Smart Sensors and Microtechnologies in the Precision Medicine Approach against Lung Cancer
Abstract
1. Introduction
2. Genetic and Biomolecular Features of Lung Cancer
2.1. Lung Cancer Genetic Drivers: Rationale for Targeted Therapy
2.2. The Basis of Immunotherapy
3. Lung Cancer Therapy: Principles, Rationale, and Novel Advances
3.1. Specific Challenges in Lung Cancer Therapy
3.2. The Importance of Multidisciplinary Expertise
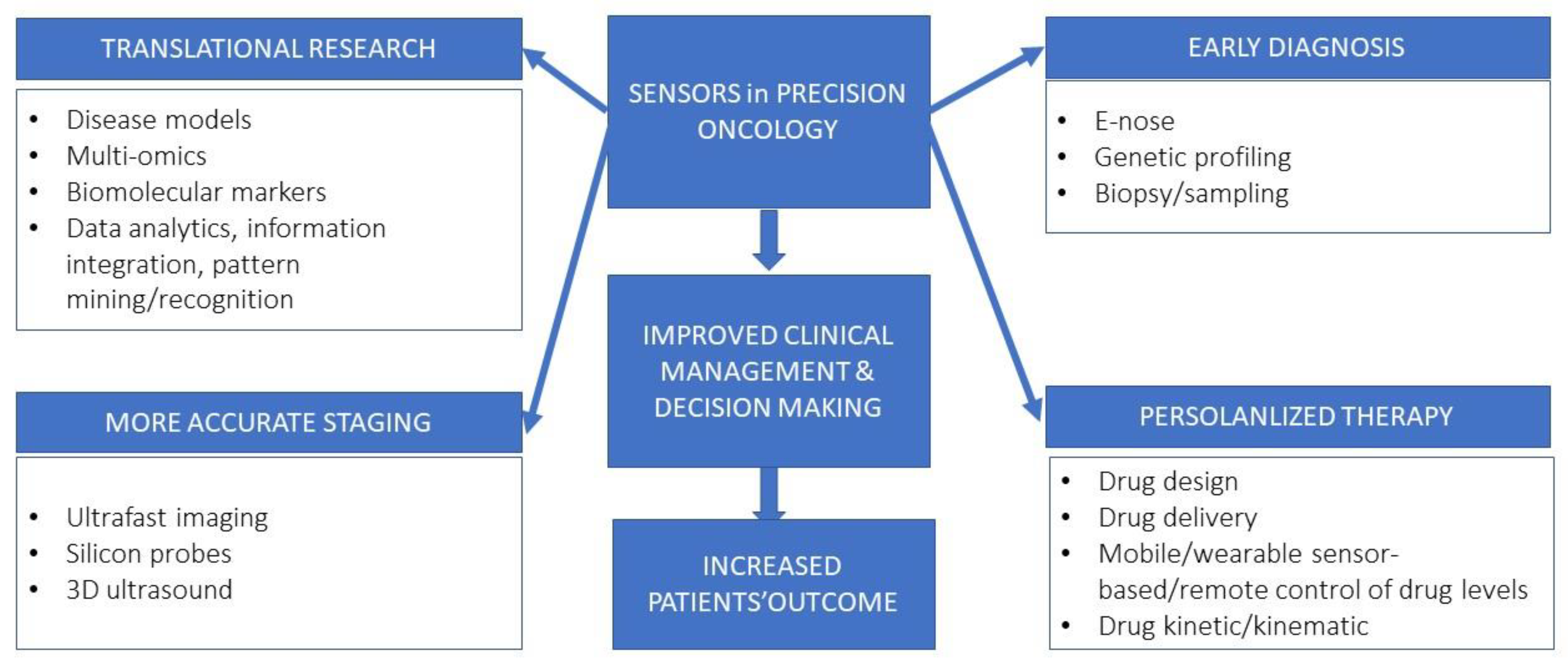
4. Impact of Micro-Technologies in Cancer Medicine
5. Micro-Technologies in Preclinical Setting
6. Frontier Technologies in Lung Cancer Clinical Management
6.1. Mutational Profiling Analysis
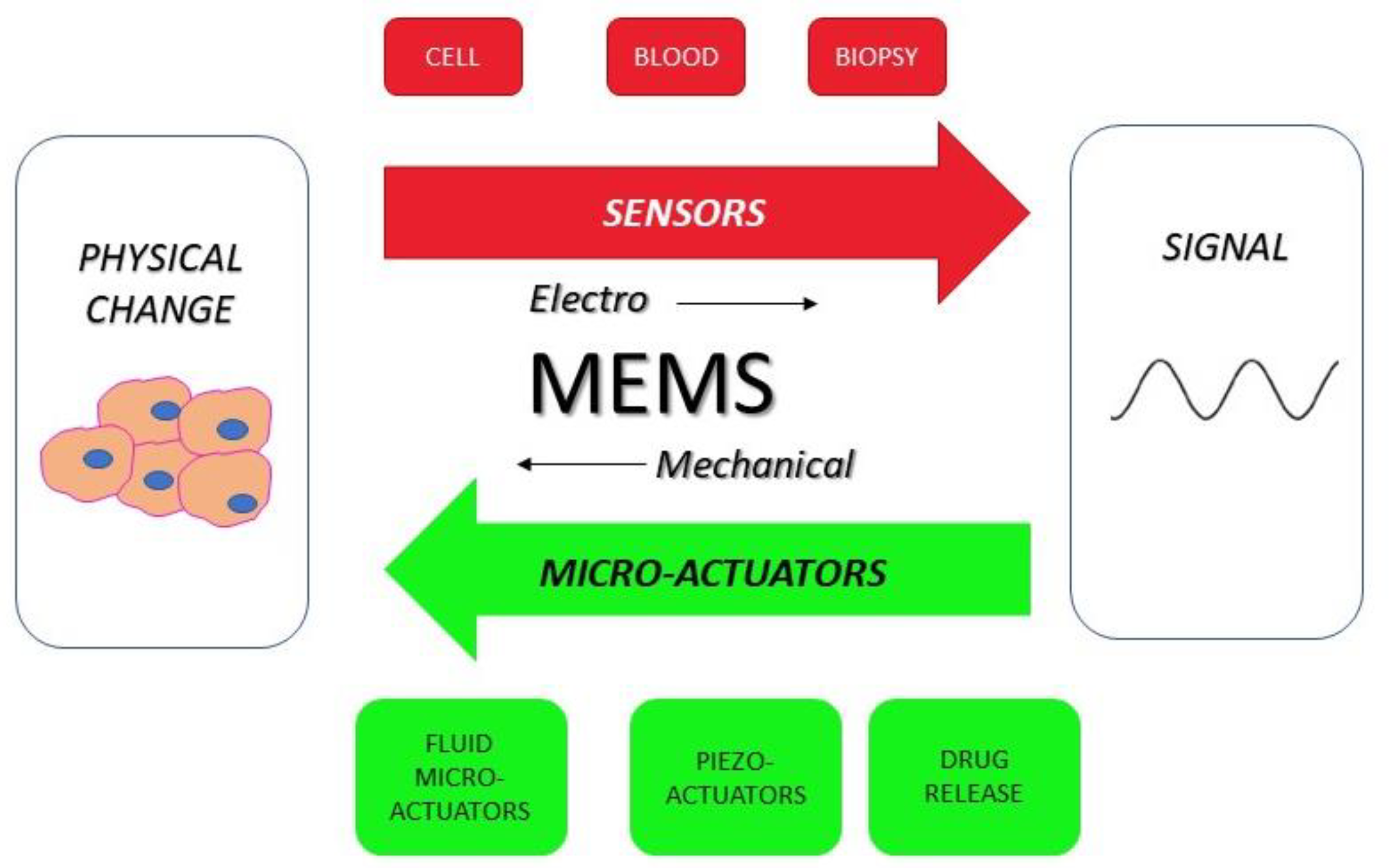
6.2. MEMS Sensor-Based Electronic Nose
6.3. Diagnostic Applications
6.3.1. Three-Dimensional Ultrasound
6.3.2. Silicon Probes
6.3.3. Ultrafast Imaging
6.3.4. Therapeutic Implications
7. Pharmaceutical Manufacturing and Drug Delivery
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, L.; Hu, H.; Scholz, A.; Feist, F.; Marques, G.C.; Kraus, S.; Bojanowski, N.M.; Blasco, E.; Barner-Kowollik, C.; Aghassi-Hagmann, J.; et al. Laser printed microelectronics. Nat. Commun. 2023, 14, 1103. [Google Scholar] [CrossRef] [PubMed]
- Zschieschang, U.; Waizmann, U.; Weis, J.; Borchert, J.W.; Klauk, H. Nanoscale flexible organic thin-film transistors. Sci. Adv. 2022, 8, eabm9845. [Google Scholar] [CrossRef]
- Fath, A.; Xia, T.; Li, W. Recent Advances in the Application of Piezoelectric Materials in Microrobotic Systems. Micromachines 2022, 13, 1422. [Google Scholar] [CrossRef] [PubMed]
- Frey, N.; Sönmez, U.M.; Minden, J.; LeDuc, P. Microfluidics for understanding model organisms. Nat. Commun. 2022, 13, 3195. [Google Scholar] [CrossRef]
- Ayuso, J.M.; Virumbrales-Muñoz, M.; Lang, J.M.; Beebe, D.J. A role for microfluidic systems in precision medicine. Nat. Commun. 2022, 13, 3086. [Google Scholar] [CrossRef] [PubMed]
- Thirunavukkarasu, M.K.; Suriya, U.; Rungrotmongkol, T.; Karuppasamy, R. In Silico Screening of Available Drugs Targeting Non-Small Cell Lung Cancer Targets: A Drug Repurposing Approach. Pharmaceutics 2021, 14, 59. [Google Scholar] [CrossRef]
- Thirunavukkarasu, M.K.; Veerappapillai, S.; Karuppasamy, R. Sequential virtual screening collaborated with machine-learning strategies for the discovery of precise medicine against non-small cell lung cancer. J. Biomol. Struct. Dyn. 2023, 30, 1–14. [Google Scholar] [CrossRef]
- Namini, A.M.; Jahangir, M.; Mohseni, M.; Kolahi, A.A.; Hassanian-Moghaddam, H.; Mazloumi, Z.; Motallebi, M.; Sheikhpour, M.; Movafagh, A. An in silico comparative transcriptome analysis identifying hub lncRNAs and mRNAs in brain metastatic small cell lung cancer (SCLC). Sci. Rep. 2022, 12, 18063. [Google Scholar] [CrossRef]
- Baldini, C.; Billeci, L.; Sansone, F.; Conte, R.; Domenici, C.; Tonacci, A. Electronic Nose as a Novel Method for Diagnosing Cancer: A Systematic Review. Biosensors 2020, 10, 84. [Google Scholar] [CrossRef]
- Chen, T.; Li, S.; Sun, H. Metamaterials Application in Sensing. Sensors 2012, 12, 2742–2765. [Google Scholar] [CrossRef]
- Romeo, M.S.; Sokolova, I.A.; Morrison, L.E.; Zeng, C.; Barón, A.E.; Hirsch, F.R.; Miller, Y.E.; Franklin, W.A.; Varella-Garcia, M. Chromosomal Abnormalities in Non-Small Cell Lung Carcinomas and in Bronchial Epithelia of High-Risk Smokers Detected by Multi-Target Interphase Fluorescence in Situ Hybridization. J. Mol. Diagn. 2003, 5, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.A.; Woo, M.S.; Getz, G.; Perner, S.; Ding, L.; Beroukhim, R.; Lin, W.M.; Province, M.A.; Kraja, A.; Johnson, L.A.; et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 2007, 450, 893–898. [Google Scholar] [CrossRef]
- Shimizu, E.; Sone, S. Tumor suppressor genes in human lung cancer. J. Med. Investig. 1997, 44, 15–24. [Google Scholar]
- Oxnard, G.R.; Binder, A.; Jänne, P.A. New Targetable Oncogenes in Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2013, 31, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Smolarz, B.; Durczyński, A.; Romanowicz, H.; Szyłło, K.; Hogendorf, P. miRNAs in Cancer (Review of Literature). Int. J. Mol. Sci. 2022, 23, 2805. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Zhong, S.; Golpon, H.; Zardo, P.; Borlak, J. miRNAs in lung cancer. A systematic review identifies predictive and prognostic miRNA candidates for precision medicine in lung cancer. Transl. Res. 2020, 230, 164–196. [Google Scholar] [CrossRef] [PubMed]
- Wani, J.A.; Majid, S.; Imtiyaz, Z.; Rehman, M.U.; Alsaffar, R.M.; Shah, N.N.; Alshehri, S.; Ghoneim, M.M.; Imam, S.S. MiRNAs in Lung Cancer: Diagnostic, Prognostic, and Therapeutic Potential. Diagnostics 2022, 12, 1610. [Google Scholar] [CrossRef]
- Ulusan, M.; Sen, S.; Yilmazer, R.; Dalay, N.; Demokan, S. The let-7 microRNA binding site variant in KRAS as a predictive biomarker for head and neck cancer patients with lymph node metastasis. Pathol. Res. Pract. 2022, 239, 154147. [Google Scholar] [CrossRef]
- Li, L.; Wang, D. MicroRNA-128-b regulates epidermal growth factor receptor expression in non-small cell lung cancer. Mol. Med. Rep. 2019, 20, 4803–4810. [Google Scholar] [CrossRef]
- Arena, S.; Benvenuti, S.; Bardelli, A. Genetic analysis of the kinome and phosphatome in cancer. Cell. Mol. Life Sci. 2005, 62, 2092–2099. [Google Scholar] [CrossRef]
- Haber, D.A.; Settleman, J. Drivers and passengers. Nature 2007, 446, 145–146. [Google Scholar] [CrossRef] [PubMed]
- Greenman, C.; Stephens, P.; Smith, R.; Dalgliesh, G.L.; Hunter, C.; Bignell, G.; Davies, H.; Teague, J.; Butler, A.; Stevens, C.; et al. Patterns of somatic mutation in human cancer genomes. Nature 2007, 446, 153–158. [Google Scholar] [CrossRef]
- Thomas, A.; Rajan, A.; Giaccone, G. Tyrosine Kinase Inhibitors in Lung Cancer. Hematol. Clin. N. Am. 2012, 26, 589–605. [Google Scholar] [CrossRef]
- Stella, G.M.; Luisetti, M.; Inghilleri, S.; Cemmi, F.; Scabini, R.; Zorzetto, M.; Pozzi, E. Targeting EGFR in non-small-cell lung cancer: Lessons, experiences, strategies. Respir. Med. 2012, 106, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Shigematsu, H.; Gazdar, A.F. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int. J. Cancer 2006, 118, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, M.G.; Di Noia, V.; D’argento, E.; Vita, E.; Damiano, P.; Cannella, A.; Ribelli, M.; Pilotto, S.; Milella, M.; Tortora, G.; et al. Oncogene-Addicted Non-Small-Cell Lung Cancer: Treatment Opportunities and Future Perspectives. Cancers 2020, 12, 1196. [Google Scholar] [CrossRef]
- Sharma, S.V.; Fischbach, M.A.; Haber, D.A.; Settleman, J. “Oncogenic Shock”: Explaining Oncogene Addiction through Differential Signal Attenuation. Clin. Cancer Res. 2006, 12, 4392s–4395s. [Google Scholar] [CrossRef]
- Tumbrink, H.L.; Heimsoeth, A.; Sos, M.L. The next tier of EGFR resistance mutations in lung cancer. Oncogene 2020, 40, 1–11. [Google Scholar] [CrossRef]
- Westover, D.; Zugazagoitia, J.; Cho, B.C.; Lovly, C.M.; Paz-Ares, L. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann. Oncol. 2018, 29, i10–i19. [Google Scholar] [CrossRef]
- Gao, J.; Li, H.-R.; Jin, C.; Jiang, J.-H.; Ding, J.-Y. Strategies to overcome acquired resistance to EGFR TKI in the treatment of non-small cell lung cancer. Clin. Transl. Oncol. 2019, 21, 1287–1301. [Google Scholar] [CrossRef]
- Rebuzzi, S.E.; Zullo, L.; Rossi, G.; Grassi, M.; Murianni, V.; Tagliamento, M.; Prelaj, A.; Coco, S.; Longo, L.; Bello, M.G.D.; et al. Novel Emerging Molecular Targets in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2021, 22, 2625. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, P.; Wei, J.; Zhang, X.; Guo, J.; Lin, Y. Recent progress in targeted therapy for non-small cell lung cancer. Front. Pharmacol. 2023, 14, 1125547. [Google Scholar] [CrossRef]
- Gridelli, C.; Ascierto, P.A.; Grossi, F.; Baldini, E.; Favaretto, A.; Garassino, M.C.; Morabito, A.; Migliorino, M.R.; Rossi, A.; de Marinis, F. Second-line Treatment of Advanced Non-small Cell Lung Cancer Non-oncogene Addicted: New Treatment Algorithm in the Era of Novel Immunotherapy. Curr. Clin. Pharmacol. 2018, 13, 76–84. [Google Scholar] [CrossRef]
- Rocco, D.; Della Gravara, L.; Franzese, N.; Maione, P.; Gridelli, C. Chemotherapy plus single/double immunotherapy in the treatment of non-oncogene addicted advanced non-small cell lung cancer: Where do we stand and where are we going? Expert Rev. Anticancer. Ther. 2022, 22, 183–189. [Google Scholar] [CrossRef]
- Hendriks, L.; Kerr, K.; Menis, J.; Mok, T.; Nestle, U.; Passaro, A.; Peters, S.; Planchard, D.; Smit, E.; Solomon, B.; et al. Non-oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023, 34, 358–376. [Google Scholar] [CrossRef] [PubMed]
