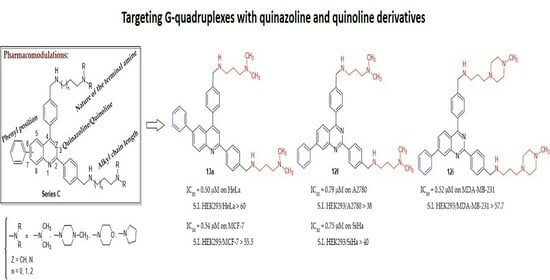
New 2,4-bis[(substituted-aminomethyl)phenyl]phenylquinazoline and 2,4-bis[(substituted-aminomethyl)phenyl]phenylquinoline Derivatives: Synthesis and Biological Evaluation as Novel Anticancer Agents by Targeting G-Quadruplex
Abstract
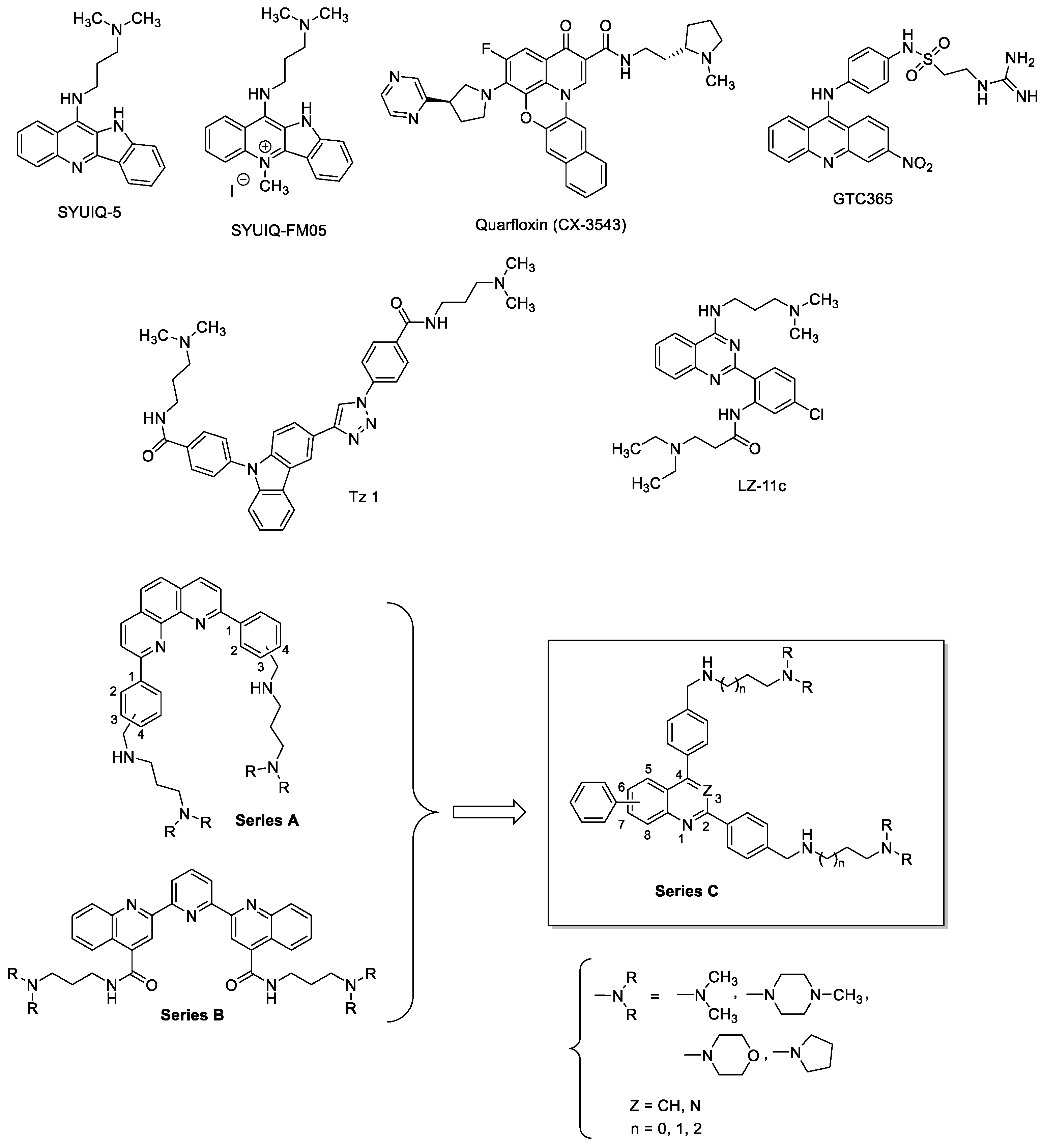
:1. Introduction
2. Results & Discussion
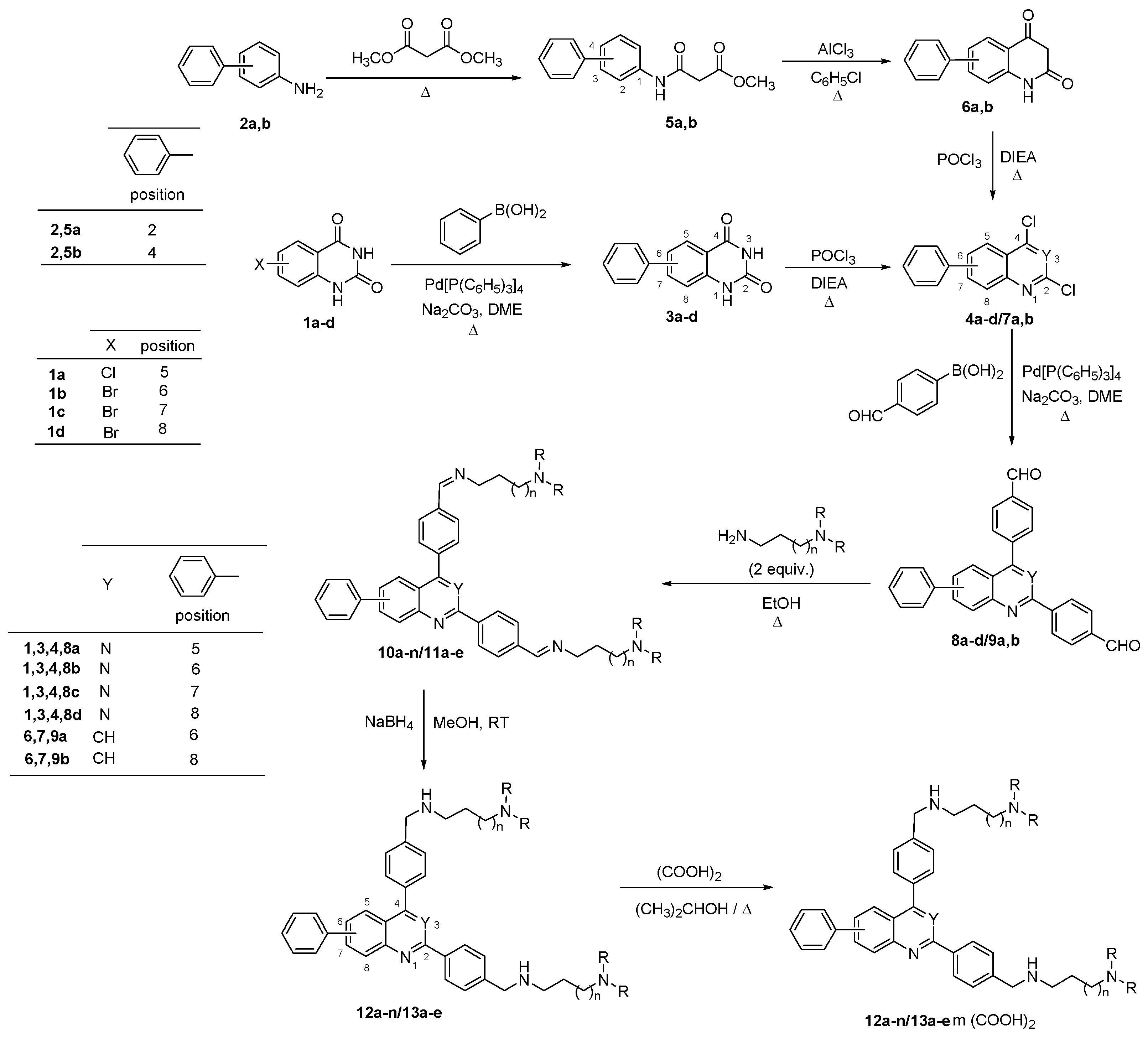
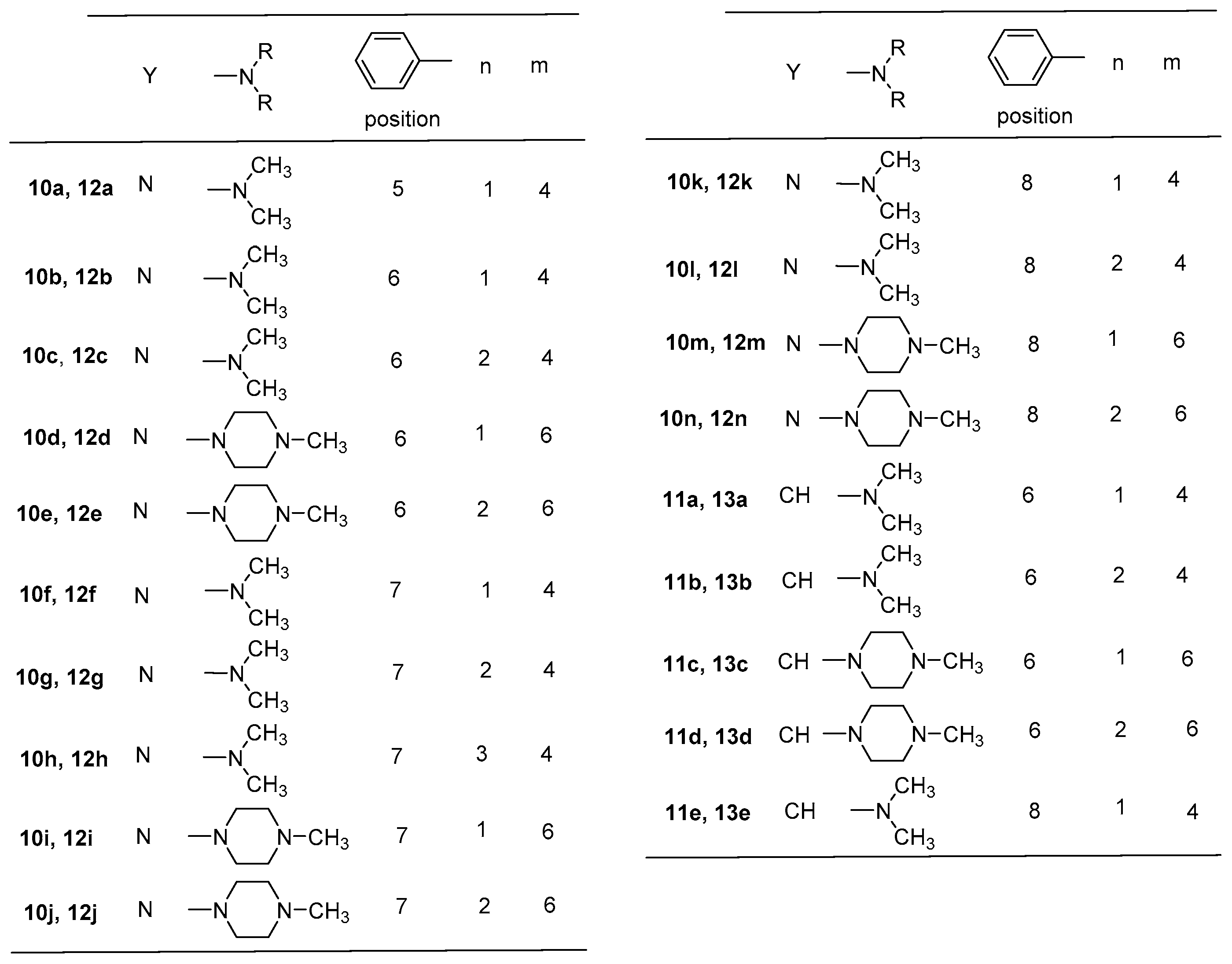
2.1. Chemistry
2.2. Biology
2.3. FRET-Melting Experiments
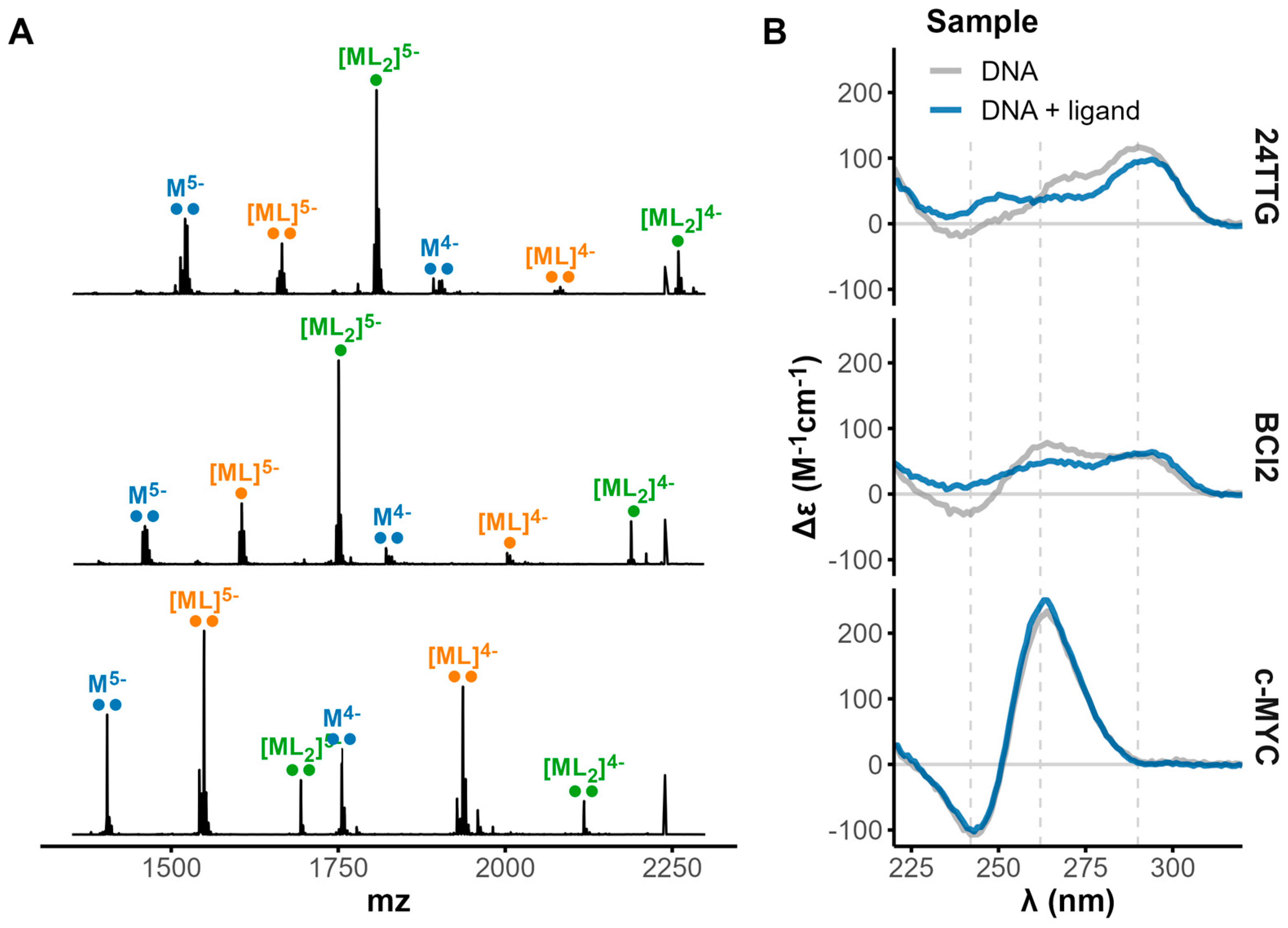
2.4. Native Electrospray Mass Spectrometry
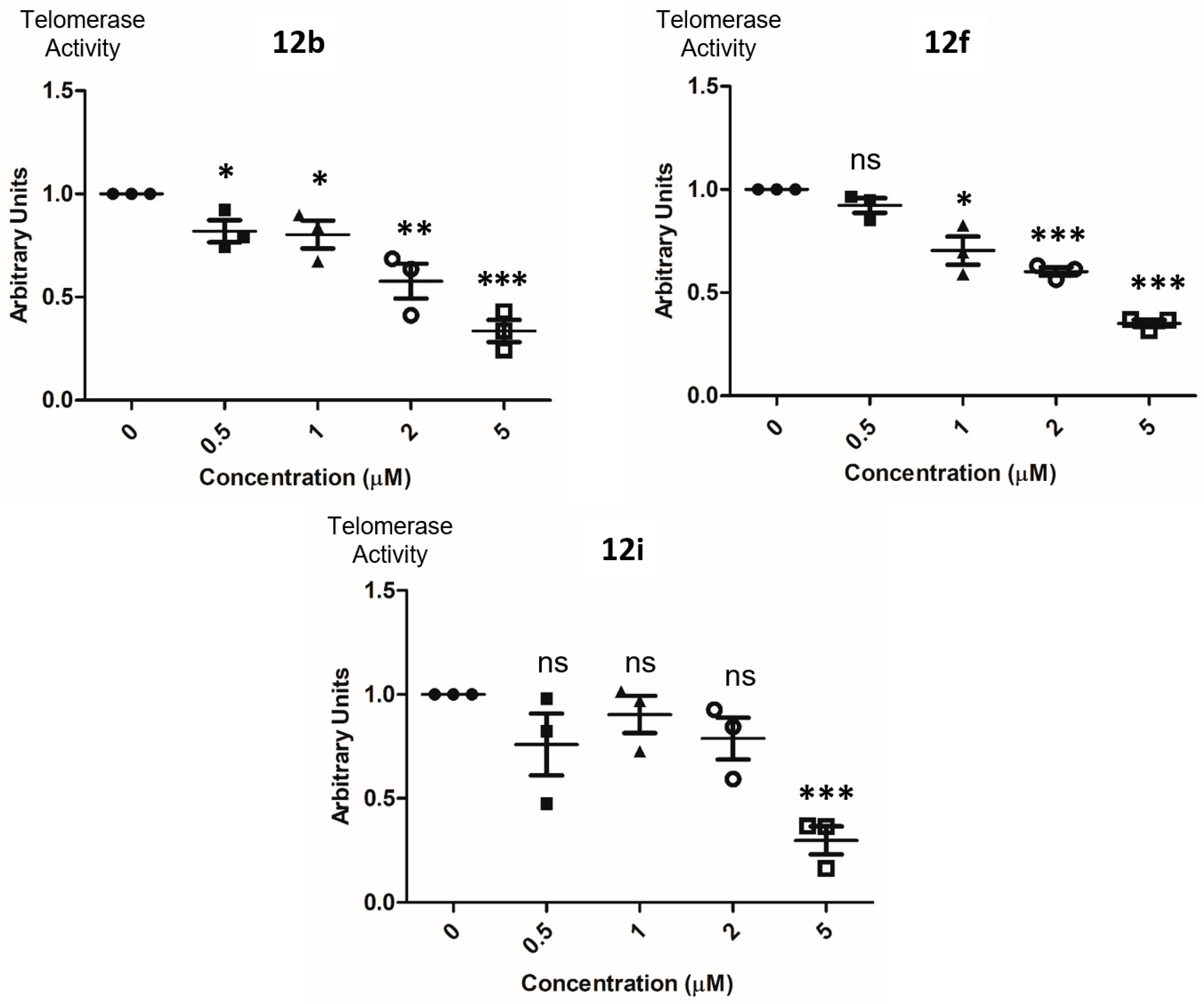
2.5. Detection of Telomerase Activity in Cell Lysates
3. Materials and Methods
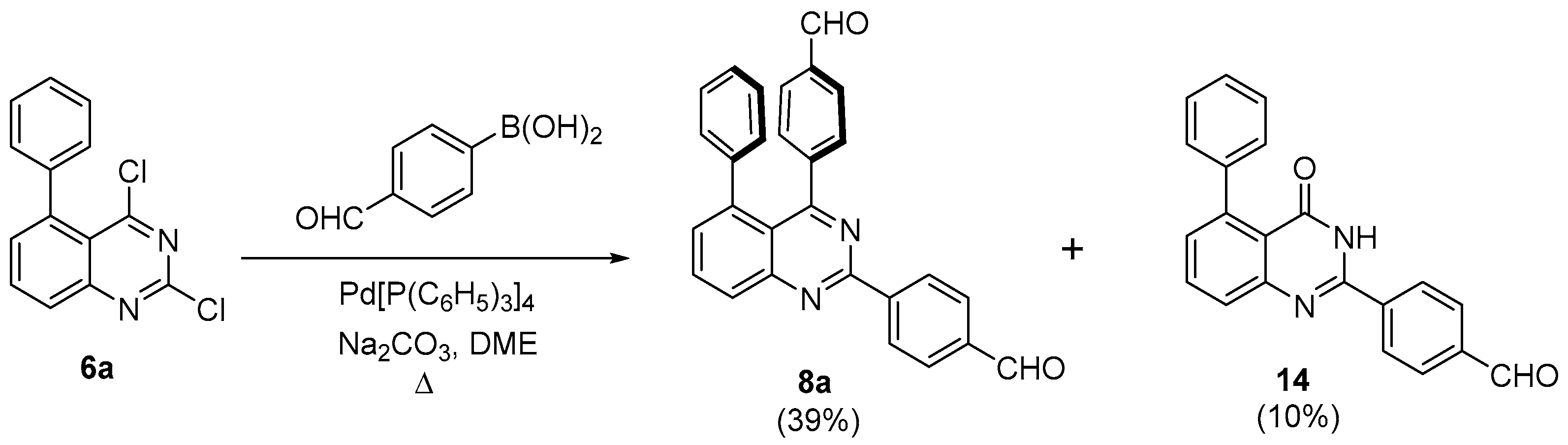
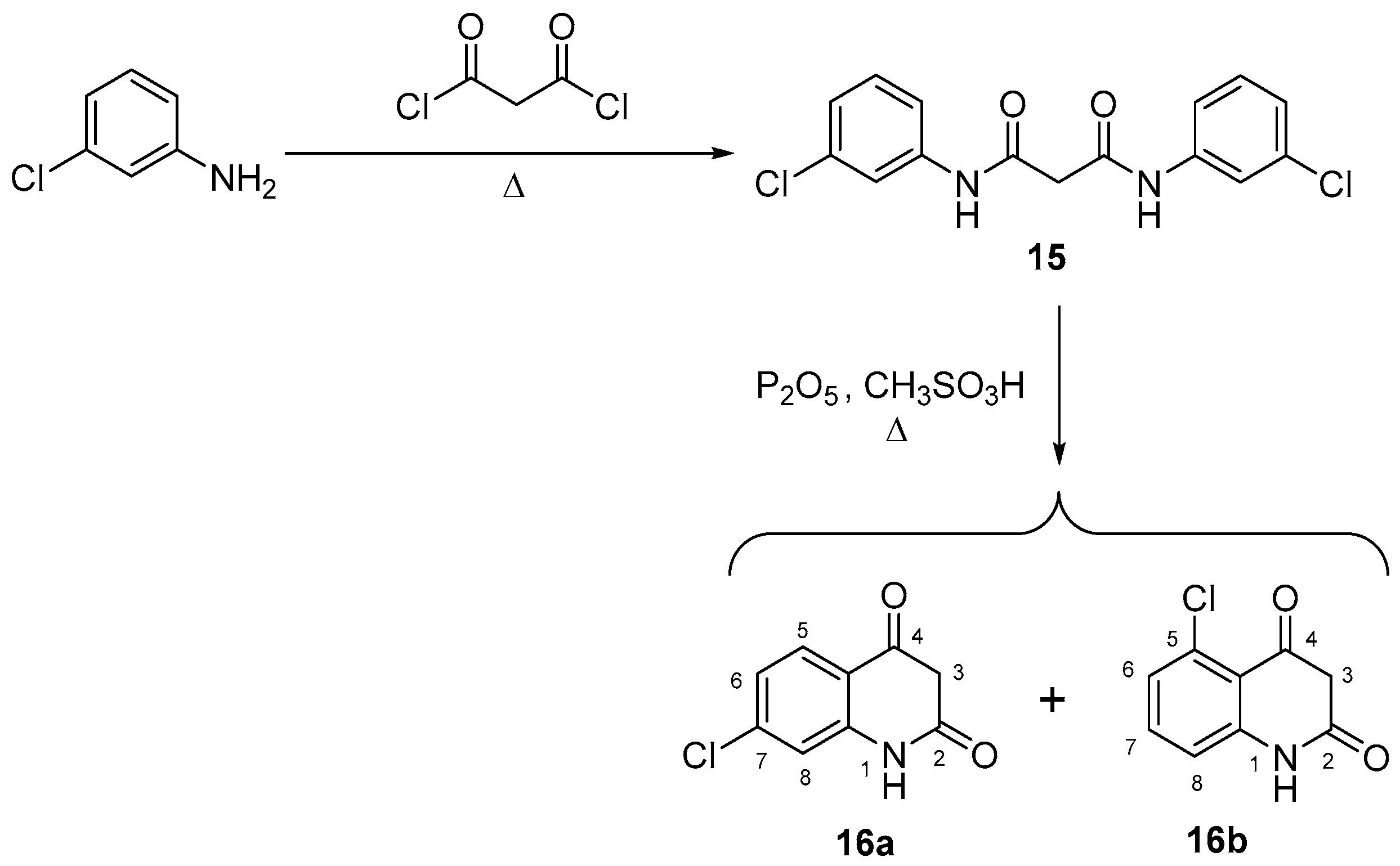
3.1. Chemistry
- General procedure for generating 2,4-bis(4-formylphenyl)-phenylquinazolines 8a–d and 2,4-bis(4-formylphenyl)-phenylquinolines 9a,b
- 2,4-bis(4-formylphenyl)-5-phenylquinazoline (8a)
- 2,4-bis(4-formylphenyl)-6-phenylquinazoline (8b)
- 2,4-bis(4-formylphenyl)-7-phenylquinazoline (8c)
- 2,4-bis(4-formylphenyl)-8-phenylquinazoline (8d)
- 2,4-bis(4-formylphenyl)-6-phenylquinoline (9a)
- 2,4-bis(4-formylphenyl)-8-phenylquinoline (9b)
- General procedure for 2,4-bis[(substituted-iminomethyl)phenyl]-phenylquinazolines 10a–n and 2,4-bis[(substituted-iminomethyl)phenyl]-phenylquinolines 11a–e
- 2,4-bis{4-[(3-dimethylaminopropyl)iminomethyl]phenyl}-5-phenylquinazoline (10a)
- 2,4-bis{4-[(3-dimethylaminopropyl)iminomethyl]phenyl}-6-phenylquinazoline (10b)
- 2,4-bis{4-[(4-dimethylaminobutyl)iminomethyl]phenyl}-6-phenylquinazoline (10c)
- 2,4-bis{4-[(3-(4-methylpiperazin-1-yl)propyl)iminomethyl]phenyl}-6-phenylquinazoline (10d)
- 2,4-bis{4-[(4-(4-methylpiperazin-1-yl)butyl)iminomethyl]phenyl}-6-phenylquinazoline (10e)
- 2,4-bis{4-[(3-dimethylaminopropyl)iminomethyl]phenyl}-7-phenylquinazoline (10f)
- 2,4-bis{4-[(4-dimethylaminobutyl)iminomethyl]phenyl}-7-phenylquinazoline (10g)
- 2,4-bis{4-[(5-dimethylaminopentyl)iminomethyl]phenyl}-7-phenylquinazoline (10h)
- 2,4-bis{4-[(3-(4-methylpiperazin-1-yl)propyl)iminomethyl]phenyl}-7-phenylquinazoline (10i)
- 2,4-bis{4-[(4-(4-methylpiperazin-1-yl)butyl)iminomethyl]phenyl}-7-phenylquinazoline (10j)
- 2,4-bis{4-[(3-dimethylaminopropyl)iminomethyl]phenyl}-8-phenylquinazoline (10k)
- 2,4-bis{4-[(4-dimethylaminobutyl)iminomethyl]phenyl}-8-phenylquinazoline (10l)
- 2,4-bis{4-[(3-(4-methylpiperazin-1-yl)propyl)iminomethyl]phenyl}-8-phenylquinazoline (10m)
- 2,4-bis{4-[(4-(4-methylpiperazin-1-yl)butyl)iminomethyl]phenyl}-8-phenylquinazoline (10n)
- 2,4-bis{4-[(3-dimethylaminopropyl)iminomethyl]phenyl}-6-phenylquinoline (11a)
