Design of Tetra-Peptide Ligands of Antibody Fc Regions Using In Silico Combinatorial Library Screening
Abstract
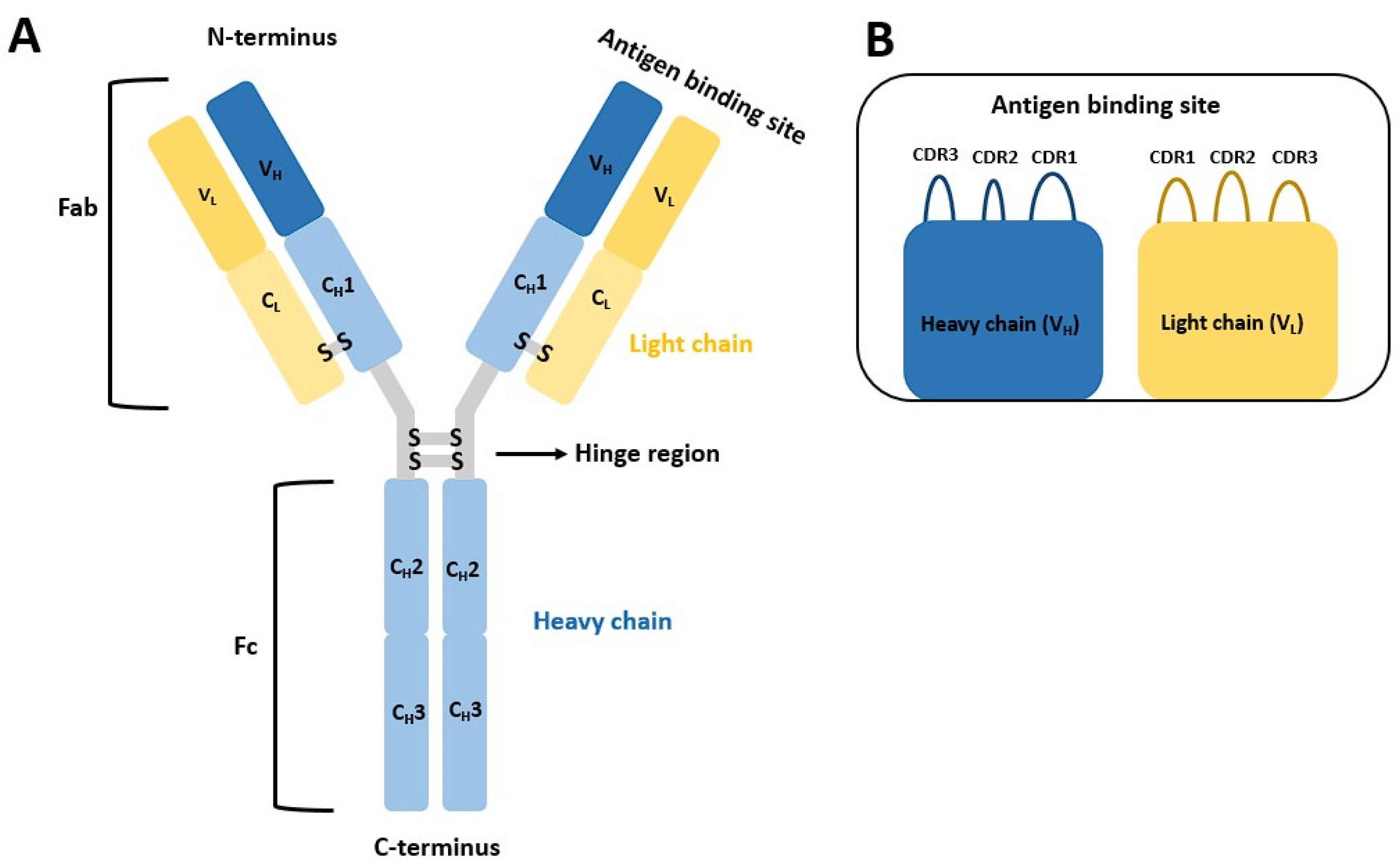
:1. Introduction
| Software Name | Type | Description |
|---|---|---|
| pepATTRACT [37] | Global docking | Web server for blind large-scale peptide–protein docking |
| MDockPeP [38] | Global docking | Protein–peptide docking server, that uses ab-initio methods to generate the peptide from sequence and docks it to the receptor structure |
| GalaxyPepDock [39] | Template-based | A protein–peptide docking tool based on interaction similarity and energy optimization |
| PIPER-FlexPepDock [40] | Global docking | High-resolution peptide–protein docking using a fragment-based approach, that is founded on the Rosseta fragment picker |
| CABS-Dock [41] | Global docking | Standalone web server for flexible docking of peptides to proteins without prior knowledge of the binding site |
| HPEPDOCK [42] | Global docking | A web server for blind protein–peptide docking through a hierarchical algorithm |
| ClusPro PeptiDock [43] | Global docking | A web server for protein–protein docking with efficient global docking of peptide recognition motifs using fast Fourier transform. |
| rDock [32] | Local docking | Small molecule docking program, suitable for docking 6–10 amino-acid residue peptides |
| CmDock | Local docking | A versatile open source fork of the small molecule docking program rDock, suitable for docking various ligands to proteins and nucleic acids |
| ZDOCK server [44] | Global docking | A protein-docking server for the prediction of protein–protein complex structures and symmetric multimers, based on the rigid-body docking programs ZDOCK and M-ZDOCK |
| FRODOCK [45] | Global docking | Flexible and fast rotational protein–protein docking |
| HawkDock [46] | Global docking | A web server * to predict and analyze a given protein–protein complex based on computational docking using the ATTRACT docking algorithm, the HawkRank scoring function and MM/GBSA free energy decomposition for key amino-acid residues. |
| DINC [47] | Local docking | Auto-dock adapted protocol for docking large ligands |
