Abstract
The continuing need for the discovery of potent antibacterial agents against antibiotic-resistant pathogens is the driving force for many researchers to design and develop such agents. Herein, we report the design, synthesis, and biological evaluation of amidine derivatives as new antibacterial agents. Compound 13d was the most active in this study against a wide range of antibiotic-resistant, and susceptible, Gram-positive, and Gram-negative bacterial strains. Time–kill assay experiments indicated that compound 13d was an effective bactericidal compound against the tested organisms at the log-phase of bacterial growth. Docking simulations were performed to assess in silico its mode of action regarding UPPS, KARI, and DNA as potential bacterial targets. Results unveiled the importance of structural features of compound 13d in its biological activity including central thiophene ring equipped with left and right pyrrolo[2,3-b]pyridine and phenyl moieties and two terminal amidines cyclized into 4,5-dihydro-1H-imidazol-2-yl functionalities. Collectively, compound 13d represents a possible hit for future development of potent antibacterial agents.
1. Introduction
Penicillin discovery and the subsequent development of several antibacterial chemotherapeutics over the 1950s to 1970s marked the ‘golden antibiotic era’ [1]. Nealy half of the known antibiotics were discovered and developed during these decades, which significantly reduced morbidity and mortality from bacterial infections. Unfortunately, the inappropriate, irresponsible, and excessive use of antibiotics resulted in the emergence and development of antimicrobial resistance (AMR) that impaired the effectiveness of antimicrobial treatment protocols [2]. Such a situation raises alarms of entry into a ‘post-antibiotic era’ in which AMR will render minor injuries and infections fatal [3]. Around 5 million deaths because of AMR were estimated in 2019 including nearly 1.3 million deaths because of bacterial AMR [4]. The global toll of AMR deaths is estimated to increase to 10 million deaths annually by 2050 [5]. It is therefore urgent to develop new agents effective against antibiotic-resistant bacteria.
As a workplan is needed to combat antimicrobial resistance, the World Health Organization (WHO) announced a global priority list of drug-resistant species that are the most urgent to be considered for development of new agents [6]. In the list, the ESKAPE bacteria that include multidrug-resistant Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp., were listed as critical and high priority organisms. Such ESKAPE organisms are a global leading cause of serious, life-threatening infections [7,8,9]. In many cases, ESKAPE pathogens are a leading cause of dangerous nosocomial infections resulting in deaths, particularly in immunocompromised patients [10]. The ability of such antibiotic-resistant bacteria to overcome the action of current drugs might be attributed to drug inactivation, modifications of the drug’s target receptor, or decreased drug accumulation within the organism either by efflux mechanisms or via decreased permeability. To foil bacterial resistance, it may be beneficial not only to address new targets but also to develop multitarget agents [11,12,13,14].
Selective toxicity is a desirable key property for the successful development of antimicrobial agents. Molecules that act on bacterial-specific targets could be toxic to bacteria but non-toxic for mammalian cells. Consequently, molecules targeting bacterial cell wall biosynthesis can selectively kill bacteria but do not impair mammalian cells. Bacterial undecaprenyl diphosphate synthase (UPPS) is an enzyme absent in mammals that is essential for the bacterial isoprenoid biosynthetic pathway of cell wall synthesis [15]. Consequently, UPPS could serve as a new promising target for the development of novel antibacterial agents. Ketol-acid reductoisomerase (KARI) is another mammals-absent key bacterial enzyme that can serve as a promising molecular target for developing novel antibacterial agents. KARI is involved in the biosynthesis of branched-chain amino acids (BCAA), which are the essential leucine, valine, and isoleucine amino acids. In contrast to bacteria and other microbes that utilize KARI for synthesis of such essential BCAA, mammals have no KARI. Accordingly, KARI inhibitors could be safe and effective antibacterial agents [16,17,18].
Minor groove binders (MGBs) are small molecules that bind within the minor groove of DNA B-form. Such binding disrupts nucleosomes resulting in suppression of DNA replication and transcription, which are essential biological processes for bacterial growth and multiplication. Such DNA replication and transcription requires promoter sequences that are sequences at which DNA unwinding is initiated. Such promoters involve either AT-rich or GC-rich repeated sequences. While human promoter sequences are mainly GC-rich, microbial promoter sequences predominantly involve AT-rich sequences [19,20,21]. Furthermore, B-DNA form of AT-rich sequences feature a narrower, deeper minor groove which is capable of establishing more hydrogen bond interactions than GC-rich sequences [22,23,24]. Such facts might offer an opportunity to develop selective and safe antimicrobials.
Diamidines are an interesting class of compounds that reported to possess interesting bioactivities [25,26,27,28]. While the literature reports show that some diamidines have antibacterial activities, studies of their antibacterial effects against antibiotic resistant bacteria, to the best of our knowledge, are scant and incomplete [29,30,31]. Interestingly, it was recently discovered that diamidines such as pentamidine and its congeners sensitize antibiotic resistant Gram-negative bacteria to Gram-positive antibiotics [32,33]. It might be intriguing to design and more thoroughly investigate the direct antibacterial effects of novel diamidines against antibiotic-resistant bacteria in comparison with antibiotic-susceptible bacteria. Consequently, we addressed, herein, the design, synthesis and assessment of small molecules incorporating the privileged amidine moiety to discover new multitarget antibacterial hit compounds against antibiotic-resistant bacteria. In this report, we present our approach and interesting results.
2. Results and Discussion
2.1. Design Approach
The structure of diamidines involves a central moiety linked to left and right aromatic moieties (Figure 1). Both left and right aromatic moieties carry an amidine moiety that might be in the form of an acyclic or cyclic moiety. Amongst interesting diamidines, the antiparasitic drug pentamidine (Figure 1) was found not only as a potential MGB but also as an inhibitor of E. coli KARI [34,35,36,37,38]. While pentamidine has a flexible central moiety of pentamidine, the rigid analog, furamidine has potential antimicrobial activity via MGB [34,37]. Compound MMV688271 (Figure 1), a furamidine analog which has chlorophenyl left and right aromatic moieties each bearing the amidine-like biguanide fragment, was found as an effective inhibitor of Mycobacterium tuberculosis KARI [18]. Compound MBX-1066 (Figure 1), which has larger left and right aromatic indole moieties combined with cyclic amidine moiety, was identified as an antibacterial agent eliciting potential UPPS inhibition that impairs bacterial cell wall synthesis in addition to its potential MGB activity [38,39]. Interestingly, a recently identified hit compound (Figure 1) possessing central indole ring combined with left and right phenyl rings each bearing acyclic amidine moiety was reported to exhibit potential activity against Gram-positive antibiotic-resistant ESKAPE bacteria [6]. However, the compound showed lower activity against Gram-negative antibiotic-resistant ESKAPE bacteria.
