The Antitubercular Activities of Natural Products with Fused-Nitrogen-Containing Heterocycles
Abstract
1. Introduction
2. Natural Products with Indole Moiety
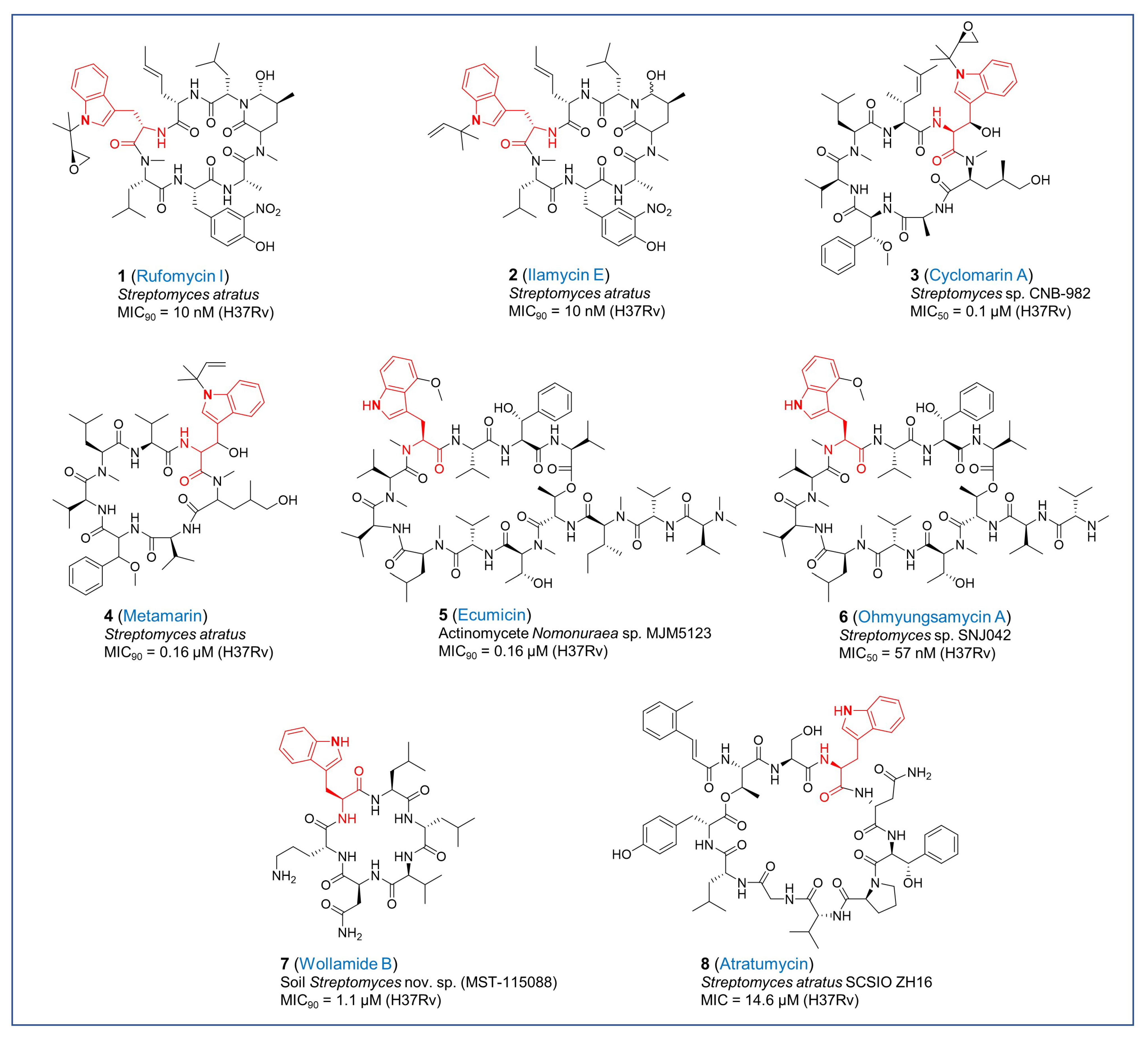
2.1. Tryptophan-Containing Cyclopeptides
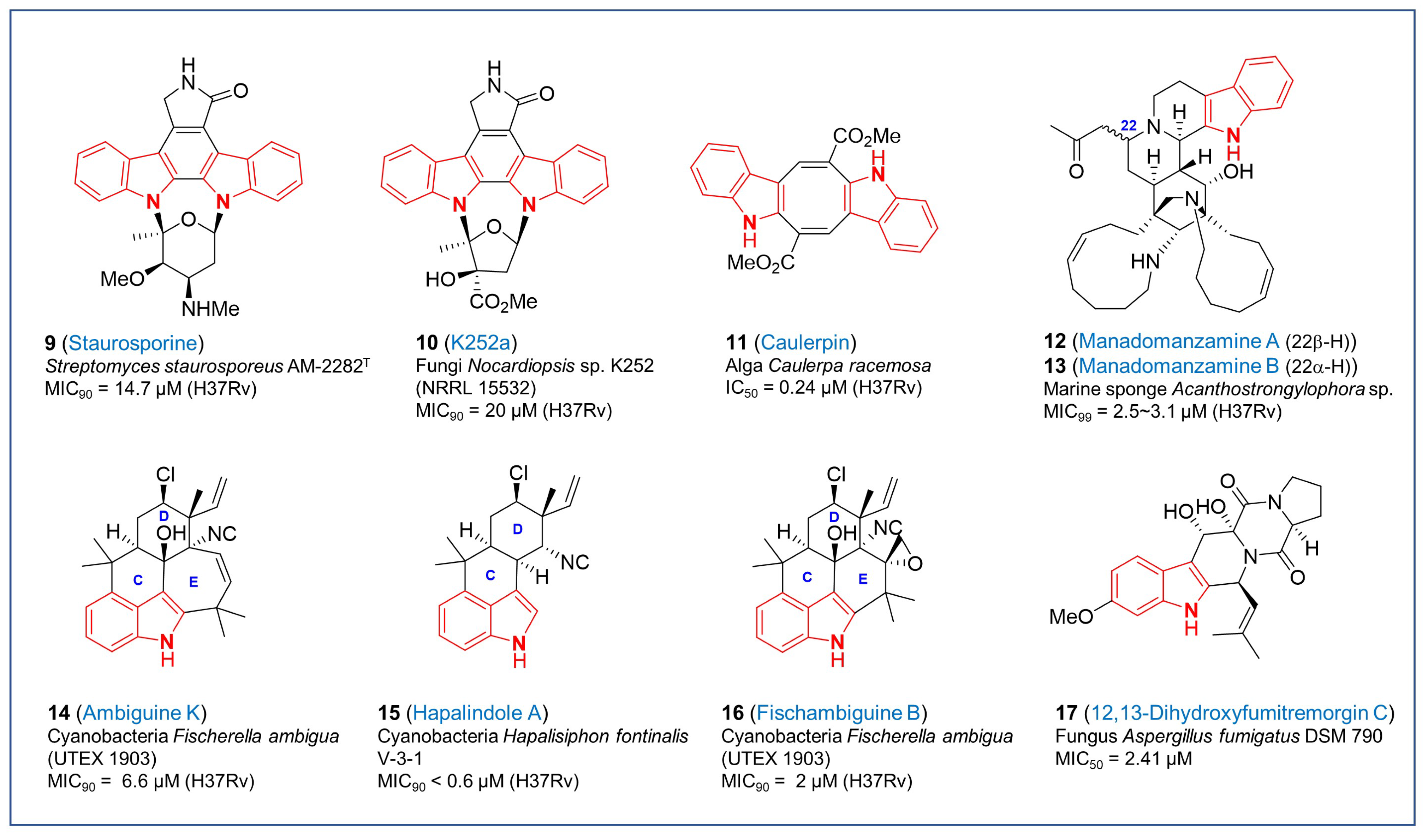
2.2. Other Indole Derivatives
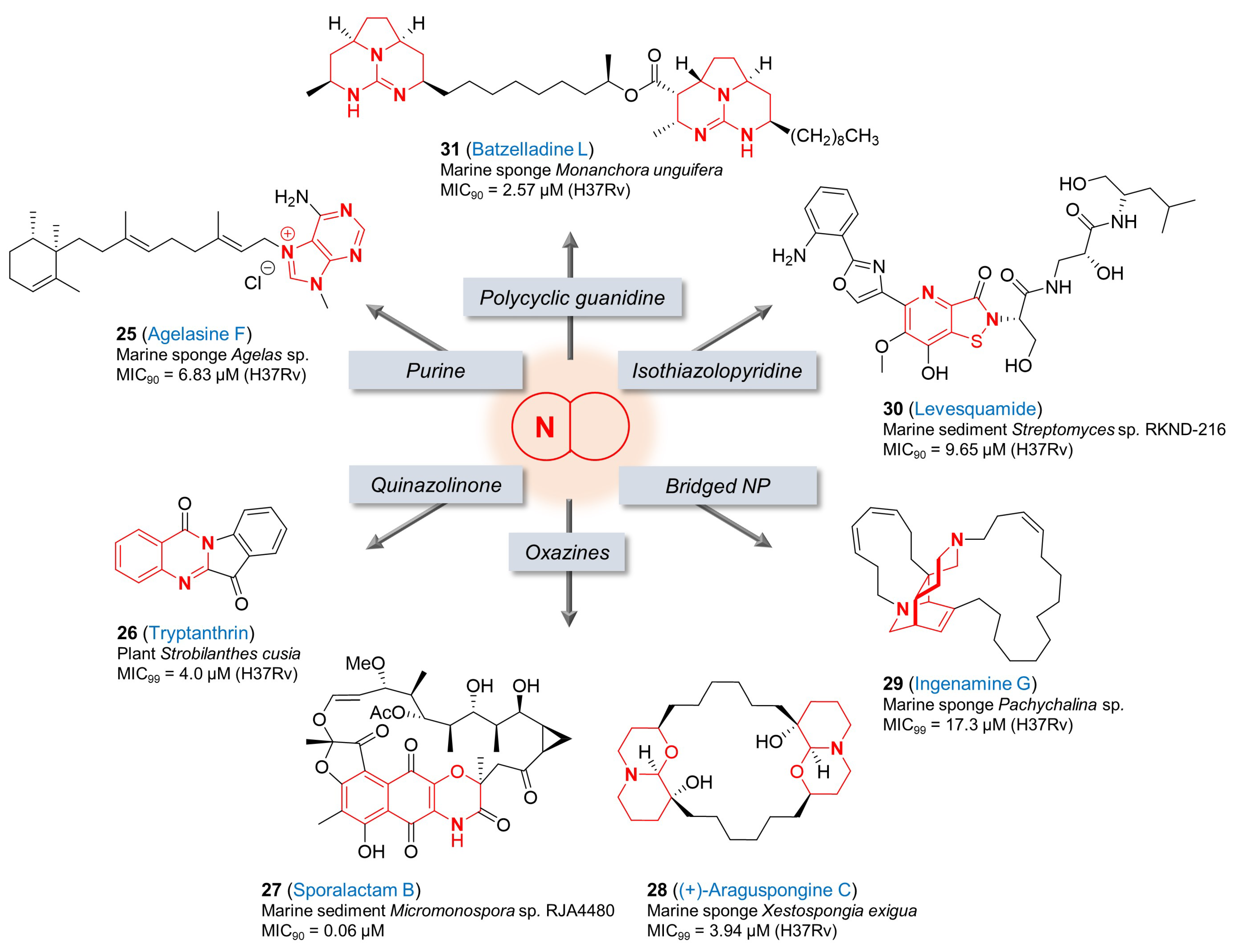
3. Natural Products with Other Fused-Nitrogen-Containing Heterocycles
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. The WHO Global Tuberculosis Report 2023; World Health Organization: Geneva, Switzerland, 2023. [Google Scholar]
- Derendinger, B.; Dippenaar, A.; de Vos, M.; Huo, S.; Alberts, R.; Tadokera, R.; Limberis, J.; Sirgel, F.; Dolby, T.; Spies, C.; et al. Bedaquiline resistance in patients with drug-resistant tuberculosis in Cape Town, South Africa: A retrospective longitudinal cohort study. Lancet Microbe 2023, 4, E972–E982. [Google Scholar] [CrossRef] [PubMed]
- Gygli, S.M.; Borrell, S.; Trauner, A.; Gagneux, S. Antimicrobial resistance in Mycobacterium tuberculosis: Mechanistic and evolutionary perspectives. FEMS Microbiol. Rev. 2017, 41, 354–373. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, G.; Midiri, A.; De Gaetano, S.; Ponzo, E.; Biondo, C. Tackling Drug-Resistant Tuberculosis: New Challenges from the Old Pathogen Mycobacterium tuberculosis. Microorganisms 2023, 11, 2277. [Google Scholar] [CrossRef] [PubMed]
- Walesch, S.; Birkelbach, J.; Jézéquel, G.; Haeckl, F.P.J.; Hegemann, J.D.; Hesterkamp, T.; Hirsch, A.K.H.; Hammann, P.; Müller, R. Fighting antibiotic resistance-strategies and (pre)clinical developments to find new antibacterials. EMBO Rep. 2023, 24, e56033. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T. Nature Builds Macrocycles and Heterocycles into Its Antimicrobial Frameworks: Deciphering Biosynthetic Strategy. ACS Infect. Dis. 2018, 4, 1283–1299. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; the International Natural Product Sciences Taskforce; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Surette, M.D.; Spanogiannopoulos, P.; Wright, G.D. The Enzymes of the Rifamycin Antibiotic Resistome. Accout. Chem. Res. 2021, 54, 2065–2075. [Google Scholar] [CrossRef]
- Skoreński, M.; Sieńczyk, M. The Fellowship of Privileged Scaffolds—One Structure to Inhibit Them All. Pharmaceuticals 2021, 14, 1164. [Google Scholar] [CrossRef]
- Bajad, N.G.; Singh, S.K.; Singh, S.K.; Singh, T.D.; Singh, M. Indole: A promising scaffold for the discovery and development of potential anti-tubercular agents. Curr. Res. Pharmacol. Drug Discov. 2022, 3, 100119. [Google Scholar] [CrossRef]
- Umer, S.M.; Solangi, M.; Khan, K.M.; Saleem, R.S.Z. Indole-Containing Natural Products 2019-2022: Isolations, Reappraisals, Syntheses, and Biological Activities. Molecules 2022, 27, 7586. [Google Scholar] [CrossRef]
- Xie, Y.; Xu, H.; Sun, C.; Yu, Y.; Chen, R. Two novel nucleosidyl-peptide antibiotics: Sansanmycin F and G produced by Streptomyces sp SS. J. Antibiot. 2010, 63, 143–146. [Google Scholar] [CrossRef]
- Tran, A.T.; Watson, E.E.; Pujari, V.; Conroy, T.; Dowman, L.J.; Giltrap, A.M.; Pang, A.; Wong, W.R.; Linington, R.G.; Mahapatra, S.; et al. Sansanmycin natural product analogues as potent and selective anti-mycobacterials that inhibit lipid I biosynthesis. Nat. Commun. 2017, 8, 14414. [Google Scholar] [CrossRef]
- Shibata, M.; Yamamoto, H.; Higashidani, E.; Nakazawa, K. Studies on Streptomycetes: Part I. Streptomyces atratus Nov. Sp., Producing New Antituberculous Antibiotics Rufomycin A and B. Agric. Biol. Chem. 1962, 26, 228–233. [Google Scholar] [CrossRef]
- Higashidani, E.; Ueyanagi, J.; Shibata, M.; Nakazawa, K.; Miyake, A.; Iwasaki, H.; Yamamoto, H. Studies on Streptomycetes: Part II. Rufomycin A and B, New Antituberculous Antibiotics. Agric. Biol. Chem. 1962, 26, 234–237. [Google Scholar] [CrossRef]
