Abstract
Little is known about the chemical and biological profiles of Dicranopteris linearis and Psychotria adenophylla. No previous studies have investigated alpha-glucosidase inhibition using extracts from D. linearis and P. adenophylla. In this paper, bioactive-guided isolation procedures were applied to the plants D. linearis and P. adenophylla based on alpha-glucosidase inhibition. From the most active fractions, 20 compounds (DL1–DL13 and PA1–PA7) were isolated. The chemical structures were elucidated using spectroscopic data and compared with those available in the literature. These compounds were evaluated for alpha-glucosidase inhibition, while a molecular docking study was performed to elucidate the mechanisms involved. Consequently, D. linearis and P. adenophylla might serve as a good potential for developing new antidiabetic preparations.
1. Introduction
Dicranopteris linearis (Burm. F.) Underw. Is a common fern belonging to the Gleicheniaceae family. This species is widely distributed in Africa and Asia, especially in the dry mountainous regions [1]. The extracts of D. linearis showed anticancer, antibacterial, antioxidant, analgesic, and anti-HIV activities [1,2,3,4,5]. This plant is commonly used as a folk medicine to treat fever (Malaysia), intestinal worms (Indochina), asthma, infertility in women (India), wounds (Papua New Guinea) [6], cough, allergies, and respiratory disorders (Mymensingh) [7]. Until now, 21 compounds have been isolated from D. linearis [1,2,3,4,5]. The major components are glycosides with aglycone parts: diterpenes, flavanols, and monoaromatic compounds. D. linearis is native to Vietnam, but alpha-glucosidase inhibition of its extracts and compounds derived from it has not been studied yet.
The genus Psychotria (Rubiaceae) comprises approximately 1700 species, popularly distributed in tropical and subtropical areas [8]. Different parts of these species (leaves, roots, and rhizomes) have been traditionally used to treat fever, bronchitis, ulcers, stomachaches, and gynecological hemorrhage in females [9]. Pharmacological studies have indicated that Psychotria plants exhibit various biological activities, including antimicrobial, antiviral, analgesic, hypoglycemic, and strong cytotoxic activities against several cancer cell lines [10,11]. The chemical data of the genus Psychotria have been comprehensively studied, indicating that the major compounds of this genus are alkaloids and terpenoids. Some alkaloids are the biomarkers of Psychotria plants [12]. Psychotria adenophylla wall is distributed in the south of Vietnam, and the phytochemical data on this plant are scarce. There has been only one report about the chemical constituents of P. adenophylla growing in India, which indicated the presence of eight sterols and triterpenes, including β-sitosterol, betulin, betulinic acid, α-amyrin, ursolic acid, friedelin, bauerenol, and bauerenol acetate [13].
There is limited information available concerning the chemical and biological profiles of D. linearis and P. adenophylla. Additionally, there have been no prior investigations into the potential of extracts from these plants to inhibit alpha-glucosidase. During our systematic research on alpha-glucosidase inhibitors from Vietnamese medicinal plants, bioactive-guided isolation procedures were applied to the plants D. linearis and P. adenophylla based on alpha-glucosidase inhibition. Twenty compounds (DL1–DL13 and PA1–PA7) were isolated from the most active fraction (Figure 1 and Figure 2). The chemical structures were elucidated using spectroscopic data and compared with those available in the literature. These compounds were evaluated for alpha-glucosidase inhibition, and a molecular docking study was performed to elucidate the mechanisms involved.
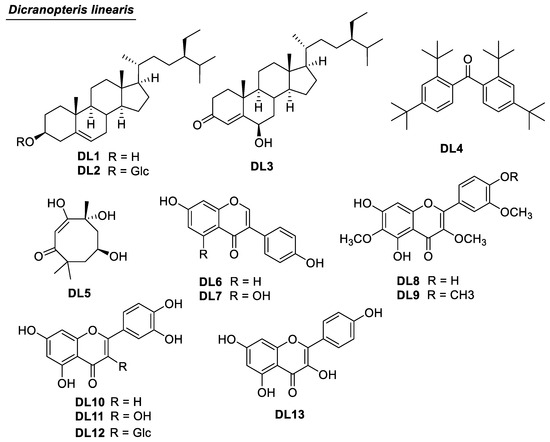
Figure 1.
Chemical structures of compounds DL1–DL13 from D. linearis.
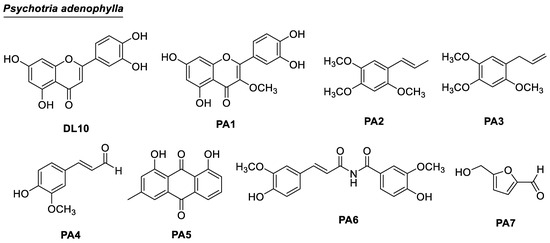
Figure 2.
Chemical structures of compounds DL10 and PA1–PA7 from P. adenophylla.
2. Results
Extracts/fractions from D. linearis and P. adenophylla were evaluated for alpha-glucosidase inhibition (Table 1). The most bioactive extract of each plant was selected for further isolation.

Table 1.
Alpha-glucosidase inhibition (IC50) by extracts and fractions.
2.1. Phytochemical Identification and Alpha-Glucosidase Inhibition of Isolated Compounds of D. linearis
Thirteen compounds were isolated from D. linearis. They were structurally elucidated as β-sitosterol (DL1) [14], daucosterol (DL2) [15], 6-hydroxystigmast-4-en-3-one (DL3) [16], 2,2′,4,4′-tetra-tert-butylbenzophenone (DL4) [17], chakyunglupulin B (DL5) [18], daidzein (DL6) [19], genistein (DL7) [20], jaceidin (DL8) [21], bonanzin (DL9) [22], luteolin (DL10) [23], quercetin (DL11) [24], isoquercetin (DL12) [25], and kaempferol (DL13) [26]. The chemical structures of all Isolated compounds were determined using 1D and 2D NMR methods. For compound DL3, the position of 6-OH was defined using the Heteronuclear Multiple Bond Correlation (HMBC) of H-4 to C-6 (Figure 3). The configuration of C-6 was defined based on the small J value of H-6 [16]. The positions of the methoxy groups of multi-oxygenated flavonoids DL8 and DL9 were defined using HMBC correlations (Figure 3).
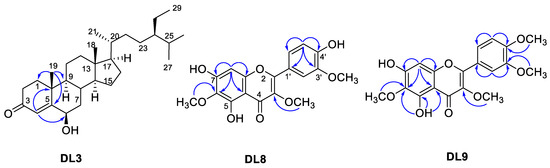
Figure 3.
Selected HMBC correlations of compounds DL3, DL8, and DL9 from P. adenophylla.
The physicochemical properties of isolated compounds are described below.
β-Sitosterol (DL1). White amorphous powder. 1H-NMR (500 MHz, acetone-d6) and 13C-NMR (125 MHz, acetone-d6) data were consistent with those reported in the literature [14].
Daucosterol (DL2). White amorphous powder. 1H-NMR (500 MHz, DMSO-d6) and 13C-NMR (125 MHz, DMSO-d6) data were consistent with those reported in the literature [15].
6-Hydroxystigmast-4-en-3-one (DL3). White amorphous powder. 1H-NMR (500 MHz, chloroform-d, δ ppm, J in Hertz): 5.82 (s, H-4), 4.35 (t, J = 2.0 Hz, H-6), 1.38 (s, H-19), 0.92 (d, J = 6.5 Hz, H-21), 0.84 (overlap, H-29), 0.83 (overlap, H-27), 0.81 (d, J = 7.0 Hz, H-26), 0.74 (s, H-18). 13C NMR (chloroform-d, 125 MHz, δ ppm):-200.6 (C-3), 168.6 (C-5), 126.5 (C-4), 73.5 (C-6), 56.2 (C-17), 56.0 (C-14), 53.8 (C-9), 46.0 (C-24), 42.7 (C-13), 39.8 (C-12), 38.7 (C-7), 38.1 (C-10), 37.3 (C-1), 36.3 (C-20), 34.4 (C-2), 34.1 (C-22), 29.9 (C-8), 29.3 (C-25), 28.3 (C-16), 26.3 (C-23), 24.3 (C-15), 23.2 (C-28), 21.1 (C-11), 20.0 (C-27), 19.7 (C-19), 19.2 (C-26), 18.9 (C-21), 12.2 (C-18), and 12.1 (C-29). These data were consistent with those reported in the literature [16].