- 2,4-bis{4-[(4-dimethylaminobutyl)iminomethyl]phenyl}-6-phenylquinoline (11b)
- 2,4-bis{4-[(3-(4-methylpiperazin-1-yl)propyl)iminomethyl]phenyl}-6-phenylquinoline (11c)
- 2,4-bis{4-[(4-(4-methylpiperazin-1-yl)butyl)iminomethyl]phenyl}-6-phenylquinoline (11d)
- 2,4-bis{4-[(3-dimethylaminopropyl)iminomethyl]phenyl}-8-phenylquinoline (11e)
- General procedure for 2,4-bis[(substituted-aminomethyl)phenyl]-phenylquinazolines 12a–n and 2,4-bis[(substituted-aminomethyl)phenyl]-phenylquinolines 13a–e
- 2,4-bis{4-[(3-dimethylaminopropyl)aminomethyl]phenyl}-5-phenylquinazoline (12a)
- 2,4-bis{4-[(3-dimethylaminopropyl)aminomethyl]phenyl}-6-phenylquinazoline (12b)
- 2,4-bis{4-[(4-dimethylaminobutyl)aminomethyl]phenyl}-6-phenylquinazoline (12c)
- 2,4-bis{4-[(4-(3-methylpiperazin-1-yl)propyl)aminomethyl]phenyl}-6-phenylquinazoline (12d)
- 2,4-bis{4-[(4-(4-methylpiperazin-1-yl)butyl)aminomethyl]phenyl}-6-phenylquinazoline (12e)
- 2,4-bis{4-[(3-dimethylaminopropyl)aminomethyl]phenyl}-7-phenylquinazoline (12f)
- 2,4-bis{4-[(4-dimethylaminobutyl)aminomethyl]phenyl}-7-phenylquinazoline (12g)
- 2,4-bis{4-[(5-dimethylaminopentyl)aminomethyl]phenyl}-7-phenylquinazoline (12h)
- 2,4-bis{4-[(4-(3-methylpiperazin-1-yl)propyl)aminomethyl]phenyl}-7-phenylquinazoline (12i)
- 2,4-bis{4-[(4-(4-methylpiperazin-1-yl)butyl)aminomethyl]phenyl}-7-phenylquinazoline (12j)
- 2,4-bis{4-[(3-dimethylaminopropyl)aminomethyl]phenyl}-8-phenylquinazoline (12k)
- 2,4-bis{4-[(4-dimethylaminobutyl)aminomethyl]phenyl}-8-phenylquinazoline (12l)
- 2,4-bis{4-[(4-(3-methylpiperazin-1-yl)propyl)aminomethyl]phenyl}-8-phenylquinazoline (12m)
- 2,4-bis{4-[(4-(4-methylpiperazin-1-yl)butyl)aminomethyl]phenyl}-8-phenylquinazoline (12n)
- 2,4-bis{4-[(3-dimethylaminopropyl)aminomethyl]phenyl}-6-phenylquinoline (13a)
- 2,4-bis{4-[(4-dimethylaminobutyl)aminomethyl]phenyl}-6-phenylquinoline (13b)
- 2,4-bis{4-[(4-(3-methylpiperazin-1-yl)propyl)aminomethyl]phenyl}-6-phenylquinoline (13c)
- 2,4-bis{4-[(4-(4-methylpiperazin-1-yl)butyl)aminomethyl]phenyl}-6-phenylquinoline (13d)
- 2,4-bis{4-[(3-dimethylaminopropyl)aminomethyl]phenyl}-8-phenylquinoline (13e)
- 2-(4-Formylphenyl)-5-phenyl-3H-quinazolin-4-one (14)
3.2. FRET-Melting Experiments
3.3. Native MS
3.4. Circular Dichroism
3.5. Determination of Antiproliferative Effects (MTT Assay)
3.6. Telomerase Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Cancer Research Fund International. Available online: https://www.wcrf.org/cancer-trends/worldwide-cancer-data/ (accessed on 1 February 2023).
- World Health Organization. Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 1 February 2023).