- Hiam-Galvez, K.J.; Allen, B.M.; Spitzer, M.H. Systemic immunity in cancer. Nat. Rev. Cancer 2021, 21, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Lasek, W. Cancer immunoediting hypothesis: History, clinical implications and controversies. Cent. Eur. J. Immunol. 2022, 47, 168–174. [Google Scholar] [CrossRef]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The Immunobiology of Cancer Immunosurveillance and Immunoediting. Immunity 2004, 21, 137–148. [Google Scholar] [CrossRef]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The Three Es of Cancer Immunoediting. Annu. Rev. Immunol. 2004, 22, 329–360. [Google Scholar] [CrossRef]
- Vesely, M.; Schreiber, R.D. Cancer immunoediting: Antigens, mechanisms, and implications to cancer immunotherapy. Ann. N. Y. Acad. Sci. 2013, 1284, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Buchbinder, E.I.; Desai, A. CTLA-4 and PD-1 Pathways: Similarities, Differences, and Implications of Their Inhibition. Am. J. Clin. Oncol. 2016, 39, 98–106. [Google Scholar] [CrossRef]
- Callahan, M.K.; Wolchok, J.D. At the Bedside: CTLA-4- and PD-1-blocking antibodies in cancer immunotherapy. J. Leukoc. Biol. 2013, 94, 41–53. [Google Scholar] [CrossRef]
- Weber, J. Immune Checkpoint Proteins: A New Therapeutic Paradigm for Cancer—Preclinical Background: CTLA-4 and PD-1 Blockade. Semin. Oncol. 2010, 37, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Seidel, J.; Otsuka, A.; Kabashima, K. Anti-PD-1 and Anti-CTLA-4 Therapies in Cancer: Mechanisms of Action, Efficacy, and Limitations. Front. Oncol. 2018, 8, 86. [Google Scholar] [CrossRef]
- Shen, X.; Huang, S.; Xiao, H.; Zeng, S.; Liu, J.; Ran, Z.; Xiong, B. Efficacy and safety of PD-1/PD-L1 plus CTLA-4 antibodies ± other therapies in lung cancer: A systematic review and meta-analysis. Eur. J. Hosp. Pharm. 2021, 30, 3–8. [Google Scholar] [CrossRef]
- Zhang, H.; Dai, Z.; Wu, W.; Wang, Z.; Zhang, N.; Zhang, L.; Zeng, W.-J.; Liu, Z.; Cheng, Q. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J. Exp. Clin. Cancer Res. 2021, 40, 184. [Google Scholar] [CrossRef]
- Wang, Q.; Lin, W.; Tang, X.; Li, S.; Guo, L.; Lin, Y.; Kwok, H.F. The Roles of microRNAs in Regulating the Expression of PD-1/PD-L1 Immune Checkpoint. Int. J. Mol. Sci. 2017, 18, 2540. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, X.; Xu, L.; Li, Y.; Zeng, C. Current insight into the regulation of PD-L1 in cancer. Exp. Hematol. Oncol. 2022, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Thandra, K.C.; Barsouk, A.; Saginala, K.; Aluru, J.S.; Barsouk, A. Epidemiology of lung cancer. Contemp. Oncol. 2021, 25, 45–52. [Google Scholar] [CrossRef]
- Strebhardt, K.; Ullrich, A. Paul Ehrlich’s magic bullet concept: 100 years of progress. Nat. Rev. Cancer 2008, 8, 473–480. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Kohman, L.J.; Gu, L.; Altorki, N.; Scalzetti, E.; Veit, L.J.; Wallen, J.M.; Wang, X. Biopsy first: Lessons learned from Cancer and Leukemia Group B (CALGB) 140503. J. Thorac. Cardiovasc. Surg. 2017, 153, 1592–1597. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.T.; Durbin, E.B.; Chen, L.; Gal, T.; Mahan, A.; Ferraris, V.; Zwischenberger, J. Nodal Upstaging During Lung Cancer Resection Is Associated With Surgical Approach. Ann. Thorac. Surg. 2015, 101, 238–245. [Google Scholar] [CrossRef]
- Paul, S.; Altorki, N.K.; Sheng, S.; Lee, P.C.; Harpole, D.H.; Onaitis, M.W.; Stiles, B.M.; Port, J.L.; D’Amico, T.A. Thoracoscopic lobectomy is associated with lower morbidity than open lobectomy: A propensity-matched analysis from the STS database. J. Thorac. Cardiovasc. Surg. 2010, 139, 366–378. [Google Scholar] [CrossRef]
- Cao, C.; Manganas, C.; Ang, S.C.; Peeceeyen, S.; Yan, T.D. Video-assisted thoracic surgery versus open thoracotomy for non-small cell lung cancer: A meta-analysis of propensity score-matched patients. Interact. Cardiovasc. Thorac. Surg. 2012, 16, 244–249. [Google Scholar] [CrossRef]
- Berry, M.F.; Hanna, J.; Tong, B.C.; Burfeind, W.R.; Harpole, D.H.; D’Amico, T.A.; Onaitis, M.W. Risk Factors for Morbidity after Lobectomy for Lung Cancer in Elderly Patients. Ann. Thorac. Surg. 2009, 88, 1093–1099. [Google Scholar] [CrossRef]
- Tandberg, D.J.; Tong, B.C.; Ackerson, B.G.; Kelsey, C.R. Surgery versus stereotactic body radiation therapy for stage I non-small cell lung cancer: A comprehensive review. Cancer 2017, 124, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Brooks, E.D.; Komaki, R.U.; Liao, Z.; Jeter, M.D.; McAleer, M.F.; Allen, P.K.; Balter, P.A.; Welsh, J.D.; O’Reilly, M.S.; et al. 7-year follow-up after stereotactic ablative radiotherapy for patients with stage I non-small cell lung cancer: Results of a phase 2 clinical trial. Cancer 2017, 123, 3031–3039. [Google Scholar] [CrossRef]
- Nagata, Y.; Hiraoka, M.; Shibata, T.; Onishi, H.; Kokubo, M.; Karasawa, K.; Shioyama, Y.; Onimaru, R.; Kozuka, T.; Kunieda, E.; et al. Prospective Trial of Stereotactic Body Radiation Therapy for Both Operable and Inoperable T1N0M0 Non-Small Cell Lung Cancer: Japan Clinical Oncology Group Study JCOG0403. Int. J. Radiat. Oncol. 2015, 93, 989–996. [Google Scholar] [CrossRef]
- Winton, T.; Livingston, R.; Johnson, D.; Rigas, J.; Johnston, M.; Butts, C.; Cormier, Y.; Goss, G.; Inculet, R.; Vallieres, E.; et al. Vinorelbine plus Cisplatin vs. Observation in Resected Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2005, 352, 2589–2597. [Google Scholar] [CrossRef]
- Strauss, G.M.; Herndon, J.E.; Maddaus, M.A.; Johnstone, D.W.; Johnson, E.A.; Harpole, D.H.; Gillenwater, H.H.; Watson, D.M.; Sugarbaker, D.J.; Schilsky, R.L.; et al. Adjuvant Paclitaxel Plus Carboplatin Compared With Observation in Stage IB Non–Small-Cell Lung Cancer: CALGB 9633 With the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J. Clin. Oncol. 2008, 26, 5043–5051. [Google Scholar] [CrossRef]
- Butts, C.A.; Ding, K.; Seymour, L.; Twumasi-Ankrah, P.; Graham, B.; Gandara, D.; Johnson, D.H.; Kesler, K.A.; Green, M.; Vincent, M.; et al. Randomized Phase III Trial of Vinorelbine Plus Cisplatin Compared With Observation in Completely Resected Stage IB and II Non–Small-Cell Lung Cancer: Updated Survival Analysis of JBR-10. J. Clin. Oncol. 2010, 28, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Douillard, J.-Y.; Tribodet, H.; Aubert, D.; Shepherd, F.A.; Rosell, R.; Ding, K.; Veillard, A.-S.; Seymour, L.; Le Chevalier, T.; Spiro, S.; et al. Adjuvant Cisplatin and Vinorelbine for Completely Resected Non-small Cell Lung Cancer: Subgroup Analysis of the Lung Adjuvant Cisplatin Evaluation. J. Thorac. Oncol. 2010, 5, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Pignon, J.-P.; Tribodet, H.; Scagliotti, G.V.; Douillard, J.-Y.; Shepherd, F.A.; Stephens, R.J.; Dunant, A.; Torri, V.; Rosell, R.; Seymour, L.; et al. Lung Adjuvant Cisplatin Evaluation: A Pooled Analysis by the LACE Collaborative Group. J. Clin. Oncol. 2008, 26, 3552–3559. [Google Scholar] [CrossRef]
- Felip, E.; Rosell, R.; Maestre, J.A.; Rodríguez-Paniagua, J.M.; Morán, T.; Astudillo, J.; Alonso, G.; Borro, J.M.; González-Larriba, J.L.; Torres, A.; et al. Preoperative Chemotherapy Plus Surgery Versus Surgery Plus Adjuvant Chemotherapy Versus Surgery Alone in Early-Stage Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2010, 28, 3138–3145. [Google Scholar] [CrossRef] [PubMed]
- Preoperative chemotherapy for non-small-cell lung cancer: A systematic review and meta-analysis of individual participant data. Lancet 2014, 383, 1561–1571. [CrossRef]
- Wakelee, H.A.; Dahlberg, S.E.; Keller, S.M.; Tester, W.J.; Gandara, D.R.; Graziano, S.L.; Adjei, A.A.; Leighl, N.B.; Aisner, S.C.; Rothman, J.M.; et al. Adjuvant chemotherapy with or without bevacizumab in patients with resected non-small-cell lung cancer (E1505): An open-label, multicentre, randomised, phase 3 trial. Lancet Oncol. 2017, 18, 1610–1623. [Google Scholar] [CrossRef]
- Kelly, K.; Altorki, N.K.; Eberhardt, W.E.E.; O’Brien, M.E.; Spigel, D.R.; Crinò, L.; Tsai, C.-M.; Kim, J.-H.; Cho, E.K.; Hoffman, P.C.; et al. Adjuvant Erlotinib Versus Placebo in Patients With Stage IB-IIIA Non–Small-Cell Lung Cancer (RADIANT): A Randomized, Double-Blind, Phase III Trial. J. Clin. Oncol. 2015, 33, 4007–4014. [Google Scholar] [CrossRef]
- Goss, G.D.; O’Callaghan, C.; Lorimer, I.; Tsao, M.-S.; Masters, G.A.; Jett, J.; Edelman, M.J.; Lilenbaum, R.; Choy, H.; Khuri, F.; et al. Gefitinib Versus Placebo in Completely Resected Non–Small-Cell Lung Cancer: Results of the NCIC CTG BR19 Study. J. Clin. Oncol. 2013, 31, 3320–3326. [Google Scholar] [CrossRef]
- Govindan, R.; Mandrekar, S.J.; Gerber, D.E.; Oxnard, G.R.; Dahlberg, S.E.; Chaft, J.; Malik, S.; Mooney, M.; Abrams, J.S.; Jänne, P.A.; et al. ALCHEMIST Trials: A Golden Opportunity to Transform Outcomes in Early-Stage Non–Small Cell Lung Cancer. Clin. Cancer Res. 2015, 21, 5439–5444. [Google Scholar] [CrossRef]