- Choules, M.P.; Wolf, N.M.; Lee, H.; Anderson, J.R.; Grzelak, E.M.; Wang, Y.; Ma, R.; Gao, W.; McAlpine, J.B.; Jin, Y.-Y.; et al. Rufomycin Targets ClpC1 Proteolysis in Mycobacterium tuberculosis and M. abscessus. Antimicrob. Agents Chemother. 2019, 63, e02204-18. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Achanta, P.S.; Shetye, G.; Chen, S.-N.; Lee, H.; Jin, Y.-Y.; Cheng, J.; Lee, M.-J.; Suh, J.-W.; Cho, S.; et al. Rufomycins or Ilamycins: Naming Clarifications and Definitive Structural Assignments. J. Nat. Prod. 2021, 84, 2644–2663. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Huang, H.; Xie, Y.; Liu, Z.; Zhao, J.; Zhang, C.; Jia, Y.; Zhang, Y.; Zhang, H.; Zhang, T.; et al. Biosynthesis of ilamycins featuring unusual building blocks and engineered production of enhanced anti-tuberculosis agents. Nat. Commun. 2017, 8, 391. [Google Scholar] [CrossRef]
- Wolf, N.M.; Lee, H.; Choules, M.P.; Pauli, G.F.; Phansalkar, R.; Anderson, J.R.; Gao, W.; Ren, J.; Santarsiero, B.D.; Lee, H.; et al. High-Resolution Structure of ClpC1-Rufomycin and Ligand Binding Studies Provide a Framework to Design and Optimize Anti-Tuberculosis Leads. ACS Infect. Dis. 2019, 5, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Shetye, G.; Yu, Y.; Santarsiero, B.D.; Klein, L.L.; Abad-Zapatero, C.; Wolf, N.M.; Cheng, J.; Jin, Y.; Lee, H.; et al. Antimycobacterial Rufomycin Analogues from Streptomyces atratus Strain MJM3502. J. Nat. Prod. 2020, 83, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Park, C.R.; Paik, S.; Kim, Y.J.; Kim, J.K.; Jeon, S.M.; Lee, S.-H.; Whang, J.; Cheng, J.; Suh, J.-W.; Cao, J.; et al. Rufomycin Exhibits Dual Effects Against Mycobacterium abscessus Infection by Inducing Host Defense and Antimicrobial Activities. Front. Microbiol. 2021, 12, 695024. [Google Scholar] [CrossRef] [PubMed]
- Lupoli, T.J.; Vaubourgeix, J.; Burns-Huang, K.; Gold, B. Targeting the Proteostasis Network for Mycobacterial Drug Discovery. ACS Infect. Dis. 2018, 4, 478–498. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Duc, N.M.; Jeong, B.-C.; Cho, S.; Shetye, G.; Cao, J.; Lee, H.; Jeong, C.; Lee, H.; Suh, J.-W. Identification of the inhibitory mechanism of ecumicin and rufomycin 4-7 on the proteolytic activity of Mycobacterium tuberculosis ClpC1/ClpP1/ClpP2 complex. Tuberculosis 2023, 138, 102298. [Google Scholar] [CrossRef] [PubMed]
- Renner, M.K.; Shen, Y.C.; Cheng, X.C.; Jensen, P.R.; Frankmoelle, W.; Kauffman, C.A.; Fenical, W.; Lobkovsky, E.; Clardy, J. Cyclomarins A–C, new antiinflammatory cyclic peptides produced by a marine bacterium (Streptomyces sp.). J. Am. Chem. Soc. 1999, 121, 11273–11276. [Google Scholar] [CrossRef]
- Schultz, A.W.; Oh, D.-C.; Carney, J.R.; Williamson, R.T.; Udwary, D.W.; Jensen, P.R.; Gould, S.J.; Fenical, W.; Moore, B.S. Biosynthesis and Structures of Cyclomarins and Cyclomarazines, Prenylated Cyclic Peptides of Marine Actinobacterial Origin. J. Am. Chem. Soc. 2008, 130, 4507–4516. [Google Scholar] [CrossRef]
- Schmitt, E.K.; Riwanto, M.; Sambandamurthy, V.; Roggo, S.; Miault, C.; Zwingelstein, C.; Krastel, P.; Noble, C.; Beer, D.; Rao, S.P.; et al. The natural product cyclomarin kills Mycobacterium tuberculosis by targeting the ClpC1 subunit of the caseinolytic protease. Angew. Chem. Int. Ed. Engl. 2011, 50, 5889–5891. [Google Scholar] [CrossRef]
- Vasudevan, D.; Rao, S.P.; Noble, C.G. Structural Basis of Mycobacterial Inhibition by Cyclomarin A. J Biol. Chem. 2013, 288, 30883–30891. [Google Scholar] [CrossRef]
- Maurer, M.; Linder, D.; Franke, K.B.; Jager, J.; Taylor, G.; Gloge, F.; Gremer, S.; Le Breton, L.; Mayer, M.P.; Weber-Ban, E.; et al. Toxic Activation of an AAA+ Protease by the Antibacterial Drug Cyclomarin A. Cell Chem. Biol. 2019, 26, 1169–1179. [Google Scholar] [CrossRef]
