RETRACTED: Design and Synthesis of Novel 5-((3-(Trifluoromethyl)piperidin-1-yl)sulfonyl)indoline-2,3-dione Derivatives as Promising Antiviral Agents: In Vitro, In Silico, and Structure–Activity Relationship Studies
Abstract
1. Introduction
2. Results and Discussion
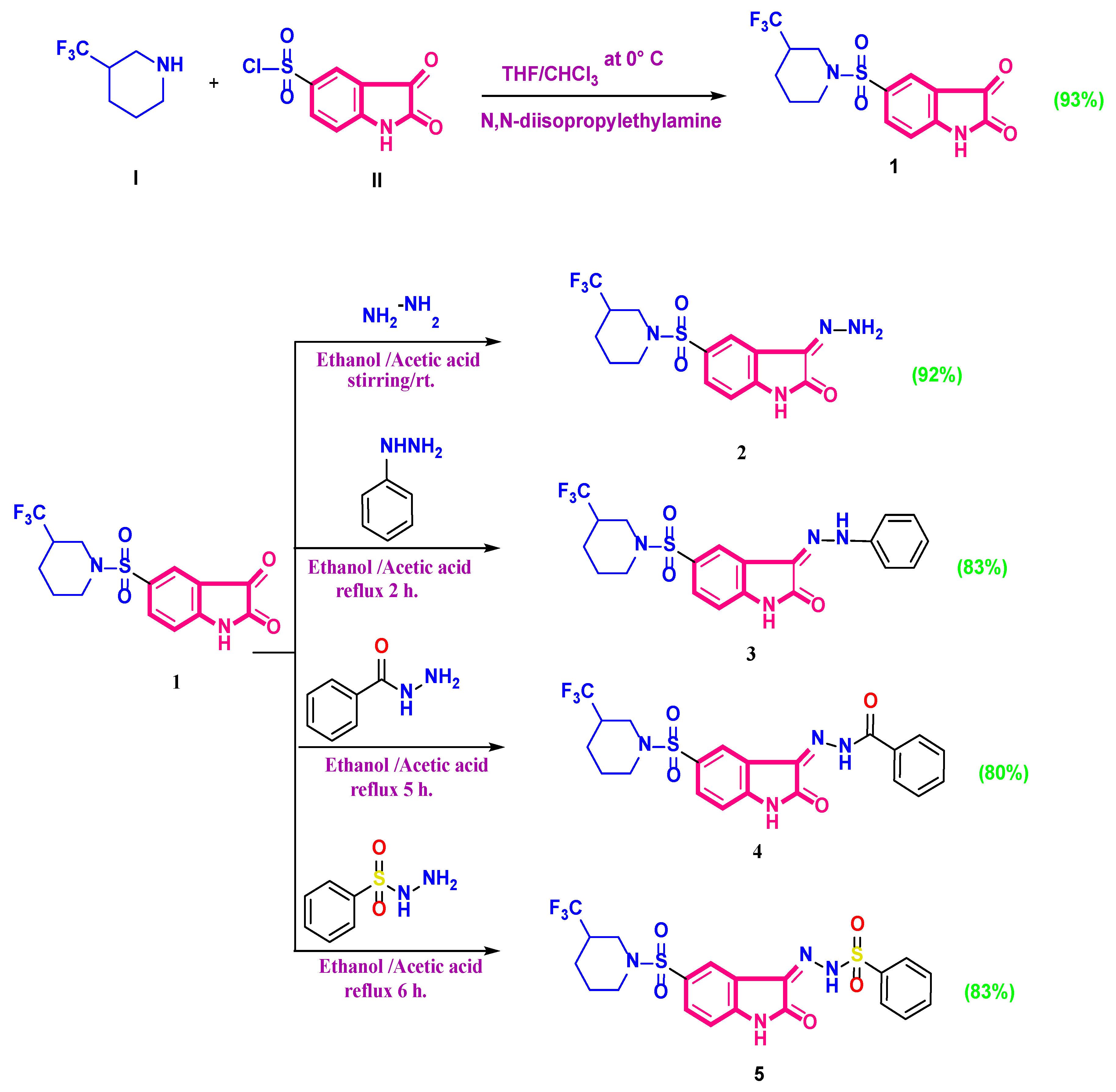
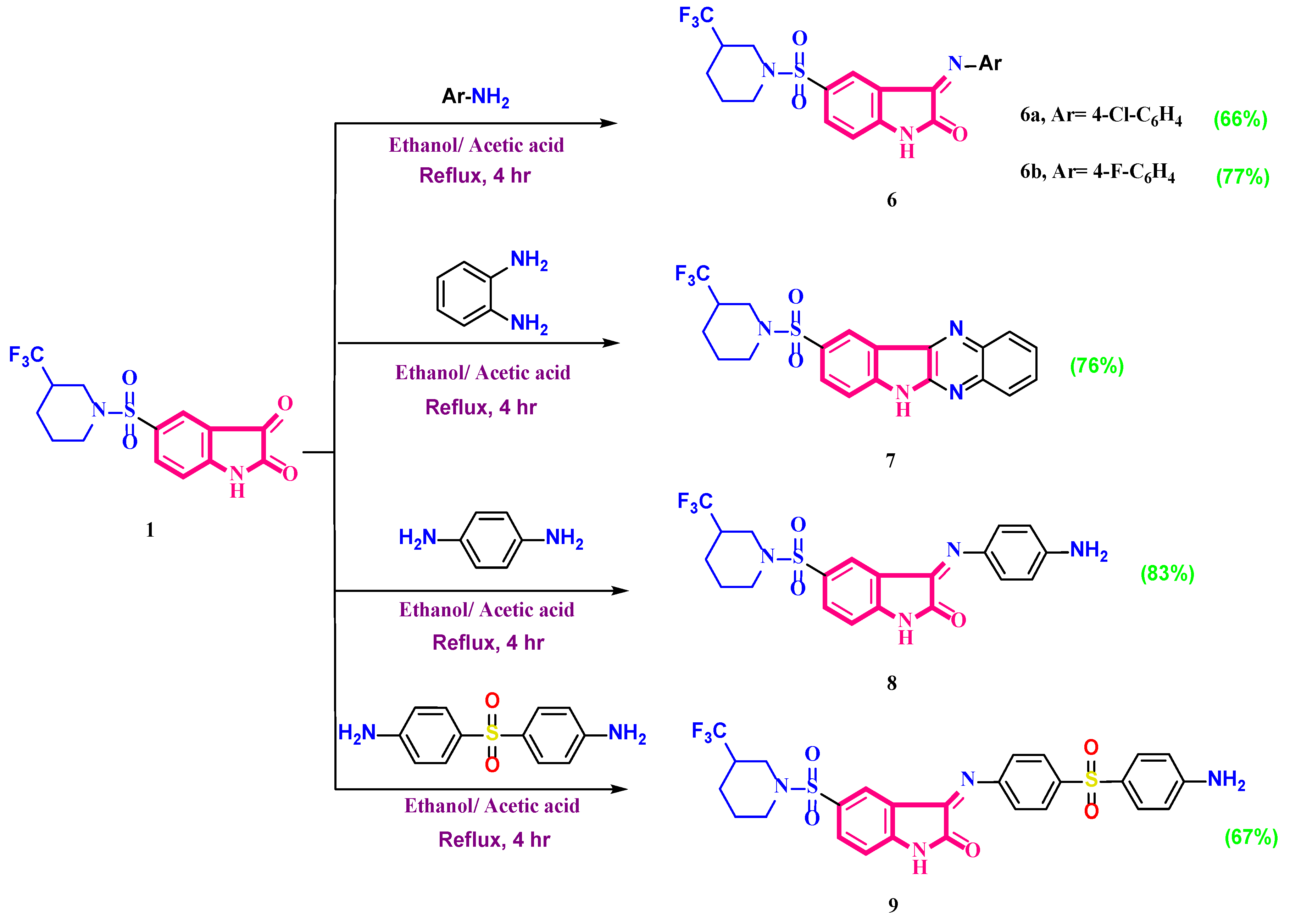
2.1. Chemistry
2.2. Biological Evaluation
2.2.1. Cytotoxicity Assay
2.2.2. The Antiviral Activity Assay (Inhibitory Concentration 50 Detection)
2.2.3. Quantitative PCR (qPCR) Assay
2.3. In Silico Studies
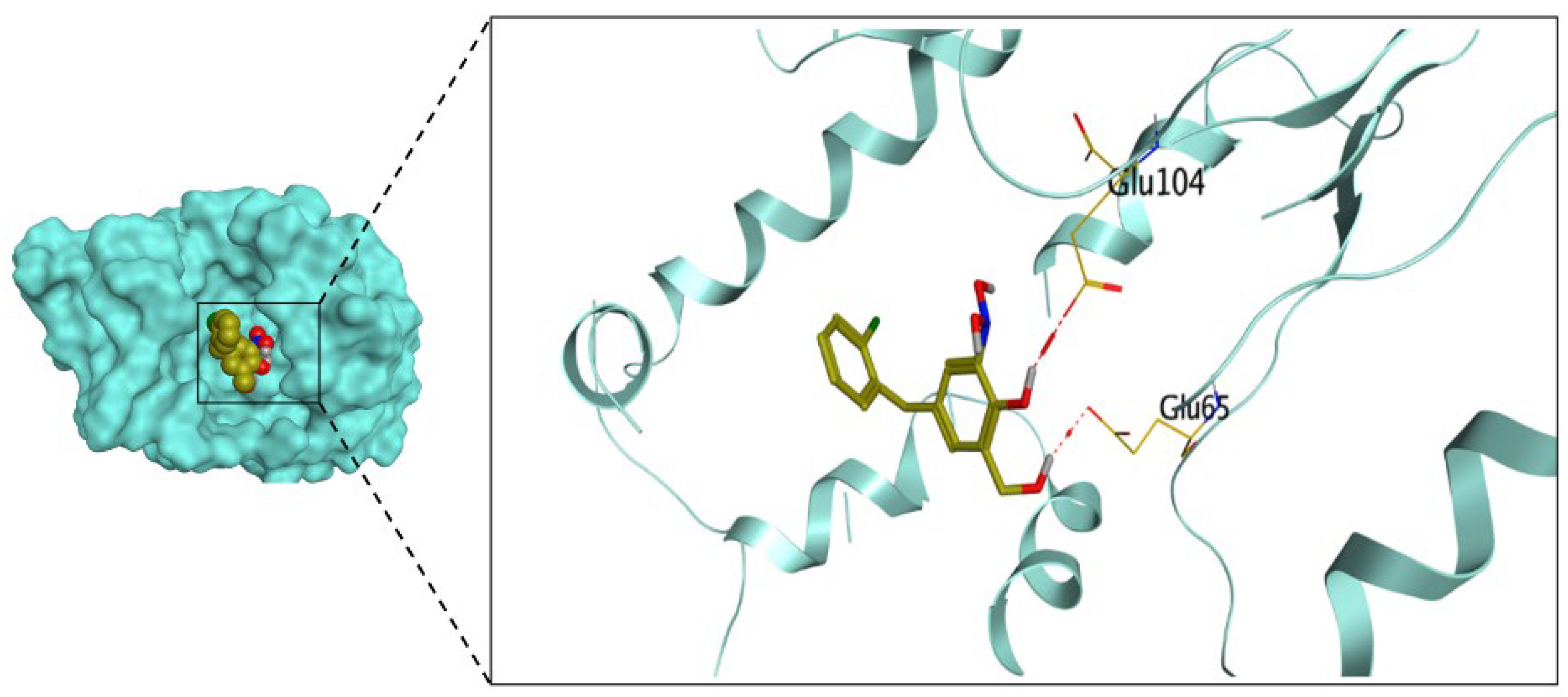
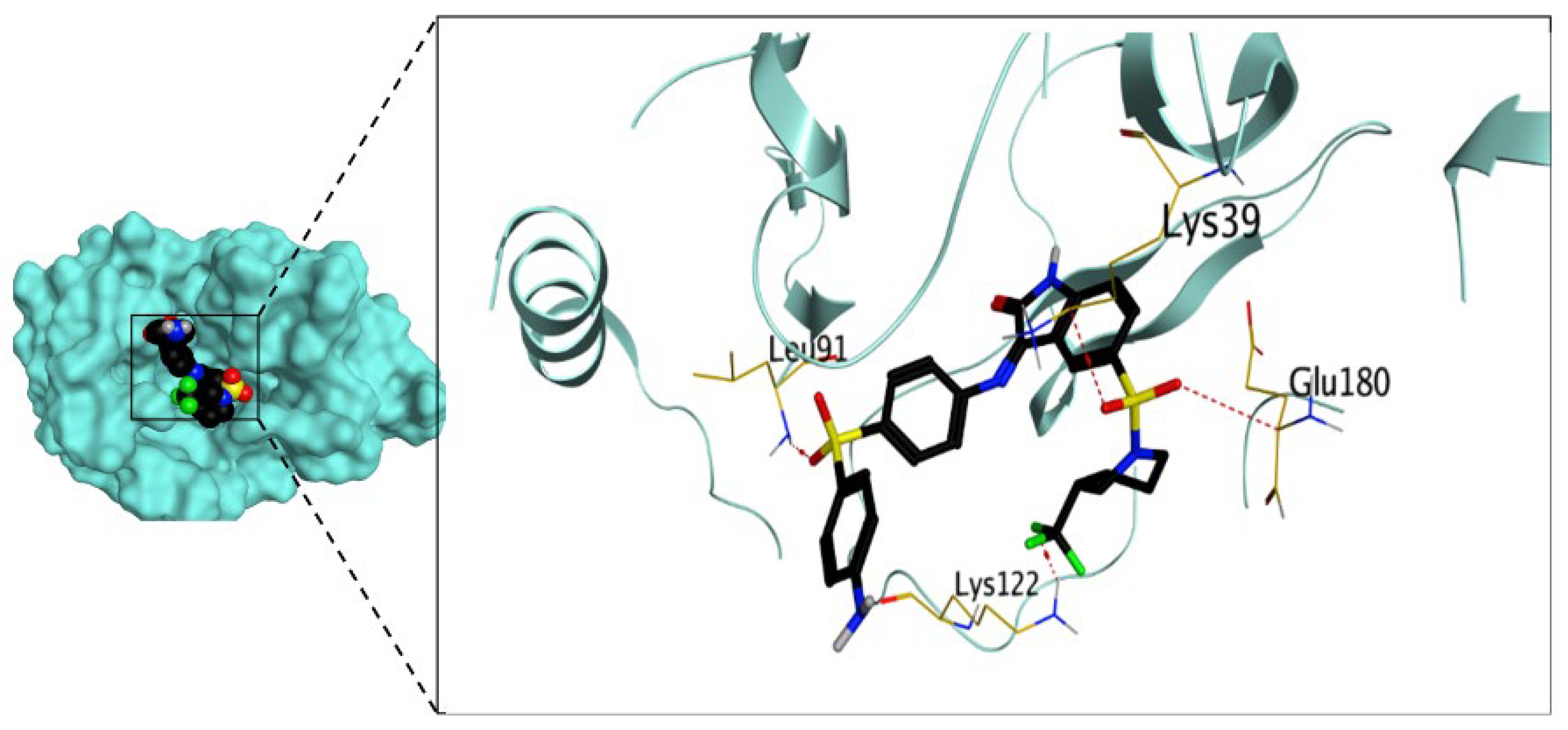
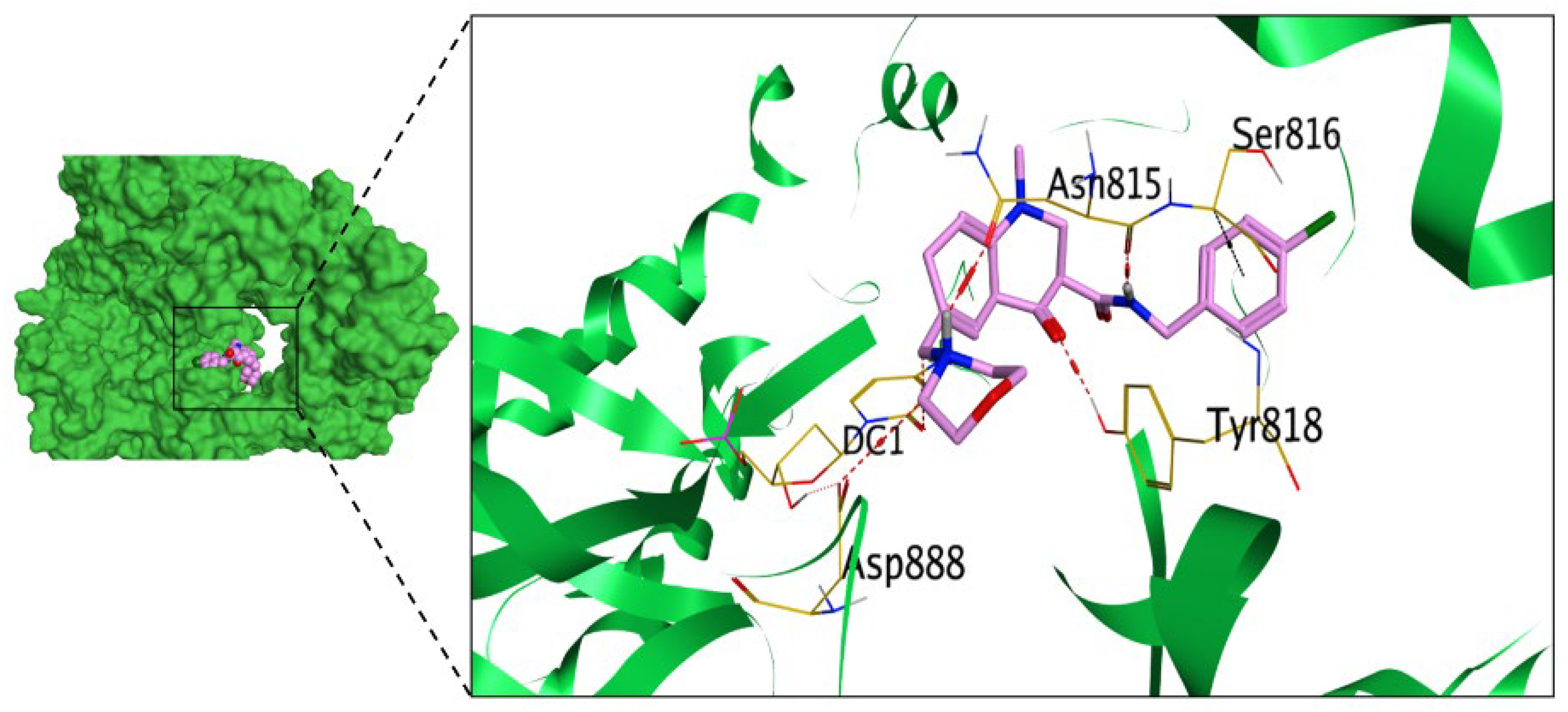
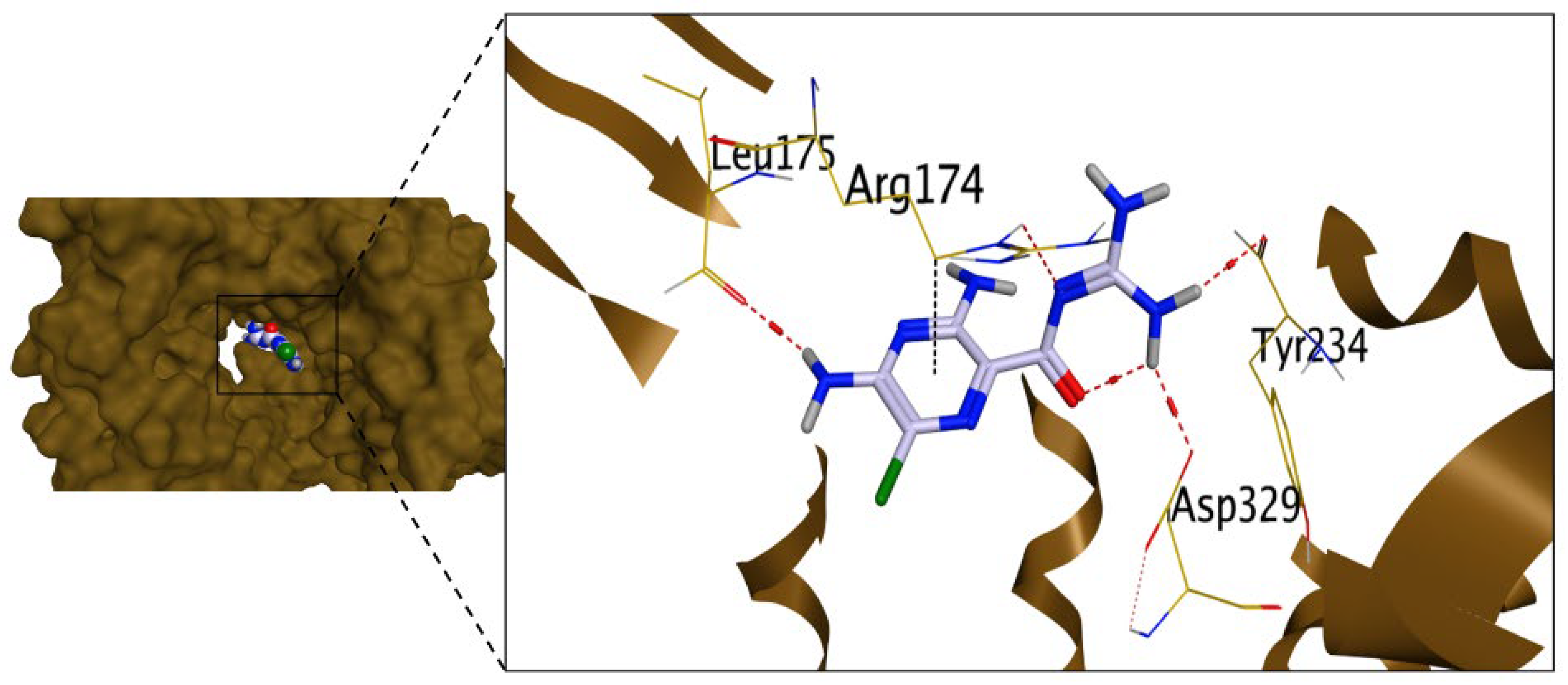
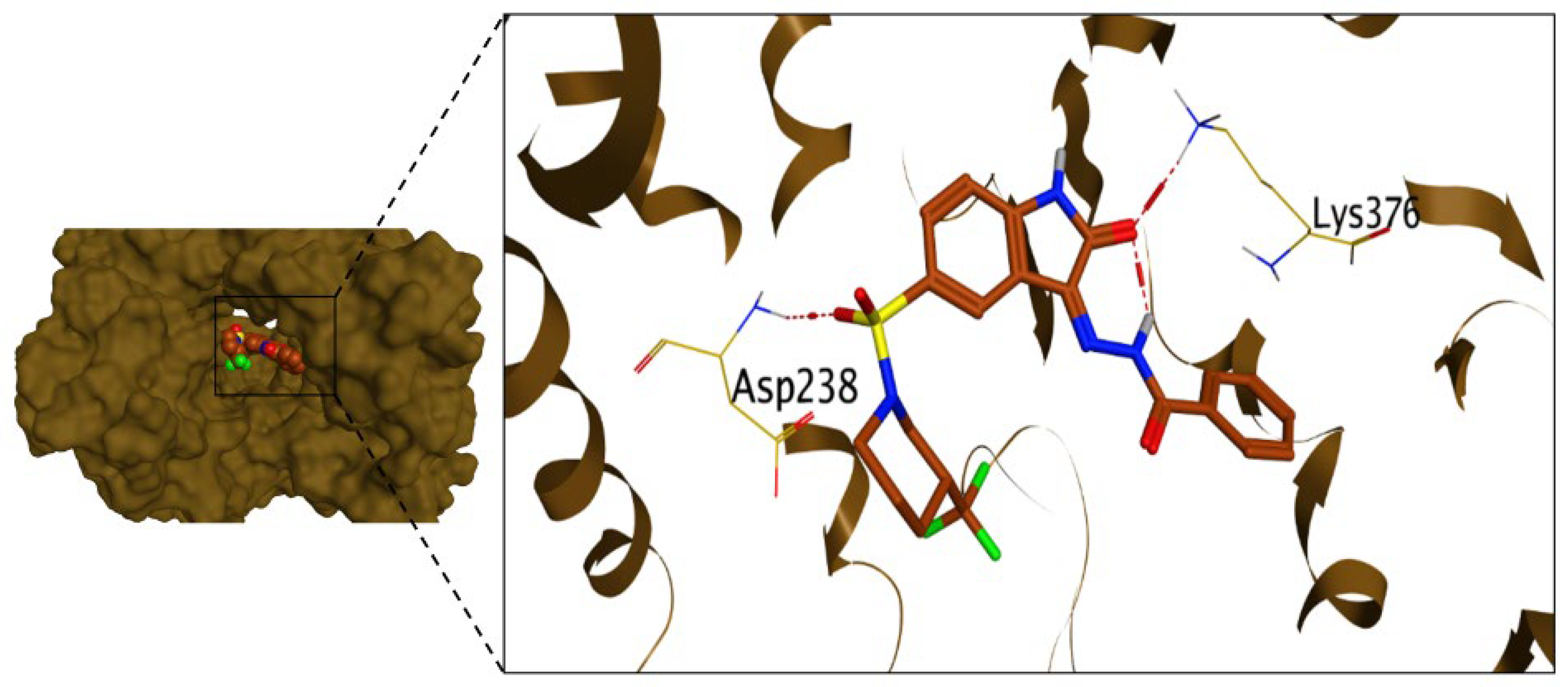
2.3.1. Molecular Docking Studies
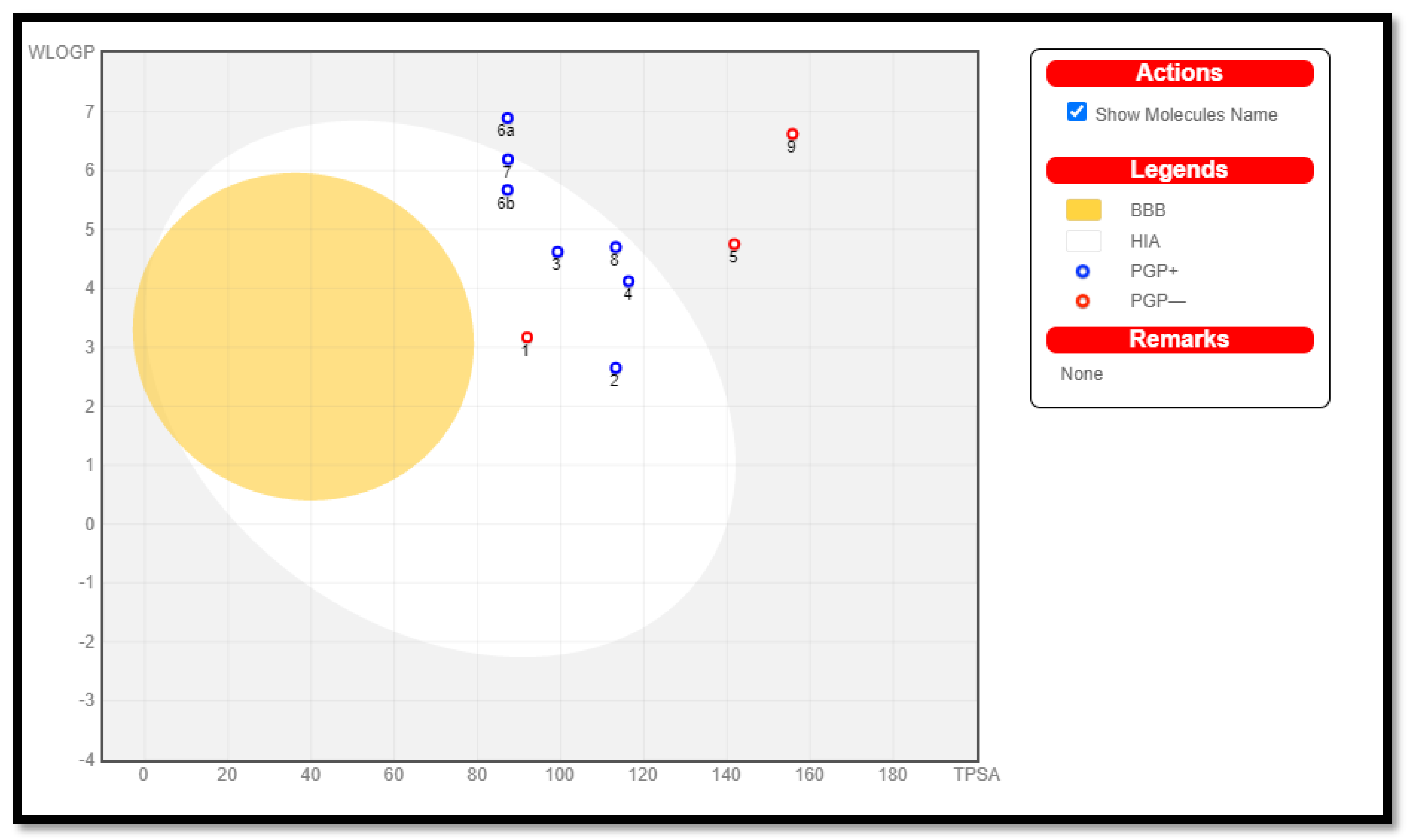
2.3.2. In Silico ADME Predictions
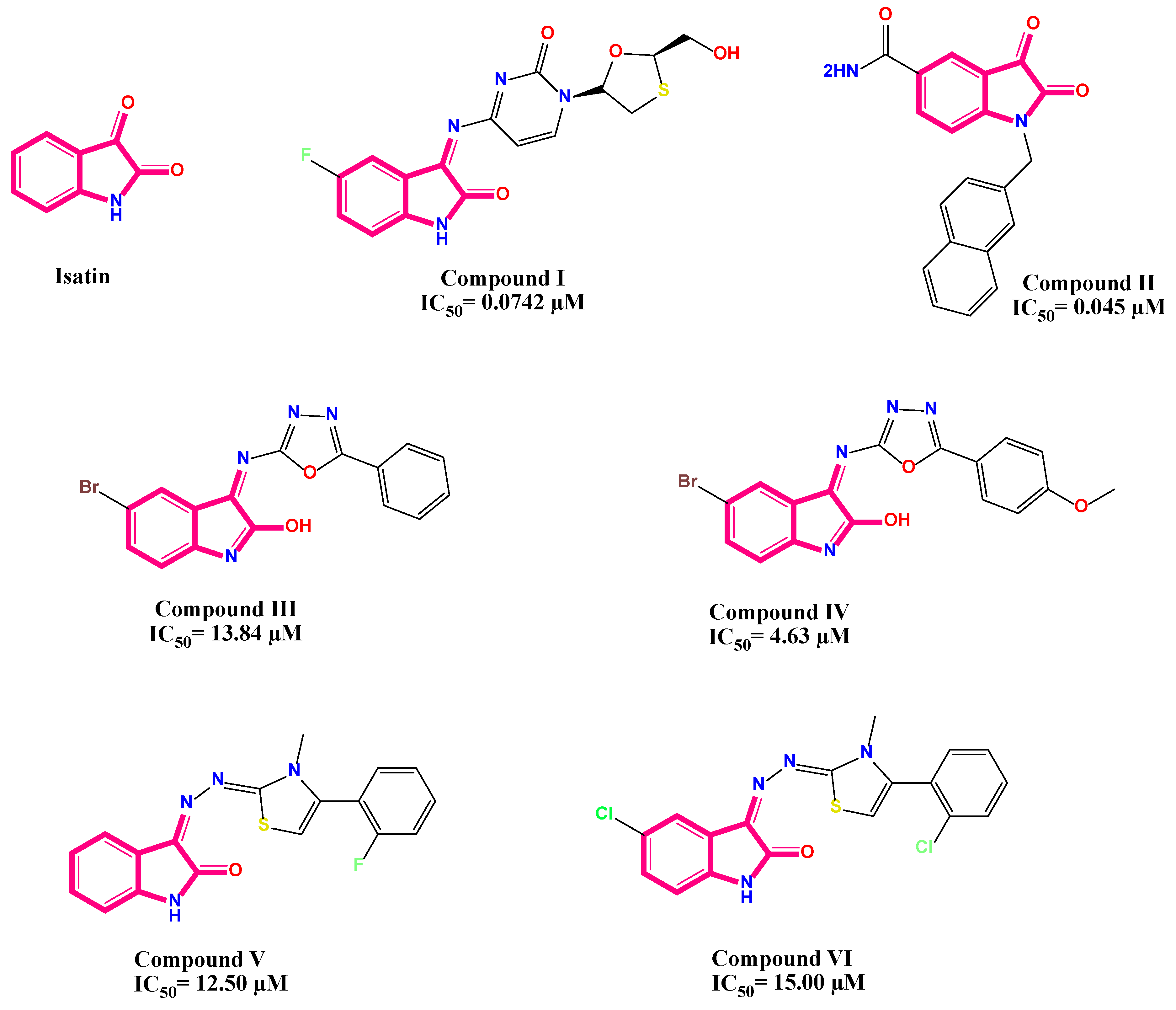
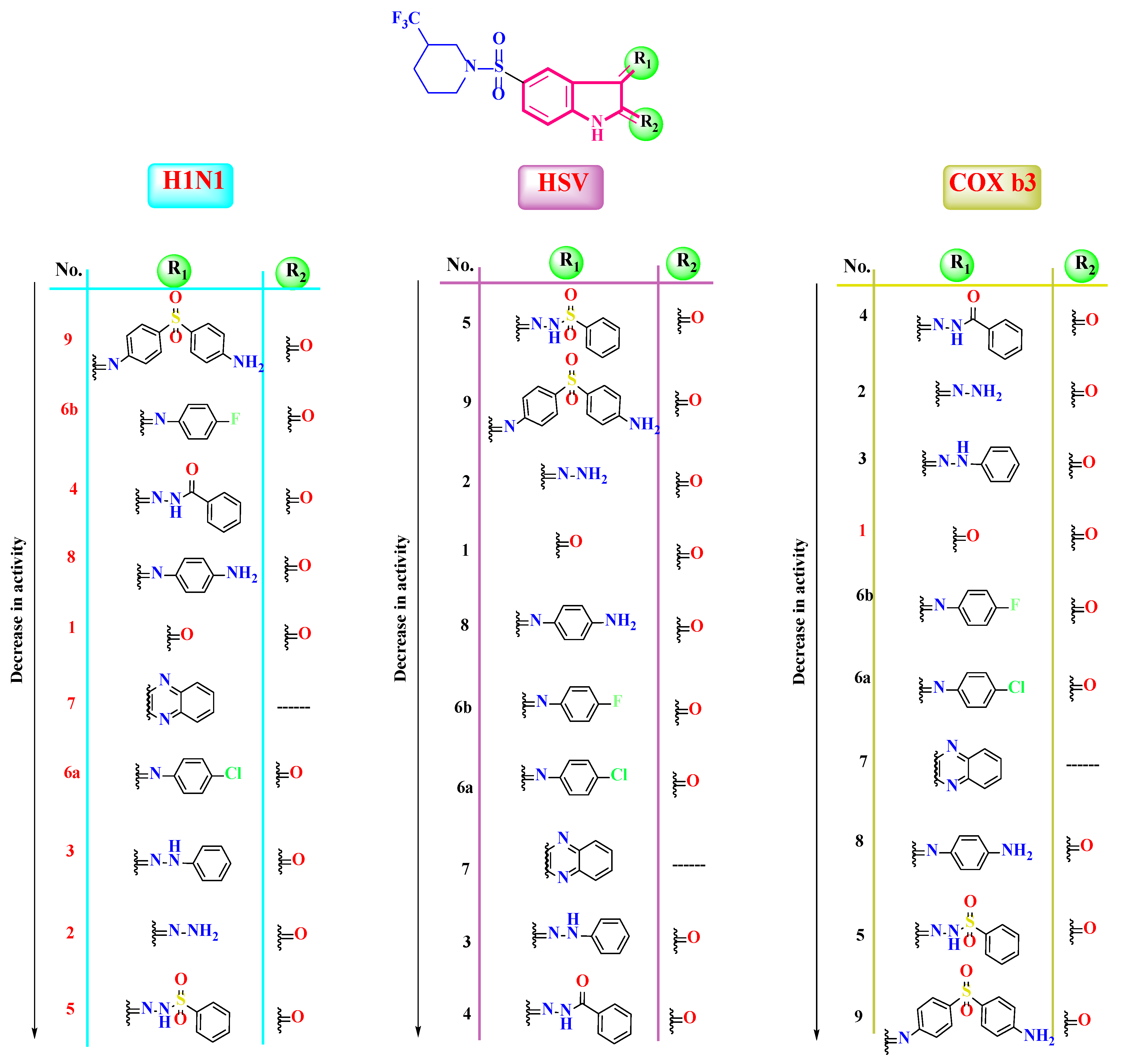
2.4. Structure–Antiviral Activity Relationship
3. Conclusions
4. Materials and Methods
4.1. Chemistry
- 5-(3-(Trifluoromethyl)piperidin-1-ylsulfonyl)isatin (1)
- 3-Hydrazono-5-(3-(trifluoromethyl)piperidin-1-ylsulfonyl)indolin-2-one (2)
- 3-(2-Phenylhydrazono)-5-(3-(trifluoromethyl)piperidin-1-ylsulfonyl)indolin-2-one (3)
- N’-(2-oxo-5-(3-(trifluoromethyl)piperidin-1-ylsulfonyl)indolin-3-ylidene)benzohydrazide (4)
- N’-(2-oxo-5-(3-(trifluoromethyl)piperidin-1-ylsulfonyl)indolin ylidene) benzenesulfonohy drazide (5)
- 3-(Arylimino)-5-(piperidin-1-ylsulfonyl)indolin-2-one (6)
- 3-((4-Chlorophenyl)imino)-5-((3-(trifluoromethyl)piperidin-1-yl)sulfonyl)indolin-2-one (6a)
- 3-((4-Fluorophenyl)imino)-5-((3-(trifluoromethyl)piperidin-1-yl)sulfonyl)indolin-2-one (6b)
- 9-((3-(Trifluoromethyl)piperidin-1-yl)sulfonyl)-6H-indolo[2,3-b]quinoxaline (7)
- 3-((4-Aminophenyl)imino)-5-((3-(trifluoromethyl)piperidin-1-yl)sulfonyl)indolin-2-one (8)
- 3-((4-((4-Aminophenyl)sulfonyl)phenyl)imino)-5-((3-(trifluoromethyl)piperidin-1-yl) sulfonyl)indolin-2-one (9)
4.2. Biological Evaluation
4.2.1. Cell Lines and Viruses
4.2.2. Cytotoxicity Assay
4.2.3. Antiviral Activity Assay (Inhibitory Concentration 50 Detection)
4.2.4. Quantitative PCR (qPCR)
4.3. In Silico Studies
4.3.1. Docking Studies
Molecular Docking
Preparation of the Synthesized Derivatives (1–9)
Preparation of the Virus Polymerase Active Site
4.3.2. In Silico ADME Prediction
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kausar, S.; Said Khan, F.; Ishaq Mujeeb Ur Rehman, M.; Akram, M.; Riaz, M.; Rasool, G.; Hamid Khan, A.; Saleem, I.; Shamim, S.; Malik, A. A review: Mechanism of action of antiviral drugs. Int. J. Immunopathol. Pharmacol. 2021, 35, 20587384211002621. [Google Scholar] [CrossRef]
- Shah, B.; Modi, P.; Sagar, S.R. In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sci. 2020, 252, 117652. [Google Scholar] [CrossRef] [PubMed]
- Harvard University. World Health Statistics 2022. Available online: https://repository.gheli.harvard.edu/repository/11008/ (accessed on 22 March 2023).
