In Silico-Motivated Discovery of Novel Potent Glycogen Synthase-3 Inhibitors: 1-(Alkyl/arylamino)-3H-naphtho[1,2,3-de]quinoline-2,7-dione Identified as a Scaffold for Kinase Inhibitor Development
Abstract
1. Introduction
2. Results and Discussion
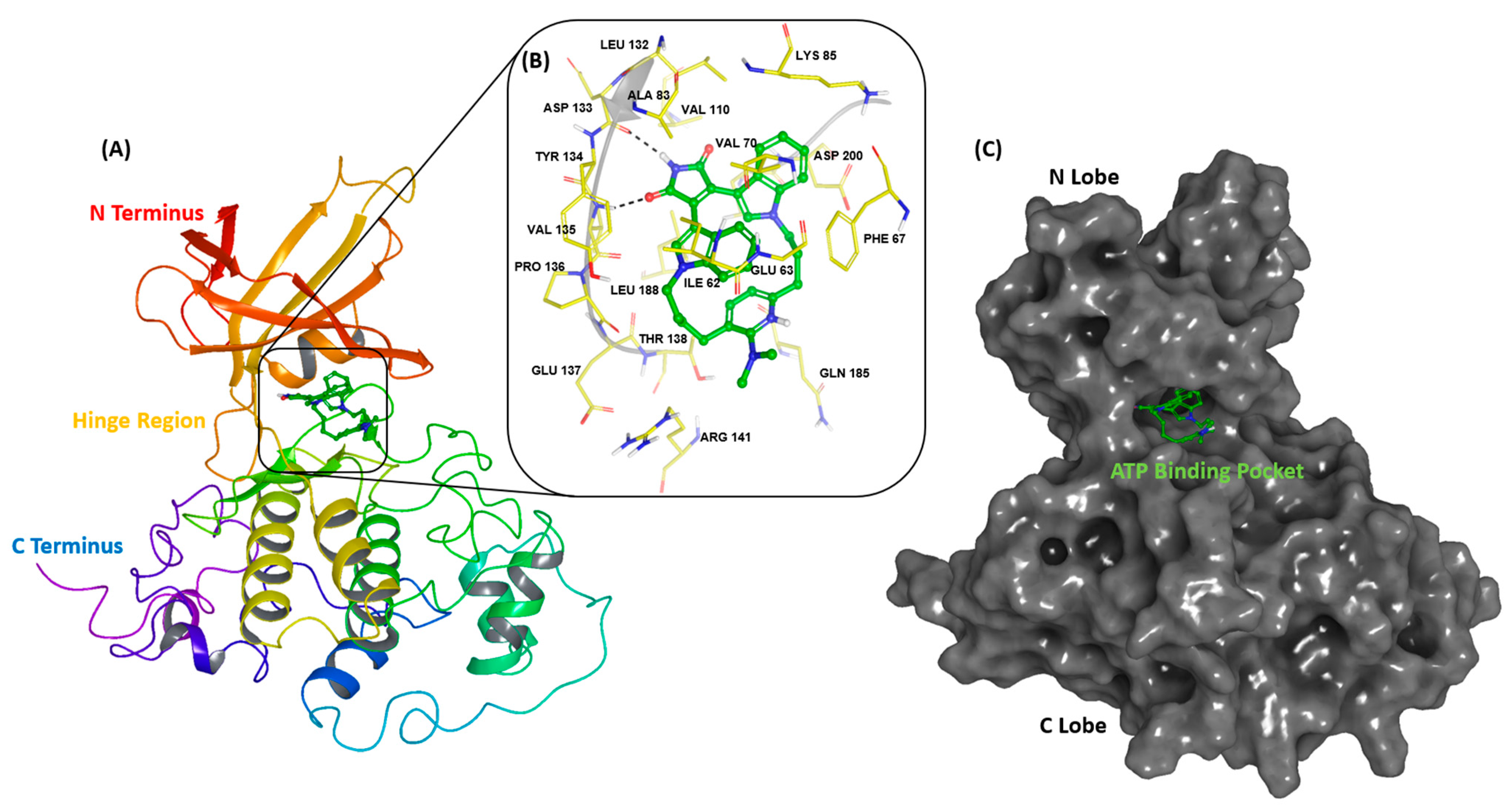
2.1. Design of In Silico Screening Protocol/Benchmarking Studies
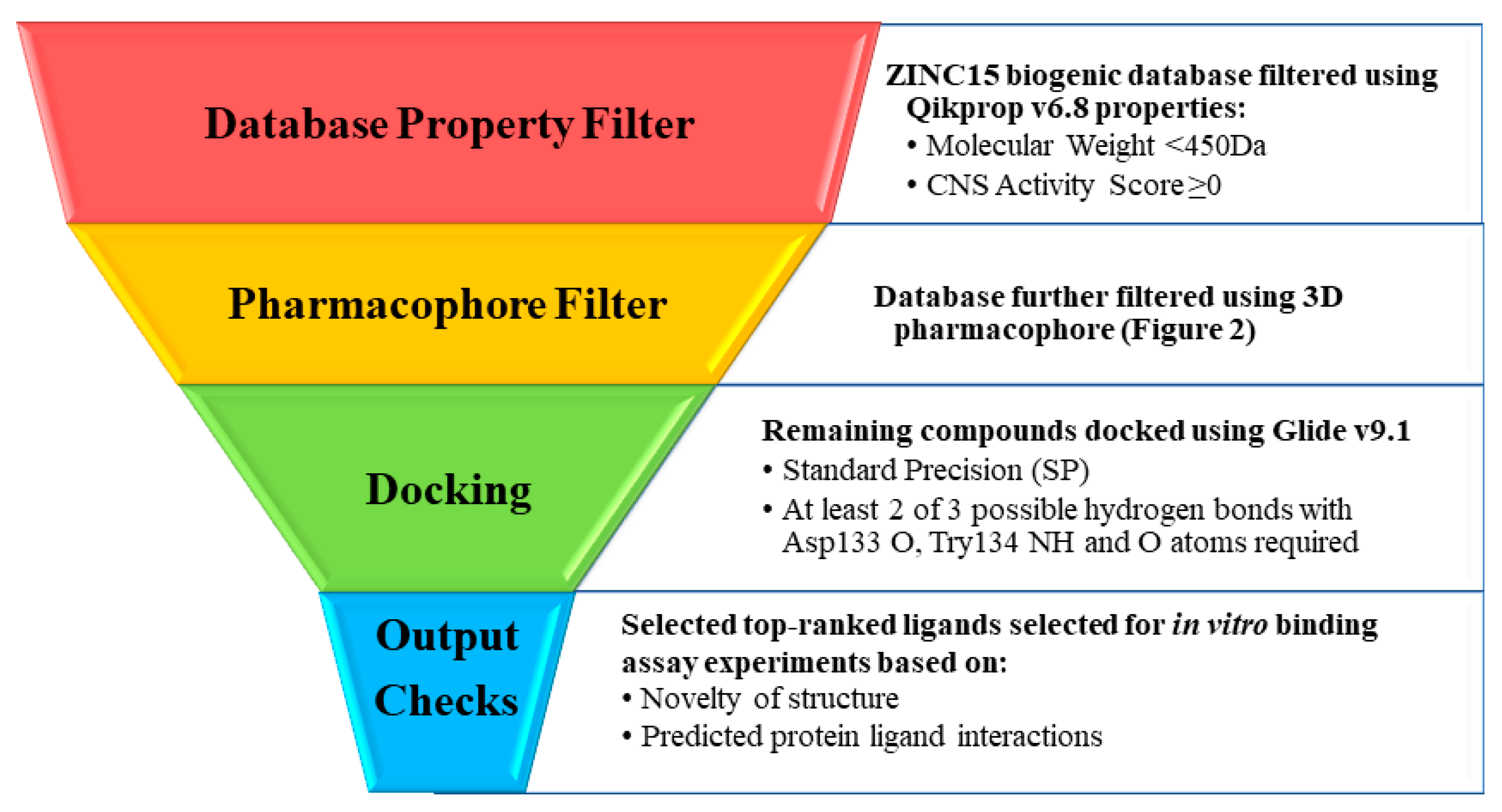
2.2. Screening of Biogenic Database
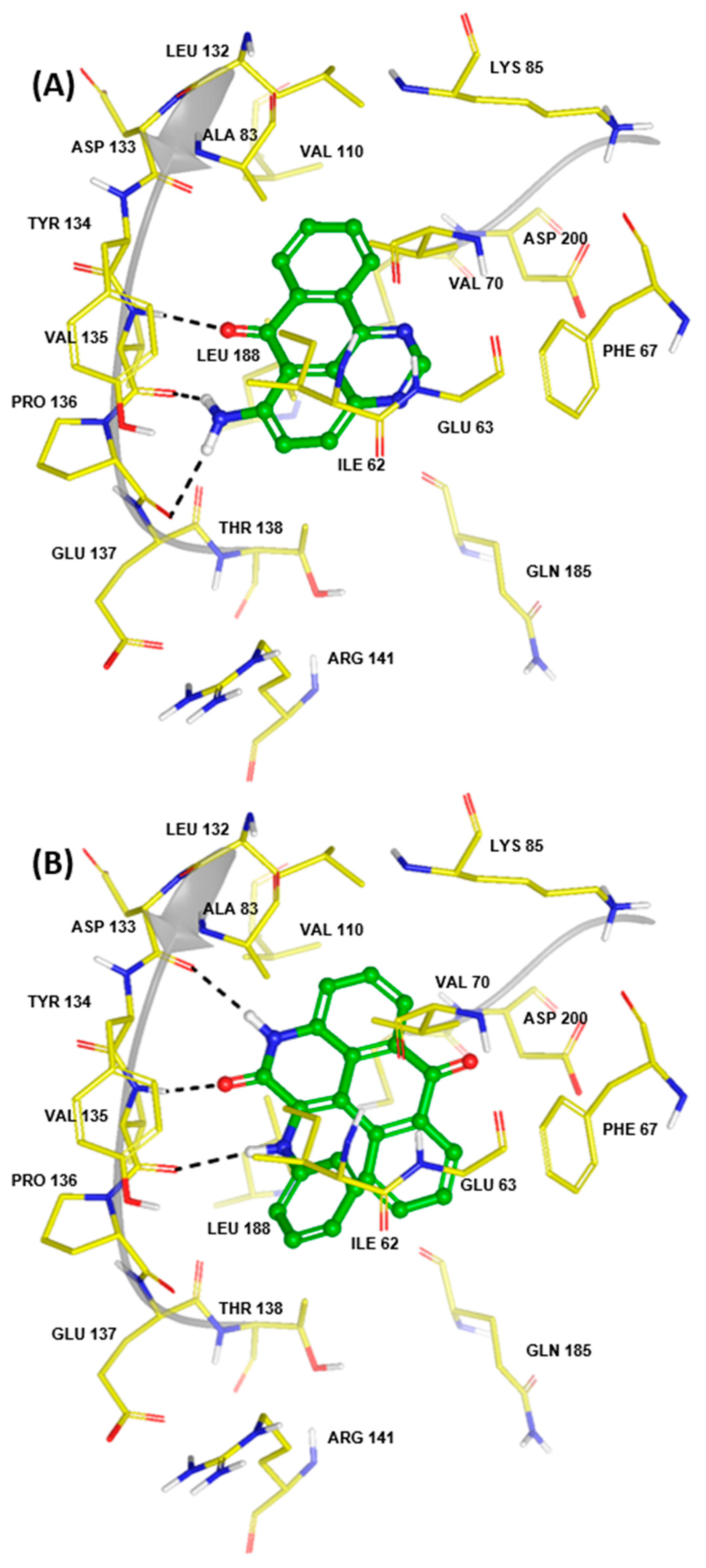
2.2.1. Generation I Selection
2.2.2. Generation II Selection
2.3. Kinase Selectivity Screening
2.4. ADME(T) Results & Analysis
3. Conclusions
4. Materials and Methods
4.1. Computational Details
4.1.1. Ligand Preparation
4.1.2. Protein Preparation
4.1.3. Docking
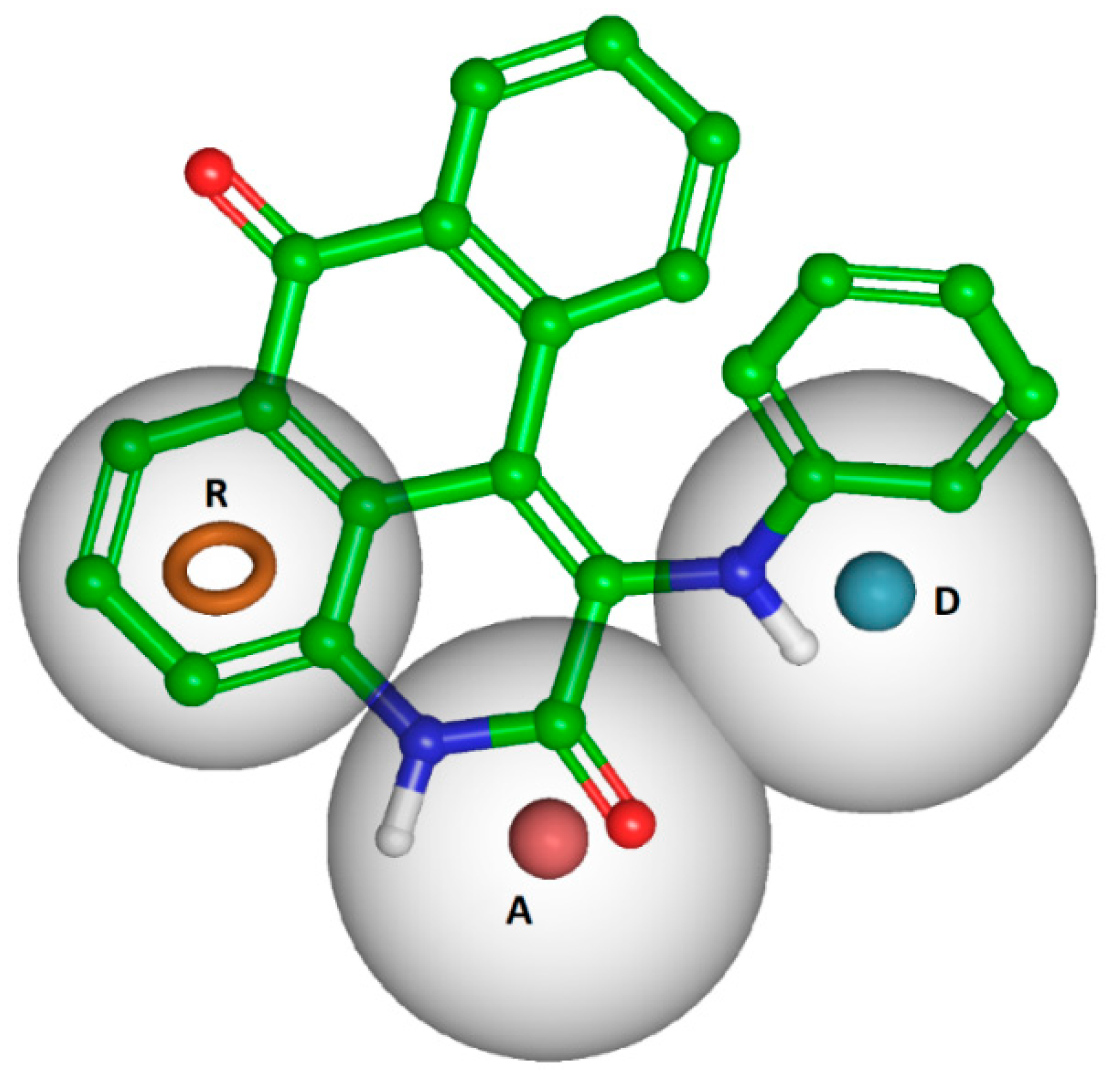
4.1.4. Pharmacophore Modelling
4.1.5. ADME(T) Calculations
4.1.6. DFT Calculations
4.2. Experimental In Vitro Binding Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kockeritz, L.; Doble, B.; Patel, S.; Woodgett, J.R. Glycogen synthase kinase-3-an overview of an over-achieving protein kinase. Curr. Drug. Targets 2006, 7, 1377–1388. [Google Scholar] [CrossRef] [PubMed]
- Embi, N.; Rylatt, D.B.; Cohen, P. Glycogen synthase kinase-3 from rabbit skeletal muscle. Separation from cyclic-AMP-dependent protein kinase and phosphorylase kinase. Eur. J. Biochem. 1980, 107, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Maurer, U.; Preiss, F.; Brauns-Schubert, P.; Schlicher, L.; Charvet, C. GSK-3—At the crossroads of cell death and survival. J. Cell Sci. 2014, 127, 1369–1378. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, C. What Are the bona fide GSK3 Substrates? Int. J. Alzheimers Dis. 2011, 2011, 505607. [Google Scholar] [CrossRef]