- Taylor, G.; Frommherz, Y.; Katikaridis, P.; Layer, D.; Sinning, I.; Carroni, M.; Weber-Ban, E.; Mogk, A. Antibacterial peptide CyclomarinA creates toxicity by deregulating the Mycobacterium tuberculosis ClpC1-ClpP1P2 protease. J. Biol. Chem. 2022, 298, 102202. [Google Scholar] [CrossRef]
- Barbie, P.; Kazmaier, U. Total Synthesis of Cyclomarin A, a Marine Cycloheptapeptide with Anti-Tuberculosis and Anti-Malaria Activity. Org. Lett. 2016, 18, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Morreale, F.E.; Kleine, S.; Leodolter, J.; Junker, S.; Hoi, D.M.; Ovchinnikov, S.; Okun, A.; Kley, J.; Kurzbauer, R.; Junk, L.; et al. BacPROTACs mediate targeted protein degradation in bacteria. Cell 2022, 185, 2338–2353. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; MacIntyre, L.W.; Ali, T.; Russo, R.; Koirala, B.; Hernandez, Y.; Brady, S.F. Biosynthetic Interrogation of Soil Metagenomes Reveals Metamarin, an Uncommon Cyclomarin Congener with Activity against Mycobacterium tuberculosis. J. Nat. Prod. 2021, 84, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Weinhaupl, K.; Gragera, M.; Bueno-Carrasco, M.T.; Arranz, R.; Krandor, O.; Akopian, T.; Soares, R.; Rubin, E.; Felix, J.; Fraga, H. Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action. J. Biol. Chem. 2022, 298, 102553. [Google Scholar] [CrossRef] [PubMed]
- Hoi, D.M.; Junker, S.; Junk, L.; Schwechel, K.; Fischel, K.; Podlesainski, D.; Hawkins, P.M.E.; van Geelen, L.; Kaschani, F.; Leodolter, J.; et al. Clp-targeting BacPROTACs impair mycobacterial proteostasis and survival. Cell 2023, 186, 2176–2192. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Kim, J.-Y.; Anderson, J.R.; Akopian, T.; Hong, S.; Jin, Y.-Y.; Kandror, O.; Kim, J.-W.; Lee, I.-A.; Lee, S.-Y.; et al. The Cyclic Peptide Ecumicin Targeting ClpC1 Is Active against Mycobacterium tuberculosis In Vivo. Antimicrob. Agents Chemother. 2015, 59, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; McAlpine, J.B.; Choules, M.P.; Napolitano, J.G.; Lankin, D.C.; Simmler, C.; Ho, N.A.; Lee, H.; Suh, J.-W.; Burton, I.W.; et al. Structural Sequencing of Oligopeptides Aided by 1H Iterative Full-Spin Analysis. J. Nat. Prod. 2017, 80, 2630–2643. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, P.M.E.; Hoi, D.M.; Cheung, C.Y.; Wang, T.; Quan, D.; Sasi, V.M.; Liu, D.Y.; Linington, R.G.; Jackson, C.J.; Oehlers, S.H.; et al. Potent Bactericidal Antimycobacterials Targeting the Chaperone ClpC1 Based on the Depsipeptide Natural Products Ecumicin and Ohmyungsamycin A. J. Med. Chem. 2022, 65, 4893–4908. [Google Scholar] [CrossRef]
- Gavrish, E.; Sit, C.S.; Cao, S.; Kandror, O.; Spoering, A.; Peoples, A.; Ling, L.; Fetterman, A.; Hughes, D.; Bissell, A.; et al. Lassomycin, a Ribosomally Synthesized Cyclic Peptide, Kills Mycobacterium tuberculosis by Targeting the ATP-Dependent Protease ClpC1P1P2. Chem. Biol. 2014, 21, 509–518. [Google Scholar] [CrossRef]
- Khalil, Z.G.; Salim, A.A.; Lacey, E.; Blumenthal, A.; Capon, R.J. Wollamides: Antimycobacterial Cyclic Hexapeptides from an Australian Soil Streptomyces. Org. Lett. 2014, 16, 5120–5123. [Google Scholar] [CrossRef]
- Asfaw, H.; Laqua, K.; Walkowska, A.M.; Cunningham, F.; Martinez-Martinez, M.S.; Cuevas-Zurita, J.C.; Ballell-Pages, L.; Imming, P. Design, synthesis and structure-activity relationship study of wollamide B; a new potential anti TB agent. PLoS ONE 2017, 12, e0176088. [Google Scholar] [CrossRef]
- Asfaw, H.; Wetzlar, T.; Martinez-Martinez, M.S.; Imming, P. An efficient synthetic route for preparation of antimycobacterial wollamides and evaluation of their in vitro and in vivo efficacy. Bioorg. Med. Chem. Lett. 2018, 28, 2899–2905. [Google Scholar] [CrossRef]
- Khalil, Z.G.; Hill, T.A.; De Leon Rodriguez, L.M.; Lohman, R.J.; Hoang, H.N.; Reiling, N.; Hillemann, D.; Brimble, M.A.; Fairlie, D.P.; Blumenthal, A.; et al. Structure-Activity Relationships of Wollamide Cyclic Hexapeptides with Activity against Drug-Resistant and Intracellular Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2019, 63, e01773-18. [Google Scholar] [CrossRef]