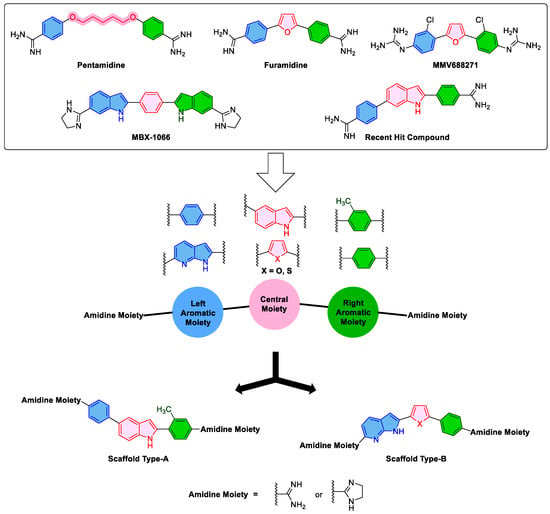
Figure 1.
The literature -reported compounds and the design of the new targeted compounds.
Molecular hybridization is well-acknowledged drug strategy that can afford bioactive multitarget compounds [40]. Aiming to discover new multitarget antibacterial agents against antibiotic-resistant ESKAPE pathogens and considering the diverse structural features of the literature-reported compounds shown in Figure 1 coupled with their antibacterial activity mediated through UPPS or KARI inhibition and MGB, new hybrid compounds were designed. As illustrated in Figure 1, designed compounds possessing scaffold A were planned to incorporate the larger central indole ring connected to left and right phenyl rings at 2- and 5-positions, each of which bears at para position either acyclic unsubstituted amidine moiety or 4,5-dihydro-1H-imidazol-2-yl as cyclic amidine moiety inherited from compound MBX-1066. Such design of scaffold A might be in part a positional isomer to the recently identified hit compound (Figure 1) in which the left aromatic ring was translocated from 6- to 5-position of the indole ring. The right phenyl ring connected to 2-position of the central indole ring was planned to bear a methyl substituent as an isosteric replacement for the chloro-substituent in compound MMV688271. Meanwhile, designed compounds conforming to scaffold B have the smaller five membered furan or thiophene rings as the central moiety connected to a left fused bicyclic pyrrolo[2,3-b]pyridine fragment as an isostere to the left indole ring of compound MBX-1066. Concurrently, the right indole ring of compound MBX-1066 was replaced in scaffold B with a phenyl ring inherited from other known compounds shown in Figure 1. Finally, the cyclic amidine moiety of compound MBX-1066 was maintained or replaced by acyclic unsubstituted amidine moiety. Such designed diverse structures of scaffolds A and B were synthesized and biologically evaluated for antibacterial activity with a particular consideration to antibiotic-resistant ESKAPE bacteria.
2.2. Chemistry
Scheme 1 shows our approach to synthesize indole-diamidines 6a and 6b. Protection of 5-bromoindole 1 was performed using Boc anhydride in presence of DMAP as a catalyst and DCM as a solvent. The Boc-protected derivative 2 was lithiated using LDA in THF at –20 °C followed by in situ conversion into organometalic stannane 3 using trimethyltin chloride [41,42]. Stille coupling of stannane derivative 3 with 4-bromo-3-methylbenzonitrile employing Pd(PPh3)4 in dioxane [41] afforded compound 4. Suzuki coupling of bromo derivative 4 [43] with 4-cyanophenylboronic acid using Pd(PPh3)4 as a catalyst in toluene afforded the dinitrile 5. The diamidine 6a was made by reacting the dinitrile 5 with lithium bis(trimethylsilyl)amide [44] in THF then subsequent desilyation using ethanolic HCl. While the imidazoline 6b was furnished by applying Pinner reaction [45,46] conditions on the dinitrile 5 employing anhydrous ethanolic HCl then reacting the formed intermediate carboximidate with ethylene diamine in ethanol.
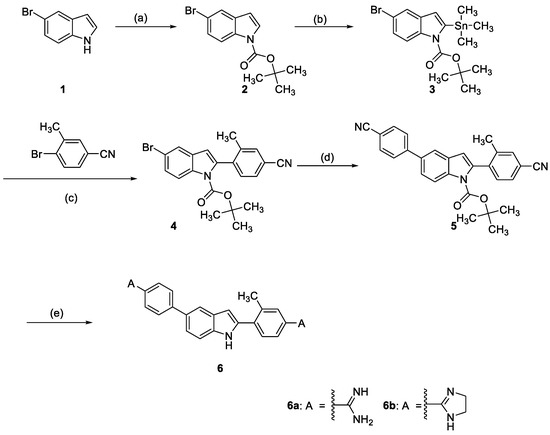
Scheme 1.
Reagents and conditions: (a) tert-Butyldicarbonate, 4-dimethylaminopyridine, CH2Cl2; (b) Trimethyltin chloride, lithium diisopropylamide/THF; (c) Palladium-tetrakis(triphenylphosphine), dioxane; (d) p-Cyanophenylboronic acid, palladium-tetrakis(triphenylphosphine, sodium carbonate, toluene; (e) For 6a: i- lithium bis(trimethylsilyl)amide/THF, ii-HCl gas/EtOH; For 6b: i-HCl gas/EtOH, ii-Ethylene diamine/EtOH.
The synthesis of the final diamidines 13a-d is outlined in Scheme 2. Applying Stille cross coupling between the stannane 7a or 7b with 4-bromobenzonitrile 8 afforded the cyano compounds 9a and 9b in high yield. Compounds 9a and 9b were converted to the bromo analogues 10a and 10b by bromination using NBS in DMF. The bisnitriles 12a and 12b were obtained after coupling of the bromo derivative 10a and 10b with the stannane compound 11 utilizing Stille cross coupling conditions. The bisnitriles 12a and 12b were converted to the final amidines either by using lithium bis(trimethylsilyl)amide in THF to afford 13a or 13b or by applying Pinner reaction condition to afford the imidazolines 13c or 13d.