| Rosseta FlexPepDock [48] | Global docking | An ab initio approach to simultaneous folding, docking and refinement of peptides onto their receptors |
| AutoDock CrankPep [49] | Local docking | Flexible peptide docking to rigid receptors based on folding and docking |
| PeptoGrid for AutoDock [50] | Local docking | Rescoring function for AutoDock based on frequency information of ligand atoms |
| DynaDock [51] | Local docking | Molecular dynamics based algorithm for flexible protein–peptide docking |
| GOLD [52] | Local docking | Docking software based on genetic algorithm for flexible ligand docking |
| Surflex [53] | Local docking | Flexible molecular docking software based on a molecular similarity-based search engine |
2. Results
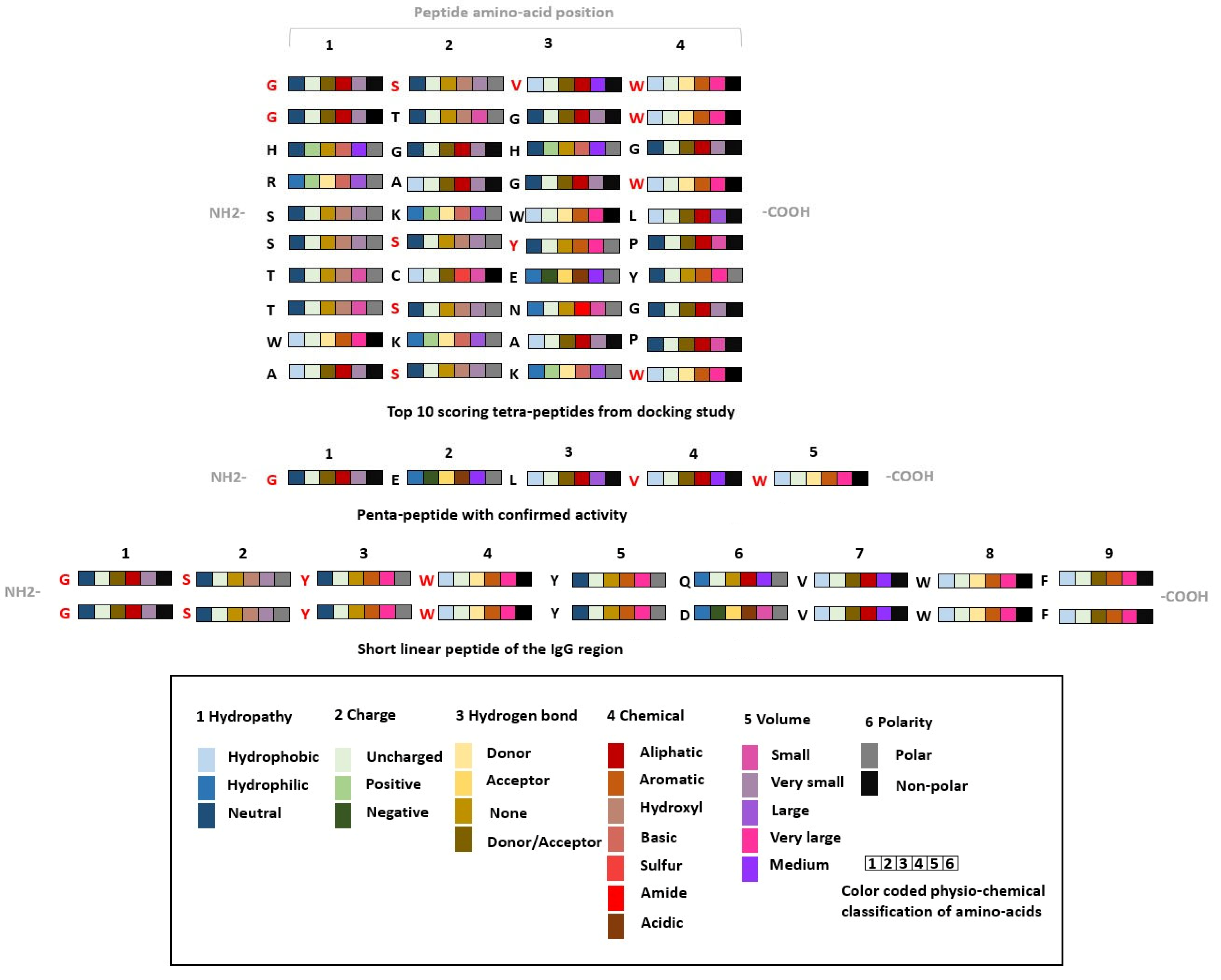
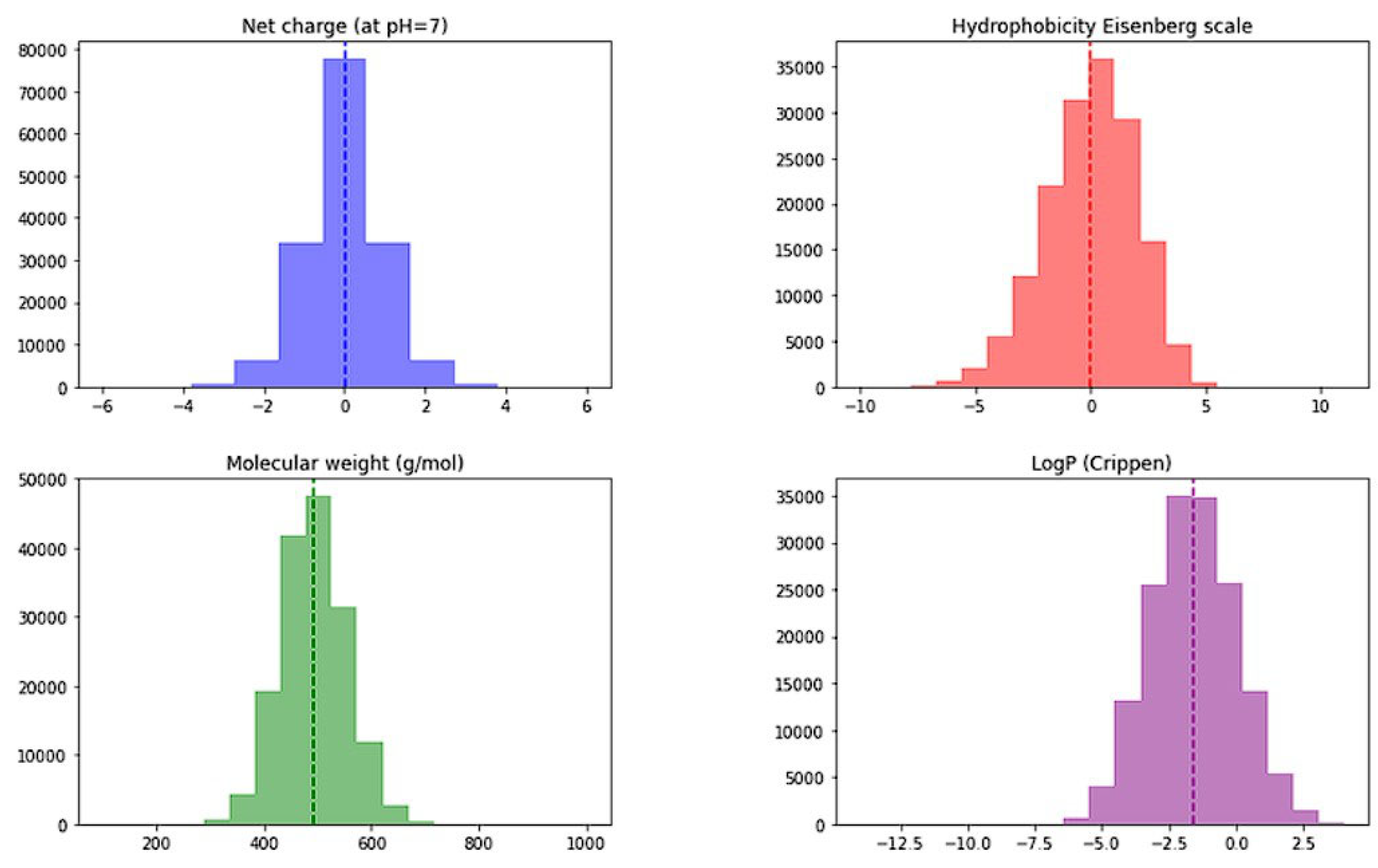
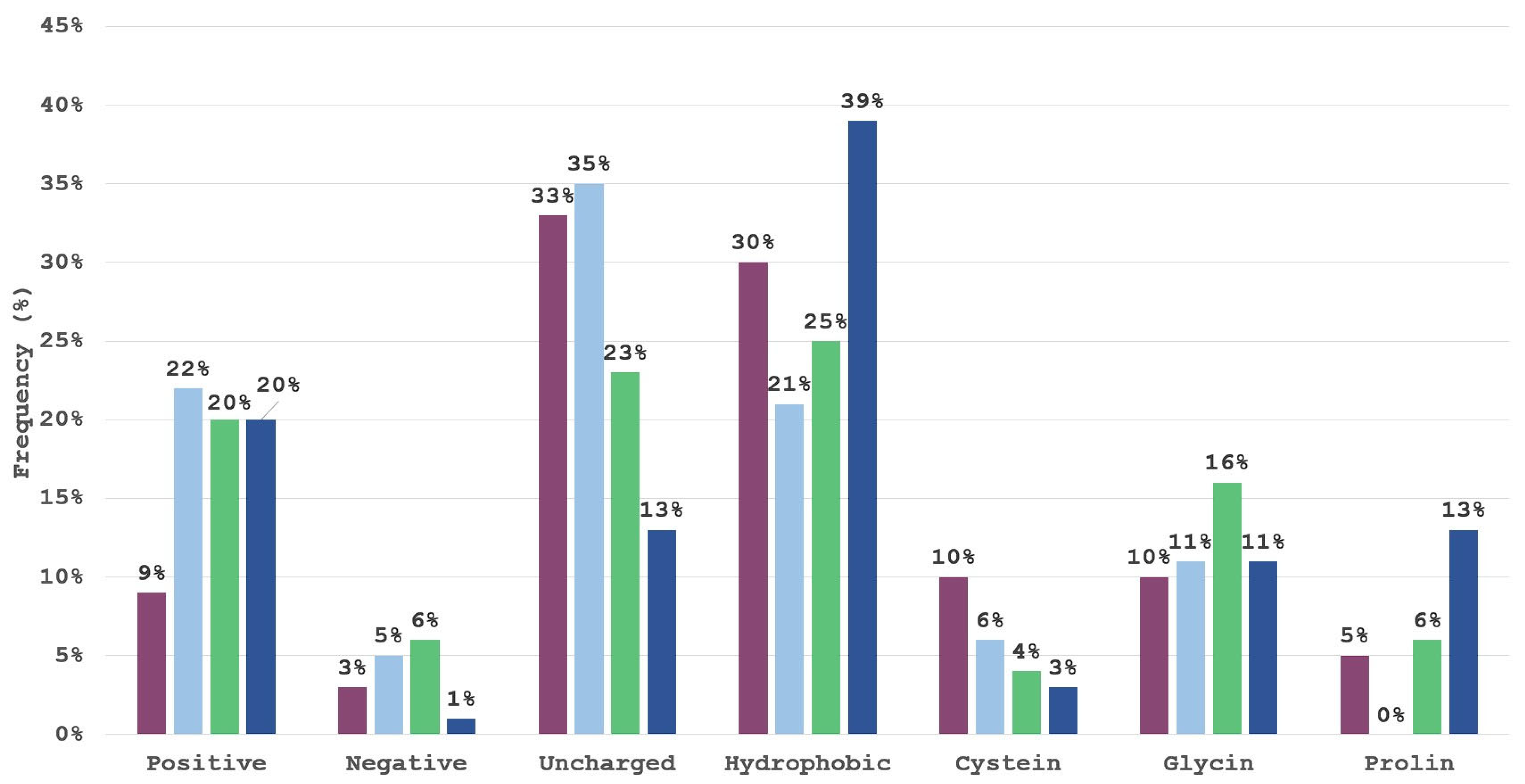
2.1. Library Preparation
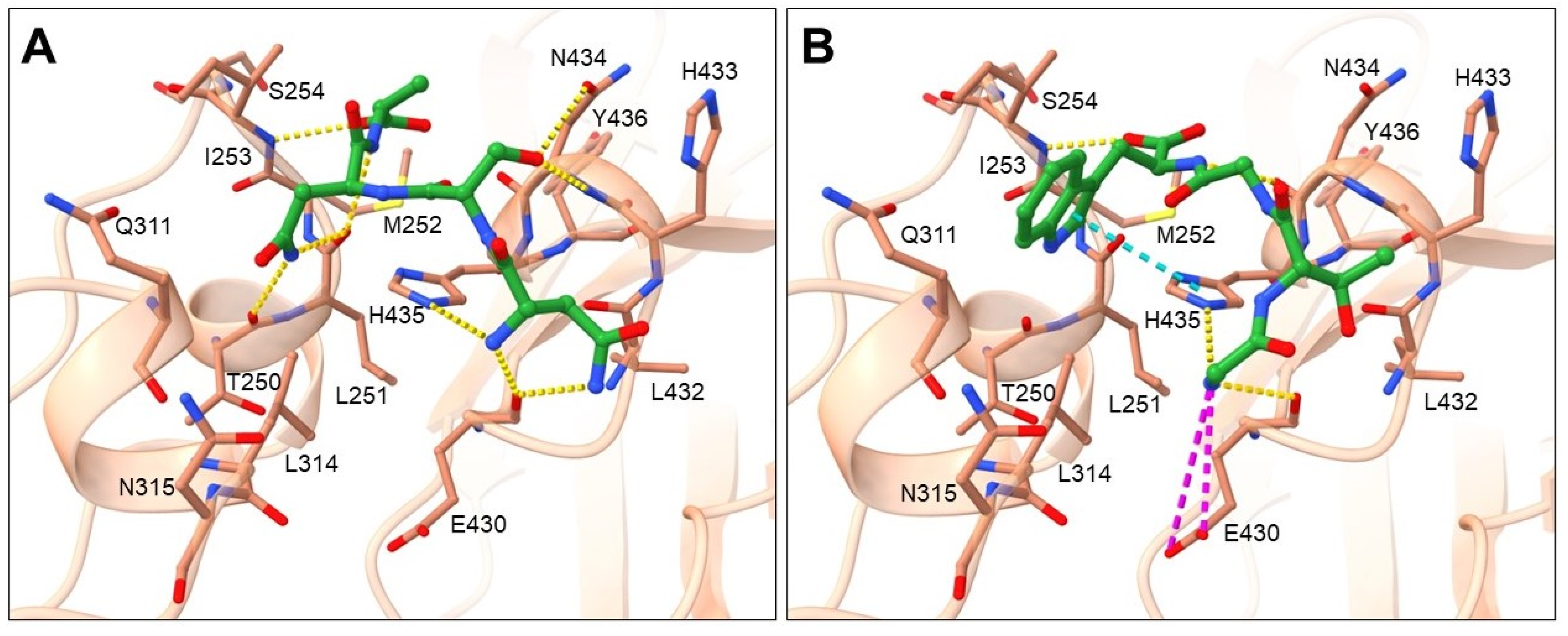
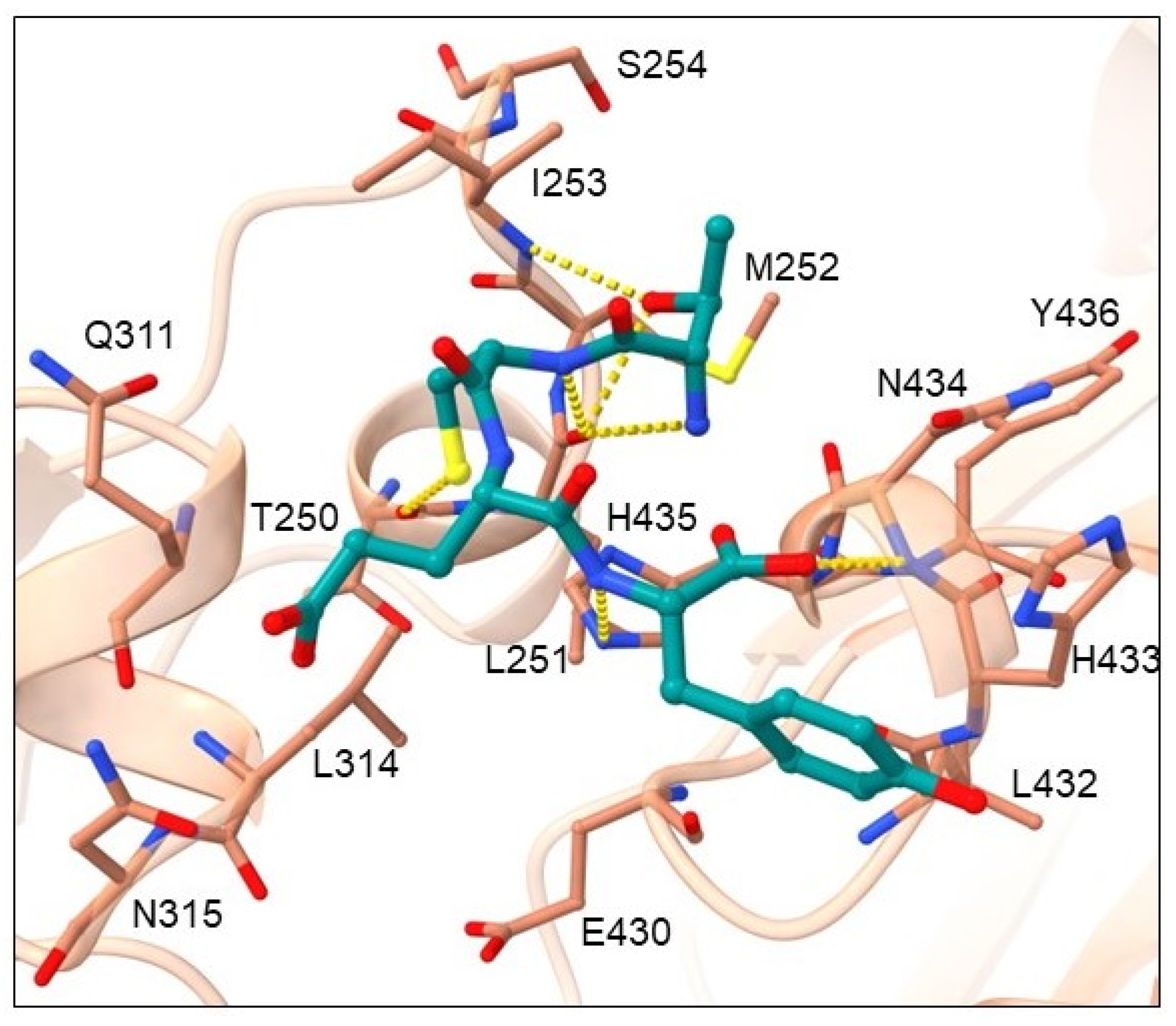
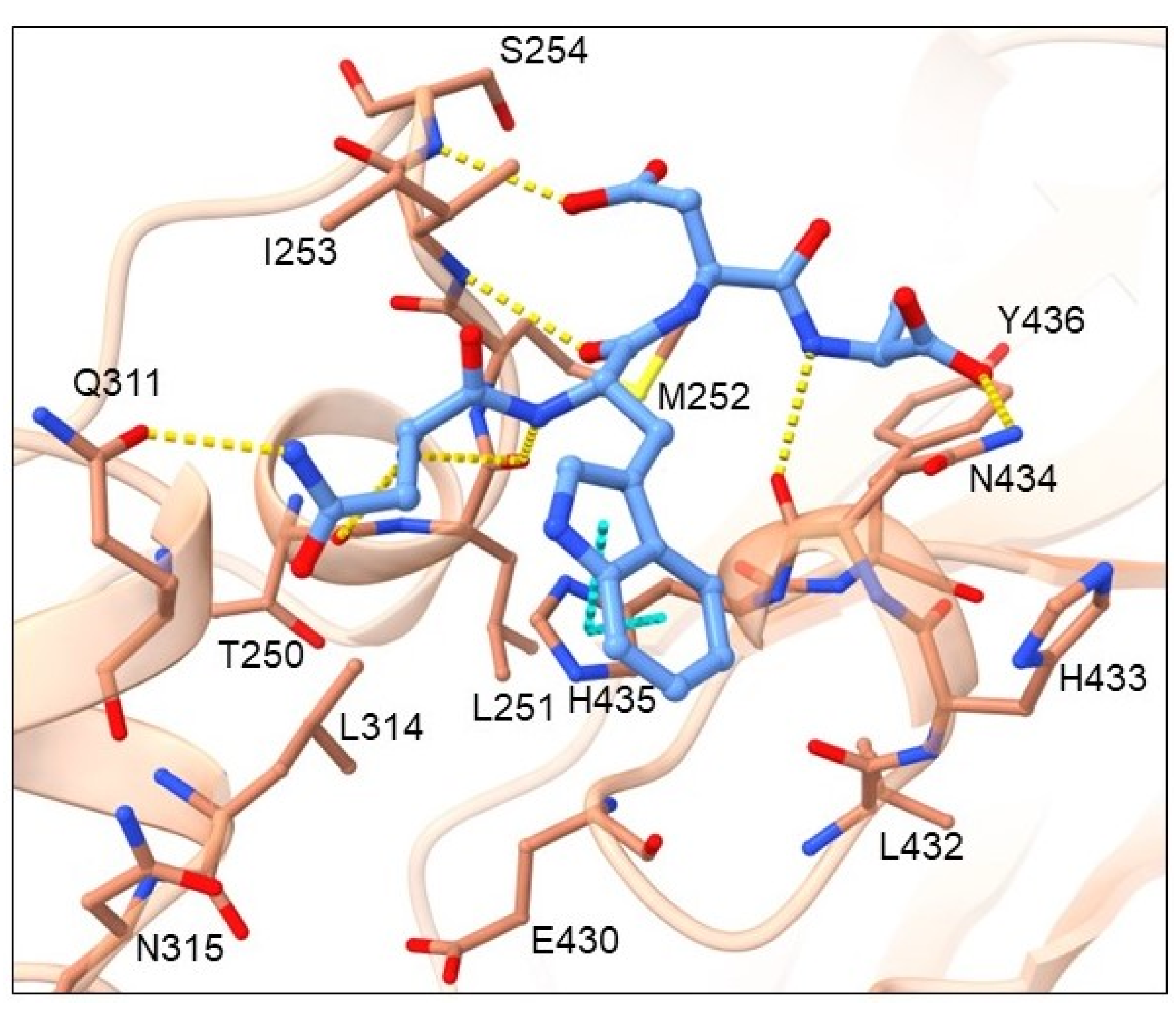
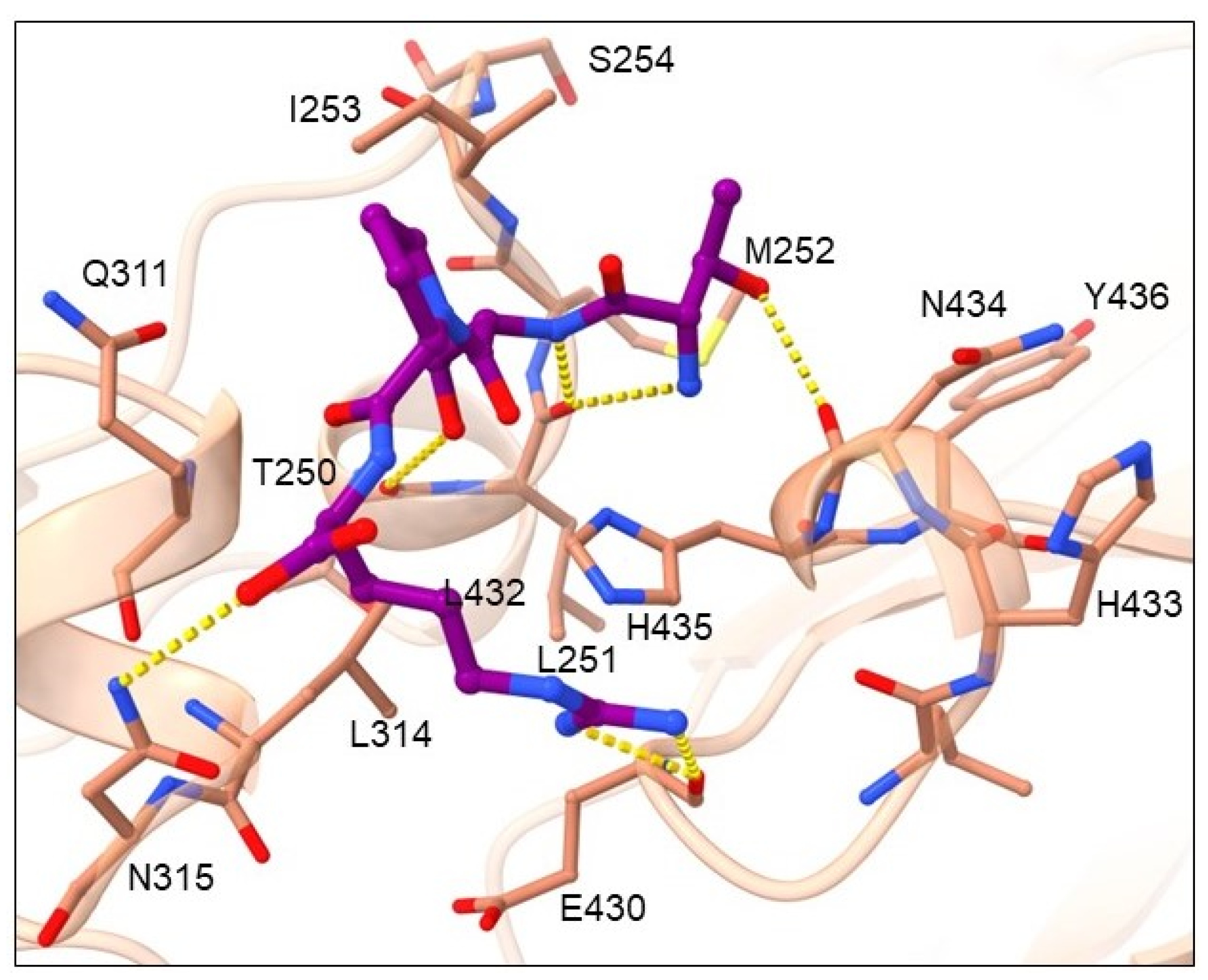
2.2. Binding Mode Analysis
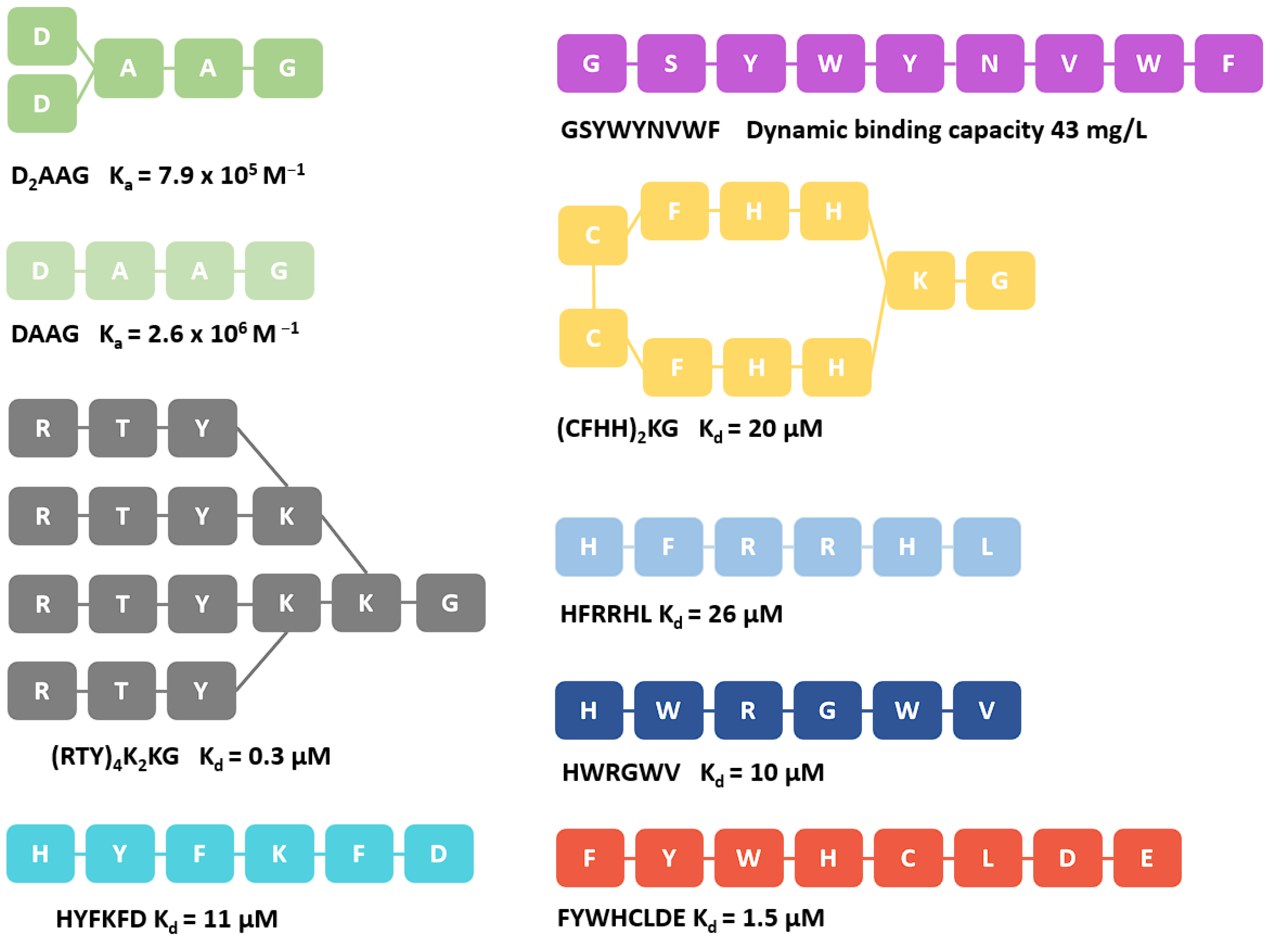
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Borrebaeck, C.A.K. Antibodies in Diagnostics—from Immunoassays to Protein Chips. Immunol. Today 2000, 21, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Price, C.P.; Newman, D.J. (Eds.) Principles and Practice of Immunoassay; Palgrave Macmillan: London, UK, 1991; ISBN 978-1-349-11236-4. [Google Scholar]
- Bozovičar, K.; Jenko Bizjan, B.; Meden, A.; Kovač, J.; Bratkovič, T. Focused Peptide Library Screening as a Route to a Superior Affinity Ligand for Antibody Purification. Sci. Rep. 2021, 11, 11650. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Xu, J.; Zhang, L.; Fu, Y.; Xu, X. Development of Novel Small Peptide Ligands for Antibody Purification. RSC Adv. 2015, 5, 67093–67101. [Google Scholar] [CrossRef]
- Chan, C.E.Z.; Chan, A.H.Y.; Hanson, B.J.; Ooi, E.E. The Use of Antibodies in the Treatment of Infectious Diseases. Singap. Med. J. 2009, 50, 663–672, quiz 673. [Google Scholar]
- Suzuki, M.; Kato, C.; Kato, A. Therapeutic Antibodies: Their Mechanisms of Action and the Pathological Findings They Induce in Toxicity Studies. J. Toxicol. Pathol. 2015, 28, 133–139. [Google Scholar] [CrossRef]