- Antonia, S.J.; Villegas, A.; Daniel, D.; Vicente, D.; Murakami, S.; Hui, R.; Yokoi, T.; Chiappori, A.; Lee, K.H.; De Wit, M.; et al. Durvalumab after Chemoradiotherapy in Stage III Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 377, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Antonia, S.J.; Villegas, A.; Daniel, D.; Vicente, D.; Murakami, S.; Hui, R.; Kurata, T.; Chiappori, A.; Lee, K.H.; De Wit, M.; et al. Overall Survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC. N. Engl. J. Med. 2018, 379, 2342–2350. [Google Scholar] [CrossRef]
- Faivre-Finn, C.; Vicente, D.; Kurata, T.; Planchard, D.; Paz-Ares, L.; Vansteenkiste, J.F.; Spigel, D.R.; Garassino, M.C.; Reck, M.; Senan, S.; et al. Four-Year Survival With Durvalumab After Chemoradiotherapy in Stage III NSCLC—An Update From the PACIFIC Trial. J. Thorac. Oncol. 2021, 16, 860–867. [Google Scholar] [CrossRef]
- SEER*Explorer: An interactive website for SEER cancer statistics [Internet]. Surveillance Research Program, National Cancer Institute. 2023. Available online: https://seer.cancer.gov/statistics-network/explorer (accessed on 16 June 2023).
- Tsao, A.S.; Scagliotti, G.V.; Bunn, P.A.; Carbone, D.P.; Warren, G.W.; Bai, C.; de Koning, H.J.; Yousaf-Khan, A.U.; McWilliams, A.; Tsao, M.S.; et al. Scientific Advances in Lung Cancer 2015. J. Thorac. Oncol. 2016, 11, 613–638. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Herbst, R.S.; Boshoff, C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat. Med. 2021, 27, 1345–1356. [Google Scholar] [CrossRef]
- Midha, A.; Dearden, S.; McCormack, R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: A systematic review and global map by ethnicity (mutMapII). Am. J. Cancer Res. 2015, 5, 2892–2911. [Google Scholar] [PubMed]
- Shigematsu, H.; Lin, L.; Takahashi, T.; Nomura, M.; Suzuki, M.; Wistuba, I.I.; Fong, K.M.; Lee, H.; Toyooka, S.; Shimizu, N.; et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J. Natl. Cancer Inst. 2005, 97, 339–346. [Google Scholar] [CrossRef]
- Ramalingam, S.S.; Vansteenkiste, J.; Planchard, D.; Cho, B.C.; Gray, J.E.; Ohe, Y.; Zhou, C.; Reungwetwattana, T.; Cheng, Y.; Chewaskulyong, B.; et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N. Engl. J. Med. 2020, 382, 41–50. [Google Scholar] [CrossRef]
- Shaw, A.T.; Yeap, B.Y.; Mino-Kenudson, M.; Digumarthy, S.R.; Costa, D.B.; Heist, R.S.; Solomon, B.; Stubbs, H.; Admane, S.; McDermott, U.; et al. Clinical Features and Outcome of Patients With Non–Small-Cell Lung Cancer Who Harbor EML4-ALK. J. Clin. Oncol. 2009, 27, 4247–4253. [Google Scholar] [CrossRef]
- Gainor, J.F.; Varghese, A.M.; Ou, S.-H.I.; Kabraji, S.; Awad, M.M.; Katayama, R.; Pawlak, A.; Mino-Kenudson, M.; Yeap, B.Y.; Riely, G.J.; et al. ALK Rearrangements Are Mutually Exclusive with Mutations in EGFR or KRAS: An Analysis of 1,683 Patients with Non–Small Cell Lung Cancer. Clin. Cancer Res. 2013, 19, 4273–4281. [Google Scholar] [CrossRef]
- Pao, W.; Wang, T.Y.; Riely, G.J.; Miller, V.A.; Pan, Q.; Ladanyi, M.; Zakowski, M.F.; Heelan, R.T.; Kris, M.G.; Varmus, H.E. KRAS Mutations and Primary Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib. PLoS Med. 2005, 2, e17. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.M.; Kim, H.R.; Lee, J.S.; Lee, K.H.; Lee, Y.G.; Min, Y.J.; Cho, E.K.; Lee, S.S.; Kim, B.S.; Choi, M.Y.; et al. Open-Label, Multicenter, Phase II Study of Ceritinib in Patients With Non-Small-Cell Lung Cancer Harboring ROS1 Rearrangement. J. Clin. Oncol. 2017, 35, 2613–2618. [Google Scholar] [CrossRef]
- Shaw, A.; Riely, G.; Bang, Y.-J.; Kim, D.-W.; Camidge, D.; Solomon, B.; Varella-Garcia, M.; Iafrate, A.; Shapiro, G.; Usari, T.; et al. Crizotinib in ROS1-rearranged advanced non-small-cell lung cancer (NSCLC): Updated results, including overall survival, from PROFILE 1001. Ann. Oncol. 2019, 30, 1121–1126. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Siena, S.; Dziadziuszko, R.; Barlesi, F.; Krebs, M.G.; Shaw, A.T.; de Braud, F.; Rolfo, C.; Ahn, M.-J.; Wolf, J.; et al. Entrectinib in ROS1 fusion-positive non-small-cell lung cancer: Integrated analysis of three phase 1–2 trials. Lancet Oncol. 2019, 21, 261–270. [Google Scholar] [CrossRef]
- Shaw, A.T.; Solomon, B.J.; Chiari, R.; Riely, G.J.; Besse, B.; Soo, R.A.; Kao, S.; Lin, C.-C.; Bauer, T.M.; Clancy, J.S.; et al. Lorlatinib in advanced ROS1-positive non-small-cell lung cancer: A multicentre, open-label, single-arm, phase 1–2 trial. Lancet Oncol. 2019, 20, 1691–1701. [Google Scholar] [CrossRef]
- Califano, R.; Abidin, A.; Tariq, N.-U.; Economopoulou, P.; Metro, G.; Mountzios, G. Beyond EGFR and ALK inhibition: Unravelling and exploiting novel genetic alterations in advanced non small-cell lung cancer. Cancer Treat. Rev. 2015, 41, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Planchard, D.; Besse, B.; Groen, H.J.M.; Souquet, P.-J.; Quoix, E.; Baik, C.S.; Barlesi, F.; Kim, T.M.; Mazieres, J.; Novello, S.; et al. Dabrafenib plus trametinib in patients with previously treated BRAFV600E-mutant metastatic non-small cell lung cancer: An open-label, multicentre phase 2 trial. Lancet Oncol. 2016, 17, 984–993. [Google Scholar] [CrossRef]
- Drilon, A.; Laetsch, T.W.; Kummar, S.; Dubois, S.G.; Lassen, U.N.; Demetri, G.D.; Nathenson, M.; Doebele, R.C.; Farago, A.F.; Pappo, A.S.; et al. Efficacy of Larotrectinib in TRK Fusion–Positive Cancers in Adults and Children. N. Engl. J. Med. 2018, 378, 731–739. [Google Scholar] [CrossRef]
- Alexander, M.; Kim, S.Y.; Cheng, H. Update 2020: Management of Non-Small Cell Lung Cancer. Lung 2020, 198, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Jänne, P.A.; Shaw, A.T.; Pereira, J.R.; Jeannin, G.; Vansteenkiste, J.; Barrios, C.; Franke, F.A.; Grinsted, L.; Zazulina, V.; Smith, P.; et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: A randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013, 14, 38–47. [Google Scholar] [CrossRef]
- Blumenschein, G.R.; Smit, E.F.; Planchard, D.; Kim, D.-W.; Cadranel, J.; De Pas, T.; Dunphy, F.; Udud, K.; Ahn, M.-J.; Hanna, N.H.; et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC). Ann. Oncol. 2015, 26, 894–901. [Google Scholar] [CrossRef]
- Lin, J.J.; Shaw, A.T. Resisting Resistance: Targeted Therapies in Lung Cancer. Trends Cancer 2016, 2, 350–364. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J., Jr.; Wu, Y.-L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1–Positive Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crinò, L.; Eberhardt, W.E.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Rizvi, N.A.; Goldman, J.W.; Gettinger, S.N.; Borghaei, H.; Brahmer, J.R.; Ready, N.E.; Gerber, D.E.; Chow, L.Q.; Juergens, R.A.; et al. Nivolumab plus ipilimumab as first-line treatment for advanced non-small-cell lung cancer (CheckMate 012): Results of an open-label, phase 1, multicohort study. Lancet Oncol. 2016, 18, 31–41. [Google Scholar] [CrossRef]
- Paz-Ares, L.; Ciuleanu, T.-E.; Cobo, M.; Schenker, M.; Zurawski, B.; Menezes, J.; Richardet, E.; Bennouna, J.; Felip, E.; Juan-Vidal, O.; et al. First-line nivolumab plus ipilimumab combined with two cycles of chemotherapy in patients with non-small-cell lung cancer (CheckMate 9LA): An international, randomised, open-label, phase 3 trial. Lancet Oncol. 2021, 22, 198–211. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Paz-Ares, L.; Bernabe Caro, R.; Zurawski, B.; Kim, S.-W.; Carcereny Costa, E.; Park, K.; Alexandru, A.; Lupinacci, L.; de la Mora Jimenez, E.; et al. Nivolumab plus Ipilimumab in Advanced Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2019, 381, 2020–2031. [Google Scholar] [CrossRef]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef]
- Zhou, C.; Li, M.; Wang, Z.; An, D.; Li, B. Adverse events of immunotherapy in non-small cell lung cancer: A systematic review and network meta-analysis. Int. Immunopharmacol. 2021, 102, 108353. [Google Scholar] [CrossRef]
- Rivera-Concepcion, J.; Uprety, D.; Adjei, A.A. Challenges in the Use of Targeted Therapies in Non–Small Cell Lung Cancer. Cancer Res. Treat. 2022, 54, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Gopal, V.; Dubashi, B.; Kayal, S.; Penumadu, P.; Rajaram, M.; Karunanithi, G.; Adithan, S.; Toi, P.C.; Ganesan, P. Challenges in the Management of Lung Cancer: Real-World Experience from a Tertiary Center in South India. South Asian J. Cancer 2021, 10, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Tsianakas, V.; Robert, G.; Maben, J.; Richardson, A.; Dale, C.; Wiseman, T. Implementing patient-centred cancer care: Using experience-based co-design to improve patient experience in breast and lung cancer services. Support. Care Cancer 2012, 20, 2639–2647. [Google Scholar] [CrossRef]
- Lahiri, A.; Maji, A.; Potdar, P.D.; Singh, N.; Parikh, P.; Bisht, B.; Mukherjee, A.; Paul, M.K. Lung cancer immunotherapy: Progress, pitfalls, and promises. Mol. Cancer 2023, 22, 40. [Google Scholar] [CrossRef]
- Hegde, P.S.; Chen, D.S. Top 10 Challenges in Cancer Immunotherapy. Immunity 2020, 52, 17–35. [Google Scholar] [CrossRef]
- Sajjad, H.; Imtiaz, S.; Noor, T.; Siddiqui, Y.H.; Sajjad, A.; Zia, M. Cancer models in preclinical research: A chronicle review of advancement in effective cancer research. Anim. Model. Exp. Med. 2021, 4, 87–103. [Google Scholar] [CrossRef]
