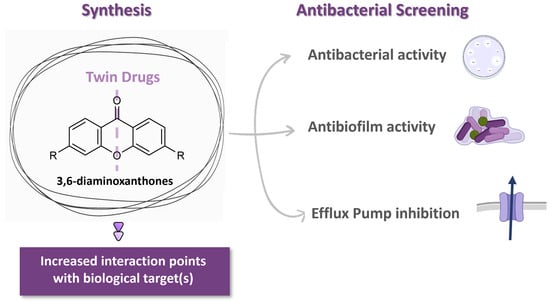
Antibacterial Potential of Symmetrical Twin-Drug 3,6-Diaminoxanthones
Abstract
1. Introduction
2. Results and Discussion
2.1. Docking Studies
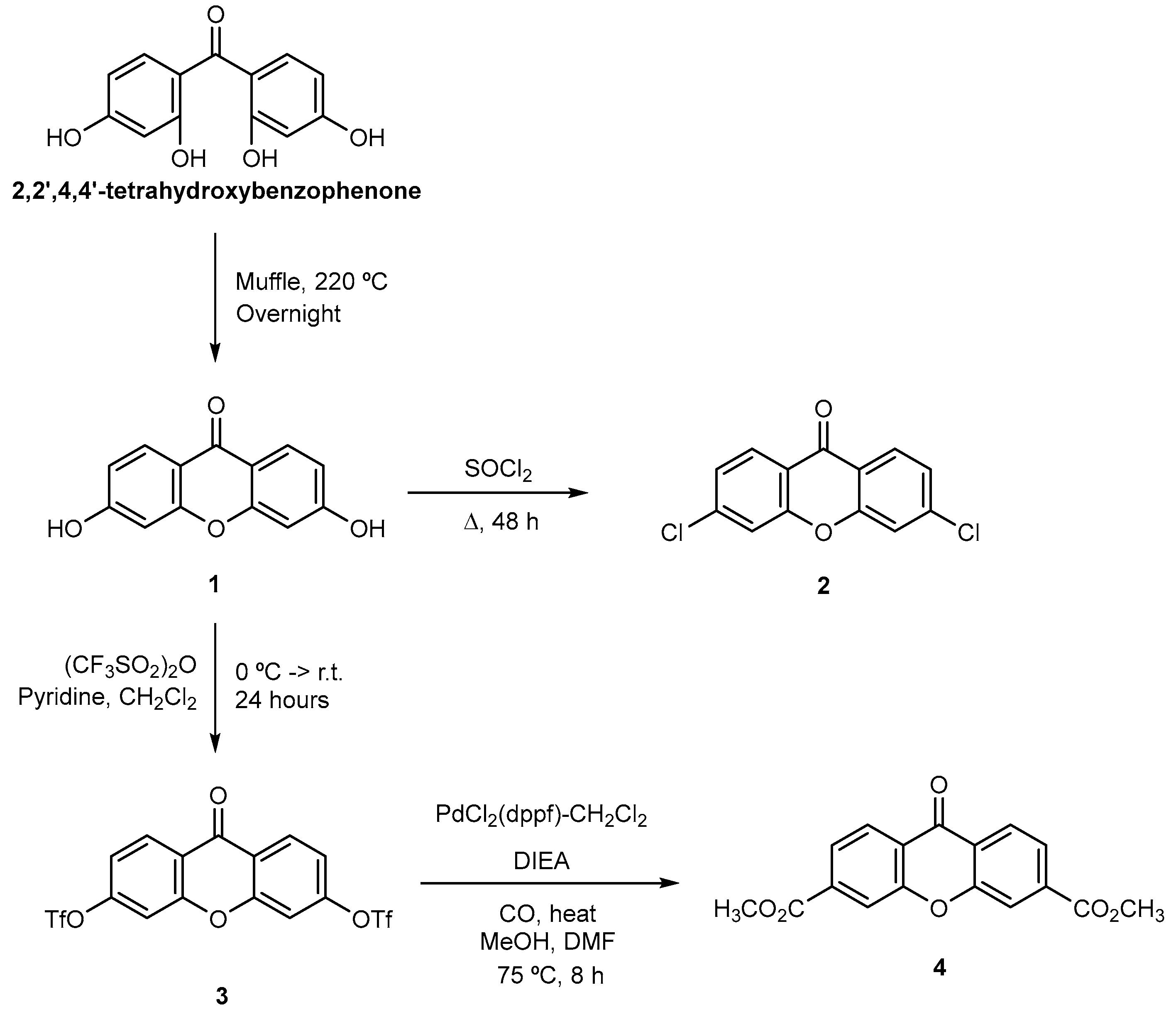
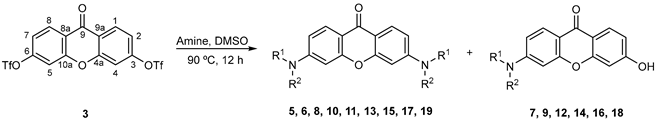
2.2. Synthesis
2.3. Antimicrobial Activity
2.4. Efflux Pump Activity Inhibition Assay
2.5. Inhibition of Biofilm Formation and Quorum-Sensing Assays
2.6. Cytotoxicity Assay
3. Materials and Methods
3.1. Chemistry
3.1.1. Reagents and Equipment
3.1.2. Synthesis
General
Synthesis of Xanthone Primary Derivatives 1–4
Synthesis of Aminated Xanthones 5–19
3.2. Microbiology
3.2.1. Culture Media and Chemicals
3.2.2. Microorganisms
3.2.3. Microbiological Analysis of Xanthone Derivatives
3.2.4. Docking Studies
3.2.5. Antibacterial Assay
3.2.6. Antifungal Activity
3.2.7. Efflux Pump Inhibition Assays
3.2.8. Inhibition of Biofilm Formation
3.2.9. Quorum Sensing Assay
3.2.10. Cell Lines and Cultures
3.2.11. Cytotoxicity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AHL | Acyl-homoserine-lactone |
| ATCC | American Type Culture Collection |
| CCCP | Carbonyl cyanide 3-chlorophenylhydrazone |
| CV026 | Chromobacterium violaceum CV026 |
| CV | Crystal violet |
| DMSO | Dimethyl sulfoxide |
| EPIs | efflux pump inhibitors |
| EB | Ethidium bromide |
| EZF | Sphingomonas paucimobilis EZF 10-17 |
| HT | Hydrophobic trap |
| IC50 | Half-maximal inhibitory concentration |
| LB-A | Luria–Bertani agar |
| LB-B | Luria–Bertani broth |
| LD | Lipoyl domain |
| MFS | Major facilitator superfamily |
| MHB II | Cation-adjusted Mueller–Hinton broth |
| MIC | Minimum inhibitory concentration |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| OD | Optical density |
| PBS | Phosphate-buffered saline |
| PMZ | Promethazine |
| QS | Quorum sensing |
| RF | Relative fluorescence |
| RFI | Relative fluorescence index |
| SBS | Substrate-binding site |
| SD | Standard deviation |
| TSA | Tryptic Soy agar |
| TSB | Tryptic Soy broth |
References
- Picot, S.; Beugnet, F.; Leboucher, G.; Bienvenu, A.-L. Drug Resistant Parasites and Fungi from a One-Health Perspective: A Global Concern That Needs Transdisciplinary Stewardship Programs. One Health 2022, 14, 100368. [Google Scholar] [CrossRef]
- Verhoef, J.; van Kessel, K.; Snippe, H. Immune Response in Human Pathology: Infections Caused by Bacteria, Viruses, Fungi, and Parasites. In Nijkamp and Parnham’s Principles of Immunopharmacology; Parnham, M.J., Nijkamp, F.P., Rossi, A.G., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 165–178. [Google Scholar]