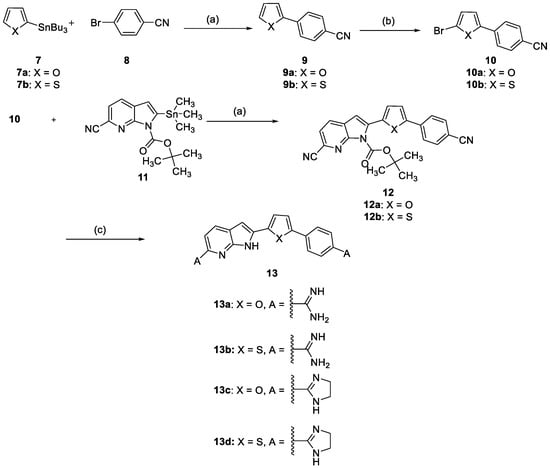
Scheme 2.
Reagents and conditions; (a) Palladium-tetrakis(triphenylphosphine, dioxane; (b) N-Bromosuccinimide/DMF; (c) For 13a and 13b: i- LiN(TMS)2/THF, ii- HCl gas/EtOH; For 13c and 13d: i- HCl gas/EtOH, ii- Ethylene diamine/EtOH.
2.3. Biological Evaluations
2.3.1. Minimum Inhibitory Concentrations and Spectrum of Activity
Minimum inhibitory concentrations (MICs) and activity spectra were determined utilizing a panel of bacterial isolates and strains consisting of two Gram-positive and two Gram-negative ESKAPE pathogens, in addition to two Gram-positive and three Gram-negative antibiotic-susceptible pathogenic bacteria. The two Gram-positive antibiotic-resistant ESKAPE isolates included the vancomycin-resistant E. faecium (VRE; Antibiotic Resistance Isolate Bank #0572) and the methicillin-resistant S. aureus (MRSA; strain COL) were used. Additionally, two Gram-positive antibiotic-susceptible bacteria were used including E. faecalis (Antibiotic Resistance Isolate Bank # 0671), and methicillin-susceptible S. aureus (MSSA ATCC 25923). The utilized Gram-negative antibiotic-resistant ESKAPE clinical isolates included a carbapenem-resistant Enterobacterale (CRE) isolate of K. pneumoniae (Clinical Isolate #015), and an isolate of carbapenem-resistant A. baumannii (CRAB; Clinical Isolate # 03-149.2). Moreover, three Gram-negative antibiotic-susceptible strains were applied namely, A. baumannii (ATCC 19606), P. aeruginosa (Antibiotic Resistance Isolate Bank # 0238), and E. coli (Antibiotic Resistance Isolate Bank # 0017). The results were compared to those of two reference standard drugs including vancomycin against Gram-positive pathogens and gentamicin against Gram-negative bacteria. The results are illustrated in Table 1.

Table 1.
Determined MIC (μg/mL) for the investigated compounds and reference drugs against antibiotic-resistant Gram-positive and Gram-negative ESKAPE pathogens and antibiotic-susceptible Gram-positive and Gram-negative bacteria.
Among the investigated compounds of scaffold type-B, compound 13d, which structurally consists of a central thiophene ring, a right phenyl ring, and a left pyrrolo[2,3-b]pyridine moiety in addition to bis-4,5-dihydro-1H-imidazol-2-yl rings as a cyclic amidine moieties, exhibited a remarkable activity. Thus, compound 13d showed a potential activity against the tested Gram-positive antibiotic-resistant bacterial strains, where it inhibited the growth of both VRE and MRSA with MIC values of 0.25 µg/mL reflecting higher potency than the reference drug, vancomycin, whose MIC values were 8 and 1–2 µg/mL. In addition, compound 13d inhibited the growth of the two investigated antibiotic-susceptible bacterial strains, E. faecalis and MSSA, at MIC values of 1 and 0.5 µg/mL, respectively. These values were also superior to the vancomycin MICs of 2 µg/mL against both strains. As for Gram-negative antibiotic-resistant bacteria, 13d inhibited the growth of CRE and CRAB at MIC values of 4 and 2 µg/mL, respectively. Again, its activity surpassed the activity of reference standard drug used against Gram-positive bacteria, gentamycin, that showed high MIC values of 64 and >128 µg/mL, respectively. Although compound 13d showed an MIC value of 0.5 µg/mL that was equal to that of gentamycin against antibiotic-susceptible E. coli, compound 13d showed intermediate to weak activity against the other antibiotic-susceptible Gram-negative bacteria A. baumannii and P. aeruginosa with MIC values of 32 and >128 µg/mL, respectively. These results suggest that compound 13d has an excellent activity against Enterobacterales, with a variable activity against Acinetobacter but poor activity against Pseudomonas. It was interesting to find out that compound 13d elicited a higher activity against antibiotic-resistant bacteria rather than against antibiotic-susceptible bacteria. This might be similar to the known phenomenon of “seesaw effect” in which bacterial resistance mechanisms decrease susceptibility to one drug class but correspondingly increase susceptibility to another drug class [47]. Additionally, it was found that resistance mutations impose complex fitness cost in case of A. baumannii [48]. Furthermore, the expression of UPPS was found to have a complex relationship with drug resistance and it is possible that bacteria with resistance to traditional antibacterial agents are more vulnerable to inhibition of UPPS [49]. Moreover, pentamidine analogues were reported as resistance breakers and therefore antibiotic-resistant bacteria might become more susceptible [50]. P. aeruginosa is intrinsically resistant to a lot of antibacterials that are active against other types of bacteria due to low permeability, so it is possible that the encountered low activity of compound 13d against P. aeruginosa is because it does not penetrate through its cell in sufficient quantities to kill the bacteria. Interestingly, maintaining all structural features of compound 13d but replacing the central thiophene ring by the isosteric furan ring afforded compound 13c that exhibited a marked decrease in the activity. Likewise, compound 13b, which maintained all structural features of compound 13d but possessed acyclic unsubstituted amidine moieties in place of the cyclic amidine moieties, exhibited much lower activity relative to compound 13d against all tested antibiotic-resistant or susceptible Gram-positive as well as Gram-negative bacteria. However, compound 13b was more active than compound 13c against antibiotic-susceptible or resistant Gram-negative bacteria. Nevertheless, compound 13b possessed decreased activity relative to compound 13c against antibiotic-resistant Gram-positive bacteria and comparable activity against antibiotic-susceptible Gram-positive bacteria. Interestingly, combining both replacement of thiophen and the cyclic amidine by furan and acyclic unsubstituted amidine afforded compound 13a with partially restored activity against antibiotic-resistant Gram-positive bacteria but not antibiotic-resistant Gram-negative bacteria. Thus, compound 13a showed moderate activity against VRE and MRSA.
