Cai, Z.; Xu, D.; et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell 2013, 154, 556–568. [Google Scholar] [CrossRef]
- Lasko, L.M.; Jakob, C.G.; Edalji, R.P.; Qiu, W.; Montgomery, D.; Digiammarino, E.L.; Hansen, T.M.; Risi, R.M.; Frey, R.; Manaves, V.; et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 2017, 550, 128–132. [Google Scholar] [CrossRef]
- Ansari, M.S.Z.; Stagni, V.; Iuzzolino, A.; Rotili, D.; Mai, A.; Del Bufalo, D.; Lavia, P.; Degrassi, F.; Trisciuoglio, D. Pharmacological targeting of CBP/p300 drives a redox/autophagy axis leading to senescence-induced growth arrest in non-small cell lung cancer cells. Cancer Gene Ther. 2023, 30, 124–136. [Google Scholar] [CrossRef]
- Cai, L.-Y.; Chen, S.-J.; Xiao, S.-H.; Sun, Q.-J.; Ding, C.-H.; Zheng, B.-N.; Zhu, X.-Y.; Liu, S.-Q.; Yang, F.; Yang, Y.-X.; et al. Targeting p300/CBP Attenuates Hepatocellular Carcinoma Progression through Epigenetic Regulation of Metabolism. Cancer Res. 2021, 81, 860–872. [Google Scholar] [CrossRef]
- Gajer, J.M.; Furdas, S.D.; Gründer, A.; Gothwal, M.; Heinicke, U.; Keller, K.; Colland, F.; Fulda, S.; Pahl, H.L.; Fichtner, I.; et al. Histone acetyltransferase inhibitors block neuroblastoma cell growth in vivo. Oncogenesis 2015, 4, e137. [Google Scholar] [CrossRef]
- Wang, Y.M.; Gu, M.L.; Meng, F.S.; Jiao, W.R.; Zhou, X.X.; Yao, H.P.; Ji, F. Histone acetyltransferase p300/CBP inhibitor C646 blocks the survival and invasion pathways of gastric cancer cell lines. Int. J. Oncol. 2017, 51, 1860–1868. [Google Scholar] [CrossRef]
- Illuzzi, G.; Staniszewska, A.D.; Gill, S.J.; Pike, A.; McWilliams, L.; Critchlow, S.E.; Cronin, A.; Fawell, S.; Hawthorne, G.; Jamal, K.; et al. Preclinical Characterization of AZD5305, A Next-Generation, Highly Selective PARP1 Inhibitor and Trapper. Clin. Cancer Res. 2022, 28, 4724–4736. [Google Scholar] [CrossRef]
- Sreekumar, S.; Zhou, D.; Mpoy, C.; Schenk, E.; Scott, J.; Arbeit, J.M.; Xu, J.; Rogers, B.E. Preclinical Efficacy of a PARP-1 Targeted Auger-Emitting Radionuclide in Prostate Cancer. Int. J. Mol. Sci. 2023, 24, 3083. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Y.-F.; Jayaraman, L.; Yang, H.; Massagué, J.; Pavletich, N.P. Crystal Structure of a Smad MH1 Domain Bound to DNA: Insights on DNA Binding in TGF-β Signaling. Cell 1998, 94, 585–594. [Google Scholar] [CrossRef]
- Zawel, L.; Le Dai, J.; Buckhaults, P.; Zhou, S.; Kinzler, K.W.; Vogelstein, B.; Kern, S.E. Human Smad3 and Smad4 Are Sequence-Specific Transcription Activators. Mol. Cell 1998, 1, 611–617. [Google Scholar] [CrossRef]
- Morikawa, M.; Koinuma, D.; Miyazono, K.; Heldin, C.H. Genome-wide mechanisms of Smad binding. Oncogene 2013, 32, 1609–1615. [Google Scholar] [CrossRef]
- Dennler, S.; Itoh, S.; Vivien, D.; ten Dijke, P.; Huet, S.; Gauthier, J.M. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J 1998, 17, 3091–3100. [Google Scholar] [CrossRef]
- Liu, L.; Liu, X.; Ren, X.; Tian, Y.; Chen, Z.; Xu, X.; Du, Y.; Jiang, C.; Fang, Y.; Liu, Z.; et al. Smad2 and Smad3 have differential sensitivity in relaying TGFβ signaling and inversely regulate early lineage specification. Sci. Rep. 2016, 6, 21602. [Google Scholar] [CrossRef]
- Yagi, K.; Goto, D.; Hamamoto, T.; Takenoshita, S.; Kato, M.; Miyazono, K. Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J. Biol. Chem. 1999, 274, 703–709. [Google Scholar] [CrossRef]
- Pyrowolakis, G.; Hartmann, B.; Müller, B.; Basler, K.; Affolter, M. A Simple Molecular Complex Mediates Widespread BMP-Induced Repression during Drosophila Development. Dev. Cell 2004, 7, 229–240. [Google Scholar] [CrossRef]
- Weiss, A.; Charbonnier, E.; Ellertsdóttir, E.; Tsirigos, A.; Wolf, C.; Schuh, R.; Pyrowolakis, G.; Affolter, M. A conserved activation element in BMP signaling during Drosophila development. Nat. Struct. Mol. Biol. 2010, 17, 69–76. [Google Scholar] [CrossRef]
- Ito, T.; Ikehara, T.; Nakagawa, T.; Kraus, W.L.; Muramatsu, M. p300-Mediated acetylation facilitates the transfer of histone H2A–H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 2000, 14, 1899–1907. [Google Scholar] [CrossRef] [PubMed]
- Kahata, K.; Hayashi, M.; Asaka, M.; Hellman, U.; Kitagawa, H.; Yanagisawa, J.; Kato, S.; Imamura, T.; Miyazono, K. Regulation of transforming growth factor-β and bone morphogenetic protein signalling by transcriptional coactivator GCN5. Genes Cells 2004, 9, 143–151. [Google Scholar] [CrossRef]
- Ross, S.; Cheung, E.; Petrakis, T.G.; Howell, M.; Kraus, W.L.; Hill, C.S. Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription. EMBO J. 2006, 25, 4490–4502. [Google Scholar] [CrossRef]
- Du, D.; Katsuno, Y.; Meyer, D.; Budi, E.H.; Chen, S.-H.; Koeppen, H.; Wang, H.; Akhurst, R.J.; Derynck, R. Smad3-mediated recruitment of the methyltransferase SETDB1/ESET controls Snail1 expression and epithelial–mesenchymal transition. EMBO Rep. 2018, 19, 135–155. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.-H.; Zhang, Y.; Wu, R.-Y.; Derynck, R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev. 1998, 12, 2153–2163. [Google Scholar] [CrossRef]
- Song, C.; Zhou, C. HOXA10 mediates epithelial-mesenchymal transition to promote gastric cancer metastasis partly via modulation of TGFB2/Smad/METTL3 signaling axis. J. Exp. Clin. Cancer Res. 2021, 40, 62. [Google Scholar] [CrossRef]