- Adebisi, Y.A.; Ogunkola, I.O. The Global Antimicrobial Resistance Response Effort Must Not Exclude Marginalised Populations. Trop. Med. Health 2023, 51, 33. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Oonsivilai, M.; Lim, C.; Niehus, R.; Cooper, B.S. Implications of Reducing Antibiotic Treatment Duration for Antimicrobial Resistance in Hospital Settings: A Modelling Study and Meta-Analysis. PLoS Med. 2023, 20, e1004013. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, G. Strategies to Overcome Antimicrobial Resistance (AMR) Making Use of Non-Essential Target Inhibitors: A Review. Int. J. Mol. Sci. 2019, 20, 5844. [Google Scholar] [CrossRef] [PubMed]
- Badiali, C.; Petruccelli, V.; Brasili, E.; Pasqua, G. Xanthones: Biosynthesis and Trafficking in Plants, Fungi and Lichens. Plants 2023, 12, 694. [Google Scholar] [CrossRef] [PubMed]
- Remali, J.; Sahidin, I.; Aizat, W.M. Xanthone Biosynthetic Pathway in Plants: A Review. Front. Plant Sci. 2022, 13, 809497. [Google Scholar] [CrossRef]
- Kuete, V.; Alibert-Franco, S.; Eyong, K.O.; Ngameni, B.; Folefoc, G.N.; Nguemeving, J.R.; Tangmouo, J.G.; Fotso, G.W.; Komguem, J.; Ouahouo, B.M.W.; et al. Antibacterial Activity of Some Natural Products against Bacteria Expressing a Multidrug-Resistant Phenotype. Int. J. Antimicrob. Agents 2011, 37, 156–161. [Google Scholar] [CrossRef]
- Fotie, J.; Bohle, S.D. Pharmacological and Biological Activities of Xanthones. Anti-Infect. Agents Med. Chem. 2006, 5, 15–31. [Google Scholar] [CrossRef]
- Resende, D.I.S.P.; Pereira-Terra, P.; Inácio, Â.S.; Costa, P.M.d.; Pinto, E.; Sousa, E.; Pinto, M.M.M. Lichen Xanthones as Models for New Antifungal Agents. Molecules 2018, 23, 2617. [Google Scholar] [CrossRef]
- Resende, D.I.S.P.; Pereira-Terra, P.; Moreira, J.; Freitas-Silva, J.; Lemos, A.; Gales, L.; Pinto, E.; de Sousa, M.E.; da Costa, P.M.; Pinto, M.M.M. Synthesis of a Small Library of Nature-Inspired Xanthones and Study of Their Antimicrobial Activity. Molecules 2020, 25, 2405. [Google Scholar] [CrossRef]
