Molecular Marvels: Small Molecules Paving the Way for Enhanced Gene Therapy
Abstract
:1. Introduction
2. Genetic Structure and Function of the CRISPR System
3. CRISPR/Cas9-Mediated Genome Editing
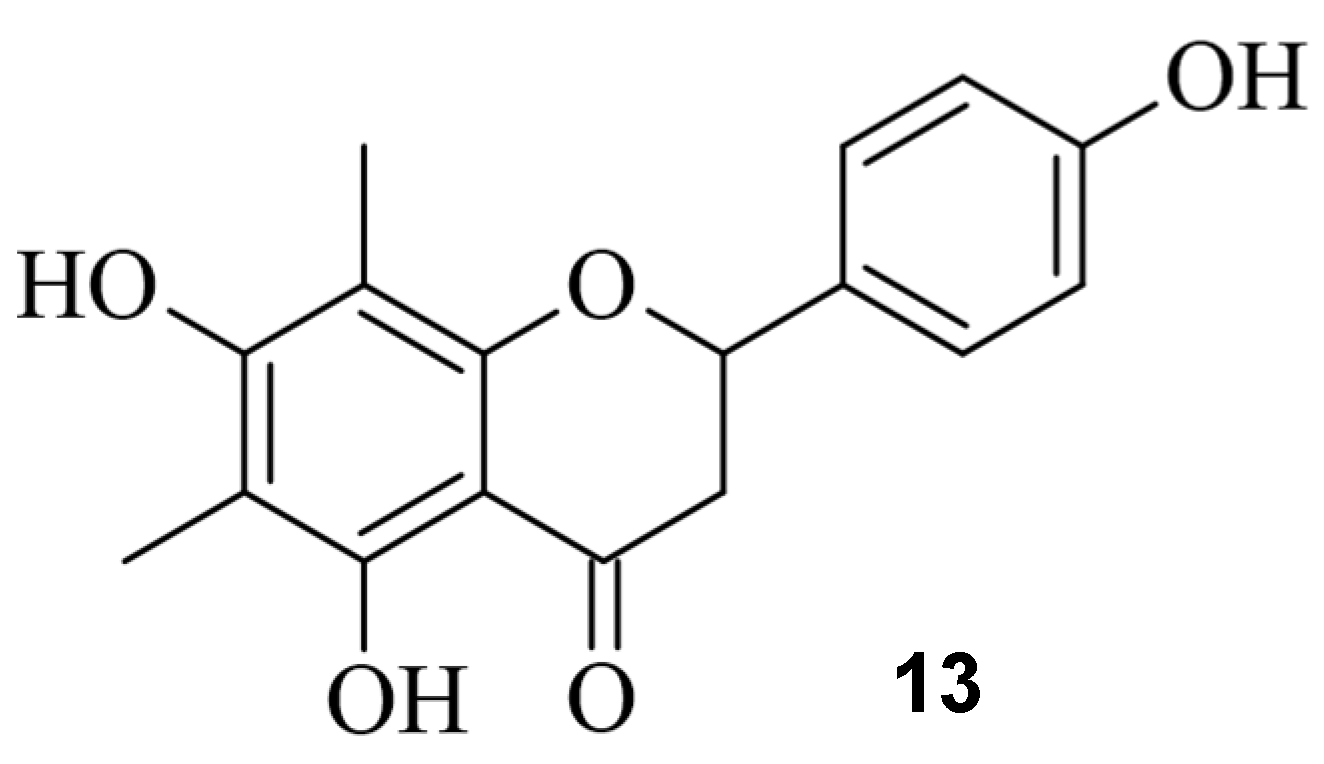
4. Small Molecules Modulate Wild-Type Cas9 Protein
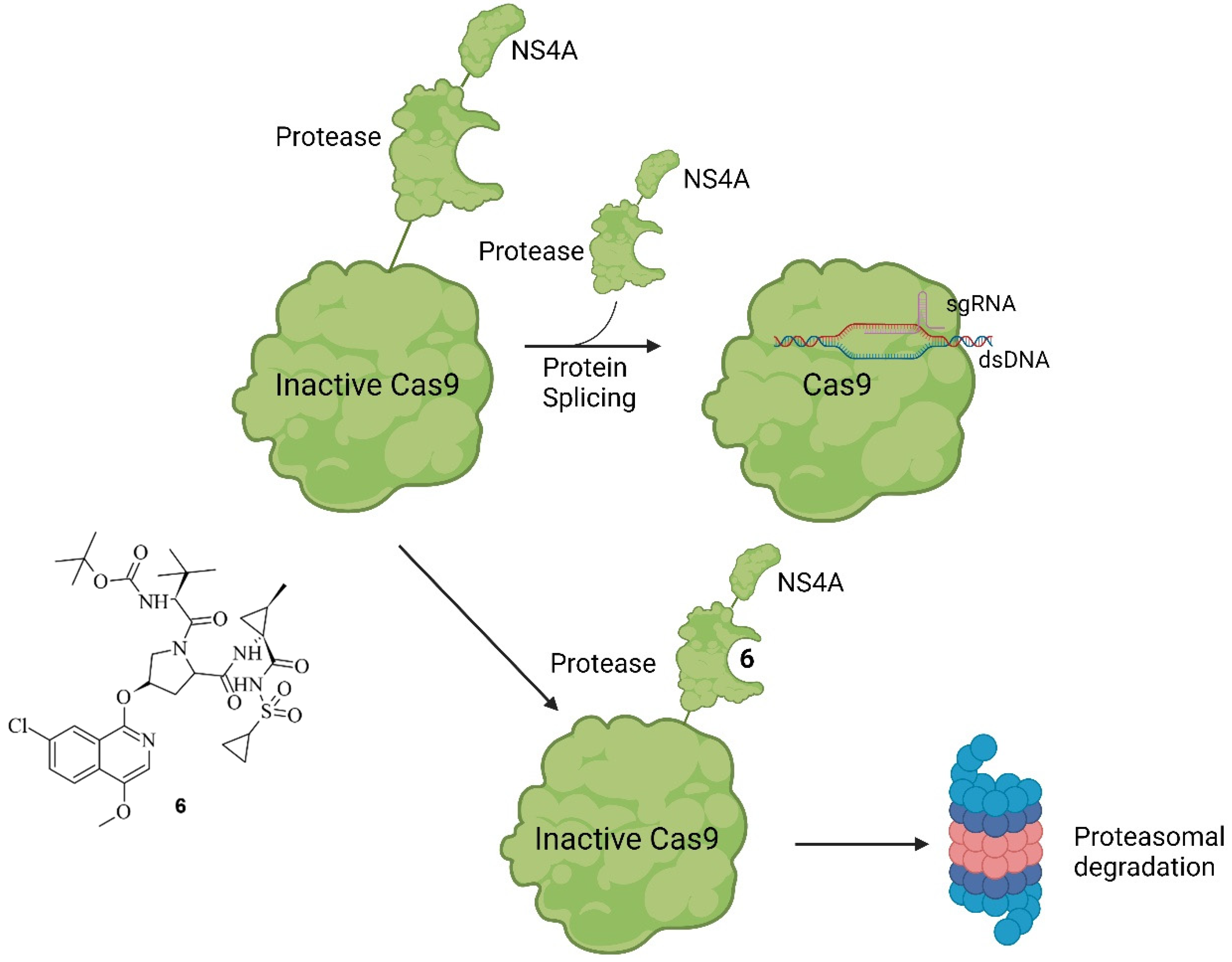
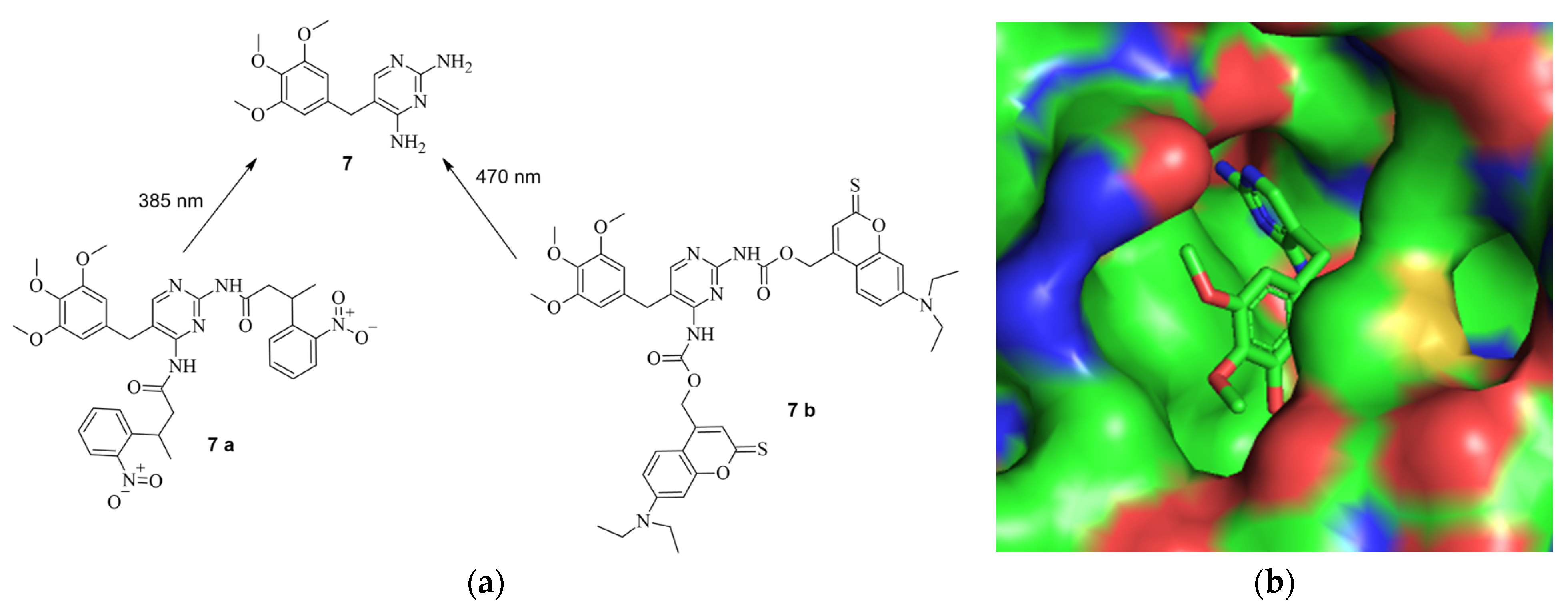
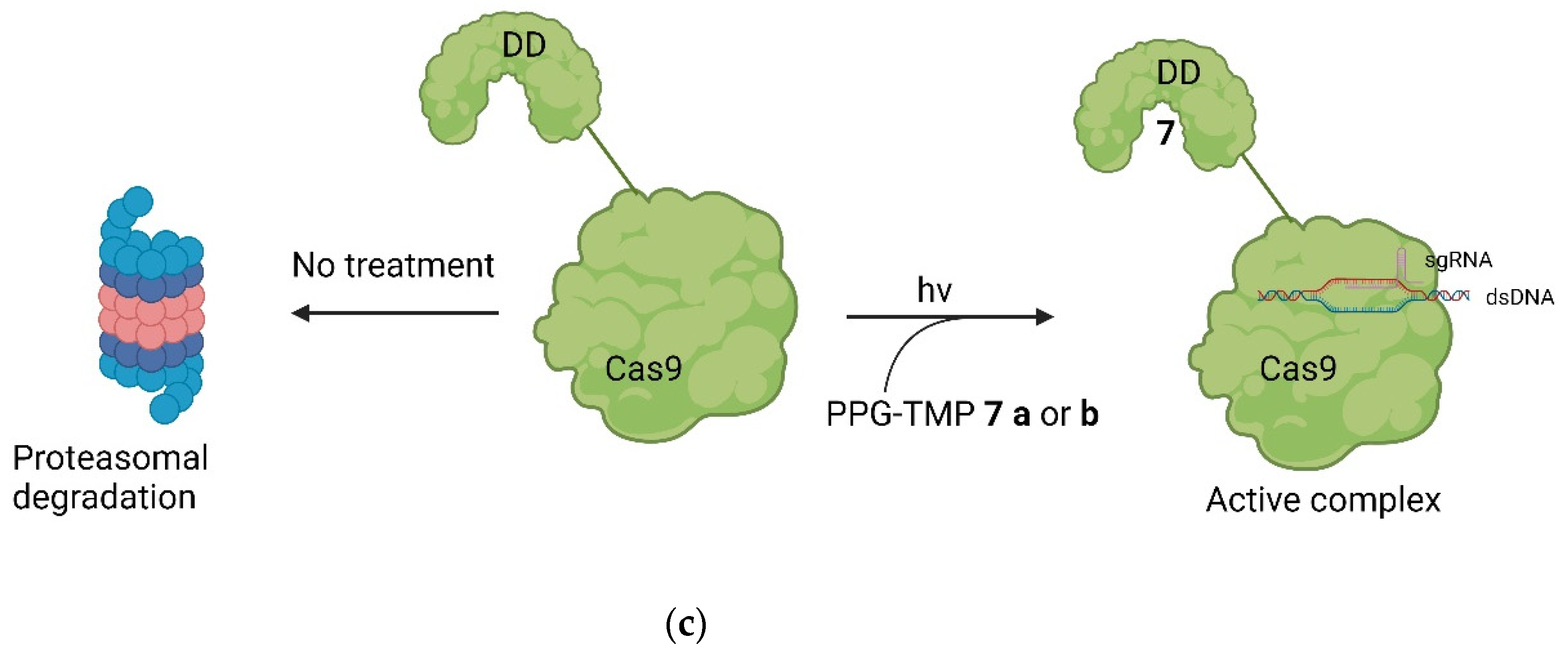
5. Small Molecules Modulate Engineered Cas9 Protein
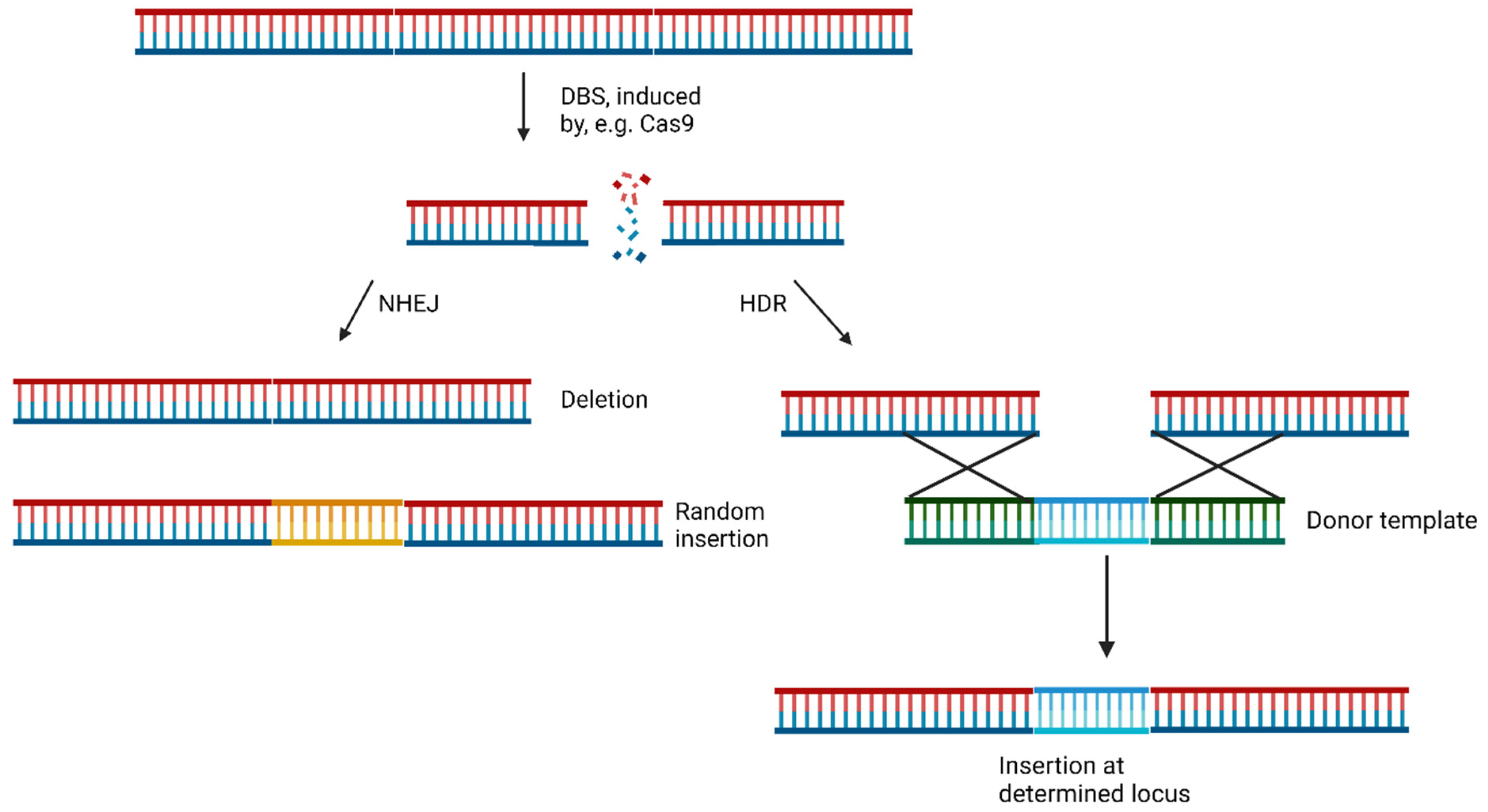
6. Small Molecules Regulate DSB Repair Mechanisms
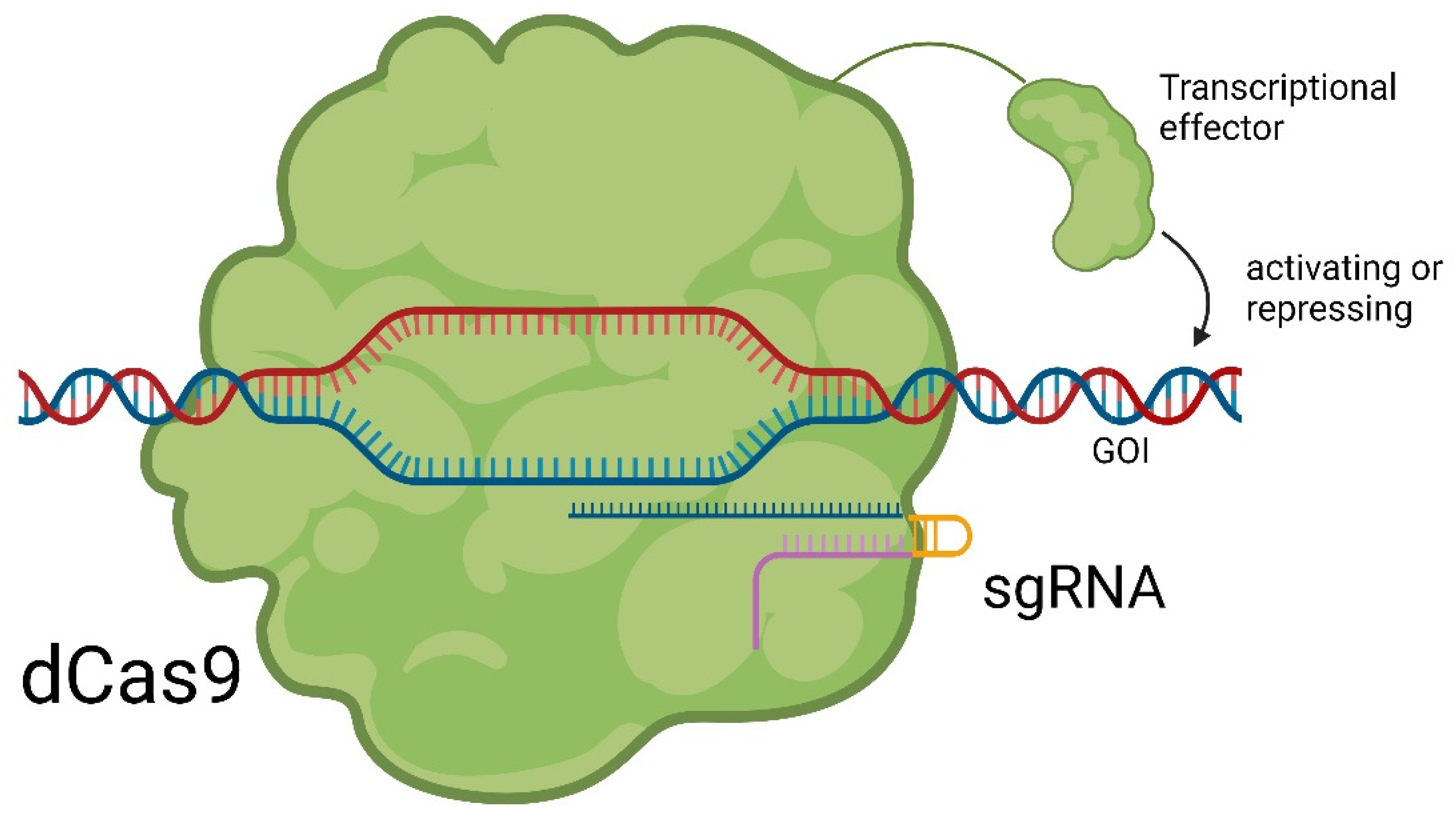
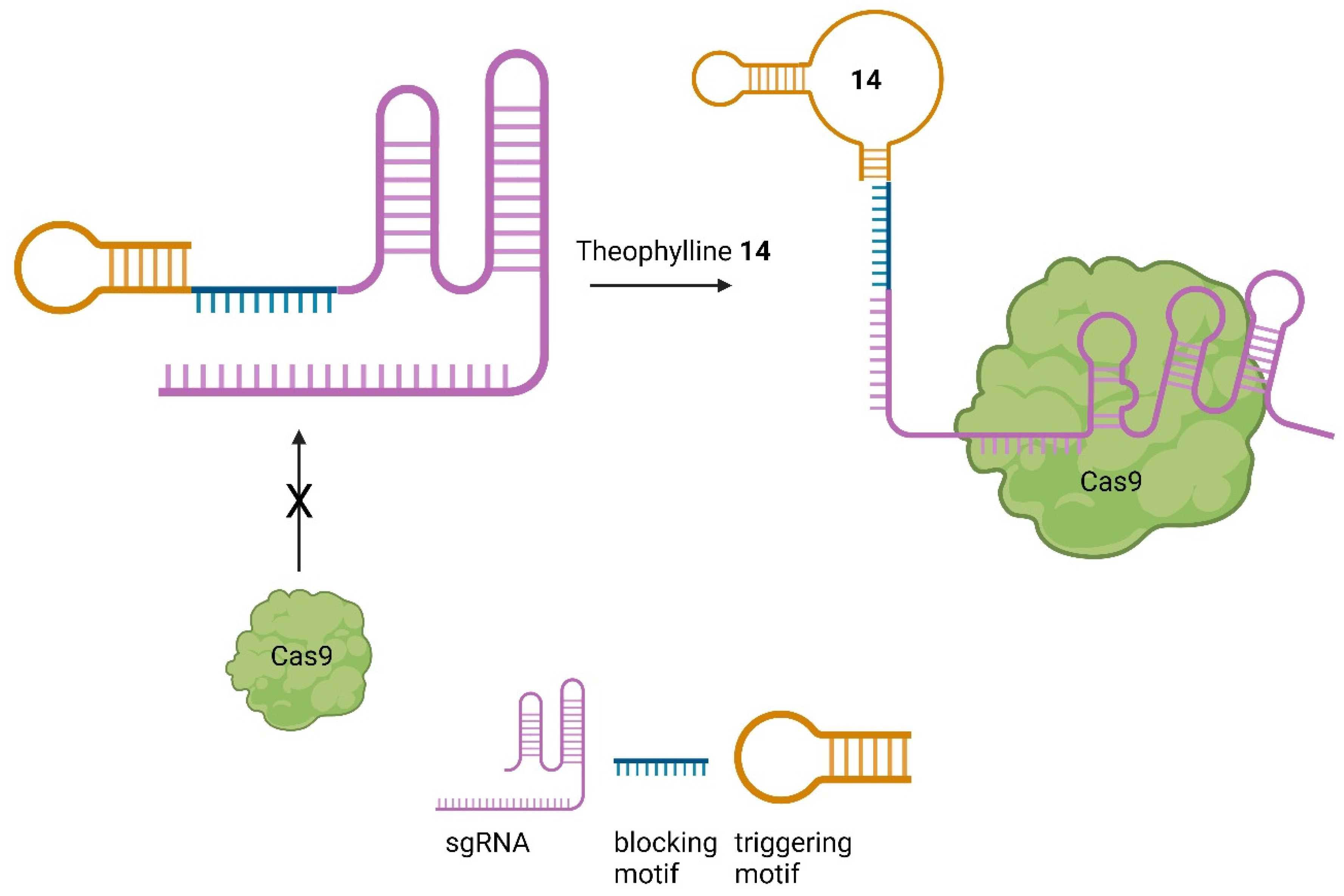
7. Small Molecules Regulate sgRNAs
8. Downsides of Small Molecules and Ethical Viewing Points
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wiedenheft, B.; Sternberg, S.H.; Doudna, J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012, 482, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, Z. CRISPR-Cas systems: Overview, innovations and applications in human disease research and gene therapy. Comput. Struct. Biotechnol. J. 2020, 18, 2401–2415. [Google Scholar] [CrossRef] [PubMed]
- Shrock, E.; Güell, M. CRISPR in Animals and Animal Models. Prog. Mol. Biol. Transl. Sci. 2017, 152, 95–114. [Google Scholar] [CrossRef] [PubMed]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Tran, K.T.; Vazquez-Uribe, R.; Gotfredsen, C.H.; Clausen, M.H.; Mendez, B.L.; Montoya, G.; Bach, A.; Sommer, M.O.A. Identification and Optimization of Novel Small-Molecule Cas9 Inhibitors by Cell-Based High-Throughput Screening. J. Med. Chem. 2022, 65, 3266–3305. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, D.; Liu, B.; Haisma, H.J. Modulating CRISPR/Cas9 genome-editing activity by small molecules. Drug Discov. Today 2022, 27, 951–966. [Google Scholar] [CrossRef]
- Shams, F.; Bayat, H.; Mohammadian, O.; Mahboudi, S.; Vahidnezhad, H.; Soosanabadi, M.; Rahimpour, A. Advance trends in targeting homology-directed repair for accurate gene editing: An inclusive review of small molecules and modified CRISPR-Cas9 systems. Bioimpacts 2022, 12, 371–391. [Google Scholar] [CrossRef]
