Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review
Abstract
1. Introduction
2. Origin of Antibiotic Resistance
3. Mechanism of Antibiotic Resistance
4. Adverse Effects of Antibiotic Resistance
5. Effects of Antibiotic Resistance on Drug Development: The Challenges
6. Latest Strategies against Drug-Resistant Microorganisms
- (i)
- New antibiotics: The development of novel antibiotics targeting drug-resistant bacteria is a critical endeavor in the face of growing antimicrobial resistance. Researchers are actively seeking new treatment options to combat these resilient pathogens. One notable example is Teixobactin, a groundbreaking antibiotic discovered in 2015. Teixobactin effectively targets a wide range of drug-resistant Gram-positive bacteria [135]. Unlike some traditional antibiotics, Teixobactin’s mechanism of action, which disrupts bacterial cell walls, makes it less likely to trigger resistance [136]. Another innovative antibiotic, Lefamulin, is a pleuromutilin that received FDA approval in 2019 [137]. Lefamulin is used to treat community-acquired bacterial pneumonia, including infections caused by drug-resistant Streptococcus pneumoniae. It inhibits bacterial protein synthesis (binding to the peptidyl transferase center of the 50S bacterial ribosome, thus preventing the binding of transfer RNA for peptide transfer), providing an alternative treatment for cases where resistance to older antibiotics has emerged [138,139]. Zoliflodacin is being studied as a novel antibiotic for drug-resistant Neisseria gonorrhoeae, the bacteria responsible for gonorrhea [140]. This antibiotic, which falls into the spiroketone class, targets DNA replication, making it a promising option for combating a sexually transmitted infection that has developed resistance to multiple drugs [141]. Cefiderocol, a siderophore cephalosporin antibiotic, has been approved in several countries. It is highly effective against many drug-resistant Gram-negative bacteria, including carbapenem-resistant Enterobacteriaceae [142]. Cefiderocol works by disrupting bacterial cell walls and facilitating iron uptake, an innovative approach to addressing antibiotic resistance [143].
- (ii)
- Bacteriophages: Bacteriophages (viruses that infect bacteria) produce lysins to break down bacterial cell walls, leading to bacterial death [144]. Engineered lysins have shown promise in targeting specific bacterial species or strains, including antibiotic-resistant ones. Lysins can be effective against a wide range of bacteria and can be developed relatively quickly. Lysins offer a potentially rapid response to bacterial infections, especially those caused by antibiotic-resistant strains [145]. However, there are challenges, such as lysin resistance and the potential for side effects due to the rapid release of bacterial toxins upon cell wall breakdown. Bacteriophages are viruses that infect and kill bacteria. They can be highly specific, targeting only certain bacterial strains. Phage therapy involves using specific phages to treat bacterial infections, including those caused by antibiotic-resistant bacteria. Phage therapy has been successfully used in some cases to treat antibiotic-resistant infections, particularly in regions where it has a history of use (e.g., Eastern Europe). However, challenges include finding the right phage for each infection, phage stability, regulatory hurdles, and the need for tailored treatments. Combining different approaches, such as antibiotics with antibodies, lysins, or phages, can enhance treatment effectiveness. This is because different strategies target different aspects of bacterial infection and resistance.
- (iii)
- Combination therapies: The use of multiple antibiotics in combination is a promising strategy to enhance treatment efficacy and prevent the development of antibiotic resistance in bacteria. This approach involves administering two or more antibiotics with distinct mechanisms of action simultaneously. By targeting different aspects of bacterial physiology, combination therapy can reduce the selective pressure that drives resistance development. Two notable examples of combination therapies are Trimethoprim-Sulfamethoxazole (TMP-SMX) and Beta-lactam/beta-lactamase inhibitor combinations [115,116]. The TMP-SMX combination therapy pairs Trimethoprim, which inhibits bacterial dihydrofolate reductase, with Sulfamethoxazole (an inhibitor of dihydropteroate synthase). Together, these drugs disrupt two essential steps in the bacterial folate synthesis pathway. TMP-SMX is highly effective against a broad range of bacterial infections, including urinary tract infections, respiratory tract infections, and opportunistic infections in immunocompromised patients [146]. This combination is particularly valuable in treating methicillin-resistant Staphylococcus aureus (MRSA) and Pneumocystis jirovecii pneumonia [147]. The Beta-lactam/beta-lactamase inhibitor combinations are those in which a beta-lactam antibiotic (such as ampicillin, piperacillin, or cefotaxime) is paired with a beta-lactamase inhibitor (like sulbactam, clavulanic acid, or tazobactam). The inhibitor prevents the enzymatic degradation of the beta-lactam antibiotic, allowing it to remain effective against a broader spectrum of bacteria, including those with beta-lactamase enzymes. Examples of such combination therapies include ampicillin/sulbactam and piperacillin/tazobactam, which are used to combat various bacterial infections, especially those caused by beta-lactamase-producing organisms [148,149].
- (iv)
- Immune modulation: Immune modulation is a strategy of boosting the immune response to enhance the clearance of bacterial infection and represents a critical strategy in addressing antibiotic resistance. The rise of drug-resistant bacteria has underscored the importance of fortifying the body’s natural defenses to complement antibiotic therapies and, in some cases, reduce the need for antibiotics. Several approaches have been explored to harness the immune system’s capabilities effectively. One of the strategies employed is the use of immunomodulatory compounds. These include cytokines such as interferons (e.g., interferon-gamma), interleukins, and colony-stimulating factors. Interferon-gamma, for instance, plays a pivotal role in activating macrophages, which are key components of the immune system. IFN-γ therapy has been utilized in treating infections like tuberculosis and non-tuberculous mycobacterial (NTM) infections [150]. By bolstering the immune response, these compounds help the body combat bacterial invaders more effectively. Vaccination is another proactive approach to stimulate the immune system. The development of vaccines targeting specific bacterial pathogens has yielded remarkable results. For instance, vaccines against drug-resistant strains of Streptococcus pneumoniae have significantly reduced the prevalence of antibiotic-resistant pneumonia [151]. Vaccination not only provides direct protection, but also curbs the spread of drug-resistant bacteria within communities. Immunostimulants, including granulocyte-colony stimulating factor (G-CSF), can be used to enhance white blood cell production [152]. This heightened immune response can assist the body in fighting off infections more effectively. While not a direct substitute for antibiotics, immunostimulants can be employed to reduce the reliance on antibiotics and support the immune system’s efforts [153].
- (v)
- Antibody therapies: The development of monoclonal antibodies (mAbs) and antibody-derived molecules to target bacterial pathogens is an innovative approach in combating bacterial infections [154]. These engineered antibodies are designed to specifically recognize and neutralize bacterial targets, offering a precise and highly targeted method of treatment. Various strategies have been employed to harness the potential of these antibodies, with promising results. One notable example is the development of mAbs targeting Clostridium difficile, a bacterium responsible for antibiotic-associated diarrhea and colitis [155]. Bezlotoxumab, a monoclonal antibody, has been approved for the prevention of recurrent C. difficile infection. Bezlotoxumab works by binding to the C. difficile toxin B, preventing its harmful effects and reducing the risk of recurrent infections in high-risk patients [156]. This approach not only provides a targeted therapy, but also reduces the reliance on antibiotics, which can exacerbate C. difficile infections. Another groundbreaking development is the creation of mAbs targeting Staphylococcus aureus. In particular, the antibody-derived molecule called Altastaph is designed to bind to the bacteria’s surface protein, effectively neutralizing the pathogen. This approach is particularly relevant in combating methicillin-resistant Staphylococcus aureus (MRSA) infections, which are notorious for their resistance to multiple antibiotics [157]. By utilizing Altastaph and similar antibodies, researchers aim to enhance the immune system’s ability to recognize and clear MRSA infections. Beyond conventional mAbs, innovative antibody-derived molecules have been engineered. A notable example is that of lysibodies, which are designed to target a broad spectrum of bacteria by attacking conserved elements on the bacterial cell wall [158]. These molecules work similarly to mAbs, but have been engineered to recognize a wide range of bacterial species, reducing the need for highly specific antibodies for each pathogen. This approach offers the potential for a versatile and effective treatment strategy against a wide variety of bacterial infections. Researchers have also explored the use of antibody-conjugates, where antibodies are coupled with toxic agents to create a potent antimicrobial. One such example is the use of antibody–toxin conjugates targeting Pseudomonas aeruginosa [159]. By using antibodies to guide the toxin to the bacterial pathogen, this approach effectively destroys the bacteria and may represent a promising alternative to conventional antibiotics, especially in cases of multidrug-resistant strains.
- (vi)
- Nanotechnology: Nanoparticles have emerged as a promising class of novel antimicrobials with diverse mechanisms of action and several advantages. One of their primary mechanisms is direct microbial inhibition, where their small size and high surface area allow them to physically disrupt microbial cell membranes. This can lead to cell lysis and the inactivation of microorganisms. Some nanoparticles, such as silver nanoparticles, release ions that interfere with microbial DNA replication and protein synthesis [160,161]. Additionally, certain nanoparticles can generate reactive oxygen species (ROS) when exposed to light. These ROS, including singlet oxygen and hydroxyl radicals, have potent antimicrobial properties which damage microbial components [162]. Nanoparticles can also inhibit biofilm formation, a crucial virulence factor in many pathogens. They achieve this by disrupting quorum sensing, preventing initial attachment, or destabilizing the biofilm matrix [163]. Moreover, nanoparticles can be designed for drug delivery, allowing the encapsulation and targeted release of antimicrobial agents directly at the infection site. This enhances drug stability and bioavailability, and minimizes systemic side effects, making them a valuable tool in antimicrobial therapy [164]. The advantages of nanoparticles as antimicrobials are manifold. They exhibit broad-spectrum activity against various microorganisms, including bacteria, fungi, and viruses. Their multiple mechanisms of action can slow down the development of antimicrobial resistance, a critical concern in contemporary medicine. Nanoparticles are also capable of enhancing the solubility of poorly soluble antimicrobial drugs, thereby increasing their therapeutic efficacy. Their ability to achieve targeted drug delivery minimizes damage to healthy tissues and maximizes the antimicrobial effect at the site of infection. Furthermore, nanoparticles offer stability to antimicrobial agents, protecting them from degradation and extending their shelf life. However, nanoparticles also come with certain limitations. Ensuring their biocompatibility with human cells and tissues is of paramount importance, particularly when considering in vivo applications. The cost of producing nanoparticles can be high, potentially limiting their widespread use, and regulatory challenges arise due to safety concerns. Achieving the specificity to target pathogens while sparing beneficial microorganisms remains a challenge. Additionally, the environmental impact of nanoparticles, especially their disposal into the environment, raises concerns regarding ecological consequences and the potential transfer of antibiotic resistance genes.
- (vii)
- Heterocyclic compounds: Heterocyclic compounds, stemming from Thiazole, Imidazole, Thiazolidinone, Oxazole, Pyrrole, Pyridine, and Pyrimidine, constitute a highly diverse and essential class of organic chemicals. They have emerged as promising contenders in the quest for novel antimicrobial agents, thanks to their ability to employ a wide array of mechanisms that enhance their efficacy against various microbial threats. This family of compounds is increasingly in demand, due to its extensive applications in both the synthetic and biological domains. Notably, derivatives like Oxazole, Pyrrole, Pyranopyrimidine, and benzimidazole-based heterocyclic compounds exhibit distinct antibacterial properties against a broad spectrum of bacteria, encompassing both Gram-positive and Gram-negative strains [121]. Their primary mode of action revolves around disrupting essential biological processes within microorganisms, effectively inhibiting their growth and survival. This action includes the inhibition of vital enzymes, interference with nucleic acid synthesis, damage to cell membranes, and disruption of energy production pathways. Heterocyclic compounds offer the advantage of a well-understood synthetic chemistry, facilitating the creation of structurally diverse derivatives, thus enabling the optimization of their antimicrobial potential. Their versatility allows for the tailored development of compounds designed to target specific microorganisms or even strains that have developed resistance. However, it is essential to acknowledge the limitations associated with heterocyclic compounds, such as the risk of resistance development and potential toxicity concerns. Extensive research is needed to ensure their safety in clinical applications. Additionally, addressing regulatory considerations and overcoming challenges in large-scale production is vital to fully harness the potential of these compounds in the ongoing battle against infectious diseases [121].
- (viii)
- Antimicrobial peptides: The exploration of naturally occurring peptides with antimicrobial properties has garnered significant attention in the search for alternative treatments against drug-resistant bacterial infections. These peptides, often referred to as antimicrobial peptides (AMPs), are naturally produced by various organisms, including humans, as part of their innate immune defense mechanisms [73]. AMPs exhibit a wide range of antimicrobial activities and have the potential to combat bacterial pathogens effectively. One remarkable example of an AMP is LL-37, which is a human host defense peptide found in various tissues and bodily fluids. LL-37 has demonstrated potent antimicrobial activity against a variety of bacteria, including drug-resistant strains [174]. It acts by disrupting bacterial cell membranes and interfering with intracellular processes. Researchers have been investigating the therapeutic potential of LL-37 in treating infections caused by multidrug-resistant bacteria, particularly in wound care and chronic skin conditions. Another group of naturally occurring peptides with antimicrobial properties is the defensins. Human defensins, such as human beta-defensins (hBDs), are small cationic peptides with broad-spectrum antimicrobial activity [175]. They can target both Gram-positive and Gram-negative bacteria, making them valuable candidates for developing novel treatments. For example, researchers are exploring the use of defensins in combination with traditional antibiotics to enhance their effectiveness against antibiotic-resistant strains. In nature, amphibians, such as frogs, produce a wide array of antimicrobial peptides as part of their defense mechanisms. Peptides like magainins and temporins have been isolated from the skin secretions of frogs and have demonstrated potent antibacterial activity [176]. These peptides function by disrupting bacterial membranes, ultimately leading to bacterial cell lysis. They have been the focus of research for their potential use in developing new antibiotics, especially for combating multidrug-resistant bacteria. Furthermore, peptides derived from various sources, including marine organisms, insects, and plants, have shown promise as antimicrobial agents. For instance, researchers have investigated the use of peptides from marine sponges and algae to combat bacterial infections. Additionally, antimicrobial peptides from bee venom, like melittin, have displayed activity against a range of pathogens [177].
- (ix)
- CRISPR-Cas system: The application of CRISPR gene-editing technology to target and eliminate drug-resistant bacteria represents a groundbreaking approach in the fight against antibiotic resistance. CRISPR, which stands for Clustered Regularly Interspaced Short Palindromic Repeats, is a revolutionary tool that allows precise modification of an organism’s DNA, including bacteria. By leveraging CRISPR, researchers can engineer specific genetic changes to disrupt drug resistance mechanisms in bacterial pathogens. One remarkable example of using CRISPR technology to combat antibiotic resistance is the development of phage therapy [178]. Phages are viruses that can infect and kill bacteria. Researchers have employed CRISPR to modify phages so that they target antibiotic-resistant bacterial strains. By introducing CRISPR-modified phages into the bacterial population, scientists can selectively eliminate resistant bacteria, restoring susceptibility to antibiotics. This approach offers a promising strategy for overcoming drug resistance and enhancing the effectiveness of conventional antibiotics. Another example involves the modification of a bacteria’s own genes to reverse antibiotic resistance. Researchers have used CRISPR technology to target and disrupt the genes responsible for antibiotic resistance in bacterial strains [125]. By doing so, they can restore the susceptibility of these bacteria to antibiotics that were previously ineffective. This approach may be particularly useful in clinical settings, where antibiotic-resistant infections can be life-threatening. Moreover, CRISPR-based gene editing has enabled the development of novel antimicrobial compounds. Scientists have used this technology to design synthetic peptides or proteins that specifically target and inhibit drug-resistant bacterial mechanisms. These customized molecules can interfere with essential bacterial processes, ultimately rendering antibiotic-resistant strains vulnerable to traditional antibiotics. This innovative approach opens up new avenues for developing antimicrobial agents. CRISPR technology’s versatility allows for the customization of strategies to combat antibiotic resistance in a highly targeted manner. Researchers can design CRISPR sequences to recognize and disrupt various resistance mechanisms, such as efflux pumps or antibiotic-modifying enzymes [179]. As a result, drug-resistant bacteria become more susceptible to existing antibiotics. The utilization of CRISPR gene-editing technology to target and eliminate drug-resistant bacteria has the potential to transform the field of antimicrobial research and clinical practice. However, there are challenges to overcome, including the need for safe and efficient delivery methods for CRISPR components. Ensuring the specificity and accuracy of CRISPR targeting is crucial to avoid unintended consequences. Nonetheless, the development and refinement of CRISPR-based strategies provide hope for combating antibiotic resistance and extending the effectiveness of existing antibiotics.