- Beurel, E.; Grieco, S.F.; Jope, R.S. Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol. Ther. 2015, 148, 114–131. [Google Scholar] [CrossRef] [PubMed]
- Ring, D.B.; Johnson, K.W.; Henriksen, E.J.; Nuss, J.M.; Goff, D.; Kinnick, T.R.; Ma, S.T.; Reeder, J.W.; Samuels, I.; Slabiak, T.; et al. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes 2003, 52, 588–595. [Google Scholar] [CrossRef]
- Ma, T. GSK3 in Alzheimer’s disease: Mind the isoforms. J. Alzheimers Dis. 2014, 39, 707–710. [Google Scholar] [CrossRef] [PubMed]
- Lovestone, S.; Killick, R.; Di Forti, M.; Murray, R. Schizophrenia as a GSK-3 dysregulation disorder. Trends Neurosci. 2007, 30, 142–149. [Google Scholar] [CrossRef]
- Lin, R.; Jones, N.C.; Kwan, P. Unravelling the Role of Glycogen Synthase Kinase-3 in Alzheimer’s Disease-Related Epileptic Seizures. Int. J. Mol. Sci. 2020, 21, 3676. [Google Scholar] [CrossRef]
- De Simone, A.; Tumiatti, V.; Andrisano, V.; Milelli, A. Glycogen Synthase Kinase 3beta: A New Gold Rush in Anti-Alzheimer’s Disease Multitarget Drug Discovery? J. Med. Chem. 2021, 64, 26–41. [Google Scholar] [CrossRef]
- Domoto, T.; Uehara, M.; Bolidong, D.; Minamoto, T. Glycogen Synthase Kinase 3beta in Cancer Biology and Treatment. Cells 2020, 9, 1388. [Google Scholar] [CrossRef] [PubMed]
- Augello, G.; Emma, M.R.; Cusimano, A.; Azzolina, A.; Montalto, G.; McCubrey, J.A.; Cervello, M. The Role of GSK-3 in Cancer Immunotherapy: GSK-3 Inhibitors as a New Frontier in Cancer Treatment. Cells 2020, 9, 1427. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.K.; DeGrado, T.R. Glycogen Synthase Kinase-3 (GSK-3)-Targeted Therapy and Imaging. Theranostics 2016, 6, 571–593. [Google Scholar] [CrossRef]