- Karginov, F.V.; Hannon, G.J. The CRISPR System: Small RNA-Guided Defense in Bacteria and Archaea. Mol. Cell 2010, 37, 7–19. [Google Scholar] [CrossRef]
- Safari, F.; Zare, K.; Negahdaripour, M.; Barekati-Mowahed, M.; Ghasemi, Y. CRISPR Cpf1 proteins: Structure, function and implications for genome editing. Cell Biosci. 2019, 9, 36. [Google Scholar] [CrossRef]
- Mengstie, M.A.; Wondimu, B.Z. Mechanism and Applications of CRISPR/Cas-9-Mediated Genome Editing. Biologics 2021, 15, 353. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Koonin, E.V. Annotation and classification of CRISPR-Cas systems. Methods Mol. Biol. 2015, 1311, 47–75. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 Structures. Mechanisms 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Weterings, E.; Chen, D.J. The endless tale of non-homologous end-joining. Cell Res. 2008, 18, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef] [PubMed]
- Filippo, J.S.; Sung, P.; Klein, H. Mechanism of Eukaryotic Homologous Recombination. Annu. Rev. Biochem. 2008, 77, 229–257. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhang, X.; Zhong, C.; Mo, J.; Quan, R.; Yang, J.; Liu, D.; Li, Z.; Yang, H.; Wu, Z. Small molecules enhance CRISPR/ Cas9-mediated homology-directed genome editing in primary cells OPEN. Sci. Rep. 2017, 7, 8943. [Google Scholar] [CrossRef]
- Wang, H.; Xu, X. Microhomology-mediated end joining: New players join the team. Cell Biosci. 2017, 7, 6. [Google Scholar] [CrossRef]
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef]
- Lee, M.; Kim, H. Therapeutic application of the CRISPR system: Current issues and new prospects. Hum. Genet. 2019, 138, 563–590. [Google Scholar] [CrossRef]