- (x)
- One Health approach: A holistic approach integrating human health, animal health, and the environment is crucial in addressing antibiotic resistance. Recognizing the interconnectedness of these elements is vital to combat this global challenge. In such an approach, human health and antibiotic use are considered in conjunction with animal health and environmental factors [180]. One example of this approach is the One Health initiative, which emphasizes the interplay between humans, animals, and the environment. It encourages collaborative efforts between healthcare professionals, veterinarians, environmental experts, and policymakers to mitigate antibiotic resistance. In the context of human health, this approach involves promoting responsible antibiotic use in healthcare settings. This includes optimizing prescription practices to reduce overuse and misuse of antibiotics [181]. In animal health, it pertains to the judicious use of antibiotics in veterinary medicine and agriculture. Implementing stricter regulations on antibiotic use in livestock and aquaculture helps minimize the risk of antibiotic-resistant bacteria entering the food chain. Furthermore, the environmental aspect involves understanding how antibiotic residues and resistant bacteria can impact ecosystems. The presence of antibiotics in the environment can promote the selection of resistant strains. This holistic approach necessitates proper disposal of pharmaceutical waste and controlling the release of antibiotics into water sources [182].
- (xi)
- Drug repurposing: Investigating existing drugs for potential activity against drug-resistant microorganisms is a promising strategy in the battle against antibiotic resistance [4,183]. This approach involves reevaluating established medications to identify whether they possess antimicrobial properties, even if that was not their primary purpose. This repurposing of drugs can provide effective treatments for drug-resistant infections. An example of this is the use of statins, commonly prescribed for managing cholesterol levels, which have shown potential antibacterial properties [184]. Statins can disrupt bacterial membranes and potentially enhance the action of traditional antibiotics. Moreover, other drugs used for non-antibiotic purposes have demonstrated antimicrobial activity. For instance, antipsychotic agents like chlorpromazine and thioridazine, which treat mental health conditions, have been explored for their ability to inhibit bacterial efflux pumps and increase the susceptibility of bacteria to antibiotics [185]. This approach maximizes the utility of existing drugs and can be particularly valuable when combating multidrug-resistant pathogens. A key advantage of investigating existing drugs is the wealth of safety and efficacy data available. These drugs have already undergone extensive testing in human patients, which can expedite their repurposing for antimicrobial applications. By identifying non-traditional antimicrobial agents among existing drugs, the antibiotic arsenal can be enhanced and new treatments for drug-resistant microorganisms can be offered.
- (xii)
- Surveillance and diagnostics: Improved monitoring and diagnostic tools are crucial for the early detection and tracking of resistance in microorganisms, ensuring more effective responses to the threat of antibiotic resistance. One example of this is the development and utilization of rapid molecular diagnostics, such as polymerase chain reaction (PCR) tests, which can quickly identify the presence of antibiotic resistance genes in bacterial samples [186]. This technology enables healthcare professionals to make informed decisions regarding the choice of antibiotics and treatment regimens, improving patient outcomes and helping contain resistance. Furthermore, advances in metagenomics, particularly next-generation sequencing techniques, have enabled researchers to monitor the entire microbial community within a given environment. It is especially valuable in tracking the emergence of new resistance mechanisms and identifying areas where antibiotic-resistant bacteria are prevalent [186]. In addition, digital health tools and telemedicine have been harnessed to enhance surveillance and monitoring. These technologies facilitate the real-time reporting of antibiotic-resistant infections, aiding healthcare authorities in responding promptly to outbreaks and implementing containment measures [187]. Efforts to improve monitoring and diagnostics also extend to the development of biosensors and microfluidic devices capable of detecting antibiotic resistance at the point of care. For instance, researchers have designed microchip-based devices that can rapidly identify drug-resistant bacteria by analyzing their responses to antibiotics [188]. These tools offer the potential for on-site diagnostics and tailored treatment decisions.
7. Safety, Efficacy and Commercial Viability of Common Alternative Therapies
8. Limitations and Challenges in the Development of New Antibiotics
- (i)
- Limited Novel Antibiotic Discovery:
- (ii)
- Antibiotic Resistance Development:
- (iii)
- Limited Spectrum of Activity:
- (iv)
- Complex Clinical Trials:
- (v)
- Need for Combination Therapies:
- (vi)
- Stewardship and Responsible Use:
9. Hurdles and Challenges in Regulatory Process for New Antibiotics
10. Modification in Regulatory Process to Facilitate Antibiotic Development
- (i)
- Adaptive Pathways and Flexible Trial Designs: The regulatory process should allow adaptive trial designs that can be adjusted based on emerging data. This flexibility can speed up the development process and accommodate the evolving nature of bacterial resistance.
- (ii)
- Streamlined Approval Process: Establishment of a streamlined regulatory pathway specifically for antibiotics that addresses their unique challenges. This could involve expedited reviews, accelerated approvals, or priority designations for antibiotics targeting urgent or unmet medical needs.
- (iii)
- Tailored Clinical Trial Endpoints: Development of clinically meaningful endpoints for antibiotic trials that reflect their unique mechanism of action and intended use. These endpoints should consider both short-term efficacy and long-term impact on antibiotic resistance.
- (iv)
- Guidance on Combination Therapies: The regulatory process should provide clear guidance on developing and testing combination therapies involving antibiotics, antibodies, probiotics, or other approaches. This guidance should address dosing, interactions, and potential synergies.
- (v)
- Biomarker Development: Investment in research to identify predictive biomarkers that can indicate early in clinical trials whether an antibiotic is effective. Biomarkers can help streamline development and reduce trial durations.
- (vi)
- Economic Incentives: Offering economic incentives such as extended market exclusivity or market entry rewards for antibiotics that target high-priority pathogens or mechanisms of resistance [205].
- (vii)
- Adaptive Licensing: Implementation of adaptive licensing strategies that allow earlier access to antibiotics based on initial safety and efficacy data, with continued monitoring and data collection post approval.
- (viii)
- Collaborative Approaches: Encouragement of collaboration between regulatory agencies, industry, academia, and public health organizations to share data, insights, and best practices for antibiotic development and regulatory evaluation.
- (ix)
- Real-World Evidence: Incorporation of real-world evidence, such as data from patient registries and observational studies, to supplement traditional clinical trial data and support post-approval evaluations.
- (x)
- Global Harmonization: Working towards global harmonization of regulatory standards for antibiotic development. This can reduce duplication of efforts and create a more efficient pathway for international approvals.
- (xi)
- Antibiotic Stewardship Education: Incorporation of antibiotic stewardship education and responsible-use recommendations into regulatory processes to ensure that new antibiotics are used appropriately to slow the development of resistance.
11. One Health Approach
- (i)
- The Netherlands’ Success in Controlling MRSA: The Netherlands adopted a One Health strategy to tackle methicillin-resistant Staphylococcus aureus (MRSA) in livestock and subsequently in humans [206]. They introduced strict regulations on antibiotic use in agriculture and implemented effective surveillance programs. This led to a significant reduction in MRSA prevalence in both animals and humans.
- (ii)
- The Danish Integrated Antimicrobial Resistance Monitoring and Research Program: Denmark has been proactive in implementing a One Health approach to address antibiotic resistance [207]. Their Integrated Antimicrobial Resistance Monitoring and Research Program (DANMAP) tracks antibiotic usage and resistance in humans, animals, and food. As a result, Denmark has seen a decrease in the use of antibiotics in animal farming and a subsequent reduction in resistance.
- (iii)
- The Case of Nipah Virus in Bangladesh: The emergence of the Nipah virus in Bangladesh was tackled through a One Health approach [208]. Health authorities coordinated with veterinary services, ecologists, and other stakeholders to investigate the source and transmission routes of the virus. This collaborative effort allowed for a better understanding of the virus, leading to more targeted public health interventions.
- (iv)
- The Swedish Strategy Against Antibiotic Resistance: Sweden implemented a One Health strategy that involves close collaboration between human and veterinary medicine [209]. This approach has resulted in low antibiotic consumption in both healthcare and agriculture, contributing to low levels of antibiotic resistance.
- (v)
- Tackling Zoonotic Diseases in Africa: Various African countries have embraced the One Health approach to combat zoonotic diseases [210]. For instance, Kenya established the Zoonotic Disease Unit (ZDU), which works to control diseases that spread between animals and humans. ZDU promotes collaborative efforts involving healthcare workers, veterinarians, and environmental experts to address diseases like rabies and anthrax.
- (vi)
- The Work of the FAO, WHO, and WOAH: The Food and Agriculture Organization (FAO), World Health Organization (WHO), and World Organization for Animal Health (WOAH) have been collaborating to address antibiotic resistance at a global level [211]. They work on guidelines for responsible antibiotic use in agriculture and the surveillance of resistance in both animals and humans.
- (vii)
- Global Initiatives on Tuberculosis: Tuberculosis (TB) is an example where the One Health approach is critical. Organizations like the Stop TB Partnership and the World Health Organization work with veterinarians to tackle TB in cattle, as it can also infect humans [212,213]. By addressing the disease in both humans and animals, they aim to reduce the overall burden of TB.
12. Economic Implications of Antibiotic Resistance and Its Impact on Drug Development
- (i)
- Increased Healthcare Costs: One of the most immediate economic consequences of antibiotic resistance is the rise in healthcare costs [215]. Resistant infections often require more extended hospital stays, complex treatments, and costly second-line antibiotics. Further, the increased financial burden on healthcare systems can limit resources available for other critical health services.
- (ii)
- Lost Productivity: Antibiotic-resistant infections can lead to extended sick leave, decreased productivity, and even disability or death, resulting in economic losses [216]. When individuals are unable to work or need prolonged medical care due to antibiotic-resistant infections, it affects not only their personal income but also overall workforce productivity.
- (iii)
- Antibiotic Research and Development Costs: Developing new antibiotics is a costly and time-consuming process. The pharmaceutical industry faces financial disincentives to invest in antibiotic research and development because the market for these drugs is limited [205]. Unlike chronic conditions where patients may take medications for an extended period, antibiotics are typically taken for short durations. Therefore, the return on investment for new antibiotics is often lower than for drugs used to treat chronic diseases.
- (iv)
- Market Dynamics: The pharmaceutical market dynamics play a significant role in the economic implications of antibiotic resistance [217]. As new antibiotics face challenges in reaching the market, existing antibiotics may experience price increases. Drug shortages, which are often linked to production and distribution issues, can lead to higher prices for antibiotics, impacting both healthcare facilities and patients.
- (v)
- Global Trade and Food Production: Antibiotic resistance can have indirect economic consequences through its impact on global trade and food production [218]. In agriculture, the use of antibiotics in animal husbandry can lead to the development of resistant bacteria and contribute to the spread of antibiotic resistance. This can disrupt food supplies, increase production costs, and potentially affect food prices, impacting both the agricultural and food industries.
- (vi)
- Tourism and Travel: Resistant infections can discourage tourism and travel to regions with higher prevalence rates [219]. Countries with reputations for antibiotic-resistant healthcare-associated infections may see a decline in medical tourism and tourism in general. This can have a direct impact on local economies that rely on tourism as a significant revenue source.
- (vii)
- Innovative Treatments: The emergence of antibiotic resistance can also drive innovation in medical treatments. Researchers and healthcare professionals are exploring alternative treatment options, such as phage therapy and monoclonal antibodies, which may have economic implications in terms of development costs and market competition [220].
- (viii)
- Global Economic Impact: Antibiotic resistance is a global issue that can have international economic repercussions. The spread of resistant strains across borders can impact trade, tourism, and international relations. Collaborative efforts to address the problem and share best practices can mitigate these economic consequences [182].
13. Healthcare Costs Associated with Antibiotic-Resistant Infections, and the Potential Return on Investment for Pharmaceutical Companies
14. Withdrawal of Major Capital Firms in Antibiotic Research: Reasons and Strategies to Re-Engage Them
15. Strategies to Overcome Economic Challenges and Low Profitability of Antibiotic Development
16. Ethical and Societal Considerations
17. Responsibilities of Healthcare Professionals
18. Role of Policy-Makers and Regulatory Bodies
19. Data Availability and Reporting
- (i)
- Under-reporting: One of the most substantial issues in surveillance systems is under-reporting. Healthcare providers and facilities may fail to report antibiotic-resistant cases due to concerns about potential consequences or the administrative burden of reporting [235]. This leads to an incomplete and inaccurate representation of the true prevalence of antibiotic resistance, hindering the ability to implement targeted interventions.
- (ii)
- Variability in Data Quality: Data on antibiotic-resistant cases can vary significantly in quality. Variations in laboratory testing methods, diagnostic capabilities, and reporting mechanisms can lead to inconsistencies in data [236]. For instance, different facilities may use diverse criteria for defining antibiotic resistance, making it difficult to compare data across regions or healthcare systems.
- (iii)
- Limited Access to Comprehensive Data: Access to comprehensive data is often limited, due to issues related to data sharing and privacy. Sharing patient-specific data while maintaining privacy is a delicate balance that can be challenging to achieve [237]. As a result, surveillance systems might not have access to complete patient histories, hindering the ability to track the progression of antibiotic resistance.
- (iv)
- Heterogeneous Reporting Standards: Reporting standards and guidelines can vary significantly from one region to another. This heterogeneity can create confusion and challenges in data interpretation and integration [238]. Harmonizing these standards can streamline data collection and reporting.
- (v)
- Data Silos: Healthcare data are often stored in isolated silos within various healthcare facilities and organizations. These silos can prevent the comprehensive sharing of data and lead to gaps in surveillance. Improved interoperability and data-sharing mechanisms are needed to facilitate data integration.
- (vi)
- Lack of Standardization in Testing Methods: The use of varying diagnostic methods and laboratory techniques for testing antibiotic resistance can affect the quality and comparability of data. The lack of standardized testing protocols can lead to discrepancies in the reported prevalence of antibiotic resistance.
- (vii)
- Delay in Reporting: A significant challenge is the delay in reporting antibiotic-resistant cases. The lag between data collection and reporting can hinder timely responses to outbreaks or emerging resistance patterns. Streamlining data collection, analysis, and reporting processes is crucial for faster response times.
- (viii)
- Data Overload: The sheer volume of data generated in healthcare settings can be overwhelming, making it challenging to extract valuable information for surveillance purposes. The development of efficient data management and analysis tools is essential for identifying trends and patterns amid the data noise.
- (ix)
- Data Security Concerns: Protecting sensitive patient data is critical, and this may limit the extent to which data can be shared among healthcare facilities and surveillance systems. Finding ways to anonymize and secure patient data while still allowing for comprehensive analysis is an ongoing challenge.
- (x)
- Global Data Sharing: Given the global nature of antibiotic resistance, sharing data and collaborating across borders is essential. However, international data sharing can be hampered by issues such as data sovereignty, legal and ethical concerns, and differing healthcare systems.
- (i)
- Standardizing Reporting Criteria: Establishing uniform reporting criteria and guidelines for defining and documenting antibiotic resistance is paramount. By ensuring that healthcare facilities, laboratories, and providers adhere to standardized definitions and protocols, data consistency and comparability can be significantly improved.
- (ii)
- Improving Data Sharing Mechanisms: Enhancing data sharing mechanisms is crucial for seamless information exchange. Developing secure, interoperable systems that allow healthcare facilities and organizations to share data while safeguarding patient privacy is essential. This might involve the implementation of anonymization techniques and secure data transfer protocols.
- (iii)
- Investing in Data Infrastructure: Investing in robust data infrastructure is fundamental for capturing, storing, and managing vast amounts of healthcare data. The development of comprehensive databases and data repositories, equipped with advanced analytical tools, can streamline data collection and analysis, making it easier to track and respond to antibiotic resistance trends.
- (iv)
- Enhancing Data Analysis Capabilities: Effective data analysis is vital for identifying patterns, trends, and emerging resistance issues. Investing in data analysis capabilities, including the use of artificial intelligence and machine learning algorithms, can help in processing large datasets rapidly, thus enabling timely responses to changing resistance patterns.
- (v)
- Promoting a Culture of Data Reporting and Transparency: Fostering a culture of data reporting and transparency within the healthcare community is crucial. Healthcare professionals, including clinicians and laboratory staff, need to be aware of the importance of reporting antibiotic-resistant cases. Education and awareness campaigns can play a significant role in encouraging data reporting as part of routine practice.
- (vi)
- Collaboration and Coordination: Collaboration among healthcare facilities, public health agencies, and regulatory bodies is essential. By coordinating efforts and sharing data and insights, a more comprehensive and accurate picture of antibiotic resistance can be achieved. This collaboration can extend beyond national borders to address global antibiotic resistance challenges.
- (vii)
- Timely Reporting: Implementing mechanisms for real-time or near-real-time reporting of antibiotic-resistant cases can significantly enhance response times. Swift reporting allows for quicker identification of outbreaks, enabling healthcare providers and policymakers to take immediate action to contain the spread of resistant strains.
- (viii)
- Interdisciplinary Approaches: Combining expertise from various disciplines, including epidemiology, microbiology, data science, and public health, can lead to more holistic and accurate data collection and analysis. Interdisciplinary teams can work together to address the multifaceted challenges posed by antibiotic resistance.