- Hur, J.; Jang, J.; Sim, J.; Son, W.S.; Ahn, H.-C.; Kim, T.S.; Shin, Y.-H.; Lim, C.; Lee, S.; An, H.; et al. Conformation-Enabled Total Syntheses of Ohmyungsamycins A and B and Structural Revision of Ohmyungsamycin B. Angew. Chem. Int. Ed. Engl. 2018, 57, 3069–3073. [Google Scholar] [CrossRef]
- Um, S.; Choi, T.J.; Kim, H.; Kim, B.Y.; Kim, S.-H.; Lee, S.K.; Oh, K.-B.; Shin, J.; Oh, D.-C. Ohmyungsamycins A and B: Cytotoxic and antimicrobial cyclic peptides produced by Streptomyces sp. from a volcanic island. J. Org. Chem. 2013, 78, 12321–12329. [Google Scholar] [CrossRef]
- Kim, T.S.; Shin, Y.-H.; Lee, H.-M.; Kim, J.K.; Choe, J.H.; Jang, J.-C.; Um, S.; Jin, H.S.; Komatsu, M.; Cha, G.-H.; et al. Ohmyungsamycins promote antimicrobial responses through autophagy activation via AMP-activated protein kinase pathway. Sci. Rep. 2017, 7, 3431. [Google Scholar] [CrossRef] [PubMed]
- Byun, W.S.; Kim, S.; Shin, Y.-H.; Kim, W.K.; Oh, D.-C.; Lee, S.K. Antitumor Activity of Ohmyungsamycin A through the Regulation of the Skp2-p27 Axis and MCM4 in Human Colorectal Cancer Cells. J. Nat. Prod. 2020, 83, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.M.; Kim, Y.J.; Nguyen, T.Q.; Cui, J.; Thi Bich Hanh, B.; Silwal, P.; Kim, J.K.; Kim, J.M.; Oh, D.-C.; Jang, J.; et al. Ohmyungsamycin promotes M1-like inflammatory responses to enhance host defense against Mycobacteroides abscessus infections. Virulence 2022, 13, 1966–1984. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yang, Z.; Zhang, C.; Liu, Z.; He, J.; Liu, Q.; Zhang, T.; Ju, J.; Ma, J. Genome Mining of Streptomyces atratus SCSIO ZH16: Discovery of Atratumycin and Identification of Its Biosynthetic Gene Cluster. Org. Lett. 2019, 21, 1453–1457. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Sun, C.; Liu, Z.; Liu, Q.; Zhang, T.; Ju, J.; Ma, J. Production of Antitubercular Depsipeptides via Biosynthetic Engineering of Cinnamoyl Units. J. Nat. Prod. 2020, 83, 1666–1673. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, V.; Moretti, C.; Sauvain, M.; Caron, C.; Porzel, A.; Massiot, G.; Richard, B.; Le Men-Olivier, L. Isolation of bis-indole alkaloids with antileishmanial and antibacterial activities from Peschiera van heurkii (syn. Tabernaemontana van heurkii). Planta Med. 1994, 60, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.-B.; Mar, W.; Kim, S.; Kim, J.-Y.; Lee, T.-H.; Kim, J.-G.; Shin, D.; Sim, C.J.; Shin, J. Antimicrobial Activity and Cytotoxicity of Bis(indole) Alkaloids from the Sponge Spongosorites sp. Biol. Pharm. Bull. 2006, 29, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, L.S.; Buchanan, M.S.; Carroll, A.R.; Feng, Y.J.; Quinn, R.J.; Avery, V.M. Flinderoles A–C: Antimalarial Bis-indole Alkaloids from Flindersia Species. Org. Lett. 2009, 11, 329–332. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, C. Anticancer activity of bisindole alkaloids derived from natural sources and synthetic bisindole hybrids. Arch. Pharm. 2020, 353, e2000092. [Google Scholar] [CrossRef]
- Ramkissoon, A.; Seepersaud, M.; Maxwell, A.; Jayaraman, J.; Ramsubhag, A. Isolation and Antibacterial Activity of Indole Alkaloids from Pseudomonas aeruginosa UWI-1. Molecules 2020, 25, 3744. [Google Scholar] [CrossRef]
- Xu, M.; Peng, R.; Min, Q.; Hui, S.; Chen, X.; Yang, G.; Qin, S. Bisindole natural products: A vital source for the development of new anticancer drugs. Eur. J. Med. Chem. 2022, 243, 114748. [Google Scholar] [CrossRef]