- Mahas, A.; Stewart, C.N.; Mahfouz, M.M. Harnessing CRISPR/Cas systems for programmable transcriptional and post-transcriptional regulation. Biotechnol. Adv. 2018, 36, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, A.A.; Lim, W.A.; Qi, L.S. Beyond editing: Repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2015, 17, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Urrutia, R. KRAB-containing zinc-finger repressor proteins. Genime Biol. 2003, 4, 231. [Google Scholar] [CrossRef]
- Casas-Mollano, J.A.; Zinselmeier, M.H.; Erickson, S.E.; Smanski, M.J. CRISPR-Cas Activators for Engineering Gene Expression in Higher Eukaryotes. CRISPR J. 2020, 3, 350–364. [Google Scholar] [CrossRef] [PubMed]
- Karlson, C.K.S.; Mohd-noor, S.N.; Nolte, N.; Tan, B.C. CRISPR/dCas9-Based Systems: Mechanisms and Applications in Plant Sciences. Plants 2021, 10, 2055. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, L.; Jiang, J.; Wu, M.; Lin, P. Applications and challenges of CRISPR-Cas gene-editing to disease treatment in clinics. Precis. Clin. Med. 2021, 4, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Ali, G.; Tariq, M.A.; Shahid, K.; Ahmad, F.J.; Akram, J. Advances in genome editing: The technology of choice for precise and efficient β-thalassemia treatment. Gene Ther. 2020, 28, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Métais, J.Y.; Doerfler, P.A.; Mayuranathan, T.; Bauer, D.E.; Fowler, S.C.; Hsieh, M.M.; Katta, V.; Keriwala, S.; Lazzarotto, C.R.; Luk, K.; et al. Genome editing of HBG1 and HBG2 to induce fetal hemoglobin. Blood Adv. 2019, 3, 3379–3392. [Google Scholar] [CrossRef]
- Liu, Q.; Yang, F.; Zhang, J.; Liu, H.; Rahman, S.; Islam, S.; Ma, W.; She, M. Application of CRISPR/Cas9 in Crop Quality Improvement. Int. J. Mol. Sci. 2021, 22, 4206. [Google Scholar] [CrossRef]
- Shen, L.; Wang, C.; Fu, Y.; Wang, J.; Liu, Q.; Zhang, X.; Yan, C.; Qian, Q.; Wang, K. QTL editing confers opposing yield performance in different rice varieties. J. Integr. Plant Biol. 2018, 60, 89–93. [Google Scholar] [CrossRef]
- Gangopadhyay, S.A.; Cox, K.J.; Manna, D.; Lim, D.; Maji, B.; Zhou, Q.; Choudhary, A. Precision Control of CRISPR-Cas9 Using Small Molecules and Light. Biochemistry 2019, 58, 234. [Google Scholar] [CrossRef] [PubMed]