- (ix)
- Feedback Loops: Establishing feedback loops for healthcare facilities and providers can enhance data quality and encourage reporting. By providing facilities with timely feedback on their data, they can improve their data collection and reporting practices.
- (x)
- Public Awareness: Educating the public about the importance of antibiotic resistance data and the role of individuals in reporting illnesses and adhering to prescribed antibiotics is crucial. A well-informed public can contribute to early reporting and more responsible antibiotic use.
20. International Collaborations or Initiatives Addressing Antibiotic Resistance
- (i)
- Global Antibiotic Research and Development Partnership (GARDP):
- (ii)
- CARB-X (Combating Antibiotic-Resistant Bacteria Biopharmaceutical Accelerator):
- (iii)
- Innovative Medicines Initiative (IMI) AMR Accelerator:
- (iv)
- ReAct—Action on Antibiotic Resistance:
- (v)
- Fleming Fund:
- (vi)
- Global Health Security Agenda (GHSA):
- (vii)
- WHO Global Action Plan on Antimicrobial Resistance:
- (viii)
- Joint Programming Initiative on Antimicrobial Resistance (JPIAMR):
- (ix)
- United Nations Interagency Coordination Group on Antimicrobial Resistance (IACG):
21. Government-Led Initiatives against Antibiotic Resistance: A Focus on the Middle East
22. Geographical Perspective of Antibiotic Resistance
23. Antibiotic Resistance in the Arabian Peninsula
- (i)
- Establish Regional Networks: Arabian Peninsula countries can establish regional networks or consortia dedicated to antibiotic resistance research and intervention. These networks can facilitate the exchange of data, research findings, and best practices. The Gulf Cooperation Council (GCC) serves as a foundation for such collaboration, and it can be expanded to include specific working groups on antibiotic resistance.
- (ii)
- Joint Research Projects: Collaborative research projects can be initiated to study antibiotic resistance patterns in the region. Researchers from different countries can work together to conduct multicenter studies, which will provide a more comprehensive understanding of regional challenges and help identify effective interventions. These projects can be funded by regional organizations or international research grants.
- (iii)
- Data Sharing and Surveillance: Establishing a regional database for antibiotic resistance data could be beneficial. Participating countries can contribute data from their surveillance programs. By sharing information, countries can track resistance trends across borders and make informed decisions on interventions.
- (iv)
- Harmonize Antibiotic Guidelines: The GCC countries have already taken steps to harmonize antibiotic guidelines. This effort can be expanded and refined to ensure consistent practices in antibiotic use, prescription policies, and resistance containment strategies. Standardized guidelines will promote effective interventions and stewardship.
- (v)
- Regional Workshops and Conferences: Regular regional workshops and conferences can bring together healthcare professionals, researchers, and policymakers to discuss antibiotic resistance challenges and solutions. These events serve as platforms for knowledge-sharing, networking, and collaboration. They can be hosted on a rotational basis by different countries in the region.
- (vi)
- Exchange Programs: Exchange programs for healthcare professionals can facilitate the transfer of knowledge and expertise. Doctors, pharmacists, and researchers can spend time working in healthcare institutions or laboratories in neighboring countries, gaining insights into different approaches to antibiotic resistance management.
- (vii)
- Telemedicine and Teleconsultation: Utilize telemedicine and teleconsultation platforms to connect healthcare professionals across borders. This technology can enable experts to provide advice and guidance on complex antibiotic resistance cases, fostering cross-border cooperation.
- (viii)
- Pharmaceutical and Industry Collaboration: Encourage pharmaceutical companies and the healthcare industry to collaborate on research and development projects targeting antibiotic resistance. Joint initiatives can lead to the development of new antibiotics and diagnostics tailored to the region’s resistance patterns.
- (ix)
- Public Awareness Campaigns: Collaborate on public awareness campaigns that educate the public about the risks of antibiotic misuse. Shared campaigns can reach a wider audience and send consistent messages about responsible antibiotic use.
24. The Intersection of Artificial Intelligence (AI) and Antibiotic Resistance
- (a)
- Predictive Analytics: Predictions that are based on historical and real-time data in order to identify patterns by machine learning techniques make up predictive analytics. AI algorithms can analyze large datasets, including genetic information from bacteria, patient health records, and environmental factors, to predict which antibiotics are likely to be effective against specific strains of bacteria. This can aid clinicians in making more informed decisions about antibiotic treatment. By analyzing historical patient data, these tools can recommend the most suitable antibiotics, dosage, and duration of treatment. They can forecast patient outcomes, such as the risk of developing antibiotic resistance, mortality rates, and the length of hospital stays. This information enables medical professionals to provide more personalized and effective care. The analysis of extensive datasets on drug interactions, pharmacokinetics, and microbial genomics, and identifying potential antibiotic candidates and predicting their effectiveness can assist in development of new antibiotics. Moreover, predictive analytics is used to investigate the synergistic effects of drug combinations. This approach can uncover new treatment strategies that enhance the effectiveness of existing antibiotics and combat resistance [262].
- (b)
- Drug Discovery: The development of antibiotics is a resource-intensive process, often requiring a long time and significant financial investments. To expedite antibiotic discovery, there is a growing need for computer-assisted exploration of innovative drugs with unique action mechanisms. Artificial intelligence (AI) has emerged as a powerful tool for accelerating antibiotic discovery. Large amounts of data can be analyzed by AI to identify novel potential drug candidates, predict their properties, and optimize their design. In this way, the drug discovery process can be quick and cheap. Virtual screening for drug discovery is one of the most promising applications of AI. Virtual screening involves using computers to simulate the interaction between potential drug candidates and bacterial targets. This can help to identify compounds that are likely to be effective against bacteria, without the need for expensive and time-consuming laboratory experiments. Another promising application of AI is drug design. AI algorithms can be used to design new antibiotics that are specifically targeted to bacterial targets [263,264].
- (c)
- Optimizing Treatment Plans: Artificial intelligence (AI) can optimize antibiotic treatment plans in several ways. For instance, Al can be used to identify the most effective antibiotic for a particular patient and infection. Medical history, lab results, and genetic information of the patient can be analyzed by Al, and the antibiotic that is most likely to be effective against the specific infection can be identified.
- (d)
- Surveillance and Early Detection: Al can analyze large amounts of data from clinical settings, laboratories, and public health records to identify patterns of antibiotic use and resistance. This information can be used to track the spread of resistant bacteria and inform targeted interventions. For example, Al can be used to identify hospitals or regions with high rates of antibiotic-resistant infections and target these areas for additional resources and education. Al can analyze genetic data to identify mutations that confer antibiotic resistance. This information can be used to track the evolution of resistance genes and develop new antibiotics that are less likely to be ineffective. For example, Al can be used to identify new mutations in the genes that code for antibiotic resistance in Staphylococcus aureus, a common cause of hospital-acquired infections. Al can develop personalized treatment plans for patients with antibiotic-resistant infections. This can be carried out by analyzing patient data, such as medical history, antibiotic use, and laboratory results. Al can also be used to identify combinations of antibiotics that are more effective against resistant bacteria [267].
- (e)
- Rapid Diagnostics: The emergence of rapid diagnostics empowered by artificial intelligence (AI) has revolutionized the fight against antibiotic resistance. These innovative tools enable healthcare professionals to swiftly identify bacterial strains and their resistance profiles, leading to faster and more precise treatment decisions. This not only improves patient outcomes but also plays a critical role in curbing the overuse of antibiotics. Early detection of pathogens is crucial for optimal treatment of infectious diseases. Conventional methods, however, often require several days for both pathogen detection and characterization, leading to delays in initiating appropriate treatment. This delay often necessitates the use of broad-spectrum antibiotics, which can contribute to the development of AMR. Al-powered rapid diagnostics have addressed this challenge by significantly reducing the turnaround time for both detection and resistance profiling. These tools can identify specific bacterial strains and their resistance patterns within hours or even minutes, allowing clinicians to select the most effective antibiotic regimen promptly [267,268].
- (f)
- Optimizing Antibiotic Stewardship: Antibiotic stewardship refers to the coordinated efforts to promote the responsible use of antibiotics in healthcare settings to combat antibiotic resistance, reduce unnecessary antibiotic prescriptions, and improve patient outcomes. Artificial intelligence (AI) can play a significant role in antibiotic stewardship by assisting healthcare providers and institutions in making more informed decisions about antibiotic use [267,269].
- (g)
- Monitoring Environmental Factors: AI can be used to monitor environmental data in real time, which can help to identify and respond to problems quickly. Environmental data can be mapped at a high spatial resolution, which can help to identify hotspots of antibiotic contamination. Data can be analyzed from water and soil samples to detect the presence of antibiotic residues and other pharmaceutical compounds. AI can identify and track antibiotic-resistant genes by processing metagenomic data from environmental samples. Machine learning models can be developed to predict and map regions where environmental factors are more conducive to the development and spread of antibiotic resistance. Data can be analyzed from various sources to trace the origin of antibiotic contamination. AI can also analyze data related to antibiotic use in agriculture and livestock farming, thereby integrating environmental data with clinical data to better understand the links between antibiotic resistance in the environment and its impact on human health. Assisting regulatory agencies in monitoring and enforcing compliance with environmental regulations related to antibiotic use, waste management, and pollution control is yet another role for AI [266,270,271].
- (h)
- Public Health Interventions: In the realm of public health, Artificial Intelligence (AI) plays a critical role in understanding and combating antibiotic resistance. By leveraging AI’s capabilities, extensive datasets are examined to gain insights into the dynamics of antibiotic resistance transmission, guiding the development of effective interventions. One noteworthy application of AI in the fight against antibiotic resistance is its integration with genomics for enhanced antimicrobial resistance (AMR) surveillance. AI-driven models monitor resistance genes, detect emerging trends, and identify new genetic variants, enhancing surveillance sensitivity and efficiency. These models focus on crucial features like resistance genes for initial monitoring, and continuously adapt as new data become available, ensuring accuracy. AI also integrates data from regions with high AMR transmission, facilitating ongoing monitoring and revealing correlations between AMR gene abundance and socioeconomic, health, and environmental factors. AI’s ability to identify emerging AMR genes accelerates the connection between surveillance data and patient care through diagnostic stewardship programs. This leads to updated treatment guidelines and improved patient outcomes. AI holds significant potential in the battle against antibiotic resistance and public health challenges [182].
- (i)
- Education and Awareness: Antibiotic resistance is a growing global threat, and AI can play a crucial role in educating healthcare professionals, researchers, and the public about this critical issue. AI-driven educational tools offer several benefits in addressing antibiotic resistance. AI-powered educational tools can continuously gather and analyze the latest data on antimicrobial use and resistance, providing up-to-date information on the evolving landscape of antibiotic resistance [272]. This real-time information access ensures that healthcare providers and the public are informed about the latest trends and developments in antibiotic resistance. AI can assist researchers in analyzing vast datasets related to antibiotic resistance, helping them identify emerging resistance patterns and potential hotspots of antimicrobial resistance [262]. This ability to analyze large datasets enables researchers to gain deeper insights into the mechanisms of antibiotic resistance and inform the development of effective strategies to combat it. AI tools contribute to a broader understanding of the antibiotic resistance issue by making complex data more accessible and facilitating informed decision-making by healthcare professionals and the public. This enhanced understanding empowers individuals to make informed choices about antibiotic use and play a role in combating antibiotic resistance.
- (j)
- Ethical Considerations: The ethical considerations of using artificial intelligence (AI) in healthcare are complex and multifaceted. There are many potential benefits to using AI in healthcare, such as improved diagnosis and treatment, but there are also risks, such as algorithmic bias and data privacy concerns. The World Health Organization (WHO) has identified six key ethical principles for the use of AI in healthcare [273,274,275]:
25. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). Priority Medicines for Europe and the World/Warren Kaplan, Richard Laing 2004. Available online: https://iris.who.int/handle/10665/68769?show=full (accessed on 25 August 2023).