- Waldmann, T.A. Monoclonal Antibodies in Diagnosis and Therapy. Science 1991, 252, 1657–1662. [Google Scholar] [CrossRef]
- Köhler, G.; Milstein, C. Continuous Cultures of Fused Cells Secreting Antibody of Predefined Specificity. Nature 1975, 256, 495–497. [Google Scholar] [CrossRef]
- Schroeder, H.W.; Cavacini, L. Structure and Function of Immunoglobulins. J. Allergy Clin. Immunol. 2010, 125, S41–S52. [Google Scholar] [CrossRef]
- Janeway, C. (Ed.) Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Publ. [u.a.]: New York, NY, USA, 2001; ISBN 978-0-8153-3642-6. [Google Scholar]
- Fernández-Quintero, M.L.; Pomarici, N.D.; Math, B.A.; Kroell, K.B.; Waibl, F.; Bujotzek, A.; Georges, G.; Liedl, K.R. Antibodies Exhibit Multiple Paratope States Influencing VH–VL Domain Orientations. Commun. Biol. 2020, 3, 589. [Google Scholar] [CrossRef]
- Vandyk, L.; Meek, K. Assembly of IgH CDR3: Mechanism, Regulation, and Influence on Antibody Diversity. Int. Rev. Immunol. 1992, 8, 123–133. [Google Scholar] [CrossRef]
- Vidarsson, G.; Dekkers, G.; Rispens, T. IgG Subclasses and Allotypes: From Structure to Effector Functions. Front. Immunol. 2014, 5, 520. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Liu, F.-F.; Dong, X.-Y.; Sun, Y. Molecular Mechanism of the Affinity Interactions between Protein A and Human Immunoglobulin G1 Revealed by Molecular Simulations. J. Phys. Chem. B 2011, 115, 4168–4176. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.L.; Goulet, D.R.; Teplyakov, A.; Gilliland, G.L. Antibody Structure and Function: The Basis for Engineering Therapeutics. Antibodies 2019, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- De Taeye, S.W.; Rispens, T.; Vidarsson, G. The Ligands for Human IgG and Their Effector Functions. Antibodies 2019, 8, 30. [Google Scholar] [CrossRef]
- Sela-Culang, I.; Kunik, V.; Ofran, Y. The Structural Basis of Antibody-Antigen Recognition. Front. Immunol. 2013, 4, 302. [Google Scholar] [CrossRef]
- Choe, W.; Durgannavar, T.; Chung, S. Fc-Binding Ligands of Immunoglobulin G: An Overview of High Affinity Proteins and Peptides. Materials 2016, 9, 994. [Google Scholar] [CrossRef]
- Necina, R.; Amatschek, K.; Jungbauer, A. Capture of Human Monoclonal Antibodies from Cell Culture Supernatant by Ion Exchange Media Exhibiting High Charge Density. Biotechnol. Bioeng. 1998, 60, 689–698. [Google Scholar] [CrossRef]
