Hybrid Molecules of Azithromycin with Chloramphenicol and Metronidazole: Synthesis and Study of Antibacterial Properties
Abstract
1. Introduction
2. Results
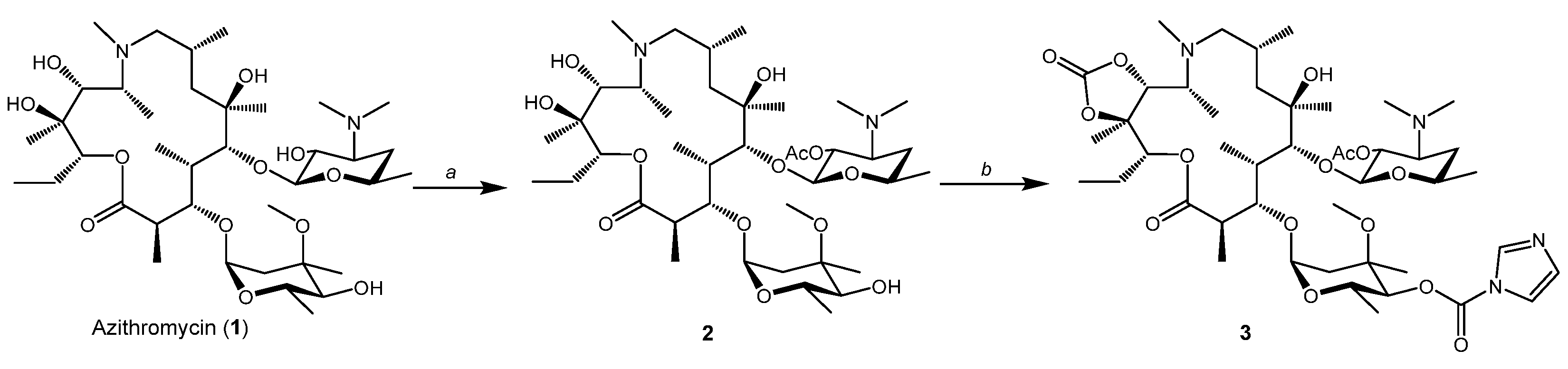
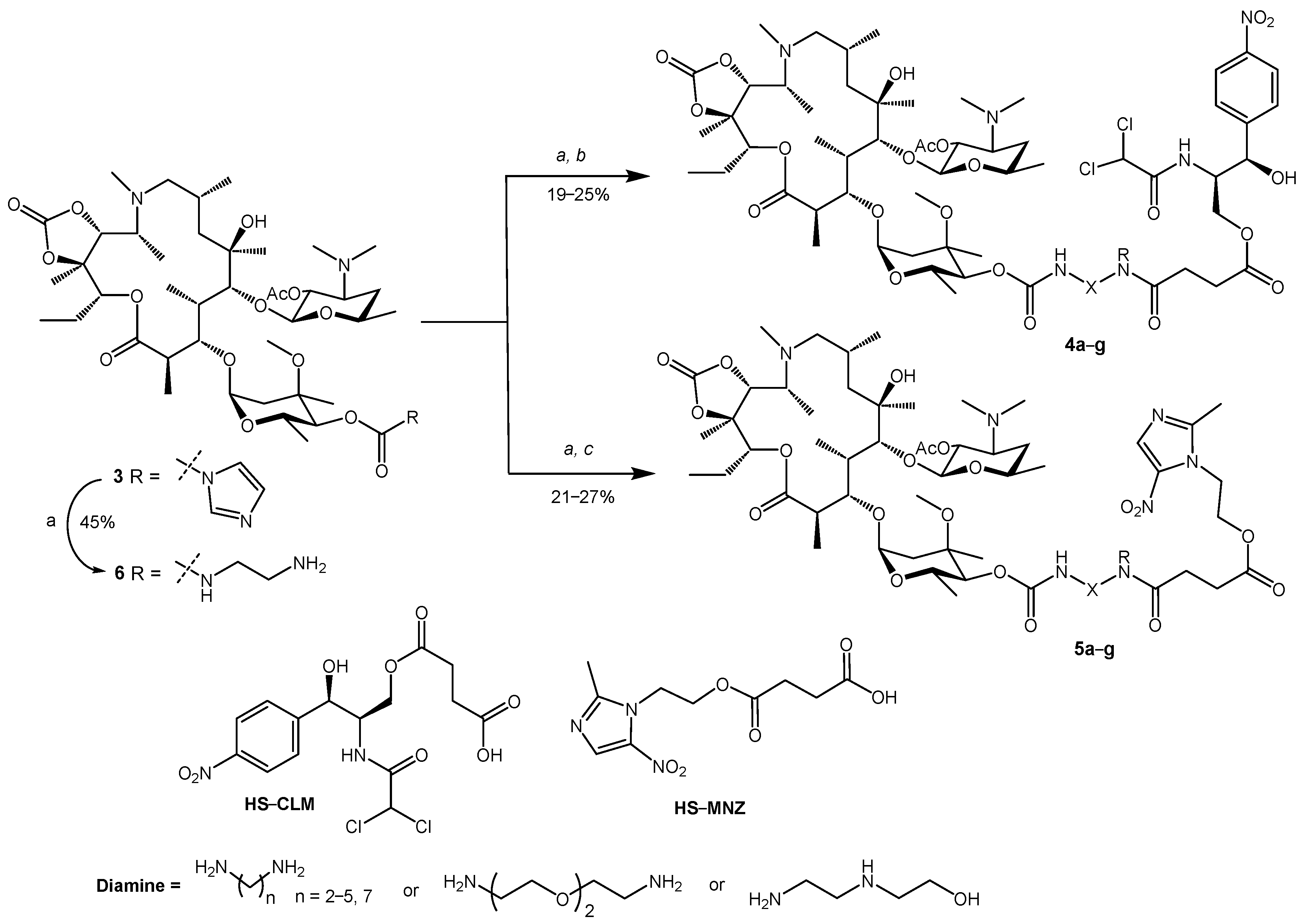
2.1. Synthesis of Hybrid Molecules
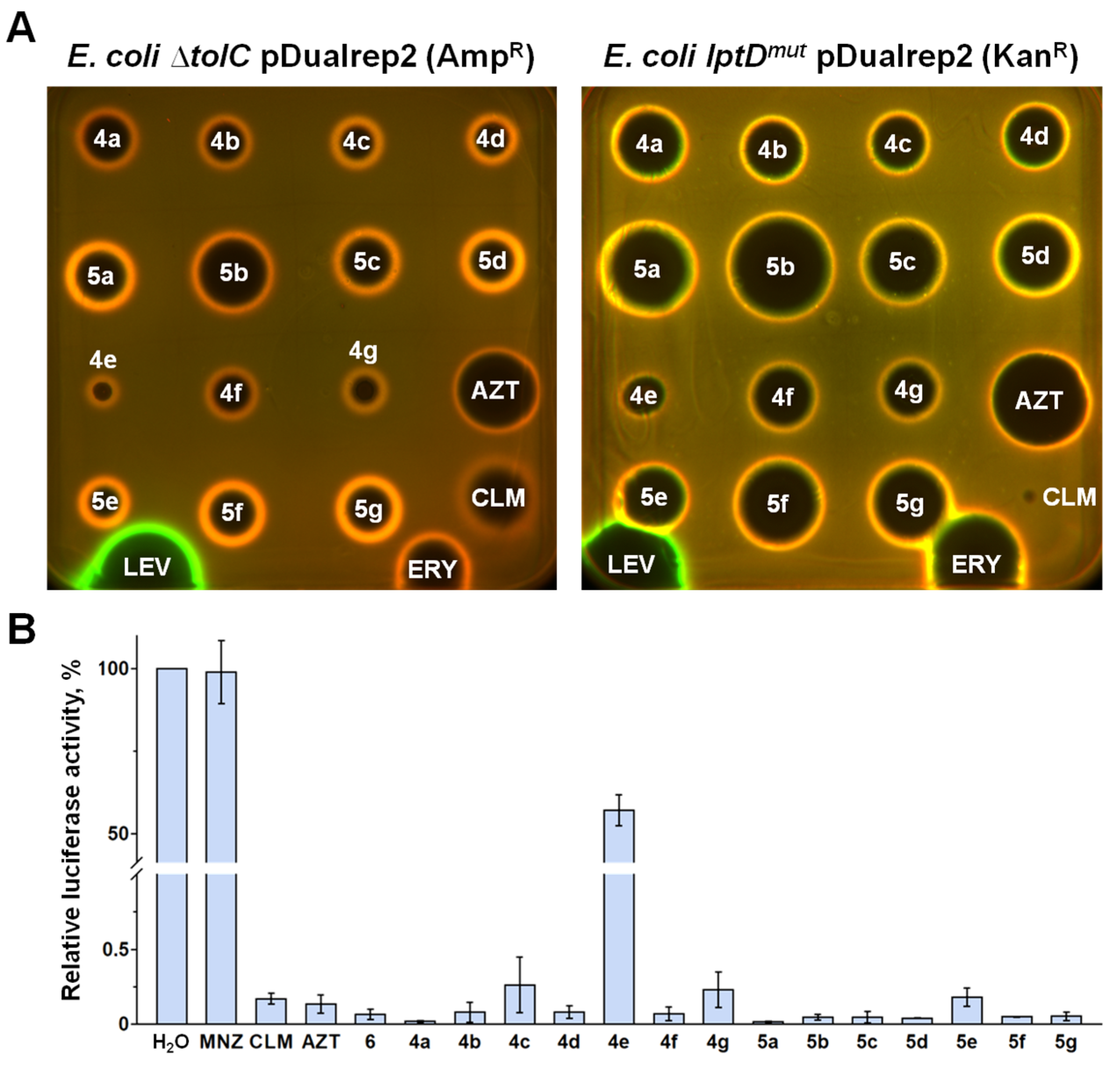
2.2. In Vitro Antimicrobial Activity
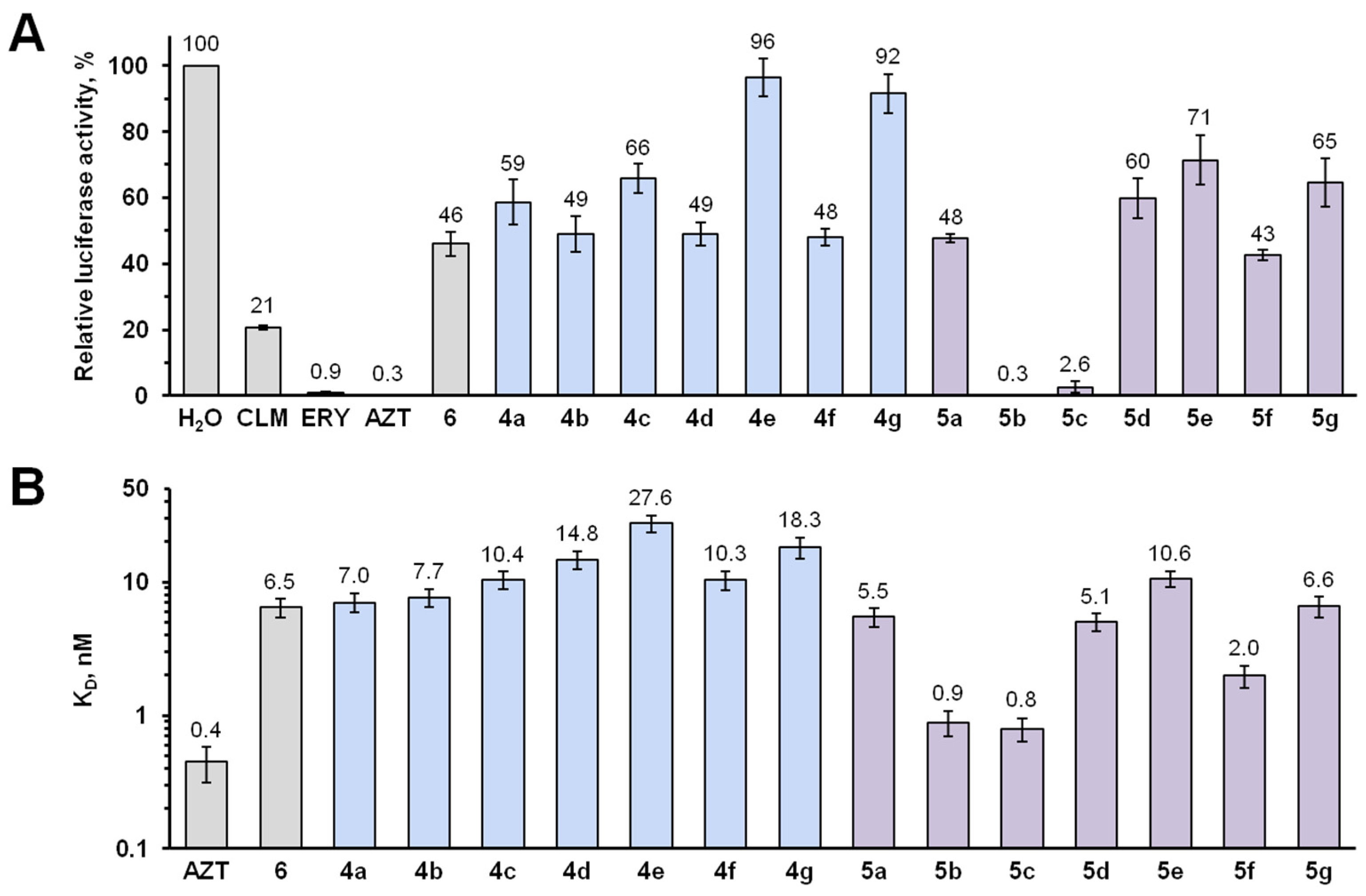
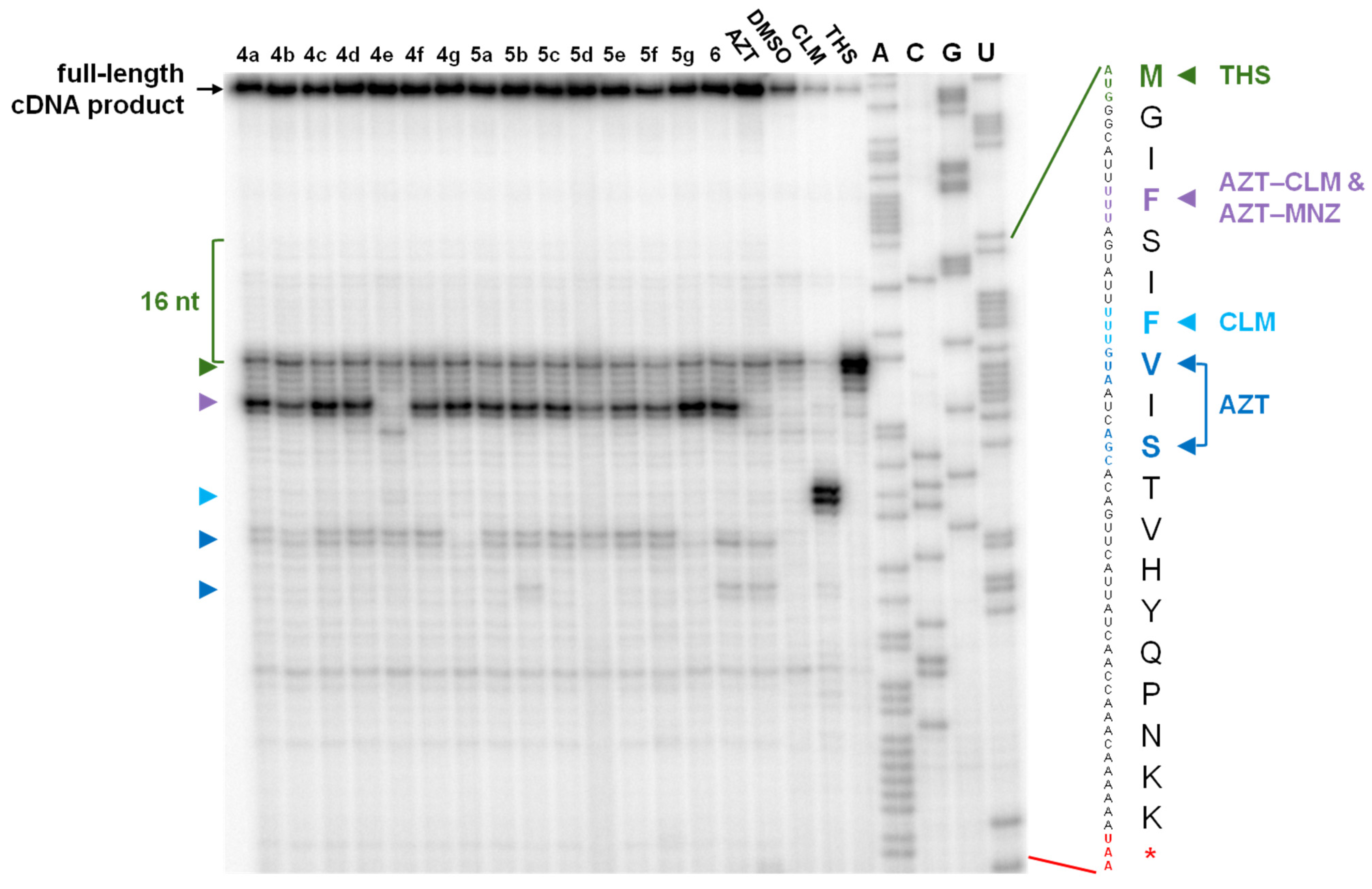
2.3. Elucidation of the Mode of Action of New Hybrid Antibiotics
3. Discussion
4. Materials and Methods
4.1. Chemistry
4.1.1. 2′-O-Acetylazithromycin (2) [17,33]
4.1.2. 2′-O-Acetyl-4″-O-acylimidazolylazithromycin-11,12-cyclic carbonate (3) [17,33]
4.1.3. 2′-O-Acetyl-4′′-O-((2-(4-((2R,3R)-2-(2,2-dichloroacetamido)-3-hydroxy-3-(4-nitrophenyl)propoxy)-4-oxobutanamido)ethyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (4a)
4.1.4. 2′-O-Acetyl-4′′-O-((3-(4-((2R,3R)-2-(2,2-dichloroacetamido)-3-hydroxy-3-(4-nitrophenyl)propoxy)-4-oxobutanamido)propyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (4b)
4.1.5. 2′-O-Acetyl-4′′-O-((4-(4-((2R,3R)-2-(2,2-dichloroacetamido)-3-hydroxy-3-(4-nitrophenyl)propoxy)-4-oxobutanamido)butyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (4c)
4.1.6. 2′-O-Acetyl-4′′-O-((5-(4-((2R,3R)-2-(2,2-dichloroacetamido)-3-hydroxy-3-(4-nitrophenyl)propoxy)-4-oxobutanamido)pentyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (4d)
4.1.7. 2′-O-Acetyl-4′′-O-((7-(4-((2R,3R)-2-(2,2-dichloroacetamido)-3-hydroxy-3-(4-nitrophenyl)propoxy)-4-oxobutanamido)heptyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (4e)
4.1.8. 2′-O-Acetyl-4′′-O-((8-(4-((2R,3R)-2-(2,2-dichloroacetamido)-3-hydroxy-3-(4-nitrophenyl)propoxy)-4-oxobutanamido)-2-(2-(2-aminoethoxy)ethoxy)ethyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (4f)
4.1.9. 2′-O-Acetyl-4′′-O-((2-(N-(2-hydroxyethyl))-2-(4-((2R,3R)-2-(2,2-dichloroacetamido)-3-hydroxy-3-(4-nitrophenyl)propoxy)-4-oxobutanamido)ethyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (4g)
4.1.10. 2′-O-Acetyl-4′′-O-((2-(4-(2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethoxy)-4-oxobutanamido)ethyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (5a)
4.1.11. 2′-O-Acetyl-4′′-O-((3-(4-(2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethoxy)-4-oxobutanamido)propyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (5b)
4.1.12. 2′-O-Acetyl-4′′-O-((4-(4-(2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethoxy)-4-oxobutanamido)butyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (5c)