- Rehman, M.T.; Faheem, M.; Khan, A.U. An insight into the biophysical characterization of different states of cefotaxime hydrolyzing β-lactamase 15 (CTX-M-15). J. Biomol. Struct. Dyn. 2015, 33, 625–638. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M. Bacterial Resistance: Origins, Epidemiology, and Impact. Clin. Infect. Dis. 2003, 36, S11–S23. [Google Scholar] [CrossRef] [PubMed]
- Muteeb, G.; Alsultan, A.; Farhan, M.; Aatif, M. Risedronate and Methotrexate Are High-Affinity Inhibitors of New Delhi Metallo-β-Lactamase-1 (NDM-1): A Drug Repurposing Approach. Molecules 2022, 27, 1283. [Google Scholar] [CrossRef]
- Khan, A.U.; Rehman, M.T. Role of Non-Active-Site Residue Trp-93 in the Function and Stability of New Delhi Metallo-β-Lactamase 1. Antimicrob. Agents Chemother. 2016, 60, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Gould, K. Antibiotics: From prehistory to the present day. J. Antimicrob. Chemother. 2016, 71, 572–575. [Google Scholar] [CrossRef]
- Clardy, J.; Fischbach, M.A.; Currie, C.R. The natural history of antibiotics. Curr. Biol. 2009, 19, R437–R441. [Google Scholar] [CrossRef]
- Fleming, A. On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to their Use in the Isolation of B. influenzæ. Br. J. Exp. Pathol. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- Hodgkin, D.C. The X-ray analysis of the structure of penicillin. Adv. Sci. 1949, 6, 85–89. [Google Scholar]
- Sheehan, J.C.; Henery-Logan, K.R. The Total Synthesis of Penicillin, V.J. Am. Chem. Soc. 1959, 81, 3089–3094. [Google Scholar] [CrossRef]
- Von Döhren, H. Antibiotics: Actions, Origins, Resistance, by C. Walsh. 2003; ASM Press: Washington, DC, USA, 2009; Volume 13, p. 345. [Google Scholar]
- Abraham, E.P.; Chain, E. An Enzyme from Bacteria able to Destroy Penicillin. Nature 1940, 146, 837. [Google Scholar] [CrossRef]
- Aminov, R.I. A Brief History of the Antibiotic Era: Lessons Learned and Challenges for the Future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef] [PubMed]
- Durand, G.A.; Raoult, D.; Dubourg, G. Antibiotic discovery: History, methods and perspectives. Int. J. Antimicrob. Agents 2019, 53, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Iskandar, K.; Murugaiyan, J.; Hammoudi Halat, D.; Hage, S.E.; Chibabhai, V.; Adukkadukkam, S.; Roques, C.; Molinier, L.; Salameh, P.; Van Dongen, M. Antibiotic Discovery and Resistance: The Chase and the Race. Antibiotics 2022, 11, 182. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.B. Drugs That Changed Society: History and Current Status of the Early Antibiotics: Salvarsan, Sulfonamides, and β-Lactams. Molecules 2021, 26, 6057. [Google Scholar] [CrossRef]
- Chait, R.; Vetsigian, K.; Kishony, R. What counters antibiotic resistance in nature? Nat. Chem. Biol. 2012, 8, 2–5. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M.; Blaser, M.; Carrs, O.; Cassell, G.; Fishman, N.; Guidos, R.; Levy, S.; Powers, J.; Norrby, R.; Tillotson, G.; et al. Discovery research: The scientific challenge of finding new antibiotics. J. Antimicrob. Chemother. 2011, 66, 1941–1944. [Google Scholar] [CrossRef]
- Saga, T.; Yamaguchi, K. History of antimicrobial agents and resistant bacteria. Japan Med. Assoc. J. 2009, 52, 103–108. [Google Scholar]
- Hwang, I.Y.; Tan, M.H.; Koh, E.; Ho, C.L.; Poh, C.L.; Chang, M.W. Reprogramming Microbes to Be Pathogen-Seeking Killers. ACS Synth. Biol. 2014, 3, 228–237. [Google Scholar] [CrossRef]
- Orfali, R.; Perveen, S.; AlAjmI, M.F.; Ghaffar, S.; Rehman, M.T.; AlanzI, A.R.; Gamea, S.B.; Essa Khwayri, M. Antimicrobial Activity of Dihydroisocoumarin Isolated from Wadi Lajab Sediment-Derived Fungus Penicillium chrysogenum: In Vitro and In Silico Study. Molecules 2022, 27, 3630. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Bartlett, J.G.; Gilbert, D.N.; Spellberg, B. Seven Ways to Preserve the Miracle of Antibiotics. Clin. Infect. Dis. 2013, 56, 1445–1450. [Google Scholar] [CrossRef] [PubMed]
- Velez, R.; Sloand, E. Combating antibiotic resistance, mitigating future threats and ongoing initiatives. J. Clin. Nurs. 2016, 25, 1886–1889. [Google Scholar] [CrossRef] [PubMed]
- D’Costa, V.M.; McGrann, K.M.; Hughes, D.W.; Wright, G.D. Sampling the Antibiotic Resistome. Science 2006, 311, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.; Kaur, P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef]
- Vega, N.M.; Gore, J. Collective antibiotic resistance: Mechanisms and implications. Curr. Opin. Microbiol. 2014, 21, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 2023, 21, 280–295. [Google Scholar] [CrossRef]
- Chellat, M.F.; Raguž, L.; Riedl, R. Targeting Antibiotic Resistance. Angew. Chemie Int. Ed. 2016, 55, 6600–6626. [Google Scholar] [CrossRef]
- Akova, M. Epidemiology of antimicrobial resistance in bloodstream infections. Virulence 2016, 7, 252–266. [Google Scholar] [CrossRef]
- Martínez, J.L.; Rojo, F. Metabolic regulation of antibiotic resistance. FEMS Microbiol. Rev. 2011, 35, 768–789. [Google Scholar] [CrossRef]
- Akiba, M.; Sekizuka, T.; Yamashita, A.; Kuroda, M.; Fujii, Y.; Murata, M.; Lee, K.-I.; Joshua, D.I.; Balakrishna, K.; Bairy, I.; et al. Distribution and Relationships of Antimicrobial Resistance Determinants among Extended-Spectrum-Cephalosporin-Resistant or Carbapenem-Resistant Escherichia coli Isolates from Rivers and Sewage Treatment Plants in India. Antimicrob. Agents Chemother. 2016, 60, 2972–2980. [Google Scholar] [CrossRef]
- Heddle, J.; Maxwell, A. Quinolone-Binding Pocket of DNA Gyrase: Role of GyrB. Antimicrob. Agents Chemother. 2002, 46, 1805–1815. [Google Scholar] [CrossRef] [PubMed]
- Romano, M.; Iovene, M.R.; Russo, M.I.; Rocco, A.; Salerno, R.; Cozzolino, D.; Pilloni, A.P.; Tufano, M.A.; Vaira, D.; Nardone, G. Failure of first-line eradication treatment significantly increases prevalence of antimicrobial-resistant Helicobacter pylori clinical isolates. J. Clin. Pathol. 2008, 61, 1112–1115. [Google Scholar] [CrossRef]
- Vester, B.; Douthwaite, S. Macrolide Resistance Conferred by Base Substitutions in 23S rRNA. Antimicrob. Agents Chemother. 2001, 45, 1–12. [Google Scholar] [CrossRef]
- Hopwood, D.A. How do antibiotic-producing bacteria ensure their self-resistance before antibiotic biosynthesis incapacitates them? Mol. Microbiol. 2007, 63, 937–940. [Google Scholar] [CrossRef]
- Tahlan, K.; Ahn, S.K.; Sing, A.; Bodnaruk, T.D.; Willems, A.R.; Davidson, A.R.; Nodwell, J.R. Initiation of actinorhodin export in Streptomyces coelicolor. Mol. Microbiol. 2007, 63, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Q&A: Antibiotic resistance: Where does it come from and what can we do about it? BMC Biol. 2010, 8, 6. [Google Scholar]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.N.; Hauryliuk, V.; Atkinson, G.C.; O’Neill, A.J. Target protection as a key antibiotic resistance mechanism. Nat. Rev. Microbiol. 2020, 18, 637–648. [Google Scholar] [CrossRef]
- Kapur, V.; Li, L.L.; Iordanescu, S.; Hamrick, M.R.; Wanger, A.; Kreiswirth, B.N.; Musser, J.M. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J. Clin. Microbiol. 1994, 32, 1095–1098. [Google Scholar] [CrossRef] [PubMed]
- Wichelhaus, T.A.; Schafer, V.; Brade, V.; Boddinghaus, B. Molecular Characterization of rpoB Mutations Conferring Cross-Resistance to Rifamycins on Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 1999, 43, 2813–2816. [Google Scholar] [CrossRef]
- Adam, M.; Murali, B.; Glenn, N.O.; Potter, S.S. Epigenetic inheritance based evolution of antibiotic resistance in bacteria. BMC Evol. Biol. 2008, 8, 52. [Google Scholar] [CrossRef]
- AlAjmi, M.F.; Rehman, M.T.; Hussain, A.; Rather, G.M. Pharmacoinformatics approach for the identification of Polo-like kinase-1 inhibitors from natural sources as anti-cancer agents. Int. J. Biol. Macromol. 2018, 116, 173–181. [Google Scholar] [CrossRef]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.A.; Dantas, G. The Shared Antibiotic Resistome of Soil Bacteria and Human Pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef]
- Colomer-Lluch, M.; Jofre, J.; Muniesa, M. Antibiotic Resistance Genes in the Bacteriophage DNA Fraction of Environmental Samples. PLoS ONE 2011, 6, e17549. [Google Scholar] [CrossRef]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef]
- Baker, K.S.; Dallman, T.J.; Field, N.; Childs, T.; Mitchell, H.; Day, M.; Weill, F.-X.; Lefèvre, S.; Tourdjman, M.; Hughes, G.; et al. Horizontal antimicrobial resistance transfer drives epidemics of multiple Shigella species. Nat. Commun. 2018, 9, 1462. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-Spectrum β-Lactamases: A Clinical Update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Mathers, A.J.; Cox, H.L.; Kitchel, B.; Bonatti, H.; Brassinga, A.K.C.; Carroll, J.; Scheld, W.M.; Hazen, K.C.; Sifri, C.D. Molecular Dissection of an Outbreak of Carbapenem-Resistant Enterobacteriaceae Reveals Intergenus KPC Carbapenemase Transmission through a Promiscuous Plasmid. mBio 2011, 2, 10–1128. [Google Scholar] [CrossRef]
- Balm, M.N.D.; Ngan, G.; Jureen, R.; Lin, R.T.P.; Teo, J.W.P. OXA-181-producing Klebsiella pneumoniae establishing in Singapore. BMC Infect. Dis. 2013, 13, 58. [Google Scholar] [CrossRef]
- Huang, T.-W.; Lauderdale, T.-L.; Liao, T.-L.; Hsu, M.-C.; Chang, F.-Y.; Chang, S.-C.; Khong, W.X.; Ng, O.T.; Chen, Y.-T.; Kuo, S.-C.; et al. Effective transfer of a 47 kb NDM-1-positive plasmid among Acinetobacter species. J. Antimicrob. Chemother. 2015, 70, 2734–2738. [Google Scholar] [CrossRef]
- Martin, J.; Phan, H.T.T.; Findlay, J.; Stoesser, N.; Pankhurst, L.; Navickaite, I.; De Maio, N.; Eyre, D.W.; Toogood, G.; Orsi, N.M.; et al. Covert dissemination of carbapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: Long- and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission. J. Antimicrob. Chemother. 2017, 72, 3025–3034. [Google Scholar] [CrossRef]
- Gorrie, C.L.; Mirceta, M.; Wick, R.R.; Judd, L.M.; Wyres, K.L.; Thomson, N.R.; Strugnell, R.A.; Pratt, N.F.; Garlick, J.S.; Watson, K.M.; et al. Antimicrobial-Resistant Klebsiella pneumoniae Carriage and Infection in Specialized Geriatric Care Wards Linked to Acquisition in the Referring Hospital. Clin. Infect. Dis. 2018, 67, 161–170. [Google Scholar] [CrossRef]
- Virolle, C.; Goldlust, K.; Djermoun, S.; Bigot, S.; Lesterlin, C. Plasmid Transfer by Conjugation in Gram-Negative Bacteria: From the Cellular to the Community Level. Genes 2020, 11, 1239. [Google Scholar] [CrossRef] [PubMed]
- Stanczak-Mrozek, K.I.; Manne, A.; Knight, G.M.; Gould, K.; Witney, A.A.; Lindsay, J.A. Within-host diversity of MRSA antimicrobial resistances. J. Antimicrob. Chemother. 2015, 70, 2191–2198. [Google Scholar] [CrossRef] [PubMed]
- Mašlaňová, I.; Stříbná, S.; Doškař, J.; Pantůček, R. Efficient plasmid transduction to Staphylococcus aureus strains insensitive to the lytic action of transducing phage. FEMS Microbiol. Lett. 2016, 363, fnw211. [Google Scholar] [CrossRef] [PubMed]
- Blahová, J.; Králiková, K.; Krčméry, V.; Ježek, P. Low-Frequency Transduction of Imipenem Resistance and High-Frequency Transduction of Ceftazidime and Aztreonam Resistance by the Bacteriophage AP-151 Isolated from a Pseudomonas aeruginosa Strain. J. Chemother. 2000, 12, 482–486. [Google Scholar] [CrossRef]
- Krahn, T.; Wibberg, D.; Maus, I.; Winkler, A.; Bontron, S.; Sczyrba, A.; Nordmann, P.; Pühler, A.; Poirel, L.; Schlüter, A. Intraspecies Transfer of the Chromosomal Acinetobacter baumannii bla NDM-1 Carbapenemase Gene. Antimicrob. Agents Chemother. 2016, 60, 3032–3040. [Google Scholar] [CrossRef]
- Zeman, M.; Mašlaňová, I.; Indráková, A.; Šiborová, M.; Mikulášek, K.; Bendíčková, K.; Plevka, P.; Vrbovská, V.; Zdráhal, Z.; Doškař, J.; et al. Staphylococcus sciuri bacteriophages double-convert for staphylokinase and phospholipase, mediate interspecies plasmid transduction, and package mecA gene. Sci. Rep. 2017, 7, 46319. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.; Coque, T.M.; Alonso, D.; Valverde, A.; Baquero, F.; Cantón, R. CTX-M-10 Linked to a Phage-Related Element Is Widely Disseminated among Enterobacteriaceae in a Spanish Hospital. Antimicrob. Agents Chemother. 2005, 49, 1567–1571. [Google Scholar] [CrossRef] [PubMed]
- Traglia, G.M.; Chua, K.; Centrón, D.; Tolmasky, M.E.; Ramírez, M.S. Whole-Genome Sequence Analysis of the Naturally Competent Acinetobacter baumannii Clinical Isolate A118. Genome Biol. Evol. 2014, 6, 2235–2239. [Google Scholar] [CrossRef] [PubMed]
- Johnston, C.; Martin, B.; Fichant, G.; Polard, P.; Claverys, J.-P. Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 2014, 12, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Griffith, F. The Significance of Pneumococcal Types. Epidemiol. Infect. 1928, 27, 113–159. [Google Scholar] [CrossRef] [PubMed]
- Dorer, M.S.; Cohen, I.E.; Sessler, T.H.; Fero, J.; Salama, N.R. Natural Competence Promotes Helicobacter pylori Chronic Infection. Infect. Immun. 2013, 81, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Kersulyte, D.; Chalkauskas, H.; Berg, D.E. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 1999, 31, 31–43. [Google Scholar] [CrossRef]
- Bello-López, J.M.; Cabrero-Martínez, O.A.; Ibáñez-Cervantes, G.; Hernández-Cortez, C.; Pelcastre-Rodríguez, L.I.; Gonzalez-Avila, L.U.; Castro-Escarpulli, G. Horizontal Gene Transfer and Its Association with Antibiotic Resistance in the Genus Aeromonas spp. Microorganisms 2019, 7, 363. [Google Scholar] [CrossRef]
- Picard, M.; Shirihai, O.S. Mitochondrial signal transduction. Cell Metab. 2022, 34, 1620–1653. [Google Scholar] [CrossRef]
- Hassett, D.J.; Ma, J.; Elkins, J.G.; McDermott, T.R.; Ochsner, U.A.; West, S.E.H.; Huang, C.; Fredericks, J.; Burnett, S.; Stewart, P.S.; et al. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 1999, 34, 1082–1093. [Google Scholar] [CrossRef]
- Michaelis, C.; Grohmann, E. Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms. Antibiotics 2023, 12, 328. [Google Scholar] [CrossRef]
- Jayaraman, R. Antibiotic resistance: An overview of mechanisms and a paradigm shift. Curr. Sci. 2009, 96, 1475–1484. [Google Scholar]
- Muteeb, G.; Rehman, M.T.; Ali, S.; Al-Shahrani, A.; Kamal, M.; Ashraf, G. Phage Display Technique: A Novel Medicinal Approach to Overcome Antibiotic Resistance by Using Peptide-Based Inhibitors Against β-Lactamases. Curr. Drug Metab. 2017, 18, 90–95. [Google Scholar] [CrossRef]
- Lai, S.; Tremblay, J.; Déziel, E. Swarming motility: A multicellular behaviour conferring antimicrobial resistance. Environ. Microbiol. 2009, 11, 126–136. [Google Scholar] [CrossRef]