- Aragón, E.; Wang, Q.; Zou, Y.; Morgani, S.M.; Ruiz, L.; Kaczmarska, Z.; Su, J.; Torner, C.; Tian, L.; Hu, J.; et al. Structural basis for distinct roles of SMAD2 and SMAD3 in FOXH1 pioneer-directed TGF-β signaling. Genes Dev. 2019, 33, 1506–1524. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, Y.; Zhang, S.; Liu, H.; He, H.; Li, N.; Gong, Y.; Zhao, S.; Jiang, J.D.; Shao, R.G. Involvement of Ras GTPase-activating protein SH3 domain-binding protein 1 in the epithelial-to-mesenchymal transition-induced metastasis of breast cancer cells via the Smad signaling pathway. Oncotarget 2015, 6, 17039–17053. [Google Scholar] [CrossRef]
- Vervoort, S.J.; de Jong, O.G.; Roukens, M.G.; Frederiks, C.L.; Vermeulen, J.F.; Lourenço, A.R.; Bella, L.; Vidakovic, A.T.; Sandoval, J.L.; Moelans, C.; et al. Global transcriptional analysis identifies a novel role for SOX4 in tumor-induced angiogenesis. Elife 2018, 7, e27706. [Google Scholar] [CrossRef]
- Seoane, J.; Le, H.-V.; Shen, L.; Anderson, S.A.; Massagué, J. Integration of Smad and Forkhead Pathways in the Control of Neuroepithelial and Glioblastoma Cell Proliferation. Cell 2004, 117, 211–223. [Google Scholar] [CrossRef] [PubMed]
- González-González, A.; Muñoz-Muela, E.; Marchal, J.A.; Cara, F.E.; Molina, M.P.; Cruz-Lozano, M.; Jiménez, G.; Verma, A.; Ramírez, A.; Qian, W.; et al. Activating Transcription Factor 4 Modulates TGFβ-Induced Aggressiveness in Triple-Negative Breast Cancer via SMAD2/3/4 and mTORC2 Signaling. Clin. Cancer Res. 2018, 24, 5697–5709. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.W.; Hui, W.W.Y.; Wang, J.J.; Yung, M.M.H.; Hui, L.M.N.; Qin, Y.; Liang, R.R.; Leung, T.H.Y.; Xu, D.; Chan, K.K.L.; et al. DLX1 acts as a crucial target of FOXM1 to promote ovarian cancer aggressiveness by enhancing TGF-β/SMAD4 signaling. Oncogene 2017, 36, 1404–1416. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.R.; Kang, Y.; Siegel, P.M.; Massagué, J. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell 2002, 110, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Verrecchia, F.; Vindevoghel, L.; Lechleider, R.J.; Uitto, J.; Roberts, A.B.; Mauviel, A. Smad3/AP-1 interactions control transcriptional responses to TGF-β in a promoter-specific manner. Oncogene 2001, 20, 3332–3340. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Wu, J.; Jiang, Q.; Cheng, H.; Han, J.-D.J.; Chen, Y.-G. CXXC5 suppresses hepatocellular carcinoma by promoting TGF-β-induced cell cycle arrest and apoptosis. J. Mol. Cell Biol. 2017, 10, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Tu, S.; Wang, J.; Luo, S.; Yan, X. CXXC5: A novel regulator and coordinator of TGF-β, BMP and Wnt signaling. J. Cell Mol. Med. 2019, 23, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Adam, P.J.; Regan, C.P.; Hautmann, M.B.; Owens, G.K. Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22alpha in vivo. J. Biol. Chem. 2000, 275, 37798–37806. [Google Scholar] [CrossRef]
- Li, H.X.; Han, M.; Bernier, M.; Zheng, B.; Sun, S.G.; Su, M.; Zhang, R.; Fu, J.R.; Wen, J.K. Krüppel-like factor 4 promotes differentiation by transforming growth factor-beta receptor-mediated Smad and p38 MAPK signaling in vascular smooth muscle cells. J. Biol. Chem. 2010, 285, 17846–17856. [Google Scholar] [CrossRef]
- Spittau, B.; Krieglstein, K. Klf10 and Klf11 as mediators of TGF-beta superfamily signaling. Cell Tissue Res. 2012, 347, 65–72. [Google Scholar] [CrossRef]
- Buck, A.; Buchholz, M.; Wagner, M.; Adler, G.; Gress, T.; Ellenrieder, V. The tumor suppressor KLF11 mediates a novel mechanism in transforming growth factor beta-induced growth inhibition that is inactivated in pancreatic cancer. Mol. Cancer Res. 2006, 4, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wu, H.; Xu, Y.; Zhang, Z.; Liu, T.; Lin, X.; Feng, X.H. Zinc finger protein 451 is a novel Smad corepressor in transforming growth factor-β signaling. J. Biol. Chem. 2014, 289, 2072–2083. [Google Scholar] [CrossRef]
- Xi, Q.; Wang, Z.; Zaromytidou, A.-I.; Zhang, X.H.F.; Chow-Tsang, L.-F.; Liu, J.X.; Kim, H.; Barlas, A.; Manova-Todorova, K.; Kaartinen, V.; et al. A Poised Chromatin Platform for TGF-β Access to Master Regulators. Cell 2011, 147, 1511–1524. [Google Scholar] [CrossRef]
- Tang, X.; Li, G.; Su, F.; Cai, Y.; Shi, L.; Meng, Y.; Liu, Z.; Sun, J.; Wang, M.; Qian, M.; et al. HDAC8 cooperates with SMAD3/4 complex to suppress SIRT7 and promote cell survival and migration. Nucleic Acids Res. 2020, 48, 2912–2923. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.S.; Hong, J.J.; Wu, J.F.; Yan, S.; Wu, D.; Liu, N.; Liu, Q.F.; Wu, Q.W.; Xie, Y.Y.; Liu, Y.J.; et al. OVOL2 antagonizes TGF-β signaling to regulate epithelial to mesenchymal transition during mammary tumor metastasis. Oncotarget 2017, 8, 39401–39416. [Google Scholar] [CrossRef]
- Stroschein, S.L.; Wang, W.; Zhou, S.; Zhou, Q.; Luo, K. Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science 1999, 286, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.; Stroschein, S.L.; Wang, W.; Chen, D.; Martens, E.; Zhou, S.; Zhou, Q. The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling. Genes Dev. 1999, 13, 2196–2206. [Google Scholar] [CrossRef]
- Wotton, D.; Lo, R.S.; Swaby, L.-A.C.; Massagué, J. Multiple Modes of Repression by the Smad Transcriptional Corepressor TGIF*. J. Biol. Chem. 1999, 274, 37105–37110. [Google Scholar] [CrossRef]
- Guca, E.; Suñol, D.; Ruiz, L.; Konkol, A.; Cordero, J.; Torner, C.; Aragon, E.; Martin-Malpartida, P.; Riera, A.; Macias, M.J. TGIF1 homeodomain interacts with Smad MH1 domain and represses TGF-β signaling. Nucleic Acids Res. 2018, 46, 9220–9235. [Google Scholar] [CrossRef]