Regarding compounds 6a and 6b that possess scaffold type-A that incorporates the larger central indole ring connected to a left phenyl moiety and a right methylphenyl moiety at 5- and 2-positions of the indole ring, respectively, they triggered moderate to good activity against antibiotic-resistant Gram-positive VRE and MRSA. However, both compounds possessed disappointing activity against antibiotic-resistant Gram-negative bacteria. Noteworthy, compound 6a possessing acyclic unsubstituted amidine moieties was more active relative to compound 6b having 4,5-dihydro-1H-imidazol-2-yl moieties as cyclic amidine fragments. Thus, compound 6a elicited MIC value of 2 µg/mL against MRSA and MIC value of 32 µg/mL against CRAB. In addition, compound 6a elicited MIC values of 2 µg/mL against antibiotic-susceptible Gram-positive MSSA as well as 32 µg/mL against Gram-negative CRAB, A. baumannii, P. aeruginosa, and E. coli. Together, these data suggest scaffold distinct structure-activity patterns where the thiophene-based scaffold type-B and 4,5-dihydro-1H-imidazol-2-yl moieties as cyclic amidine moieties are the best structural feature combinations to afford an active compound while in indole-based-scaffold type-A the acyclic unsubstituted amidine moieties affords better active compounds.
2.3.2. Time–Kill Assay
As compound 13d emerged as potential agent against tested antibiotic-resistant Gram-positive and Gram-negative bacteria, it was interesting to explore the pharmacodynamic interactions as a function of both time and concentration [51]. Hence, four different bacterial strains including antibiotic-resistant Gram-positive COL (MRSA) and E. faecium AR Bank # 0572 (VRE), in addition to antibiotic-resistant Gram-negative CRAB 03-149.2 (polymyxin-resistant A. baumannii) and antibiotic-susceptible Gram-negative E. coli AR Bank # 0017, were utilized in 24-h time–killing experiments employing compound 13d (Figure 2A) and the first order growth rate constants were calculated (Figure 2B). Initially, bacterial growth follows an exponential function characterizing the log phase (rapidly dividing cells phase). After a certain time, the bacterial growth enters a stationary phase (slowly dividing cells phase). In fact, the majority of antibiotics show most of their efficacy during the log-phase of bacterial growth rather than other phases [52]. Accordingly, the action of compound 13d on different growth phases was assessed. The growth control experiments showed that all of these tested four bacteria were in the initial growth log phase over the first four to six hours, after which the growth rate declined as the bacteria entered the stationary phase. As illustrated in Figure 2, the results indicated that 1X and 2X MIC concentrations of compound 13d decreased but did not completely stop the growth of VRE, MRSA, and CRAB over the growth log phase. Meanwhile, the 4X and 8X MIC concentrations of compound 13d not only completely inhibited the growth of VRE, MRSA, and CRAB over the growth log phase, but also achieved sustained killing against all of these antibiotic-resistant bacteria. The calculated first-order growth rate constants for 4X and 8X MIC concentrations were negative over the whole experiment duration indicating that the total number of living bacteria, measured as colony-forming units, remained less than the initial inoculum confirming that the bacteria–kill activity maintained by these concentration of compounds 13d over log and stationary growth phases. Calculation of the slope of the first-order growth rate showed more negative values over the log-phase rather than the stationary phase, which suggests compound 13d is more effective in killing VRE, MRSA, and CRAB during the log-phase. Nevertheless, it has a considerable killing effect at the stationary phase as well, indicated by negative first-order growth rate constants over the whole time of each experiment for 4X and 8X MIC concentrations. Interestingly, the measured time–kill dynamics showed that as low as 2X MIC concentration of compound 13d was effective in killing the antibiotic-susceptible Gram-negative E. coli. As the numbers of CFU/mL at almost all sampling times were lower than the initial number of CFU/mL, the calculated growth rate constant of E. Coli at 2X MIC concentration showed negative values over almost all follow time (Figure 2B). Together, these results indicate that compound 13d has potential antibacterial activity against antibiotic-resistant Gram-positive and Gram-negative bacteria as well as antibiotic-susceptible bacteria and might be subject to further development.
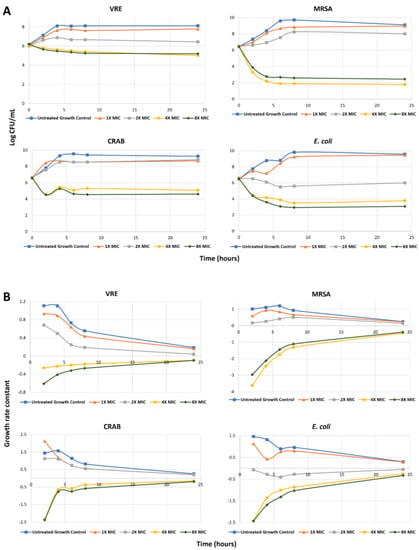
Figure 2.
(A) Time–kill curve for variable concentrations of compound 13d against VRE, MRSA, CRAB, and E. coli over 24 h; (B) Calculated growth rate constants for variable concentrations of 13d against VRE, MRSA, CRAB, and E. coli over 24 h.
2.4. Docking Study
As compound 13d was discovered as a promising agent, it was subjected to docking studies to explore the possible mode of action considering the three molecular targets, viz., UPPS, KARI, and DNA.