4.1.13. 2′-O-Acetyl-4′′-O-((5-(4-(2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethoxy)-4-oxobutanamido)pentyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (5d)
4.1.14. 2′-O-Acetyl-4′′-O-((7-(4-(2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethoxy)-4-oxobutanamido)heptyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (5e)
4.1.15. 2′-O-Acetyl-4′′-O-((8-(4-(2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethoxy)-4-oxobutanamido)-2-(2-(2-aminoethoxy)ethoxy)ethyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (5f)
4.1.16. 2′-O-Acetyl-4′′-O-((2-(N-(2-hydroxyethyl))-2-(4-(2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethoxy)-4-oxobutanamido)ethyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (5g)
4.1.17. 2′-O-Acetyl-4′′-O-((2-aminoethyl)carbamoyl)-11,12-cyclic carbonate of azithromycin (6)
4.2. Plasmids and Cloning
4.3. Bacterial Strains and Media
4.4. In Vitro Antimicrobial Activity
4.5. Dual Reporter Assay on Agar Plates
4.6. In Vitro Translation in a Cell-Free Bacterial System
4.7. In Vitro Competition-Binding Assay with E. coli Ribosomes
4.8. Toe-Printing Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marston, H.D.; Dixon, D.M.; Knisely, J.M.; Palmore, T.N.; Fauci, A.S. Antimicrobial Resistance. JAMA 2016, 316, 1193–1204. [Google Scholar] [CrossRef]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed]
- Hofer, U. The cost of antimicrobial resistance. Nat. Rev. Microbiol. 2019, 17, 3. [Google Scholar] [CrossRef]
- Lungu, I.A.; Moldovan, O.L.; Biris, V.; Rusu, A. Fluoroquinolones Hybrid Molecules as Promising Antibacterial Agents in the Fight against Antibacterial Resistance. Pharmaceutics 2022, 14, 1749. [Google Scholar] [CrossRef]
- Muhammad, A.; Simcha, W.; Rawish, F.; Sabih, R.; Albert, E.; Ali, N. Cadazolid vs Vancomycin for the Treatment of Clostridioides difficile Infection: Systematic Review with Meta-analysis. Curr. Clin. Pharmacol. 2020, 15, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.U.; Dalhoff, A.; Weintraub, A.; Nord, C.E. In vitro activity of MCB3681 against Clostridium difficile strains. Anaerobe 2014, 28, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Blais, J.; Lewis, S.R.; Krause, K.M.; Benton, B.M. Antistaphylococcal activity of TD-1792, a multivalent glycopeptide-cephalosporin antibiotic. Antimicrob. Agents Chemother. 2012, 56, 1584–1587. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Lynch, A.S. Development of a Dual-Acting Antibacterial Agent (TNP-2092) for the Treatment of Persistent Bacterial Infections. J. Med. Chem. 2016, 59, 6645–6657. [Google Scholar] [CrossRef]