- Vasan, N.; Baselga, J.; Hyman, D.M. A View on Drug Resistance in Cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, S.; Kumar, M.S.; Peters, G.J.; Mayur, Y.C. Targeting telomerase for its advent in cancer therapeutics. Med. Res. Rev. 2020, 40, 1871–1919. [Google Scholar] [CrossRef] [PubMed]
- Dolinnaya, N.G.; Ogloblina, A.M.; Yakubovskaya, M.G. Structure, properties, and biological relevance of the DNA and RNA G-quadruplexes: Overview 50 years after their discovery. Biochemistry 2016, 81, 1602–1649. [Google Scholar] [CrossRef] [PubMed]
- Roxo, C.; Kotkowiak, W.; Pasternak, A. G-quadruplex forming aptamers—Characteristics, applications, and perspectives. Molecules 2019, 24, 3781. [Google Scholar] [CrossRef] [PubMed]
- Ivancich, M.; Schrank, Z.; Wojdyla, L.; Leviskas, B.; Kuckovic, A.; Sanjali, A.; Puri, N. Treating Cancer by Targeting Telomeres and Telomerase. Antioxidants 2017, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Mengual Gomez, D.L.; Armando, R.G.; Cerrudo, C.S.; Ghiringhelli, P.D.; Gomez, D.E. Telomerase as a Cancer Target. Development of New Molecules. Curr. Med. Chem. 2016, 16, 2432–2440. [Google Scholar] [CrossRef] [PubMed]
- Lipinska, N.; Romaniuk, A.; Paszel-Jaworska, A.; Toton, E.; Kopczynski, P.; Rubis, B. Telomerase and drug resistance in cancer. Cell. Mol. Life Sci. 2017, 74, 4121–4132. [Google Scholar] [CrossRef]
- Zidanloo, S.G.; Colagar, A.H.; Ayatollahi, H.; Bagheryan, Z. G-quadruplex forming region within WT1 promoter is selectively targeted by daunorubicin and mitoxantrone: A possible mechanism for anti-leukemic effect of drugs. J. Biosci. 2019, 44, 12. [Google Scholar]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy. Molecules 2019, 24, 429. [Google Scholar] [CrossRef]
- Sun, Z.-Y.; Wang, X.-N.; Cheng, S.-Q.; Su, X.-X.; Ou, T.-M. Developing Novel G-Quadruplex Ligands: From Interaction with Nucleic Acids to Interfering with Nucleic Acid-Protein Interaction. Molecules 2019, 24, 396. [Google Scholar] [CrossRef]
- Che, T.; Wang, Y.-Q.; Huang, Z.L.; Tan, J.-H.; Huang, Z.-S.; Chen, S.B. Natural Alkaloids and Heterocycles as G-Quadruplex Ligands and Potential Anticancer Agents. Molecules 2018, 23, 493. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.R.; Cadoni, E.; Ressurreição, A.S.; Moreira, R.; Paulo, A. Design of Modular G-quadruplex Ligands. ChemMedChem 2018, 13, 869–893. [Google Scholar] [CrossRef]
- Ilyinsky, N.S.; Varizhuk, A.M.; Beniaminov, A.D.; Puzanov, M.A.; Shchyolkina, A.K.; Kaluzhny, D.N. G-quadruplex ligands: Mechanisms of anticancer action and target binding. Mol. Biol. 2014, 48, 778–794. [Google Scholar] [CrossRef]