- Boon, R.A.; Fledderus, J.O.; Volger, O.L.; van Wanrooij, E.J.; Pardali, E.; Weesie, F.; Kuiper, J.; Pannekoek, H.; ten Dijke, P.; Horrevoets, A.J. KLF2 suppresses TGF-beta signaling in endothelium through induction of Smad7 and inhibition of AP-1. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Nakao, A.; Afrakhte, M.; Morén, A.; Nakayama, T.; Christian, J.L.; Heuchel, R.; Itoh, S.; Kawabata, M.; Heldin, N.E.; Heldin, C.H.; et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature 1997, 389, 631–635. [Google Scholar] [CrossRef]
- Itoh, S.; Landström, M.; Hermansson, A.; Itoh, F.; Heldin, C.-H.; Heldin, N.-E.; ten Dijke, P. Transforming Growth Factor β1 Induces Nuclear Export of Inhibitory Smad7*. J. Biol. Chem. 1998, 273, 29195–29201. [Google Scholar] [CrossRef] [PubMed]
- Elkouris, M.; Kontaki, H.; Stavropoulos, A.; Antonoglou, A.; Nikolaou, K.C.; Samiotaki, M.; Szantai, E.; Saviolaki, D.; Brown, P.J.; Sideras, P.; et al. SET9-Mediated Regulation of TGF-β Signaling Links Protein Methylation to Pulmonary Fibrosis. Cell Rep. 2016, 15, 2733–2744. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Sixt, K.M.; Gao, S.; Xu, X.; Huang, J.; Weigert, R.; Zhou, M.; Zhang, Y.E. Direct Regulation of Alternative Splicing by SMAD3 through PCBP1 Is Essential to the Tumor-Promoting Role of TGF-β. Mol. Cell 2016, 64, 549–564. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.N.; Hilyard, A.C.; Lagna, G.; Hata, A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 2008, 454, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.N.; Hilyard, A.C.; Nguyen, P.H.; Lagna, G.; Hata, A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol. Cell 2010, 39, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Xiong, X.; Chen, Y.-G. Feedback regulation of TGF-β signaling. Acta Biochim. Biophys. Sin. 2017, 50, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Adylova, A.; Mukhanbetzhanovna, A.A.; Attar, R.; Yulaevna, I.M.; Farooqi, A.A. Regulation of TGFβ/SMAD signaling by long non-coding RNAs in different cancers: Dark Knight in the Castle of molecular oncology. Noncoding RNA Res. 2021, 6, 23–28. [Google Scholar] [CrossRef]
- Bertero, A.; Brown, S.; Madrigal, P.; Osnato, A.; Ortmann, D.; Yiangou, L.; Kadiwala, J.; Hubner, N.C.; de Los Mozos, I.R.; Sadée, C.; et al. The SMAD2/3 interactome reveals that TGFβ controls m(6)A mRNA methylation in pluripotency. Nature 2018, 555, 256–259. [Google Scholar] [CrossRef]
- Bassi, Z.I.; Fillmore, M.C.; Miah, A.H.; Chapman, T.D.; Maller, C.; Roberts, E.J.; Davis, L.C.; Lewis, D.E.; Galwey, N.W.; Waddington, K.E.; et al. Modulating PCAF/GCN5 Immune Cell Function through a PROTAC Approach. ACS Chem. Biol. 2018, 13, 2862–2867. [Google Scholar] [CrossRef]
- Collins, H.M.; Abdelghany, M.K.; Messmer, M.; Yue, B.; Deeves, S.E.; Kindle, K.B.; Mantelingu, K.; Aslam, A.; Winkler, G.S.; Kundu, T.K.; et al. Differential effects of garcinol and curcumin on histone and p53 modifications in tumour cells. BMC Cancer 2013, 13, 37. [Google Scholar] [CrossRef]
- Mota, M.; Sweha, S.R.; Pun, M.; Natarajan, S.K.; Ding, Y.; Chung, C.; Hawes, D.; Yang, F.; Judkins, A.R.; Samajdar, S.; et al. Targeting SWI/SNF ATPases in H3.3K27M diffuse intrinsic pontine gliomas. Proc. Natl. Acad. Sci. USA 2023, 120, e2221175120. [Google Scholar] [CrossRef]
- Lee, D.; Lee, D.Y.; Hwang, Y.S.; Seo, H.R.; Lee, S.A.; Kwon, J. The Bromodomain Inhibitor PFI-3 Sensitizes Cancer Cells to DNA Damage by Targeting SWI/SNF. Mol. Cancer Res. 2021, 19, 900–912. [Google Scholar] [CrossRef]
- Papillon, J.P.N.; Nakajima, K.; Adair, C.D.; Hempel, J.; Jouk, A.O.; Karki, R.G.; Mathieu, S.; Möbitz, H.; Ntaganda, R.; Smith, T.; et al. Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers. J. Med. Chem. 2018, 61, 10155–10172. [Google Scholar] [CrossRef]
- Federico, A.; Steinfass, T.; Larribère, L.; Novak, D.; Morís, F.; Núñez, L.E.; Umansky, V.; Utikal, J. Mithramycin A and Mithralog EC-8042 Inhibit SETDB1 Expression and Its Oncogenic Activity in Malignant Melanoma. Mol. Ther. Oncolytics 2020, 18, 83–99. [Google Scholar] [CrossRef]
- Lee, J.K.; Kim, K.C. DZNep, inhibitor of S-adenosylhomocysteine hydrolase, down-regulates expression of SETDB1 H3K9me3 HMTase in human lung cancer cells. Biochem. Biophys. Res. Commun. 2013, 438, 647–652. [Google Scholar] [CrossRef]
- Shao, Y.; Song, X.; Jiang, W.; Chen, Y.; Ning, Z.; Gu, W.; Jiang, J. MicroRNA-621 Acts as a Tumor Radiosensitizer by Directly Targeting SETDB1 in Hepatocellular Carcinoma. Mol. Ther. 2019, 27, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.H.; Gmachl, M.; Ramharter, J.; Savarese, F.; Gerlach, D.; Marszalek, J.R.; Sanderson, M.P.; Kessler, D.; Trapani, F.; Arnhof, H.; et al. BI-3406, a Potent and Selective SOS1-KRAS Interaction Inhibitor, Is Effective in KRAS-Driven Cancers through Combined MEK Inhibition. Cancer Discov. 2021, 11, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Park, D.; Magis, A.T.; Zhang, J.; Zhou, W.; Sica, G.L.; Ramalingam, S.S.; Curran, W.J.; Deng, X. Small Molecule KRAS Agonist for Mutant KRAS Cancer Therapy. Mol. Cancer 2019, 18, 85. [Google Scholar] [CrossRef]
- Shima, F.; Yoshikawa, Y.; Ye, M.; Araki, M.; Matsumoto, S.; Liao, J.; Hu, L.; Sugimoto, T.; Ijiri, Y.; Takeda, A.; et al. In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras–effector interaction. Proc. Natl. Acad. Sci. USA 2013, 110, 8182–8187. [Google Scholar] [CrossRef] [PubMed]
- Janes, M.R.; Zhang, J.; Li, L.-S.; Hansen, R.; Peters, U.; Guo, X.; Chen, Y.; Babbar, A.; Firdaus, S.J.; Darjania, L.; et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018, 172, 578–589.e17. [Google Scholar] [CrossRef] [PubMed]