- Reese, H.R.; Xiao, X.; Shanahan, C.C.; Chu, W.; Van Den Driessche, G.A.; Fourches, D.; Carbonell, R.G.; Hall, C.K.; Menegatti, S. Novel Peptide Ligands for Antibody Purification Provide Superior Clearance of Host Cell Protein Impurities. J. Chromatogr. A 2020, 1625, 461237. [Google Scholar] [CrossRef]
- Kruljec, N.; Molek, P.; Hodnik, V.; Anderluh, G.; Bratkovič, T. Development and Characterization of Peptide Ligands of Immunoglobulin G Fc Region. Bioconjugate Chem. 2018, 29, 2763–2775. [Google Scholar] [CrossRef]
- Gong, Y.; Zhang, L.; Li, J.; Feng, S.; Deng, H. Development of the Double Cyclic Peptide Ligand for Antibody Purification and Protein Detection. Bioconjugate Chem. 2016, 27, 1569–1573. [Google Scholar] [CrossRef]
- Fassina, G.; Verdoliva, A.; Odierna, M.R.; Ruvo, M.; Cassini, G. Protein a Mimetic Peptide Ligand for Affinity Purification of Antibodies. J. Mol. Recognit. 1996, 9, 564–569. [Google Scholar] [CrossRef]
- Verdoliva, A.; Marasco, D.; De Capua, A.; Saporito, A.; Bellofiore, P.; Manfredi, V.; Fattorusso, R.; Pedone, C.; Ruvo, M. A New Ligand for Immunoglobulin G Subdomains by Screening of a Synthetic Peptide Library. ChemBioChem 2005, 6, 1242–1253. [Google Scholar] [CrossRef] [PubMed]
- Fassina, G.; Ruvo, M.; Palombo, G.; Verdoliva, A.; Marino, M. Novel Ligands for the Affinity-Chromatographic Purification of Antibodies. J. Biochem. Biophys. Methods 2001, 49, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Lund, L.N.; Gustavsson, P.-E.; Michael, R.; Lindgren, J.; Nørskov-Lauritsen, L.; Lund, M.; Houen, G.; Staby, A.; St. Hilaire, P.M. Novel Peptide Ligand with High Binding Capacity for Antibody Purification. J. Chromatogr. A 2012, 1225, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Verdoliva, A.; Pannone, F.; Rossi, M.; Catello, S.; Manfredi, V. Affinity Purification of Polyclonal Antibodies Using a New All-D Synthetic Peptide Ligand: Comparison with Protein A and Protein G. J. Immunol. Methods 2002, 271, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Gurgel, P.V.; Carbonell, R.G. Hexamer Peptide Affinity Resins That Bind the Fc Region of Human Immunoglobulin G: Fc-Binding Hexamer Peptide Resins. J. Pept. Res. 2008, 66, 120–137. [Google Scholar] [CrossRef]