- Bhullar, K.; Waglechner, N.; Pawlowski, A.; Koteva, K.; Banks, E.D.; Johnston, M.D.; Barton, H.A.; Wright, G.D. Antibiotic Resistance Is Prevalent in an Isolated Cave Microbiome. PLoS ONE 2012, 7, e34953. [Google Scholar] [CrossRef]
- Kümmerer, K.; Henninger, A. Promoting resistance by the emission of antibiotics from hospitals and households into effluent. Clin. Microbiol. Infect. 2003, 9, 1203–1214. [Google Scholar] [CrossRef]
- Li, B.; Webster, T.J. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 2017, 36, 22–32. [Google Scholar] [CrossRef]
- von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef]
- Mulvey, M.R.; Simor, A.E. Antimicrobial resistance in hospitals: How concerned should we be? Can. Med. Assoc. J. 2009, 180, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Cycoń, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment—Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef]
- Felis, E.; Kalka, J.; Sochacki, A.; Kowalska, K.; Bajkacz, S.; Harnisz, M.; Korzeniewska, E. Antimicrobial pharmaceuticals in the aquatic environment—Occurrence and environmental implications. Eur. J. Pharmacol. 2020, 866, 172813. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: Part 2: Management strategies and new agents. Pharm. Ther. 2015, 40, 344–352. [Google Scholar]
- Arciola, C.R.; Campoccia, D.; Montanaro, L. Implant infections: Adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 2018, 16, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-K.; Cheng, N.-C.; Cheng, C.-M. Biofilms in Chronic Wounds: Pathogenesis and Diagnosis. Trends Biotechnol. 2019, 37, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J. The crisis of no new antibiotics—What is the way forward? Lancet Infect. Dis. 2012, 12, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-S.; Gupta, A.; Rotello, V.M. Nanomaterials for the Treatment of Bacterial Biofilms. ACS Infect. Dis. 2016, 2, 3–4. [Google Scholar] [CrossRef]
- Wright, G.D. Something old, something new: Revisiting natural products in antibiotic drug discovery. Can. J. Microbiol. 2014, 60, 147–154. [Google Scholar] [CrossRef]
- Gajdács, M. The Concept of an Ideal Antibiotic: Implications for Drug Design. Molecules 2019, 24, 892. [Google Scholar] [CrossRef] [PubMed]
- Gould, I.M.; Bal, A.M. New antibiotic agents in the pipeline and how they can help overcome microbial resistance. Virulence 2013, 4, 185. [Google Scholar] [CrossRef]
- Minandri, F.; Bonchi, C.; Frangipani, E.; Imperi, F.; Visca, P. Promises and failures of gallium as an antibacterial agent. Future Microbiol. 2014, 9, 379–397. [Google Scholar] [CrossRef]
- Lemire, J.A.; Harrison, J.J.; Turner, R.J. Antimicrobial activity of metals: Mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 2013, 11, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Giedraitienė, A.; Vitkauskienė, A.; Naginienė, R.; Pavilonis, A. Antibiotic Resistance Mechanisms of Clinically Important Bacteria. Medicina 2011, 47, 19. [Google Scholar] [CrossRef]
- Flamm, R.K.; Rhomberg, P.R.; Simpson, K.M.; Farrell, D.J.; Sader, H.S.; Jones, R.N. In Vitro Spectrum of Pexiganan Activity When Tested against Pathogens from Diabetic Foot Infections and with Selected Resistance Mechanisms. Antimicrob. Agents Chemother. 2015, 59, 1751–1754. [Google Scholar] [CrossRef]
- Reardon, S. Antibiotic alternatives rev up bacterial arms race. Nature 2015, 521, 402–403. [Google Scholar] [CrossRef]
- Sengupta, S.; Chattopadhyay, M.K.; Grossart, H.-P. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front. Microbiol. 2013, 4, 47. [Google Scholar] [CrossRef]
- Spellberg, B.; Gilbert, D.N. The Future of Antibiotics and Resistance: A Tribute to a Career of Leadership by John Bartlett. Clin. Infect. Dis. 2014, 59, S71–S75. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Recover the lost art of drug discovery. Nature 2012, 485, 439–440. [Google Scholar] [CrossRef] [PubMed]
- Baym, M.; Lieberman, T.D.; Kelsic, E.D.; Chait, R.; Gross, R.; Yelin, I.; Kishony, R. Spatiotemporal microbial evolution on antibiotic landscapes. Science 2016, 353, 1147–1151. [Google Scholar] [CrossRef] [PubMed]
- Lyddiard, D.; Jones, G.L.; Greatrex, B.W. Keeping it simple: Lessons from the golden era of antibiotic discovery. FEMS Microbiol. Lett. 2016, 363, fnw084. [Google Scholar] [CrossRef]
- Parmanik, A.; Das, S.; Kar, B.; Bose, A.; Dwivedi, G.R.; Pandey, M.M. Current Treatment Strategies Against Multidrug-Resistant Bacteria: A Review. Curr. Microbiol. 2022, 79, 388. [Google Scholar] [CrossRef]
- Colson, A.R.; Morton, A.; Årdal, C.; Chalkidou, K.; Davies, S.C.; Garrison, L.P.; Jit, M.; Laxminarayan, R.; Megiddo, I.; Morel, C.; et al. Antimicrobial Resistance: Is Health Technology Assessment Part of the Solution or Part of the Problem? Value Health 2021, 24, 1828–1834. [Google Scholar] [CrossRef]
- Ndagi, U.; Falaki, A.A.; Abdullahi, M.; Lawal, M.M.; Soliman, M.E. Antibiotic resistance: Bioinformatics-based understanding as a functional strategy for drug design. RSC Adv. 2020, 10, 18451–18468. [Google Scholar] [CrossRef] [PubMed]
- Levy, S. Antibiotic resistance—The problem intensifies. Adv. Drug Deliv. Rev. 2005, 57, 1446–1450. [Google Scholar] [CrossRef]
- Faheem, M.; Rehman, M.T.; Danishuddin, M.; Khan, A.U. Biochemical Characterization of CTX-M-15 from Enterobacter cloacae and Designing a Novel Non-β-Lactam-β-Lactamase Inhibitor. PLoS ONE 2013, 8, e56926. [Google Scholar] [CrossRef]
- Harbarth, S.; Samore, M.H. Antimicrobial Resistance Determinants and Future Control. Emerg. Infect. Dis. 2005, 11, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Summers, A.O. Generally Overlooked Fundamentals of Bacterial Genetics and Ecology. Clin. Infect. Dis. 2002, 34, S85–S92. [Google Scholar] [CrossRef] [PubMed]
- Lipsitch, M.; Bergstrom, C.T.; Levin, B.R. The epidemiology of antibiotic resistance in hospitals: Paradoxes and prescriptions. Proc. Natl. Acad. Sci. USA 2000, 97, 1938–1943. [Google Scholar] [CrossRef] [PubMed]
- Goossens, H.; Ferech, M.; Vander Stichele, R.; Elseviers, M. Outpatient antibiotic use in Europe and association with resistance: A cross-national database study. Lancet 2005, 365, 579–587. [Google Scholar] [CrossRef]
- Cárdenas, L.L.; Merchán, M.A.; López, D.P. New antibiotics against bacterial resistance. Infectio 2019, 23, 382. [Google Scholar] [CrossRef]
- Cui, X.; Lü, Y.; Yue, C. Development and Research Progress of Anti-Drug Resistant Bacteria Drugs. Infect. Drug Resist. 2021, 14, 5575–5593. [Google Scholar] [CrossRef]
- Abouelhassan, Y.; Garrison, A.T.; Yang, H.; Chávez-Riveros, A.; Burch, G.M.; Huigens, R.W. Recent Progress in Natural-Product-Inspired Programs Aimed To Address Antibiotic Resistance and Tolerance. J. Med. Chem. 2019, 62, 7618–7642. [Google Scholar] [CrossRef]
- Wang, Y.; Subedi, D.; Li, J.; Wu, J.; Ren, J.; Xue, F.; Dai, J.; Barr, J.J.; Tang, F. Phage Cocktail Targeting STEC O157:H7 Has Comparable Efficacy and Superior Recovery Compared with Enrofloxacin in an Enteric Murine Model. Microbiol. Spectr. 2022, 10, e0023222. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, H.; Ryu, S.; Jeon, B. Inhibition of Antimicrobial-Resistant Escherichia coli Using a Broad Host Range Phage Cocktail Targeting Various Bacterial Phylogenetic Groups. Front. Microbiol. 2021, 12, 699630. [Google Scholar] [CrossRef]
- Yahav, D.; Giske, C.G.; Grāmatniece, A.; Abodakpi, H.; Tam, V.H.; Leibovici, L. New β-Lactam–β-Lactamase Inhibitor Combinations. Clin. Microbiol. Rev. 2020, 34, e00115–e00120. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Hagihara, M.; Asai, N.; Umemura, T.; Shibata, Y.; Hirai, J.; Yamagishi, Y.; Iwamoto, T.; Mikamo, H. Efficacy of Trimethoprim–Sulfamethoxazole in Combination with an Echinocandin as a First-Line Treatment Option for Pneumocystis Pneumonia: A Systematic Review and Meta-Analysis. Antibiotics 2022, 11, 719. [Google Scholar] [CrossRef]
- Bergman, P.; Raqib, R.; Rekha, R.S.; Agerberth, B.; Gudmundsson, G.H. Host Directed Therapy Against Infection by Boosting Innate Immunity. Front. Immunol. 2020, 11, 01209. [Google Scholar] [CrossRef]
- Vacca, F.; Sala, C.; Rappuoli, R. Monoclonal Antibodies for Bacterial Pathogens: Mechanisms of Action and Engineering Approaches for Enhanced Effector Functions. Biomedicines 2022, 10, 2126. [Google Scholar] [CrossRef]
- Yeh, Y.-C.; Huang, T.-H.; Yang, S.-C.; Chen, C.-C.; Fang, J.-Y. Nano-Based Drug Delivery or Targeting to Eradicate Bacteria for Infection Mitigation: A Review of Recent Advances. Front. Chem. 2020, 8, 00286. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227–1249. [Google Scholar] [CrossRef] [PubMed]
- Aatif, M.; Raza, M.A.; Javed, K.; Nashre-ul-Islam, S.M.; Farhan, M.; Alam, M.W. Potential Nitrogen-Based Heterocyclic Compounds for Treating Infectious Diseases: A Literature Review. Antibiotics 2022, 11, 1750. [Google Scholar] [CrossRef]
- Zhang, Q.-Y.; Yan, Z.-B.; Meng, Y.-M.; Hong, X.-Y.; Shao, G.; Ma, J.-J.; Cheng, X.-R.; Liu, J.; Kang, J.; Fu, C.-Y. Antimicrobial peptides: Mechanism of action, activity and clinical potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef] [PubMed]
- Branco, P.; Maurício, E.M.; Costa, A.; Ventura, D.; Roma-Rodrigues, C.; Duarte, M.P.; Fernandes, A.R.; Prista, C. Exploring the Multifaceted Potential of a Peptide Fraction Derived from Saccharomyces cerevisiae Metabolism: Antimicrobial, Antioxidant, Antidiabetic, and Anti-Inflammatory Properties. Antibiotics 2023, 12, 1332. [Google Scholar] [CrossRef]
- Tao, S.; Chen, H.; Li, N.; Liang, W. The Application of the CRISPR-Cas System in Antibiotic Resistance. Infect. Drug Resist. 2022, 15, 4155–4168. [Google Scholar] [CrossRef] [PubMed]
- Mayorga-Ramos, A.; Zúñiga-Miranda, J.; Carrera-Pacheco, S.E.; Barba-Ostria, C.; Guamán, L.P. CRISPR-Cas-Based Antimicrobials: Design, Challenges, and Bacterial Mechanisms of Resistance. ACS Infect. Dis. 2023, 9, 1283–1302. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Ousterout, D.G. Repurposing CRISPR-Cas systems as DNA-based smart antimicrobials. Cell Gene Ther. Insights 2017, 3, 63–72. [Google Scholar] [CrossRef]
- Sleeman, J.M.; DeLiberto, T.; Nguyen, N. Optimization of human, animal, and environmental health by using the One Health approach. J. Vet. Sci. 2017, 18, 263. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.-Y.; Feng, J.-X.; Ai, L.; Xue, J.-B.; Liu, J.-S.; Zhang, X.-X.; Cao, C.-L.; Xu, J.; Xia, S.; Zhou, X.-N.; et al. Assessment of integrated patterns of human-animal-environment health: A holistic and stratified analysis. Infect. Dis. Poverty 2023, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Schein, C.H. Repurposing approved drugs on the pathway to novel therapies. Med. Res. Rev. 2020, 40, 586–605. [Google Scholar] [CrossRef]
- Barbarossa, A.; Rosato, A.; Corbo, F.; Clodoveo, M.L.; Fracchiolla, G.; Carrieri, A.; Carocci, A. Non-Antibiotic Drug Repositioning as an Alternative Antimicrobial Approach. Antibiotics 2022, 11, 816. [Google Scholar] [CrossRef]
- Zulak, K.G.; Cox, B.A.; Tucker, M.A.; Oliver, R.P.; Lopez-Ruiz, F.J. Improved Detection and Monitoring of Fungicide Resistance in Blumeria graminis f. sp. hordei With High-Throughput Genotype Quantification by Digital PCR. Front. Microbiol. 2018, 9, 00706. [Google Scholar] [CrossRef]
- Gupta, E.; Saxena, J.; Kumar, S.; Sharma, U.; Rastogi, S.; Srivastava, V.K.; Kaushik, S.; Jyoti, A. Fast Track Diagnostic Tools for Clinical Management of Sepsis: Paradigm Shift from Conventional to Advanced Methods. Diagnostics 2023, 13, 277. [Google Scholar] [CrossRef]
- Kaprou, G.D.; Bergšpica, I.; Alexa, E.A.; Alvarez-Ordóñez, A.; Prieto, M. Rapid Methods for Antimicrobial Resistance Diagnostics. Antibiotics 2021, 10, 209. [Google Scholar] [CrossRef] [PubMed]
- Kali, A. Teixobactin: A Novel Antibiotic in Treatment of Gram Positive Bacterial Infections. J. Clin. Diagn. Res. 2015, 9, DL01. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.; Karas, J.A.; Schneider-Futschik, E.K.; Chen, F.; Swarbrick, J.; Paulin, O.K.A.; Hoyer, D.; Baker, M.; Zhu, Y.; Li, J.; et al. The Killing Mechanism of Teixobactin against Methicillin-Resistant Staphylococcus aureus: An Untargeted Metabolomics Study. Msystems 2020, 5, e00077-20. [Google Scholar] [CrossRef]
- Adhikary, S.; Duggal, M.K.; Nagendran, S.; Chintamaneni, M.; Tuli, H.S.; Kaur, G. Lefamulin: A New Hope in the Field of Community-Acquired Bacterial Pneumonia. Curr. Pharmacol. Reports 2022, 8, 418–426. [Google Scholar] [CrossRef]
- Eraikhuemen, N.; Julien, D.; Kelly, A.; Lindsay, T.; Lazaridis, D. Treatment of Community-Acquired Pneumonia: A Focus on Lefamulin. Infect. Dis. Ther. 2021, 10, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Veve, M.P.; Wagner, J.L. Lefamulin: Review of a Promising Novel Pleuromutilin Antibiotic. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2018, 38, 935–946. [Google Scholar] [CrossRef]
- Bradford, P.A.; Miller, A.A.; O’Donnell, J.; Mueller, J.P. Zoliflodacin: An Oral Spiropyrimidinetrione Antibiotic for the Treatment of Neisseria gonorrheae, Including Multi-Drug-Resistant Isolates. ACS Infect. Dis. 2020, 6, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Jacobsson, S.; Golparian, D.; Oxelbark, J.; Franceschi, F.; Brown, D.; Louie, A.; Drusano, G.; Unemo, M. Pharmacodynamic Evaluation of Zoliflodacin Treatment of Neisseria gonorrhoeae Strains With Amino Acid Substitutions in the Zoliflodacin Target GyrB Using a Dynamic Hollow Fiber Infection Model. Front. Pharmacol. 2022, 13, 874176. [Google Scholar] [CrossRef]
- Ong’uti, S.; Czech, M.; Robilotti, E.; Holubar, M. Cefiderocol: A New Cephalosporin Stratagem Against Multidrug-Resistant Gram-Negative Bacteria. Clin. Infect. Dis. 2022, 74, 1303–1312. [Google Scholar] [CrossRef]
- Domingues, S.; Lima, T.; Saavedra, M.J.; Da Silva, G.J. An Overview of Cefiderocol’s Therapeutic Potential and Underlying Resistance Mechanisms. Life 2023, 13, 1427. [Google Scholar] [CrossRef] [PubMed]
- Ghose, C.; Euler, C.W. Gram-Negative Bacterial Lysins. Antibiotics 2020, 9, 74. [Google Scholar] [CrossRef]
- Fischetti, V.A. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 2008, 11, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Wróbel, A.; Arciszewska, K.; Maliszewski, D.; Drozdowska, D. Trimethoprim and other nonclassical antifolates an excellent template for searching modifications of dihydrofolate reductase enzyme inhibitors. J. Antibiot. 2020, 73, 5–27. [Google Scholar] [CrossRef]