- Khan, N.A.; Kaur, N.; Owens, P.; Thomas, O.P.; Boyd, A. Bis-Indole Alkaloids Isolated from the Sponge Spongosorites calcicola Disrupt Cell Membranes of MRSA. Int. J. Mol. Sci. 2022, 23, 1991. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Gutiérrez, S.L.; Silva-Miranda, M.; Krengel, F.; Huerta-Salazar, E.; León-Santiago, M.; Díaz-Cantón, J.K.; Espitia Pinzón, C.; Reyes-Chilpa, R. Antimycobacterial Activity of Alkaloids and Extracts from Tabernaemontana alba and T. arborea. Planta Med. 2022, 88, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Omura, S.; Iwai, Y.; Hirano, A.; Nakagawa, A.; Awaya, J.; Tsuchya, H.; Takahashi, Y.; Masuma, R. A new alkaloid AM-2282 of Streptomyces origin. Taxonomy, fermentation, isolation and preliminary characterization. J. Antibiot. 1977, 30, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Nakano, H.; Omura, S. Chemical biology of natural indolocarbazole products: 30 years since the discovery of staurosporine. J. Antibiot. 2009, 62, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Tamaoki, T.; Nomoto, H.; Takahashi, I.; Kato, Y.; Morimoto, M.; Tomita, F. Staurosporine, a potent inhibitor of phospholipid/Ca++dependent protein kinase. Biochem. Biophys. Res. Commun. 1986, 135, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Kase, H.; Iwahashi, K.; Matsuda, Y. K-252a, a potent inhibitor of protein kinase C from microbial origin. J. Antibiot. 1986, 39, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Tapley, P.; Lamballe, F.; Barbacid, M. K252a is a selective inhibitor of the tyrosine protein kinase activity of the trk family of oncogenes and neurotrophin receptors. Oncogene 1992, 7, 371–381. [Google Scholar]
- Lawrie, A.M.; Noble, M.E.; Tunnah, P.; Brown, N.R.; Johnson, L.N.; Endicott, J.A. Protein kinase inhibition by staurosporine revealed in details of the molecular interaction with CDK2. Nat. Struct. Biol. 1997, 4, 796–801. [Google Scholar] [CrossRef]
- Zhao, B.; Bower, M.J.; McDevitt, P.J.; Zhao, H.; Davis, S.T.; Johanson, K.O.; Green, S.M.; Concha, N.O.; Zhou, B.B. Structural basis for Chk1 inhibition by UCN-01. J. Biol. Chem. 2002, 277, 46609–46615. [Google Scholar] [CrossRef]
- Atwell, S.; Adams, J.M.; Badger, J.; Buchanan, M.D.; Feil, I.K.; Froning, K.J.; Gao, X.; Hendle, J.; Keegan, K.; Leon, B.C.; et al. A Novel Mode of Gleevec Binding Is Revealed by the Structure of Spleen Tyrosine Kinase. J. Biol. Chem. 2004, 279, 55827–55832. [Google Scholar] [CrossRef]
- Tanramluk, D.; Schreyer, A.; Pitt, W.R.; Blundell, T.L. On the Origins of Enzyme Inhibitor Selectivity and Promiscuity: A Case Study of Protein Kinase Binding to Staurosporine. Chem. Biol. Drug Des. 2009, 74, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Tipparaju, S.K.; Pegan, S.D.; Wan, B.; Mo, S.; Orjala, J.; Mesecar, A.D.; Franzblau, S.G.; Kozikowski, A.P. Natural Product Leads for Drug Discovery: Isolation, Synthesis and Biological Evaluation of 6-Cyano-5-Methoxyindolo[2,3-a]carbazole Based Ligands as Antibacterial Agents. Bioorg. Med. Chem. 2009, 17, 7126–7130. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fernandez, P.; Saint-Joanis, B.; Barilone, N.; Jackson, M.; Gicquel, B.; Cole, S.T.; Alzari, P.M. The Ser/Thr protein kinase PknB is essential for sustaining mycobacterial growth. J. Bacteriol. 2006, 188, 7778–7784. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Sammartino, J.C.; Costantino, L.; Gelain, A.; Meneghetti, F.; Villa, S.; Chiarelli, L.R. An Overview on the Potential Antimycobacterial Agents Targeting Serine/Threonine Protein Kinases from Mycobacterium tuberculosis. Curr. Top. Med. Chem. 2019, 19, 646–661. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Santos, G. Caulerpin, a new red pigment from green algae of the genus Caulerpa. J. Chem. Soc. Perkin Trans. 1970, 6, 842–843. [Google Scholar] [CrossRef] [PubMed]
- Vidal, J.P.; Laurent, D.; Kabore, S.A.; Rechencq, E.; Boucard, M.; Girard, J.P.; Escale, R.; Rossi, J.C. Caulerpin, Caulerpicin, Caulerpa scalpelliformis: Comparative Acute Toxicity Study. Bot. Mar. 1984, 27, 533–537. [Google Scholar] [CrossRef]