- Afaloniati, H.; Angelopoulou, K.; Giakoustidis, A.; Hardas, A.; Pseftogas, A.; Makedou, K.; Gargavanis, A.; Goulopoulos, T.; Iliadis, S.; Papadopoulos, V.; et al. HDAC1/2 Inhibitor Romidepsin Suppresses DEN-Induced Hepatocellular Carcinogenesis in Mice. Onco Targets Ther. 2020, 13, 5575–5588. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C.; Kiesslich, T.; Erber, S.; Bekric, D.; Dobias, H.; Beyreis, M.; Ritter, M.; Jäger, T.; Neumayer, B.; Winkelmann, P.; et al. HDAC Screening Identifies the HDAC Class I Inhibitor Romidepsin as a Promising Epigenetic Drug for Biliary Tract Cancer. Cancers 2021, 13, 3862. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-H.; Zhang, X.-B.; Han, X.-Q.; Feng, C.-R.; Wang, F.S.; Wang, P.G.; Shen, J.; Shi, Y.-K. Antitumor effects of a novel histone deacetylase inhibitor NK-HDAC-1 on breast cancer. Oncol. Rep. 2013, 30, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Park, S.B.; Kim, S.A.; Kwon, S.K.; Cha, H.; Lee, D.Y.; Ro, S.; Cho, J.M.; Song, S.Y. A novel HDAC inhibitor, CG200745, inhibits pancreatic cancer cell growth and overcomes gemcitabine resistance. Sci. Rep. 2017, 7, 41615. [Google Scholar] [CrossRef]
- Chun, S.-M.; Lee, J.-Y.; Choi, J.; Lee, J.-H.; Hwang, J.J.; Kim, C.-S.; Suh, Y.-A.; Jang, S.J. Epigenetic Modulation with HDAC Inhibitor CG200745 Induces Anti-Proliferation in Non-Small Cell Lung Cancer Cells. PLoS ONE 2015, 10, e0119379. [Google Scholar] [CrossRef] [PubMed]
- Sandor, V.; Bakke, S.; Robey, R.W.; Kang, M.H.; Blagosklonny, M.V.; Bender, J.; Brooks, R.; Piekarz, R.L.; Tucker, E.; Figg, W.D.; et al. Phase I trial of the histone deacetylase inhibitor, depsipeptide (FR901228, NSC 630176), in patients with refractory neoplasms. Clin. Cancer Res. 2002, 8, 718–728. [Google Scholar] [PubMed]
- Sandor, V.; Senderowicz, A.; Mertins, S.; Sackett, D.; Sausville, E.; Blagosklonny, M.V.; Bates, S.E. P21-dependent G1arrest with downregulation of cyclin D1 and upregulation of cyclin E by the histone deacetylase inhibitor FR901228. Br. J. Cancer 2000, 83, 817–825. [Google Scholar] [CrossRef]
- Nian, H.; Bisson, W.H.; Dashwood, W.-M.; Pinto, J.T.; Dashwood, R.H. α-Keto acid metabolites of organoselenium compounds inhibit histone deacetylase activity in human colon cancer cells. Carcinogenesis 2009, 30, 1416–1423. [Google Scholar] [CrossRef]
- Suzuki, T.; Muto, N.; Bando, M.; Itoh, Y.; Masaki, A.; Ri, M.; Ota, Y.; Nakagawa, H.; Iida, S.; Shirahige, K.; et al. Design, Synthesis, and Biological Activity of NCC149 Derivatives as Histone Deacetylase 8-Selective Inhibitors. ChemMedChem 2014, 9, 657–664. [Google Scholar] [CrossRef]
- Rettig, I.; Koeneke, E.; Trippel, F.; Mueller, W.C.; Burhenne, J.; Kopp-Schneider, A.; Fabian, J.; Schober, A.; Fernekorn, U.; von Deimling, A.; et al. Selective inhibition of HDAC8 decreases neuroblastoma growth in vitro and in vivo and enhances retinoic acid-mediated differentiation. Cell Death Dis. 2015, 6, e1657. [Google Scholar] [CrossRef]
- Du, Y.; Yuan, Y.; Xu, L.; Zhao, F.; Wang, W.; Xu, Y.; Tian, X. Discovery of METTL3 Small Molecule Inhibitors by Virtual Screening of Natural Products. Front Pharmacol 2022, 13, 878135. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Zhao, R.; Meng, W.; Liao, Y. Effects and translatomics characteristics of a small-molecule inhibitor of METTL3 against non-small cell lung cancer. J. Pharm. Anal. 2023, 13, 625–639. [Google Scholar] [CrossRef] [PubMed]
- Barsyte-Lovejoy, D.; Li, F.; Oudhoff, M.J.; Tatlock, J.H.; Dong, A.; Zeng, H.; Wu, H.; Freeman, S.A.; Schapira, M.; Senisterra, G.A.; et al. (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proc. Natl. Acad. Sci. USA 2014, 111, 12853–12858. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.K.; Pardoux, C.; Hall, M.C.; Lee, P.S.; Warburton, D.; Qing, J.; Smith, S.M.; Derynck, R. TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. Embo J. 2007, 26, 3957–3967. [Google Scholar] [CrossRef] [PubMed]
- Zuo, W.; Chen, Y.G. Specific activation of mitogen-activated protein kinase by transforming growth factor-beta receptors in lipid rafts is required for epithelial cell plasticity. Mol. Biol. Cell 2009, 20, 1020–1029. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, A.; Thakur, N.; Grimsby, S.; Marcusson, A.; von Bulow, V.; Schuster, N.; Zhang, S.; Heldin, C.-H.; Landström, M. The type I TGF-β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat. Cell Biol. 2008, 10, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, M.; Fatyol, K.; Jin, C.; Wang, X.; Liu, Z.; Zhang, Y.E. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol. Cell 2008, 31, 918–924. [Google Scholar] [CrossRef] [PubMed]
- Yumoto, K.; Thomas, P.S.; Lane, J.; Matsuzaki, K.; Inagaki, M.; Ninomiya-Tsuji, J.; Scott, G.J.; Ray, M.K.; Ishii, M.; Maxson, R.; et al. TGF-β-activated kinase 1 (Tak1) mediates agonist-induced Smad activation and linker region phosphorylation in embryonic craniofacial neural crest-derived cells. J. Biol. Chem. 2013, 288, 13467–13480. [Google Scholar] [CrossRef]
- Edlund, S.; Bu, S.; Schuster, N.; Aspenström, P.; Heuchel, R.; Heldin, N.E.; ten Dijke, P.; Heldin, C.H.; Landström, M. Transforming growth factor-beta1 (TGF-beta)-induced apoptosis of prostate cancer cells involves Smad7-dependent activation of p38 by TGF-beta-activated kinase 1 and mitogen-activated protein kinase kinase 3. Mol. Biol. Cell 2003, 14, 529–544. [Google Scholar] [CrossRef]
