Anthocyanin Oligomers Induce Apoptosis and Autophagy by Inhibiting the mTOR Signaling Pathway in Human Breast Cancer Cells
Abstract
:1. Introduction
2. Results
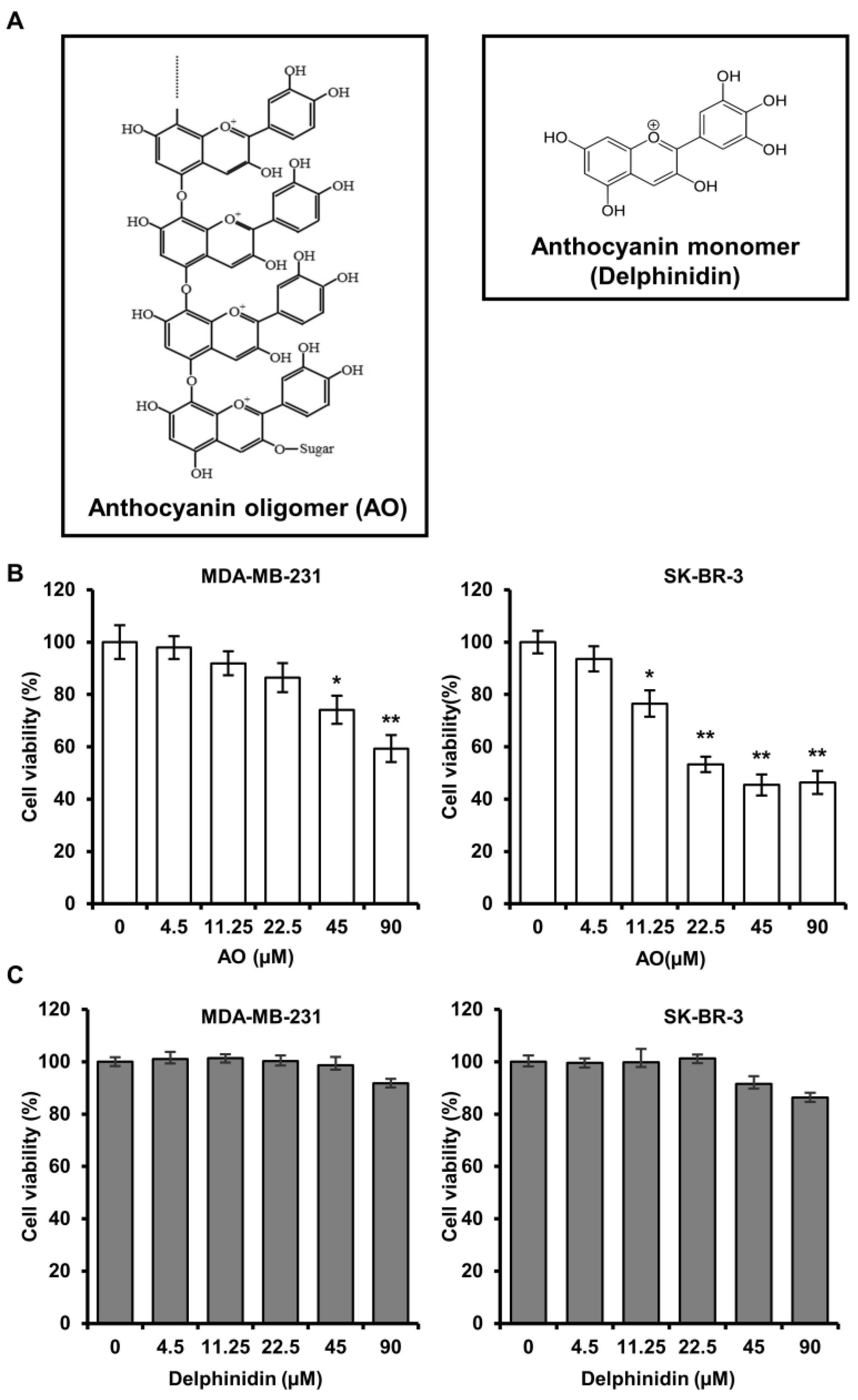
2.1. AO Significantly Induced the Cytotoxicity of MDA-MB-231 and SK-BR-3 Cells
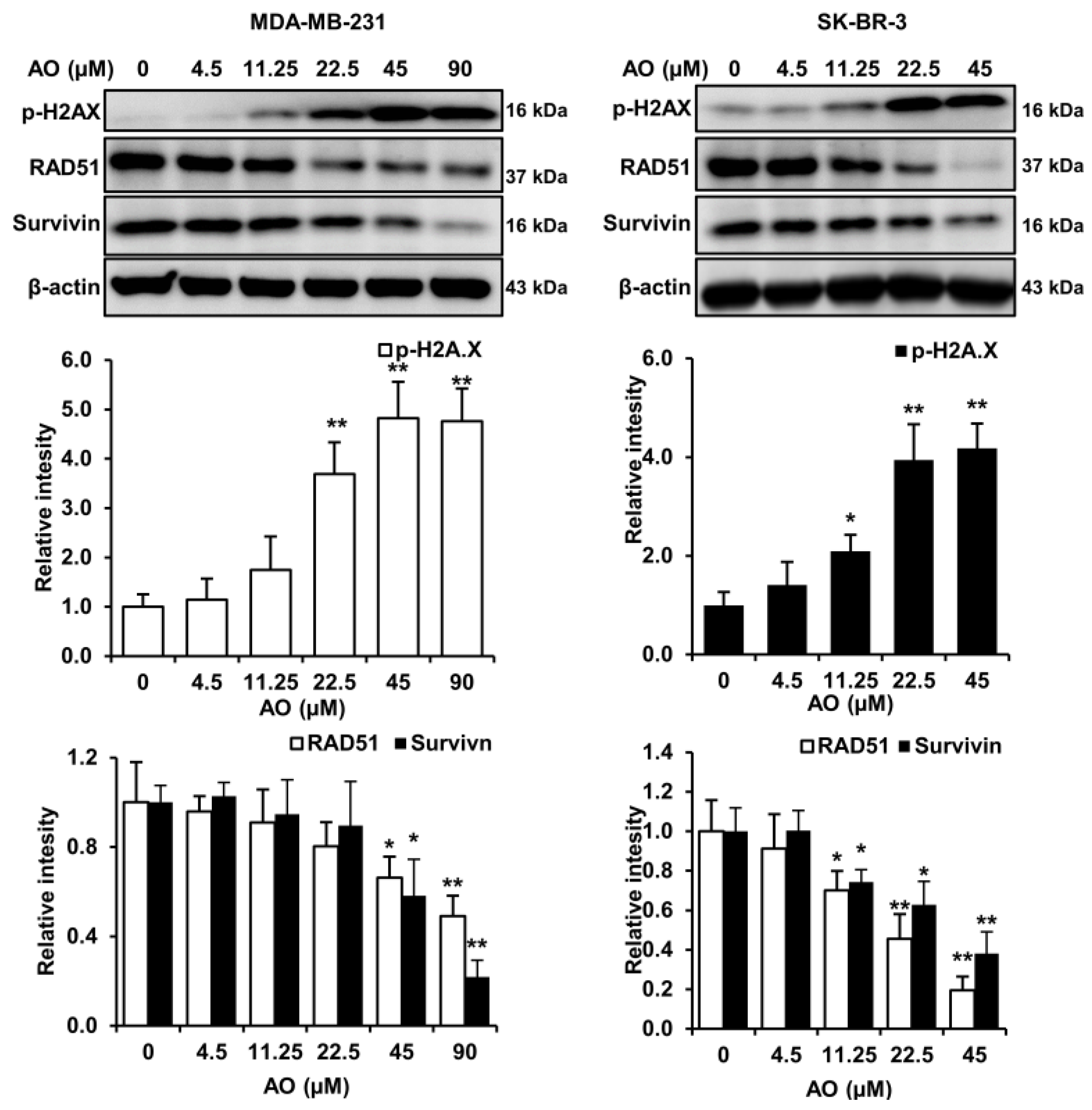
2.2. AO Caused DNA Damage and Attenuated DNA Repair in MDA-MB-231 and SK-BR-3 Cells
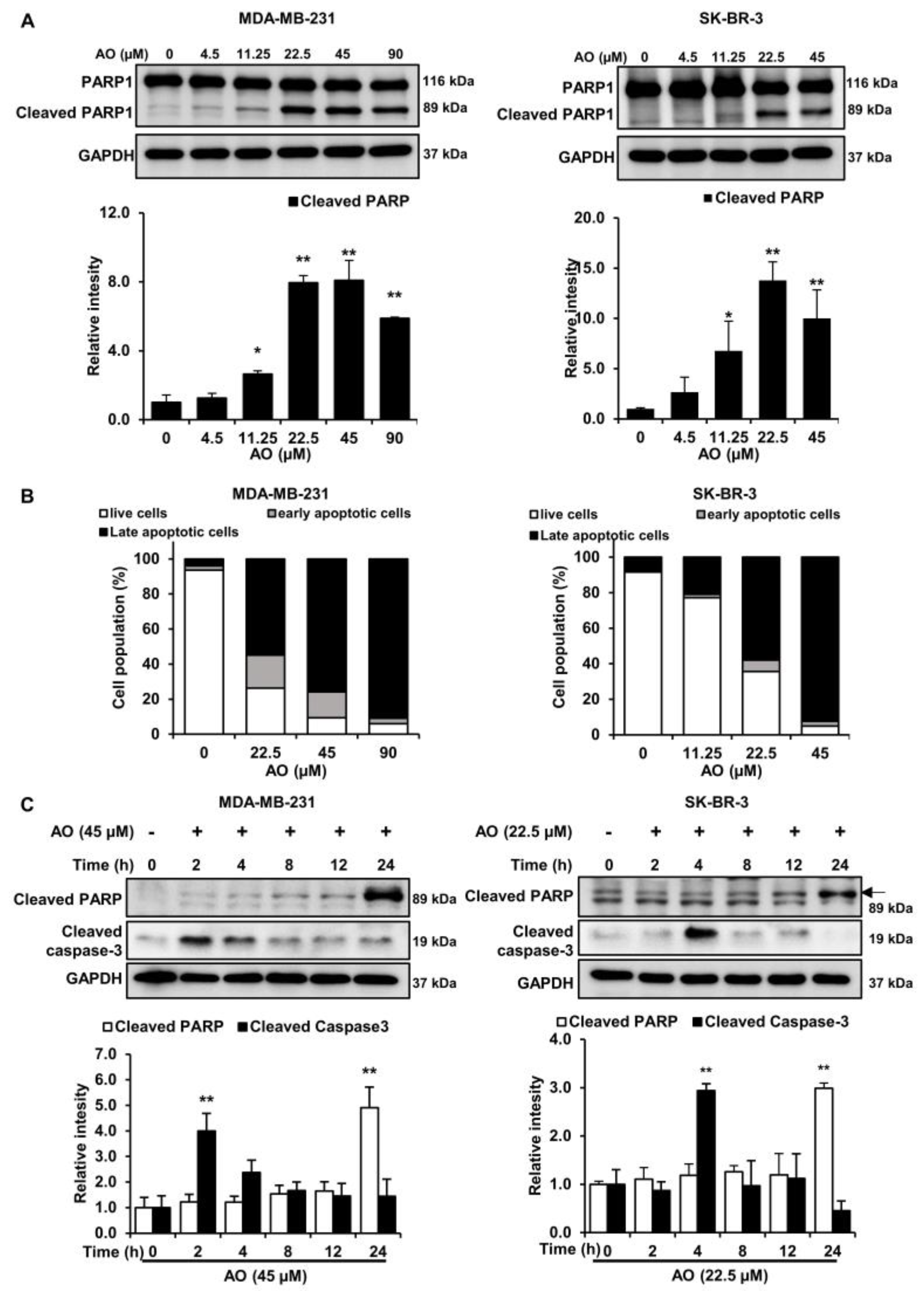
2.3. AO Induced Apoptosis via Caspase-3 Dependent PARP1 Cleavage in MDA-MB-231 and SK-BR-3 Cells
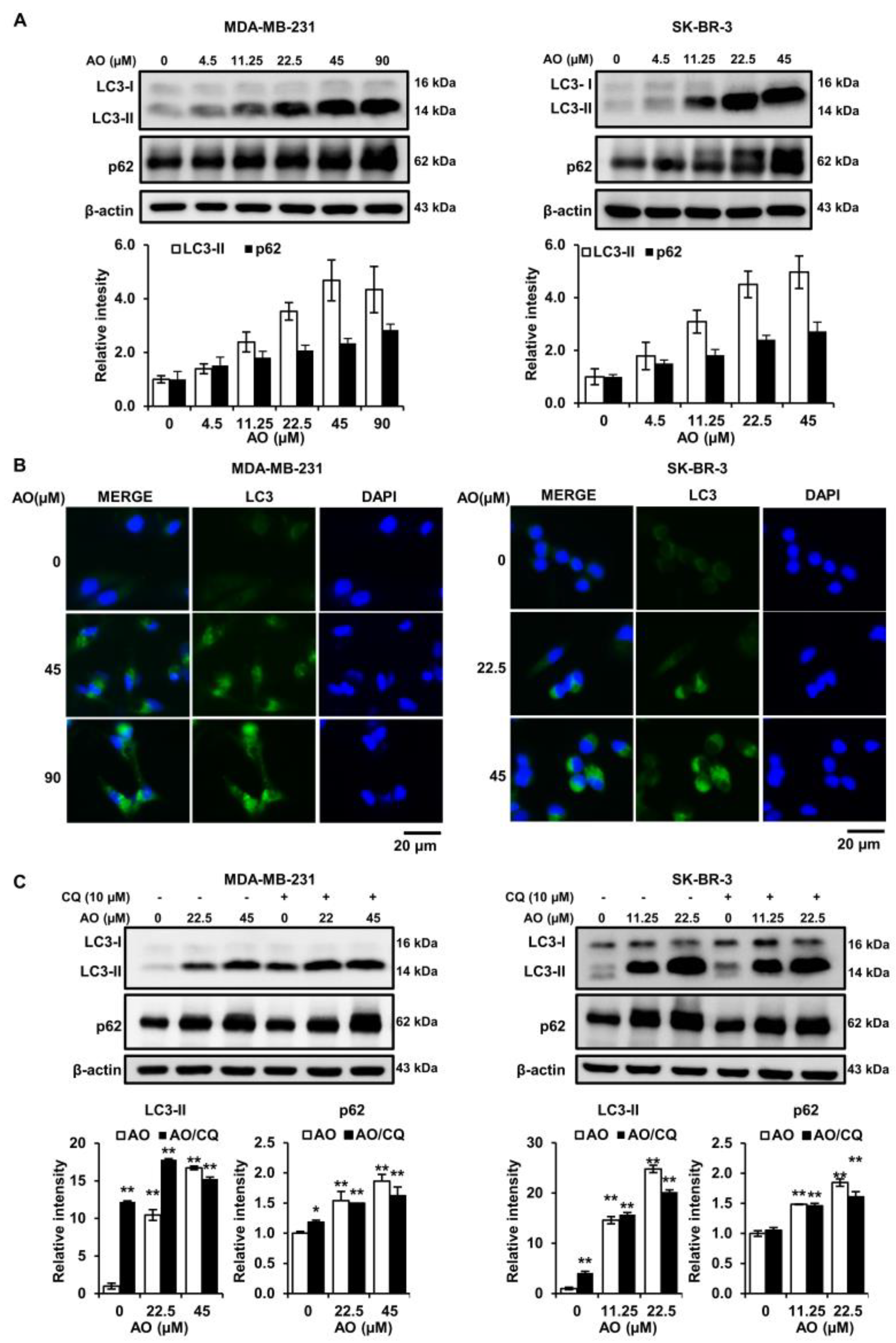
2.4. AO Significantly Increased LC3-II and p62 Levels in the Two Cell Lines
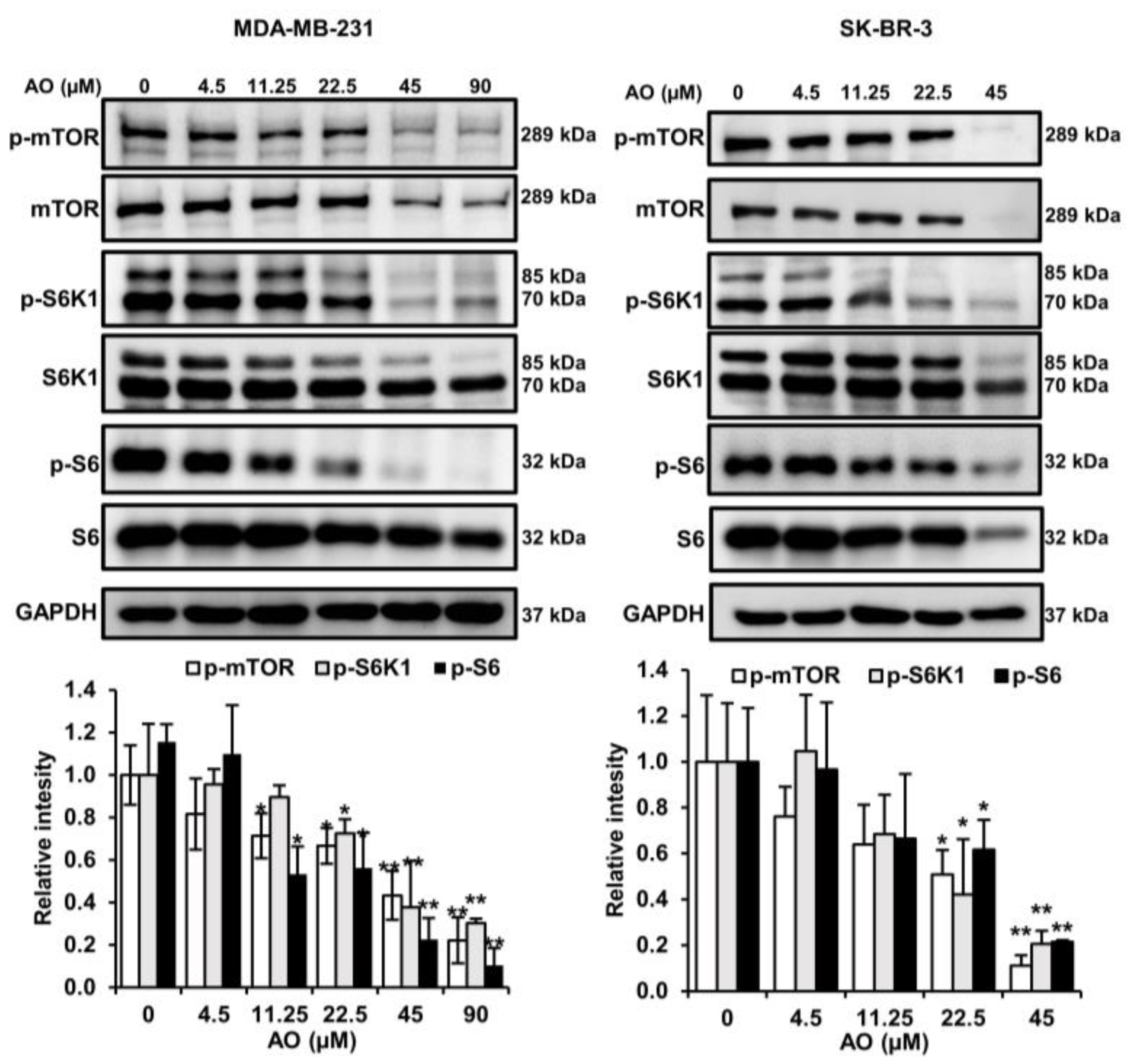
2.5. AO Inhibited mTOR Pathways in MDA-MB-231 and SK-BR-3 Cells
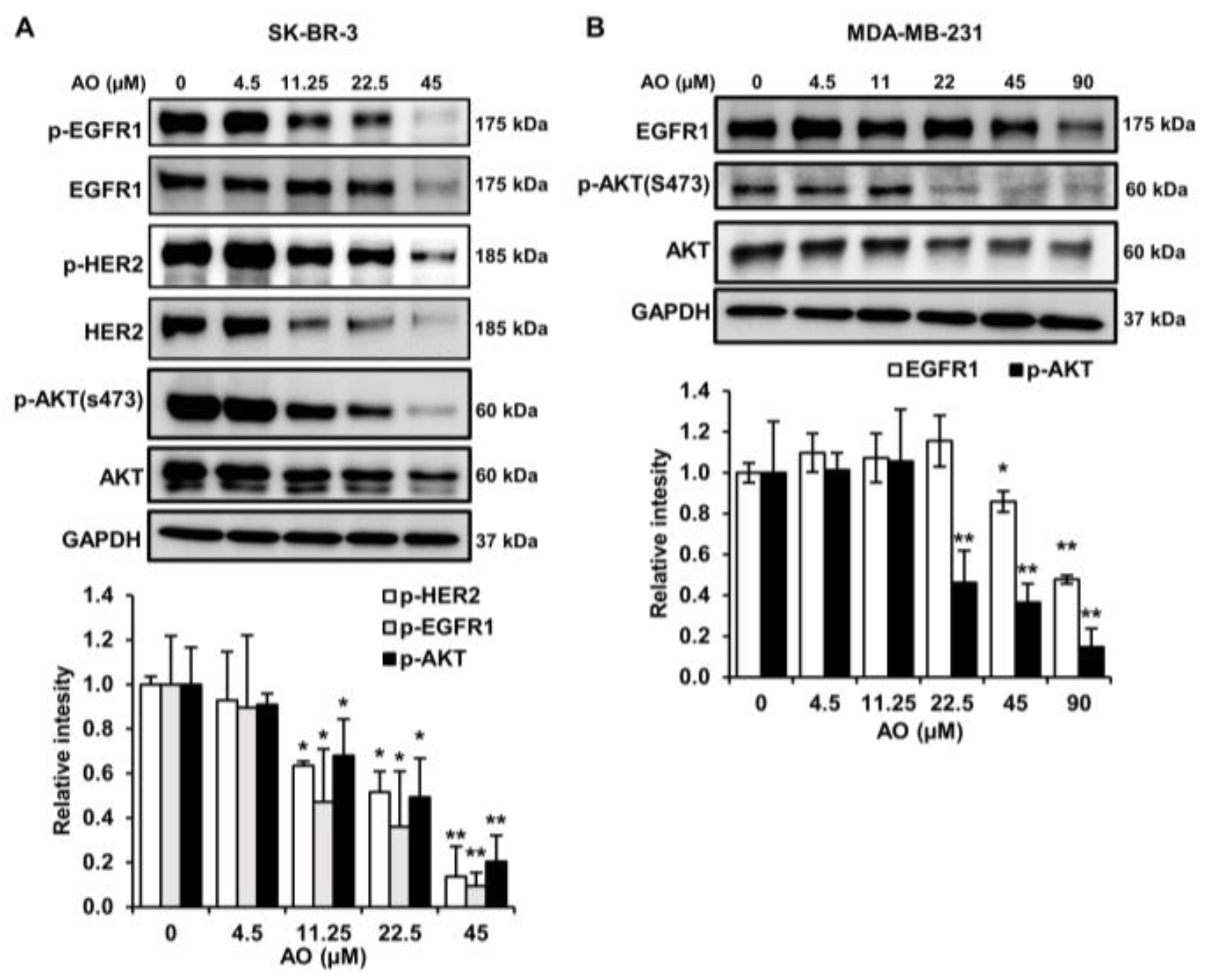
2.6. AO Inhibited the mTOR Pathway by Suppressing the HER2/EGFR1, and AKT Pathways in SK-BR-3 Cells
2.7. AO Inhibited the mTOR Pathway by Suppressing the EGFR1and AKT Pathways in MDA-MB-231 Cells
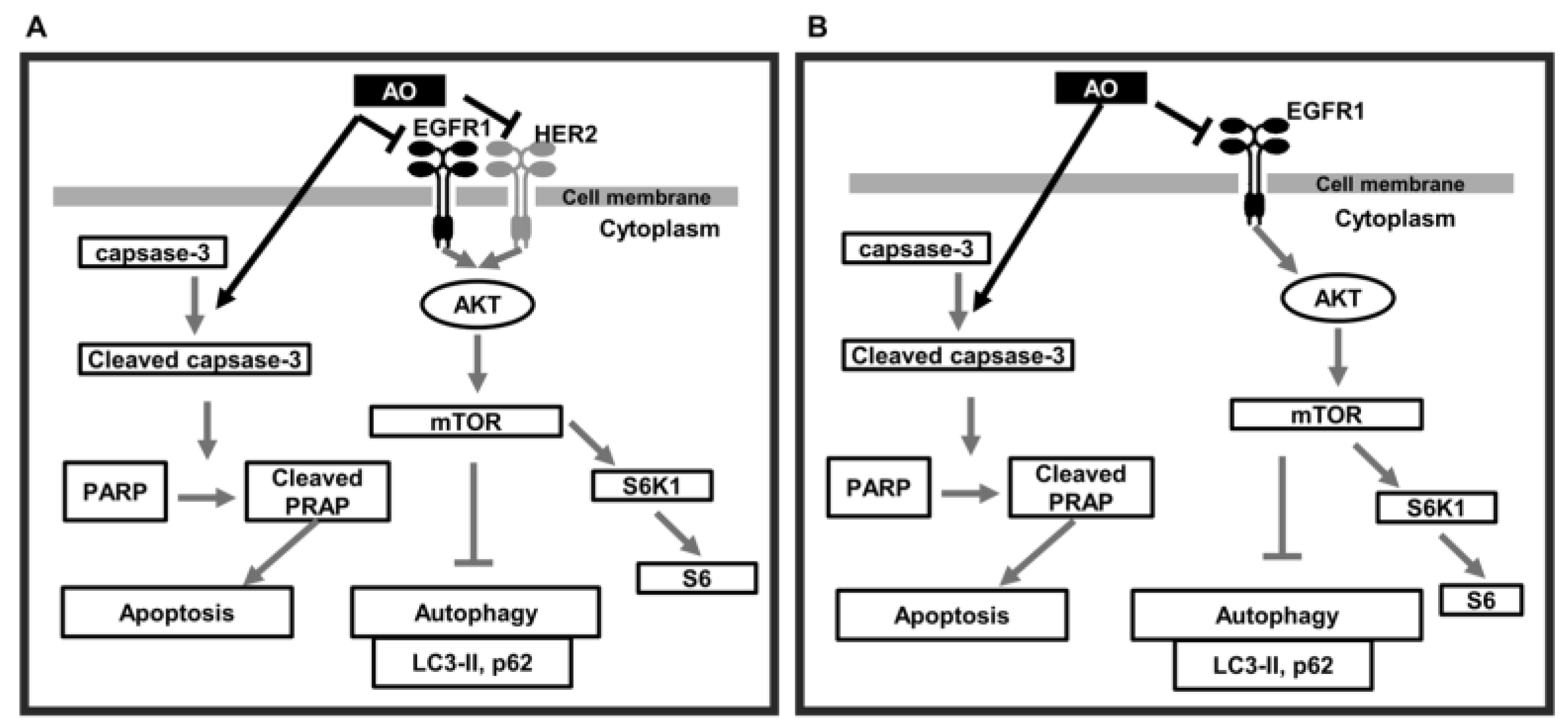
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Sulforhodamine B (SRB) Cell Viability Assay
4.4. Western Blotting
4.5. Fluorescent Immunocytochemistry
4.6. Apoptosis Detection through 7-AAD/FITC-Conjugated Annexin V Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, H.; Rhee, W.J. Exosome-mediated Let7c-5p Delivery for Breast Cancer Therapeutic Development. Biotechnol. Bioproc. E 2020, 25, 513–520. [Google Scholar] [CrossRef]