- Liu, J.N.; Guo, J.F.; Zhou, J.M.; Feng, G.K.; Huang, Z.S.; Gu, L.Q.; Zeng, Y.X.; Zhu, X.F. Inhibition of myc promoter and telomerase activity and induction of delayed apoptosis by SYUIQ-5, a novel G-quadruplex interactive agent in leukemia cells. Leukemia 2007, 21, 1300–1302. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, A.-C.; Kuang, G.-T.; Wang, S.-K.; Ou, T.-M.; Tan, J.-H.; Li, D.; Huang, Z.-S. Design, synthesis and biological evaluation of 4-anilinoquinazoline derivatives as new c-myc G-quadruplex lignads. Eur. J. Med. Chem. 2016, 122, 264–279. [Google Scholar] [CrossRef] [PubMed]
- Navneetha, O.; Deepthi, K.; Rao, A.M.; Jyostna, T.S. A review on chemotherapeutic activities of quinoline. Int. J. Pharm. Chem. Biol. Sci. 2017, 7, 364–372. [Google Scholar]
- Man, R.-J.; Jeelani, N.; Zhou, C.; Yang, Y.-S. Recent progress in the development of quinoline derivatives for the exploitation of anti-cancer agents. Anticancer Med. Chem. 2021, 21, 825–838. [Google Scholar] [CrossRef]
- Hussaini, S.M.A. Therapeutic significance of quinolines: A patent review (2013–2015). Expert Opin. Ther. Pat. 2016, 26, 1201–1221. [Google Scholar] [CrossRef]
- Wdowiak, P.; Matysiak, J.; Kuszta, P.; Czarnek, K.; Niezabitowska, E.; Baj, T. Quinazoline derivatives as potential therapeutic agents in urinary bladder cancer therapy. Front. Chem. 2021, 9, 765552. [Google Scholar] [CrossRef]
- Desplat, V.; Vincenzi, M.; Lucas, R.; Moreau, S.; Savrimoutou, S.; Pinaud, N.; Lesbordes, J.; Peyrilles, E.; Marchivie, M.; Routier, S.; et al. Synthesis and evaluation of the cytotoxic activity of novel ethyl 4-[4-(4-substitutedpiperidin-1-yl)]benzyl-phenylpyrrolo[1,2-a]quinoxaline-carboxylate derivatives in myeloid and lymphoid leukemia cell lines. Eur. J. Med. Chem. 2016, 113, 214–227. [Google Scholar] [CrossRef]
- Desplat, V.; Vincenzi, M.; Lucas, R.; Moreau, S.; Savrimoutou, S.; Rubio, S.; Pinaud, N.; Bigat, D.; Enriquez, E.; Marchivie, M.; et al. Synthesis and Antiproliferative Effect of Ethyl 4-[4-(4-Substituted Piperidin-1-yl)]benzylpyrrolo[1,2-a]quinoxalinecarboxylate Derivatives on Human Leukemia Cells. ChemMedChem 2017, 12, 940–953. [Google Scholar] [CrossRef]
- Guillon, J.; Vincenzi, M.; Pinaud, N.; Ronga, L.; Rossi, F.; Savrimoutou, S.; Moreau, S.; Desplat, V.; Marchivie, M. Synthesis and Crystal Structure of 3-{4-[(4-(2-Oxo-2,3-dihydro-1H-benzimidazol- 1-yl)piperidin-1-yl)benzyl]}-2-phenylindole. Struct. Chem. Crystallogr. Commun. 2016, 2, 8. [Google Scholar]
- Guillon, J.; Savrimoutou, S.; Rubio, S.; Desplat, V. 1-Methyl-3-{4-[(4-(2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)piperidin-1-yl)benzyl]}-2-phenylindole. Molbank 2018, 2018, M1023. [Google Scholar] [CrossRef]