- Gomes, C.; Ruiz-Roldán, L.; Mateu, J.; Ochoa, T.J.; Ruiz, J. Azithromycin resistance levels and mechanisms in Escherichia coli. Sci. Rep. 2019, 9, 6089. [Google Scholar] [CrossRef]
- Derbie, A.; Mekonnen, D.; Woldeamanuel, Y.; Abebe, T. Azithromycin resistant gonococci: A literature review. Antimicrob. Resist. Infect. Control 2020, 9, 138. [Google Scholar] [CrossRef]
- Doan, T.; Worden, L.; Hinterwirth, A.; Arzika, A.M.; Maliki, R.; Abdou, A.; Zhong, L.; Chen, C.; Cook, C.; Lebas, E.; et al. Macrolide and Nonmacrolide Resistance with Mass Azithromycin Distribution. N. Engl. J. Med. 2020, 383, 1941–1950. [Google Scholar] [CrossRef]
- El Ashkar, S.; Osman, M.; Rafei, R.; Mallat, H.; Achkar, M.; Dabboussi, F.; Hamze, M. Molecular detection of genes responsible for macrolide resistance among Streptococcus pneumoniae isolated in North Lebanon. J. Infect. Public Health 2017, 10, 745–748. [Google Scholar] [CrossRef]
- Tevyashova, A.N.; Bychkova, E.N.; Korolev, A.M.; Isakova, E.B.; Mirchink, E.P.; Osterman, I.A.; Erdei, R.; Szucs, Z.; Batta, G. Synthesis and evaluation of biological activity for dual-acting antibiotics on the basis of azithromycin and glycopeptides. Bioorg. Med. Chem. Lett. 2019, 29, 276–280. [Google Scholar] [CrossRef]
- Pavlovic, D.; Mutak, S. Discovery of 4″-ether linked azithromycin-quinolone hybrid series: Influence of the central linker on the antibacterial activity. ACS Med. Chem. Lett. 2011, 2, 331–336. [Google Scholar] [CrossRef]
- Pavlovic, D.; Mutak, S. Synthesis and antibacterial evaluation of novel 4″-glycyl linked quinolyl-azithromycins with potent activity against macrolide-resistant pathogens. Bioorg. Med. Chem. 2016, 24, 1255–1267. [Google Scholar] [CrossRef]
- Fajdetic, A.; Vinter, A.; Paljetak, H.C.; Padovan, J.; Jakopovic, I.P.; Kapic, S.; Alihodzic, S.; Filic, D.; Modric, M.; Kosutic-Hulita, N.; et al. Synthesis, activity and pharmacokinetics of novel antibacterial 15-membered ring macrolones. Eur. J. Med. Chem. 2011, 46, 3388–3397. [Google Scholar] [CrossRef]
- Tevyashova, A.N.; Korolev, A.M.; Mirchink, E.P.; Isakova, E.B.; Osterman, I.A. Synthesis and evaluation of biological activity of benzoxaborole derivatives of azithromycin. J. Antibiot. 2019, 72, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, S.A.; Li, X.; He, S.; Yu, M.; Wu, M.; Vederas, J.C. Synthesis of Tridecaptin-Antibiotic Conjugates with in Vivo Activity against Gram-Negative Bacteria. J. Med. Chem. 2015, 58, 9779–9785. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cong, C.; Chai, W.C.; Dong, R.; Jia, L.; Song, D.; Zhou, Z.; Ma, S. Synthesis and antibacterial activity of novel 4″-O-(1-aralkyl-1,2,3-triazol-4-methyl-carbamoyl) azithromycin analogs. Bioorg. Med. Chem. Lett. 2017, 27, 3872–3877. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Xu, L.; Wang, Y.; Wan, J.; Li, Q.; Wang, R. Synthesis and antibacterial activity of 11, 12-cyclic carbonate 4″-O-aralkylacetylhydrazineacyl azithromycin derivatives. Bioorg. Chem. 2020, 94, 103475. [Google Scholar] [CrossRef] [PubMed]
- Brook, I. Antimicrobials therapy of anaerobic infections. J. Chemother. 2016, 28, 143–150. [Google Scholar] [CrossRef]
- Erslev, A.J.; Iossifides, I.A. In vitro action of chloramphenicol and chloramphenicol-analogues on the metabolism of human immature red blood cells. Acta Haematol. 1962, 28, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Dinos, G.P.; Athanassopoulos, C.M.; Missiri, D.A.; Giannopoulou, P.C.; Vlachogiannis, I.A.; Papadopoulos, G.E.; Papaioannou, D.; Kalpaxis, D.L. Chloramphenicol Derivatives as Antibacterial and Anticancer Agents: Historic Problems and Current Solutions. Antibiotics 2016, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Tevyashova, A.N. Recent Trends in Synthesis of Chloramphenicol New Derivatives. Antibiotics 2021, 10, 370. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, D. Binding of chloramphenicol to ribosomes. The effect of a number of antibiotics. Biochim. Biophys. Acta 1966, 114, 277–288. [Google Scholar] [CrossRef]
- Bulkley, D.; Innis, C.A.; Blaha, G.; Steitz, T.A. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl. Acad. Sci. USA 2010, 107, 17158–17163. [Google Scholar] [CrossRef] [PubMed]
- Schlunzen, F.; Zarivach, R.; Harms, J.; Bashan, A.; Tocilj, A.; Albrecht, R.; Yonath, A.; Franceschi, F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 2001, 413, 814–821. [Google Scholar] [CrossRef]
- Brook, I. Treatment of anaerobic infection. Expert Rev. Anti. Infect. Ther. 2007, 5, 991–1006. [Google Scholar] [CrossRef]
- Mouton, C.; Dextraze, L.; Mayrand, D. Susceptibility of potential periodontopathic bacteria to metronidazole, spiramycin and their combination. J. Biol. Buccale 1984, 12, 17–26. [Google Scholar]
- Cederbrant, G.; Kahlmeter, G.; Schalen, C.; Kamme, C. Additive effect of clarithromycin combined with 14-hydroxy clarithromycin, erythromycin, amoxycillin, metronidazole or omeprazole against Helicobacter pylori. J. Antimicrob. Chemother. 1994, 34, 1025–1029. [Google Scholar] [CrossRef]
- Wang, J.; Cooper, D.L.; Zhan, W.; Wu, D.; He, H.; Sun, S.; Lovett, S.T.; Xu, B. Diglycine Enables Rapid Intrabacterial Hydrolysis for Activating Anbiotics against Gram-negative Bacteria. Angew. Chem. Int. Ed. Engl. 2019, 58, 10631–10634. [Google Scholar] [CrossRef]