- Masoud, V.; Pages, G. Targeted therapies in breast cancer: New challenges to fight against resistance. World J. Clin. Oncol. 2017, 8, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Newton, E.E.; Mueller, L.E.; Treadwell, S.M.; Morris, C.A.; Machado, H.L. Molecular Targets of Triple-Negative Breast Cancer: Where Do We Stand? Cancers 2022, 14, 482. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, H.; Merkher, Y.; Chen, L.; Liu, N.; Leonov, S.; Chen, Y. Recent advances in therapeutic strategies for triple-negative breast cancer. J. Hematol. Oncol. 2022, 15, 121. [Google Scholar] [CrossRef] [PubMed]
- Israel, B.B.; Tilghman, S.L.; Parker-Lemieux, K.; Payton-Stewart, F. Phytochemicals: Current strategies for treating breast cancer. Oncol. Lett. 2018, 15, 7471–7478. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Sharifi-Rad, J.; Cappellini, F.; Reiner, A.; Zorzan, D.; Imran, M.; Sener, B.; Kilic, M.; El-Shazly, M.; Fahmy, N.M.; et al. The Therapeutic Potential of Anthocyanins: Current Approaches Based on Their Molecular Mechanism of Action. Front. Pharmacol. 2020, 11, 1300. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.W.; Natarajan, S.B.; Kim, Y.S.; Kim, E.K.; Lee, J.W.; Moon, S.H.; Jeon, B.T.; Park, P.J. Biosynthesis of Oligomeric Anthocyanins from Grape Skin Extracts. Molecules 2017, 22, 497. [Google Scholar] [CrossRef]