- Das, R.N.; Chevret, E.; Desplat, V.; Rubio, S.; Mergny, J.-L.; Guillon, J. Design, Synthesis and Biological Evaluation of New Substituted Diquinolinyl-Pyridine Ligands as Anticancer Agents by Targeting G-Quadruplex. Molecules 2018, 23, 81. [Google Scholar] [CrossRef]
- Guillon, J.; Savrimoutou, S.; Rubio, S.; Moreau, S.; Pinaud, N.; Marchivie, M.; Desplat, V. 1-Phenyl-8-[[4-(pyrrolo[1,2-a]quinoxalin-4-yl)phenyl]methyl]-1,3,8-triazaspiro[4.5]decan-4-one: Synthesis, Crystal Structure and Anti-Leukemic Activity. Molbank 2020, 2020, M1113. [Google Scholar] [CrossRef]
- Gueddouda, N.M.; Hurtado, M.R.; Moreau, S.; Ronga, L.; Das, R.N.; Savrimoutou, S.; Rubio, S.; Marchand, A.; Mendoza, O.; Marchivie, M.; et al. Design, Synthesis, and Evaluation of 2,9-Bis[(substituted-aminomethyl)phenyl]-1,10-phenanthroline Derivatives as G-Quadruplex Ligands. ChemMedChem 2017, 12, 146–160. [Google Scholar] [CrossRef]
- Guillon, J.; Denevault-Sabourin, C.; Chevret, E.; Brachet-Botineau, M.; Milano, V.; Guedin-Beaurepaire, A.; Moreau, S.; Ronga, L.; Savrimoutou, S.; Rubio, S.; et al. Design, synthesis, and antiproliferative effect of 2,9-bis[4-(pyridinylalkylaminomethyl)phenyl]-1,10-phenanthroline derivatives on human leukemic cells by targeting G-quadruplex. Arch. Pharm. (Weinheim) 2021, 354, e2000450. [Google Scholar] [CrossRef]
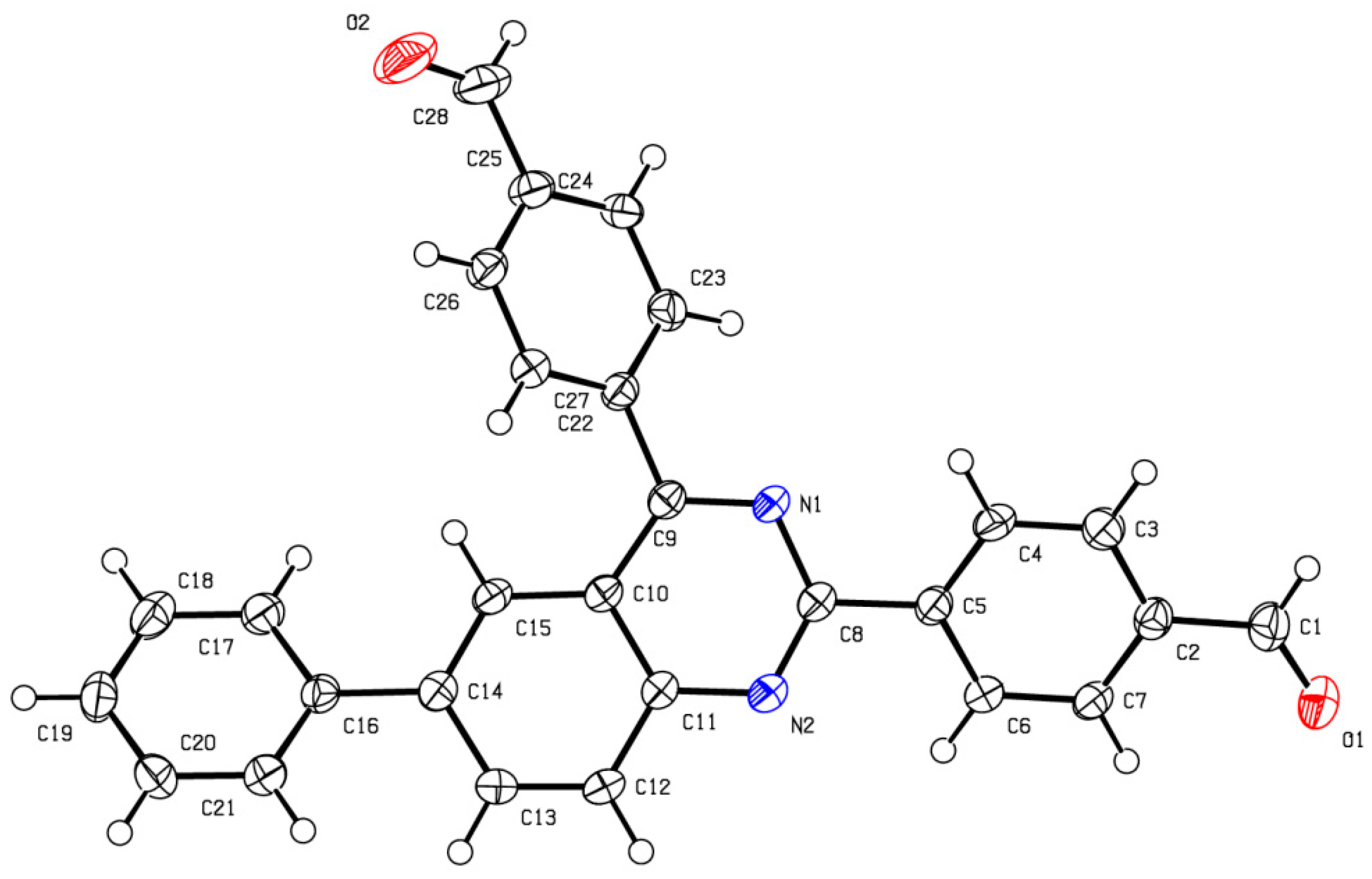
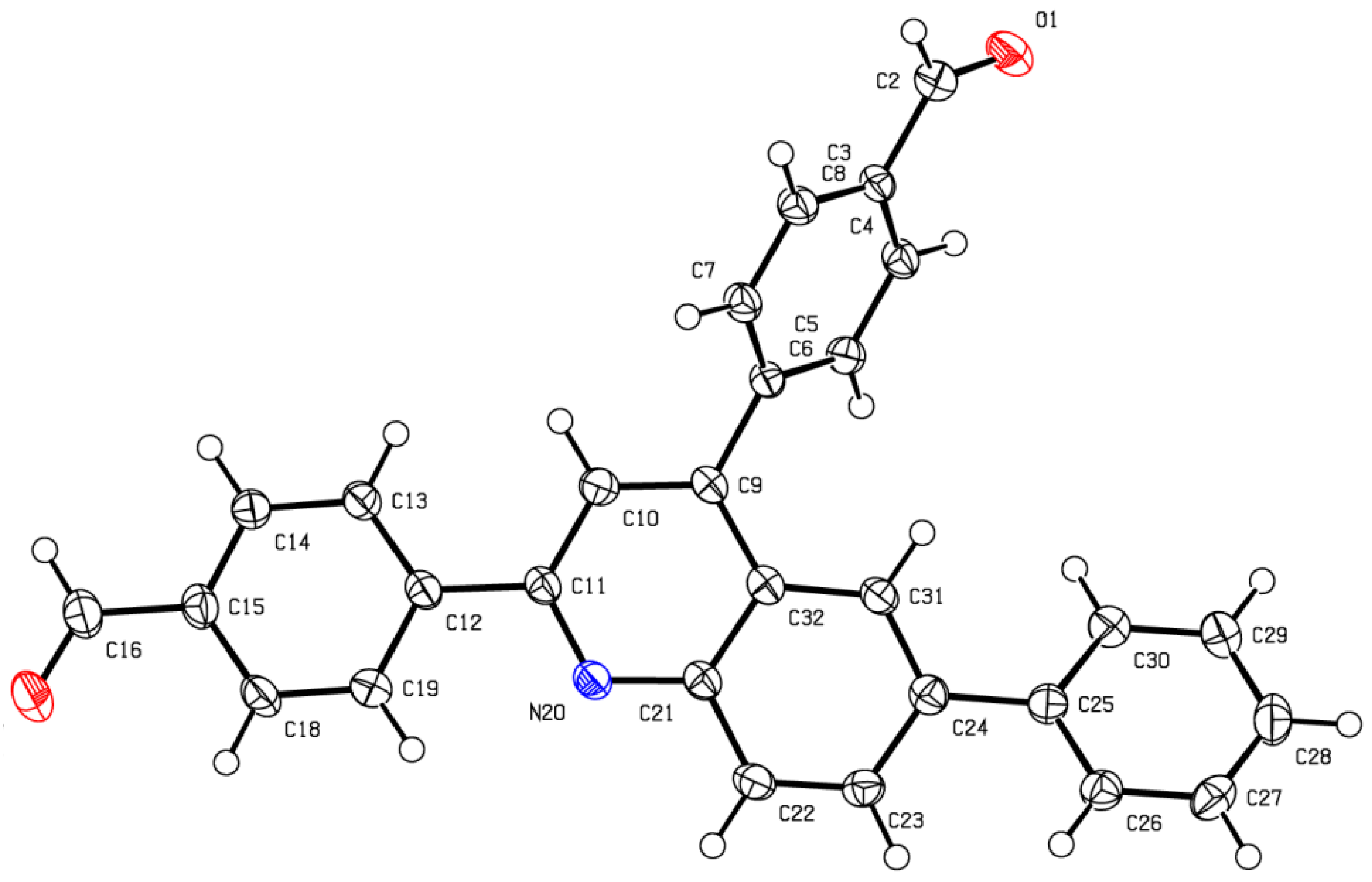
- CCDC-2238804 and CCDC-2238806 Contain the Supplementary Crystallographic Data for This Paper. These Data Can Be Obtained Free of Charge from the Cambridge Crystallographic Data Centre, University Chemical Lab, Lensfield Road, Cambridge, CB2 1EW, UK. Available online: https://www.ccdc.cam.ac.uk/ (accessed on 30 January 2023).
- Liu, X.; Fu, H.; Jiang, Y.; Zhao, Y. A Simple and Highly Efficient Approach to Quinazolinones under Mild Copper-Catalyzed Conditions. Angew. Chem. Int. Ed. 2009, 48, 348–351. [Google Scholar] [CrossRef]
- Kappe, T.; Karem, A.S.; Stadlbauer, W. Synthesis of benzo-halogenated 4-hydroxy-2(1H)-quinolones. J. Heterocycl. Chem. 1988, 25, 857–862. [Google Scholar] [CrossRef]
- Largy, E.; König, A.; Ghosh, A.; Ghosh, D.; Benabou, S.; Rosu, F.; Gabelica, V. Mass Spectrometry of Nucleic Acid Noncovalent Complexes. Chem. Rev. 2022, 122, 7720–7839. [Google Scholar] [CrossRef]
- Rosu, F.; De Pauw, E.; Gabelica, V. Electrospray mass spectrometry to study drug-nucleic acids interactions. Biochimie 2008, 90, 1074–1087. [Google Scholar] [CrossRef] [PubMed]
- Largy, E.; Marchand, A.; Amrane, S.; Gabelica, V.; Mergny, J.-L. Quadruplex Turncoats: Cation-Dependent Folding and Stability of Quadruplex-DNA Double Switches. J. Am. Chem. Soc. 2016, 138, 2780–2792. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Trajkovski, M.; Teulade-Fichou, M.-P.; Gabelica, V.; Plavec, J. Phen-DC3 Induces Refolding of Human Telomeric DNA into a Chair-Type Antiparallel G-Quadruplex through Ligand Intercalation. Angew. Chem. 2022, 134, e202207. [Google Scholar] [CrossRef]