- Manna, D.; Maji, B.; Gangopadhyay, S.A.; Cox, K.J.; Zhou, Q.; Law, B.K.; Mazitschek, R.; Choudhary, A. A Singular System with Precise Dosing and Spatiotemporal Control of CRISPR-Cas9. Angew. Chem. Int. Ed. 2019, 58, 6285–6289. [Google Scholar] [CrossRef]
- Bondy-Denomy, J. Protein inhibitors of CRISPR-Cas9. ACS Chem. Biol. 2018, 13, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Vyas, P.; Harish. Anti-CRISPR proteins as a therapeutic agent against drug-resistant bacteria. Microbiol. Res. 2022, 257, 126963. [Google Scholar] [CrossRef] [PubMed]
- Franklin, S.E.; Mayfield, S.P. Expert Opinion on Biological Therapy Recent developments in the production of human therapeutic proteins in eukaryotic algae Recent developments in the production of human therapeutic proteins in eukaryotic algae. Expert. Opin. Biol. Ther. 2005, 5, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Beck, H.; Härter, M.; Haß, B.; Schmeck, C.; Baerfacker, L. Small molecules and their impact in drug discovery: A perspective on the occasion of the 125th anniversary of the Bayer Chemical Research Laboratory. Drug Discov. Today 2022, 27, 1560–1574. [Google Scholar] [CrossRef]
- Maji, B.; Gangopadhyay, S.A.; Lee, M.; Shi, M.; Wu, P.; Heler, R.; Mok, B.; Lim, D.; Siriwardena, S.U.; Paul, B.; et al. A High-Throughput Platform to Identify Small-Molecule Inhibitors of CRISPR-Cas9. Cell 2019, 177, 1067–1079.e19. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, D.; Wan, F.; Chen, B.; Wu, G.; Li, F.; Ren, Y.; Liang, P.; Wan, J.; Songyang, Z. Identification and Analysis of Small Molecule Inhibitors of CRISPR-Cas9 in Human Cells. Cells 2022, 11, 3574. [Google Scholar] [CrossRef]
- Cheng, X. Valproic Acid Thermally Destabilizes and Inhibits SpyCas9 Activity. Mol. Ther. 2020, 28, 2635–2641. [Google Scholar] [CrossRef]
- Crowley, W.R. BIBO3304. Xpharm: Compr. Pharmacol. Ref. 2007, 1, 1–3. [Google Scholar] [CrossRef]
- Davis, K.M.; Pattanayak, V.; Thompson, D.B.; Zuris, J.A.; Liu, D.R. Small molecule–triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 2015, 11, 316–318. [Google Scholar] [CrossRef] [PubMed]