- Domoto, T.; Pyko, I.V.; Furuta, T.; Miyashita, K.; Uehara, M.; Shimasaki, T.; Nakada, M.; Minamoto, T. Glycogen synthase kinase-3beta is a pivotal mediator of cancer invasion and resistance to therapy. Cancer Sci. 2016, 107, 1363–1372. [Google Scholar] [CrossRef]
- Papadopoli, D.; Pollak, M.; Topisirovic, I. The role of GSK3 in metabolic pathway perturbations in cancer. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 119059. [Google Scholar] [CrossRef]
- Duda, P.; Akula, S.M.; Abrams, S.L.; Steelman, L.S.; Martelli, A.M.; Cocco, L.; Ratti, S.; Candido, S.; Libra, M.; Montalto, G.; et al. Targeting GSK3 and Associated Signaling Pathways Involved in Cancer. Cells 2020, 9, 1110. [Google Scholar] [CrossRef] [PubMed]
- Kotliarova, S.; Pastorino, S.; Kovell, L.C.; Kotliarov, Y.; Song, H.; Zhang, W.; Bailey, R.; Maric, D.; Zenklusen, J.C.; Lee, J.; et al. Glycogen synthase kinase-3 inhibition induces glioma cell death through c-MYC, nuclear factor-kappaB, and glucose regulation. Cancer Res. 2008, 68, 6643–6651. [Google Scholar] [CrossRef]
- Woodgett, J.R. Molecular cloning and expression of glycogen synthase kinase-3/factor A. EMBO J. 1990, 9, 2431–2438. [Google Scholar] [CrossRef]
- Yao, H.B.; Shaw, P.C.; Wong, C.C.; Wan, D.C. Expression of glycogen synthase kinase-3 isoforms in mouse tissues and their transcription in the brain. J. Chem. Neuroanat. 2002, 23, 291–297. [Google Scholar] [CrossRef]
- Pandey, G.N.; Dwivedi, Y.; Rizavi, H.S.; Teppen, T.; Gaszner, G.L.; Roberts, R.C.; Conley, R.R. GSK-3beta gene expression in human postmortem brain: Regional distribution, effects of age and suicide. Neurochem. Res. 2009, 34, 274–285. [Google Scholar] [CrossRef]