2.4.1. Docking Simulation of Compound 13d to UPPS
Interestingly, the crystal structure of UPPS shows a tunnel-like elongated core, which is enclosed with two α-helices and four β-sheets, with four possible binding sites (Figure 3A). UPPS may adopt two conformations for substrate binding and product release, either opened or closed [53,54]. Accordingly, it was mandatory to carry out docking simulations against both forms of UPPS (PDB IDs: 2E98 and 1X06; respectively) to predict the binding mode, if present, of compound 13d to UPPS conformer(s) in which active site(s). Results unveiled that our potential antibacterial hit compound could dock successfully to the open conformation of UPPS, specifically, into biding site number four of 2E98 crystal structure as illustrated in Figure 3B. The calculated binding score was −7.9218 Kcal/mol and the predicted binding poses unveiled that the tested compound established a network of favourable binding interactions (Figure 3C,D). The incorporation of two amidine moieties into dihydroimidazole rings was proved to cause a remarkable increase in the potency of compound 13d and this might be explained by docking simulation where the predicted binding mode unveiled that one of the moieties formed hydrogen bond with Ser55 amino acid residue in the binding site of 2E98 and the other formed carbon hydrogen interaction with Ala142. In addition, the central thiophene ring formed two π-alkyl interactions with Val50 and Arg51. The right phenyl ring established favourable π-sigma interaction with Ile141 and π-alkyl interaction with Val50 while the left pyrrolo[2,3-b]pyridine formed various π-alkyl and π-sigma interactions with Arg51, Val54, and Leu100. Collectively, it is anticipated that compound 13d docked into the open conformation of UPPS specifically in binding site 4 (Figure 3).
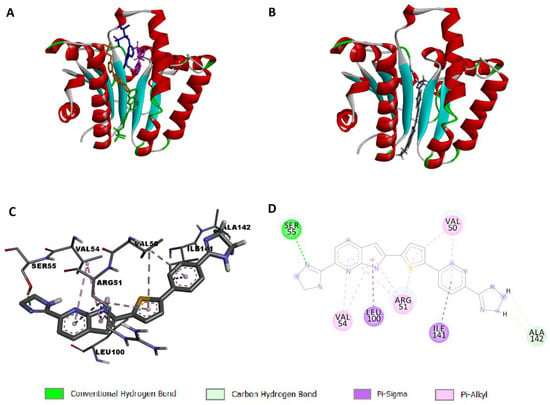
Figure 3.
Docking simulation of compound 13d to 2E98: (A) open conformation of the enzyme showing four binding sites with the co-crystallized ligand represented in four different colors in each binding site; (B) predicted pose showing compound 13d docked into site 4 of 2E98; (C) predicted 3D pose; (D) predicted 2D pose of compound 13d to 2E98.
2.4.2. Docking Simulation of Compound 13d to KARI
It is reported that there are two classes of KARI enzymes namely, class I and II. They differ in the length of polypeptide chain, where class I has about 340 amino acid residues while class II has extra polypeptide segment. Moreover, class I enzymes are usually found within microorganisms while those belonging to class II are typically found in plants. Enzymes from both classes are composed of a dimeric structure with two intertwined C-domains with the active site is located between these two domains [55]. Activity of KARI is essentially dependent on two Mg2+ ions and NADPH cofactor. Interestingly, the location of these ions is highly dependent upon whether the enzyme is NADPH-bound or not. This suggested that the two ions have non-significant role in the catalysis process of KARI. Moreover, it was observed that binding of KARI and its cofactors aids widening the active site for substrate binding. Consequently, KARI inhibitors might bind to either NADPH-bound or unbound forms of the enzyme [56]. Accordingly, a docking experiment was carried out in the presence and absence of NADPH to explore whether compound 13d would dock into the bound or unbound enzyme. Calculations unveiled that compound 13d docked successfully into the NADPH-unbound form of the enzyme with respectable binding score of −7.5505 Kcal/mol. The predicted 3D-pose of the docking simulation clarified that the dihydroimidazole moiety attached to the left pyrrolo[2,3-b]pyridine ring is located near the interwind C-domain of the second monomeric chain (Figure 4). Additionally, this side of the ligand was found to form favorable hydrogen bonds between the NH of the dihydroimidazole moiety and Ser249 and another hydrogen bond between NH of the pyrrolo[2,3-b]pyridine and Ile250. Additionally, the first monomeric chain was involved in a network of favorable binding interactions with the docked molecule including carbon hydrogen and π-alkyl interactions between the central thiophene and the phenyl rings with Ser27, Leu80, and Pro81. These results ascertained that compound 13d acts as KARI inhibitor via binding to the NADPH-unbound form of the enzyme.
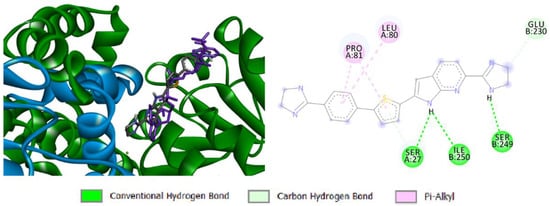
Figure 4.
Docking simulation of compound 13d to dimeric 7KH7: 3D representation showing the first monomeric chain in green color, the intertwined C-domain of the other chain in blue color, and NADPH in violet color (left) and bonding interactions of the docked compound (right).
2.4.3. Docking Simulation of Compound 13d to Bacterial DNA
Docking simulation of compound 13d to DNA of S. aureus was performed to explore the potential selective binding to bacterial DNA. Accordingly, a 105-mer sequence of S. aureus TY4, ETB plasmid DNA was obtained from GenBank (accession No. AP003088, sequence 19,540–19,644). A preliminary docking study was conducted to broadly identify the possible sites for binding, followed by another more accurate run utilizing the identified possible sequences in the first experiment. It was found that compound 13d docked into the minor groove of B-DNA, not the A-conformation, with calculated binding scores ranging from −9.2059 to −9.0894 Kcal/mol for the top ten poses of the docking experiment. In fact, six of the top ten poses within the docking simulation were bound to the sequence TTTTTT 19,582–19,587, three poses were predicted to bind with sequence TAATTA 19,553–19,556, and only one pose was bound to sequence CATTA 19,622–19,626 as illustrated in Figure 5. These results suggested that compound 13d is a potential MGB that possibly binds mainly to sequence TTTTTT 19,582–19,587.