- Buskirk, A.R.; Ong, Y.C.; Gartner, Z.J.; Liu, D.R. Directed evolution of ligand dependence: Small-molecule-activated protein splicing. Proc. Natl. Acad. Sci. USA 2004, 101, 10505–10510. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yang, L.; Chang, T.; Kandeel, F.; Yee, J.K. A Small Molecule Controlled Cas9 Repressible Sysytem. Mol. Ther. Nucleic Acids 2020, 19, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.C.; Popp, N.A.; Richardson, C.D.; Stephany, J.J.; Mathieu, J.; Wei, C.T.; Corn, J.E.; Maly, D.J.; Fowler, D.M. Suppression of unwanted CRISPR-Cas9 editing by co-administration of catalytically inactivating truncated guide RNAs. Nat. Commun. 2020, 11, 2697. [Google Scholar] [CrossRef] [PubMed]
- Sekine, R.; Kawata, T.; Muramoto, T. CRISPR/Cas9 mediated targeting of multiple genes in Dictyostelium. Sci. Rep. 2018, 8, 8471. [Google Scholar] [CrossRef] [PubMed]
- Weinstain, R.; Slanina, T.T.; Kand, D.; Kla, P.K. Visible-to-NIR-Light Activated Release: From Small Molecules to Nanomaterials. Chem. Rev. 2020, 120, 13135–13272. [Google Scholar] [CrossRef]
- Maji, B.; Moore, C.L.; Zetsche, B.; Volz, S.E.; Zhang, F.; Shoulders, M.D.; Choudhary, A. Multidimensional chemical control of CRISPR–Cas9. Nat. Chem. Biol. 2016, 13, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Krucinska, J.; Lombardo, M.N.; Erlandsen, H.; Estrada, A.; Si, D.; Viswanathan, K.; Wright, D.L. Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens. Commun. Biol. 2022, 5, 459. [Google Scholar] [CrossRef]
- Gama-Brambila, R.A.; Chen, J.; Dabiri, Y.; Tascher, G.; Němec, V.; Münch, C.; Song, G.; Knapp, S.; Cheng, X. A Chemical Toolbox for Labeling and Degrading Engineered Cas Proteins. JACS Au 2021, 1, 777–785. [Google Scholar] [CrossRef]
- Zhang, C.; Welborn, M.; Zhu, T.; Yang, N.J.; Santos, M.S.; Van Voorhis, T.; Pentelute, B.L. π-Clamp-mediated cysteine conjugation. Nat. Chem. 2016, 8, 120–128. [Google Scholar] [CrossRef]