- Durães, F.; Resende, D.I.S.P.; Palmeira, A.; Szemerédi, N.; Pinto, M.M.M.; Spengler, G.; Sousa, E. Xanthones Active against Multidrug Resistance and Virulence Mechanisms of Bacteria. Antibiotics 2021, 10, 600. [Google Scholar] [CrossRef] [PubMed]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of Bacterial Efflux Pumps in Biofilm Formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Luo, Y. Bacterial Quorum-Sensing Systems and Their Role in Intestinal Bacteria-Host Crosstalk. Front. Microbiol. 2021, 12, 611413. [Google Scholar] [CrossRef] [PubMed]
- Tomassoli, I.; Gündisch, D. The Twin Drug Approach for Novel Nicotinic Acetylcholine Receptor Ligands. Bioorganic Med. Chem. 2015, 23, 4375–4389. [Google Scholar] [CrossRef] [PubMed]
- Fujisaki, F.; Aki, H.; Naito, A.; Fukami, E.; Kashige, N.; Miake, F.; Sumoto, K. Synthesis of New 5-Substituted Hydantoins and Symmetrical Twin-Drug Type Hydantoin Derivatives. Chem. Pharm. Bull. 2014, 62, 429–438. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eicher, T.; Cha, H.-J.; Seeger, M.A.; Brandstätter, L.; El-Delik, J.; Bohnert, J.A.; Kern, W.V.; Verrey, F.; Grütter, M.G.; Diederichs, K.; et al. Transport of Drugs by the Multidrug Transporter Acrb Involves an Access and a Deep Binding Pocket That Are Separated by a Switch-Loop. Proc. Natl. Acad. Sci. USA 2012, 109, 5687. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Wang, Z.; James, N.R.; Voss, J.E.; Klimont, E.; Ohene-Agyei, T.; Venter, H.; Chiu, W.; Luisi, B.F. Structure of the Acrab–Tolc Multidrug Efflux Pump. Nature 2014, 509, 512–515. [Google Scholar] [CrossRef]
- Aron, Z.; Opperman, T.J. The Hydrophobic Trap—The Achilles Heel of Rnd Efflux Pumps. Res. Microbiol. 2018, 169, 393–400. [Google Scholar] [CrossRef]
- Rosa, G.P.; Palmeira, A.; Resende, D.I.S.P.; Almeida, I.F.; Kane-Pagès, A.; Barreto, M.C.; Sousa, E.; Pinto, M.M.M. Xanthones for Melanogenesis Inhibition: Molecular Docking and Qsar Studies to Understand Their Anti-Tyrosinase Activity. Bioorganic Med. Chem. 2021, 29, 115873. [Google Scholar] [CrossRef]
- Wu, L.; Burgess, K. Synthesis and Spectroscopic Properties of Rosamines with Cyclic Amine Substituents. J. Org. Chem. 2008, 73, 8711–8718. [Google Scholar] [CrossRef] [PubMed]
- Dax, S.; DeCorte, B.; Liu, L.; McDonnell, M.; McNally, J. Tricyclic-Bridged Piperidinylidene Derivatives as δ-Opioid Modulators. U.S. Patent 7553850B2, 30 June 2009. [Google Scholar]
- Durães, F.; Cravo, S.; Freitas-Silva, J.; Szemerédi, N.; Martins-da-Costa, P.; Pinto, E.; Tiritan, M.E.; Spengler, G.; Fernandes, C.; Sousa, E.; et al. Enantioselectivity of Chiral Derivatives of Xanthones in Virulence Effects of Resistant Bacteria. Pharmaceuticals 2021, 14, 1141. [Google Scholar] [CrossRef] [PubMed]