- Kondel, R.; Shafiq, N.; Kaur, I.P.; Singh, M.P.; Pandey, A.K.; Ratho, R.K.; Malhotra, S. Effect of acyclovir solid lipid nanoparticles for the treatment of herpes simplex virus (HSV) infection in an animal model of HSV-1 infection. Pharm. Nanotechnol. 2019, 7, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Waisner, H.; Kalamvoki, M. The ICP0 protein of herpes simplex virus 1 (HSV-1) downregulates major autophagy adaptor proteins sequestosome 1 and optineurin during the early stages of HSV-1 infection. J. Virol. 2019, 93, e01258-19. [Google Scholar] [CrossRef]
- Tuddenham, S.; Hamill, M.M.; Ghanem, K.G. Diagnosis and treatment of sexually transmitted infections: A review. JAMA 2022, 327, 161–172. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Park, S.M.; Kim, B.H. Synthesis of 5-isoxazol-5-yl-2′-deoxyuridines exhibiting antiviral activity against HSV and several RNA viruses. Bioorganic Med. Chem. Lett. 2009, 19, 1126–1128. [Google Scholar] [CrossRef]
- Biswas, B.K.; Shin, J.S.; Malpani, Y.R.; Hwang, D.; Jung, E.; Han, S.B.; Vishakantegowda, A.G.; Jung, Y.-S. Enteroviral replication inhibition by N-Alkyl triazolopyrimidinone derivatives through a non-capsid binding mode. Bioorganic Med. Chem. Lett. 2022, 64, 128673. [Google Scholar] [CrossRef] [PubMed]
- Kaga, A.; Katata, Y.; Suzuki, A.; Otani, K.; Watanabe, H.; Kitaoka, S.; Kumaki, S. Perinatal coxsackievirus B3 infection with transient thrombocytopenia. Tohoku J. Exp. Med. 2016, 239, 135–138. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Obadawo, B.; Oyeneyin, O.; Anifowose, M.; Fagbohungbe, K.; Amoko, J. QSAR evaluation of C-8-tert-butyl substituted as potent anti-enterovirus agents. Sci. Lett. 2020, 8, 28–35. [Google Scholar]
- De Clercq, E. Strategies in the design of antiviral drugs. Nat. Rev. Drug Discov. 2002, 1, 13–25. [Google Scholar] [CrossRef]
- Brisse, M.; Vrba, S.M.; Kirk, N.; Liang, Y.; Ly, H. Emerging concepts and technologies in vaccine development. Front. Immunol. 2020, 11, 583077. [Google Scholar] [CrossRef]
- Salas, J.H.; Urbanowicz, R.A.; Guest, J.D.; Frumento, N.; Figueroa, A.; Clark, K.E.; Keck, Z.; Cowton, V.M.; Cole, S.J.; Patel, A.H. An antigenically diverse, representative panel of envelope glycoproteins for hepatitis C virus vaccine development. Gastroenterology 2022, 162, 562–574. [Google Scholar] [CrossRef]
- Heida, R.; Hinrichs, W.L.; Frijlink, H.W. Inhaled vaccine delivery in the combat against respiratory viruses: A 2021 overview of recent developments and implications for COVID-19. Expert Rev. Vaccines 2022, 21, 957–974. [Google Scholar] [CrossRef] [PubMed]
- Chiaretti, A.; Pulitanò, S.; Barone, G.; Ferrara, P.; Romano, V.; Capozzi, D.; Riccardi, R. IL-1β and IL-6 upregulation in children with H1N1 influenza virus infection. Mediat. Inflamm. 2013, 2013, 495848. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.D.; Hu, X.; Ko, E.-J.; Park, S.; Lee, Y.-T.; Orr, M.; Fernandes, J.; Uppal, K.; Kang, S.-M.; Jones, D.P. Metabolic pathways of lung inflammation revealed by high-resolution metabolomics (HRM) of H1N1 influenza virus infection in mice. Am. J. Physiol. -Regul. Integr. Comp. Physiol. 2016, 311, R906–R916. [Google Scholar] [CrossRef] [PubMed]