- Nagappan, T.; Vairappan, C.S. Nutritional and bioactive properties of three edible species of green algae, genus Caulerpa (Caulerpaceae). J. Appl Phycol. 2014, 26, 1019–1027. [Google Scholar] [CrossRef]
- Canché Chay, C.I.; Gómez Cansino, R.; Espitia Pinzón, C.I.; Torres-Ochoa, R.O.; Martínez, R. Synthesis and Anti-Tuberculosis Activity of the Marine Natural Product Caulerpin and Its Analogues. Mar. Drugs 2014, 12, 1757–1772. [Google Scholar] [CrossRef]
- Peng, J.; Hu, J.-F.; Kazi, A.B.; Li, Z.; Avery, M.; Peraud, O.; Hill, R.T.; Franzblau, S.G.; Zhang, F.; Schinazi, R.F.; et al. Manadomanzamines A and B: A Novel Alkaloid Ring System with Potent Activity against Mycobacteria and HIV-1. J. Am. Chem. Soc. 2003, 125, 13382–13386. [Google Scholar] [CrossRef] [PubMed]
- Mo, S.; Krunic, A.; Chlipala, G.; Orjala, J. Antimicrobial ambiguine isonitriles from the cyanobacterium Fischerella ambigua. J. Nat. Prod. 2009, 72, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.E.; Cheuk, C.; Patterson, G.M.L. Hapalindoles: New Alkaloids from the Blue-Green-Alga Hapalosiphon Fontinalis. J. Am. Chem. Soc. 1984, 106, 6456–6457. [Google Scholar] [CrossRef]
- Kim, H.; Lantvit, D.; Hwang, C.H.; Kroll, D.J.; Swanson, S.M.; Franzblau, S.G.; Orjala, J. Indole alkaloids from two cultured cyanobacteria, Westiellopsis sp. and Fischerella muscicola. Bioorg. Med. Chem. 2012, 20, 5290–5295. [Google Scholar] [CrossRef] [PubMed]
- Mo, S.; Krunic, A.; Santarsiero, B.D.; Franzblau, S.G.; Orjala, J. Hapalindole-related alkaloids from the cultured cyanobacterium Fischerella ambigua. Phytochemistry 2010, 71, 2116–2123. [Google Scholar] [CrossRef] [PubMed]
- Abraham, W.-R.; Arfmann, H.A. 12,13-Dihydroxy-fumitremorgin-C from Aspergillus-fumigatus. Phytochemistry 1990, 29, 1025–1026. [Google Scholar] [CrossRef]
- Li, S.-M. Genome mining and biosynthesis of fumitremorgin-type alkaloids in ascomycetes. J. Antibiot. 2011, 64, 45–49. [Google Scholar] [CrossRef][Green Version]
- Luo, X.; Zhou, X.; Lin, X.; Qin, X.; Zhang, T.; Wang, J.; Tu, Z.; Yang, B.; Liao, S.; Tian, Y.; et al. Antituberculosis compounds from a deep-sea-derived fungus Aspergillus sp. SCSIO Ind09F01. Nat. Prod. Res. 2017, 31, 1958–1962. [Google Scholar] [CrossRef]
- Macabeo, A.P.; Vidar, W.S.; Chen, X.; Decker, M.; Heilmann, J.; Wan, B.; Franzblau, S.G.; Galvez, E.V.; Aguinaldo, M.A.; Cordell, G.A. Mycobacterium tuberculosis and cholinesterase inhibitors from Voacanga globosa. Eur. J. Med. Chem. 2011, 46, 3118–3123. [Google Scholar] [CrossRef]
- Kwon-Chung, K.J.; Sugui, J.A. What do we know about the role of gliotoxin in the pathobiology of Aspergillus fumigatus? Med. Mycol. 2009, 47, S97–S103. [Google Scholar] [CrossRef]
- Fu, J.; Luo, X.; Lin, M.; Xiao, Z.; Huang, L.; Wang, J.; Zhu, Y.; Liu, Y.; Huaming Tao, H. Marine-Fungi-Derived Gliotoxin Promotes Autophagy to Suppress Mycobacteria tuberculosis Infection in Macrophage. Mar. Drugs 2023, 21, 616. [Google Scholar] [CrossRef] [PubMed]
- Stanley, S.A.; Grant, S.S.; Kawate, T.; Iwase, N.; Shimizu, M.; Wivagg, C.; Silvis, M.; Kazyanskaya, E.; Aquadro, J.; Golas, A.; et al. Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening. ACS Chem. Biol. 2012, 7, 1377–1384. [Google Scholar] [CrossRef] [PubMed]
- Chinworrungsee, M.; Kittakoop, P.; Saenboonrueng, J.; Kongsaeree, P.; Thebtaranonth, Y. Bioactive compounds from the seed fungus Menisporopsis theobromae BCC 3975. J. Nat. Prod. 2006, 69, 1404–1410. [Google Scholar] [CrossRef]
- Murali Krishna Kumar, M.; Devilal Naik, J.; Satyavathi, K.; Ramana, H.; Raghuveer Varma, P.; Purna Nagasree, K.; Smitha, D.; Venkata Rao, D. Denigrins A–C: New antitubercular 3,4-diarylpyrrole alkaloids from Dendrilla nigra. Nat. Prod. Res. 2014, 28, 888–894. [Google Scholar] [CrossRef]
- Wada, Y.; Fujioka, H.; Kita, Y. Synthesis of the Marine Pyrroloiminoquinone Alkaloids, Discorhabdins. Mar. Drugs 2010, 8, 1394–1416. [Google Scholar] [CrossRef] [PubMed]