- de Górgolas, M.; Avilés, P.; Verdejo, C.; Fernández Guerrero, M.L. Treatment of experimental endocarditis due to methicillin-susceptible or methicillin-resistant Staphylococcus aureus with trimethoprim-sulfamethoxazole and antibiotics that inhibit cell wall synthesis. Antimicrob. Agents Chemother. 1995, 39, 953–957. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nomura, S.; Hanaki, H.; Nagayama, A. Tazobactam-Piperacillin Compared with Sulbactam-Ampicillin, Clavulanic Acid-Ticarcillin, Sulbactam-Cefoperazone, and Piperacillin for Activity against Beta-Lactamase-Producing Bacteria Isolated from Patients with Complicated Urinary Tract Infections. J. Chemother. 1997, 9, 89–94. [Google Scholar] [CrossRef]
- Rafailidis, P.I.; Ioannidou, E.N.; Falagas, M.E. Ampicillin/Sulbactam. Drugs 2007, 67, 1829–1849. [Google Scholar] [CrossRef] [PubMed]
- Ghanavi, J.; Farnia, P.; Farnia, P.; Velayati, A. The role of interferon-gamma and interferon-gamma receptor in tuberculosis and nontuberculous mycobacterial infections. Int. J. Mycobacteriol. 2021, 10, 349. [Google Scholar] [PubMed]
- Cillóniz, C.; Garcia-Vidal, C.; Ceccato, A.; Torres, A. Antimicrobial Resistance Among Streptococcus Pneumoniae. In Antimicrobial Resistance in the 21st Century; Springer International Publishing: Cham, Switzerland, 2018; pp. 13–38. [Google Scholar]
- Mathias, B.; Szpila, B.E.; Moore, F.A.; Efron, P.A.; Moldawer, L.L. A Review of GM-CSF Therapy in Sepsis. Medicine 2015, 94, e2044. [Google Scholar] [CrossRef]
- Schneider, C.M.; Daschner, F.D. Colony-stimulating factors and antibiotics—A new prospect in treating infectious diseases? Clin. Microbiol. Infect. 1998, 4, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Motley, M.P.; Banerjee, K.; Fries, B.C. Monoclonal antibody-based therapies for bacterial infections. Curr. Opin. Infect. Dis. 2019, 32, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, M.; Debnath, A.K.; Todi, S.K. Clostridium difficile and Antibiotic-associated Diarrhea. Indian. J. Crit. Care Med. 2020, 24, S162–S167. [Google Scholar] [CrossRef]
- Navalkele, B.D.; Chopra, T. Bezlotoxumab: An emerging monoclonal antibody therapy for prevention of recurrent Clostridium difficile infection. Biol. Targets Ther. 2018, 12, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Park, S.; Thompson, C.D.; Li, X.; Lee, J.C. Antibodies to Staphylococcus aureus capsular polysaccharides 5 and 8 perform similarly in vitro but are functionally distinct in vivo. Virulence 2017, 8, 859–874. [Google Scholar] [CrossRef] [PubMed]
- Raz, A.; Serrano, A.; Lawson, C.; Thaker, M.; Alston, T.; Bournazos, S.; Ravetch, J.V.; Fischetti, V.A. Lysibodies are IgG Fc fusions with lysin binding domains targeting Staphylococcus aureus wall carbohydrates for effective phagocytosis. Proc. Natl. Acad. Sci. USA 2017, 114, 4781–4786. [Google Scholar] [CrossRef]
- Kajihara, K.K.; Pantua, H.; Hernandez-Barry, H.; Hazen, M.; Deshmukh, K.; Chiang, N.; Ohri, R.; Castellanos, E.R.; Martin, L.; Matsumoto, M.L.; et al. Potent Killing of Pseudomonas aeruginosa by an Antibody-Antibiotic Conjugate. mBio 2021, 12, 10–1128. [Google Scholar] [CrossRef]
- Yin, I.X.; Zhang, J.; Zhao, I.S.; Mei, M.L.; Li, Q.; Chu, C.H. The Antibacterial Mechanism of Silver Nanoparticles and Its Application in Dentistry. Int. J. Nanomed. 2020, 15, 2555–2562. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Alajmi, M.F.; Khan, M.A.; Pervez, S.A.; Ahmed, F.; Amir, S.; Husain, F.M.; Khan, M.S.; Shaik, G.M.; Hassan, I.; et al. Biosynthesized silver nanoparticle (AgNP) from pandanus odorifer leaf extract exhibits anti-metastasis and anti-biofilm potentials. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Memar, M.Y.; Ghotaslou, R.; Samiei, M.; Adibkia, K. Antimicrobial use of reactive oxygen therapy: Current insights. Infect. Drug Resist. 2018, 11, 567–576. [Google Scholar] [CrossRef]
- Sahli, C.; Moya, S.E.; Lomas, J.S.; Gravier-Pelletier, C.; Briandet, R.; Hémadi, M. Recent advances in nanotechnology for eradicating bacterial biofilm. Theranostics 2022, 12, 2383–2405. [Google Scholar] [CrossRef]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef] [PubMed]
- Abdelhameed, R.M.; Abu-Elghait, M.; El-Shahat, M. Engineering titanium-organic framework decorated silver molybdate and silver vanadate as antimicrobial, anticancer agents, and photo-induced hydroxylation reactions. J. Photochem. Photobiol. A Chem. 2022, 423, 113572. [Google Scholar] [CrossRef]
- Zhang, X.; Peng, F.; Wang, D. MOFs and MOF-Derived Materials for Antibacterial Application. J. Funct. Biomater. 2022, 13, 215. [Google Scholar] [CrossRef]
- Abdelhameed, R.M.; Abu-Elghait, M.; El-Shahat, M. Hybrid three MOFs composites (ZIF-67@ZIF-8@MIL-125-NH2): Enhancement the biological and visible—Light photocatalytic activity. J. Environ. Chem. Eng. 2020, 8, 104107. [Google Scholar] [CrossRef]
- Liu, J.; Wu, D.; Zhu, N.; Wu, Y.; Li, G. Antibacterial mechanisms and applications of metal-organic frameworks and their derived nanomaterials. Trends Food Sci. Technol. 2021, 109, 413–434. [Google Scholar] [CrossRef]
- Emam, H.E.; El-Shahat, M.; Allayeh, A.K.; Ahmed, H.B. Functionalized starch for formulation of graphitic carbon nanodots as viricidal/anticancer laborers. Biocatal. Agric. Biotechnol. 2023, 47, 102577. [Google Scholar] [CrossRef]
- Ahmed, H.B.; El-Shahat, M.; Allayeh, A.K.; Emam, H.E. Maillard reaction for nucleation of polymer quantum dots from chitosan-glucose conjugate: Antagonistic for cancer and viral diseases. Int. J. Biol. Macromol. 2023, 224, 858–870. [Google Scholar] [CrossRef] [PubMed]
- Muteeb, G. Nanotechnology—A Light of Hope for Combating Antibiotic Resistance. Microorganisms 2023, 11, 1489. [Google Scholar] [CrossRef]
- Ghirardello, M.; Ramos-Soriano, J.; Galan, M.C. Carbon Dots as an Emergent Class of Antimicrobial Agents. Nanomaterials 2021, 11, 1877. [Google Scholar] [CrossRef]
- Wu, Y.; Li, C.; van der Mei, H.C.; Busscher, H.J.; Ren, Y. Carbon Quantum Dots Derived from Different Carbon Sources for Antibacterial Applications. Antibiotics 2021, 10, 623. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.; Cho, Y.; Dinh, N.N.; Waring, A.J.; Lehrer, R.I. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob. Agents Chemother. 1998, 42, 2206–2214. [Google Scholar] [CrossRef]
- Joly, S.; Maze, C.; McCray, P.B.; Guthmiller, J.M. Human β-Defensins 2 and 3 Demonstrate Strain-Selective Activity against Oral Microorganisms. J. Clin. Microbiol. 2004, 42, 1024–1029. [Google Scholar] [CrossRef]
- Wang, G. Bioinformatic Analysis of 1000 Amphibian Antimicrobial Peptides Uncovers Multiple Length-Dependent Correlations for Peptide Design and Prediction. Antibiotics 2020, 9, 491. [Google Scholar] [CrossRef]
- Choi, J.H.; Jang, A.Y.; Lin, S.; Lim, S.; Kim, D.; Park, K.; Han, S.-M.; Yeo, J.-H.; Seo, H.S. Melittin, a honeybee venom-derived antimicrobial peptide, may target methicillin-resistant Staphylococcus aureus. Mol. Med. Rep. 2015, 12, 6483–6490. [Google Scholar] [CrossRef]
- Gholizadeh, P.; Köse, Ş.; Dao, S.; Ganbarov, K.; Tanomand, A.; Dal, T.; Aghazadeh, M.; Ghotaslou, R.; Ahangarzadeh Rezaee, M.; Yousefi, B.; et al. How CRISPR-Cas System Could Be Used to Combat Antimicrobial Resistance. Infect. Drug Resist. 2020, 13, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Cao, H.; Zhang, L.-H.; Xu, Z. Harnessing the CRISPR-Cas Systems to Combat Antimicrobial Resistance. Front. Microbiol. 2021, 12, e716064. [Google Scholar] [CrossRef]
- Velazquez-Meza, M.E.; Galarde-López, M.; Carrillo-Quiróz, B.; Alpuche-Aranda, C.M. Antimicrobial resistance: One Health approach. Vet. World 2022, 15, 743–749. [Google Scholar] [CrossRef] [PubMed]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6, 521–547. [Google Scholar] [CrossRef] [PubMed]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, e771510. [Google Scholar] [CrossRef] [PubMed]
- Boyd, N.K.; Teng, C.; Frei, C.R. Brief Overview of Approaches and Challenges in New Antibiotic Development: A Focus on Drug Repurposing. Front. Cell. Infect. Microbiol. 2021, 11, 684515. [Google Scholar] [CrossRef] [PubMed]
- Masadeh, M.; Mhaidat, N.; Alzoubi, K.; Al-azzam, S.; Alnasser, Z. Antibacterial activity of statins: A comparative study of Atorvastatin, Simvastatin, and Rosuvastatin. Ann. Clin. Microbiol. Antimicrob. 2012, 11, 13. [Google Scholar] [CrossRef]
- Rácz, B.; Spengler, G. Repurposing Antidepressants and Phenothiazine Antipsychotics as Efflux Pump Inhibitors in Cancer and Infectious Diseases. Antibiotics 2023, 12, 137. [Google Scholar] [CrossRef] [PubMed]
- Galhano, B.S.P.; Ferrari, R.G.; Panzenhagen, P.; de Jesus, A.C.S.; Conte-Junior, C.A. Antimicrobial Resistance Gene Detection Methods for Bacteria in Animal-Based Foods: A Brief Review of Highlights and Advantages. Microorganisms 2021, 9, 923. [Google Scholar] [CrossRef] [PubMed]
- Pierce, J.; Stevens, M.P. The Emerging Role of Telehealth in Antimicrobial Stewardship: A Systematic Review and Perspective. Curr. Treat. Options Infect. Dis. 2021, 13, 175–191. [Google Scholar] [CrossRef]
- Magnano San Lio, R.; Barchitta, M.; Maugeri, A.; La Rosa, M.C.; Favara, G.; Agodi, A. Updates on developing and applying biosensors for the detection of microorganisms, antimicrobial resistance genes and antibiotics: A scoping review. Front. Public. Health 2023, 11, e1240584. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhang, J.; Tian, L.; Xin, L.; Liang, C.; Ren, X.; Li, M. A Comprehensive Overview of the Antibiotics Approved in the Last Two Decades: Retrospects and Prospects. Molecules 2023, 28, 1762. [Google Scholar] [CrossRef]
- Ye, J.; Chen, X. Current Promising Strategies against Antibiotic-Resistant Bacterial Infections. Antibiotics 2022, 12, 67. [Google Scholar] [CrossRef] [PubMed]
- Zavišić, G.; Popović, M.; Stojkov, S.; Medić, D.; Gusman, V.; Jovanović Lješković, N.; Jovanović Galović, A. Antibiotic Resistance and Probiotics: Knowledge Gaps, Market Overview and Preliminary Screening. Antibiotics 2023, 12, 1281. [Google Scholar] [CrossRef]
- Murray, E.; Draper, L.A.; Ross, R.P.; Hill, C. The Advantages and Challenges of Using Endolysins in a Clinical Setting. Viruses 2021, 13, 680. [Google Scholar] [CrossRef]
- Chung, K.M.; Nang, S.C.; Tang, S.S. The Safety of Bacteriophages in Treatment of Diseases Caused by Multidrug-Resistant Bacteria. Pharmaceuticals 2023, 16, 1347. [Google Scholar] [CrossRef] [PubMed]
- Stan, D.; Enciu, A.-M.; Mateescu, A.L.; Ion, A.C.; Brezeanu, A.C.; Stan, D.; Tanase, C. Natural Compounds with Antimicrobial and Antiviral Effect and Nanocarriers Used for Their Transportation. Front. Pharmacol. 2021, 12, e723233. [Google Scholar] [CrossRef]
- Silver, L.L. Challenges of Antibacterial Discovery. Clin. Microbiol. Rev. 2011, 24, 71–109. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, M.A.; Walsh, C.T. Antibiotics for Emerging Pathogens. Science 2009, 325, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C. Where will new antibiotics come from? Nat. Rev. Microbiol. 2003, 1, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Chinemerem Nwobodo, D.; Ugwu, M.C.; Oliseloke Anie, C.; Al-Ouqaili, M.T.S.; Chinedu Ikem, J.; Victor Chigozie, U.; Saki, M. Antibiotic resistance: The challenges and some emerging strategies for tackling a global menace. J. Clin. Lab. Anal. 2022, 36, e24655. [Google Scholar] [CrossRef] [PubMed]
- Serwecińska, L. Antimicrobials and Antibiotic-Resistant Bacteria: A Risk to the Environment and to Public Health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Sakamaki, K.; Uemura, Y.; Shimizu, Y. Definitions and elements of endpoints in phase III randomized trials for the treatment of COVID-19: A cross-sectional analysis of trials registered in ClinicalTrials.gov. Trials 2021, 22, 788. [Google Scholar] [CrossRef] [PubMed]
- Terreni, M.; Taccani, M.; Pregnolato, M. New Antibiotics for Multidrug-Resistant Bacterial Strains: Latest Research Developments and Future Perspectives. Molecules 2021, 26, 2671. [Google Scholar] [CrossRef]
- Majumder, M.A.A.; Rahman, S.; Cohall, D.; Bharatha, A.; Singh, K.; Haque, M.; Gittens-St Hilaire, M. Antimicrobial Stewardship: Fighting Antimicrobial Resistance and Protecting Global Public Health. Infect. Drug Resist. 2020, 13, 4713–4738. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.R.; Haste, N.M.; Gluckstein, D.P. The Role of Antibiotic Stewardship in Promoting Appropriate Antibiotic Use. Am. J. Lifestyle Med. 2019, 13, 376–383. [Google Scholar] [CrossRef]
- Dhingra, S.; Rahman, N.A.A.; Peile, E.; Rahman, M.; Sartelli, M.; Hassali, M.A.; Islam, T.; Islam, S.; Haque, M. Microbial Resistance Movements: An Overview of Global Public Health Threats Posed by Antimicrobial Resistance, and How Best to Counter. Front. Public. Health 2020, 8, 535668. [Google Scholar] [CrossRef] [PubMed]
- Dutescu, I.A.; Hillier, S.A. Encouraging the Development of New Antibiotics: Are Financial Incentives the Right Way Forward? A Systematic Review and Case Study. Infect. Drug Resist. 2021, 14, 415–434. [Google Scholar] [CrossRef] [PubMed]
- Westgeest, A.C.; Schippers, E.F.; Sijbom, M.; Visser, L.G.; de Boer, M.G.J.; Numans, M.E.; Lambregts, M.M.C. Exploring the Barriers in the Uptake of the Dutch MRSA ‘Search and Destroy’ Policy Using the Cascade of Care Approach. Antibiotics 2022, 11, 1216. [Google Scholar] [CrossRef] [PubMed]
- Hammerum, A.M.; Heuer, O.E.; Emborg, H.-D.; Bagger-Skjøt, L.; Jensen, V.F.; Rogues, A.-M.; Skov, R.L.; Agersø, Y.; Brandt, C.T.; Seyfarth, A.M.; et al. Danish Integrated Antimicrobial Resistance Monitoring and Research Program. Emerg. Infect. Dis. 2007, 13, 1633–1639. [Google Scholar] [CrossRef] [PubMed]
- Nazmunnahar; Ahmed, I.; Roknuzzaman, A.S.M.; Islam, M.R. The recent Nipah virus outbreak in Bangladesh could be a threat for global public health: A brief report. Health Sci. Rep. 2023, 6, e1423. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, J.; Björkman, I.; Röing, M.; Essack, S.Y.; Stålsby Lundborg, C. Exploring the One Health Perspective in Sweden’s Policies for Containing Antibiotic Resistance. Antibiotics 2021, 10, 526. [Google Scholar] [CrossRef] [PubMed]
- Alimi, Y.; Wabacha, J. Strengthening coordination and collaboration of one health approach for zoonotic diseases in Africa. One Health Outlook 2023, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- de la Rocque, S.; Errecaborde, K.M.M.; Belot, G.; Brand, T.; Shadomy, S.; von Dobschuetz, S.; Aguanno, R.; Carron, M.; Caya, F.; Ding, S.; et al. One health systems strengthening in countries: Tripartite tools and approaches at the human-animal-environment interface. BMJ Glob. Health 2023, 8, e011236. [Google Scholar] [CrossRef] [PubMed]