- Clarke, M.A.; Fisher, J. Executable cancer models: Successes and challenges. Nat. Rev. Cancer 2020, 20, 343–354. [Google Scholar] [CrossRef]
- Neufeld, L.; Yeini, E.; Pozzi, S.; Satchi-Fainaro, R. 3D bioprinted cancer models: From basic biology to drug development. Nat. Rev. Cancer 2022, 22, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Patkar, V.; Acosta, D.; Davidson, T.; Jones, A.; Fox, J.; Keshtgar, M. Cancer Multidisciplinary Team Meetings: Evidence, Challenges, and the Role of Clinical Decision Support Technology. Int. J. Breast Cancer 2011, 2011, 831605. [Google Scholar] [CrossRef]
- Chaturvedi, V.K.; Singh, A.; Singh, V.K.; Singh, M.P. Cancer Nanotechnology: A New Revolution for Cancer Diagnosis and Therapy. Curr. Drug Metab. 2019, 20, 416–429. [Google Scholar] [CrossRef]
- Zhang, Q.; Hou, K.; Chen, H.; Zeng, N.; Wu, Y. Nanotech Probes: A Revolution in Cancer Diagnosis. Front. Oncol. 2022, 12, 933125. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Rodrigues, M.; Andrade, I.; Cruz, R. Current Point-of-Care testing in cancer and future perspectives: A systematic review. Eur. J. Public Health 2020, 30, ckaa040-033. [Google Scholar] [CrossRef]
- Syedmoradi, L.; Norton, M.L.; Omidfar, K. Point-of-care cancer diagnostic devices: From academic research to clinical translation. Talanta 2020, 225, 122002. [Google Scholar] [CrossRef]
- Arshavsky-Graham, S.; Segal, E. Lab-on-a-Chip Devices for Point-of-Care Medical Diagnostics. Adv. Biochem. Eng. Biotechnol. 2022, 179, 247–265. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef]
- Malhotra, B.D.; Srivastava, S.; Ali, A.; Singh, C. Nanomaterial-Based Biosensors for Food Toxin Detection. Appl. Biochem. Biotechnol. 2014, 174, 880–896. [Google Scholar] [CrossRef] [PubMed]
- Shanbhag, P.P.; Patil, N.S. BioMicroelectromechanical systems: A novel approach for drug targeting in chronic diseases. Eur. J. Mol. Clin. Med. 2017, 3, 265. [Google Scholar] [CrossRef]
- Rafiee, P.; Khatibi, G.; Zehetbauer, M. A review of the most important failure, reliability and nonlinearity aspects in the development of microelectromechanical systems (MEMS). Microelectron. Int. 2017, 34, 9–21. [Google Scholar] [CrossRef]
- Herrera-May, A.L.; Soler-Balcazar, J.C.; Vázquez-Leal, H.; Martínez-Castillo, J.; Vigueras-Zuñiga, M.O.; Aguilera-Cortés, L.A. Recent Advances of MEMS Resonators for Lorentz Force Based Magnetic Field Sensors: Design, Applications and Challenges. Sensors 2016, 16, 1359. [Google Scholar] [CrossRef] [PubMed]
- Chircov, C.; Grumezescu, A.M. Microelectromechanical Systems (MEMS) for Biomedical Applications. Micromachines 2022, 13, 164. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Fan, Z.H. Thermoplastic microfluidic devices and their applications in protein and DNA analysis. Analyst 2011, 136, 1288–1297. [Google Scholar] [CrossRef]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar] [CrossRef]
- Iqbal, M.J.; Javed, Z.; Herrera-Bravo, J.; Sadia, H.; Anum, F.; Raza, S.; Tahir, A.; Shahwani, M.N.; Sharifi-Rad, J.; Calina, D.; et al. Biosensing chips for cancer diagnosis and treatment: A new wave towards clinical innovation. Cancer Cell Int. 2022, 22, 354. [Google Scholar] [CrossRef]
- Žvirblytė, J.; Mažutis, L. Microfluidics for Cancer Biomarker Discovery, Research, and Clinical Application. Adv. Exp. Med. Biol. 2022, 1379, 499–524. [Google Scholar] [CrossRef]
- Yang, Y.; Kannisto, E.; Yu, G.; Reid, M.E.; Patnaik, S.K.; Wu, Y. An Immuno-Biochip Selectively Captures Tumor-Derived Exosomes and Detects Exosomal RNAs for Cancer Diagnosis. ACS Appl. Mater. Interfaces 2018, 10, 43375–43386. [Google Scholar] [CrossRef]
- Parihar, A.; Singhal, A.; Kumar, N.; Khan, R.; Khan, M.A.; Srivastava, A.K. Next-Generation Intelligent MXene-Based Electrochemical Aptasensors for Point-of-Care Cancer Diagnostics. Nano-Micro Lett. 2022, 14, 100. [Google Scholar] [CrossRef] [PubMed]
- Sadiq, M.; Pang, L.; Johnson, M.; Sathish, V.; Zhang, Q.; Wang, D. 2D Nanomaterial, Ti3C2 MXene-Based Sensor to Guide Lung Cancer Therapy and Management. Biosensors 2021, 11, 40. [Google Scholar] [CrossRef]
- Yang, X.; Xu, Y.; Wang, T.; Shu, D.; Guo, P.; Miskimins, K.; Qian, S.Y. Inhibition of cancer migration and invasion by knocking down delta-5-desaturase in COX-2 overexpressed cancer cells. Redox Biol. 2017, 11, 653–662. [Google Scholar] [CrossRef]
- Saghir, Z.; Dirksen, A.; Ashraf, H.; Bach, K.S.; Brodersen, J.; Clementsen, P.F.; Døssing, M.; Hansen, H.; Kofoed, K.F.; Larsen, K.R.; et al. CT screening for lung cancer brings forward early disease. The randomised Danish Lung Cancer Screening Trial: Status after five annual screening rounds with low-dose CT. Thorax 2012, 67, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Oudkerk, M.; Devaraj, A.; Vliegenthart, R.; Henzler, T.; Prosch, H.; Heussel, C.P.; Bastarrika, G.; Sverzellati, N.; Mascalchi, M.; Delorme, S.; et al. European position statement on lung cancer screening. Lancet Oncol. 2017, 18, e754–e766. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gole, J.; Gore, A.; He, Q.; Lu, M.; Min, J.; Yuan, Z.; Yang, X.; Jiang, Y.; Zhang, T.; et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat. Commun. 2020, 11, 3475. [Google Scholar] [CrossRef] [PubMed]
- Fiala, C.; Diamandis, E.P. Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection. BMC Med. 2018, 16, 166. [Google Scholar] [CrossRef]
- Liang, W.; Zhao, Y.; Huang, W.; Gao, Y.; Xu, W.; Tao, J.; Yang, M.; Li, L.; Ping, W.; Shen, H.; et al. Non-invasive diagnosis of early-stage lung cancer using high-throughput targeted DNA methylation sequencing of circulating tumor DNA (ctDNA). Theranostics 2019, 9, 2056–2070. [Google Scholar] [CrossRef]
- Zhang, B.; Niu, X.; Zhang, Q.; Wang, C.; Liu, B.; Yue, D.; Li, C.; Giaccone, G.; Li, S.; Gao, L.; et al. Circulating tumor DNA detection is correlated to histologic types in patients with early-stage non-small-cell lung cancer. Lung Cancer 2019, 134, 108–116. [Google Scholar] [CrossRef]
- Knight, S.B.; Phil, A.; Crosbie, P.A.; Balata, H.; Chudziak, J.; Hussell, T.; Dive, C. Progress and prospects of early detection in lung cancer. Open Biol. 2017, 7, 170070. [Google Scholar] [CrossRef]
- Marmor, H.N.; Zorn, J.T.; Deppen, S.A.; Massion, P.P.; Grogan, E.L. Biomarkers in lung cancer screening: A narrative review. Curr. Chall. Thorac. Surg. 2023, 5, 5. [Google Scholar] [CrossRef]
- Xing, W.; Sun, H.; Yan, C.; Zhao, C.; Wang, D.; Li, M.; Ma, J. A prediction model based on DNA methylation biomarkers and radiological characteristics for identifying malignant from benign pulmonary nodules. BMC Cancer 2021, 21, 263. [Google Scholar] [CrossRef]
- Voigt, W.; Prosch, H.; Silva, M. Clinical Scores, Biomarkers and IT Tools in Lung Cancer Screening—Can an Integrated Approach Overcome Current Challenges? Cancers 2023, 15, 1218. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Dave, S.; Nie, S.; True, L.; Gao, X. Multicolor quantum dots for molecular diagnostics of cancer. Expert Rev. Mol. Diagn. 2006, 6, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Pietryga, J.M.; Schaller, R.D.; Werder, D.; Stewart, M.H.; Klimov, V.I.; Hollingsworth, J.A. Pushing the Band Gap Envelope: Mid-Infrared Emitting Colloidal PbSe Quantum Dots. J. Am. Chem. Soc. 2004, 126, 11752–11753. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Feng, Y.; Knoll, W.; Han, M. Alloyed ZnxCd1-xS Nanocrystals with Highly Narrow Luminescence Spectral Width. J. Am. Chem. Soc. 2003, 125, 13559–13563. [Google Scholar] [CrossRef]
- Tade, R.S.; More, M.P.; Nangare, S.N.; Patil, P.O. Graphene quantum dots (GQDs) nanoarchitectonics for theranostic application in lung cancer. J. Drug Target. 2021, 30, 269–286. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Xu, T.; Zhang, X. Graphene-Based Biosensors for Detection of Biomarkers. Micromachines 2020, 11, 60. [Google Scholar] [CrossRef]
- Mastroianni, L.; Vajglová, Z.; Eränen, K.; Peurla, M.; Di Serio, M.; Murzin, D.Y.; Russo, V.; Salmi, T. Microreactor technology in experimental and modelling study of alcohol oxidation on nanogold. Chem. Eng. Sci. 2022, 260, 117920. [Google Scholar] [CrossRef]
- Šalić, A.; Tušek, A.; Zelić, B. Application of microreactors in medicine and biomedicine. J. Appl. Biomed. 2012, 10, 137–153. [Google Scholar] [CrossRef]
- Moro, M.; Bertolini, G.; Tortoreto, M.; Pastorino, U.; Sozzi, G.; Roz, L. Patient-Derived Xenografts of Non Small Cell Lung Cancer: Resurgence of an Old Model for Investigation of Modern Concepts of Tailored Therapy and Cancer Stem Cells. J. Biomed. Biotechnol. 2012, 2012, 568567. [Google Scholar] [CrossRef]
- Baschnagel, A.M.; Kaushik, S.; Durmaz, A.; Goldstein, S.; Ong, I.M.; Abel, L.; Clark, P.A.; Gurel, Z.; Leal, T.; Buehler, D.; et al. Development and characterization of patient-derived xenografts from non-small cell lung cancer brain metastases. Sci. Rep. 2021, 11, 2520. [Google Scholar] [CrossRef]