- Novoa, R.R.; Calderita, G.; Arranz, R.; Fontana, J.; Granzow, H.; Risco, C. Virus factories: Associations of cell organelles for viral replication and morphogenesis. Biol. Cell 2005, 97, 147–172. [Google Scholar] [CrossRef]
- Jones, J.E.; Le Sage, V.; Lakdawala, S.S. Viral and host heterogeneity and their effects on the viral life cycle. Nature Reviews Microbiology 2021, 19, 272–282. [Google Scholar] [CrossRef]
- Staufer, O.; Gantner, G.; Platzman, I.; Tanner, K.; Berger, I.; Spatz, J.P. Bottom-up assembly of viral replication cycles. Nat. Commun. 2022, 13, 6530. [Google Scholar] [CrossRef]
- Reis, E.V.; Damas, B.M.; Mendonça, D.C.; Abrahão, J.S.; Bonjardim, C.A. In-Depth Characterization of the Chikungunya Virus Replication Cycle. J. Virol. 2022, 96, e01732-21. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Cortese, M.; Haselmann, U.; Tabata, K.; Romero-Brey, I.; Funaya, C.; Schieber, N.L.; Qiang, Y.; Bartenschlager, M.; Kallis, S. Spatiotemporal coupling of the hepatitis C virus replication cycle by creating a lipid droplet-proximal membranous replication compartment. Cell Rep. 2019, 27, 3602–3617.e5. [Google Scholar] [CrossRef]
- Pathania, S.; Rawal, R.K.; Singh, P.K. RdRp (RNA-dependent RNA polymerase): A key target providing anti-virals for the management of various viral diseases. J. Mol. Struct. 2022, 1250, 131756. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Chen, C.Z.; Gorshkov, K.; Xu, M.; Lo, D.C.; Zheng, W. RNA-dependent RNA polymerase as a target for COVID-19 drug discovery. SLAS Discov. Adv. Sci. Drug Discov. 2020, 25, 1141–1151. [Google Scholar] [CrossRef]
- Andersen, P.I.; Ianevski, A.; Lysvand, H.; Vitkauskiene, A.; Oksenych, V.; Bjørås, M.; Telling, K.; Lutsar, I.; Dumpis, U.; Irie, Y. Discovery and development of safe-in-man broad-spectrum antiviral agents. Int. J. Infect. Dis. 2020, 93, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Sk, M.F.; Sonawane, A.; Kar, P.; Sadhukhan, S. Plant-derived natural polyphenols as potential antiviral drugs against SARS-CoV-2 via RNA-dependent RNA polymerase (RdRp) inhibition: An in-silico analysis. J. Biomol. Struct. Dyn. 2021, 39, 6249–6264. [Google Scholar] [CrossRef]
- Mouffouk, C.; Mouffouk, S.; Mouffouk, S.; Hambaba, L.; Haba, H. Flavonols as potential antiviral drugs targeting SARS-CoV-2 proteases (3CLpro and PLpro), spike protein, RNA-dependent RNA polymerase (RdRp) and angiotensin-converting enzyme II receptor (ACE2). Eur. J. Pharmacol. 2021, 891, 173759. [Google Scholar] [CrossRef] [PubMed]
- Elsaman, T.; Mohamed, M.S.; Eltayib, E.M.; Abdel-Aziz, H.A.; Abdalla, A.E.; Munir, M.U.; Mohamed, M.A. Isatin derivatives as broad-spectrum antiviral agents: The current landscape. Med. Chem. Res. 2022, 31, 244–273. [Google Scholar] [CrossRef]
- Ilyas, M.; Muhammad, S.; Iqbal, J.; Amin, S.; Al-Sehemi, A.G.; Algarni, H.; Alarfaji, S.S.; Alshahrani, M.Y.; Ayub, K. Insighting isatin derivatives as potential antiviral agents against NSP3 of COVID-19. Chem. Pap. 2022, 76, 6271–6285. [Google Scholar] [CrossRef]
- El-Masry, R.M.; Al-Karmalawy, A.A.; Alnajjar, R.; Mahmoud, S.H.; Mostafa, A.; Kadry, H.H.; Abou-Seri, S.M.; Taher, A.T. Newly synthesized series of oxoindole–oxadiazole conjugates as potential anti-SARS-CoV-2 agents: In silico and in vitro studies. New J. Chem. 2022, 46, 5078–5090. [Google Scholar] [CrossRef]