2,2′,4,4′-Tetra-tert-butylbenzophenone (DL4). Colorless oil. 1H-NMR (500 MHz, chloroform-d, δ ppm, J in Hertz): 7.53 (d, J = 9.0 Hz, H-6), 7.35 (t, J = 2.3 Hz, H-4), 7.12 (d, J = 8.5, 2.5 Hz, H-5), 1.33 (s, H-10), 1.28 (s, H-9). 13C NMR (125 MHz, chloroform-d, δ ppm): 196.8 (C=O), 147.2 (C-1), 141.1 (C-2), 138.5 (C-3), 124.6 (C-4), 124.1 (C-5), 119.3 (C-6), 35.8 (C-7), 35.0 (C-8), 31.6 (C-9), and 30.3 (C-10). These data were consistent with those reported in the literature [17].
Chakyunglupulin B (DL5). Colorless oil. 1H-NMR (500 MHz, chloroform-d, δ ppm, J in Hertz): 5.69 (s, H-2), 4.33 (m, H-6), 2.46 (dt, J = 14.0, 2.8 Hz, H-5β), 1.97 (dt, J = 14.5, 2.8 Hz, H-7β), 1.78 (overlap, H-5α), 1.78 (s, H-11), 1.53 (dd, J = 14.5, 4.0 Hz, H-7α), 1.47 (s, H-9), 1.27 (s, H-10). 13C NMR (125 MHz, chloroform-d, δ ppm):182.5 (C-1), 171.2 (C-3), 113.1 (C-2), 86.8 (C-4), 67.0 (C-6), 47.5 (C-7), 45.8 (C-5), 36.1 (C-8), 30.8 (C-10), 27.2 (C-11), and 26.6 (C-9). These data were consistent with those reported in the literature [18].
Daidzein (DL6). Yellow amorphous powder. 1H-NMR (500 MHz, DMSO-d6, δ ppm, J in Hertz): 9.52 (s, 4′-OH), 8.29 (s, H-2), 7.97 (d, J = 8.5 Hz, H-5), 7.38 (d, J = 8.5 Hz, H-2′), 7.38 (d, J = 8.5 Hz, H-6′), 6.94 (dd, J = 8.5, 2.0 Hz, H-6), 6.87 (d, J = 2.5 Hz, H-8), 6.81 (d, J = 8.5 Hz, H-3′), 6.81 (d, J = 8.5 Hz, H-5′). 13C NMR (125 MHz, DMSO-d6, δ ppm): 174.6 (C-4), 162.3 (C-7), 157.4 (C-9), 157.4 (C-4′), 152.9 (C-2), 129.9 (C-2′), 129.9 (C-6′), 127.0 (C-5), 123.5 (C-3), 122.6 (C-1′), 116.6 (C-10), 115.1 (C-6), 114.9 (C-3′), 114.9 (C-5′), and 101.7 (C-8). These data were consistent with those reported in the literature [19].
Genistein (DL7). Yellow amorphous powder. 1H-NMR (500 MHz, Acetone-d6, δ ppm, J in Hertz): 13.03 (s, 5-OH), 8.16 (s, H-2), 7.45 (d, J = 8.5 Hz, H-2′), 7.45 (d, J = 8.5 Hz, H-6′), 6.90 (d, J = 8.5 Hz, H-3′), 6.90 (d, J = 8.5 Hz, H-3′), 6.42 (d, J = 2.5 Hz, H-8) and 6.28 (d, J = 2.5 Hz, H-6). 13C NMR (125 MHz, Acetone-d6, δ ppm): 181.9 (C-4), 165.3 (C-7), 163.9 (C-5), 159.2 (C-9), 158.6 (C-4′), 154.3 (C-2), 131.2 (C-2′), 131.2 (C-6′), 124.3 (C-3), 123.8 (C-1′), 116.0 (C-3′), 116.0 (C-5′), 106.2 (C-10), 99.9 (C-6), and 94.5 (C-8). These data were consistent with those reported in the literature [20].
Jaceidin (DL8). Yellow amorphous powder. 1H-NMR (500 MHz, Acetone-d6, δ ppm, J in Hertz): 7.71 (d, J = 2.0, H-2′), 7.60 (dd, J = 9.0, 2.0, H-6′), 6.99 (d, J = 9.0, H-5′), 6.80 (s, H-8), 3.98 (s, 3′-OCH3), 3.87 (s, 6-OCH3) and 3.80 (s, 3-OCH3). 13C NMR (125 MHz, Acetone-d6, δ ppm): 177.3 (C-4), 160.1 (C-7), 156.8 (C-2), 153.2 (C-9), 151.8 (C-5), 149.3 (C-4′), 146.1 (C-3′), 136.0 (C-3), 133.2 (C-6), 122.9 (C-1′), 122.2 (C-6′), 116.4 (C-5′), 112.1 (C-2′), 107.1 (C-10), 97.1 (C-8), 60.5 (6-OCH3), 60.1 (3-OCH3), and 56.8 (3′-OCH3). These data were consistent with those reported in the literature [21].
Bonanzin (DL9). Yellow amorphous powder. 1H-NMR (500 MHz, Acetone-d6, δ ppm, J in Hertz): 12.7 (s, 5-OH), 7.71 (dd, J = 8.8, 2.3 Hz, H-6′), 7.66 (d, J = 2.0 Hz, H-2′), 7.13 (d, J = 8.5 Hz, H-5′), 6.85 (s, H-8), 3.99 (s, 4′-OCH3), 3.96 (s, 4′-OCH3), 3.89 (s, 3-OCH3) and 3.80 (s, 6-OCH3). 13C-NMR (125 MHz, Acetone-d6, δ ppm, J in Hertz: 180.7 (C-4), 160.3 (C-4′), 158.9 (C-7), 155.4 (C-9), 153.7 (C-5), 152.2 (C-2), 151.3 (C-3′), 139.3 (C-3), 133.2 (C-6), 124.2 (C-6′), 121.9 (C-1′), 115.8 (C-2′), 112.2 (C-5′), 107.1 (C-10), 91.8 (C-8), 60.6 (3-OCH3), 60.2 (6-OCH3), 56.9 (3′-OCH3), and 56.3 (4′-OCH3). These data were consistent with those reported in the literature [22].
Luteolin (DL10). Yellow amorphous powder. 1H-NMR (500 MHz, acetone-d6) and 13C-NMR (125 MHz, acetone-d6). These data were consistent with those reported in the literature [23].
Quercetin (DL11). Yellow amorphous powder. 1H-NMR (500 MHz, acetone-d6) and 13C-NMR (125 MHz, acetone-d6). These data were consistent with those reported in the literature [24].
Isoquercetin (DL12). Yellow powder. 1H-NMR (500 MHz, DMSO-d6) and 13C-NMR (125 MHz, DMSO-d6). These data were consistent with those reported in the literature [25].
Kaempferol (DL13). Yellow amorphous powder. 1H-NMR (500 MHz, acetone-d6) and 13C-NMR (125 MHz, acetone-d6). These data were consistent with those reported in the literature [26].
2.2. Phytochemical Identification and Alpha-Glucosidase Inhibition of Isolated Compounds of P. adenophylla
Eight compounds were isolated and structurally elucidated. They were luteolin (DL10) [23], 3-O-methylquercetin (PA1) [27], α-asarone (PA2) [28], γ-asarone (PA3) [29], coniferyl aldehyde (PA4) [30], chrysophanol (PA5) [31], tribulusimide D (PA6) [32], and 5-hydroxymethylfurfural (PA7) [33].
3-O-methylquercetin (PA1). Yellow amorphous powder. 1H-NMR (500 MHz, acetone-d6) and 13C-NMR (125 MHz, acetone-d6). These data were consistent with those reported in the literature [27].
α-Asarone (PA2). Colorless oil. 1H-NMR (500 MHz, acetone-d6, δ ppm, J in Hertz): 7.04 (1H, s, H-6), 6.67 (1H, s, H-3), 6.64 (1H, dd, 16.0, 2.0, H-1′), 6.13 (1H, dq, 16.0, 6.5, H-2′), 3.84 (3H, s, 2-OCH3), 3.82 (3H, s, 3-OCH3), 3.78 (3H, s, 5-OCH3), 1.84 (3H, dd, 6.5, 2.0, H-3′). 13C-NMR (125 MHz, acetone-d6, δ ppm): 151.1 (C-4), 149.6 (C-2), 143.7 (C-5), 125.3 (C-2′), 122.9 (C-1′), 118.3 (C-1), 110.7 (C-6), 98.5 (C-3), 56.3, 56.1, 55.7 (4-OCH3, 2-OCH3, and 5-OCH3 signals could be interchanged), and 17.8 (C-3′). These data were consistent with those reported in the literature [28].