- Jung, S.M.; Lee, J.H.; Park, J.; Oh, Y.S.; Lee, S.K.; Park, J.S.; Lee, Y.S.; Kim, J.H.; Lee, J.Y.; Bae, Y.S.; et al. Smad6 inhibits non-canonical TGF-β1 signaling by recruiting the deubiquitinase A20 to TRAF6. Nat. Commun. 2013, 4, 2562. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, F.; García de Vinuesa, A.; de Kruijf, E.M.; Mesker, W.E.; Hui, L.; Drabsch, Y.; Li, Y.; Bauer, A.; Rousseau, A.; et al. TRAF4 Promotes TGF-β Receptor Signaling and Drives Breast Cancer Metastasis. Mol. Cell 2013, 51, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Owen, K.L.; Brockwell, N.K.; Parker, B.S. JAK-STAT Signaling: A Double-Edged Sword of Immune Regulation and Cancer Progression. Cancers 2019, 11, 2002. [Google Scholar] [CrossRef]
- Tang, L.Y.; Heller, M.; Meng, Z.; Yu, L.R.; Tang, Y.; Zhou, M.; Zhang, Y.E. Transforming Growth Factor-β (TGF-β) Directly Activates the JAK1-STAT3 Axis to Induce Hepatic Fibrosis in Coordination with the SMAD Pathway. J. Biol. Chem. 2017, 292, 4302–4312. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.Y.; Shin, I.; Arteaga, C.L. Type I Transforming Growth Factor β Receptor Binds to and Activates Phosphatidylinositol 3-Kinase*. J. Biol. Chem. 2005, 280, 10870–10876. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, A.; Song, J.; Thakur, N.; Itoh, S.; Marcusson, A.; Bergh, A.; Heldin, C.H.; Landström, M. TGF-β promotes PI3K-AKT signaling and prostate cancer cell migration through the TRAF6-mediated ubiquitylation of p85α. Sci. Signal 2017, 10, aal4186. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Connolly, E.; Smyth, J.W.; Akhurst, R.J.; Derynck, R. TGF-β-induced activation of mTOR complex 2 drives epithelial-mesenchymal transition and cell invasion. J. Cell Sci. 2012, 125 Pt 5, 1259–1273. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Derynck, R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J. Cell Biol. 2007, 178, 437–451. [Google Scholar] [CrossRef]
- Mu, Y.; Sundar, R.; Thakur, N.; Ekman, M.; Gudey, S.K.; Yakymovych, M.; Hermansson, A.; Dimitriou, H.; Bengoechea-Alonso, M.T.; Ericsson, J.; et al. TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer. Nat. Commun. 2011, 2, 330. [Google Scholar] [CrossRef]
- Gudey, S.K.; Sundar, R.; Mu, Y.; Wallenius, A.; Zang, G.; Bergh, A.; Heldin, C.H.; Landström, M. TRAF6 stimulates the tumor-promoting effects of TGFβ type I receptor through polyubiquitination and activation of presenilin 1. Sci. Signal 2014, 7, ra2. [Google Scholar] [CrossRef]
- Sundar, R.; Gudey, S.K.; Heldin, C.-H.; Landström, M. TRAF6 promotes TGFβ-induced invasion and cell-cycle regulation via Lys63-linked polyubiquitination of Lys178 in TGFβ type I receptor. Cell Cycle 2015, 14, 554–565. [Google Scholar] [CrossRef]
- Bhowmick, N.A.; Ghiassi, M.; Bakin, A.; Aakre, M.; Lundquist, C.A.; Engel, M.E.; Arteaga, C.L.; Moses, H.L. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol. Biol. Cell 2001, 12, 27–36. [Google Scholar] [CrossRef]
- Edlund, S.; Landström, M.; Heldin, C.-H.; Aspenström, P. Transforming Growth Factor-β–induced Mobilization of Actin Cytoskeleton Requires Signaling by Small GTPases Cdc42 and RhoA. Mol. Biol. Cell 2002, 13, 902–914. [Google Scholar] [CrossRef] [PubMed]
- Ozdamar, B.; Bose, R.; Barrios-Rodiles, M.; Wang, H.-R.; Zhang, Y.; Wrana, J.L. Regulation of the Polarity Protein Par6 by TGFß Receptors Controls Epithelial Cell Plasticity. Science 2005, 307, 1603–1609. [Google Scholar] [CrossRef] [PubMed]
- Wilkes, M.C.; Murphy, S.J.; Garamszegi, N.; Leof, E.B. Cell-Type-Specific Activation of PAK2 by Transforming Growth Factor β Independent of Smad2 and Smad3. Mol. Cell. Biol. 2003, 23, 8878–8889. [Google Scholar] [CrossRef] [PubMed]
- Ferrigno, O.; Lallemand, F.; Verrecchia, F.; L’Hoste, S.; Camonis, J.; Atfi, A.; Mauviel, A. Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-β/Smad signaling. Oncogene 2002, 21, 4879–4884. [Google Scholar] [CrossRef] [PubMed]
- Alarcón, C.; Zaromytidou, A.-I.; Xi, Q.; Gao, S.; Yu, J.; Fujisawa, S.; Barlas, A.; Miller, A.N.; Manova-Todorova, K.; Macias, M.J.; et al. Nuclear CDKs Drive Smad Transcriptional Activation and Turnover in BMP and TGF-β Pathways. Cell 2009, 139, 757–769. [Google Scholar] [CrossRef] [PubMed]
- Nallet-Staub, F.; Yin, X.; Gilbert, C.; Marsaud, V.; Ben Mimoun, S.; Javelaud, D.; Leof, E.B.; Mauviel, A. Cell Density Sensing Alters TGF-β Signaling in a Cell-Type-Specific Manner, Independent from Hippo Pathway Activation. Dev. Cell 2015, 32, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Szeto, S.G.; Narimatsu, M.; Lu, M.; He, X.; Sidiqi, A.M.; Tolosa, M.F.; Chan, L.; De Freitas, K.; Bialik, J.F.; Majumder, S.; et al. YAP/TAZ Are Mechanoregulators of TGF-β-Smad Signaling and Renal Fibrogenesis. J. Am. Soc. Nephrol. 2016, 27, 3117–3128. [Google Scholar] [CrossRef]
- Lee, D.-H.; Park, J.O.; Kim, T.-S.; Kim, S.-K.; Kim, T.-h.; Kim, M.-c.; Park, G.S.; Kim, J.-H.; Kuninaka, S.; Olson, E.N.; et al. LATS-YAP/TAZ controls lineage specification by regulating TGFβ signaling and Hnf4α expression during liver development. Nat. Commun. 2016, 7, 11961. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, Y.; Sharma, K. Transforming Growth Factor-β1 Stimulates Protein Kinase A in Mesangial Cells*. J. Biol. Chem. 1998, 273, 8522–8527. [Google Scholar] [CrossRef]
- Yang, H.; Li, G.; Wu, J.J.; Wang, L.; Uhler, M.; Simeone, D.M. Protein kinase A modulates transforming growth factor-β signaling through a direct interaction with Smad4 protein. J. Biol. Chem. 2013, 288, 8737–8749. [Google Scholar] [CrossRef]