- Kumar, R.; Gideon, D.A.; Mariadasse, R.; Nirusimhan, V.; Castin, J.; Jeyakanthan, J.; Dhayabaran, V.V. In silico evaluation of isatin-based derivatives with RNA-dependent RNA polymerase of the novel coronavirus SARS-CoV-2. J. Biomol. Struct. Dyn. 2021, 40, 6710–6724. [Google Scholar]
- Konkel, M.J.; Lagu, B.; Boteju, L.W.; Jimenez, H.; Noble, S.; Walker, M.W.; Chandrasena, G.; Blackburn, T.P.; Nikam, S.S.; Wright, J.L. 3-arylimino-2-indolones are potent and selective galanin GAL3 receptor antagonists. J. Med. Chem. 2006, 49, 3757–3758. [Google Scholar] [CrossRef]
- Meleddu, R.; Distinto, S.; Corona, A.; Tramontano, E.; Bianco, G.; Melis, C.; Cottiglia, F.; Maccioni, E. Isatin thiazoline hybrids as dual inhibitors of HIV-1 reverse transcriptase. J. Enzym. Inhib. Med. Chem. 2017, 32, 130–136. [Google Scholar] [CrossRef]
- Pawar, V.; Lokwani, D.; Bhandari, S.; Mitra, D.; Sabde, S.; Bothara, K.; Madgulkar, A. Design of potential reverse transcriptase inhibitor containing Isatin nucleus using molecular modeling studies. Bioorganic Med. Chem. 2010, 18, 3198–3211. [Google Scholar] [CrossRef] [PubMed]
- Sriram, D.; Bal, T.; Yogeeswari, P. Newer aminopyrimidinimino isatin analogues as non-nucleoside HIV-1 reverse transcriptase inhibitors for HIV and other opportunistic infections of AIDS: Design, synthesis and biological evaluation. Il Farm. 2005, 60, 377–384. [Google Scholar] [CrossRef]
- El-Demerdash, A.; Al-Karmalawy, A.A.; Abdel-Aziz, T.M.; Elhady, S.S.; Darwish, K.M.; Hassan, A.H.E. Investigating the structure–activity relationship of marine natural polyketides as promising SARS-CoV-2 main protease inhibitors. RSC Adv. 2021, 11, 31339–31363. [Google Scholar] [CrossRef] [PubMed]
- Elebeedy, D.; Elkhatib, W.F.; Kandeil, A.; Ghanem, A.; Kutkat, O.; Alnajjar, R.; Saleh, M.A.; Abd El Maksoud, A.I.; Badawy, I.; Al-Karmalawy, A.A. Anti-SARS-CoV-2 activities of tanshinone IIA, carnosic acid, rosmarinic acid, salvianolic acid, baicalein, and glycyrrhetinic acid between computational and in vitro insights. RSC Adv. 2021, 11, 29267–29286. [Google Scholar] [CrossRef] [PubMed]
- Elmaaty, A.A.; Alnajjar, R.; Hamed, M.I.; Khattab, M.; Khalifa, M.M.; Al-Karmalawy, A.A. Revisiting activity of some glucocorticoids as a potential inhibitor of SARS-CoV-2 main protease: Theoretical study. RSC Adv. 2021, 11, 10027–10042. [Google Scholar] [CrossRef] [PubMed]
- Khattab, M.; Al-Karmalawy, A.A. Computational repurposing of benzimidazole anthelmintic drugs as potential colchicine binding site inhibitors. Future Med. Chem. 2021, 13, 1623–1638. [Google Scholar] [CrossRef]
- Krämer, S.D.; Wunderli-Allenspach, H. Physicochemical properties in pharmacokinetic lead optimization. Il Farm. 2001, 56, 145–148. [Google Scholar] [CrossRef]
- Neervannan, S. Preclinical formulations for discovery and toxicology: Physicochemical challenges. Expert Opin. Drug Metab. Toxicol. 2006, 2, 715–731. [Google Scholar] [CrossRef]
- Müller, J.; Martins, A.; Csábi, J.; Fenyvesi, F.; Könczöl, Á.; Hunyadi, A.; Balogh, G.T. BBB penetration-targeting physicochemical lead selection: Ecdysteroids as chemo-sensitizers against CNS tumors. Eur. J. Pharm. Sci. 2017, 96, 571–577. [Google Scholar] [CrossRef]