γ-Asarone (PA3). Colorless oil. 1H-NMR (500 MHz, acetone-d6, δ ppm, J in Hertz): 6.76 (1H, s, H-6), 6.69 (1H, s, H-3), 5.95 (1H, m, H-2′), 5.02 (1H, m, H-3′a), 4.97 (1H, m, H-3′b), 3.81 (3H, s, 2-OCH3), 3.83 (3H, s, 3-OCH3), 3.74 (3H, s, 5-OCH3), 3.29 (2H, d, 6.5, H-1′). 13C-NMR (125 MHz, acetone-d6, δ ppm): 151.5 (C-2), 148.6 (C-4), 143.2 (C-5), 138.1 (C-2′), 119.6 (C-1), 115.4 (C-6), 114.3 (C-3′), 98.7 (C-3), 55.8, 55.7, 55.5 (4-OCH3, 2-OCH3, and 5-OCH3 signals could be interchanged), and 33.4 (C-1′). These data were consistent with those reported in the literature [29].
Coniferyl aldehyde (PA4). White amorphous solid. The 1H-NMR (400 MHz, methanol-d4) data were consistent with those reported in the literature [30].
Chrysophanol (PA5). Yellow amorphous solid. 1H-NMR (500 MHz, methanol-d4, δ ppm, J in Hertz): 12.05 (1H, s, 8-OH), 11.95 (1H, s, 1-OH), 7.83 (1H, t, 8.0, H-6), 7.79 (1H, dd, 8.0, 1.0, H-5), 7.63 (1H, d, 1.0, H-4), 7.36 (1H, dd, 8.0, 1.0, H-7), 7.20 (1H, brs, H-2), 2.50 (3H, s, H-11). 13C-NMR (125 MHz, methanol-d4, δ ppm): 163.3 (C-1), 124.9 (C-2), 150.7 (C-3), 121.7 (C-4), 115.0 (C-4a), 120.3 (C-5), 138.3 (C-6), 125.2 (C-7), 163.7 (C-8), 116.7 (C-8a), 190.0 (C-9), 114.7 (C-9a), 182.4 (C-10), 134.8 (C-10a), and 20.1 (C-11). These data were consistent with those reported in the literature [31].
Tribulusimide D (PA6). Colorless oil. 1H-NMR (500 MHz, acetone-d6, δ ppm, J in Hertz): 7.33 (brs; H-2), 6.87 (d, J = 8.0; H-5), 7.26 (dd, J = 6.5, 9.0; H-6), 7.59 (d, J = 16.0; H-7), 6.39 (d, J = 16.0; H-8), 7.57 (d, J = 6.5; H-2′), 7.14 (d, J = 8.0; H-5′), 7.47 (brs; H-6′), 3.92 (s; 3-OCH3), 3,72 (s; 3′-OCH3). 13C-NMR (125 MHz, acetone-d6, δ ppm): 171.4 (C-9), 168.8 (C-7′), 150.4 (C-4′), 149.7 (C-4), 147.9 (C-3), 147.8 (C-3′), 145.7 (C-7), 126.6 (C-1), 125.5 (C-6′), 124.9 (C-6), 123.9 (C-5), 122.6 (C-1′), 119.9 (C-5′), 116.1 (C-8), 115.6 (C-2′), 111.4 (C-2), 56.4 (3-OCH3), and 51.5 (3′-OCH3). The NMR data were consistent with those reported previously [32].
5-Hydroxymethylfurfural (PA7). Colorless oil. 1H-NMR (500 MHz, acetone-d6) and 13C-NMR (125 MHz, acetone-d6). These data were consistent with those reported in the literature [33].
2.3. Alpha-Glucosidase Inhibition of Extracts, Fractions, and Compounds from D. linearis and P. adenophylla
The results of alpha-glucosidase inhibition for both extracts and fractions obtained from D. linearis and P. adenophylla are presented in Table 1. Additionally, the evaluation of the isolated compounds for their alpha-glucosidase inhibitory activity is presented in Table 2. From D. linearis, compounds DL3, DL5-DL8, and DL10-13 exhibited good inhibition with IC50 values ranging from 67.1 to 282.1 µM, whereas others were inactive. From P. adenophylla, compounds DL10 and PA4-PA6 showed potent inhibition with IC50 values of 67.1, 249.7, 98.2, and 76.2 µM, respectively.

Table 2.
Alpha-glucosidase inhibition (IC50) of isolated compounds.
The molecular docking models of DL1 [34], DL2 [35], DL6 [36], DL10 [37], DL11 [38], DL12 [39], DL13 [39], and PA4 [40] were investigated extensively, so no in silico analysis of those compounds were performed in the current study. Molecular docking studies were applied to compounds DL3, DL5, DL8, PA5, and PA6. The results are shown in Table 3 and Figure 4 and Figure 5.

Table 3.
The XP-IFD docking scores, the experimental/estimated binding affinities, and the number of H-bonds networks in the ligand–protein complex.
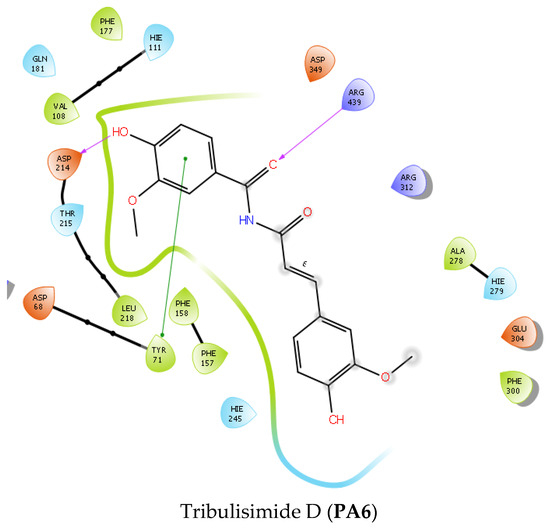
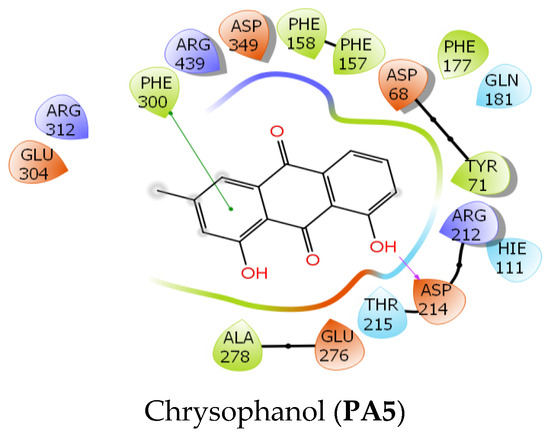
Figure 4.
The 2D interactions of two ligand–protein complexes: PA5 and PA6.
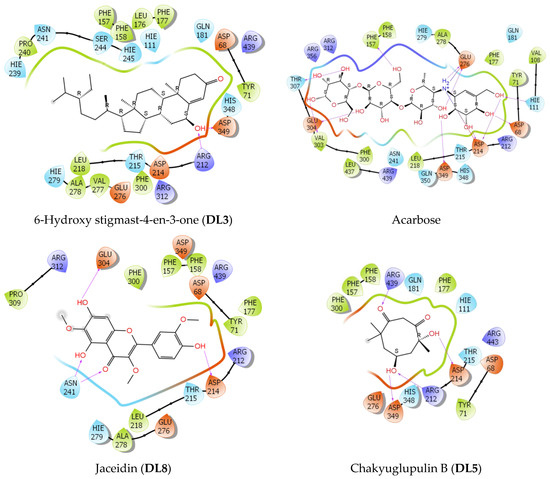
Figure 5.
The 2D interactions of three ligand–protein complexes (DL3, DL5, and DL8) in comparison with acarbose.
3. Discussion
3.1. Chemical Composition of D. linearis and P. adenophylla
Until now, 21 compounds have been reported previously for D. linearis (see Figure S21). Most of them are glycosides with 1–3 sugar units, and they appear in polar fractions. From the Vietnamese D. linearis, three steroids (DL1–DL3), eight flavonoids (DL6–DL13), one diphenylketone (DL4), and an 8-member-ring compound (DL5) were isolated. In 2021, Zakaria and co-workers undertook multiple analytic methods to determine phenolic compounds and triterpenes from the Malaysian Dicranopteris linearis leaves without isolation [41]. To the best of our knowledge, compounds DL1 and DL3–DL10 were reported for the first time in the genus Dicranopteris.