- London, N.; Raveh, B.; Schueler-Furman, O. Modeling Peptide–Protein Interactions. In Homology Modeling; Orry, A.J.W., Abagyan, R., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2011; Volume 857, pp. 375–398. ISBN 978-1-61779-587-9. [Google Scholar]
- Weng, G.; Gao, J.; Wang, Z.; Wang, E.; Hu, X.; Yao, X.; Cao, D.; Hou, T. Comprehensive Evaluation of Fourteen Docking Programs on Protein–Peptide Complexes. J. Chem. Theory Comput. 2020, 16, 3959–3969. [Google Scholar] [CrossRef]
- Ruiz-Carmona, S.; Alvarez-Garcia, D.; Foloppe, N.; Garmendia-Doval, A.B.; Juhos, S.; Schmidtke, P.; Barril, X.; Hubbard, R.E.; Morley, S.D. RDock: A Fast, Versatile and Open Source Program for Docking Ligands to Proteins and Nucleic Acids. PLoS Comput. Biol. 2014, 10, e1003571. [Google Scholar] [CrossRef]
- Soler, D.; Westermaier, Y.; Soliva, R. Extensive Benchmark of RDock as a Peptide-Protein Docking Tool. J. Comput. Aided Mol. Des. 2019, 33, 613–626. [Google Scholar] [CrossRef]
- Tubert-Brohman, I.; Sherman, W.; Repasky, M.; Beuming, T. Improved Docking of Polypeptides with Glide. J. Chem. Inf. Model. 2013, 53, 1689–1699. [Google Scholar] [CrossRef]
- Clackson, T.; Wells, J.A. A Hot Spot of Binding Energy in a Hormone-Receptor Interface. Science 1995, 267, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Ultsch, M.; Braisted, A.; Maun, H.R.; Eigenbrot, C. 3-2-1: Structural Insights from Stepwise Shrinkage of a Three-Helix Fc-Binding Domain to a Single Helix. Protein Eng. Des. Sel. 2017, 30, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Kotlyar, M.; Rossos, A.E.M.; Jurisica, I. Prediction of Protein-Protein Interactions. Curr. Protoc. Bioinform. 2017, 60, 8.2.1–8.2.14. [Google Scholar] [CrossRef] [PubMed]
- de Vries, S.J.; Rey, J.; Schindler, C.E.M.; Zacharias, M.; Tuffery, P. The PepATTRACT Web Server for Blind, Large-Scale Peptide–Protein Docking. Nucleic Acids Res. 2017, 45, W361–W364. [Google Scholar] [CrossRef]
- Xu, X.; Yan, C.; Zou, X. MDockPeP: An Ab-initio Protein–Peptide Docking Server. J. Comput. Chem. 2018, 39, 2409–2413. [Google Scholar] [CrossRef]