- Mahfouz, N.M.; Aboul-Fadl, T.; Diab, A.K. Metronidazole twin ester prodrugs: Synthesis, physicochemical properties, hydrolysis kinetics and antigiardial activity. Eur. J. Med. Chem. 1998, 33, 675–683. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, L.; Wang, R.; Cao, J.; Liu, C.; Fang, Y.; Wang, J.; Ma, S. Novel C-4″ modified azithromycin analogs with remarkably enhanced activity against erythromycin-resistant Streptococcus pneumoniae: The synthesis and antimicrobial evaluation. Eur. J. Med. Chem. 2011, 46, 5196–5205. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Liu, Z.; Song, H.; Jiang, R.; He, F.; Ma, S. Synthesis and antibacterial activity of novel 11,12-cyclic carbonate azithromycin 4″-O-carbamate derivatives. J. Antibiot. 2010, 63, 3–8. [Google Scholar] [CrossRef]
- Maravic, G. Macrolide resistance based on the Erm-mediated rRNA methylation. Curr. Drug Targets Infect. Disord. 2004, 4, 193–202. [Google Scholar] [CrossRef]
- Svetlov, M.S.; Syroegin, E.A.; Aleksandrova, E.V.; Atkinson, G.C.; Gregory, S.T.; Mankin, A.S.; Polikanov, Y.S. Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance. Nat. Chem. Biol. 2021, 17, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Weisblum, B. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 1995, 39, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Miklasinska-Majdanik, M. Mechanisms of Resistance to Macrolide Antibiotics among Staphylococcus aureus. Antibiotics 2021, 10, 1406. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Sothiselvam, S.; Vazquez-Laslop, N.; Mankin, A.S. Deregulation of translation due to post-transcriptional modification of rRNA explains why erm genes are inducible. Nat. Commun. 2013, 4, 1984. [Google Scholar] [CrossRef]
- Vazquez-Laslop, N.; Thum, C.; Mankin, A.S. Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell. 2008, 30, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Osterman, I.A.; Komarova, E.S.; Shiryaev, D.I.; Korniltsev, I.A.; Khven, I.M.; Lukyanov, D.A.; Tashlitsky, V.N.; Serebryakova, M.V.; Efremenkova, O.V.; Ivanenkov, Y.A.; et al. Sorting Out Antibiotics’ Mechanisms of Action: A Double Fluorescent Protein Reporter for High-Throughput Screening of Ribosome and DNA Biosynthesis Inhibitors. Antimicrob. Agents Chemother. 2016, 60, 7481–7489. [Google Scholar] [CrossRef]
- Sharff, A.; Fanutti, C.; Shi, J.; Calladine, C.; Luisi, B. The role of the TolC family in protein transport and multidrug efflux. From stereochemical certainty to mechanistic hypothesis. Eur. J. Biochem. 2001, 268, 5011–5026. [Google Scholar] [CrossRef]