- Hwang, J.W.; Kim, E.K.; Lee, S.J.; Kim, Y.S.; Moon, S.H.; Jeon, B.T.; Sung, S.H.; Kim, E.T.; Park, P.J. Antioxidant activity and protective effect of anthocyanin oligomers on H(2)O(2)-triggered G2/M arrest in retinal cells. J. Agric. Food Chem. 2012, 60, 4282–4288. [Google Scholar] [CrossRef]
- Fan, M.; Kim, S.A.; Choi, Y.J.; Tang, Y.; Yang, H.P.; Kim, E.K. Anthocyanin oligomer (grape skin extract) administration improves dry eye disease: A randomised, double-blind, placebo-controlled study. Clin. Exp. Ophthalmol. 2023, 51, 122–130. [Google Scholar] [CrossRef]
- Choi, M.; Mukherjee, S.; Yun, J.W. Anthocyanin oligomers stimulate browning in 3T3-L1 white adipocytes via activation of the beta3-adrenergic receptor and ERK signaling pathway. Phytother. Res. 2021, 35, 6281–6294. [Google Scholar] [CrossRef]
- Zhou, J.; Zhu, Y.F.; Chen, X.Y.; Han, B.; Li, F.; Chen, J.Y.; Peng, X.L.; Luo, L.P.; Chen, W.; Yu, X.P. Black rice-derived anthocyanins inhibit HER-2-positive breast cancer epithelial-mesenchymal transition-mediated metastasis in vitro by suppressing FAK signaling. Int. J. Mol. Med. 2017, 40, 1649–1656. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Tollefsbol, T.O. Combinational Proanthocyanidins and Resveratrol Synergistically Inhibit Human Breast Cancer Cells and Impact Epigenetic(-)Mediating Machinery. Int. J. Mol. Sci. 2018, 19, 2204. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A Comprehensive Review of Their Chemical Properties and Health Effects on Cardiovascular and Neurodegenerative Diseases. Molecules 2020, 25, 3809. [Google Scholar] [CrossRef] [PubMed]