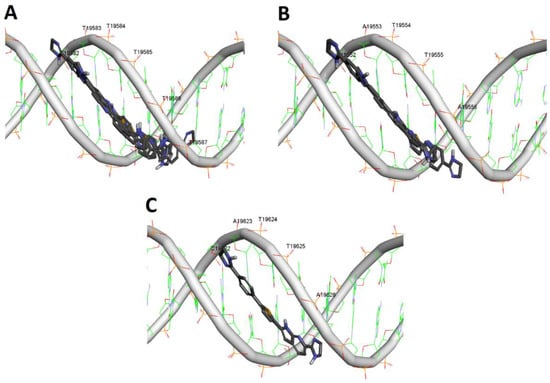
Figure 5.
Docking simulation of compound 13d to DNA sequence of S. aureus (19,540–19,644): (A) Anticipated poses of compound 13d to TTTTTT sequence 19,582–19,587; (B) Anticipated poses of compound 13d to TATTA sequence 19,552–19,556; (C) Anticipated poses of compound 13d to CATTA sequence 19,622–19,626.
3. Materials and Methods
3.1. Chemistry
Synthesis and compounds characterization are as detailed in the supplementary materials.
3.2. Biological Evaluations
Biological evaluations were performed following standard protocols [6,57] as detailed in the supplementary materials.
3.3. Docking Simulations
The docking simulations were carried out adopting reported procedures as detailed in the supplementary materials.
4. Conclusions
Towards the discovery of novel antibacterial agents against antibiotic-resistant pathogens, novel hybrid molecules of two scaffold types were designed, synthesized, and evaluated. Compound 13d, which possesses a type-B scaffold, was the most promising compound and elicited potential activity against Gram-positive and Gram-negative bacteria with various resistance profiles including VRE, MRSA, CRE, CRAB, MSSA, E. faecalis, E. coli, A. baumannii, and P. aeruginosa. Study of time–kill activity of compound 13d showed that it achieved sustained killing against tested bacteria at 4X MIC concentrations with maximum killing observed during the log phase of bacterial growth. In silico simulations suggested compound 13d is a multifunctional molecule that possibly interacts with bacterial UPPS, KARI, and DNA as potential targets. Overall, compound 13d represents a promising hit for future development of potential antibacterial agents against antibiotic-resistant as well as antibiotic-susceptible bacteria.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph16071040/s1, Chemical synthesis procedures and characterization data; Biological evaluations protocols; In silico calculations methods.
Author Contributions
Conceptualization, A.A.F. and A.H.E.H.; methodology, A.A.F., A.H.E.H. and J.R.L.; validation, A.A.F., J.R.L. and A.H.E.H.; formal analysis, A.A.F., A.H.E.H. and J.R.L.; investigation, S.M.E.-S., S.A.A. and K.G.; resources, A.A.F., A.H.E.H. and J.R.L.; data curation, A.A.F., A.H.E.H. and J.R.L.; writing—original draft preparation, S.M.E.-S. and A.H.E.H.; writing—review and editing, A.A.F., A.H.E.H. and J.R.L.; visualization, S.M.E.-S. and S.A.A.; supervision, A.A.F. and A.H.E.H.; project administration, A.A.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data is contained within the article or supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Davies, J. Where Have all the Antibiotics Gone? Can. J. Infect. Dis. Med. Microbiol. 2006, 17, 707296. [Google Scholar] [CrossRef]
- Theuretzbacher, U.; Outterson, K.; Engel, A.; Karlén, A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2020, 18, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Kåhrström, C.T. Entering a post-antibiotic era? Nat. Rev. Microbiol. 2013, 11, 146. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Kwon, J.H.; Powderly, W.G. The post-antibiotic era is here. Science 2021, 373, 471. [Google Scholar] [CrossRef]
- Gulia, K.; Hassan, A.H.E.; Lenhard, J.R.; Farahat, A.A. Escaping ESKAPE resistance: In vitro and in silico studies of multifunctional carbamimidoyl-tethered indoles against antibiotic-resistant bacteria. R. Soc. Open Sci. 2023, 10, 230020. [Google Scholar] [CrossRef] [PubMed]
- Denissen, J.; Reyneke, B.; Waso-Reyneke, M.; Havenga, B.; Barnard, T.; Khan, S.; Khan, W. Prevalence of ESKAPE pathogens in the environment: Antibiotic resistance status, community-acquired infection and risk to human health. Int. J. Hyg. Environ. Health. 2022, 244, 114006. [Google Scholar] [CrossRef] [PubMed]
- Zhen, X.; Lundborg, C.S.; Sun, X.; Hu, X.; Dong, H. Economic burden of antibiotic resistance in ESKAPE organisms: A systematic review. Antimicrob. Resist. Infect. Control. 2019, 8, 137. [Google Scholar] [CrossRef]
- Ayobami, O.; Brinkwirth, S.; Eckmanns, T.; Markwart, R. Antibiotic resistance in hospital-acquired ESKAPE-E infections in low- and lower-middle-income countries: A systematic review and meta-analysis. Emerg. Microbes Infect. 2022, 11, 443. [Google Scholar] [CrossRef]
- Idris, F.N.; Nadzir, M.M. Multi-drug resistant ESKAPE pathogens and the uses of plants as their antimicrobial agents. Arch. Microbiol. 2023, 205, 115. [Google Scholar] [CrossRef] [PubMed]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.M.P.D.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Bharath Prasad, A.S.; Mehta, C.H.; Nayak, U.Y. Antimicrobial peptide polymers: No escape to ESKAPE pathogens—A review. World J. Microbiol. Biotechnol. 2020, 36, 131. [Google Scholar] [CrossRef]
- Suckling, C.J.; Hunter, I.S.; Scott, F.J. Multitargeted anti-infective drugs: Resilience to resistance in the antimicrobial resistance era. Future Drug Discov. 2022, 4, FDD73. [Google Scholar] [CrossRef]
- Jukič, M.; Rožman, K.; Sova, M.; Barreteau, H.; Gobec, S. Anthranilic Acid Inhibitors of Undecaprenyl Pyrophosphate Synthase (UppS), an Essential Enzyme for Bacterial Cell Wall Biosynthesis. Front. Microbiol. 2019, 9, 3322. [Google Scholar] [CrossRef] [PubMed]