- Wu, T.; McCandlish, A.C.; Gronenberg, L.S.; Chng, S.S.; Silhavy, T.J.; Kahne, D. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 2006, 103, 11754–11759. [Google Scholar] [CrossRef]
- Orelle, C.; Carlson, S.; Kaushal, B.; Almutairi, M.M.; Liu, H.; Ochabowicz, A.; Quan, S.; Pham, V.C.; Squires, C.L.; Murphy, B.T.; et al. Tools for characterizing bacterial protein synthesis inhibitors. Antimicrob. Agents Chemother. 2013, 57, 5994–6004. [Google Scholar] [CrossRef]
- Sampson, B.A.; Misra, R.; Benson, S.A. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics 1989, 122, 491–501. [Google Scholar] [CrossRef]
- Schwarz, S.; Kehrenberg, C.; Doublet, B.; Cloeckaert, A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 2004, 28, 519–542. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Hunt, E.; Berge, J.; May, E.; Copeland, R.A.; Gontarek, R.R. Fluorescence polarization method to characterize macrolide-ribosome interactions. Antimicrob. Agents Chemother. 2005, 49, 3367–3372. [Google Scholar] [CrossRef] [PubMed]
- Tereshchenkov, A.G.; Shishkina, A.V.; Karpenko, V.V.; Chertkov, V.A.; Konevega, A.L.; Kasatsky, P.S.; Bogdanov, A.A.; Sumbatyan, N.V. New Fluorescent Macrolide Derivatives for Studying Interactions of Antibiotics and Their Analogs with the Ribosomal Exit Tunnel. Biochemistry 2016, 81, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.R.; Gohara, D.W.; Yap, M.N. Sequence selectivity of macrolide-induced translational attenuation. Proc. Natl. Acad. Sci. USA 2014, 111, 15379–15384. [Google Scholar] [CrossRef]
- Marks, J.; Kannan, K.; Roncase, E.J.; Klepacki, D.; Kefi, A.; Orelle, C.; Vazquez-Laslop, N.; Mankin, A.S. Context-specific inhibition of translation by ribosomal antibiotics targeting the peptidyl transferase center. Proc. Natl. Acad. Sci. USA 2016, 113, 12150–12155. [Google Scholar] [CrossRef]
- Troup, R.I.; Fallan, C.; Baud, M.G.J. Current strategies for the design of PROTAC linkers: A critical review. Explor. Target. Antitumor Ther. 2020, 1, 273–312. [Google Scholar] [CrossRef]
- Osterman, I.A.; Wieland, M.; Maviza, T.P.; Lashkevich, K.A.; Lukianov, D.A.; Komarova, E.S.; Zakalyukina, Y.V.; Buschauer, R.; Shiriaev, D.I.; Leyn, S.A.; et al. Tetracenomycin X inhibits translation by binding within the ribosomal exit tunnel. Nat. Chem. Biol. 2020, 16, 1071–1077. [Google Scholar] [CrossRef]