- Na, M.; Ding, Y.; Wang, B.; Tekwani, B.L.; Schinazi, R.F.; Franzblau, S.; Kelly, M.; Stone, R.; Li, X.-C.; Ferreira, D.; et al. Anti-infective discorhabdins from a deep-water alaskan sponge of the genus Latrunculia. J. Nat. Prod. 2010, 73, 383–387. [Google Scholar] [CrossRef]
- Suwanborirux, K.; Charupant, K.; Amnuoypol, S.; Pummangura, S.; Kubo, A.; Saito, N. Ecteinascidins 770 and 786 from the Thai tunicate Ecteinascidia thurstoni. J. Nat. Prod. 2002, 65, 935–937. [Google Scholar] [CrossRef]
- Le, V.H.; Inai, M.; Williams, R.M.; Kan, T. Ecteinascidins. A Review of the Chemistry, Biology and Clinical Utility of Potent Tetrahydroisoquinoline Antitumor Antibiotics. Nat. Prod. Rep. 2015, 32, 328–347. [Google Scholar] [CrossRef]
- Kanokmedhakul, S.; Kanokmedhakul, K.; Lekphrom, R. Bioactive Constituents of the Roots of Polyalthia cerasoides. J. Nat. Prod. 2007, 70, 1536–1538. [Google Scholar] [CrossRef]
- Mangalindan, G.C.; Talaue, M.T.; Cruz, L.J.; Franzblau, S.G.; Adams, L.B.; Richardson, A.D.; Ireland, C.M.; Concepcion, G.P. Agelasine F from a Philippine Agelas sp. Sponge Exhibits in vitro Antituberculosis Activity. Planta Med. 2000, 66, 364–365. [Google Scholar] [CrossRef]
- Bakkestuen, A.K.; Gundersen, L.L.; Petersen, D.; Utenova, B.T.; Vik, A. Synthesis and antimycobacterial activity of agelasine E and analogs. Org. Biomol. Chem. 2005, 3, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Vik, A.; Hedner, E.; Charnock, C.; Samuelsen, O.; Larsson, R.; Gundersen, L.L.; Bohlin, L. (+)-Agelasine D: Improved Synthesis and Evaluation of Antibacterial and Cytotoxic Activities. J. Nat. Prod. 2006, 69, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Duca, G.; Pogrebnoi, S.; Boldescu, V.; Aksakal, F.; Uncu, A.; Valica, V.; Uncu, L.; Negres, S.; Nicolescu, F.; Macaev, F. Tryptanthrin Analogues as Inhibitors of Enoyl-acyl Carrier Protein Reductase: Activity against Mycobacterium tuberculosis, Toxicity, Modeling of Enzyme Binding. Curr. Top. Med. Chem. 2019, 19, 609–619. [Google Scholar] [CrossRef]
- Hwang, J.-M.; Oh, T.; Kaneko, T.; Upton, A.M.; Franzblau, S.G.; Ma, Z.; Cho, S.-N.; Kim, P. Design, Synthesis, and Structure–Activity Relationship Studies of Tryptanthrins as Antitubercular Agents. J. Nat. Prod. 2013, 76, 354–367. [Google Scholar] [CrossRef]
- Tripathi, A.; Wadia, N.; Bindal, D.; Jana, T. Docking studies on novel alkaloid tryptanthrin and its analogues against enoyl-acyl carrier protein reductase (InhA) of Mycobacterium tuberculosis. Indian J. Biochem. Biophys. 2012, 49, 435–441. [Google Scholar] [PubMed]
- Frolova, S.G.; Klimina, K.M.; Kumar, R.; Vatlin, A.A.; Salunke, D.B.; Kendrekar, P.; Danilenko, V.N.; Maslov, D.A. Identification of Mutations Conferring Tryptanthrin Resistance to Mycobacterium smegmatis. Antibiotics 2020, 10, 6. [Google Scholar] [CrossRef]
- Williams, D.E.; Dalisay, D.S.; Chen, J.; Polishchuck, E.A.; Patrick, B.O.; Narula, G.; Ko, M.; Av-Gay, Y.; Li, H.; Magarvey, N.; et al. Aminorifamycins and Sporalactams Produced in Culture by a Micromonospora sp. Isolated from a Northeastern-Pacific Marine Sediment Are Potent Antibiotics. Org. Lett. 2017, 19, 766–769. [Google Scholar] [CrossRef]
- Althagbi, H.I.; Alarif, W.M.; Al-Footy, K.O.; Abdel-Lateff, A. Marine-Derived Macrocyclic Alkaloids (MDMAs): Chemical and Biological Diversity. Mar. Drugs 2020, 18, 368. [Google Scholar] [CrossRef]
- Orabi, K.Y.; El Sayed, K.A.; Hamann, M.T.; Dunbar, D.C.; Al-Said, M.S.; Higa, T.; Kelly, M. Araguspongines K and L, new bioactive bis-1-oxaquinolizidine N-oxide alkaloids from Red Sea specimens of Xestospongia exigua. J. Nat. Prod. 2002, 65, 1782–1785. [Google Scholar] [CrossRef]
- Ismatullah, H.; Jabeen, I.; Muhammad Tariq Saeed, M.T. Biological Regulatory Network (BRN) Analysis and Molecular Docking Simulations to Probe the Modulation of IP3R Mediated Ca2+ Signaling in Cancer. Genes 2021, 12, 34. [Google Scholar] [CrossRef]