- Lagunas-Rangel, F.A.; Bermudez-Cruz, R.M. Natural Compounds That Target DNA Repair Pathways and Their Therapeutic Potential to Counteract Cancer Cells. Front. Oncol. 2020, 10, 598174. [Google Scholar] [CrossRef]
- Chun, S.Y.; Nam, K.S.; Lee, K.S. Proton Beam Induces P53-mediated Cell Cycle Arrest in HepG2 Hepatocellular Carcinoma Cells. Biotechnol. Bioproc. E 2020, 25, 141–148. [Google Scholar] [CrossRef]
- Collins, P.L.; Purman, C.; Porter, S.I.; Nganga, V.; Saini, A.; Hayer, K.E.; Gurewitz, G.L.; Sleckman, B.P.; Bednarski, J.J.; Bassing, C.H.; et al. DNA double-strand breaks induce H2Ax phosphorylation domains in a contact-dependent manner. Nat. Commun. 2020, 11, 3158. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Wang, Q.; Shuryak, I.; Brenner, D.J.; Turner, H.C. Development of a high-throughput gamma-H2AX assay based on imaging flow cytometry. Radiat. Oncol. 2019, 14, 150. [Google Scholar] [CrossRef]
- Martinez-Pastor, B.; Silveira, G.G.; Clarke, T.L.; Chung, D.; Gu, Y.; Cosentino, C.; Davidow, L.S.; Mata, G.; Hassanieh, S.; Salsman, J.; et al. Assessing kinetics and recruitment of DNA repair factors using high content screens. Cell Rep. 2021, 37, 110176. [Google Scholar] [CrossRef]