Only six compounds were reported previously for P. adenophylla (see Figure S22). All of them are common compounds found in many plants. Using bioactive-guided isolation on the Vietnamese P. adenophylla, eight compounds were successfully isolated. To the best of our knowledge, compounds PA1–PA6 were reported for the first time in the genus Psychotria. Many previous phytochemical investigations on the Psychotria plants focused on alkaloids using an alkaloid-isolation procedure [42,43,44,45]. Interestingly, the alkaloid tribulusimide D (PA6) was detected in P. adenophylla, representing a new alkaloid type within the genus Psychotria.
3.2. Alpha-Glucosidase Inhibition of Extracts, Fractions, and Compounds from D. linearis and P. adenophylla
Our literature review showed that crude extracts of both D. linearis and P. adenophylla have not been evaluated for alpha-glucosidase inhibition. Very little is known about the alpha-glucosidase inhibitory activity of the Dicranopteris and Psychotria plants. Recent studies regarding the alpha-glucosidase inhibition of the crude extracts from other Dicranopteris and Psychotria plants, D. caudata, P. malayana, and P viridiflora, were performed, indicating the potent inhibition of the extracts of these plants [46,47,48].
Although the crude methanol extract of D. linearis showed good activity, its derived extracts and fractions showed weaker alpha-glucosidase inhibition (Table 1). This indicated that the combination of all components of D. linearis might increase the activity. Further antidiabetic investigation of this plant should be conducted on the crude extract.
As seen in Table 1, the ethyl acetate extract of D. linearis had an IC50 value of 124.1 µg/mL. Therefore, it was further fractionated to obtain five fractions (EA1–EA5). Fractions EA2–EA4 were chosen for further isolation based on their good alpha-glucosidase inhibition. Compounds DL7 and DL10–DL13 were isolated from these fractions. Two isoflavones, DL6 and DL7, were isolated from the extract HEA. Those mentioned flavonoids are well-known alpha-glucosidase inhibitors that have been comprehensively studied. Particularly, the consistency between in vitro alpha-glucosidase inhibitory activity and in vivo data of luteolin (DL10) and daizein (DL6) was confirmed by various reports [37,49,50]. Quercetin (DL11), isoquercitin (DL12), and kaempferol (DL13) were potent inhibitors, and the two formers were non-competitive types [39,51]. Two isoflavones, genistein (DL5) and daizein (DL6), also inhibited alpha-glucosidase, which is consistent with previously published reports [36,52]. These compounds are abundant in the ethyl acetate extract of D. linearis, indicating that they determine the activity of the extract EA. Multi-oxygenated flavonoids DL8 and DL9 are less potent than the above-mentioned flavonoids. Their low inhibition might be affected by the presence of the 3-oMe group, which was reported previously by Nguyen et al., 2023 [53].
The EA extract and its derived fractions of P. adenophylla showed potent activities, with IC50 values ranging from 1.7 to 26.6 µg/mL, much lower than that of the crude MeOH extract (Table 1). However, the isolation of bioactive components from these extracts and fractions is limited due to their high lignin content. The detection of the mixture of undefined lignins was determined using NMR and HPLC methods (Figure S20.1–20.3). Such macromolecules were known to be potent alpha-glucosidase inhibitors [54]. Only three compounds, PA5–PA7, were isolated from the most active fraction, EA5, but these compounds do not reflect the activity of the starting fraction. These compounds showed moderate activity with IC50 values in the range of 76.2–249.7 µM. Tribulusimide D (PA6) was previously found in Euphorbia dracunculoides [55] and Tribuli fructus [32], showing significant hepatoprotective activity with an EC50 value of 13.46 µM.
The docking results showed that 5/7 ligands were able to bind to residues within the binding site of alpha-glucosidase. The steric effect with bulky groups prevented complexation with proteins of 2,2′,4,4′-tetra-tert-butyl benzophenone and 6-hydroxy stigmast-4-en-3-one. In order of docking score, acarbose had the lowest value of −17.3 kcal/mol while the studied compounds ranged from −7.4 to −4.3 kcal/mol. In terms of energy estimates using MM-GBSA, acarbose showed the lowest value at −128.2 kcal/mol, followed by 6-hydroxy stigmast-4-en-3-one (DL3) at -65.4 kcal/mol, and tribulisimide D (PA6) and jaceidin (DL8) in the same range at −43 kcal/mol. Chakyuglupulin B (DL4) exhibited a higher value of −29.3 kcal/mol. Chrysophanol (PA5) had the highest energy estimate at −24 kcal/mol. These results show some deviation from the experimental values, but overall, the experimental values of all the studied compounds fall within the range of −5.5 to −4.5 kcal/mol. This indicates that these compounds exhibit moderate inhibition of Saccharomyces cerevisiae alpha-glucosidase.
In terms of interactions, acarbose stood out due to its extensive occupancy of the chemical space and the significant presence of hydrogen bonding within the interaction networks. The inhibitory mechanism of acarbose ignited with the H-bond between the hydroxyl group of acarbose and Asp 214, whereas Glu 276, the residue responsible for catalyzing the hydrolysis of the normal 1,4-alpha-gluco bond, was engaged with two hydrogen bonds, one from acarbose’s nitrogen and one from the hydroxyl group. In addition, a salt bridge was also formed between the cationic ammonium and the carboxylate group of Glu 276. This observation was only visible when the nitrogen of acarbose was protonated in a physiological environment (pH = 7 ± 2), which was achieved using Ligprep. Therefore, Glu 276 could not hydrolyze the C-N bond between the glucose molecules of acarbose. Additionally, Asp 349, a residue considered a transition state stabilizer of diose or triose degradation, could also form a hydrogen bond with acarbose. According to the described mechanism, Figure 4 and Figure 5 and Table 3 indicate that among the three key residues (Asp 214, Glu 276, and Asp 349), all the studied compounds formed hydrogen bonds with either Asp 214 or both Asp 214 and Glu 276, except for compound B, which only formed a H-bond with the key residue Asp 349. This provides compelling evidence of their potential inhibitory effect on the enzyme’s activity, potentially impeding polysaccharide hydrolysis by Saccharomyces cerevisiae alpha-glucosidase.
The moderate activity of these ligands may be attributed to the specific structure of each substance, which prevents them from optimally filling the chemical space within the alpha-glucosidase binding site. As a result, there are easily accessible solvent regions deep inside the binding site, which reduces their inhibitory activity.
4. Materials and Methods
4.1. Source of the Plant Material
D. linearis leaves were collected in Ba Ria-Vung Tau Province, Vietnam, from June to July 2022. The scientific name was identified as Dicranopteris linearis (Burm. F.) Underw. by Dr. Dang Van Son (deposited as No UE-P017).
The leaves of P. adenophylla were collected in Ba Ria-Vung Tau Province, Vietnam, from May to July 2022. The scientific name of the material was identified as P. adenophylla by Dr. Dang Van Son. A voucher specimen (No UE-P018) was deposited in the herbarium of the Department of Organic Chemistry, Faculty of Chemistry, Ho Chi Minh University of Education, Ho Chi Minh City, Vietnam.
4.2. Isolation of Compounds DL1-DL13 from D. linearis
Dried and ground materials (5.0 kg) were exhaustedly extracted with methanol (3 × 30 L) at room temperature to provide a crude methanol extract (280.0 g). Liquid–liquid partition was applied to this extract using consecutive solvents n-hexane, n-hexane: EtOAc (1:1, v/v), EtOAc to give H (27.44 g), HEA (34.79 g), EA (23.00 g), and MeOH (60.03 g) extracts, respectively (Scheme 1).
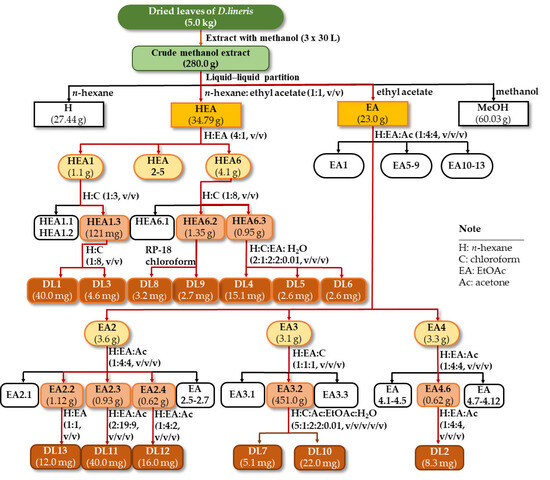
Scheme 1.