- Soutar, M.P.; Kim, W.Y.; Williamson, R.; Peggie, M.; Hastie, C.J.; McLauchlan, H.; Snider, W.D.; Gordon-Weeks, P.R.; Sutherland, C. Evidence that glycogen synthase kinase-3 isoforms have distinct substrate preference in the brain. J. Neurochem. 2010, 115, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Lovestone, S.; Boada, M.; Dubois, B.; Hull, M.; Rinne, J.O.; Huppertz, H.J.; Calero, M.; Andres, M.V.; Gomez-Carrillo, B.; Leon, T.; et al. A Phase II Trial of Tideglusib in Alzheimer’s Disease. J. Alzheimers Dis. 2015, 45, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Arciniegas Ruiz, S.M.; Eldar-Finkelman, H. Glycogen Synthase Kinase-3 Inhibitors: Preclinical and Clinical Focus on CNS-A Decade Onward. Front. Mol. Neurosci. 2021, 14, 792364. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, J.A.; Thieffine, S.; Vulpetti, A.; Cristiani, C.; Valsasina, B.; Knapp, S.; Kalisz, H.M.; Flocco, M. Structural characterization of the GSK-3beta active site using selective and non-selective ATP-mimetic inhibitors. J. Mol. Biol. 2003, 333, 393–407. [Google Scholar] [CrossRef]
- Zhang, H.C.; Bonaga, L.V.; Ye, H.; Derian, C.K.; Damiano, B.P.; Maryanoff, B.E. Novel bis(indolyl)maleimide pyridinophanes that are potent, selective inhibitors of glycogen synthase kinase-3. Bioorg. Med. Chem. Lett. 2007, 17, 2863–2868. [Google Scholar] [CrossRef]
- Fu, G.; Sivaprakasam, P.; Dale, O.R.; Manly, S.P.; Cutler, S.J.; Doerksen, R.J. Pharmacophore Modeling, Ensemble Docking, Virtual Screening, and Biological Evaluation on Glycogen Synthase Kinase-3beta. Mol. Inf. 2014, 33, 610–626. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15-Ligand Discovery for Everyone. J. Chem. In.f Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Schrödinger Release 2020-4; Schrödinger, LLC: New York, NY, USA, 2020.