- Wun, S.J.; Johnson, L.A.; You, L.; McGeary, R.P.; Brueck, T.; Schenk, G.; Guddat, L.W. Inhibition studies of ketol-acid reductoisomerases from pathogenic microorganisms. Arch. Biochem. Biophys. 2020, 692, 108516. [Google Scholar] [CrossRef]
- Tyagi, R.; Duquerroy, S.; Navaza, J.; Guddat, L.W.; Duggleby, R.G. The crystal structure of a bacterial Class II ketol-acid reductoisomerase: Domain conservation and evolution. Protein Sci. 2005, 14, 3089. [Google Scholar] [CrossRef]
- Bayaraa, T.; Kurz, J.L.; Patel, K.M.; Hussein, W.M.; Bilyj, J.K.; West, N.P.; Schenk, G.; McGeary, R.P.; Guddat, L.W. Discovery, Synthesis and Evaluation of a Ketol-Acid Reductoisomerase Inhibitor. Chem. Eur. J. 2020, 26, 8958–8968. [Google Scholar] [CrossRef]
- Boeva, V. Analysis of Genomic Sequence Motifs for Deciphering Transcription Factor Binding and Transcriptional Regulation in Eukaryotic Cells. Front. Genet. 2016, 7, 24. [Google Scholar] [CrossRef]
- Rajewska, M.; Wegrzyn, K.; Konieczny, I. AT-rich region and repeated sequences—The essential elements of replication origins of bacterial replicons. FEMS Microbiol. Rev. 2012, 36, 408. [Google Scholar] [CrossRef]
- Rahman, A.; O’Sullivan, P.; Rozas, I. Recent developments in compounds acting in the DNA minor groove. Medchemcomm 2019, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Barrett, M.P.; Gemmell, C.G.; Suckling, C.J. Minor groove binders as anti-infective agents. Pharmacol. Ther. 2013, 139, 12. [Google Scholar] [CrossRef] [PubMed]
- Larsen, T.A.; Goodsell, D.S.; Cascio, D.; Grzeskowiak, K.; Dickerson, R.E. The Structure of DAPI Bound to DNA. J. Biomol. Struct. Dyn. 1989, 7, 477. [Google Scholar] [CrossRef] [PubMed]
- Tanious, F.A.; Spychala, J.; Kumar, A.; Greene, K.; Boykin, D.W.; Wilson, W.D. Different Binding Mode in AT and GC Sequences for Unfused-Aromatic Dications. J. Biomol. Struct. 1994, 11, 1063. [Google Scholar] [CrossRef]
- Hassan, A.H.E.; El-Sayed, S.M.; Yamamoto, M.; Gohda, J.; Matsumoto, T.; Shirouzu, M.; Inoue, J.-i.; Kawaguchi, Y.; Mansour, R.M.A.; Anvari, A.; et al. In Silico and In Vitro Evaluation of Some Amidine Derivatives as Hit Compounds towards Development of Inhibitors against Coronavirus Diseases. Viruses 2023, 15, 1171. [Google Scholar] [CrossRef]
- Farahat, A.A.; Kumar, A.; Wenzler, T.; Brun, R.; Paul, A.; Guo, P.; Wilson, W.D.; Boykin, D.W. Investigation of the effect of structure modification of furamidine on the DNA minor groove binding and antiprotozoal activity. Eur. J. Med. Chem. 2023, 252, 115287. [Google Scholar] [CrossRef]
- Depauw, S.; Lambert, M.; Jambon, S.; Paul, A.; Peixoto, P.; Nhili, R.; Marongiu, L.; Figeac, M.; Dassi, C.; Paul-Constant, C.; et al. Heterocyclic Diamidine DNA Ligands as HOXA9 Transcription Factor Inhibitors: Design, Molecular Evaluation, and Cellular Consequences in a HOXA9-Dependant Leukemia Cell Model. J. Med. Chem. 2019, 62, 1306–1329. [Google Scholar] [CrossRef]
- Simone, R.; Balendra, R.; Moens, T.G.; Preza, E.; Wilson, K.M.; Heslegrave, A.; Woodling, N.S.; Niccoli, T.; Gilbert-Jaramillo, J.; Abdelkarim, S.; et al. G-quadruplex-binding small molecules ameliorate C9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol. Med. 2018, 10, 22–31. [Google Scholar] [CrossRef]
- Hu, L.; Kully, M.L.; Boykin, D.W.; Abood, N. Synthesis and in vitro activity of dicationic bis-benzimidazoles as a new class of anti-MRSA and anti-VRE agents. Bioorg. Med. Chem. Lett. 2009, 19, 1292–1295. [Google Scholar] [CrossRef]
- Hu, L.; Kully, M.L.; Boykin, D.W.; Abood, N. Optimization of the central linker of dicationic bis-benzimidazole anti-MRSA and anti-VRE agents. Bioorg. Med. Chem. Lett. 2009, 19, 3374–3377. [Google Scholar] [CrossRef]
- Hu, L.; Kully, M.L.; Boykin, D.W.; Abood, N. Synthesis and structure–activity relationship of dicationic diaryl ethers as novel potent anti-MRSA and anti-VRE agents. Bioorg. Med. Chem. Lett. 2009, 19, 4626–4629. [Google Scholar] [CrossRef] [PubMed]
- Stokes, J.M.; MacNair, C.R.; Ilyas, B.; French, S.; Côté, J.-P.; Bouwman, C.; Farha, M.A.; Sieron, A.O.; Whitfield, C.; Coombes, B.K.; et al. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol. 2017, 2, 17028. [Google Scholar] [CrossRef] [PubMed]
- Wesseling, C.M.J.; Slingerland, C.J.; Veraar, S.; Lok, S.; Martin, N.I. Structure–Activity Studies with Bis-Amidines That Potentiate Gram-Positive Specific Antibiotics against Gram-Negative Pathogens. ACS Infect. Dis. 2021, 7, 3314–3335. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Guo, P.; Boykin, D.W.; Wilson, W.D. A New Generation of Minor-Groove-Binding—Heterocyclic Diamidines That Recognize G·C Base Pairs in an AT Sequence Context. Molecules 2019, 24, 946. [Google Scholar] [CrossRef]
- Donkor, I.O.; Clark, A.M. In vitro antimicrobial activity of aromatic diamidines and diimidazolines related to pentamidine. Eur. J. Med. Chem. 1999, 34, 639. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, X.; Wu, Y.; Zhang, W.; Chen, X.; You, X.; Hu, L. Synthesis and structure-activity relationship of novel bisindole amidines active against MDR Gram-positive and Gram-negative bacteria. Eur. J. Med. Chem. 2018, 150, 771. [Google Scholar] [CrossRef]