- Zimmer, J.; Tacconi, E.M.C.; Folio, C.; Badie, S.; Porru, M.; Klare, K.; Tumiati, M.; Markkanen, E.; Halder, S.; Ryan, A.; et al. Targeting BRCA1 and BRCA2 Deficiencies with G-Quadruplex-Interacting Compounds. Mol. Cell 2016, 61, 449–460. [Google Scholar] [CrossRef]
- Jaiswal, P.K.; Goel, A.; Mittal, R.D. Survivin: A molecular biomarker in cancer. Indian J. Med. Res. 2015, 141, 389–397. [Google Scholar]
- Vequaud, E.; Desplanques, G.; Jezequel, P.; Juin, P.; Barille-Nion, S. Survivin contributes to DNA repair by homologous recombination in breast cancer cells. Breast Cancer Res. Treat. 2016, 155, 53–63. [Google Scholar] [CrossRef]
- Roos, W.P.; Kaina, B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006, 12, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Lopez, K.E.; Bouchier-Hayes, L. Lethal and Non-Lethal Functions of Caspases in the DNA Damage Response. Cells 2022, 11, 1887. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J.; Khandre, N.S.; Sarin, A. Caspase-3 activation is an early event and initiates apoptotic damage in a human leukemia cell line. Apoptosis 2003, 8, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Scheel-Toellner, D.; Raza, K.; Assi, L.; Pilling, D.; Ross, E.J.; Lee, W.Y.; Curnow, S.J.; Buckley, C.D.; Akbar, A.N.; Lord, J.M.; et al. Differential regulation of nuclear and mitochondrial Bcl-2 in T cell apoptosis. Apoptosis 2008, 13, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Surova, O.; Zhivotovsky, B. Various modes of cell death induced by DNA damage. Oncogene 2013, 32, 3789–3797. [Google Scholar] [CrossRef]