- Felici, A.; Wurthner, J.U.; Parks, W.T.; Ruh-yu Giam, L.; Reiss, M.; Karpova, T.S.; McNally, J.G.; Roberts, A.B. TLP, a novel modulator of TGF-β signaling, has opposite effects on Smad2- and Smad3-dependent signaling. EMBO J. 2003, 22, 4465–4477. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, B.; Liang, H.; Lu, Y.; Ai, X.; Zhang, B.; Chen, X. JNK inhibitor SP600125 enhances TGF-β-induced apoptosis of RBE human cholangiocarcinoma cells in a Smad-dependent manner. Mol. Med. Rep. 2013, 8, 1623–1629. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.Y.; Chen, T.S.; Wang, X.P.; Qu, J.L.; Chen, M. The JNK inhibitor SP600125 enhances dihydroartemisinin-induced apoptosis by accelerating Bax translocation into mitochondria in human lung adenocarcinoma cells. FEBS Lett. 2010, 584, 4019–4026. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, Y.; Chen, L.; Yu, F.; Li, X.; Dan, T.; Zhao, J.; Zhou, S. Polyphyllin I induces G2/M phase arrest and apoptosis in U251 human glioma cells via mitochondrial dysfunction and the JNK signaling pathway. Acta Biochim. Biophys. Sin. 2017, 49, 479–486. [Google Scholar] [CrossRef]
- Cerbone, A.; Toaldo, C.; Pizzimenti, S.; Pettazzoni, P.; Dianzani, C.; Minelli, R.; Ciamporcero, E.; Roma, G.; Dianzani, M.U.; Canaparo, R.; et al. AS601245, an Anti-Inflammatory JNK Inhibitor, and Clofibrate Have a Synergistic Effect in Inducing Cell Responses and in Affecting the Gene Expression Profile in CaCo-2 Colon Cancer Cells. PPAR Res. 2012, 2012, 269751. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yang, X.; Kang, Q.; Lu, J.; Denzinger, M.; Kornmann, M.; Traub, B. JNK inhibitor IX restrains pancreatic cancer through p53 and p21. Front. Oncol. 2022, 12, 1006131. [Google Scholar] [CrossRef]
- Ebelt, N.D.; Kaoud, T.S.; Edupuganti, R.; Van Ravenstein, S.; Dalby, K.N.; Van, C.L. A c-Jun N-terminal kinase inhibitor, JNK-IN-8, sensitizes triple negative breast cancer cells to lapatinib. Oncotarget 2017, 8, 104894. [Google Scholar] [CrossRef] [PubMed]
- Campbell, R.M.; Anderson, B.D.; Brooks, N.A.; Brooks, H.B.; Chan, E.M.; De Dios, A.; Gilmour, R.; Graff, J.R.; Jambrina, E.; Mader, M.; et al. Characterization of LY2228820 Dimesylate, a Potent and Selective Inhibitor of p38 MAPK with Antitumor Activity. Mol. Cancer Ther. 2014, 13, 364–374. [Google Scholar] [CrossRef]
- Jin, X.; Mo, Q.; Zhang, Y.; Gao, Y.; Wu, Y.; Li, J.; Hao, X.; Ma, D.; Gao, Q.; Chen, P. The p38 MAPK inhibitor BIRB796 enhances the antitumor effects of VX680 in cervical cancer. Cancer Biol. Ther. 2016, 17, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, Y.; Xu, Y.; Sun, Q.; Liu, H.; Chen, Q.; Liu, B. BIRB796, an Inhibitor of p38 Mitogen-Activated Protein Kinase, Inhibits Proliferation and Invasion in Glioblastoma Cells. ACS Omega 2021, 6, 11466–11473. [Google Scholar] [CrossRef]
- Kim, M.N.; Lee, S.M.; Kim, J.S.; Hwang, S.G. Preclinical efficacy of a novel dual PI3K/mTOR inhibitor, CMG002, alone and in combination with sorafenib in hepatocellular carcinoma. Cancer Chemother. Pharmacol. 2019, 84, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Gao, B.; Gao, Y.; Yang, X.; Cheng, X.; Ou, Y. Phycocyanin Inhibits Tumorigenic Potential of Pancreatic Cancer Cells: Role of Apoptosis and Autophagy. Sci. Rep. 2016, 6, 34564. [Google Scholar] [CrossRef] [PubMed]
- Rafiq, R.A.; Quadri, A.; Nazir, L.A.; Peerzada, K.; Ganai, B.A.; Tasduq, S.A. A Potent Inhibitor of Phosphoinositide 3-Kinase (PI3K) and Mitogen Activated Protein (MAP) Kinase Signalling, Quercetin (3, 3′, 4′, 5, 7-Pentahydroxyflavone) Promotes Cell Death in Ultraviolet (UV)-B-Irradiated B16F10 Melanoma Cells. PLoS ONE 2015, 10, e0131253. [Google Scholar] [CrossRef] [PubMed]
- Raina, K.; Agarwal, C.; Wadhwa, R.; Serkova, N.J.; Agarwal, R. Energy deprivation by silibinin in colorectal cancer cells. Autophagy 2013, 9, 697–713. [Google Scholar] [CrossRef] [PubMed]
- Nanta, R.; Shrivastava, A.; Sharma, J.; Shankar, S.; Srivastava, R.K. Inhibition of sonic hedgehog and PI3K/Akt/mTOR pathways cooperate in suppressing survival, self-renewal and tumorigenic potential of glioblastoma-initiating cells. Mol. Cell. Biochem. 2019, 454, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Li, C.F.; Chen, J.Y.; Ho, Y.H.; Hsu, W.H.; Wu, L.C.; Lan, H.Y.; Hsu, D.S.; Tai, S.K.; Chang, Y.C.; Yang, M.H. Snail-induced claudin-11 prompts collective migration for tumour progression. Nat. Cell Biol. 2019, 21, 251–262. [Google Scholar] [CrossRef]
- Qi, Y.; Zhao, X.; Chen, J.; Pradipta, A.R.; Wei, J.; Ruan, H.; Zhou, L.; Hsung, R.P.; Tanaka, K. In vitro and in vivo cancer cell apoptosis triggered by competitive binding of Cinchona alkaloids to the RING domain of TRAF6. Biosci. Biotechnol. Biochem. 2019, 83, 1011–1026. [Google Scholar] [CrossRef]
- Shang, X.; Marchioni, F.; Sipes, N.; Evelyn, C.R.; Jerabek-Willemsen, M.; Duhr, S.; Seibel, W.; Wortman, M.; Zheng, Y. Rational Design of Small Molecule Inhibitors Targeting RhoA Subfamily Rho GTPases. Chem. Biol. 2012, 19, 699–710. [Google Scholar] [CrossRef]
- Shang, X.; Marchioni, F.; Evelyn, C.R.; Sipes, N.; Zhou, X.; Seibel, W.; Wortman, M.; Zheng, Y. Small-molecule inhibitors targeting G-protein-coupled Rho guanine nucleotide exchange factors. Proc. Natl. Acad. Sci. USA 2013, 110, 3155–3160. [Google Scholar] [CrossRef]