- Katale, B.Z.; Mbugi, E.V.; Keyyu, J.D.; Fyumagwa, R.D.; Rweyemamu, M.M.; van Helden, P.D.; Dockrell, H.M.; Matee, M.I. One Health approach in the prevention and control of mycobacterial infections in Tanzania: Lessons learnt and future perspectives. One Health Outlook 2019, 1, 2. [Google Scholar] [CrossRef]
- Ramanujam, H.; Palaniyandi, K. Bovine tuberculosis in India: The need for One Health approach and the way forward. One Health 2023, 16, 100495. [Google Scholar] [CrossRef] [PubMed]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed]
- Founou, R.C.; Founou, L.L.; Essack, S.Y. Clinical and economic impact of antibiotic resistance in developing countries: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0189621. [Google Scholar] [CrossRef] [PubMed]
- Larsson, S.; Svensson, M.; Ternhag, A. Production loss and sick leave caused by antibiotic resistance: A register-based cohort study. BMC Public Health 2022, 22, 527. [Google Scholar] [CrossRef] [PubMed]
- Hermsen, E.D.; Sibbel, R.L.; Holland, S. The Role of Pharmaceutical Companies in Antimicrobial Stewardship: A Case Study. Clin. Infect. Dis. 2020, 71, 677–681. [Google Scholar] [CrossRef]
- Lekshmi, M.; Ammini, P.; Kumar, S.; Varela, M.F. The Food Production Environment and the Development of Antimicrobial Resistance in Human Pathogens of Animal Origin. Microorganisms 2017, 5, 11. [Google Scholar] [CrossRef]
- Bokhary, H.; Pangesti, K.N.A.; Rashid, H.; Abd El Ghany, M.; Hill-Cawthorne, G.A. Travel-Related Antimicrobial Resistance: A Systematic Review. Trop. Med. Infect. Dis. 2021, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Łusiak-Szelachowska, M.; Międzybrodzki, R.; Drulis-Kawa, Z.; Cater, K.; Knežević, P.; Winogradow, C.; Amaro, K.; Jończyk-Matysiak, E.; Weber-Dąbrowska, B.; Rękas, J.; et al. Bacteriophages and antibiotic interactions in clinical practice: What we have learned so far. J. Biomed. Sci. 2022, 29, 23. [Google Scholar] [CrossRef]
- Thorpe, K.E.; Joski, P.; Johnston, K.J. Antibiotic-Resistant Infection Treatment Costs Have Doubled Since 2002, Now Exceeding $2 Billion Annually. Health Aff. 2018, 37, 662–669. [Google Scholar] [CrossRef]
- Power, E. Impact of antibiotic restrictions: The pharmaceutical perspective. Clin. Microbiol. Infect. 2006, 12, 25–34. [Google Scholar] [CrossRef] [PubMed]
- van der Gronde, T.; Uyl-de Groot, C.A.; Pieters, T. Addressing the challenge of high-priced prescription drugs in the era of precision medicine: A systematic review of drug life cycles, therapeutic drug markets and regulatory frameworks. PLoS ONE 2017, 12, e0182613. [Google Scholar]
- Guidos, R.J. Combating Antimicrobial Resistance: Policy Recommendations to Save Lives. Clin. Infect. Dis. 2011, 52, S397–S428. [Google Scholar]
- Littmann, J.; Viens, A. The Ethical Significance of Antimicrobial Resistance. Public Health Ethics 2015, 8, 209–224. [Google Scholar] [CrossRef] [PubMed]
- Papadimou, D.; Malmqvist, E.; Ancillotti, M. Socio-cultural determinants of antibiotic resistance: A qualitative study of Greeks’ attitudes, perceptions and values. BMC Public Health 2022, 22, 1439. [Google Scholar] [CrossRef] [PubMed]
- Ashiru-Oredope, D.; Hopkins, S.; Vasandani, S.; Umoh, E.; Oloyede, O.; Nilsson, A.; Kinsman, J.; Elsert, L.; Monnet, D.L. Healthcare workers’ knowledge, attitudes and behaviours with respect to antibiotics, antibiotic use and antibiotic resistance across 30 EU/EEA countries in 2019. Eurosurveillance 2021, 26, 1900633. [Google Scholar] [CrossRef]
- Adebisi, Y.A. Balancing the risks and benefits of antibiotic use in a globalized world: The ethics of antimicrobial resistance. Global. Health 2023, 19, 27. [Google Scholar] [CrossRef] [PubMed]
- Leekha, S.; Terrell, C.L.; Edson, R.S. General Principles of Antimicrobial Therapy. Mayo Clin. Proc. 2011, 86, 156–167. [Google Scholar] [CrossRef]
- Rocha-Pereira, N.; Castro Sanchez, E.; Nathwani, D. How Can Multi-Professional Education Support Better Stewardship? Infect. Dis. Rep. 2017, 9, 6917. [Google Scholar] [CrossRef] [PubMed]
- Kållberg, C.; Mathiesen, L.; Gopinathan, U.; Salvesen Blix, H. The role of drug regulatory authorities and health technology assessment agencies in shaping incentives for antibiotic R&D: A qualitative study. J. Pharm. Policy Pract. 2023, 16, 53. [Google Scholar] [CrossRef] [PubMed]
- Metlay, J.P.; Powers, J.H.; Dudley, M.N.; Christiansen, K.; Finch, R.G. Antimicrobial Drug Resistance, Regulation, and Research1. Emerg. Infect. Dis. 2006, 12, 183–190. [Google Scholar] [CrossRef]
- MacDougall, C.; Polk, R.E. Antimicrobial Stewardship Programs in Health Care Systems. Clin. Microbiol. Rev. 2005, 18, 638–656. [Google Scholar] [CrossRef]
- Barlam, T.F.; Cosgrove, S.E.; Abbo, L.M.; MacDougall, C.; Schuetz, A.N.; Septimus, E.J.; Srinivasan, A.; Dellit, T.H.; Falck-Ytter, Y.T.; Fishman, N.O.; et al. Implementing an Antibiotic Stewardship Program: Guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin. Infect. Dis. 2016, 62, e51–e77. [Google Scholar] [CrossRef]
- Kazerooni, P.A.; Nejat, M.; Akbarpoor, M.; Sedaghat, Z.; Fararouei, M. Underascertainment, underreporting, representativeness and timeliness of the Iranian communicable disease surveillance system for tuberculosis. Public Health 2019, 171, 50–56. [Google Scholar] [CrossRef]
- Iskandar, K.; Molinier, L.; Hallit, S.; Sartelli, M.; Hardcastle, T.C.; Haque, M.; Lugova, H.; Dhingra, S.; Sharma, P.; Islam, S.; et al. Surveillance of antimicrobial resistance in low- and middle-income countries: A scattered picture. Antimicrob. Resist. Infect. Control 2021, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Geiderman, J.M.; Moskop, J.C.; Derse, A.R. Privacy and Confidentiality in Emergency Medicine: Obligations and Challenges. Emerg. Med. Clin. N. Am. 2006, 24, 633–656. [Google Scholar] [CrossRef]
- Torab-Miandoab, A.; Samad-Soltani, T.; Jodati, A.; Rezaei-Hachesu, P. Interoperability of heterogeneous health information systems: A systematic literature review. BMC Med. Inform. Decis. Mak. 2023, 23, 18. [Google Scholar] [CrossRef]
- Balasegaram, M.; Piddock, L.J.V. The Global Antibiotic Research and Development Partnership (GARDP) Not-for-Profit Model of Antibiotic Development. ACS Infect. Dis. 2020, 6, 1295–1298. [Google Scholar] [CrossRef]
- Piddock, L.J.V. The global antibiotic research and development partnership (GARDP): Researching and developing new antibiotics to meet global public health needs. Medchemcomm 2019, 10, 1227–1230. [Google Scholar] [CrossRef]
- Alm, R.A.; Gallant, K. Innovation in Antimicrobial Resistance: The CARB-X Perspective. ACS Infect. Dis. 2020, 6, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.; Panteli, D.; van Kessel, R.; Ljungqvist, G.; Colombo, F.; Mossialos, E. Challenges and opportunities for incentivising antibiotic research and development in Europe. Lancet Reg. Health-Eur. 2023, 33, 100705. [Google Scholar] [CrossRef] [PubMed]
- Cars, O.; Nordberg, P. Antibiotic resistance-The faceless threat. Int. J. Risk Saf. Med. 2005, 17, 103–110. [Google Scholar]
- Cars, O.; Chandy, S.J.; Mpundu, M.; Peralta, A.Q.; Zorzet, A.; So, A.D. Resetting the agenda for antibiotic resistance through a health systems perspective. Lancet Glob. Health 2021, 9, e1022–e1027. [Google Scholar] [CrossRef] [PubMed]
- Thabit, A.K.; Alabbasi, A.Y.; Alnezary, F.S.; Almasoudi, I.A. An Overview of Antimicrobial Resistance in Saudi Arabia (2013–2023) and the Need for National Surveillance. Microorganisms 2023, 11, 2086. [Google Scholar] [CrossRef]
- Talaat, M.; Zayed, B.; Tolba, S.; Abdou, E.; Gomaa, M.; Itani, D.; Hutin, Y.; Hajjeh, R. Increasing Antimicrobial Resistance in World Health Organization Eastern Mediterranean Region, 2017–2019. Emerg. Infect. Dis. 2022, 28, 717–724. [Google Scholar] [CrossRef]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Chokshi, A.; Sifri, Z.; Cennimo, D.; Horng, H. Global contributors to antibiotic resistance. J. Glob. Infect. Dis. 2019, 11, 36. [Google Scholar] [PubMed]
- Samreen; Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental antimicrobial resistance and its drivers: A potential threat to public health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Mölstad, S.; Erntell, M.; Hanberger, H.; Melander, E.; Norman, C.; Skoog, G.; Lundborg, C.S.; Söderström, A.; Torell, E.; Cars, O. Sustained reduction of antibiotic use and low bacterial resistance: 10-year follow-up of the Swedish Strama programme. Lancet Infect. Dis. 2008, 8, 125–132. [Google Scholar] [CrossRef]
- Moura, P.; Sandberg, M.; Høg, B.B.; Niza-Ribeiro, J.; Nielsen, E.O.; Alban, L. Characterisation of antimicrobial usage in Danish pigs in 2020. Front. Vet. Sci. 2023, 10, 1155811. [Google Scholar] [CrossRef]
- Louh, I.K.; Greendyke, W.G.; Hermann, E.A.; Davidson, K.W.; Falzon, L.; Vawdrey, D.K.; Shaffer, J.A.; Calfee, D.P.; Furuya, E.Y.; Ting, H.H. Clostridium Difficile Infection in Acute Care Hospitals: Systematic Review and Best Practices for Prevention. Infect. Control Hosp. Epidemiol. 2017, 38, 476–482. [Google Scholar] [CrossRef]
- Pandey, R.P.; Mukherjee, R.; Chang, C.-M. Antimicrobial resistance surveillance system mapping in different countries. Drug Target Insights 2022, 16, 36–48. [Google Scholar] [CrossRef]
- Pollack, L.A.; van Santen, K.L.; Weiner, L.M.; Dudeck, M.A.; Edwards, J.R.; Srinivasan, A. Antibiotic Stewardship Programs in U.S. Acute Care Hospitals: Findings From the 2014 National Healthcare Safety Network Annual Hospital Survey. Clin. Infect. Dis. 2016, 63, 443–449. [Google Scholar] [CrossRef]
- Voss, A.; Ghafur, A. “The Chennai declaration”—Indian doctors’ fight against antimicrobial resistance. Antimicrob. Resist. Infect. Control 2013, 2, 7. [Google Scholar] [CrossRef]
- Obakiro, S.B.; Kiyimba, K.; Paasi, G.; Napyo, A.; Anthierens, S.; Waako, P.; Van Royen, P.; Iramiot, J.S.; Goossens, H.; Kostyanev, T. Prevalence of antibiotic-resistant bacteria among patients in two tertiary hospitals in Eastern Uganda. J. Glob. Antimicrob. Resist. 2021, 25, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Zowawi, H.M. Antimicrobial resistance in Saudi Arabia. Saudi Med. J. 2016, 37, 935–940. [Google Scholar] [CrossRef] [PubMed]
- El-Lababidi, R.M.; Atallah, B.; Abdel-Razig, S. Design and Implementation of an Antimicrobial Stewardship Certificate Program in the United Arab Emirates. Int. Med. Educ. 2023, 2, 41–48. [Google Scholar] [CrossRef]
- Awad, A.I.; Aboud, E.A. Knowledge, Attitude and Practice towards Antibiotic Use among the Public in Kuwait. PLoS ONE 2015, 10, e0117910. [Google Scholar] [CrossRef]
- Khamis, F.; Al Awaidy, S.; Al Salmi, Z.; Zayed, B. Antimicrobial Stewardship Opportunities: An Example from Oman. Oman Med. J. 2023, 38, e498. [Google Scholar] [CrossRef] [PubMed]
- Enani, M.A. The antimicrobial stewardship program in Gulf Cooperation Council (GCC) states: Insights from a regional survey. J. Infect. Prev. 2016, 17, 16–20. [Google Scholar] [CrossRef]
- Ali, T.; Ahmed, S.; Aslam, M. Artificial Intelligence for Antimicrobial Resistance Prediction: Challenges and Opportunities towards Practical Implementation. Antibiotics 2023, 12, 523. [Google Scholar] [CrossRef]
- Melo, M.C.R.; Maasch, J.R.M.A.; de la Fuente-Nunez, C. Accelerating antibiotic discovery through artificial intelligence. Commun. Biol. 2021, 4, 1050. [Google Scholar] [CrossRef]
- David, L.; Brata, A.M.; Mogosan, C.; Pop, C.; Czako, Z.; Muresan, L.; Ismaiel, A.; Dumitrascu, D.I.; Leucuta, D.C.; Stanculete, M.F.; et al. Artificial Intelligence and Antibiotic Discovery. Antibiotics 2021, 10, 1376. [Google Scholar] [CrossRef]
- Amin, D.; Garzόn-Orjuela, N.; Garcia Pereira, A.; Parveen, S.; Vornhagen, H.; Vellinga, A. Artificial Intelligence to Improve Antibiotic Prescribing: A Systematic Review. Antibiotics 2023, 12, 1293. [Google Scholar] [CrossRef]
- Lv, J.; Deng, S.; Zhang, L. A review of artificial intelligence applications for antimicrobial resistance. Biosaf. Health 2021, 3, 22–31. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Alhumaid, S.; Mutair, A.A.; Garout, M.; Abulhamayel, Y.; Halwani, M.A.; Alestad, J.H.; Bshabshe, A.A.; Sulaiman, T.; AlFonaisan, M.K.; et al. Application of Artificial Intelligence in Combating High Antimicrobial Resistance Rates. Antibiotics 2022, 11, 784. [Google Scholar] [CrossRef]
- Wang, H.; Jia, C.; Li, H.; Yin, R.; Chen, J.; Li, Y.; Yue, M. Paving the way for precise diagnostics of antimicrobial resistant bacteria. Front. Mol. Biosci. 2022, 9, 976705. [Google Scholar] [CrossRef] [PubMed]
- Cavallaro, M.; Moran, E.; Collyer, B.; McCarthy, N.D.; Green, C.; Keeling, M.J. Informing antimicrobial stewardship with explainable AI. PLOS Digit. Health 2023, 2, e0000162. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Abramova, A.; Berendonk, T.U.; Coelho, L.P.; Forslund, S.K.; Gschwind, R.; Heikinheimo, A.; Jarquín-Díaz, V.H.; Khan, A.A.; Klümper, U.; et al. Towards monitoring of antimicrobial resistance in the environment: For what reasons, how to implement it, and what are the data needs? Environ. Int. 2023, 178, 108089. [Google Scholar] [CrossRef]
- Hart, A.; Warren, J.; Wilkinson, H.; Schmidt, W. Environmental surveillance of antimicrobial resistance (AMR), perspectives from a national environmental regulator in 2023. Eurosurveillance 2023, 28, 2200367. [Google Scholar] [CrossRef] [PubMed]
- Seaton, J. AI Could Quickly Screen Thousands of Antibiotics to Tackle Superbugs: 2023, Scientific American. Available online: https://www.scientificamerican.com/article/ai-could-quickly-screen-thousands-of-antibiotics-to-tackle-superbugs/ (accessed on 29 October 2023).
- Díaz-Rodríguez, N.; Del Ser, J.; Coeckelbergh, M.; de Prado, M.L.; Herrera-Viedma, E.; Herrera, F. Connecting the dots in trustworthy Artificial Intelligence: From AI principles, ethics, and key requirements to responsible AI systems and regulation. Inf. Fusion 2023, 99, 101896. [Google Scholar] [CrossRef]
- Mustard, A. WHO Calls for Safe and Ethical AI for Health’ 2023, Departmental News. Available online: https://www.who.int/news/item/16-05-2023-who-calls-for-safe-and-ethical-ai-for-health (accessed on 29 October 2023).
- WHO Guidance. Ethics and Governance of Artificial Intelligence for Health. Available online: https://www.who.int/publications/i/item/9789240029200 (accessed on 28 June 2021).