- Perola, E. Minimizing false positives in kinase virtual screens. Proteins 2006, 64, 422–435. [Google Scholar] [CrossRef]
- Huggins, D.J.; Sherman, W.; Tidor, B. Rational Approaches to Improving Selectivity in Drug Design. J. Med. Chem. 2012, 55, 1424–1444. [Google Scholar] [CrossRef]
- Hayes, J.M.; Skamnaki, V.T.; Archontis, G.; Lamprakis, C.; Sarrou, J.; Bischler, N.; Skaltsounis, A.L.; Zographos, S.E.; Oikonomakos, N.G. Kinetics, in silico docking, molecular dynamics, and MM-GBSA binding studies on prototype indirubins, KT5720, and staurosporine as phosphorylase kinase ATP-binding site inhibitors: The role of water molecules examined. Proteins-Struct. Funct. Bioinform. 2011, 79, 703–719. [Google Scholar] [CrossRef]
- Tosovic, J.; Fijan, D.; Jukic, M.; Bren, U. Conserved Water Networks Identification for Drug Design Using Density Clustering Approaches on Positional and Orientational Data. J. Chem. Inf. Model. 2022, 62, 6105–6117. [Google Scholar] [CrossRef] [PubMed]
- Hagar, M.; Chaieb, K.; Parveen, S.; Ahmed, H.A.; Alnoman, R.B. N-alkyl 2-pyridone versus O-alkyl 2-pyridol: Ultrasonic synthesis, DFT, docking studies and their antimicrobial evaluation. J. Mol. Struct. 2020, 1199, 126926. [Google Scholar] [CrossRef]
- Barr, D.; Szennyes, E.; Bokor, E.; Al-Oanzi, Z.H.; Moffatt, C.; Kun, S.; Docsa, T.; Sipos, A.; Davies, M.P.; Mathomes, R.T.; et al. Identification of C-beta-d-Glucopyranosyl Azole-Type Inhibitors of Glycogen Phosphorylase That Reduce Glycogenolysis in Hepatocytes: In Silico Design, Synthesis, in Vitro Kinetics, and ex Vivo Studies. ACS Chem. Biol. 2019, 14, 1460–1470. [Google Scholar] [CrossRef] [PubMed]
- Kun, S.; Mathomes, R.T.; Docsa, T.; Somsak, L.; Hayes, J.M. Design and Synthesis of 3-(β-D-Glucopyranosyl)-4-amino/4-guanidino Pyrazole Derivatives and Analysis of Their Glycogen Phosphorylase Inhibitory Potential. Molecules 2023, 28, 3005. [Google Scholar] [CrossRef]
- Goller, A.H. Reliable gas-phase tautomer equilibria of drug-like molecule scaffolds and the issue of continuum solvation. J. Comput. Aid. Mol. Des. 2022, 36, 805–824. [Google Scholar] [CrossRef] [PubMed]
- Marenich, A.V.; Olson, R.M.; Kelly, C.P.; Cramer, C.J.; Truhlar, D.G. Self-consistent reaction field model for aqueous and nonaqueous solutions based on accurate polarized partial charges. J. Chem. Theory Comput. 2007, 3, 2011–2033. [Google Scholar] [CrossRef]
- Volynets, G.P.; Chekanov, M.O.; Synyugin, A.R.; Golub, A.G.; Kukharenko, O.P.; Bdzhola, V.G.; Yarmoluk, S.M. Identification of 3H-Naphtho[1,2,3-de]quinoline-2,7-diones as Inhibitors of Apoptosis Signal-Regulating Kinase 1 (ASK1). J. Med. Chem. 2011, 54, 2680–2686. [Google Scholar] [CrossRef]