- Evelyn, C.R.; Wade, S.M.; Wang, Q.; Wu, M.; Iñiguez-Lluhí, J.A.; Merajver, S.D.; Neubig, R.R. CCG-1423: A small-molecule inhibitor of RhoA transcriptional signaling. Mol. Cancer Ther. 2007, 6, 2249–2260. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.I.; Watanabe, B.; Nakagawa, Y.; Minami, S.; Morita, T. RPEL Proteins Are the Molecular Targets for CCG-1423, an Inhibitor of Rho Signaling. PLoS ONE 2014, 9, e89016. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.C.; Huang, T.H.; Chang, C.S.; Tsai, Y.R.; Lin, R.H.; Lee, P.W.; Hsu, M.F.; Huang, L.J.; Wang, J.P. Signaling mechanisms of inhibition of phospholipase D activation by CHS-111 in formyl peptide-stimulated neutrophils. Biochem. Pharmacol. 2011, 81, 269–278. [Google Scholar] [CrossRef]
- Hoy, J.J.; Salinas Parra, N.; Park, J.; Kuhn, S.; Iglesias-Bartolome, R. Protein kinase A inhibitor proteins (PKIs) divert GPCR-Gαs-cAMP signaling toward EPAC and ERK activation and are involved in tumor growth. Faseb J. 2020, 34, 13900–13917. [Google Scholar] [CrossRef] [PubMed]
- Zynda, E.R.; Matveev, V.; Makhanov, M.; Chenchik, A.; Kandel, E.S. Protein kinase A type II-α regulatory subunit regulates the response of prostate cancer cells to taxane treatment. Cell Cycle 2014, 13, 3292–3301. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cassoni, P.; Sapino, A.; Fortunati, N.; Munaron, L.; Chini, B.; Bussolati, G. Oxytocin inhibits the proliferation of MDA-MB231 human breast-cancer cells via cyclic adenosine monophosphate and protein kinase A. Int. J. Cancer 1997, 72, 340–344. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, W.E.; Zhang, X.; Li, Y.; Chen, B.; Liu, C.; Ai, X.; Zhang, X.; Tian, Y.; Zhang, C.; et al. Cardiomyocyte PKA Ablation Enhances Basal Contractility While Eliminates Cardiac β-Adrenergic Response Without Adverse Effects on the Heart. Circ. Res. 2019, 124, 1760–1777. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Hong, W.; Wei, X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J. Hematol. Oncol. 2022, 15, 129. [Google Scholar] [CrossRef]
- Jan, R.; Chaudhry, G.E. Understanding Apoptosis and Apoptotic Pathways Targeted Cancer Therapeutics. Adv. Pharm. Bull. 2019, 9, 205–218. [Google Scholar] [CrossRef]
- Ramesh, S.; Wildey, G.M.; Howe, P.H. Transforming growth factor beta (TGFbeta)-induced apoptosis: The rise & fall of Bim. Cell Cycle 2009, 8, 11–17. [Google Scholar] [CrossRef]
- Zhang, Y.; Alexander, P.B.; Wang, X.F. TGF-β Family Signaling in the Control of Cell Proliferation and Survival. Cold Spring Harb. Perspect. Biol. 2017, 9, a022145. [Google Scholar] [CrossRef]
- Meulmeester, E.; ten Dijke, P. The dynamic roles of TGF-β in cancer. J. Pathol. 2011, 223, 206–219. [Google Scholar] [CrossRef]
- Yu, L.; Hébert, M.C.; Zhang, Y.E. TGF-beta receptor-activated p38 MAP kinase mediates Smad-independent TGF-beta responses. EMBO J. 2002, 21, 3749–3759. [Google Scholar] [CrossRef]
- Chen, B.; Mu, C.; Zhang, Z.; He, X.; Liu, X. The Love-Hate Relationship Between TGF-β Signaling and the Immune System During Development and Tumorigenesis. Front. Immunol. 2022, 13, 891268. [Google Scholar] [CrossRef]
- Schrantz, N.; Auffredou, M.T.; Bourgeade, M.F.; Besnault, L.; Leca, G.; Vazquez, A. Zinc-mediated regulation of caspases activity: Dose-dependent inhibition or activation of caspase-3 in the human Burkitt lymphoma B cells (Ramos). Cell Death Differ. 2001, 8, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Hu, Y.; Fan, X.; Wu, X.; Mao, Y.; Hu, B.; Guo, H.; Wen, L.; Tang, F. Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis. Genome Biol. 2018, 19, 31. [Google Scholar] [CrossRef]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e1624. [Google Scholar] [CrossRef]
- Aiello, N.M.; Maddipati, R.; Norgard, R.J.; Balli, D.; Li, J.; Yuan, S.; Yamazoe, T.; Black, T.; Sahmoud, A.; Furth, E.E.; et al. EMT Subtype Influences Epithelial Plasticity and Mode of Cell Migration. Dev. Cell 2018, 45, 681–695.e684. [Google Scholar] [CrossRef] [PubMed]
- Medici, D.; Kalluri, R. Endothelial–mesenchymal transition and its contribution to the emergence of stem cell phenotype. Semin. Cancer Biol. 2012, 22, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Piera-Velazquez, S.; Jimenez, S.A. Endothelial to Mesenchymal Transition: Role in Physiology and in the Pathogenesis of Human Diseases. Physiol. Rev. 2019, 99, 1281–1324. [Google Scholar] [CrossRef]
- Derynck, R.; Weinberg, R.A. EMT and Cancer: More Than Meets the Eye. Dev. Cell 2019, 49, 313–316. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Kajita, M.; McClinic, K.N.; Wade, P.A. Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol. Cell Biol. 2004, 24, 7559–7566. [Google Scholar] [CrossRef] [PubMed]
- Qiao, B.; Johnson, N.W.; Gao, J. Epithelial-mesenchymal transition in oral squamous cell carcinoma triggered by transforming growth factor-beta1 is Snail family-dependent and correlates with matrix metalloproteinase-2 and -9 expressions. Int. J. Oncol. 2010, 37, 663–668. [Google Scholar] [CrossRef]
- Li, W.; Shen, M.; Jiang, Y.Z.; Zhang, R.; Zheng, H.; Wei, Y.; Shao, Z.M.; Kang, Y. Deubiquitinase USP20 promotes breast cancer metastasis by stabilizing SNAI2. Genes Dev. 2020, 34, 1310–1315. [Google Scholar] [CrossRef]
- Fan, L.; Lei, H.; Zhang, S.; Peng, Y.; Fu, C.; Shu, G.; Yin, G. Non-canonical signaling pathway of SNAI2 induces EMT in ovarian cancer cells by suppressing miR-222-3p transcription and upregulating PDCD10. Theranostics 2020, 10, 5895–5913. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Gross, K.M.; Kuperwasser, C. Molecular regulation of Snail2 in development and disease. J. Cell Sci. 2019, 132, 235127. [Google Scholar] [CrossRef]