- Lee, H.; Heo, L.; Lee, M.S.; Seok, C. GalaxyPepDock: A Protein–Peptide Docking Tool Based on Interaction Similarity and Energy Optimization. Nucleic Acids Res. 2015, 43, W431–W435. [Google Scholar] [CrossRef]
- Alam, N.; Goldstein, O.; Xia, B.; Porter, K.A.; Kozakov, D.; Schueler-Furman, O. High-Resolution Global Peptide-Protein Docking Using Fragments-Based PIPER-FlexPepDock. PLoS Comput. Biol. 2017, 13, e1005905. [Google Scholar] [CrossRef]
- Kurcinski, M.; Jamroz, M.; Blaszczyk, M.; Kolinski, A.; Kmiecik, S. CABS-Dock Web Server for the Flexible Docking of Peptides to Proteins without Prior Knowledge of the Binding Site. Nucleic Acids Res. 2015, 43, W419–W424. [Google Scholar] [CrossRef]
- Zhou, P.; Jin, B.; Li, H.; Huang, S.-Y. HPEPDOCK: A Web Server for Blind Peptide–Protein Docking Based on a Hierarchical Algorithm. Nucleic Acids Res. 2018, 46, W443–W450. [Google Scholar] [CrossRef]
- Porter, K.A.; Xia, B.; Beglov, D.; Bohnuud, T.; Alam, N.; Schueler-Furman, O.; Kozakov, D. ClusPro PeptiDock: Efficient Global Docking of Peptide Recognition Motifs Using FFT. Bioinformatics 2017, 33, 3299–3301. [Google Scholar] [CrossRef]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.-H.; Vreven, T.; Weng, Z. ZDOCK Server: Interactive Docking Prediction of Protein-Protein Complexes and Symmetric Multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef]
- Garzon, J.I.; Lopéz-Blanco, J.R.; Pons, C.; Kovacs, J.; Abagyan, R.; Fernandez-Recio, J.; Chacon, P. FRODOCK: A New Approach for Fast Rotational Protein–Protein Docking. Bioinformatics 2009, 25, 2544–2551. [Google Scholar] [CrossRef] [PubMed]
- Weng, G.; Wang, E.; Wang, Z.; Liu, H.; Zhu, F.; Li, D.; Hou, T. HawkDock: A Web Server to Predict and Analyze the Protein–Protein Complex Based on Computational Docking and MM/GBSA. Nucleic Acids Res. 2019, 47, W322–W330. [Google Scholar] [CrossRef] [PubMed]
- Dhanik, A.; McMurray, J.S.; Kavraki, L.E. DINC: A New AutoDock-Based Protocol for Docking Large Ligands. BMC Struct. Biol. 2013, 13, S11. [Google Scholar] [CrossRef] [PubMed]
- Raveh, B.; London, N.; Zimmerman, L.; Schueler-Furman, O. Rosetta FlexPepDock Ab-Initio: Simultaneous Folding, Docking and Refinement of Peptides onto Their Receptors. PLoS ONE 2011, 6, e18934. [Google Scholar] [CrossRef]