- Balthasart, F.; Plavec, J.; Gabelica, V. Ammonium Ion Binding to DNA G-Quadruplexes: Do Electrospray Mass Spectra Faithfully Reflect the Solution-Phase Species? J. Am. Soc. Mass Spectrom. 2013, 24, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Zhang, G.; Tang, Z.; Fan, S.; Li, C.; Li, Y.; Liu, W.; Long, X.; Zhang, W.; Zhang, Y.; Li, Z.; et al. Synthesis and biological assessment of indole derivatives containing penta-heterocycles scaffold as novel anticancer agents towards A549 and K562 cells. J. Enzyme Inhib. Med. Chem. 2023, 38, 2163393. [Google Scholar] [CrossRef]
- Cawthon, R.M. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009, 37, e21. [Google Scholar] [CrossRef]
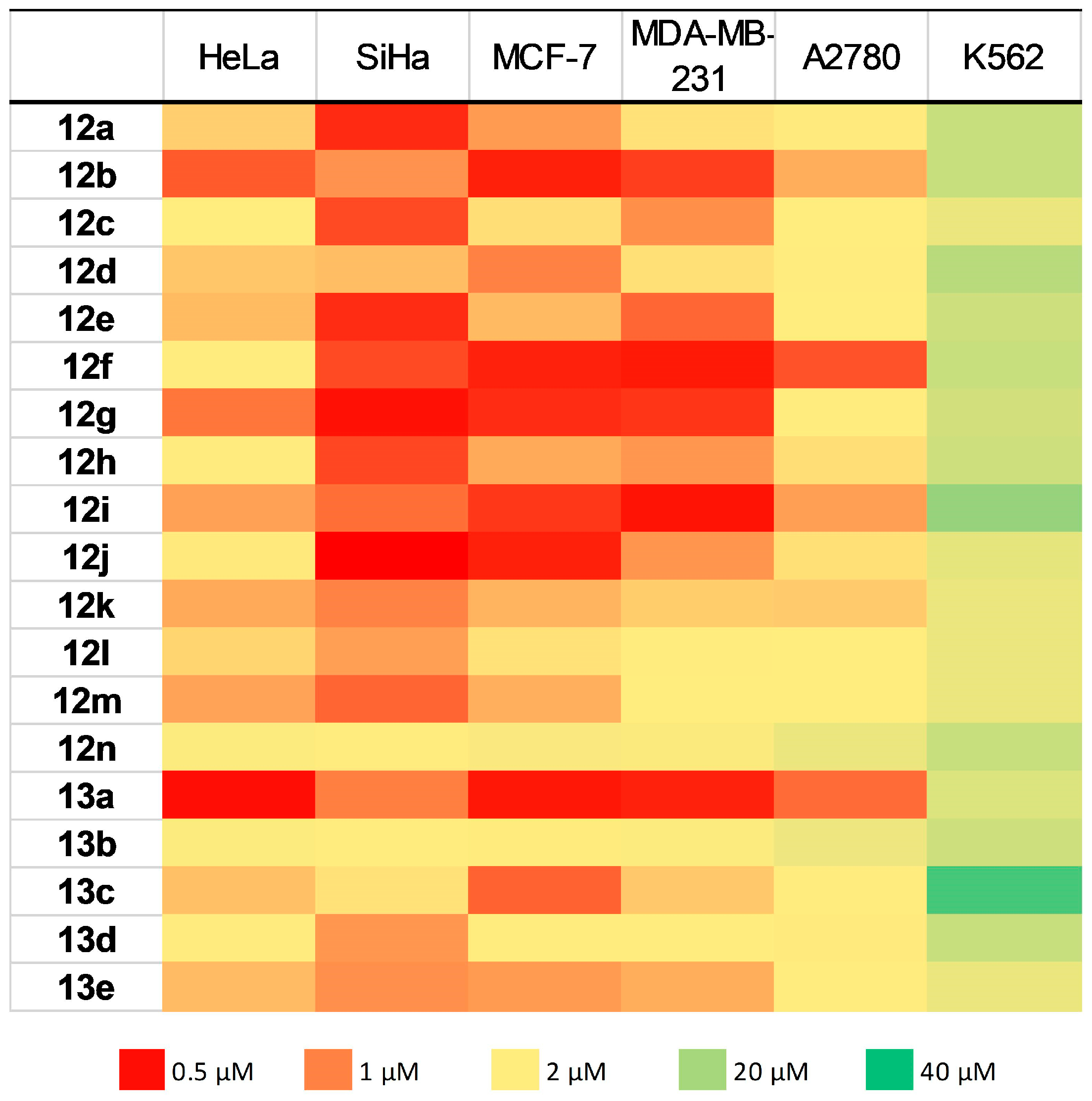
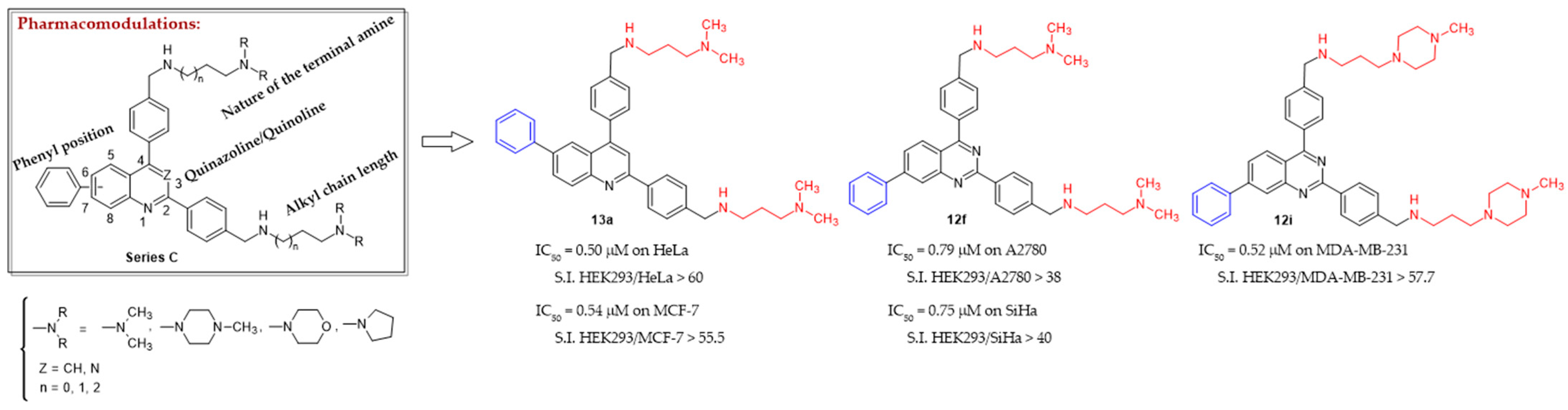
| Compound | IC50 Values (μM) ± SEM a | ||||||
|---|---|---|---|---|---|---|---|
| HeLa | SiHa | MCF-7 | MDA-MB-231 | A2780 | K562 | HEK293 | |
| 12a | 1.36 ± 0.04 | 0.62 ± 0.12 | 1.12 ± 0.13 | 1.45 ± 0.14 | 2.60 ± 0.03 | 14.0 ± 0.2 | n.d. b |
| 12b | 0.82 ± 0.31 | 1.08 ± 0.10 | 0.58 ± 0.02 | 0.70 ± 0.16 | 1.20 ± 0.27 | 14.0 ± 0.3 | >30 |
| 12c | 2.21 ± 0.86 | 0.76 ± 0.01 | 1.44 ± 0.03 | 1.07 ± 0.42 | 2.04 ± 0.54 | 7.0 ± 0.2 | n.d. b |
| 12d | 1.32 ± 0.02 | 1.28 ± 0.27 | 1.01 ± 0.30 | 1.44 ± 0.17 | 1.74 ± 0.11 | 17.0 ± 0.4 | n.d. b |
| 12e | 1.27 ± 0.02 | 0.63 ± 0.05 | 1.27 ± 0.01 | 0.89 ± 0.46 | 2.19 ± 0.41 | 13.0 ± 0.2 | n.d. b |
| 12f | 1.51 ± 0.09 | 0.75 ± 0.10 | 0.58 ± 0.08 | 0.55 ± 0.01 | 0.79 ± 0.19 | 14.0 ± 0.3 | >30 |
| 12g | 0.95 ± 0.51 | 0.52 ± 0.22 | 0.63 ± 0.35 | 0.67 ± 0.35 | 1.74 ± 0.89 | 12.0 ± 0.3 | 3.98 ± 0.12 |
| 12h | 2.31 ± 0.76 | 0.75 ± 0.01 | 1.19 ± 0.27 | 1.10 ± 0.38 | 1.43 ± 0.07 | 13.0 ± 0.2 | n.d. b |
| 12i | 1.15 ± 0.36 | 0.92 ± 0.24 | 0.67 ± 0.31 | 0.52 ± 0.02 | 1.14 ± 0.16 | 23.0 ± 0.5 | >30 |
| 12j | 1.48 ± 0.80 | 0.33 ± 0.08 | 0.58 ± 0.25 | 1.10 ± 0.15 | 1.44 ± 0.10 | 8.0 ± 0.2 | 4.90 ± 0.15 |
| 12k | 1.19 ± 0.24 | 1.01 ± 0.27 | 1.24 ± 0.14 | 1.36 ± 0.06 | 1.34 ± 0.14 | 7.0 ± 0.1 | n.d. b |
| 12l | 1.39 ± 0.16 | 1.14 ± 0.51 | 1.45 ± 0.02 | 1.55 ± 0.12 | 2.08 ± 0.48 | 7.0 ± 0.2 | n.d. b |
| 12m | 1.17 ± 0.23 | 0.88 ± 0.14 | 1.22 ± 0.19 | 2.00 ± 0.45 | 2.10 ± 0.48 | 7.0 ± 0.2 | n.d. b |
| 12n | 3.51 ± 0.17 | 1.89 ± 0.74 | 4.17 ± 0.50 | 3.94 ± 0.54 | 7.10 ± 1.15 | 14.0 ± 0.3 | n.d. b |
| 13a | 0.50 ± 0.01 | 0.99 ± 0.15 | 0.54 ± 0.01 | 0.58 ± 0.02 | 0.90 ± 0.04 | 10.0 ± 0.4 | >30 |
| 13b | 3.58 ± 0.18 | 1.76 ± 0.61 | 3.03 ± 0.64 | 3.68 ± 0.80 | 6.49 ± 1.76 | 13.0 ± 0.4 | n.d. b |