- Scott, F.J.; Khalaf, A.I.; Giordani, F.; Wong, P.E.; Duffy, S.; Barrett, M.; Avery, V.M.; Suckling, C.J. An evaluation of Minor Groove Binders as anti-Trypanosoma brucei brucei therapeutics. Eur. J. Med. Chem. 2016, 116, 116. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, Y.; Li, K.; Gao, J.; Huang, C.-H.; Chen, C.-C.; Ko, T.-P.; Zhang, Y.; Guo, R.-T.; Oldfield, E. Antibacterial Drug Leads: DNA and Enzyme Multitargeting. J. Med. Chem. 2015, 58, 1215–1227. [Google Scholar] [CrossRef]
- Zhu, W.; Zhang, Y.; Sinko, W.; Hensler, M.E.; Olson, J.; Molohon, K.J.; Lindert, S.; Cao, R.; Li, K.; Wang, K.; et al. Antibacterial drug leads targeting isoprenoid biosynthesis. Proc. Natl. Acad. Sci. USA 2013, 110, 123. [Google Scholar] [CrossRef]
- Decker, M. Introduction. In Design of Hybrid Molecules for Drug Development; Decker, M., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 1–3. [Google Scholar] [CrossRef]
- Farahat, A.A.; Kumar, A.; Say, M.; Barghash, A.E.-D.M.; Goda, F.E.; Eisa, H.M.; Wenzler, T.; Brun, R.; Liu, Y.; Mickelson, L.; et al. Synthesis, DNA binding, fluorescence measurements and antiparasitic activity of DAPI related diamidines. Bioorg. Med. Chem. 2010, 18, 557–566. [Google Scholar] [CrossRef]
- Kumar, A.; Say, M.; Boykin, D.W. Synthesis of New Substituted 2-(Trimethylstannyl)indoles. Synthesis 2008, 2008, 707–710. [Google Scholar] [CrossRef]
- Guo, P.; Farahat, A.A.; Paul, A.; Harika, N.K.; Boykin, D.W.; Wilson, W.D. Compound Shape Effects in Minor Groove Binding Affinity and Specificity for Mixed Sequence DNA. J. Am. Chem. Soc. 2018, 140, 14761–14769. [Google Scholar] [CrossRef]
- Guo, P.; Paul, A.; Kumar, A.; Farahat, A.A.; Kumar, D.; Wang, S.; Boykin, D.W.; Wilson, W.D. The Thiophene “Sigma-Hole” as a Concept for Preorganized, Specific Recognition of G⋅C Base Pairs in the DNA Minor Groove. Chem. Eur. J. 2016, 22, 15404–15412. [Google Scholar] [CrossRef]
- Laughlin, S.; Wang, S.; Kumar, A.; Farahat, A.A.; Boykin, D.W.; Wilson, W.D. Resolution of Mixed Site DNA Complexes with Dimer-Forming Minor-Groove Binders by Using Electrospray Ionization Mass Spectrometry: Compound Structure and DNA Sequence Effects. Chem. Eur. J. 2015, 21, 5528–5539. [Google Scholar] [CrossRef]
- Paul, A.; Kumar, A.; Nanjunda, R.; Farahat, A.A.; Boykin, D.W.; Wilson, W.D. Systematic synthetic and biophysical development of mixed sequence DNA binding agents. Org. Biomol. Chem. 2017, 15, 827–835. [Google Scholar] [CrossRef]
- Jiang, S.; Zhuang, H.; Zhu, F.; Wei, X.; Zhang, J.; Sun, L.; Ji, S.; Wang, H.; Wu, D.; Zhao, F.; et al. The Role of mprF Mutations in Seesaw Effect of Daptomycin-Resistant Methicillin-Resistant Staphylococcus aureus Isolates. Antimicrob. Agents Chemother. 2022, 66, e01295–e01221. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, G.J.; Domingues, S. Interplay between Colistin Resistance, Virulence and Fitness in Acinetobacter baumannii. Antibiotics 2017, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Lee Yong, H.; Helmann John, D. Reducing the Level of Undecaprenyl Pyrophosphate Synthase Has Complex Effects on Susceptibility to Cell Wall Antibiotics. Antimicrob. Agents Chemother. 2013, 57, 4267–4275. [Google Scholar] [CrossRef]
- Dhanda, G.; Acharya, Y.; Haldar, J. Antibiotic Adjuvants: A Versatile Approach to Combat Antibiotic Resistance. ACS Omega 2023, 8, 10757–10783. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rojas, A.; Rolff, J. Antimicrobial activity of cationic antimicrobial peptides against stationary phase bacteria. Front. Microbiol. 2022, 13, 1029084. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.-Y.; Ko, T.-P.; Liang, P.-H.; Wang, A.H.J. Catalytic Mechanism Revealed by the Crystal Structure of Undecaprenyl Pyrophosphate Synthase in Complex with Sulfate, Magnesium, and Triton. J. Biol. Chem. 2003, 278, 29298. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.-Y.; Ko, T.-P.; Chen, A.P.-C.; Wang, A.H.-J.; Liang, P.-H. Substrate binding mode and reaction mechanism of undecaprenyl pyrophosphate synthase deduced from crystallographic studies. J. Mol. Biol. 2004, 13, 971. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.-H.; Lonhienne, T.G.A.; Winzor, D.J.; Schenk, G.; Guddat, L.W. Bacterial and Plant Ketol-Acid Reductoisomerases Have Different Mechanisms of Induced Fit during the Catalytic Cycle. J. Mol. Biol. 2012, 424, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Kurz, J.L.; Patel, K.M.; Wun, S.J.; Hussein, W.M.; Lonhienne, T.; West, N.P.; McGeary, R.P.; Schenk, G.; Guddat, L.W. Discovery of a Pyrimidinedione Derivative with Potent Inhibitory Activity against Mycobacterium tuberculosis Ketol–Acid Reductoisomerase. Chem. Eur. J. 2021, 27, 3130–3141. [Google Scholar] [CrossRef] [PubMed]
- The Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Fifteenth Informational Supplement. CLSI/NCCLS Document M100-S15; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2005. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).