- Hers, I.; Vincent, E.E.; Tavare, J.M. Akt signalling in health and disease. Cell. Signal. 2011, 23, 1515–1527. [Google Scholar] [CrossRef]
- Goode, N.; Hughes, K.; Woodgett, J.R.; Parker, P.J. Differential regulation of glycogen synthase kinase-3 beta by protein kinase C isotypes. J. Biol. Chem. 1992, 267, 16878–16882. [Google Scholar] [CrossRef]
- Davies, M.P.; Benitez, R.; Perez, C.; Jakupovic, S.; Welsby, P.; Rzepecka, K.; Alder, J.; Davidson, C.; Martinez, A.; Hayes, J.M. Structure-Based Design of Potent Selective Nanomolar Type-II Inhibitors of Glycogen Synthase Kinase-3 beta. J. Med. Chem. 2021, 64, 1497–1509. [Google Scholar] [CrossRef]
- Wei, J.; Wang, J.; Zhang, J.; Yang, J.; Wang, G.; Wang, Y. Development of inhibitors targeting glycogen synthase kinase-3beta for human diseases: Strategies to improve selectivity. Eur. J. Med. Chem. 2022, 236, 114301. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Lv, X.; Zhang, J. Exploiting polypharmacology for improving therapeutic outcome of kinase inhibitors (KIs): An update of recent medicinal chemistry efforts. Eur. J. Med. Chem. 2018, 143, 449–463. [Google Scholar] [CrossRef] [PubMed]
- van de Waterbeemd, H.; Camenisch, G.; Folkers, G.; Chretien, J.R.; Raevsky, O.A. Estimation of blood-brain barrier crossing of drugs using molecular size and shape, and H-bonding descriptors. J. Drug. Target 1998, 6, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.K.; Herbertz, T.; Hudkins, R.L.; Dorsey, B.D.; Mallamo, J.P. Knowledge-Based, Central Nervous System (CNS) Lead Selection and Lead Optimization for CNS Drug Discovery. ACS Chem. Neurosci. 2012, 3, 50–68. [Google Scholar] [CrossRef] [PubMed]
- Luco, J.M. Prediction of the brain-blood distribution of a large set of drugs from structurally derived descriptors using partial least-squares (PLS) modeling. J. Chem. Inf. Comput. Sci. 1999, 39, 396–404. [Google Scholar] [CrossRef]
- Carpenter, T.S.; Kirshner, D.A.; Lau, E.Y.; Wong, S.E.; Nilmeier, J.P.; Lightstone, F.C. A method to predict blood-brain barrier permeability of drug-like compounds using molecular dynamics simulations. Biophys. J. 2014, 107, 630–641. [Google Scholar] [CrossRef]
- Kralj, S.; Jukic, M.; Bren, U. Comparative Analyses of Medicinal Chemistry and Cheminformatics Filters with Accessible Implementation in Konstanz Information Miner (KNIME). Int. J. Mol. Sci. 2022, 23, 5727. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Duffy, E.M. Prediction of drug solubility from Monte Carlo simulations. Bioorg. Med. Chem. Lett. 2000, 10, 1155–1158. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Duffy, E.M. Prediction of drug solubility from structure. Adv. Drug Deliv. Rev. 2002, 54, 355–366. [Google Scholar] [CrossRef]