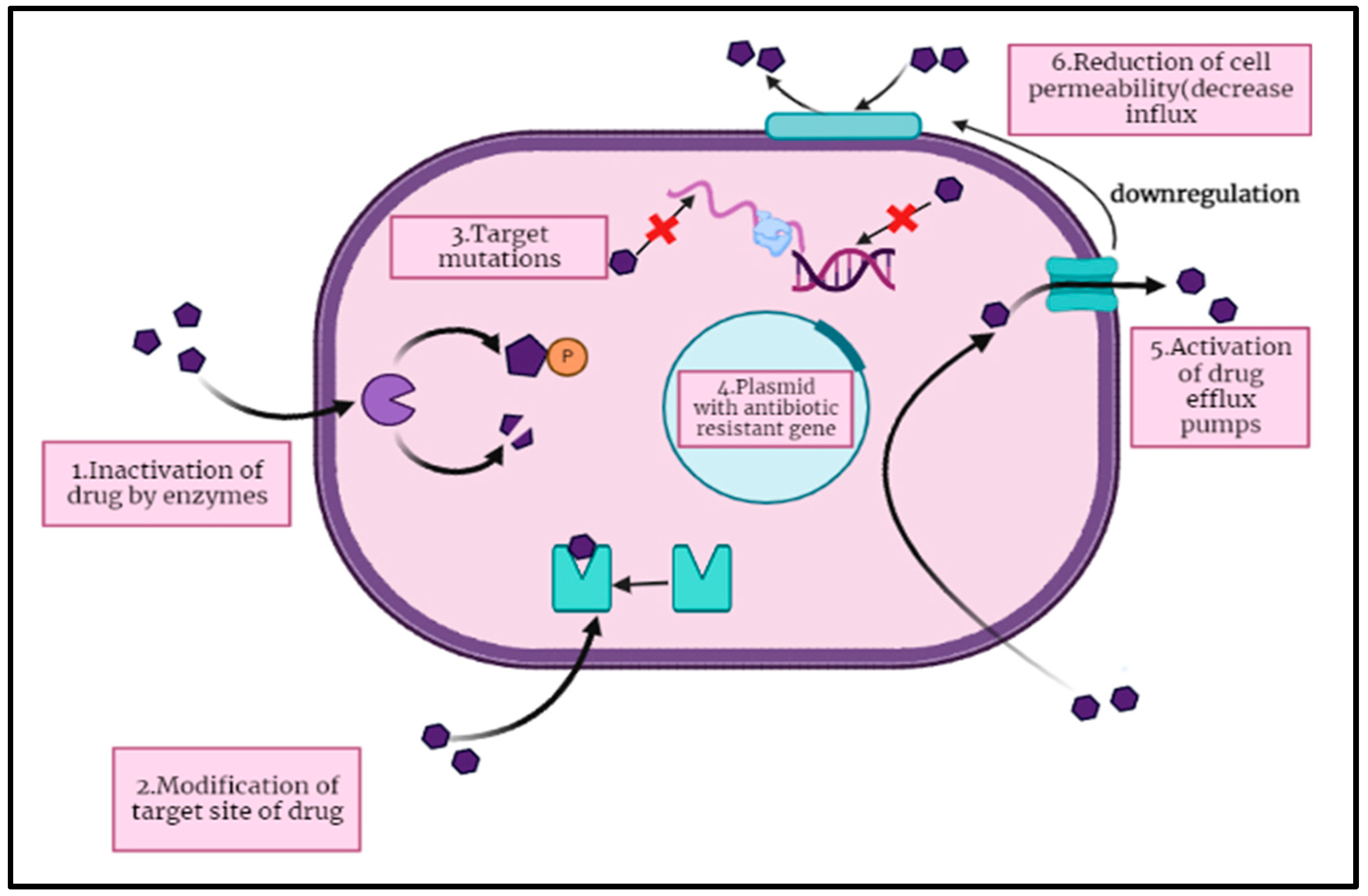
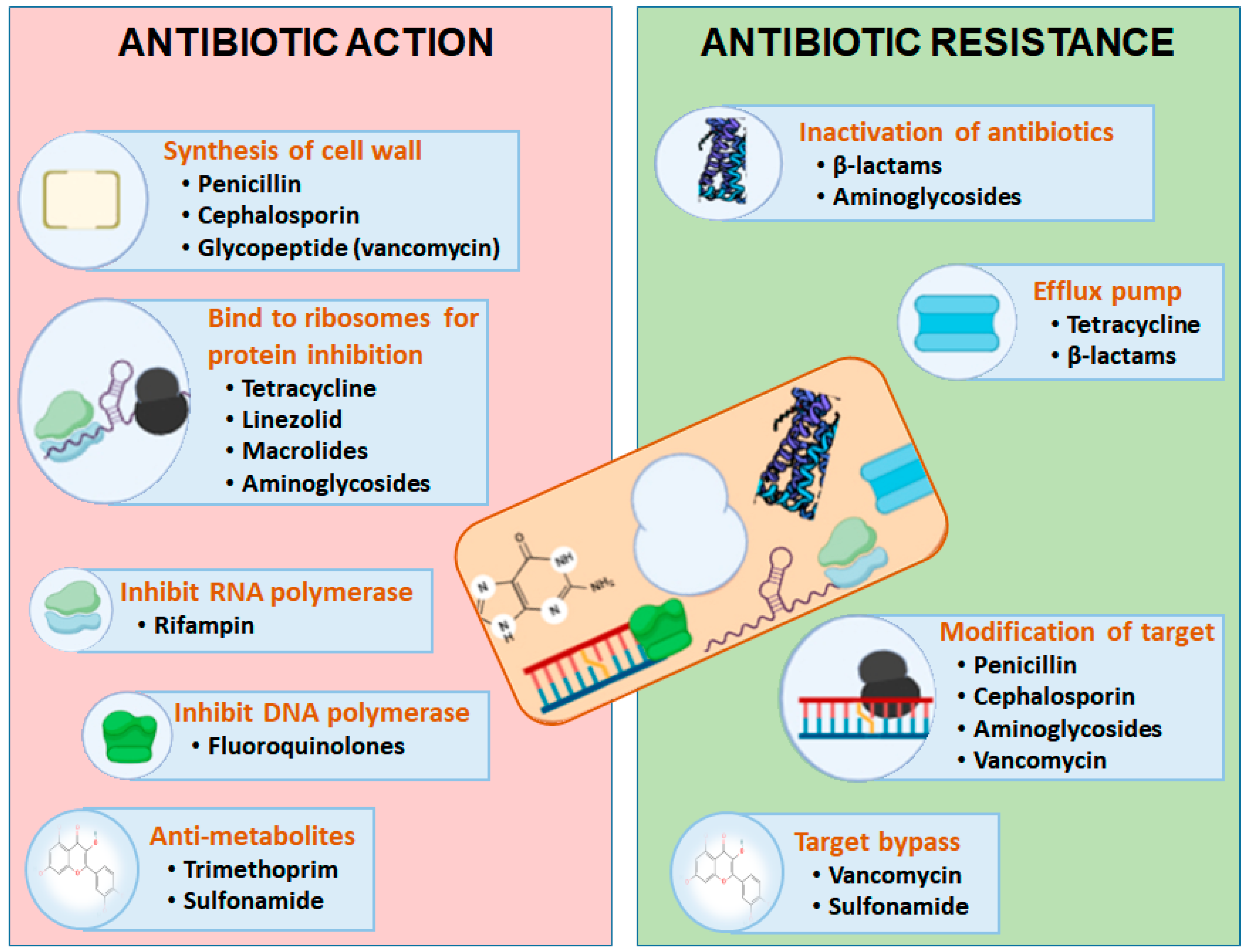
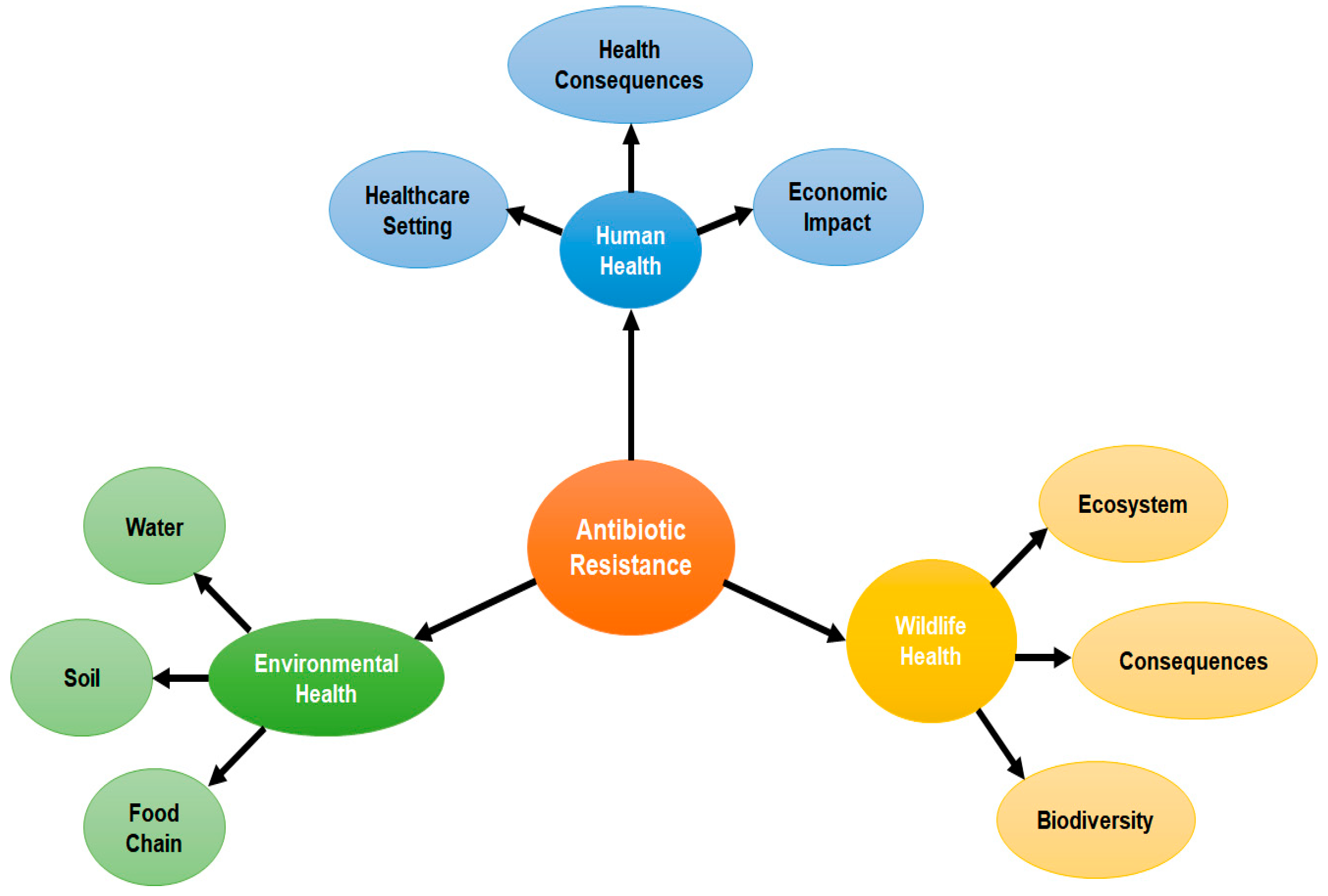
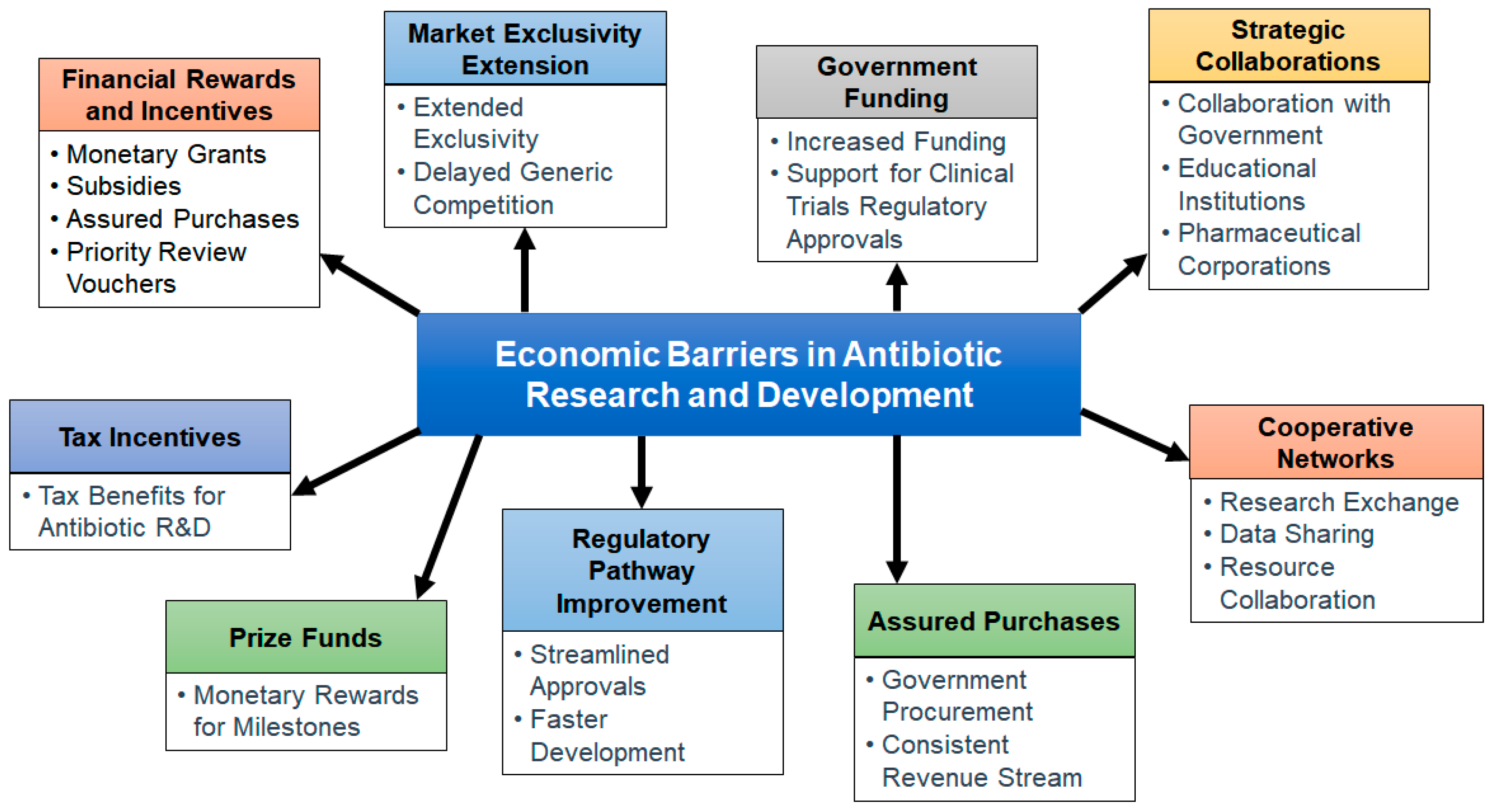
| Strategy | Description | Examples | Ref. |
|---|---|---|---|
| New Antibiotics | Development of novel antibiotics targeting drug-resistance bacteria | Teixobactin, Lefamulin, Zoliflodacin, Cefiderocol, Eravacycline | [109,110,111] |
| Bacteriophage Therapy | Utilization of bacteriophages to target and kill bacteria | Phage cocktails targeting antibiotic-resistant strains | [112,113,114] |
| Combination Therapies | Use of multiple antibiotics in combination to enhance and prevent resistance | Trimethoprim-Sulfamethoxazole, Beta-lactam/beta-lactamase inhibitor combinations | [115,116] |
| Immune Modulation | Boosting the immune response to enhance clearance of bacterial infections | Interferons, interleukins, colony-stimulating factors, etc. | [117] |
| Antibody Therapies | Development of monoclonal antibodies or antibody-derived molecules to target bacterial pathogens | Bezlotoxumab, altastaph, lysibodies, antibody-toxin conjugates | [118] |
| Nanotechnology | Use of nanoparticles for drug delivery or disruption of bacterial membranes | Metal–organic frameworks, carbon quantum dots | [119,120] |
| Heterocyclic compounds | Use of heterocyclic compounds as novel antimicrobial agents | Thiazole, Imidazole, Thiazolidinone, Oxazole, Pyrrole, Pyridine, Pyrimidine | [121] |
| Antimicrobial Peptides | Exploration of naturally occurring peptides with antimicrobial properties | LL-37, defensins, magainins, temporins, melittin | [122,123,124] |
| CRISPR-Cas Systems | Using CRISPR gene-editing technology to target and eliminate drug-resistant bacteria | CRISPR-Cas systems for selective targeting of bacterial DNA | [125,126,127] |
| One Health Approach | Holistic approach integrating human health, animal health, and the environment | Coordinated efforts to combat antimicrobial resistance across various sectors | [128,129] |
| Drug Repurposing | Investigating existing drugs for potential activity against drug-resistant microorganisms | Statins, chlorpromazine, thioridazine | [130,131] |
| Surveillance and Diagnostics | Improved monitoring and diagnostic tools for early detection and tracking of resistance | PCR, next-generation sequencing, telemedicine, biosensors, microfluidic devices | [132,133,134] |
| Strategies | Safety | Efficacy | Commercial Viability | Ref. |
|---|---|---|---|---|
| Antibiotics | Traditional antibiotics can have a broad spectrum of activity, affecting both harmful and beneficial bacteria in the body. This can lead to disruptions in the microbiome and potential side effects. | Traditional antibiotics have a history of widespread use and can be effective against a wide range of bacterial infections. However, efficacy can decrease due to resistance. | Traditional antibiotics have a well-established market and regulatory pathways. However, increased resistance, lengthy development times, and low profit margins have reduced their commercial viability. | [189] |
| Antimicrobial peptides | Generally safe; Minimal toxicity; Concerns in high doses. | Effective against some bacterial strains; Ongoing development | Potential commercial viability with research and development. | [73] |
| Monoclonal antibodies | Highly safe profile; Minimal side effects. | Effective in targeting specific pathogens; Reduce infection risks. | Costly research and development; Potential regulatory approval challenges. | [190] |
| Probiotics | Generally safe; Risk in individuals with compromised immune systems. | Effective for certain infections; Efficacy varies by strain, infection type, and host factors. | Gained commercial traction in the dietary supplement and functional food industries; Need more investment for specific bacterial infections. | [191] |
| Bacteriophages | Generally safe; Minimal side effects. | Variable, efficacy may be phage-specific. | Potential viability; Ongoing research. | [192] |
| Phage therapy | Generally safe; Minimal side effects. | Variable, depending on specificity, research and matching phages. | Increasing interest and investment with ongoing studies. | [193] |
| Essential oils and plant extracts | Generally safe. | Challenges in standardization of their components. | Potential viability; Ongoing research. | [194] |
| Nanoparticles | Long-term use needs evaluation. | Effective against some bacterial strains; Ongoing development. | High-quality nanoparticle production can be costly. | [171] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals 2023, 16, 1615. https://doi.org/10.3390/ph16111615
Muteeb G, Rehman MT, Shahwan M, Aatif M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals. 2023; 16(11):1615. https://doi.org/10.3390/ph16111615
Chicago/Turabian StyleMuteeb, Ghazala, Md Tabish Rehman, Moayad Shahwan, and Mohammad Aatif. 2023. "Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review" Pharmaceuticals 16, no. 11: 1615. https://doi.org/10.3390/ph16111615
APA StyleMuteeb, G., Rehman, M. T., Shahwan, M., & Aatif, M. (2023). Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals, 16(11), 1615. https://doi.org/10.3390/ph16111615