- Lee, H.W.; Lee, J.-I.; Lee, S.J.; Cho, H.J.; Song, H.J.; Jeong, D.E.; Seo, Y.J.; Shin, S.; Joung, J.-G.; Kwon, Y.-J.; et al. Patient-Derived Xenografts from Non–Small Cell Lung Cancer Brain Metastases Are Valuable Translational Platforms for the Development of Personalized Targeted Therapy. Clin. Cancer Res. 2015, 21, 1172–1182. [Google Scholar] [CrossRef]
- Wang, D.; Pham, N.-A.; Tong, J.; Sakashita, S.; Allo, G.; Kim, L.; Yanagawa, N.; Raghavan, V.; Wei, Y.; To, C.; et al. Molecular heterogeneity of non-small cell lung carcinoma patient-derived xenografts closely reflect their primary tumors. Int. J. Cancer 2016, 140, 662–673. [Google Scholar] [CrossRef]
- Abdolahi, S.; Ghazvinian, Z.; Muhammadnejad, S.; Saleh, M.; Aghdaei, H.A.; Baghaei, K. Patient-derived xenograft (PDX) models, applications and challenges in cancer research. J. Transl. Med. 2022, 20, 206. [Google Scholar] [CrossRef]
- Huang, Y.; Huang, Z.; Tang, Z.; Chen, Y.; Huang, M.; Liu, H.; Huang, W.; Ye, Q.; Jia, B. Research Progress, Challenges, and Breakthroughs of Organoids as Disease Models. Front. Cell Dev. Biol. 2021, 9, 740574. [Google Scholar] [CrossRef] [PubMed]
- Hoarau-Véchot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and Animal Models: Are 3D Cultures the Ideal Tool to Study Cancer-Microenvironment Interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef] [PubMed]
- Voskoglou-Nomikos, T.; Pater, J.L.; Seymour, L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin. Cancer Res. 2003, 9, 4227–4239. [Google Scholar]
- Katt, M.E.; Placone, A.L.; Wong, A.D.; Xu, Z.S.; Searson, P.C. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front. Bioeng. Biotechnol. 2016, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Monteduro, A.G.; Rizzato, S.; Caragnano, G.; Trapani, A.; Giannelli, G.; Maruccio, G. Organs-on-chips technologies—A guide from disease models to opportunities for drug development. Biosens. Bioelectron. 2023, 231, 115271. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Flecknell, P. Replacement, reduction and refinement. Altex 2002, 19, 73–78. [Google Scholar]
- Ronaldson-Bouchard, K.; Vunjak-Novakovic, G. Organs-on-a-Chip: A Fast Track for Engineered Human Tissues in Drug Development. Cell Stem Cell 2018, 22, 310–324. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Li, E.; Guo, Z.; Yu, R.; Hao, H.; Xu, Y.; Sun, Z.; Li, X.; Lyu, J.; Wang, Q. Design and Construction of a Multi-Organ Microfluidic Chip Mimicking the in vivo Microenvironment of Lung Cancer Metastasis. ACS Appl. Mater. Interfaces 2016, 8, 25840–25847. [Google Scholar] [CrossRef]
- Kang, D.; Park, J.A.; Kim, W.; Kim, S.; Lee, H.; Kim, W.; Yoo, J.; Jung, S. All-Inkjet-Printed 3D Alveolar Barrier Model with Physiologically Relevant Microarchitecture. Adv. Sci. 2021, 8, 2004990. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Lee, Y.; Kang, D.; Kwak, T.; Lee, H.-R.; Jung, S. 3D Inkjet-Bioprinted Lung-on-a-Chip. ACS Biomater. Sci. Eng. 2023, 9, 2806–2815. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fang, J.; Huang, S.; Wu, X.; Xie, X.; Wang, J.; Liu, F.; Zhang, M.; Peng, Z.; Hu, N. Tumor-on-a-chip: From bioinspired design to biomedical application. Microsystems Nanoeng. 2021, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef]
- Long, C.; Finch, C.; Esch, M.; Anderson, W.; Shuler, M.; Hickman, J. Design Optimization of Liquid-Phase Flow Patterns for Microfabricated Lung on a Chip. Ann. Biomed. Eng. 2012, 40, 1255–1267. [Google Scholar] [CrossRef]
- Hassell, B.A.; Goyal, G.; Lee, E.; Sontheimer-Phelps, A.; Levy, O.; Chen, C.S.; Ingber, D.E. Human Organ Chip Models Recapitulate Orthotopic Lung Cancer Growth, Therapeutic Responses, and Tumor Dormancy In Vitro. Cell Rep. 2017, 21, 508–516. [Google Scholar] [CrossRef]
- Yang, X.; Li, K.; Zhang, X.; Liu, C.; Guo, B.; Wen, W.; Gao, X. Nanofiber membrane supported lung-on-a-chip microdevice for anti-cancer drug testing. Lab a Chip 2018, 18, 486–495. [Google Scholar] [CrossRef]
- Park, S.; Kim, T.H.; Kim, S.H.; You, S.; Jung, Y. Three-Dimensional Vascularized Lung Cancer-on-a-Chip with Lung Extracellular Matrix Hydrogels for In Vitro Screening. Cancers 2021, 13, 3930. [Google Scholar] [CrossRef]
- Khalid, M.A.U.; Kim, Y.S.; Ali, M.; Lee, B.G.; Cho, Y.-J.; Choi, K.H. A lung cancer-on-chip platform with integrated biosensors for physiological monitoring and toxicity assessment. Biochem. Eng. J. 2020, 155, 107469. [Google Scholar] [CrossRef]
- Liao, H.-J.; Chieh, J.-A.; Chen, Y.-C.; Lee, K.-Y.; Chan, Y.-F.; Ho, S.-C.; Sun, W.-L.; Wang, Y.-S.; Huang, W.-C.; Chang, W.-C.; et al. Lung Cancer On Chip for Testing Immunotherapy. In Proceedings of the 21st International Conference on Solid-State Sensors, Actuators and Microsystems (Transducers), Orlando, FL, USA, 20–24 June 2021; pp. 1032–1035. [Google Scholar] [CrossRef]
- Carvalho, Â.; Ferreira, G.; Seixas, D.; Guimarães-Teixeira, C.; Henrique, R.; Monteiro, F.J.; Jerónimo, C. Emerging Lab-on-a-Chip Approaches for Liquid Biopsy in Lung Cancer: Status in CTCs and ctDNA Research and Clinical Validation. Cancers 2021, 13, 2101. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. Cancer genes and the pathways they control. Nat. Med. 2004, 10, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Kuijjer, M.L.; Paulson, J.N.; Salzman, P.; Ding, W.; Quackenbush, J. Cancer subtype identification using somatic mutation data. Br. J. Cancer 2018, 118, 1492–1501. [Google Scholar] [CrossRef]
- Waarts, M.R.; Stonestrom, A.J.; Park, Y.C.; Levine, R.L. Targeting mutations in cancer. J. Clin. Investig. 2022, 132, e154943. [Google Scholar] [CrossRef] [PubMed]
- Rodak, O.; Peris-Díaz, M.D.; Olbromski, M.; Podhorska-Okołów, M.; Dzięgiel, P. Current Landscape of Non-Small Cell Lung Cancer: Epidemiology, Histological Classification, Targeted Therapies, and Immunotherapy. Cancers 2021, 13, 4705. [Google Scholar] [CrossRef]
- Tafe, L.J.; Pierce, K.J.; Peterson, J.D.; de Abreu, F.; Memoli, V.A.; Black, C.C.; Pettus, J.R.; Marotti, J.D.; Gutmann, E.J.; Liu, X.; et al. Clinical Genotyping of Non–Small Cell Lung Cancers Using Targeted Next-Generation Sequencing: Utility of Identifying Rare and Co-mutations in Oncogenic Driver Genes. Neoplasia 2016, 18, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Beasley, M.B.; Chitale, D.A.; Dacic, S.; Giaccone, G.; Jenkins, R.B.; Kwiatkowski, D.J.; Saldivar, J.-S.; Squire, J.; et al. Molecular Testing Guideline for Selection of Lung Cancer Patients for EGFR and ALK Tyrosine Kinase Inhibitors. J. Mol. Diagn. 2013, 15, 415–453. [Google Scholar] [CrossRef]
- McCoach, C.E.; Bivona, T.G.; Blakely, C.M.; Doebele, R.C. Neoadjuvant Oncogene-Targeted Therapy in Early Stage Non–Small-Cell Lung Cancer as a Strategy to Improve Clinical Outcome and Identify Early Mechanisms of Resistance. Clin. Lung Cancer 2016, 17, 466–469. [Google Scholar] [CrossRef]
- Reyes, R.; Reguart, N. Neoadjuvant treatment of stage IIIA-N2 in EGFR-Mutant/ALK-rearranged non-small cell lung cancer. Transl. Lung Cancer Res. 2021, 10, 607–621. [Google Scholar] [CrossRef] [PubMed]
- de Scordilli, M.; Michelotti, A.; Bertoli, E.; De Carlo, E.; Del Conte, A.; Bearz, A. Targeted Therapy and Immunotherapy in Early-Stage Non-Small Cell Lung Cancer: Current Evidence and Ongoing Trials. Int. J. Mol. Sci. 2022, 23, 7222. [Google Scholar] [CrossRef] [PubMed]
- Loda, M. Polymerase chain reaction-based methods for the detection of mutations in oncogenes and tumor suppressor genes. Hum. Pathol. 1994, 25, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.; Pham, H.-A.T.; Tran, V.-U.; Tran, T.-T.; Dang, A.-T.H.; Le, D.-T.; Nguyen, S.-L.; Nguyen, N.-V.; Nguyen, T.-V.; Vo, B.T.; et al. Ultra-deep massively parallel sequencing with unique molecular identifier tagging achieves comparable performance to droplet digital PCR for detection and quantification of circulating tumor DNA from lung cancer patients. PLoS ONE 2019, 14, e0226193. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Tran, D.H.; Ngo, Q.D.; Pham, H.-A.T.; Tran, T.-T.; Tran, V.-U.; Pham, T.-V.N.; Le, T.K.; Le, N.-A.T.; Nguyen, N.M.; et al. Evaluation of a Liquid Biopsy Protocol using Ultra-Deep Massive Parallel Sequencing for Detecting and Quantifying Circulation Tumor DNA in Colorectal Cancer Patients. Cancer Investig. 2020, 38, 85–93. [Google Scholar] [CrossRef]
- Cainap, C.; Balacescu, O.; Cainap, S.S.; Pop, L.-A. Next Generation Sequencing Technology in Lung Cancer Diagnosis. Biology 2021, 10, 864. [Google Scholar] [CrossRef]
- Ahmad, R.; Wolfbeis, O.S.; Hahn, Y.-B.; Alshareef, H.N.; Torsi, L.; Salama, K.N. Deposition of nanomaterials: A crucial step in biosensor fabrication. Mater. Today Commun. 2018, 17, 289–321. [Google Scholar] [CrossRef]