- Doogue, M.P.; Polasek, T.M. The ABCD of clinical pharmacokinetics. Ther. Adv. Drug Saf. 2013, 4, 5–7. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. Prediction of hydrophobic (lipophilic) properties of small organic molecules using fragmental methods: An analysis of ALOGP and CLOGP methods. J. Phys. Chem. A 1998, 102, 3762–3772. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Egan, W.J.; Lauri, G. Prediction of intestinal permeability. Adv. Drug Deliv. Rev. 2002, 54, 273–289. [Google Scholar] [CrossRef] [PubMed]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Daina, A.; Blatter, M.-C.; Baillie Gerritsen, V.; Palagi, P.M.; Marek, D.; Xenarios, I.; Schwede, T.; Michielin, O.; Zoete, V. Drug Design Workshop: A web-based educational tool to introduce computer-aided drug design to the general public. J. Chem. Educ. 2017, 94, 335–344. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. iLOGP: A simple, robust, and efficient description of n-octanol/water partition coefficient for drug design using the GB/SA approach. J. Chem. Inf. Model. 2014, 54, 3284–3301. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 2016, 11, 1117. [Google Scholar] [CrossRef] [PubMed]
- Members, S.S.I.o.B. The SIB Swiss Institute of Bioinformatics’ resources: Focus on curated databases. Nucleic Acids Res. 2016, 44, D27–D37. [Google Scholar]
- Azam, F.; Madi, A.M.; Ali, H.I. Molecular Docking and Prediction of Pharmacokinetic Properties of Dual Mechanism Drugs that Block MAO-B and Adenosine A2A Receptors for the Treatment of Parkinson’s Disease. J. Young Pharm. 2012, 4, 184–192. [Google Scholar] [CrossRef]
- Zhao, Y.H.; Abraham, M.H.; Le, J.; Hersey, A.; Luscombe, C.N.; Beck, G.; Sherborne, B.; Cooper, I. Rate-limited steps of human oral absorption and QSAR studies. Pharm. Res. 2002, 19, 1446–1457. [Google Scholar] [CrossRef]
- Geinoz, S.; Guy, R.H.; Testa, B.; Carrupt, P.-A. Quantitative structure-permeation relationships (QSPeRs) to predict skin permeation: A critical evaluation. Pharm. Res. 2004, 21, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Delaney, J.S. ESOL: Estimating aqueous solubility directly from molecular structure. J. Chem. Inf. Comput. Sci. 2004, 44, 1000–1005. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Feoktistova, M.; Geserick, P.; Leverkus, M. Crystal violet assay for determining viability of cultured cells. Cold Spring Harb. Protoc. 2016, 2016, prot087379. [Google Scholar] [CrossRef]
- Mostafa, A.; Kandeil, A.; AMM Elshaier, Y.; Kutkat, O.; Moatasim, Y.; Rashad, A.A.; Shehata, M.; Gomaa, M.R.; Mahrous, N.; Mahmoud, S.H. FDA-approved drugs with potent in vitro antiviral activity against severe acute respiratory syndrome coronavirus 2. Pharmaceuticals 2020, 13, 443. [Google Scholar] [CrossRef]
- Aurelius, E.; Johansson, B.; Sköldenberg, B.; Forsgren, M. Encephalitis in immunocompetent patients due to herpes simplex virus type 1 or 2 as determined by type-specific polymerase chain reaction and antibody assays of cerebrospinal fluid. J. Med. Virol. 1993, 39, 179–186. [Google Scholar] [CrossRef]