Isolation procedure of compounds DL1–DL13 from D. linearis.
The HEA extract (34.79 g) was subjected to a silica gel column chromatography (CC) using a mobile phase of n-hexane-EtOAc (4:1, v/v) to obtain six fractions (HEA1-HEA6). Fraction HEA1 (1.1 g) was applied to a silica gel CC using a gradient system of n-hexane: chloroform (1:3, v/v) to yield three subfractions (HEA1.1-HEA1.3). Subfraction HEA1.3 (121 mg) was subjected to a silica gel CC using a gradient system of n-hexane: chloroform (1:8, v/v) to afford compounds DL1 (40.0 mg) and DL3 (4.6 mg). Fraction HEA6 (4.1 g) was chromatographed on a silica gel CC and eluted with n-hexane: chloroform (1:9, v/v) to obtain three fractions (HEA6.1-HEA6.3). Fraction HEA6.2 (1.35 g) was rechromatographed on a silica gel CC and eluted with chloroform to afford compounds DL8 (3.2 mg) and DL9 (2.7 mg). Fraction HEA6.3 (0.95 g) was rechromatographed on a silica gel CC and eluted with n-hexane: chloroform: EtOAc: acetone: water (2:1:2:2:0.01, v/v/v/v/v) to yield compounds DL4 (15.1 mg), DL5 (2.6 mg), and DL6 (2.6 mg).
Fractionation of the EA extract was performed on a silica gel CC and then eluted with n-hexane: EtOAc: acetone (1:4:4, v/v/v) to yield 13 fractions (EA1-EA13). Fraction EA2 (3.6 g) was subjected to a silica gel CC using a mobile phase as n-hexane: EtOAc: acetone (1:4:4, v/v/v) to obtain seven fractions (EA2.1-EA2.7). Fraction EA2.2 (1.12 g) was rechromatographed and then eluted with n-hexane: EtOAc (1:1, v/v) to yield compound DL13 (12.0 mg). Fraction EA2.3 (0.93 g) was fractionated on a silica gel CC and eluted with n-hexane: EtOAc: acetone (2:19:9, v/v/v) to afford compound DL11 (40.0 mg). Fraction EA2.4 (0.62 g) was purified on a silica gel CC and eluted with n-hexane: EtOAc: acetone (1:4:2, v/v/v) to obtain compound DL12 (16.0 mg). Fraction EA3 (3.1 g) was applied to a silica gel CC using n-hexane: EtOAc: acetone (1:1:1, v/v/v) as a mobile phase to yield three fractions (EA3.1-EA3.3). Fraction EA3.2 (451 mg) was rechromatographed and then eluted with n-hexane: chloroform: acetone: EtOAc: water (5:1:2:2:0.01, v/v/v/v/v) to afford compounds DL10 (22.0 mg) and DL7 (5.1 mg). Fraction EA4 (3.3 g) was rechromatographed using a solvent system of n-hexane: EtOAc: acetone (1:4:4, v/v/v) to yield 12 fractions (EA4.1-EA4.12). Fraction EA4.6 (0.33 g) was chromatographed on a silica gel CC using n-hexane: EtOAc (1:3, v/v) as an eluent to yield compound DL2 (8.3 mg).
4.3. Isolation of Compounds PA1-PA7 from P. adenophylla
Dried leaves of P. adenophylla (3.0 kg) were extracted using methanol (3 × 10 L) at room temperature. The filtrated solution was evaporated at reduced pressure to obtain a crude extract (650 g). This extract was separated into n-hexane extract (22.2 g, H), n-hexane: EtOAc extract (67 g, HEA), and EtOAc (190.0 g, EA) via liquid–liquid partition. The EA extract was applied to silica gel CC and eluted with n-hexane: EtOAc (1:4, v/v) to afford five fractions (EA1-EA5) (Scheme 2).
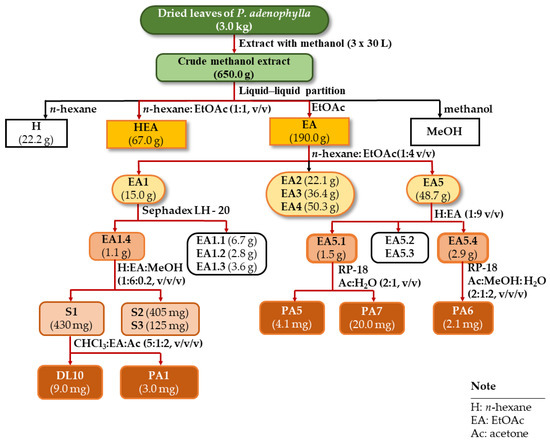
Scheme 2.
Isolation procedure of compounds DL10, PA1, and PA5-7 from P. adenophylla.
Fraction EA1 (15.0 g) was further subjected to a Sephadex LH-20 gel CC and eluted with MeOH to obtain four subfractions (EA1.1-EA1.4). Next, subfraction EA1.4 (1.1 g) was separated into three subfractions (EA1.4.1-EA1.4.3) using a silica gel CC and eluted with n-hexane: EtOAc: MeOH (1:6:0.2, v/v/v). Subfraction EA1.4.1 (430 mg) was purified using a silica gel CC with the solvent system of chloroform: EtOAc: acetone (5:1:2, v/v/v), resulting in the isolation of compounds DL10 (9.0 mg) and PA1 (3.0 mg). Fraction EA5 (48.7 g) was applied to a silica gel CC and eluted with n-hexane: EtOAc (1:9, v/v) to afford four subfractions (EA5.1-EA5.4). Subfraction EA5.1 (1.5 g) was continually chromatographed on a C-18 reverse-phase silica gel CC and eluted with acetone: water (2:1, v/v) to yield PA5 (4.1 mg) and PA7 (20 mg). Subfraction EA5.4 (2.9 g) was subjected to a C-18 reverse-phase silica gel CC and eluted with acetone: MeOH: water (2:1:2, v/v/v) to obtain compound PA6 (2.1 mg).
The H extract was applied to silica gel CC and eluted with n-hexane: EtOAc: acetone (9:1:1, v/v/v) to afford 11 fractions (H1–H11). Fraction H4 (2.1 g) was further subjected to a silica gel CC and eluted with the solvent system of n-hexane: EtOAc: acetone (9:1:1, v/v/v) to yield seven subfractions (H4.1–H4.7). Next, subfraction H4.1 (123 mg) was separated into three fractions (S1–S3) on a silica gel CC and eluted with the same solvent system. Purifying fraction S1 (31 mg) on silica gel CC obtained compound PA4 (3.5 mg). Subfraction H4.5 (67 mg) was subjected to a silica gel CC, using a solvent system of n-hexane: chloroform: MeOH: water (10:1:0.2:0.01, v/v/v/v) to afford three fractions (R1–R3). The same manner was applied to fraction R3 to afford a mixture of the compounds PA2 and PA3 (17.2 mg).
4.4. Alpha-Glucosidase Inhibition Assay
The enzyme alpha-glucosidase was derived from Saccharomyces cerevisiae (E.C 3.2.1.20) and compounds [acarbose, and 4-nitrophenyl β-D-glucopyranoside (pNPG)] were obtained from Sigma-Aldrich Co. (Saint Louis, MO, USA). The conditions followed that of our previous report with small modifications [56].