- Parai, D.; Banerjee, M.; Dey, P.; Mukherjee, S.K. Reserpine Attenuates Biofilm Formation and Virulence of Staphylococcus Aureus. Microb. Pathog. 2020, 138, 103790. [Google Scholar] [CrossRef]
- Kaur, B.; Gupta, J.; Sharma, S.; Sharma, D.; Sharma, S. Focused Review on Dual Inhibition of Quorum Sensing and Efflux Pumps: A Potential Way to Combat Multi Drug Resistant Staphylococcus Aureus Infections. Int. J. Biol. Macromol. 2021, 190, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Benomar, S.; Evans, K.C.; Unckless, R.L.; Chandler, J.R. Efflux Pumps in Chromobacterium Species Increase Antibiotic Resistance and Promote Survival in a Coculture Competition Model. Appl. Environ. Microbiol. 2019, 85, e00908-19. [Google Scholar] [CrossRef] [PubMed]
- Sussman, J.L.; Lin, D.; Jiang, J.; Manning, N.O.; Prilusky, J.; Ritter, O.; Abola, E.E. Protein Data Bank (Pdb): Database of Three-Dimensional Structural Information of Biological Macromolecules. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998, 54, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. Autodock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (Ed.) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Clinical and Laboratory Standards Institute (Ed.) Clinical and Laboratory Standards Institute: Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard-CLSI Document M27-A3; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Clinical and Laboratory Standards Institute (Ed.) Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi; Approved Standard—Second Edition. CLSI Document M38-A2; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Gajdács, M.; Spengler, G. The Role of Drug Repurposing in the Development of Novel Antimicrobial Drugs: Non-Antibiotic Pharmacological Agents as Quorum Sensing-Inhibitors. Antibiotics 2019, 8, 270. [Google Scholar] [CrossRef]
- Ferreira, R.J.; Kincses, A.; Gajdács, M.; Spengler, G.; dos Santos, D.J.V.A.; Molnár, J.; Ferreira, M.-J.U. Terpenoids from Euphorbia Pedroi as Multidrug-Resistance Reversers. J. Nat. Prod. 2018, 81, 2032–2040. [Google Scholar] [CrossRef]
- Kaatz Glenn, W.; Moudgal Varsha, V.; Seo Susan, M.; Kristiansen Jette, E. Phenothiazines and Thioxanthenes Inhibit Multidrug Efflux Pump Activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 2003, 47, 719–726. [Google Scholar] [CrossRef]