- Zhang, Y.; Sanner, M.F. AutoDock CrankPep: Combining Folding and Docking to Predict Protein–Peptide Complexes. Bioinformatics 2019, 35, 5121–5127. [Google Scholar] [CrossRef]
- Zalevsky, A.; Zlobin, A.; Gedzun, V.; Reshetnikov, R.; Lovat, M.; Malyshev, A.; Doronin, I.; Babkin, G.; Golovin, A. PeptoGrid—Rescoring Function for AutoDock Vina to Identify New Bioactive Molecules from Short Peptide Libraries. Molecules 2019, 24, 277. [Google Scholar] [CrossRef]
- Antes, I. DynaDock: A New Molecular Dynamics-Based Algorithm for Protein-Peptide Docking Including Receptor Flexibility. Proteins 2010, 78, 1084–1104. [Google Scholar] [CrossRef]
- Verdonk, M.L.; Berdini, V.; Hartshorn, M.J.; Mooij, W.T.M.; Murray, C.W.; Taylor, R.D.; Watson, P. Virtual Screening Using Protein−Ligand Docking: Avoiding Artificial Enrichment. J. Chem. Inf. Comput. Sci. 2004, 44, 793–806. [Google Scholar] [CrossRef]
- Jain, A.N. Surflex: Fully Automatic Flexible Molecular Docking Using a Molecular Similarity-Based Search Engine. J. Med. Chem. 2003, 46, 499–511. [Google Scholar] [CrossRef]
- Ochoa, R.; Cossio, P. PepFun: Open Source Protocols for Peptide-Related Computational Analysis. Molecules 2021, 26, 1664. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Vriend, G. New Ways to Boost Molecular Dynamics Simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- Canutescu, A.A.; Shelenkov, A.A.; Dunbrack, R.L. A Graph-Theory Algorithm for Rapid Protein Side-Chain Prediction. Protein Sci. 2003, 12, 2001–2014. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Lee, J.; Worrall, L.J.; Vuckovic, M.; Rosell, F.I.; Gentile, F.; Ton, A.-T.; Caveney, N.A.; Ban, F.; Cherkasov, A.; Paetzel, M.; et al. Crystallographic Structure of Wild-Type SARS-CoV-2 Main Protease Acyl-Enzyme Intermediate with Physiological C-Terminal Autoprocessing Site. Nat. Commun. 2020, 11, 5877. [Google Scholar] [CrossRef] [PubMed]
- Tange, O. Gnu parallel-the command-line power tool. Usenix Mag. 2011, 36, 42. [Google Scholar]
- Jukič, M.; Škrlj, B.; Tomšič, G.; Pleško, S.; Podlipnik, Č.; Bren, U. Prioritisation of Compounds for 3CLpro Inhibitor Development on SARS-CoV-2 Variants. Molecules 2021, 26, 3003. [Google Scholar] [CrossRef]