- Leparc, I.; Aymard, M.; Fuchs, F. Acute, chronic and persistent enterovirus and poliovirus infections: Detection of viral genome by seminested PCR amplification in culture-negative samples. Mol. Cell. Probes 1994, 8, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chemical Computing Group Inc. Molecular Operating Environment (MOE). 2016. Available online: https://www.chemcomp.com (accessed on 20 March 2023).
- Elmaaty, A.A.; Darwish, K.M.; Khattab, M.; Elhady, S.S.; Salah, M.; Hamed, M.I.; Al-Karmalawy, A.A.; Saleh, M.M. In a search for potential drug candidates for combating COVID-19: Computational study revealed salvianolic acid B as a potential therapeutic targeting 3CLpro and spike proteins. J. Biomol. Struct. Dyn. 2021, 40, 8866–8893. [Google Scholar] [CrossRef]
- Abo Elmaaty, A.; Hamed, M.I.; Ismail, M.I.; Elkaeed, E.B.; Abulkhair, H.S.; Khattab, M.; Al-Karmalawy, A.A. Computational insights on the potential of some NSAIDs for treating COVID-19: Priority set and lead optimization. Molecules 2021, 26, 3772. [Google Scholar] [CrossRef]
- Hamed, M.I.; Darwish, K.M.; Soltane, R.; Chrouda, A.; Mostafa, A.; Shama, N.M.A.; Elhady, S.S.; Abulkhair, H.S.; Khodir, A.E.; Elmaaty, A.A. β-Blockers bearing hydroxyethylamine and hydroxyethylene as potential SARS-CoV-2 Mpro inhibitors: Rational based design, in silico, in vitro, and SAR studies for lead optimization. RSC Adv. 2021, 11, 35536–35558. [Google Scholar] [CrossRef]
- Elmaaty, A.A.; Darwish, K.M.; Chrouda, A.; Boseila, A.A.; Tantawy, M.A.; Elhady, S.S.; Shaik, A.B.; Mustafa, M.; Al-Karmalawy, A.A. In silico and in vitro studies for benzimidazole anthelmintics repurposing as VEGFR-2 antagonists: Novel mebendazole-loaded mixed micelles with enhanced dissolution and anticancer activity. ACS Omega 2021, 7, 875–899. [Google Scholar] [CrossRef] [PubMed]
- Elebeedy, D.; Badawy, I.; Elmaaty, A.A.; Saleh, M.M.; Kandeil, A.; Ghanem, A.; Kutkat, O.; Alnajjar, R.; Abd El Maksoud, A.I.; Al-Karmalawy, A.A. In vitro and computational insights revealing the potential inhibitory effect of Tanshinone IIA against influenza A virus. Comput. Biol. Med. 2022, 141, 105149. [Google Scholar] [CrossRef] [PubMed]
- Hammoud, M.M.; Nageeb, A.S.; Morsi, M.; Gomaa, E.A.; Elmaaty, A.A.; Al-Karmalawy, A.A. Design, synthesis, biological evaluation, and SAR studies of novel cyclopentaquinoline derivatives as DNA intercalators, topoisomerase II inhibitors, and apoptotic inducers. New J. Chem. 2022, 46, 11422–11436. [Google Scholar] [CrossRef]
- Hammouda, M.M.; Elmaaty, A.A.; Nafie, M.S.; Abdel-Motaal, M.; Mohamed, N.S.; Tantawy, M.A.; Belal, A.; Alnajjar, R.; Eldehna, W.M.; Al-Karmalawy, A.A. Design and Synthesis of Novel Benzoazoninone Derivatives as Potential CBSIs and Apoptotic Inducers: In Vitro, In Vivo, Molecular Docking, Molecular Dynamics, and SAR Studies. Bioorganic Chem. 2022, 127, 105995. [Google Scholar] [CrossRef]
- Fudo, S.; Yamamoto, N.; Nukaga, M.; Odagiri, T.; Tashiro, M.; Hoshino, T. Two distinctive binding modes of endonuclease inhibitors to the N-terminal region of influenza virus polymerase acidic subunit. Biochemistry 2016, 55, 2646–2660. [Google Scholar] [CrossRef]