- Békés, M.; Langley, D.R.; Crews, C.M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Bubeck, F.; Hoffmann, M.D.; Harteveld, Z.; Aschenbrenner, S.; Bietz, A.; Waldhauer, M.C.; Börner, K.; Fakhiri, J.; Schmelas, C.; Dietz, L.; et al. Engineered anti-CRISPR proteins for optogenetic control of CRISPR–Cas9. Nat. Methods 2018, 15, 924–927. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, M.; Nambiar, M.; Sharma, S.; Karki, S.S.; Goldsmith, G.; Hegde, M.; Kumar, S.; Pandey, M.; Singh, R.K.; Ray, P.; et al. An Inhibitor of Nonhomologous End-Joining Abrogates Double-Strand Break Repair and Impedes Cancer Progression. Cell 2012, 151, 1474–1487. [Google Scholar] [CrossRef] [PubMed]
- Bermudez-Cabrera, H.C.; Culbertson, S.; Barkal, S.; Holmes, B.; Shen, M.W.; Zhang, S.; Gifford, D.K.; Sherwood, R.I. Small molecule inhibition of ATM kinase increases CRISPR-Cas9 1-bp insertion frequency. Nat. Commun. 2021, 12, 5111. [Google Scholar] [CrossRef] [PubMed]
- Ueno, S.; Sudo, T.; Hirasawa, A. ATM: Functions of ATM Kinase and Its Relevance to Hereditary Tumors. Int. J. Mol. Sci. 2022, 23, 523. [Google Scholar] [CrossRef] [PubMed]
- Lombard, D.B.; Chua, K.F.; Mostoslavsky, R.; Franco, S.; Gostissa, M.; Alt, F.W. DNA Repair, Genome Stability, and Aging. Cell 2005, 120, 497–512. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair. 2008, 7, 1765–1771. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, H.; Xu, Z.; Tang, H.; Geng, A.; Cai, B.; Su, T.; Shi, J.; Jiang, C.; Tian, X.; et al. A PARP1-BRG1-SIRT1 axis promotes HR repair by reducing nucleosome density at DNA damage sites. Nucleic Acids Res. 2019, 47, 8563–8580. [Google Scholar] [CrossRef]
- Wassing, I.E.; Graham, E.; Saayman, X.; Rampazzo, L.; Ralf, C.; Bassett, A.; Esashi, F. The RAD51 recombinase protects mitotic chromatin in human cells. Nat. Commun. 2021, 12, 5380. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, Y.; Yang, J.; Zhang, J.; Yu, J.; Wang, M.; Zhao, X.; Wei, K.; Wan, X.; Xu, X.; et al. A high-throughput small molecule screen identifies farrerol as a potentiator of CRISPR/Cas9-mediated genome editing. eLife 2020, 9, e56008. [Google Scholar] [CrossRef] [PubMed]