4.5. Molecular Docking Studies
The PDB structures of proteins (4j5t and 1t2p) were downloaded from the Protein Data Bank (PDB), while the 3D structures of ligands were modeled via the website chemicalize.com. After the conversion from PDB files into PDBQT format using AutodockTools, the docking study was designated on AutoDock4.2 using the Lamarckian genetic algorithm with 250 runs; the maximum number of evals was 25,000,000 (long) for each ligand–protein complex. The configurations with the most repetitions were employed to extract the estimated free energy as a scoring function for predicting the binding affinities to the macromolecular targets. Docking was carried out using the Maestro 12.5 software of Schrodinger Suites (Schrödinger Release 2020-3: Maestro, Schrödinger, LLC, New York, NY, USA, 2020). The protein of Saccharomyces cerevisiae alpha-glucosidase was obtained from the Uniprot database (P53341·MAL12_YEAST) and prepared using the Protein Preparation Wizard protocol. Next, seven reagents and an acarbose reference were generated and prepared using Ligprep [57]. As the protein itself does not contain ligands, the grid generation was based on Sitemaps detection’s suggestion, incorporating key residues such as Asp 214, Glu 276, and Asp 349 in the binding sites, with the highest site score of 1.1 [58]. In addition, the absence of a co-crystallized ligand in a binding site of the protein may not accurately represent the appropriate chemical space for ligand entry. Therefore, rigid docking can potentially lead to misinterpretation of ligand binding modes and inhibitory capacities. In contrast, Induced Fit docking (IFD) [59] allows the binding residues to dynamically adjust their positions to accommodate the ligands, allowing them to penetrate deeper into the binding sites. In the IFD process, specific restraints are implemented to prioritize the formation of h-bonds between the ligands and Asp 214, Glu 276, or Asp 349. After a glide docking SP (standard precision) with IFD, fine-tuning was performed using XP (extra precision)—IFD. The MM-GBSA method [60] was used to calculate the top two poses with the lowest docking scores for each ligand, and the result was chosen based on the best experimental fit. The experimental binding affinity was calculated based on the following equation:
is the concentration of a drug or inhibitor needed to inhibit a biological process or response by 50%.
4.6. Structure Elucidation of the Compounds
Gravity column chromatography was performed on silica gel 60 (0.040–0.063 mm, Merck, Darmstadt, Germany). Thin-layer chromatography (TLC) for checking the chromatographic patterns of fractions and isolated compounds was carried out on silica gel 60 F254 (Merck, Darmstadt, Germany) and the spots were visualized by spraying with 10% H2SO4 solution followed by heating. Specific rotations were obtained on a Jasco P-1010 polarimeter (Oklahoma City, OK, USA). The high-resolution electrospray mass spectra (HR-ESI-MS) were recorded using a MicrOTOF-Q mass spectrometer (Bruker, MA, USA). The Nuclear Magnetic Resonance (NMR) spectra were measured using a Bruker Avance 500 MHz spectrometer (Bruker, MA, USA).
4.7. HPLC Experiments Detected the Presence of Lignins in P. adenophylla
High-performance liquid chromatography (HPLC Agilent 1260 Infinity II) using the detector Diode Array detector (DAD) was employed for the analysis. A total of 35 µL of each sample (at the concentration of 1 mg/mL) was injected separately. A gradient system ofIN and water was used during the 60 min analysis: 5% to 10% ACN in 5 min, 10% to 30% ACN in 15 min, 30% to 80% ACN in 10 min, 80% to 100% ACN in 5 min, and 100% A in 5 min. A Luna C18 column (Phenomenex, 150 mm × 4.6 mm. i.d., 5 µm) and a C18 guard column (Phenomenex, Torrance, CA, USA) were employed for this analysis.
5. Conclusions
Twenty compounds were isolated from D. linearis and P. adenophylla plants using a bioactive-guided isolation procedure. Compounds DL1, DL3–DL10, and PA1–PA6 were reported for the first time in the genera Dicranopteris and Psychotria. The isolated compounds were evaluated for alpha-glucosidase inhibition. Compounds DL3, DL5–DL8, DL10–DL13, and PA4–PA6 showed potent inhibition with IC50 values ranging from 67.1 to 282.1 µM. A molecular docking study was applied to compounds DL3, DL5, DL8, PA5, and PA6 for elucidating the mechanisms of alpha-glucosidase inhibition. Consequently, this study illustrated that D. linearis and P. adenophylla might be good potential natural sources to develop new antidiabetic preparations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph16091253/s1. Figure S1. NMR spectra of DL1. Figure S2. NMR spectra of DL2. Figure S3. NMR spectra of DL3. Figure S4. NMR spectra of DL4. Figure S5. NMR spectra of DL5. Figure S6. NMR spectra of DL6. Figure S7. NMR spectra of DL7. Figure S8. NMR spectra of DL8. Figure S9. NMR spectra of DL9. Figure S10. NMR spectra of DL10. Figure S11. NMR spectra of DL11. Figure S12. NMR spectra of DL12. Figure S13. NMR spectra of DL13. Figure S14. NMR spectra of PA1. Figure S15. NMR spectra of PA2 and PA3. Figure S16. NMR spectra PA4. Figure S17. NMR spectra PA5. Figure S18. MS and NMR spectra of PA6. Figure S19. NMR spectra PA7. Figure S20. NMR spectra and HPLC chromatogram of the mixture of lignins. Figure S21. Chemical structures of compounds previously reported from D. linearis. Figure S22. Chemical structures of compounds previously reported from P. adenophylla.
Author Contributions
Conceptualization, T.-H.D., H.T.N., N.P.L. and T.-T.N.; methodology, Y.T.V. and H.T.N.; software, Y.T.V. and C.-B.D.; validation, N.-H.-N.P., N.-K.-T.P. and J.S.; formal analysis, T.-H.D., N.P.L., Y.T.V. and H.T.N.; investigation, V.-S.D., T.-H.D. and Y.T.V.; resources, V.-S.D., N.-K.-T.P., J.S. and N.-K.-D.K.; data curation, N.-K.-T.P., J.S. and N.-K.-D.K.; writing—original draft preparation, T.-H.D. and H.T.N.; writing—review and editing, T.-H.D., T.-T.N. and H.T.N.; visualization, J.S.; supervision, T.-H.D. and T.-T.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research is funded by Ho Chi Minh City University of Education Foundation for Science and Technology under grant number CS.2022.19.12TĐ.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and supplementary material.
Acknowledgments
This study was supported by the Thammasat University Research Unit in Natural Products Chemistry and Bioactivities (chemicals and NMR recording).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, J.; Chen, J.-J.; Gao, K. Chemical Constituents and Biological Activities of Dicranopteris linearis. Chem. Nat. Compd. 2014, 49, 1129–1131. [Google Scholar] [CrossRef]
- Li, X.-L.; Cheng, X.; Yang, L.-M.; Wang, R.-R.; Zheng, Y.-T.; Xiao, W.-L.; Zhao, Y.; Xu, G.; Lu, Y.; Chang, Y.; et al. Dichotomains A and B: Two New Highly Oxygenated Phenolic Derivatives from Dicranopteris dichotoma. Org. Lett. 2006, 8, 1937–1940. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-L.; Tu, L.; Zhao, Y.; Peng, L.-Y.; Xu, G.; Cheng, X.; Zhao, Q.-S. Terpenoids from Two Dicranopteris Species. Helv. Chim. Acta 2008, 91, 856–861. [Google Scholar] [CrossRef]