| 13c | 1.30 ± 0.06 | 1.45 ± 0.13 | 0.86 ± 0.38 | 1.33 ± 0.13 | 1.58 ± 0.10 | 34.0 ± 0.7 | n.d. b |
| 13d | 2.27 ± 0.89 | 1.11 ± 0.37 | 1.62 ± 0.09 | 1.64 ± 0.11 | 2.10 ± 0.09 | 14.0 ± 0.3 | n.d. b |
| 13e | 1.28 ± 0.21 | 1.07 ± 0.45 | 1.12 ± 0.10 | 1.21 ± 0.03 | 1.50 ± 0.34 | 7.0 ± 0.1 | n.d. b |
| Compound | Selectivity Index a | |||||
|---|---|---|---|---|---|---|
| HEK293/HeLa | HEK293/SiHa | HEK293/MCF-7 | HEK293/MDA-MB-231 | HEK293/A2780 | HEK293/K562 | |
| 12b | >36.6 | >27.8 | >51.7 | >42.8 | >25.0 | >2.14 |
| 12f | >19.9 | >40.0 | >51.7 | >54.5 | >38.0 | >2.14 |
| 12g | 4.19 | 7.65 | 6.32 | 5.94 | 2.29 | 0.33 |
| 12i | >26.1 | >32.6 | >44.8 | >57.7 | >26.3 | >1.30 |
| 12j | 3.31 | 14.85 | 8.45 | 4.45 | 3.40 | 0.61 |
| 13a | >60.0 | >30.3 | >55.5 | >51.7 | >33.3 | >3.0 |
| Compound | ΔT1/2 (°C) a | ||||
|---|---|---|---|---|---|
| Fc-MYCT | FK-RAST | FBCL-2T | F21T | FdxT | |
| PhenDC3 | 11.5 ± 0.3 | 16.9 ± 0.4 | 13.1 ± 0.1 | 21.2 ± 0.5 | −0.6 ± 0.1 |
| 12a | 9.3 ± 0.3 | 10.6 ± 0.6 | 7.4 ± 0.9 | 6.8 ± 0.8 | 0.9 ± 0.2 |
| 12b | 5.8 ± 0.9 | 17.4 ± 0.6 | 8.9 ± 0.7 | 23.7 ± 1.7 | 0.4 ± 0.3 |
| 12c | 17.7 ± 0.8 | 24.5 ± 0.7 | 16.4 ± 0.4 | 28.0 ± 0.4 | 3.8 ± 0.8 |
| 12d | 8.4 ± 0.9 | 15.5 ± 0.5 | 9.7 ± 0.06 | 15.8 ± 1.2 | 0.5 ± 0.3 |
| 12e | 19.5 ± 1.0 | 33.1 ± 2.1 | 22.7 ± 0.4 | 33.9 ± 0.5 | 5.4 ± 1.6 |
| 12f | 9.1 ± 1.4 | 20.1 ± 0.3 | 12.6 ± 0.6 | 16.3 ± 1.5 | 2.4 ± 0.1 |
| 12g | 14.7 ± 0.8 | 24.3 ± 1.1 | 17.5 ± 0.6 | 19.2 ± 1.0 | 3.2 ± 0.3 |
| 12h | 18.7 ± 0.2 | 28.6 ± 2.1 | 18.2 ± 1.7 | 28.1 ± 2.3 | 2.7 ± 1.0 |
| 12i | 12.5 ± 0.7 | 20.2 ± 0.5 | 14.8 ± 0.7 | 18.6 ± 0.2 | 1.9 ± 0.1 |
| 12j | 14.1 ± 0.3 | 18.7 ± 1.6 | 9.8 ± 1.0 | 14.5 ± 1.2 | 1.5 ± 0.4 |
| 12k | 9.9 ± 1.4 | 13.0 ± 0.4 | 9.5 ± 0.2 | 17.7 ± 0.3 | 0.8 ± 0.1 |
| 12l | 14.2 ± 1.1 | 18.7 ± 0.8 | 17.2 ± 1.2 | 24.9 ± 0.3 | 2.9 ± 0.2 |
| 12m | 14.6 ± 0.9 | 18.2 ± 0.6 | 15.5 ± 0.3 | 23.3 ± 0.7 | 2.2 ± 0.2 |
| 12n | 17.0 ± 2.0 | 18.0 ± 2.5 | 12.0 ± 2.0 | 20.7 ± 3.1 | 2.4 ± 0.7 |
| 13a | 7.3 ± 1.0 | 15.0 ± 0.3 | 7.7 ± 0.6 | 21.5 ± 0.4 | 0.4 ± 0.1 |
| 13b | 13.8 ± 3.0 | 19.6 ± 3.5 | 11.1 ± 2.6 | 13.6 ± 2.9 | 1.5 ± 0.6 |
| 13c | 6.8 ± 1.4 | 14.5 ± 0.8 | 7.5 ± 0.5 | 11.3 ± 0.9 | 0.1 ± 0.2 |
| 13d | 10.6 ± 0.7 | 12.7 ± 2.2 | 5.0 ± 1.1 | 8.0 ± 1.5 | 0.8 ± 0.2 |
| 13e | 5.5 ± 0.9 | 11.6 ± 1.4 | 8.3 ± 0.1 | 15.9 ± 0.4 | 0.2 ± 0.1 |
| Name | Sequences a | Topology |
|---|---|---|
| FK-RAST | FAM-A GGG C GG TGT GGG AAGA GGG A-TAMRA | Parallel |
| FBCL-2T | FAM-GGG CGC GGG A GG AAG GGG GC GGG-TAMRA | Parallel |
| F21T | FAM-GGG TTA GGG TTA GGG TTA GGG-TAMRA | Hybrid |
| Fc-MYCT | FAM-GGG T GGG TA GGG T GGG TAA-TAMRA | Parallel |
| FdxT | FAM-TAT AGC TAT A-hexaethylene glycol-T ATA GCT ATA-TAMRA | Hairpin duplex |