- Meek, K.N.; Rangel, A.E.; Heemstra, J.M. Enhancing aptamer function and stability via in vitro selection using modified nucleic acids. Methods 2016, 106, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, R.S.; Ozdilek, B.A.; Garst, A.D.; Choudhury, A.; Batey, R.T. Small molecule regulated sgRNAs enable control of genome editing in E. coli by Cas9. Nat. Commun. 2020, 11, 1394. [Google Scholar] [CrossRef] [PubMed]
- Wrist, A.; Sun, W.; Summers, R.M. The Theophylline Aptamer: 25 Years as an Important Tool in Cellular Engineering Research. ACS Synth. Biol. 2020, 9, 682–697. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Zhao, L.; Pardi, A.; Xia, T. Ultrafast dynamics show that the theophylline and 3-methylxanthine aptamers employ a conformational capture mechanism for binding their ligands. Biochemistry 2010, 49, 2943–2951. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; An, Y.; Meng, L.; Zhang, H.; Song, J.; Zhu, Z.; Liu, W.; Song, Y.; Yang, C. Control of CRISPR-Cas9 with small molecule-activated allosteric aptamer regulating sgRNAs. Chem. Commun. 2019, 55, 12223. [Google Scholar] [CrossRef] [PubMed]
- La Manna, S.; Di Natale, C.; Florio, D.; Marasco, D. Peptides as Therapeutic Agents for Inflammatory-Related Diseases. Int. J. Mol. Sci. 2018, 19, 2714. [Google Scholar] [CrossRef]
- Sishc, B.J.; Davis, A.J. The Role of the Core Non-Homologous End Joining Factors in Carcinogenesis and Cancer. Cancers 2017, 9, 81. [Google Scholar] [CrossRef]
- Rehman, W.; Arfons, L.M.; Lazarus, H.M. The Rise, Fall and Subsequent Triumph of Thalidomide: Lessons Learned in Drug Development. Ther. Adv. Hematol. 2011, 2, 291. [Google Scholar] [CrossRef]
- Jahangirian, H.; Kalantari, K.; Izadiyan, Z.; Rafiee-Moghaddam, R.; Shameli, K.; Webster, T.J. A review of small molecules and drug delivery applications using gold and iron nanoparticles. Int. J. Nanomed. 2019, 14, 1633–1657. [Google Scholar] [CrossRef]