- Das, G.; Shravage, B.V.; Baehrecke, E.H. Regulation and function of autophagy during cell survival and cell death. Cold Spring Harb. Perspect. Biol. 2012, 4, a008813. [Google Scholar] [CrossRef]
- Jung, S.; Jeong, H.; Yu, S.W. Autophagy as a decisive process for cell death. Exp. Mol. Med. 2020, 52, 921–930. [Google Scholar] [CrossRef]
- Jiang, P.; Mizushima, N. LC3- and p62-based biochemical methods for the analysis of autophagy progression in mammalian cells. Methods 2015, 75, 13–18. [Google Scholar] [CrossRef]
- Lin, Y.C.; Lin, J.F.; Wen, S.I.; Yang, S.C.; Tsai, T.F.; Chen, H.E.; Chou, K.Y.; Hwang, T.I. Chloroquine and hydroxychloroquine inhibit bladder cancer cell growth by targeting basal autophagy and enhancing apoptosis. Kaohsiung J. Med. Sci. 2017, 33, 215–223. [Google Scholar] [CrossRef]
- Xie, Z.; Xie, Y.; Xu, Y.; Zhou, H.; Xu, W.; Dong, Q. Bafilomycin A1 inhibits autophagy and induces apoptosis in MG63 osteosarcoma cells. Mol. Med. Rep. 2014, 10, 1103–1107. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, H.; Yuan, M.; Fan, H.; Cai, Z. Role of AMPK in autophagy. Front. Physiol. 2022, 13, 1015500. [Google Scholar] [CrossRef]