- Lagorce, D.; Bouslama, L.; Becot, J.; Miteva, M.A.; Villoutreix, B.O. FAF-Drugs4: Free ADME-tox filtering computations for chemical biology and early stages drug discovery. Bioinformatics 2017, 33, 3658–3660. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.; Nowotka, M.; Papadatos, G.; Dedman, N.; Gaulton, A.; Atkinson, F.; Bellis, L.; Overington, J.P. ChEMBL web services: Streamlining access to drug discovery data and utilities. Nucleic Acids Res. 2015, 43, W612–W620. [Google Scholar] [CrossRef] [PubMed]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef] [PubMed]
- Sondergaard, C.R.; Olsson, M.H.M.; Rostkowski, M.; Jensen, J.H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pK(a) Values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, G.A.; Friesner, R.A.; Tirado-Rives, J.; Jorgensen, W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar] [CrossRef]
- Hehre, W.J.; Ditchfie, R.; Pople, J.A. Self-Consistent Molecular-Orbital Methods .12. Further Extensions of Gaussian-Type Basis Sets for Use in Molecular-Orbital Studies of Organic-Molecules. J. Chem. Phys. 1972, 56, 2257–2261. [Google Scholar] [CrossRef]
- Francl, M.M.; Pietro, W.J.; Hehre, W.J.; Binkley, J.S.; Gordon, M.S.; Defrees, D.J.; Pople, J.A. Self-Consistent Molecular-Orbital Methods .23. A Polarization-Type Basis Set for 2nd-Row Elements. J. Chem. Phys. 1982, 77, 3654–3665. [Google Scholar] [CrossRef]

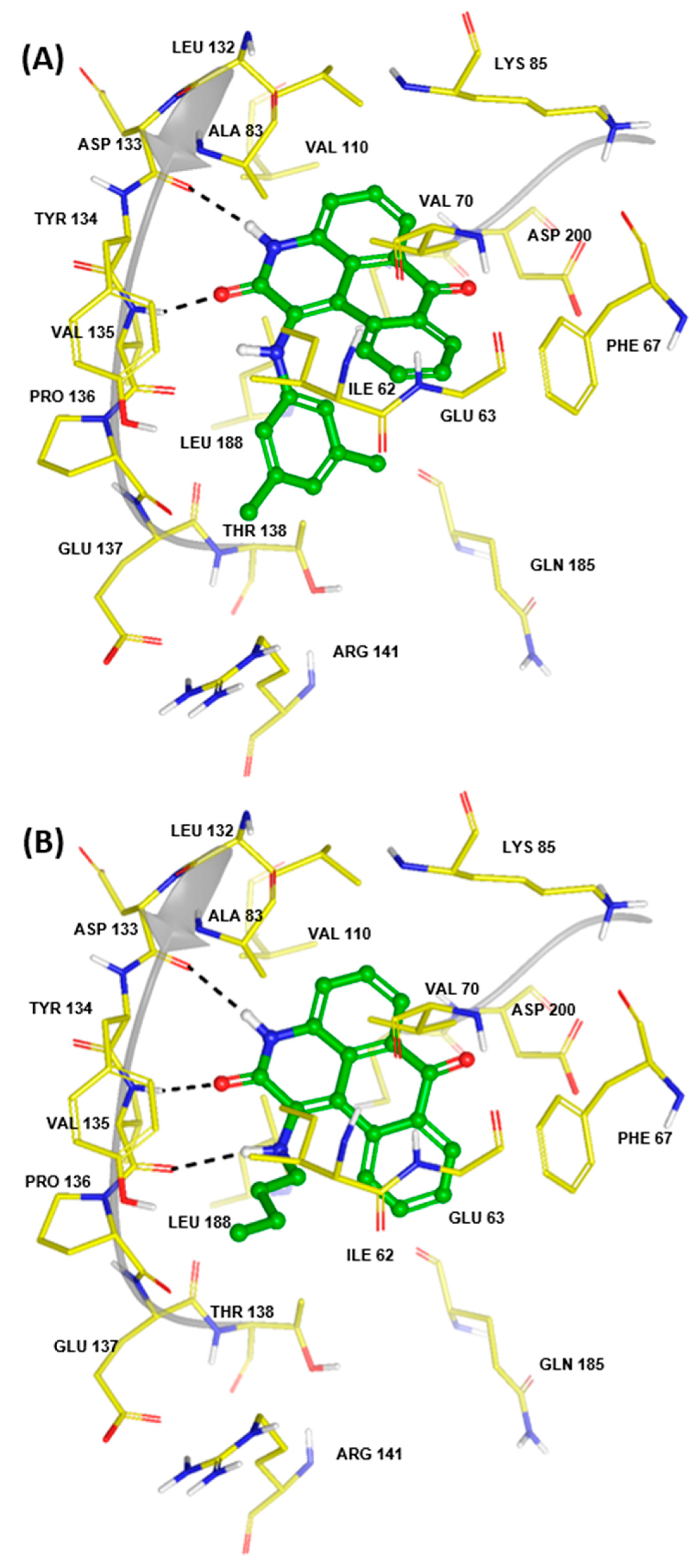
| Enrichment Statistics | Protocol 1 | Protocol 2 | Protocol 3 |
|---|---|---|---|
| # Actives recovered a | 50 | 42 | 41 |
| AU-ROC | 0.8 | 0.76 | 0.77 |
| Top-ranked 1% Statistics | |||
| # Actives in top 1% | 10 | 15 | 19 |
| EF—1% b | 16 | 28 | 34 |
| Top-ranked 2% Statistics | |||
| # Actives in top 2% | 16 | 16 | 22 |
| EF—2% b | 15 | 16 | 22 |
| Top-ranked 5% Statistics | |||
| # Actives in top 5% | 22 | 22 | 26 |
| EF—5% b | 8.8 | 8.8 | 10 |
| Compound | Computational a | Experimental | ||
|---|---|---|---|---|
| Docking Score | Log BB | PSA (Å2) | IC50 (µM) (% Inhibition at 50 µM) | |
| 1 | −8.21 | −0.55 | 73.6 | 1.63 ± 0.30 |
| 2 | −8.19 | −0.66 | 74.1 | 20.55 ± 3.80 |
| 3 | −8.84 | −0.32 | 69.2 | 63.73 ± 3.3 |
| 4 | −8.37 | −0.41 | 55.1 | 87.51 ± 9.17 |
| 5 | −8.29 | −0.35 | 66.6 | (20%) |
| 6 | −8.37 | −0.29 | 89.7 | (11%) |
| 7 | −8.31 | −0.63 | 95.4 | (10%) |
| 8 | −8.35 | −0.50 | 66.0 | (10%) |
| 9 | −8.41 | −0.46 | 71.5 | (9%) |
| 10 | −8.19 | −0.63 | 138.7 | (6%) |
| 11 | −8.92 | −0.52 | 117.2 | (2%) |