- Ponnusamy, Y.; Chear, N.J.-Y.; Ramanathan, S.; Lai, C.-S. Polyphenols Rich Fraction of Dicranopteris linearis Promotes Fibroblast Cell Migration and Proliferation in Vitro. J. Ethnopharmacol. 2015, 168, 305–314. [Google Scholar] [CrossRef]
- Zakaria, Z.A.; Kamisan, F.H.; Omar, M.H.; Mahmood, N.D.; Othman, F.; Abdul Hamid, S.S.; Abdullah, M.N.H. Methanol Extract of Dicranopteris linearis L. Leaves Impedes Acetaminophen-Induced Liver Intoxication Partly by Enhancing the Endogenous Antioxidant System. BMC Complement. Altern. Med. 2017, 17, 271. [Google Scholar] [CrossRef]
- Kamisan, F.H.; Yahya, F.; Mamat, S.S.; Kamarolzaman, M.F.F.; Mohtarrudin, N.; Kek, T.L.; Salleh, M.Z.; Hussain, M.K.; Zakaria, Z.A. Effect of Methanol Extract of Dicranopteris linearis against Carbon Tetrachloride- Induced Acute Liver Injury in Rats. BMC Complement. Altern. Med. 2014, 14, 123. [Google Scholar] [CrossRef]
- Sarker, S.K.; Hossain, A.B.M.E. Pteridophytes of Greater Mymensingh District of Bangladesh Used as Vegetables and Medicines. Bangladesh J. Plant Taxon. 1970, 16, 47–56. [Google Scholar] [CrossRef]
- Davis, A.P.; Bridson, D.; Jarvis, C.; Govaerts, R. The Typification and Characterization of the Genus Psychotria L. (Rubiaceae). Bot. J. Linn. Soc. 2001, 135, 35–42. [Google Scholar] [CrossRef]
- Calixto, N.O.; Pinto, M.E.F.; Ramalho, S.D.; Burger, M.C.M.; Bobey, A.F.; Young, M.C.M.; Bolzani, V.S.; Pinto, A.C. The Genus Psychotria: Phytochemistry, Chemotaxonomy, Ethnopharmacology and Biological Properties. J. Braz. Chem. Soc. 2016, 27, 1355–1378. [Google Scholar] [CrossRef]
- Benevides, P.J.C.; Young, M.C.M.; Bolzani, V.d.S. Biological Activities of Constituents from Psychotria spectabilis. Pharm. Biol. 2005, 42, 565–569. [Google Scholar] [CrossRef]
- Pimenta, A.T.A.; Uchôa, D.E.A.; Braz-Filho, R.; Silveira, E.R.; Lima, M.A.S. Alkaloid and Other Chemical Constituents from Psychotria stachyoides Benth. J. Braz. Chem. Soc. 2011, 22, 2216–2219. [Google Scholar] [CrossRef]
- De Carvalho, A.R.; De Carvalho, M.G.; Braz-Filho, R.; Vieira, I.J.C. Chapter 7—Psychotria Genus: Chemical Constituents, Biological Activities, and Synthetic Studies. Stud. Nat. Prod. Chem. 2016, 48, 231–261. [Google Scholar] [CrossRef]
- Dan, S.; Dan, S.S. Phytochemical Study of Adansonia digitata, Coccoloba excoriata, Psychotria adenophylla and Schleichera oleosa. Fitoterapia 1986, 57, 445–446. [Google Scholar]
- Chaturvedula, V.S.P.; Prakash, I. Isolation of Stigmasterol and β-Sitosterol from the Dichloromethane Extract of Rubus suavissimus. Int. Curr. Pharm. J. 2012, 1, 239–242. [Google Scholar] [CrossRef]
- Katja, D.G.; Harneti, D.; Mayanti, T.; Nurlelasari, N.; Maharani, R.; Shiono, Y.; Supratman, U. Cytotoxic Steroids From The Stembak of Chisocheton celebicus KOORD. J. Kim. Valensi 2019, 5, 143–148. [Google Scholar] [CrossRef]
- Wu, S.-Y.; Huang, G.-L.; Zhou, Z.-L.; Chen, Z.-M. Chemical Constituents from the Solid Culture of the Edible Mushroom Flammulina velutipes. Chem. Nat. Compd. 2022, 58, 981–983. [Google Scholar] [CrossRef]
- Olah, G.A.; Wu, A.; Farooq, O. Ring tert-Butylation of Benzophenones and Benzaldehyde with tert-Butyllithium and Thionyl Chloride. Synthesis 1991, 1991, 1179–1182. [Google Scholar] [CrossRef]
- Kim, K.H.; Clardy, J.; Senger, D.; Cao, S. Chakyunglupulins A and B, Two Novel 4,8,8-Trimethylcyclooct-2-Enone Derivatives from Barleria lupulina. Tetrahedron Lett. 2015, 56, 2732–2734. [Google Scholar] [CrossRef]
- Coward, L.; Barnes, N.C.; Setchell, K.D.R.; Barnes, S. Genistein, Daidzein, and Their Beta-Glycoside Conjugates: Antitumor Isoflavones in Soybean Foods from American and Asian Diets. J. Agric. Food Chem. 1993, 41, 1961–1967. [Google Scholar] [CrossRef]
- He, J.; Fan, P.; Feng, S.; Shao, P.; Sun, P. Isolation and Purification of Two Isoflavones from Hericium erinaceum Mycelium by High-Speed Counter-Current Chromatography. Molecules 2018, 23, 560. [Google Scholar] [CrossRef]
- Long, C.; Sauleau, P.; David, B.; Lavaud, C.; Cassabois, V.; Ausseil, F.; Massiot, G. Bioactive Flavonoids of Tanacetum parthenium Revisited. Phytochemistry 2003, 64, 567–569. [Google Scholar] [CrossRef]
- Hamed, A.R.; Mohamed, T.A.; Tawfik, W.A.; Hassan, E.M.; El-Toumy, S.A.; Dinkova-Kostova, A.T. Bioactive Polymethoxylated Flavonoids from Chiliadenus montanus. J. Chem. Pharm. Res. 2016, 8, 788–793. [Google Scholar]
- Lin, L.-C.; Pai, Y.-F.; Tsai, T.-H. Isolation of Luteolin and Luteolin-7-O-Glucoside from Dendranthema morifolium Ramat Tzvel and Their Pharmacokinetics in Rats. J. Agric. Food Chem. 2015, 63, 7700–7706. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, D.; Yang, L.; Zhou, D.; Zhang, J. Purification and Characterization of Flavonoids from the Leaves of Zanthoxylum bungeanum and Correlation between Their Structure and Antioxidant Activity. PLoS ONE 2014, 9, e105725. [Google Scholar] [CrossRef] [PubMed]
- Kazuma, K. Malonylated Flavonol Glycosides from the Petals of Clitoria ternatea. Phytochemistry 2003, 62, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.P.; Wu, H.K.; Wu, T.; Shi, H.; Hang, B.; Aisa, H.A. Kaempferol and Quercetin Flavonoids from Rosa rugosa. Chem. Nat. Compd. 2006, 42, 736–737. [Google Scholar] [CrossRef]
- Yamauchi, K.; Mitsunaga, T.; Inagaki, M.; Suzuki, T. Synthesized Quercetin Derivatives Stimulate Melanogenesis in B16 Melanoma Cells by Influencing the Expression of Melanin Biosynthesis Proteins MITF and P38 MAPK. Bioorg. Med. Chem. 2014, 22, 3331–3340. [Google Scholar] [CrossRef]
- Zuo, H.L.; Yang, F.Q.; Zhang, X.M.; Xia, Z.N. Separation of Cis- and Trans-Asarone from Acorus tatarinowii by Preparative Gas Chromatography. J. Anal. Methods Chem. 2012, 2012, e402081. [Google Scholar] [CrossRef]
- Sinha, A.K.; Acharya, R.; Joshi, B.P. A mild and convenient procedure for the conversion of toxic β-asarone into rare phenylpropanoids: 2,4,5-Trimethoxycinnamaldehyde and γ-asarone. J. Nat. Prod. 2002, 65, 764–765. [Google Scholar] [CrossRef]
- Lim, E.-K.; Jackson, R.G.; Bowles, D.J. Identification and Characterisation of Arabidopsis Glycosyltransferases Capable of Glucosylating Coniferyl Aldehyde and Sinapyl Aldehyde. FEBS Lett. 2005, 579, 2802–2806. [Google Scholar] [CrossRef]
- Zhang, H.; Guo, Z.; Wu, N.; Xu, W.; Han, L.; Li, N.; Han, Y. Two Novel Naphthalene Glucosides and an Anthraquinone Isolated from Rumex dentatus and Their Antiproliferation Activities in Four Cell Lines. Molecules 2012, 17, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Byun, E.; Jeong, G.-S.; An, R.-B.; Min, T.S.; Kim, Y.-C. Tribuli Fructus Constituents Protect against Tacrine-Induced Cytotoxicity in HepG2 Cells. Arch. Pharm. Res. 2010, 33, 67–70. [Google Scholar] [CrossRef]
- Li, Y.-X.; Li, Y.; Zhong, Q.; Kim, M.-M.; Kim, S.-K. In Vitro Antioxidant Activity of 5-HMF Isolated from Marine Red Alga Laurencia undulata in Free Radical Mediated Oxidative Systems. J. Microbiol. Biotechnol. 2009, 19, 1319–1327. [Google Scholar] [CrossRef] [PubMed]