- Hayes, R.P.; Heo, M.R.; Mason, M.; Reid, J.; Burlein, C.; Armacost, K.A.; Tellers, D.M.; Raheem, I.; Shaw, A.W.; Murray, E. Structural understanding of non-nucleoside inhibition in an elongating herpesvirus polymerase. Nat. Commun. 2021, 12, 3040. [Google Scholar] [CrossRef]
- Campagnola, G.; Weygandt, M.; Scoggin, K.; Peersen, O. Crystal structure of coxsackievirus B3 3Dpol highlights the functional importance of residue 5 in picornavirus polymerases. J. Virol. 2008, 82, 9458–9464. [Google Scholar] [CrossRef] [PubMed]
- Gazina, E.V.; Smidansky, E.D.; Holien, J.K.; Harrison, D.N.; Cromer, B.A.; Arnold, J.J.; Parker, M.W.; Cameron, C.E.; Petrou, S. Amiloride is a competitive inhibitor of coxsackievirus B3 RNA polymerase. J. Virol. 2011, 85, 10364–10374. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Saleh, M.A.; Elmaaty, A.A.; El Saeed, H.S.; Saleh, M.M.; Salah, M.; Eldin, R.R.E. Structure based design and synthesis of 3-(7-nitro-3-oxo-3, 4-dihydroquinoxalin-2-yl) propanehydrazide derivatives as novel bacterial DNA-gyrase inhibitors: In-vitro, In-vivo, In-silico and SAR studies. Bioorganic Chem. 2022, 129, 106186. [Google Scholar] [CrossRef] [PubMed]


| Samples | Gene Being Tested Experimental (TE) (CT) | Gene Being Tested Control (TC) (CT) | House-Keeping Gene Experimental (HE) (CT) | House-Keeping Gene Control (HC) (CT) | ΔCt Values for the Experimental (ΔCTE) | ΔCt Values for the Control (ΔCTC) | Delta Ct Value (ΔΔCt) | 2−ΔΔCt (Expression Fold Change) Fold Expression Level in the Experimental Condition the Expression as in the Control Condition | |
|---|---|---|---|---|---|---|---|---|---|
| H1N1 | V H1N1 | 27 | 22.4 | 26.9 | 21.2 | 0.1 | 1.2 | −1.1 | 2.1 |
| C H1N1 | 28 | 22.4 | 26.8 | 21.2 | 1.2 | 1.2 | 0 | 1.0 | |
| Comp. 1 | 27 | 22.4 | 26.7 | 21.2 | 0.3 | 1.2 | −0.9 | 1.9 | |
| Comp. 2 | 27.1 | 22.4 | 26.7 | 21.2 | 0.4 | 1.2 | −0.8 | 1.7 | |
| Comp. 6b | 27.1 | 22.4 | 26.9 | 21.2 | 0.2 | 1.2 | −1 | 2.0 | |
| Comp. 6a | 27.3 | 22.4 | 26.7 | 21.2 | 0.6 | 1.2 | −0.6 | 1.5 | |
| HSV-1 | V HSV | 27.4 | 22.6 | 26.7 | 21 | 0.7 | 1.6 | −0.9 | 1.9 |
| C HSV | 28 | 22.6 | 26.4 | 21 | 1.6 | 1.6 | 0 | 1.0 | |
| Comp. 1 | 27.5 | 22.6 | 26.7 | 21 | 0.8 | 1.6 | −0.8 | 1.7 | |
| Comp. 2 | 27.6 | 22.6 | 26.6 | 21 | 1 | 1.6 | −0.6 | 1.5 | |
| Comp. 6b | 27.4 | 22.6 | 26.6 | 21 | 0.8 | 1.6 | −0.8 | 1.7 | |
| Comp. 6a | 27.7 | 22.6 | 26.5 | 21 | 1.2 | 1.6 | −0.4 | 1.3 | |
| COX-B3 | V COX | 24.4 | 22 | 24.6 | 21.4 | −0.2 | 0.6 | −0.8 | 1.7 |
| C COX | 25.6 | 22 | 25 | 21.4 | 0.6 | 0.6 | 0 | 1.0 | |
| Comp. 1 | 24.8 | 22 | 24.7 | 21.4 | 0.1 | 0.6 | −0.5 | 1.4 | |
| Comp. 2 | 24.4 | 22 | 24.4 | 21.4 | 0 | 0.6 | −0.6 | 1.5 | |
| Comp. 6b | 24.5 | 22 | 24.5 | 21.4 | 0 | 0.6 | −0.6 | 1.5 | |
| Comp. 6a | 24.9 | 22 | 24.6 | 21.4 | 0.3 | 0.6 | −0.3 | 1.2 | |
| Comp. No. | iLog P | Molar Refractivity | Log S | WLOGP | TPSA (Ǻ2) | % ABS | GI Absorption | P-gp Substrate | Log Kp |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1.56 | 84.17 | MS | 3.17 | 91.93 | 77.28 | High | No | −7.29 |
| 2 | 1.64 | 90.14 | S | 2.65 | 113.24 | 69.93 | High | Yes | −7.03 |
| 3 | 1.95 | 116.26 | PS | 4.62 | 99.25 | 74.75 | High | Yes | −6.31 |