- Gao, G.Y.; Chen, W.W.; Yan, M.J.; Liu, J.S.; Luo, H.L.; Wang, C.; Yang, P. Rapamycin regulates the balance between cardiomyocyte apoptosis and autophagy in chronic heart failure by inhibiting mTOR signaling. Int. J. Mol. Med. 2020, 45, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Cordover, E.; Minden, A. Signaling pathways downstream to receptor tyrosine kinases: Targets for cancer treatment. J. Cancer Metastasis Treat. 2020, 6, 45. [Google Scholar] [CrossRef]
- Chadha, R.; Meador-Woodruff, J.H. Downregulated AKT-mTOR signaling pathway proteins in dorsolateral prefrontal cortex in Schizophrenia. Neuropsychopharmacol 2020, 45, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Lauring, J.; Park, B.H.; Wolff, A.C. The Phosphoinositide-3-Kinase-Akt-mTOR Pathway as a Therapeutic Target in Breast Cancer. J. Natl. Compr. Cancer Netw. 2013, 11, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Liu, Y.; Li, X. The interaction mechanism between autophagy and apoptosis in colon cancer. Transl. Oncol. 2020, 13, 100871. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, H.; Yang, D.; Yu, X.; Irwin, D.M.; Niu, G.; Tan, H. Excessive Autophagy Activation and Increased Apoptosis Are Associated with Palmitic Acid-Induced Cardiomyocyte Insulin Resistance. J. Diabetes Res. 2017, 2017, 2376893. [Google Scholar] [CrossRef]
- Lee, M.G.; Lee, K.S.; Nam, K.S. Arctigenin-mediated cell death of SK-BR-3 cells is caused by HER2 inhibition and autophagy-linked apoptosis. Pharmacol. Rep. 2021, 73, 629–641. [Google Scholar] [CrossRef]