- Eawsakul, K.; Panichayupakaranant, P.; Ongtanasup, T.; Warinhomhoun, S.; Noonong, K.; Bunluepuech, K. Computational Study and in Vitro Alpha-Glucosidase Inhibitory Effects of Medicinal Plants from a Thai Folk Remedy. Heliyon 2021, 7, e08078. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Yang, X.; Shang, C.; Thao, T.T.P.; Koyama, T. Inhibitory Properties of Saponin from Eleocharis dulcis Peel against α-Glucosidase. RSC Adv. 2021, 11, 15400–15409. [Google Scholar] [CrossRef]
- Seong, S.H.; Roy, A.; Jung, H.A.; Jung, H.J.; Choi, J.S. Protein Tyrosine Phosphatase 1B and α-Glucosidase Inhibitory Activities of Pueraria lobata Root and Its Constituents. J. Ethnopharmacol. 2016, 194, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Kahksha; Alam, O.; Al-Keridis, L.A.; Khan, J.; Naaz, S.; Alam, A.; Ashraf, S.A.; Alshammari, N.; Adnan, M.; Beg, M.A. Evaluation of Antidiabetic Effect of Luteolin in STZ Induced Diabetic Rats: Molecular Docking, Molecular Dynamics, In Vitro and In Vivo Studies. J. Funct. Biomater. 2023, 14, 126. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.J.; Zhao, Y.M.; Lu, H.N.; Wang, X.E.; Jin, N.Z. Exploring Molecular Flexibility and the Interactions of Quercetin Derivatives in the Active Site of α-Glucosidase Using Molecular Docking and Charge Density Analysis. Comput. Theor. Chem. 2016, 1094, 55–68. [Google Scholar] [CrossRef]
- Hua, F.; Zhou, P.; Wu, H.-Y.; Chu, G.-X.; Xie, Z.-W.; Bao, G.-H. Inhibition of α-Glucosidase and α-Amylase by Flavonoid Glycosides from Lu’an GuaPian Tea: Molecular Docking and Interaction Mechanism. Food Funct. 2018, 9, 4173–4183. [Google Scholar] [CrossRef]
- Arvindekar, A.; More, T.; Payghan, P.V.; Laddha, K.; Ghoshal, N.; Arvindekar, A. Evaluation of Anti-Diabetic and Alpha Glucosidase Inhibitory Action of Anthraquinones from Rheum Emodi. Food Funct. 2015, 6, 2693–2700. [Google Scholar] [CrossRef]
- Zakaria, Z.A.; Sahmat, A.; Azmi, A.H.; Nur Zainol, A.S.; Omar, M.H.; Balan, T.; Sulistyorini, L.; Azizah, R.; Abdullah, M.N.H. Polyphenolics and Triterpenes Presence in Chloroform Extract of Dicranopteris linearis Leaves Attenuated Paracetamol-Induced Liver Intoxication in Rat. BMC Complement. Med. Ther. 2021, 21, 35. [Google Scholar] [CrossRef] [PubMed]
- Guéritte-Voegelein, F.; Sévenet, T.; Pusset, J.; Adeline, M.-T.; Gillet, B.; Beloeil, J.-C.; Guénard, D.; Potier, P.; Rasolonjanahary, R.; Kordon, C. Alkaloids from Psychotria oleoides with Activity on Growth Hormone Release. J. Nat. Prod. 1992, 55, 923–930. [Google Scholar] [CrossRef]
- Li, X.-N.; Zhang, Y.; Cai, X.-H.; Feng, T.; Liu, Y.-P.; Li, Y.; Ren, J.; Zhu, H.-J.; Luo, X.-D. Psychotripine: A New Trimeric Pyrroloindoline Derivative from Psychotria pilifera. Org. Lett. 2011, 13, 5896–5899. [Google Scholar] [CrossRef] [PubMed]
- Takayama, H.; Mori, I.; Kitajima, M.; Aimi, N.; Lajis, N.H. New Type of Trimeric and Pentameric Indole Alkaloids from Psychotria rostrata. Org. Lett. 2004, 6, 2945–2948. [Google Scholar] [CrossRef] [PubMed]
- Lajis, N.H.; Mahmud, Z.; Toia, R.F. The Alkaloids of Psychotria rostrata. Planta Med. 1993, 59, 383–384. [Google Scholar] [CrossRef]
- Mala, P.; Khan, G.A.; Gopalan, R.; Gedefaw, D.; Soapi, K. Fijian Medicinal Plants and Their Role in the Prevention of Type 2 Diabetes Mellitus. Biosci. Rep. 2022, 42, BSR20220461. [Google Scholar] [CrossRef]
- Nipun, T.S.; Khatib, A.; Ahmed, Q.U.; Redzwan, I.E.; Ibrahim, Z.; Khan, A.Y.F.; Primaharinastiti, R.; Khalifa, S.A.M.; El-Seedi, H.R. Alpha-Glucosidase Inhibitory Effect of Psychotria malayana Jack Leaf: A Rapid Analysis Using Infrared Fingerprinting. Molecules 2020, 25, 4161. [Google Scholar] [CrossRef]
- Chen, Q.; Toy, J.Y.H.; Seta, C.; Yeo, T.C.; Huang, D. Inhibition Effect of Extract of Psychotria viridiflora Stem on α-Amylase and α-Glucosidase and Its Application in Lowering the Digestibility of Noodles. Front. Nutr. 2021, 8, 701114. [Google Scholar] [CrossRef]
- Li, M.; Bao, X.; Zhang, X.; Ren, H.; Cai, S.; Hu, X.; Yi, J. Exploring the Phytochemicals and Inhibitory Effects against α-Glucosidase and Dipeptidyl Peptidase-IV in Chinese Pickled Chili Pepper: Insights into Mechanisms by Molecular Docking Analysis. LWT 2022, 162, 113467. [Google Scholar] [CrossRef]
- Park, M.-H.; Ju, J.-W.; Park, M.; Han, J. Daidzein Inhibits Carbohydrate Digestive Enzymes in Vitro and Alleviates Postprandial Hyperglycemia in Diabetic Mice. Eur. J. Pharmacol. 2013, 712, 48–52. [Google Scholar] [CrossRef]
- Şöhretoğlu, D.; Sari, S.; Barut, B.; Özel, A. Discovery of Potent α-Glucosidase Inhibitor Flavonols: Insights into Mechanism of Action through Inhibition Kinetics and Docking Simulations. Bioorg. Chem. 2018, 79, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Ning, Z.; Zhai, L.; Huang, T.; Peng, J.; Hu, D.; Xiao, H.; Wen, B.; Lin, C.; Zhao, L.; Bian, Z. Identification of α-Glucosidase Inhibitors from Cyclocarya paliurus Tea Leaves Using UF-UPLC-Q/TOF-MS/MS and Molecular Docking. Food Funct. 2019, 10, 1893–1902. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.; Duong, T.; Truong Nguyen, H.; Vu, Y.T.; Tran, T.; Ho, T.; Mai, C.; Mai, D.; Nguyen, H.; Thuy Le, H.; et al. New Halogenated Flavonoids from Adenosma bracteosum and Vitex negundo and Their α-Glucosidase Inhibition. Chem. Biodivers. 2023, 20, e202300390. [Google Scholar] [CrossRef]
- Qi, S.; Jiang, B.; Huang, C.; Jin, Y. Dual Regulation of Sulfonated Lignin to Prevent and Treat Type 2 Diabetes Mellitus. Biomacromolecules 2023, 24, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, Y.-T.; Sun, Q.-Y.; Zang, Z.; Yang, F.-M.; Liu, J.-P.; Jiang, J.-H.; Huang, S.-X.; Zhao, Y. A New Lathyrane Diterpenoid Ester from Euphorbia dracunculoides. Chem. Nat. Compd. 2016, 52, 1037–1040. [Google Scholar] [CrossRef]
- Nguyen, N.-H.; Tran, N.-M.-A.; Duong, T.-H.; Vo, G.V. α-Glucosidase Inhibitory Activities of Flavonoid Derivatives Isolated from Bouea macrophylla: In Vitro and in Silico Studies. RSC Adv. 2023, 13, 8190–8201. [Google Scholar] [CrossRef]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.F.; Honig, B.; Shaw, D.E.; Friesner, R.A. A Hierarchical Approach to All-Atom Protein Loop Prediction. Proteins Struct. Funct. 2004, 55, 351–367. [Google Scholar] [CrossRef]
- Wang, G.; Chen, M.; Wang, J.; Peng, Y.; Li, L.; Xie, Z.; Deng, B.; Chen, S.; Li, W. Synthesis, Biological Evaluation and Molecular Docking Studies of Chromone Hydrazone Derivatives as α-Glucosidase Inhibitors. Bioorg. Med. Chem. Lett. 2017, 27, 2957–2961. [Google Scholar] [CrossRef]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel Procedure for Modeling Ligand/Receptor Induced Fit Effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef]
- Guimarães, C.R.W.; Cardozo, M. MM-GB/SA Rescoring of Docking Poses in Structure-Based Lead Optimization. J. Chem. Inf. Model. 2008, 48, 958–970. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).