- Chaffer, C.L.; Marjanovic, N.D.; Lee, T.; Bell, G.; Kleer, C.G.; Reinhardt, F.; D’Alessio, A.C.; Young, R.A.; Weinberg, R.A. Poised Chromatin at the ZEB1 Promoter Enables Breast Cancer Cell Plasticity and Enhances Tumorigenicity. Cell 2013, 154, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Krebs, A.M.; Mitschke, J.; Lasierra Losada, M.; Schmalhofer, O.; Boerries, M.; Busch, H.; Boettcher, M.; Mougiakakos, D.; Reichardt, W.; Bronsert, P.; et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017, 19, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Chen, B.; Zhu, Z.; Ye, W.; Zeng, J.; Liu, G.; Wang, S.; Gao, J.; Xu, G.; Huang, Z. Prognostic value of ZEB-1 in solid tumors: A meta-analysis. BMC Cancer 2019, 19, 635. [Google Scholar] [CrossRef]
- Zhang, G.J.; Zhou, T.; Tian, H.P.; Liu, Z.L.; Xia, S.S. High expression of ZEB1 correlates with liver metastasis and poor prognosis in colorectal cancer. Oncol. Lett. 2013, 5, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, C.L.T.; Forsare, C.; Bendahl, P.O.; Falck, A.K.; Fernö, M.; Lövgren, K.; Aaltonen, K.; Rydén, L. Expression of epithelial-mesenchymal transition-related markers and phenotypes during breast cancer progression. Breast Cancer Res. Treat. 2020, 181, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Kurahara, H.; Takao, S.; Maemura, K.; Mataki, Y.; Kuwahata, T.; Maeda, K.; Ding, Q.; Sakoda, M.; Iino, S.; Ishigami, S.; et al. Epithelial–mesenchymal transition and mesenchymal–epithelial transition via regulation of ZEB-1 and ZEB-2 expression in pancreatic cancer. J. Surg. Oncol. 2012, 105, 655–661. [Google Scholar] [CrossRef]
- Yang, M.H.; Wu, M.Z.; Chiou, S.H.; Chen, P.M.; Chang, S.Y.; Liu, C.J.; Teng, S.C.; Wu, K.J. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat. Cell Biol. 2008, 10, 295–305. [Google Scholar] [CrossRef]
- Meng, J.; Chen, S.; Han, J.X.; Qian, B.; Wang, X.R.; Zhong, W.L.; Qin, Y.; Zhang, H.; Gao, W.F.; Lei, Y.Y.; et al. Twist1 Regulates Vimentin through Cul2 Circular RNA to Promote EMT in Hepatocellular Carcinoma. Cancer Res. 2018, 78, 4150–4162. [Google Scholar] [CrossRef]
- Yang, J.; Mani, S.A.; Donaher, J.L.; Ramaswamy, S.; Itzykson, R.A.; Come, C.; Savagner, P.; Gitelman, I.; Richardson, A.; Weinberg, R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004, 117, 927–939. [Google Scholar] [CrossRef]
- Lee, T.K.; Poon, R.T.; Yuen, A.P.; Ling, M.T.; Kwok, W.K.; Wang, X.H.; Wong, Y.C.; Guan, X.Y.; Man, K.; Chau, K.L.; et al. Twist overexpression correlates with hepatocellular carcinoma metastasis through induction of epithelial-mesenchymal transition. Clin. Cancer Res. 2006, 12, 5369–5376. [Google Scholar] [CrossRef]
- Pattabiraman, D.R.; Weinberg, R.A. Tackling the cancer stem cells—What challenges do they pose? Nat. Rev. Drug Discov. 2014, 13, 497–512. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Scheel, C.; Eaton, E.N.; Li, S.H.; Chaffer, C.L.; Reinhardt, F.; Kah, K.J.; Bell, G.; Guo, W.; Rubin, J.; Richardson, A.L.; et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 2011, 145, 926–940. [Google Scholar] [CrossRef]
- McCormack, N.; O’Dea, S. Regulation of epithelial to mesenchymal transition by bone morphogenetic proteins. Cell Signal. 2013, 25, 2856–2862. [Google Scholar] [CrossRef] [PubMed]
- Giampieri, S.; Manning, C.; Hooper, S.; Jones, L.; Hill, C.S.; Sahai, E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat. Cell Biol. 2009, 11, 1287–1296. [Google Scholar] [CrossRef] [PubMed]
- Katsuno, Y.; Derynck, R. Epithelial plasticity, epithelial-mesenchymal transition, and the TGF-β family. Dev. Cell 2021, 56, 726–746. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Turley, S.J.; Akhurst, R.J. TGFβ biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol. 2021, 18, 9–34. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef]
- Cheung, K.J.; Gabrielson, E.; Werb, Z.; Ewald, A.J. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 2013, 155, 1639–1651. [Google Scholar] [CrossRef]
- Katsuno, Y.; Meyer, D.S.; Zhang, Z.; Shokat, K.M.; Akhurst, R.J.; Miyazono, K.; Derynck, R. Chronic TGF-β exposure drives stabilized EMT, tumor stemness, and cancer drug resistance with vulnerability to bitopic mTOR inhibition. Sci. Signal 2019, 12, aau8544. [Google Scholar] [CrossRef] [PubMed]
- Ruscetti, M.; Quach, B.; Dadashian, E.L.; Mulholland, D.J.; Wu, H. Tracking and Functional Characterization of Epithelial-Mesenchymal Transition and Mesenchymal Tumor Cells during Prostate Cancer Metastasis. Cancer Res. 2015, 75, 2749–2759. [Google Scholar] [CrossRef]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef]
- Gomis, R.R.; Gawrzak, S. Tumor cell dormancy. Mol. Oncol. 2017, 11, 62–78. [Google Scholar] [CrossRef]
- Yeh, A.C.; Ramaswamy, S. Mechanisms of Cancer Cell Dormancy--Another Hallmark of Cancer? Cancer Res. 2015, 75, 5014–5022. [Google Scholar] [CrossRef] [PubMed]
- Harper, K.L.; Sosa, M.S.; Entenberg, D.; Hosseini, H.; Cheung, J.F.; Nobre, R.; Avivar-Valderas, A.; Nagi, C.; Girnius, N.; Davis, R.J.; et al. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature 2016, 540, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Rhim, A.D.; Mirek, E.T.; Aiello, N.M.; Maitra, A.; Bailey, J.M.; McAllister, F.; Reichert, M.; Beatty, G.L.; Rustgi, A.K.; Vonderheide, R.H.; et al. EMT and dissemination precede pancreatic tumor formation. Cell 2012, 148, 349–361. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).