- Duan, L.; Ouyang, K.; Xu, X.; Xu, L.; Wen, C.; Zhou, X.; Qin, Z.; Xu, Z.; Sun, W.; Liang, Y. Nanoparticle Delivery of CRISPR/Cas9 for Genome Editing. Front. Genet. 2021, 12, 673286. [Google Scholar] [CrossRef] [PubMed]
- Gaohua, L.; Miao, X.; Dou, L. Crosstalk of physiological pH and chemical pKa under the umbrella of physiologically based pharmacokinetic modeling of drug absorption, distribution, metabolism, excretion, and toxicity. Expert. Opin. Drug Metab. Toxicol. 2021, 17, 1103–1124. [Google Scholar] [CrossRef] [PubMed]
- Gómez, R.R.; Valencia, P.R. In vitro-in vivo pharmacokinetic correlation model for quality assurance of antiretroviral drugs. Colomb. Med. 2015, 46, 109–116. [Google Scholar] [CrossRef]
- Preeti; Sambhakar, S.; Saharan, R.; Narwal, S.; Malik, R.; Gahlot, V.; Khalid, A.; Najmi, A.; Zoghebi, K.; Halawi, M.A.; et al. Exploring LIPIDs for their potential to improves bioavailability of lipophilic drugs candidates: A review. Saudi Pharm. J. 2023, 31, 101870. [Google Scholar] [CrossRef]
- Osuna-Marco, M.P.; López-Barahona, M.; López-Ibor, B.; Tejera, Á.M. Ten Reasons Why People With Down Syndrome are Protected From the Development of Most Solid Tumors—A Review. Front. Genet. 2021, 12, 749480. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasselbeck, S.; Cheng, X. Molecular Marvels: Small Molecules Paving the Way for Enhanced Gene Therapy. Pharmaceuticals 2024, 17, 41. https://doi.org/10.3390/ph17010041
Hasselbeck S, Cheng X. Molecular Marvels: Small Molecules Paving the Way for Enhanced Gene Therapy. Pharmaceuticals. 2024; 17(1):41. https://doi.org/10.3390/ph17010041
Chicago/Turabian StyleHasselbeck, Sebastian, and Xinlai Cheng. 2024. "Molecular Marvels: Small Molecules Paving the Way for Enhanced Gene Therapy" Pharmaceuticals 17, no. 1: 41. https://doi.org/10.3390/ph17010041
APA StyleHasselbeck, S., & Cheng, X. (2024). Molecular Marvels: Small Molecules Paving the Way for Enhanced Gene Therapy. Pharmaceuticals, 17(1), 41. https://doi.org/10.3390/ph17010041