- Fuchs, A.; Jeanson, H.; Claustre, P.; Gruss, J.; Revol-Cavalier, F.; Caillat, P.; Mastromatteo, U.; Scurati, M.; Villa, F.; Barlocchi, G.; et al. A silicon lab-on-chip for integrated sample preparation by PCR and DNA analysis by hybridization. In Proceedings of the 2nd Annual International IEEE-EMBS Special Topic Conference on Microtechnologies in Medicine and Biology. Proceedings (Cat. No.02EX578), Madison, WI, USA, 2–4 May 2002; pp. 227–231. [Google Scholar] [CrossRef]
- Ventimiglia, G.; Pesaturo, M.; Malcolm, A.; Petralia, S. A Miniaturized Silicon Lab-on-Chip for Integrated PCR and Hybridization Microarray for High Multiplexing Nucleic Acids Analysis. Biosensors 2022, 12, 563. [Google Scholar] [CrossRef]
- Li, H.; Sun, Z.; Zhong, W.; Hao, N.; Xu, D.; Chen, H.-Y. Ultrasensitive Electrochemical Detection For DNA Arrays Based on Silver Nanoparticle Aggregates. Anal. Chem. 2010, 82, 5477–5483. [Google Scholar] [CrossRef]
- Moulder, S.L.; Yakes, F.M.; Muthuswamy, S.K.; Bianco, R.; Simpson, J.F.; Arteaga, C.L. Epidermal growth factor receptor (HER1) tyrosine kinase inhibitor ZD1839 (Iressa) inhibits HER2/neu (erbB2)-overexpressing breast cancer cells in vitro and in vivo. Cancer Res. 2001, 61, 8887–8895. [Google Scholar]
- Gohring, J.T.; Dale, P.S.; Fan, X. Detection of HER2 breast cancer biomarker using the opto-fluidic ring resonator biosensor. Sens. Actuators B Chem. 2010, 146, 226–230. [Google Scholar] [CrossRef]
- Wu, Z.-S.; Sun, Y.; Tan, Y.-Z.; Yang, S.; Feng, X.; Müllen, K. Three-Dimensional Graphene-Based Macro- and Mesoporous Frameworks for High-Performance Electrochemical Capacitive Energy Storage. J. Am. Chem. Soc. 2012, 134, 19532–19535. [Google Scholar] [CrossRef]
- Hun, X.; Xie, G.; Luo, X. Scaling up an electrochemical signal with a catalytic hairpin assembly coupling nanocatalyst label for DNA detection. Chem. Commun. 2015, 51, 7100–7103. [Google Scholar] [CrossRef]
- Elhakim, H.K.; Azab, S.M.; Fekry, A.M. A novel simple biosensor containing silver nanoparticles/propolis (bee glue) for microRNA let-7a determination. Mater. Sci. Eng. C 2018, 92, 489–495. [Google Scholar] [CrossRef]
- Boots, A.W.; van Berkel, J.J.B.N.; Dallinga, J.W.; Smolinska, A.; Wouters, E.F.; van Schooten, F.J. The versatile use of exhaled volatile organic compounds in human health and disease. J. Breath Res. 2012, 6, 027108. [Google Scholar] [CrossRef] [PubMed]
- Heffler, E.; Carpagnano, G.E.; Favero, E.; Guida, G.; Maniscalco, M.; Motta, A.; Paoletti, G.; Rolla, G.; Baraldi, E.; Pezzella, V.; et al. Fractional Exhaled Nitric Oxide (FENO) in the management of asthma: A position paper of the Italian Respiratory Society (SIP/IRS) and Italian Society of Allergy, Asthma and Clinical Immunology (SIAAIC). Multidiscip. Respir. Med. 2020, 15, 36. [Google Scholar] [CrossRef] [PubMed]
- Gardner, J.W.; Bartlett, P.N. A brief history of electronic noses. Sens. Actuators B Chem. 1994, 18, 210–211. [Google Scholar] [CrossRef]
- Behera, B.; Chandra, S. A MEMS based acetone sensor incorporating ZnO nanowires synthesized by wet oxidation of Zn film. J. Micromech. Microeng. 2014, 25, 15007. [Google Scholar] [CrossRef]
- Behera, B.; Chandra, S. An innovative gas sensor incorporating ZnO–CuO nanoflakes in planar MEMS technology. Sens. Actuators B Chem. 2016, 229, 414–424. [Google Scholar] [CrossRef]
- Scott, S.M.; James, D.; Ali, Z. Data analysis for electronic nose systems. Microchim. Acta 2006, 156, 183–207. [Google Scholar] [CrossRef]
- Machado, R.F.; Laskowski, D.; Deffenderfer, O.; Burch, T.; Zheng, S.; Mazzone, P.J.; Mekhail, T.; Jennings, C.; Stoller, J.K.; Pyle, J.; et al. Detection of Lung Cancer by Sensor Array Analyses of Exhaled Breath. Am. J. Respir. Crit. Care Med. 2005, 171, 1286–1291. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.; Cataneo, R.N.; Cummin, A.R.; Gagliardi, A.J.; Gleeson, K.; Greenberg, J.; Maxfield, R.A.; Rom, W.N. Detection of Lung Cancer With Volatile Markers in the Breath. Chest 2003, 123, 2115–2123. [Google Scholar] [CrossRef] [PubMed]
- Horvath, I.; Lazar, Z.; Gyulai, N.; Kollai, M.; Losonczy, G. Exhaled biomarkers in lung cancer. Eur. Respir. J. 2009, 34, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Singh, T.S.; Yadava, R.D.S. MEMS sensor array-based electronic nose for breath analysis—A simulation study. J. Breath Res. 2018, 13, 016003. [Google Scholar] [CrossRef]
- Behera, B.; Joshi, R.; Vishnu, G.K.A.; Bhalerao, S.; Pandya, H.J. Electronic nose: A non-invasive technology for breath analysis of diabetes and lung cancer patients. J. Breath Res. 2019, 13, 024001. [Google Scholar] [CrossRef]
- Filipiak, W.; Mochalski, P.; Filipiak, A.; Ager, C.; Cumeras, R.; Davis, C.E.; Agapiou, A.; Unterkofler, K.; Troppmair, J. A Compendium of Volatile Organic Compounds (VOCs) Released By Human Cell Lines. Curr. Med. Chem. 2016, 23, 2112–2131. [Google Scholar] [CrossRef] [PubMed]
- Shlomi, D.; Abud, M.; Liran, O.; Bar, J.; Gai-Mor, N.; Ilouze, M.; Onn, A.; Ben-Nun, A.; Haick, H.; Peled, N. Detection of Lung Cancer and EGFR Mutation by Electronic Nose System. J. Thorac. Oncol. 2017, 12, 1544–1551. [Google Scholar] [CrossRef]
- Tirzïte, M.; Bukovskis, M.; Strazda, G.; Jurka, N.; Taivans, I. Detection of lung cancer with electronic nose and logistic regression analysis. J. Breath Res. 2018, 13, 016006. [Google Scholar] [CrossRef]
- Tran, V.H.; Chan, H.P.; Thurston, M.; Jackson, P.; Lewis, C.; Yates, D.; Bell, G.; Thomas, P.S. Breath Analysis of Lung Cancer Patients Using an Electronic Nose Detection System. IEEE Sens. J. 2010, 10, 1514–1518. [Google Scholar] [CrossRef]
- Bruins, M.; Gerritsen, J.W.; van de Sande, W.W.; van Belkum, A.; Bos, A. Enabling a transferable calibration model for metal-oxide type electronic noses. Sens. Actuators B Chem. 2013, 188, 1187–1195. [Google Scholar] [CrossRef]
- Kort, S.; Brusse-Keizer, M.; Gerritsen, J.-W.; van der Palen, J. Data analysis of electronic nose technology in lung cancer: Generating prediction models by means of Aethena. J. Breath Res. 2017, 11, 026006. [Google Scholar] [CrossRef]
- Kort, S.; Brusse-Keizer, M.; Schouwink, H.; Citgez, E.; de Jongh, F.H.; van Putten, J.W.; Borne, B.v.D.; Kastelijn, E.A.; Stolz, D.; Schuurbiers, M.; et al. Diagnosing Non-Small Cell Lung Cancer by Exhaled Breath Profiling Using an Electronic Nose. Chest 2022, 163, 697–706. [Google Scholar] [CrossRef]
- Chen, M.; Cui, D.; Haick, H.; Tang, N. Artificial Intelligence-Based Medical Sensors for Healthcare System. Adv. Sens. Res. 2023, 2300009. [Google Scholar] [CrossRef]
- Kammer, M.N.; Paez, R. It Doesn’t Smell Like Cancer to Me. Chest 2023, 163, 479–480. [Google Scholar] [CrossRef]
- Narula, J.; Chandrashekhar, Y.; Braunwald, E. Time to Add a Fifth Pillar to Bedside Physical Examination. JAMA Cardiol. 2018, 3, 346–350. [Google Scholar] [CrossRef]
- Price, A.; Long, B. Fibonacci Spiral Arranged Ultrasound Phased Array for Mid-Air Haptics. In Proceedings of the 2018 IEEE International Ultrasonics Symposium (IUS), Kobe, Japan, 22–25 October 2018; pp. 1–4. [Google Scholar] [CrossRef]
- Choi, H. Prelinearized Class-B Power Amplifier for Piezoelectric Transducers and Portable Ultrasound Systems. Sensors 2019, 19, 287. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ma, Y.; Yang, H.; Jiang, H.; Ding, Y.; Xie, H. MEMS Ultrasound Transducers for Endoscopic Photoacoustic Imaging Applications. Micromachines 2020, 11, 928. [Google Scholar] [CrossRef] [PubMed]
- Schabowicz, K. Ultrasonic tomography—The latest nondestructive technique for testing concrete members—Description, test methodology, application example. Arch. Civ. Mech. Eng. 2014, 14, 295–303. [Google Scholar] [CrossRef]
- Takiguchi, T. Ultrasonic Tomographic Technique and Its Applications. Appl. Sci. 2019, 9, 1005. [Google Scholar] [CrossRef]
- He, Y.; Wan, H.; Jiang, X.; Peng, C. Piezoelectric Micromachined Ultrasound Transducer Technology: Recent Advances and Applications. Biosensors 2022, 13, 55. [Google Scholar] [CrossRef]
- Birjis, Y.; Swaminathan, S.; Nazemi, H.; Raj, G.C.A.; Munirathinam, P.; Abu-Libdeh, A.; Emadi, A. Piezoelectric Micromachined Ultrasonic Transducers (PMUTs): Performance Metrics, Advancements, and Applications. Sensors 2022, 22, 9151. [Google Scholar] [CrossRef]
- Jiao, P.; Egbe, K.-J.I.; Xie, Y.; Nazar, A.M.; Alavi, A.H. Piezoelectric Sensing Techniques in Structural Health Monitoring: A State-of-the-Art Review. Sensors 2020, 20, 3730. [Google Scholar] [CrossRef] [PubMed]
- Muralt, P.; Ledermann, N.; Baborowski, J.; Barzegar, A.; Gentil, S.; Belgacem, B.; Petitgrand, S.; Bosseboeuf, A.; Setter, N. Piezoelectric micromachined ultrasonic transducers based on PZT thin films. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2005, 52, 2276–2288. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.-H.; Liu, H.-J. Piezoelectric Micromachined Ultrasonic Transducers with a Cost-Effective Bottom-Up Fabrication Scheme for Millimeter-Scale Range Finding. Sensors 2019, 19, 4696. [Google Scholar] [CrossRef]