- Ma, S.; Ma, R.; Liu, Z.; Ma, C.; Shen, X. Synthesis and antibacterial activity of novel 15-membered macrolide derivatives: 4″-carbamate, 11,12-cyclic carbonate-4″-carbamate and 11,4″-di-O-arylcarbamoyl analogs of azithromycin. Eur. J. Med. Chem. 2009, 44, 4010–4020. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Laslop, N.; Ramu, H.; Klepacki, D.; Kannan, K.; Mankin, A.S. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 2010, 29, 3108–3117. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.; Chettiath, T.; Mankin, A.S. Induction of erm(C) expression by noninducing antibiotics. Antimicrob. Agents Chemother. 2008, 52, 866–874. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 10th ed.; CLSI Document M07-A10; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Antimicrobial Susceptibility of Anaerobic Bacteria, 9th ed.; CLSI document M11-A09; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Milon, P.; Konevega, A.L.; Peske, F.; Fabbretti, A.; Gualerzi, C.O.; Rodnina, M.V. Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Methods Enzym. 2007, 430, 1–30. [Google Scholar] [CrossRef]
- Wang, Z.X. An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett. 1995, 360, 111–114. [Google Scholar] [CrossRef]
- Orelle, C.; Szal, T.; Klepacki, D.; Shaw, K.J.; Vazquez-Laslop, N.; Mankin, A.S. Identifying the targets of aminoacyl-tRNA synthetase inhibitors by primer extension inhibition. Nucleic Acids Res. 2013, 41, e144. [Google Scholar] [CrossRef]

| AZT–CLM derivatives | |||||||
| Cmpd | 4a | 4b | 4c | 4d | 4e | 4f | 4g |
| R | H | H | H | H | H | H | (CH2)2OH |
| X | (CH2)2 | (CH2)3 | (CH2)4 | (CH2)5 | (CH2)7 | ((CH2)2O)2(CH2)2 | (CH2)2 |
| Yield, % | 20 | 21 | 24 | 25 | 25 | 22 | 19 |
| AZT–MZN derivatives | |||||||
| Cmpd | 5a | 5b | 5c | 5d | 5e | 5f | 5g |
| R | H | H | H | H | H | H | (CH2)2OH |
| X | (CH2)2 | (CH2)3 | (CH2)4 | (CH2)5 | (CH2)7 | ((CH2)2O)2(CH2)2 | (CH2)2 |
| Yield, % | 24 | 26 | 25 | 27 | 26 | 23 | 21 |
| Strain | CLM | AZT | 4a | 4b | 4c | 4d | 4e | 4f | 4g | 6 |
|---|---|---|---|---|---|---|---|---|---|---|
| S. pneumoniae ATCC 49619 | 4 | 0.06 | 0.5 | 0.25 | 0.25 | 0.25 | 0.5 | 0.125 | 0.5 | >32 |
| S. aureus ATCC 29213 | 8 | 0.5 | 32 | 32 | 32 | 16 | 32 | 16 | >32 | 32 |
| E. coli ATCC 25922 | 8 | 2 | 32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| Strain | MNZ | AZT | 5a | 5b | 5c | 5d | 5e | 5f | 5g |
|---|---|---|---|---|---|---|---|---|---|
| S. pneumoniae ATCC 49619 | >32 | 0.06 | 0.25 | 0.125 | 0.125 | 0.125 | 0.125 | 0.06 | 0.25 |
| S. agalactiae 1Cp | >32 | 0.015 | 1–2 | 1 | 1–2 | 1 | 0.5 | 0.5 | 1 |
| S. aureus ATCC 29213 | >32 | 0.5 | 8–16 | 8 | 16 | 4 | 8 | 4 | 32 |
| C. sporogenes ATCC 19404 | 8 | 4 | 0.25 | 4 | 8 | 8 | 4 | 4 | 16 |