- Wang, Z.-J.; Zhao, F.; Wang, C.-F.; Zhang, X.-M.; Xiao, Y.; Zhou, F.; Wu, M.-N.; Zhang, J.; Qi, J.-S.; Yang, W. Xestospongin C, a Reversible IP3 Receptor Antagonist, Alleviates the Cognitive and Pathological Impairments in APP/PS1 Mice of Alzheimer’s Disease. J. Alzheimers Dis. 2019, 72, 1217–1231. [Google Scholar] [CrossRef] [PubMed]
- Akl, M.R.; Ayoub, N.M.; Ebrahim, H.Y.; Mohyeldin, M.M.; Orabi, K.Y.; Foudah, A.I.; El Sayed, K.A. Araguspongine C induces autophagic death in breast cancer cells through suppression of c-Met and HER2 receptor tyrosine kinase signaling. Mar. Drugs 2015, 13, 288–311. [Google Scholar] [CrossRef]
- de Oliveira, J.H.; Grube, A.; Kock, M.; Berlinck, R.G.; Macedo, M.L.; Ferreira, A.G.; Hajdu, E. Ingenamine G and Cyclostellettamines G-I, K, and L from the New Brazilian Species of Marine Sponge Pachychalina sp. J. Nat. Prod. 2004, 67, 1685–1689. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, J.E.; Whitehead, R.C. On the Biosynthesis of Manzamines. Tetrahedron Lett. 1992, 33, 2059–2062. [Google Scholar] [CrossRef]
- Meng, Z.; Fürstner, A. Total Synthesis Provides Strong Evidence: Xestocyclamine A is the Enantiomer of Ingenamine. J. Am. Chem. Soc. 2020, 142, 11703–11708. [Google Scholar] [CrossRef]
- Meng, Z.; Spohr, S.M.; Tobegen, S.; Fares, C.; Fürstner, A. A Unified Approach to Polycyclic Alkaloids of the Ingenamine Estate: Total Syntheses of Keramaphidin B, Ingenamine, and Nominal Njaoamine I. J. Am. Chem. Soc. 2021, 143, 14402–14414. [Google Scholar] [CrossRef]
- Liang, L.; Haltli, B.; Marchbank, D.H.; Fischer, M.; Kirby, C.W.; Correa, H.; Clark, T.N.; Gray, C.A.; Kerr, R.G. Discovery of an Isothiazolinone-Containing Antitubercular Natural Product Levesquamide. J. Org. Chem. 2020, 85, 6450–6462. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liu, X.; Li, W.; Jiao, X.; Xie, P. Total Synthesis of (−)-Levesquamide. J. Org. Chem. 2023, 88, 3981–3986. [Google Scholar] [CrossRef]
- Hua, H.M.; Peng, J.; Dunbar, D.C.; Schinazi, R.F.; Andrews, A.G.D.C.; Cuevas, C.; Garcia-Fernandez, L.F.; Kelly, M.; Hamann, M.T. Batzelladine alkaloids from the caribbean sponge Monanchora unguifera and the significant activities against HIV-1 and AIDS opportunistic infectious pathogens. Tetrahedron 2007, 63, 11179–11188. [Google Scholar] [CrossRef]
- Abd Rani, N.Z.; Lee, Y.K.; Ahmad, S.; Meesala, R.; Abdullah, I. Fused Tricyclic Guanidine Alkaloids: Insights into Their Structure, Synthesis and Bioactivity. Mar. Drugs 2022, 20, 579. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.newtbdrugs.org/pipeline/clinical (accessed on 3 February 2024).
- Lee, R.E.; Hurdle, J.G.; Liu, J.; Bruhn, D.F.; Matt, T.; Scherman, M.S.; Vaddady, P.K.; Zheng, Z.; Qi, J.; Akbergenov, R.; et al. Spectinamides: A New Class of Semisynthetic Anti-Tuberculosis Agents that Overcome Native Drug Efflux. Nat. Med. 2014, 20, 152–158. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boshoff, H.I.; Malhotra, N.; Barry, C.E., III; Oh, S. The Antitubercular Activities of Natural Products with Fused-Nitrogen-Containing Heterocycles. Pharmaceuticals 2024, 17, 211. https://doi.org/10.3390/ph17020211
Boshoff HI, Malhotra N, Barry CE III, Oh S. The Antitubercular Activities of Natural Products with Fused-Nitrogen-Containing Heterocycles. Pharmaceuticals. 2024; 17(2):211. https://doi.org/10.3390/ph17020211
Chicago/Turabian StyleBoshoff, Helena I., Neha Malhotra, Clifton E. Barry, III, and Sangmi Oh. 2024. "The Antitubercular Activities of Natural Products with Fused-Nitrogen-Containing Heterocycles" Pharmaceuticals 17, no. 2: 211. https://doi.org/10.3390/ph17020211
APA StyleBoshoff, H. I., Malhotra, N., Barry, C. E., III, & Oh, S. (2024). The Antitubercular Activities of Natural Products with Fused-Nitrogen-Containing Heterocycles. Pharmaceuticals, 17(2), 211. https://doi.org/10.3390/ph17020211