- Pan, J.; Bai, C.; Zheng, Q.; Xie, H. Review of Piezoelectric Micromachined Ultrasonic Transducers for Rangefinders. Micromachines 2023, 14, 374. [Google Scholar] [CrossRef]
- Rothberg, J.M.; Ralston, T.S.; Rothberg, A.G.; Martin, J.; Zahorian, J.S.; Alie, S.A.; Sanchez, N.J.; Chen, K.; Chen, C.; Thiele, K.; et al. Ultrasound-on-chip platform for medical imaging, analysis, and collective intelligence. Proc. Natl. Acad. Sci. USA 2021, 118, e2019339118. [Google Scholar] [CrossRef]
- Song, E.; Huang, Y.; Huang, N.; Mei, Y.; Yu, X.; Rogers, J.A. Recent advances in microsystem approaches for mechanical characterization of soft biological tissues. Microsyst. Nanoeng. 2022, 8, 77. [Google Scholar] [CrossRef]
- Qiu, Y.; Gigliotti, J.V.; Wallace, M.; Griggio, F.; DeMore, C.E.M.; Cochran, S.; Trolier-McKinstry, S. Piezoelectric Micromachined Ultrasound Transducer (PMUT) Arrays for Integrated Sensing, Actuation and Imaging. Sensors 2015, 15, 8020–8041. [Google Scholar] [CrossRef]
- Peralta, L.; Gomez, A.; Luan, Y.; Kim, B.-H.; Hajnal, J.V.; Eckersley, R.J. Coherent Multi-Transducer Ultrasound Imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2019, 66, 1316–1330. [Google Scholar] [CrossRef] [PubMed]
- Peralta, L.; Ramalli, A.; Reinwald, M.; Eckersley, R.J.; Hajnal, J.V. Impact of Aperture, Depth, and Acoustic Clutter on the Performance of Coherent Multi-Transducer Ultrasound Imaging. Appl. Sci. 2020, 10, 7655. [Google Scholar] [CrossRef]
- Montaldo, G.; Tanter, M.; Bercoff, J.; Benech, N.; Fink, M. Coherent plane-wave compounding for very high frame rate ultrasonography and transient elastography. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2009, 56, 489–506. [Google Scholar] [CrossRef]
- Tanter, M.; Fink, M. Ultrafast imaging in biomedical ultrasound. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2014, 61, 102–119. [Google Scholar] [CrossRef]
- Denarie, B.; Tangen, T.A.; Ekroll, I.K.; Rolim, N.; Torp, H.; Bjåstad, T.; Lovstakken, L. Coherent Plane Wave Compounding for Very High Frame Rate Ultrasonography of Rapidly Moving Targets. IEEE Trans. Med Imaging 2013, 32, 1265–1276. [Google Scholar] [CrossRef]
- Foiret, J.; Cai, X.; Bendjador, H.; Park, E.-Y.; Kamaya, A.; Ferrara, K.W. Improving plane wave ultrasound imaging through real-time beamformation across multiple arrays. Sci. Rep. 2022, 12, 13386. [Google Scholar] [CrossRef] [PubMed]
- Moghimirad, E.; Bamber, J.C.; Harris, E.J. Plane wave versus focused transmissions for contrast enhanced ultrasound imaging: The role of parameter settings and the effects of flow rate on contrast measurements. Phys. Med. Biol. 2019, 64, 095003. [Google Scholar] [CrossRef] [PubMed]
- Schiefler, J.N.T.; Maia, J.M.; Schneider, F.K.; Zimbico, A.J.; Assef, A.A.; Costa, E.T. Generation and Analysis of Ultrasound Images Using Plane Wave and Sparse Arrays Techniques. Sensors 2018, 18, 3660. [Google Scholar] [CrossRef] [PubMed]
- Saad, Y. Parallel Iterative Methods for Sparse Linear Systems. In Studies in Computational Mathematics; Elsevier: Amsterdam, The Netherlands, 2001; Volume 8, pp. 423–440. [Google Scholar] [CrossRef]
- Garcia, D.; Le Tarnec, L.; Muth, S.; Montagnon, E.; Porée, J.; Cloutier, G. Stolt’s f-k migration for plane wave ultrasound imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2013, 60, 1853–1867. [Google Scholar] [CrossRef]
- Tomizawa, M.; Shinozaki, F.; Motoyoshi, Y.; Sugiyama, T.; Yamamoto, S.; Sueishi, M. Sonoporation: Gene transfer using ultrasound. World J. Methodol. 2013, 3, 39–44. [Google Scholar] [CrossRef]
- Carovac, A.; Smajlovic, F.; Junuzovic, D. Application of Ultrasound in Medicine. Acta Inform. Medica 2011, 19, 168–171. [Google Scholar] [CrossRef]
- Heikkola, E.; Laitinen, M. Model-based optimization of ultrasonic transducers. Ultrason. Sonochem. 2005, 12, 53–57. [Google Scholar] [CrossRef]
- Fröhlich, E.; Salar-Behzadi, S. Oral inhalation for delivery of proteins and peptides to the lungs. Eur. J. Pharm. Biopharm. 2021, 163, 198–211. [Google Scholar] [CrossRef]
- Carvalho, T.C.; McConville, J.T. The function and performance of aqueous aerosol devices for inhalation therapy. J. Pharm. Pharmacol. 2016, 68, 556–578. [Google Scholar] [CrossRef]
- van Rooij, T.; Skachkov, I.; Beekers, I.; Lattwein, K.R.; Voorneveld, J.D.; Kokhuis, T.J.; Bera, D.; Luan, Y.; van der Steen, A.F.; de Jong, N.; et al. Viability of endothelial cells after ultrasound-mediated sonoporation: Influence of targeting, oscillation, and displacement of microbubbles. J. Control. Release 2016, 238, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, L.; Folliero, V.; Palomba, L.; Zannella, C.; Isticato, R.; Di Francia, R.; Berretta, M.; de Sio, I.; Adinolfi, L.E.; Morelli, G.; et al. Sonoporation by microbubbles as gene therapy approach against liver cancer. Oncotarget 2018, 9, 32182–32190. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hu, Z.; Sukumar, U.K.; Bose, R.J.; Telichko, A.; Dahl, J.J.; Paulmurugan, R. Ultrasound-Guided Microbubble-Mediated Locoregional Delivery of Multiple MicroRNAs Improves Chemotherapy in Hepatocellular Carcinoma. Nanotheranostics 2022, 6, 62–78. [Google Scholar] [CrossRef] [PubMed]
- Feichtinger, G.A.; Hofmann, A.T.; Slezak, P.; Schuetzenberger, S.; Kaipel, M.; Schwartz, E.; Neef, A.; Nomikou, N.; Nau, T.; van Griensven, M.; et al. Sonoporation Increases Therapeutic Efficacy of Inducible and Constitutive BMP2/7 In Vivo Gene Delivery. Hum. Gene Ther. Methods 2014, 25, 57–71. [Google Scholar] [CrossRef]
- Ling, T.-Y.; Liu, Y.-L.; Huang, Y.-K.; Gu, S.-Y.; Chen, H.-K.; Ho, C.-C.; Tsao, P.-N.; Tung, Y.-C.; Chen, H.-W.; Cheng, C.-H.; et al. Differentiation of lung stem/progenitor cells into alveolar pneumocytes and induction of angiogenesis within a 3D gelatin—Microbubble scaffold. Biomaterials 2014, 35, 5660–5669. [Google Scholar] [CrossRef]
- Petrikaite, V.; Paškevičiūtė, M.; Raišutis, R.; Sakalauskienė, K. Application of sonoporation to increase anticancer drug efficacy in 2D and 3D NSCLC cell cultures. Ann. Oncol. 2019, 30, v4–v5. [Google Scholar] [CrossRef]
- Castle, J.; Kotopoulis, S.; Forsberg, F. Sonoporation for Augmenting Chemotherapy of Pancreatic Ductal Adenocarcinoma. Methods Mol Biol. 2019, 2059, 191–205. [Google Scholar] [CrossRef]
- Hirabayashi, F.; Iwanaga, K.; Okinaga, T.; Takahashi, O.; Ariyoshi, W.; Suzuki, R.; Sugii, M.; Maruyama, K.; Tominaga, K.; Nishihara, T. Epidermal growth factor receptor-targeted sonoporation with microbubbles enhances therapeutic efficacy in a squamous cell carcinoma model. PLoS ONE 2017, 12, e0185293. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, G.; Liu, L.; Li, Z. Emerging biosensing technologies for improved diagnostics of COVID-19 and future pandemics. Talanta 2020, 225, 121986. [Google Scholar] [CrossRef]
- Mahshid, S.S.; Flynn, S.E.; Mahshid, S. The potential application of electrochemical biosensors in the COVID-19 pandemic: A perspective on the rapid diagnostics of SARS-CoV-2. Biosens. Bioelectron. 2020, 176, 112905. [Google Scholar] [CrossRef]
- Velten, T.; Ruf, H.; Barrow, D.; Aspragathos, N.; Lazarou, P.; Jung, E.; Malek, C.; Richter, M.; Kruckow, J.; Wackerle, M. Packaging of bio-MEMS: Strategies, technologies, and applications. IEEE Trans. Adv. Packag. 2005, 28, 533–546. [Google Scholar] [CrossRef]
- Laurencin, C.T.; Langer, R. Polymeric Controlled Release Systems: New Methods for Drug Delivery. Clin. Lab. Med. 1987, 7, 301–324. [Google Scholar] [CrossRef] [PubMed]
- Robitzki, A.A.; Kurz, R. Biosensing and Drug Delivery at the Microscale. Handb. Exp. Pharmacol. 2010, 197, 87–112. [Google Scholar] [CrossRef]
- Ravi Kumar, M.N. Nano and microparticles as controlled drug delivery devices. J. Pharm. Pharm. Sci. 2000, 3, 234–358. [Google Scholar] [PubMed]
- Wolinsky, J.B.; Colson, Y.L.; Grinstaff, M.W. Local drug delivery strategies for cancer treatment: Gels, nanoparticles, polymeric films, rods, and wafers. J. Control. Release 2012, 159, 14–26. [Google Scholar] [CrossRef]
- Altorki, N.K.; Wang, X.; Wigle, D.; Gu, L.; Darling, G.; Ashrafi, A.S.; Landrenau, R.; Miller, D.; Liberman, M.; Jones, D.R.; et al. Perioperative mortality and morbidity after sublobar versus lobar resection for early-stage non-small-cell lung cancer: Post-hoc analysis of an international, randomised, phase 3 trial (CALGB/Alliance 140503). Lancet Respir. Med. 2018, 6, 915–924. [Google Scholar] [CrossRef]
- Piloni, D.; Bertuccio, F.R.; Primiceri, C.; Rinaldi, P.; Chino, V.; Abbott, D.M.; Sottotetti, F.; Bortolotto, C.; Agustoni, F.; Saddi, J.; et al. Smoking Habit and Respiratory Function Predict Patients’ Outcome after Surgery for Lung Cancer, Irrespective of Histotype and Disease Stage. J. Clin. Med. 2023, 12, 1561. [Google Scholar] [CrossRef]
- Harper, E.; Dang, W.; Lapidus, R.G.; I Garver, R. Enhanced efficacy of a novel controlled release paclitaxel formulation (PACLIMER delivery system) for local-regional therapy of lung cancer tumor nodules in mice. Clin. Cancer Res. 1999, 5, 4242–4248. [Google Scholar]