| Compound | Docking Score | Compound | Docking Score | ||
|---|---|---|---|---|---|
| SBS | HT | SBS | HT | ||
| 1 | −6.8 | −6.1 | 13 | −9.1 | 5.0 |
| 2 | −6.7 | −5.4 | 14 | −8.4 | −5.4 |
| 3 | −9.9 | −6.6 | 15 | −9.2 | 15.0 |
| 4 | −7.2 | −5.5 | 16 | −8.3 | −0.2 |
| 5 | −8.7 | −4.3 | 17 | −9.4 | 12.8 |
| 6 | −6.7 | −4.2 | 18 | −8.2 | −2.9 |
| 7 | −6.8 | −5.4 | 19 | −10.4 | 11.8 |
| 8 | −9.0 | −3.9 | D13-9001 | −9.7 | 26.5 |
| 9 | −7.6 | −6.2 | Doxorubicin | −8.9 | 15.4 |
| 10 | −8.6 | −3.6 | MBX-3132 | −7.9 | 2.9 |
| 11 | −8.2 | −5.2 | Minocycline | −8.7 | 26.7 |
| 12 | −7.5 | −6.0 | PAβN | −7.1 | −4.7 |
| Entry | Compound | Yield, % | Compound | Yield, % |
|---|---|---|---|---|
| 1 | 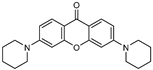 | 5 (82) | - | - |
| 2 | 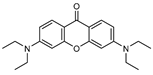 | 6 (29) |  | 7 (50) |
| 3 | 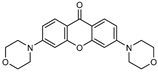 | 8 (18) | 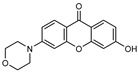 | 9 (52) |
| 4 | 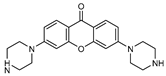 | 10 (64) | - | - |
| 5 | 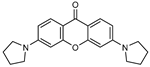 | 11 (24) | 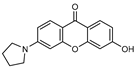 | 12 (23) |
| 6 | 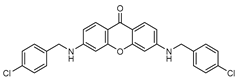 | 13 (48) | 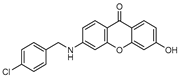 | 14 (29) |
| 7 | 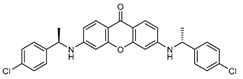 | 15 (13) | 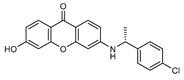 | 16 (31) |
| 8 | 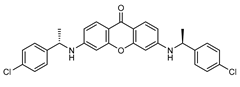 | 17 (23) |  | 18 (35) |
| 9 | 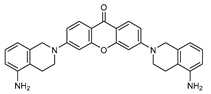 | 19 (26) | - | - |
| Compound | MIC in µM (µg/mL) | |||
|---|---|---|---|---|
| E. coli ATCC 25922 | S. aureus ATCC 25923 | S. aureus 272123 | E. faecalis ATCC 29212 | |
| 1–4 | >200 | >200 | >200 | >200 |
| 10 | 176 (64 µg/mL) | >200 | >200 | >200 |
| 15 | >200 | >200 | >200 | 127 (64 µg/mL) |
| 16 | >200 | 11 (4 µg/mL) | 25 (9 µg/mL) | 11 (4 µg/mL) |
| 17 | >200 | >200 | 100 (50 µg/mL) | 127 (64 µg/mL) |
| 18 | >200 | 44 (16 µg/mL) | 50 (18 µg/mL) | 22 (8 µg/mL) |
| Relative Fluorescence Index (RFI) | Relative Fluorescence Index (RFI) | ||||
|---|---|---|---|---|---|
| Compound | S. aureus 272123 | S. enterica Typhimurium SL1344 | Compound | S. aureus 272123 | S. enterica Typhimurium SL1344 |
| 1 | −0.23 ± 0.04 | −0.17 ± 0.02 | 12 | 0.88 ± 0.07 | 2.24 ± 0.05 |
| 2 | −0.09 ± 0.04 | 0.02 ± 0.04 | 13 | 2.12 ± 0.07 | 3.52 ± 0.64 |
| 3 | −0.05 ± 0.07 | 0.09 ± 0.02 | 14 | 0.06 ± 0.05 | 0.17 ± 0.03 |
| 4 | −0.21 ± 0.04 | −0.05 ± 0.03 | 15 | −0.08 ± 0.09 | 0.06 ± 0.02 |