| 4 | 1.90 | 119.95 | PS | 4.12 | 116.32 | 68.86 | High | Yes | −6.59 |
| 5 | 1.84 | 122.85 | PS | 4.75 | 141.77 | 60.08 | Low | No | −7.03 |
| 6a | 2.88 | 117.49 | PS | 5.76 | 87.22 | 78.90 | High | No | −6.20 |
| 6b | 2.12 | 112.44 | PS | 5.67 | 87.22 | 78.90 | High | Yes | −6.48 |
| 7 | 2.78 | 110.92 | PS | 6.19 | 87.33 | 78.87 | High | Yes | −6.04 |
| 8 | 2.18 | 116.89 | PS | 4.70 | 113.24 | 69.93 | High | Yes | −7.02 |
| 9 | 2.61 | 148.82 | PS | 6.62 | 155.76 | 55.26 | Low | No | −7.30 |
| Comp. No | Mol. Weight [g/mol] | Lipophilicity (MLogP) | H-Bond Donors | H-Bond Acceptors | Lipinski Violations | Ghose Viol. | Veber Viol. | Egan Viol. | Muegge Viol. | Bioavailability Score |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 362.32 | 0.70 | 1 | 8 | Yes; 0 | Yes | Yes | Yes | Yes | 0.55 |
| 2 | 376.35 | 0.72 | 2 | 8 | Yes; 0 | Yes | Yes | Yes | Yes | 0.55 |
| 3 | 452.45 | 2.15 | 2 | 8 | Yes; 0 | Yes | Yes | Yes | Yes | 0.55 |
| 4 | 480.46 | 1.92 | 2 | 9 | Yes; 0 | No; 1 | Yes | Yes | Yes | 0.55 |
| 5 | 516.51 | 1.26 | 2 | 10 | Yes; 1 | No; 1 | No; 1 | No; 1 | Yes | 0.55 |
| 6a | 471.88 | 2.63 | 1 | 8 | Yes; 0 | No; 1 | Yes | Yes | Yes | 0.55 |
| 6b | 455.43 | 2.52 | 1 | 9 | Yes; 0 | No; 1 | Yes | Yes | Yes | 0.55 |
| 7 | 434.43 | 2.80 | 1 | 8 | Yes; 0 | No; 1 | Yes | No; 1 | Yes | 0.55 |
| 8 | 452.45 | 1.61 | 2 | 8 | Yes; 0 | Yes | Yes | Yes | Yes | 0.55 |
| 9 | 592.61 | 2.29 | 2 | 10 | Yes; 1 | No; 3 | No; 1 | No; 2 | No; 1 | 0.55 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ezz Eldin, R.R.; Saleh, M.A.; Alwarsh, S.A.; Rushdi, A.; Althoqapy, A.A.; El Saeed, H.S.; Abo Elmaaty, A. RETRACTED: Design and Synthesis of Novel 5-((3-(Trifluoromethyl)piperidin-1-yl)sulfonyl)indoline-2,3-dione Derivatives as Promising Antiviral Agents: In Vitro, In Silico, and Structure–Activity Relationship Studies. Pharmaceuticals 2023, 16, 1247. https://doi.org/10.3390/ph16091247
Ezz Eldin RR, Saleh MA, Alwarsh SA, Rushdi A, Althoqapy AA, El Saeed HS, Abo Elmaaty A. RETRACTED: Design and Synthesis of Novel 5-((3-(Trifluoromethyl)piperidin-1-yl)sulfonyl)indoline-2,3-dione Derivatives as Promising Antiviral Agents: In Vitro, In Silico, and Structure–Activity Relationship Studies. Pharmaceuticals. 2023; 16(9):1247. https://doi.org/10.3390/ph16091247
Chicago/Turabian StyleEzz Eldin, Rogy R., Marwa A. Saleh, Sefat A. Alwarsh, Areej Rushdi, Azza Ali Althoqapy, Hoda S. El Saeed, and Ayman Abo Elmaaty. 2023. "RETRACTED: Design and Synthesis of Novel 5-((3-(Trifluoromethyl)piperidin-1-yl)sulfonyl)indoline-2,3-dione Derivatives as Promising Antiviral Agents: In Vitro, In Silico, and Structure–Activity Relationship Studies" Pharmaceuticals 16, no. 9: 1247. https://doi.org/10.3390/ph16091247
APA StyleEzz Eldin, R. R., Saleh, M. A., Alwarsh, S. A., Rushdi, A., Althoqapy, A. A., El Saeed, H. S., & Abo Elmaaty, A. (2023). RETRACTED: Design and Synthesis of Novel 5-((3-(Trifluoromethyl)piperidin-1-yl)sulfonyl)indoline-2,3-dione Derivatives as Promising Antiviral Agents: In Vitro, In Silico, and Structure–Activity Relationship Studies. Pharmaceuticals, 16(9), 1247. https://doi.org/10.3390/ph16091247