- Lee, K.S.; Lee, M.G.; Nam, K.S. Evaluation of the antimetastatic and anticancer activities of morin in HER2-overexpressing breast cancer SK-BR-3 cells. Oncol. Rep. 2021, 46, 126. [Google Scholar] [CrossRef]
- Fan, M.; Choi, Y.J.; Tang, Y.; Bae, S.M.; Yang, H.P.; Kim, E.K. Efficacy and Mechanism of Polymerized Anthocyanin from Grape-Skin Extract on High-Fat-Diet-Induced Nonalcoholic Fatty Liver Disease. Nutrients 2019, 11, 2586. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.-G.; Hong, H.-J.; Nam, K.-S. Anthocyanin Oligomers Induce Apoptosis and Autophagy by Inhibiting the mTOR Signaling Pathway in Human Breast Cancer Cells. Pharmaceuticals 2024, 17, 24. https://doi.org/10.3390/ph17010024
Lee M-G, Hong H-J, Nam K-S. Anthocyanin Oligomers Induce Apoptosis and Autophagy by Inhibiting the mTOR Signaling Pathway in Human Breast Cancer Cells. Pharmaceuticals. 2024; 17(1):24. https://doi.org/10.3390/ph17010024
Chicago/Turabian StyleLee, Min-Gu, Hyun-Jin Hong, and Kyung-Soo Nam. 2024. "Anthocyanin Oligomers Induce Apoptosis and Autophagy by Inhibiting the mTOR Signaling Pathway in Human Breast Cancer Cells" Pharmaceuticals 17, no. 1: 24. https://doi.org/10.3390/ph17010024
APA StyleLee, M.-G., Hong, H.-J., & Nam, K.-S. (2024). Anthocyanin Oligomers Induce Apoptosis and Autophagy by Inhibiting the mTOR Signaling Pathway in Human Breast Cancer Cells. Pharmaceuticals, 17(1), 24. https://doi.org/10.3390/ph17010024