| P. acnes 55 | >32 | <0.06 | 4 | 4 | 2 | 2 | 0.25 | 8 | 4 |
| Strain | MNZ | CLM | AZT | ERY | 4a | 4b | 4c | 4d | 4e | 4f | 4g | 5a | 5b | 5c | 5d | 5e | 5f | 5g | 6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli ΔtolC pErmC | >200 | 1.6 | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| E. coli ΔtolC pErmCL-ErmC | >200 | 1.6 | 3.1 | 50 | 12.5 | 25 | 12.5 | 25 | >200 | 25 | 100 | 12.5 | 6.3 | 12.5 | 12.5 | 12.5 | 12.5 | 25 | 6.3 |
| E. coli ΔtolC | >200 | 1.6 | 0.8 | 3.1 | 12.5 | 12.5 | 12.5 | 12.5 | >200 | 12.5 | 50 | 12.5 | 6.3 | 6.3 | 6.3 | 6.3 | 12.5 | 25 | 3.1 |
| E. coli ΔtolC pERMZα | >200 | 1.6 | 0.8 | 1.6 | 12.5 | 12.5 | 12.5 | 12.5 | >200 | 12.5 | 50 | 12.5 | 6.3 | 6.3 | 6.3 | 12.5 | 12.5 | 25 | 3.1 |
| Ratio * | - | 1:1 | 1:4 | 1:16 | 1:1 | 1:2 | 1:1 | 1:2 | - | 1:2 | 1:2 | 1:1 | 1:1 | 1:2 | 1:2 | 1:2 | 1:1 | 1:1 | 1:2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volynkina, I.A.; Bychkova, E.N.; Karakchieva, A.O.; Tikhomirov, A.S.; Zatonsky, G.V.; Solovieva, S.E.; Martynov, M.M.; Grammatikova, N.E.; Tereshchenkov, A.G.; Paleskava, A.; et al. Hybrid Molecules of Azithromycin with Chloramphenicol and Metronidazole: Synthesis and Study of Antibacterial Properties. Pharmaceuticals 2024, 17, 187. https://doi.org/10.3390/ph17020187
Volynkina IA, Bychkova EN, Karakchieva AO, Tikhomirov AS, Zatonsky GV, Solovieva SE, Martynov MM, Grammatikova NE, Tereshchenkov AG, Paleskava A, et al. Hybrid Molecules of Azithromycin with Chloramphenicol and Metronidazole: Synthesis and Study of Antibacterial Properties. Pharmaceuticals. 2024; 17(2):187. https://doi.org/10.3390/ph17020187
Chicago/Turabian StyleVolynkina, Inna A., Elena N. Bychkova, Anastasiia O. Karakchieva, Alexander S. Tikhomirov, George V. Zatonsky, Svetlana E. Solovieva, Maksim M. Martynov, Natalia E. Grammatikova, Andrey G. Tereshchenkov, Alena Paleskava, and et al. 2024. "Hybrid Molecules of Azithromycin with Chloramphenicol and Metronidazole: Synthesis and Study of Antibacterial Properties" Pharmaceuticals 17, no. 2: 187. https://doi.org/10.3390/ph17020187
APA StyleVolynkina, I. A., Bychkova, E. N., Karakchieva, A. O., Tikhomirov, A. S., Zatonsky, G. V., Solovieva, S. E., Martynov, M. M., Grammatikova, N. E., Tereshchenkov, A. G., Paleskava, A., Konevega, A. L., Sergiev, P. V., Dontsova, O. A., Osterman, I. A., Shchekotikhin, A. E., & Tevyashova, A. N. (2024). Hybrid Molecules of Azithromycin with Chloramphenicol and Metronidazole: Synthesis and Study of Antibacterial Properties. Pharmaceuticals, 17(2), 187. https://doi.org/10.3390/ph17020187