| 12b | 12e | 13a | 13d | |
|---|---|---|---|---|
| BCL-2 | ||||
| Bound DNA (%) | 79 | 79 | 92 | 33 |
| Kd1 | 5.6 | 6.2 | 1.6 | 53 |
| Kd2 | 2.8 | 2.2 | 0.75 | 20 |
| Kd1/Kd2 | 2.0 | 2.8 | 2.1 | 2.6 |
| 24TTG | ||||
| Bound DNA (%) | 77 | 72 | 81 | 39 |
| Kd1 | 9.5 | 14 | 3.7 | 29 |
| Kd2 | 1.5 | 1.9 | 7.1 | 83 |
| Kd1/Kd2 | 6.3 | 7.2 | 0.5 | 0.4 |
| c-MYC | ||||
| Bound DNA (%) | 70 | 75 | 78 | 44 |
| Kd1 | 5.8 | 4.3 | 3.8 | 20 |
| Kd2 | 100 | 74 | 36 | 440 |
| Kd1/Kd2 | 0.1 | 0.1 | 0.1 | 0.1 |
| ds26 | ||||
| Bound DNA (%) | 48 | 36 | 53 | 18 |
| Kd1 | 17 | 30 | 14 | 85 |
| Kd2 | 130 | 280 | 95 | 460 |
| Kd1/Kd2 | 0.1 | 0.1 | 0.2 | 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guillon, J.; Le Borgne, M.; Milano, V.; Guédin-Beaurepaire, A.; Moreau, S.; Pinaud, N.; Ronga, L.; Savrimoutou, S.; Albenque-Rubio, S.; Marchivie, M.; et al. New 2,4-bis[(substituted-aminomethyl)phenyl]phenylquinazoline and 2,4-bis[(substituted-aminomethyl)phenyl]phenylquinoline Derivatives: Synthesis and Biological Evaluation as Novel Anticancer Agents by Targeting G-Quadruplex. Pharmaceuticals 2024, 17, 30. https://doi.org/10.3390/ph17010030
Guillon J, Le Borgne M, Milano V, Guédin-Beaurepaire A, Moreau S, Pinaud N, Ronga L, Savrimoutou S, Albenque-Rubio S, Marchivie M, et al. New 2,4-bis[(substituted-aminomethyl)phenyl]phenylquinazoline and 2,4-bis[(substituted-aminomethyl)phenyl]phenylquinoline Derivatives: Synthesis and Biological Evaluation as Novel Anticancer Agents by Targeting G-Quadruplex. Pharmaceuticals. 2024; 17(1):30. https://doi.org/10.3390/ph17010030
Chicago/Turabian StyleGuillon, Jean, Marc Le Borgne, Vittoria Milano, Aurore Guédin-Beaurepaire, Stéphane Moreau, Noël Pinaud, Luisa Ronga, Solène Savrimoutou, Sandra Albenque-Rubio, Mathieu Marchivie, and et al. 2024. "New 2,4-bis[(substituted-aminomethyl)phenyl]phenylquinazoline and 2,4-bis[(substituted-aminomethyl)phenyl]phenylquinoline Derivatives: Synthesis and Biological Evaluation as Novel Anticancer Agents by Targeting G-Quadruplex" Pharmaceuticals 17, no. 1: 30. https://doi.org/10.3390/ph17010030
APA StyleGuillon, J., Le Borgne, M., Milano, V., Guédin-Beaurepaire, A., Moreau, S., Pinaud, N., Ronga, L., Savrimoutou, S., Albenque-Rubio, S., Marchivie, M., Kalout, H., Walker, C., Chevallier, L., Buré, C., Largy, E., Gabelica, V., Mergny, J.-L., Baylot, V., Ferrer, J., ... Zupkó, I. (2024). New 2,4-bis[(substituted-aminomethyl)phenyl]phenylquinazoline and 2,4-bis[(substituted-aminomethyl)phenyl]phenylquinoline Derivatives: Synthesis and Biological Evaluation as Novel Anticancer Agents by Targeting G-Quadruplex. Pharmaceuticals, 17(1), 30. https://doi.org/10.3390/ph17010030