| 12 | −8.58 | −0.30 | 54.5 | (NI) b |
| Skeletal Structure | 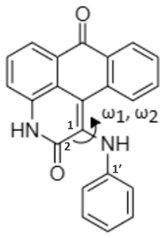 | 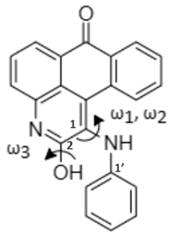 |
| Global minimum conformation | 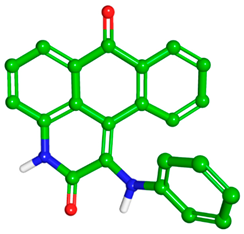 |  |
| t1 | t2 | |
| Energy (kcal/mol) a | ||
| Relative GPE | 0.0 (0.0) | 7.1 (6.8) |
| Relative SPE | 0.0 | 6.3 |
| Dihedral Angle (°) b | ||
| ω1 [C(2)-C(1)-N-H] | 9.3 | 15.7 |
| ω2 [C(2)-C(1)-N-C(1′)] | −138.6 | −128.9 |
| ω3 [C(1)-C(2)-O-H] | - | −179.1 |
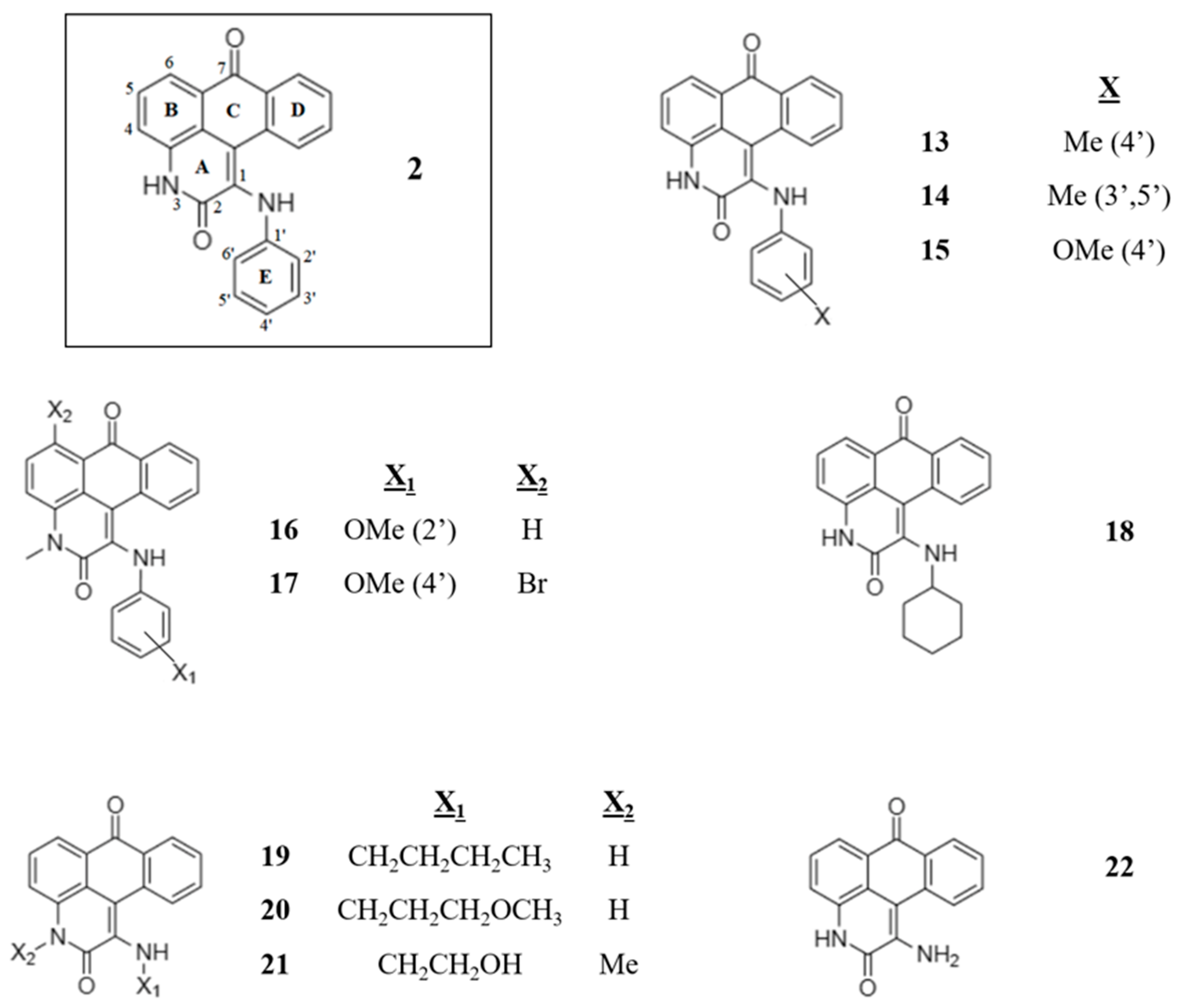
| Compound | Computational a | Experimental | ||
|---|---|---|---|---|
| Docking Score | Log BB | PSA (Å2) | IC50 (µM) | |
| 2 | −8.19 | −0.66 | 74.1 | 20.55 ± 3.80 |
| 13 | −8.32 | −0.70 | 74.1 | 9.4 ± 0.5 |
| 14 | −8.44 | −0.74 | 74.5 | 9.1 ± 0.2 |
| 15 | −8.31 | −0.76 | 82.4 | 11.4 ± 0.6 |
| 16 | −4.79 | −0.41 | 70.0 | 27.1 ± 1.1 |
| 17 | −7.68 | −0.21 | 69.0 | 14.0 ± 3.1 |
| 18 | −8.41 | −0.60 | 73.3 | 25.3 ± 3.5 |
| 19 | −7.54 | −0.76 | 73.0 | 4.9 ± 0.2 |
| 20 | −7.89 | −0.83 | 81.8 | 9.1 ± 1.7 |
| 21 | −7.91 | −0.85 | 87.7 | 17.0 ± 2.7 |
| 22 | −8.40 | −0.85 | 89.4 | 36.7 ± 5.7 |
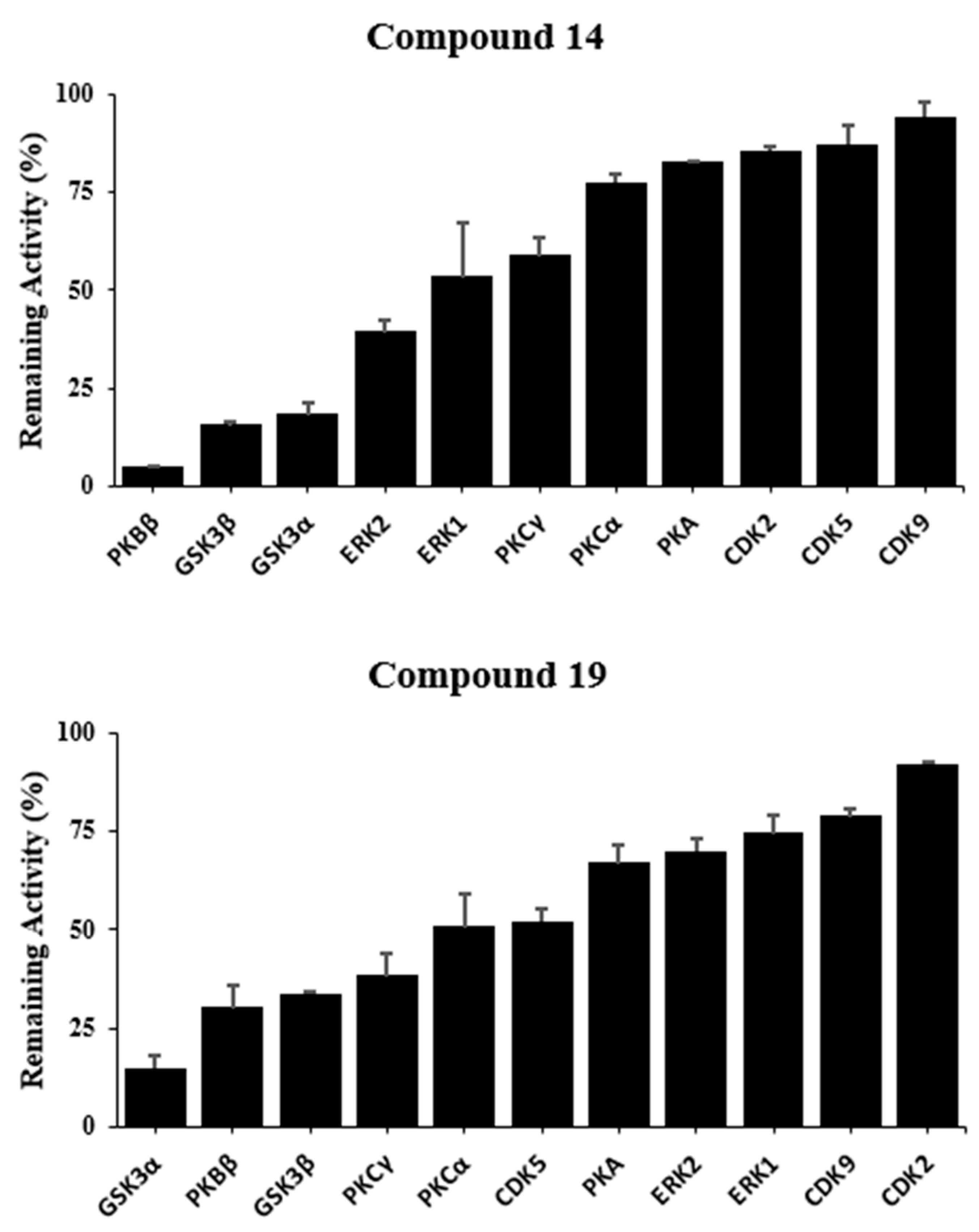
| Kinase | Compound IC50 (µM) | |
|---|---|---|
| 14 | 19 | |
| GSK-3α | 2.3 ± 0.4 | 1.7 ± 0.1 |
| GSK-3β | 9.1 ± 2.4 | 4.9 ± 0.2 |
| PKBβ | 14.9 ± 1.5 | 24.6 ± 0.2 |
| ERK2 | 21. 3 ± 2.3 | - |
| PKCγ | - | 11.0 ± 1.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Emmerich, T.D.; Hayes, J.M. In Silico-Motivated Discovery of Novel Potent Glycogen Synthase-3 Inhibitors: 1-(Alkyl/arylamino)-3H-naphtho[1,2,3-de]quinoline-2,7-dione Identified as a Scaffold for Kinase Inhibitor Development. Pharmaceuticals 2023, 16, 661. https://doi.org/10.3390/ph16050661
Emmerich TD, Hayes JM. In Silico-Motivated Discovery of Novel Potent Glycogen Synthase-3 Inhibitors: 1-(Alkyl/arylamino)-3H-naphtho[1,2,3-de]quinoline-2,7-dione Identified as a Scaffold for Kinase Inhibitor Development. Pharmaceuticals. 2023; 16(5):661. https://doi.org/10.3390/ph16050661
Chicago/Turabian StyleEmmerich, Thomas D., and Joseph M. Hayes. 2023. "In Silico-Motivated Discovery of Novel Potent Glycogen Synthase-3 Inhibitors: 1-(Alkyl/arylamino)-3H-naphtho[1,2,3-de]quinoline-2,7-dione Identified as a Scaffold for Kinase Inhibitor Development" Pharmaceuticals 16, no. 5: 661. https://doi.org/10.3390/ph16050661
APA StyleEmmerich, T. D., & Hayes, J. M. (2023). In Silico-Motivated Discovery of Novel Potent Glycogen Synthase-3 Inhibitors: 1-(Alkyl/arylamino)-3H-naphtho[1,2,3-de]quinoline-2,7-dione Identified as a Scaffold for Kinase Inhibitor Development. Pharmaceuticals, 16(5), 661. https://doi.org/10.3390/ph16050661