- DeLano, W.L.; Ultsch, M.H.; De, A.M.; Vos; Wells, J. A. Convergent Solutions to Binding at a Protein-Protein Interface. Science 2000, 287, 1279–1283. [Google Scholar] [CrossRef]
- Fang, Y.-M.; Lin, D.-Q.; Yao, S.-J. Tetrapeptide Ligands Screening for Antibody Separation and Purification by Molecular Simulation and Experimental Verification. Biochem. Eng. J. 2021, 176, 108213. [Google Scholar] [CrossRef]
- Hartshorn, M.J.; Verdonk, M.L.; Chessari, G.; Brewerton, S.C.; Mooij, W.T.M.; Mortenson, P.N.; Murray, C.W. Diverse, High-Quality Test Set for the Validation of Protein−Ligand Docking Performance. J. Med. Chem. 2007, 50, 726–741. [Google Scholar] [CrossRef]
- Morley, S.D.; Afshar, M. Validation of an Empirical RNA-Ligand Scoring Function for Fast Flexible Docking Using RiboDock®. J. Comput. Aided Mol. Des. 2004, 18, 189–208. [Google Scholar] [CrossRef] [PubMed]
- Berthold, M.R.; Cebron, N.; Dill, F.; Gabriel, T.R.; Kötter, T.; Meinl, T.; Ohl, P.; Thiel, K.; Wiswedel, B. KNIME—The Konstanz Information Miner: Version 2.0 and Beyond. SIGKDD Explor. Newsl. 2009, 11, 26–31. [Google Scholar] [CrossRef]
- Yoshida, S.; Uehara, S.; Kondo, N.; Takahashi, Y.; Yamamoto, S.; Kameda, A.; Kawagoe, S.; Inoue, N.; Yamada, M.; Yoshimura, N.; et al. Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides. J. Med. Chem. 2022, 65, 10655–10673. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jukič, M.; Kralj, S.; Kolarič, A.; Bren, U. Design of Tetra-Peptide Ligands of Antibody Fc Regions Using In Silico Combinatorial Library Screening. Pharmaceuticals 2023, 16, 1170. https://doi.org/10.3390/ph16081170
Jukič M, Kralj S, Kolarič A, Bren U. Design of Tetra-Peptide Ligands of Antibody Fc Regions Using In Silico Combinatorial Library Screening. Pharmaceuticals. 2023; 16(8):1170. https://doi.org/10.3390/ph16081170
Chicago/Turabian StyleJukič, Marko, Sebastjan Kralj, Anja Kolarič, and Urban Bren. 2023. "Design of Tetra-Peptide Ligands of Antibody Fc Regions Using In Silico Combinatorial Library Screening" Pharmaceuticals 16, no. 8: 1170. https://doi.org/10.3390/ph16081170
APA StyleJukič, M., Kralj, S., Kolarič, A., & Bren, U. (2023). Design of Tetra-Peptide Ligands of Antibody Fc Regions Using In Silico Combinatorial Library Screening. Pharmaceuticals, 16(8), 1170. https://doi.org/10.3390/ph16081170