| 5 | 1.12 ± 0.02 | 0.82 ± 0.02 | 16 | 0.74 ± 0.01 | 4.30 ± 0.01 |
| 6 | 0.32 ± 0.05 | 0.04 ± 0.04 | 17 | 0.19 ± 0.01 | −0.06 ± 0.02 |
| 7 | 0.76 ± 0.04 | 0.28 ± 0.14 | 18 | 0.36 ± 0.05 | 0.57 ± 0.11 |
| 8 | 0.12 ± 0.03 | 0.03 ± 0.08 | 19 | −0.27 ± 0.04 | 0.49 ± 0.04 |
| 9 | −0.58 ± 0.06 | −0.26 ± 0.10 | Reserpine | 0.50 ± 0.04 | - |
| 10 | 0.06 ± 0.07 | −0.11 ± 0.02 | CCCP | - | 0.40 ± 0.07 |
| 11 | −0.01 ± 0.18 | 0.01 ± 0.06 | |||
| Compound | Inhibition of Biofilm Formation (%) | |
|---|---|---|
| S. aureus ATCC 25923 | S. aureus 272123 | |
| 5 | 0 | 13.1 ± 2.65 |
| 7 | 0 | 8.16 ± 0.93 |
| 18 | 0 | 47.1 ± 0.04 |
| 19 | 56.1 ± 0.05 | 93.6 ± 0.03 |
| Reserpine | 22.3 ± 5.10 | 63.1 ± 2.33 |
| Compound | Quorum Sensing Inhibition (mm) | ||
|---|---|---|---|
| S. marcescens | wt85 | EZF + CV026 | |
| 5 | 0 | 0 | 21 ± 0.5 |
| 7 | 0 | 0 | 53 ± 0.1 |
| 18 | 0 | 0 | 59 ± 0.5 |
| 19 | 0 | 0 | 29 ± 0.5 |
| PMZ | 18 ± 0.8 | 40 ± 0.1 | 41 ± 0.5 |
| Compound | IC50 (µM) ± SD | IC50 (µg/mL) ± SD |
|---|---|---|
| 5 | 82.03 ± 0.27 | 29.73 ± 0.10 |
| 7 | >100 | >28.31 |
| 18 | >100 | >36.58 |
| 19 | 64.22 ± 1.09 | 31.38 ± 0.53 |
| Doxorubicin | 12.05 ± 0.81 | 6.55 ± 0.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Resende, D.I.S.P.; Durães, F.; Zubarioglu, S.; Freitas-Silva, J.; Szemerédi, N.; Pinto, M.; Pinto, E.; Martins da Costa, P.; Spengler, G.; Sousa, E. Antibacterial Potential of Symmetrical Twin-Drug 3,6-Diaminoxanthones. Pharmaceuticals 2024, 17, 209. https://doi.org/10.3390/ph17020209
Resende DISP, Durães F, Zubarioglu S, Freitas-Silva J, Szemerédi N, Pinto M, Pinto E, Martins da Costa P, Spengler G, Sousa E. Antibacterial Potential of Symmetrical Twin-Drug 3,6-Diaminoxanthones. Pharmaceuticals. 2024; 17(2):209. https://doi.org/10.3390/ph17020209
Chicago/Turabian StyleResende, Diana I. S. P., Fernando Durães, Sidika Zubarioglu, Joana Freitas-Silva, Nikoletta Szemerédi, Madalena Pinto, Eugénia Pinto, Paulo Martins da Costa, Gabriella Spengler, and Emília Sousa. 2024. "Antibacterial Potential of Symmetrical Twin-Drug 3,6-Diaminoxanthones" Pharmaceuticals 17, no. 2: 209. https://doi.org/10.3390/ph17020209
APA StyleResende, D. I. S. P., Durães, F., Zubarioglu, S., Freitas-Silva, J., Szemerédi, N., Pinto, M., Pinto, E., Martins da Costa, P., Spengler, G., & Sousa, E. (2024). Antibacterial Potential of Symmetrical Twin-Drug 3,6-Diaminoxanthones. Pharmaceuticals, 17(2), 209. https://doi.org/10.3390/ph17020209