- Shen, H.; Hu, D.; Du, J.; Wang, X.; Liu, Y.; Wang, Y.; Wei, J.-M.; Ma, D.; Wang, P.; Li, L. Paclitaxel–octreotide conjugates in tumor growth inhibition of A549 human non-small cell lung cancer xenografted into nude mice. Eur. J. Pharmacol. 2008, 601, 23–29. [Google Scholar] [CrossRef]
- Goel, A.; Sahni, J.; Ali, J.; Baboota, S. Exploring targeted pulmonary delivery for treatment of lung cancer. Int. J. Pharm. Investig. 2013, 3, 8–14. [Google Scholar] [CrossRef]
- Storti, C.; Le Noci, V.; Sommariva, M.; Tagliabue, E.; Balsari, A.; Sfondrini, L. Aerosol delivery in the treatment of lung cancer. Curr. Cancer Drug Targets 2015, 15, 604–612. [Google Scholar] [CrossRef]
- Gagnadoux, F.; Hureaux, J.; Vecellio, L.; Urban, T.; Valo, I.; Montharu, J.; Leblond, V.; Boisdron-Celle, M.; Lerondel, S.; Majoral, C.; et al. Aerosolized Chemotherapy. J. Aerosol Med. Pulm. Drug Deliv. 2008, 21, 61–70. [Google Scholar] [CrossRef]
- Laube, B.L. The expanding role of aerosols in systemic drug delivery, gene therapy, and vaccination. Respir. Care 2005, 50, 1161–1176. [Google Scholar] [CrossRef]
- Rao, S.N.M.A.P.M.A.R.D.; Markovic, S.; Anderson, P. Aerosol Therapy for Malignancy Involving the Lungs. Curr. Cancer Drug Targets 2003, 3, 239–250. [Google Scholar] [CrossRef]
- Densmore, C.L. The Re-Emergence of Aerosol Gene Delivery: A Viable Approach to Lung Cancer Therapy. Curr. Cancer Drug Targets 2003, 3, 275–286. [Google Scholar] [CrossRef]
- Chen, L.; Chen, F.; Li, J.; Pu, Y.; Yang, C.; Wang, Y.; Lei, Y.; Huang, Y. CAR-T cell therapy for lung cancer: Potential and perspective. Thorac. Cancer 2022, 13, 889–899. [Google Scholar] [CrossRef]
- Xiao, B.-F.; Zhang, J.-T.; Zhu, Y.-G.; Cui, X.-R.; Lu, Z.-M.; Yu, B.-T.; Wu, N. Chimeric Antigen Receptor T-Cell Therapy in Lung Cancer: Potential and Challenges. Front. Immunol. 2021, 12, 782775. [Google Scholar] [CrossRef] [PubMed]
- Lisini, D.; Lettieri, S.; Nava, S.; Accordino, G.; Frigerio, S.; Bortolotto, C.; Lancia, A.; Filippi, A.R.; Agustoni, F.; Pandolfi, L.; et al. Local Therapies and Modulation of Tumor Surrounding Stroma in Malignant Pleural Mesothelioma: A Translational Approach. Int. J. Mol. Sci. 2021, 22, 9014. [Google Scholar] [CrossRef] [PubMed]
- Sutradhar, K.B.; Sumi, C.D. Implantable microchip: The futuristic controlled drug delivery system. Drug Deliv. 2014, 23, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Uvarov, I.; Lemekhov, V.; Melenev, S.S.; Naumov, A.E.; Koroleva, V.V.; Izyumov, M.O.; Svetovoy, V.B. A simple electrochemical micropump: Design and fabrication. J. Phys. Conf. Ser. 2016, 741, 012167. [Google Scholar] [CrossRef]
- Uvarov, I.; Lemekhov, V.; Melenev, S.S.; Svetovoy, V.B. A micropump driven by electrochemically produced short-lived bubbles. J. Phys. Conf. Ser. 2016, 757, 012008. [Google Scholar] [CrossRef]
- Zhang, H.; Jackson, J.K.; Chiao, M. Microfabricated Drug Delivery Devices: Design, Fabrication, and Applications. Adv. Funct. Mater. 2017, 27, 1703606. [Google Scholar] [CrossRef]
- Ashraf, M.W.; Tayyaba, S.; Afzulpurkar, N. Micro Electromechanical Systems (MEMS) Based Microfluidic Devices for Biomedical Applications. Int. J. Mol. Sci. 2011, 12, 3648–3704. [Google Scholar] [CrossRef] [PubMed]
- Mohith, S.; Karanth, P.N.; Kulkarni, S. Recent trends in mechanical micropumps and their applications: A review. Mechatronics 2019, 60, 34–55. [Google Scholar] [CrossRef]
- Meng, E.; Hoang, T. MEMS-enabled implantable drug infusion pumps for laboratory animal research, preclinical, and clinical applications. Adv. Drug Deliv. Rev. 2012, 64, 1628–1638. [Google Scholar] [CrossRef]
- Jonas, O.; Landry, H.M.; Fuller, J.E.; Santini, J.T., Jr.; Baselga, J.; Tepper, R.I.; Cima, M.J.; Langer, R. An implantable microdevice to perform high-throughput in vivo drug sensitivity testing in tumors. Sci. Transl. Med. 2015, 7, 284ra57. [Google Scholar] [CrossRef]
- Nathan, M. Microbattery Technologies for Miniaturized Implantable Medical Devices. Curr. Pharm. Biotechnol. 2010, 11, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Chung, A.J.; Huh, Y.S.; Erickson, D. A robust, electrochemically driven microwell drug delivery system for controlled vasopressin release. Biomed. Microdevices 2009, 11, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, D.W.; Al-Sarawi, S.; Abbott, D. Modelling and simulation of wirelessly and securely interrogated low-powered actuators for bio-MEMS. Smart Mater. Struct. 2010, 20, 015025. [Google Scholar] [CrossRef]
- Defrère, S.; Mestagdt, M.; Riva, R.; Krier, F.; Van Langendonckt, A.; Drion, P.; Jérôme, C.; Evrard, B.; Dehoux, J.-P.; Foidart, J.-M.; et al. In Vivo Biocompatibility of Three Potential Intraperitoneal Implants. Macromol. Biosci. 2011, 11, 1336–1345. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Sun, W.; Fang, J.; Lee, K.; Li, S.; Gu, Z.; Dokmeci, M.R.; Khademhosseini, A. Biodegradable Gelatin Methacryloyl Microneedles for Transdermal Drug Delivery. Adv. Health Mater. 2018, 8, e1801054. [Google Scholar] [CrossRef]
- Mendoza, L.A.V.; Scilletta, N.A.; Bellino, M.G.; DeSimone, M.F.; Catalano, P.N. Recent Advances in Micro-Electro-Mechanical Devices for Controlled Drug Release Applications. Front. Bioeng. Biotechnol. 2020, 8, 827. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Terry, R.N.; Tang, J.; Feng, M.R.; Schwendeman, S.P.; Prausnitz, M.R. Rapidly separable microneedle patch for the sustained release of a contraceptive. Nat. Biomed. Eng. 2019, 3, 220–229. [Google Scholar] [CrossRef]
- Al-Zu’bi, M.; Mohan, A. Modelling of combination therapy using implantable anticancer drug delivery with thermal ablation in solid tumor. Sci. Rep. 2020, 10, 19366. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Ruan, W.; Qin, M.; Long, Y.; Wan, T.; Yu, K.; Zhai, Y.; Wu, C.; Xu, Y. Intradermal delivery of STAT3 siRNA to treat melanoma via dissolving microneedles. Sci. Rep. 2018, 8, 1117. [Google Scholar] [CrossRef]
- Kim, K.; Sung, H.K.; Lee, K.; Park, S.K. Semiconductor Work, Leukemia, and Cancer Risk: A Systematic Review and Meta-Analysis. Int. J. Environ. Res. Public Health 2022, 19, 14733. [Google Scholar] [CrossRef]
- Mundt, K.A. Cancer risk in the semiconductor industry: Responding to the call for action. Occup. Environ. Med. 2006, 64, 5–6. [Google Scholar] [CrossRef]
- Kim, M.-H.; Kim, H.; Paek, D. The health impacts of semiconductor production: An epidemiologic review. Int. J. Occup. Environ. Health 2013, 20, 95–114. [Google Scholar] [CrossRef]
| Study Focus | Device/Sensor | Study Scope | Reference |
|---|---|---|---|
| Preclinical | Patients’ derived organoids based on 3D nanomatrices and 3D tumors Microfluidic device Microfluidic device | Evaluation of tumor functional vasculature Purification of circulating tumor cells Analysis of EGFR-mutated circulating tumor cells | NCT04826913 NCT04957602 NCT01193829 |
| Diagnosis | MEMS e-nose MEMS e-nose Acoustic sensors array (e-stethoscope) Wireless palpatory Electrosensing antibody probing system | Mutation detection Analysis of cancerogenic VOCs after surgery for cancer Mapping chest sound propagation Detection of subpleural tumors To improve costs and save time for mutation detection | [9] NCT0803137 NCT03043898 NCT03521615 NCT01359436 |
| Disease Monitoring | Mobile sensor technology MEMS mixed approach | To help in assessing symptoms, response to therapy and quality of life Adherence to therapy and persistence in relation to clinical outcome | NCT04465214 NCT 04347161 |
| Treatment | MEMS magnetic field sensor-based Artificial intelligence/Machine learning | Radiotherapy: more accurate breathing signal graph with lower measurement error and higher spatial resolution than conventional Prediction of efficacy of immunotherapy | [10] NCT05537922 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stella, G.M.; Lettieri, S.; Piloni, D.; Ferrarotti, I.; Perrotta, F.; Corsico, A.G.; Bortolotto, C. Smart Sensors and Microtechnologies in the Precision Medicine Approach against Lung Cancer. Pharmaceuticals 2023, 16, 1042. https://doi.org/10.3390/ph16071042
Stella GM, Lettieri S, Piloni D, Ferrarotti I, Perrotta F, Corsico AG, Bortolotto C. Smart Sensors and Microtechnologies in the Precision Medicine Approach against Lung Cancer. Pharmaceuticals. 2023; 16(7):1042. https://doi.org/10.3390/ph16071042
Chicago/Turabian StyleStella, Giulia Maria, Sara Lettieri, Davide Piloni, Ilaria Ferrarotti, Fabio Perrotta, Angelo Guido Corsico, and Chandra Bortolotto. 2023. "Smart Sensors and Microtechnologies in the Precision Medicine Approach against Lung Cancer" Pharmaceuticals 16, no. 7: 1042. https://doi.org/10.3390/ph16071042
APA StyleStella, G. M., Lettieri, S., Piloni, D., Ferrarotti, I., Perrotta, F., Corsico, A. G., & Bortolotto, C. (2023). Smart Sensors and Microtechnologies in the Precision Medicine Approach against Lung Cancer. Pharmaceuticals, 16(7), 1042. https://doi.org/10.3390/ph16071042







