Pharmacokinetics in Pharmacometabolomics: Towards Personalized Medication
Abstract
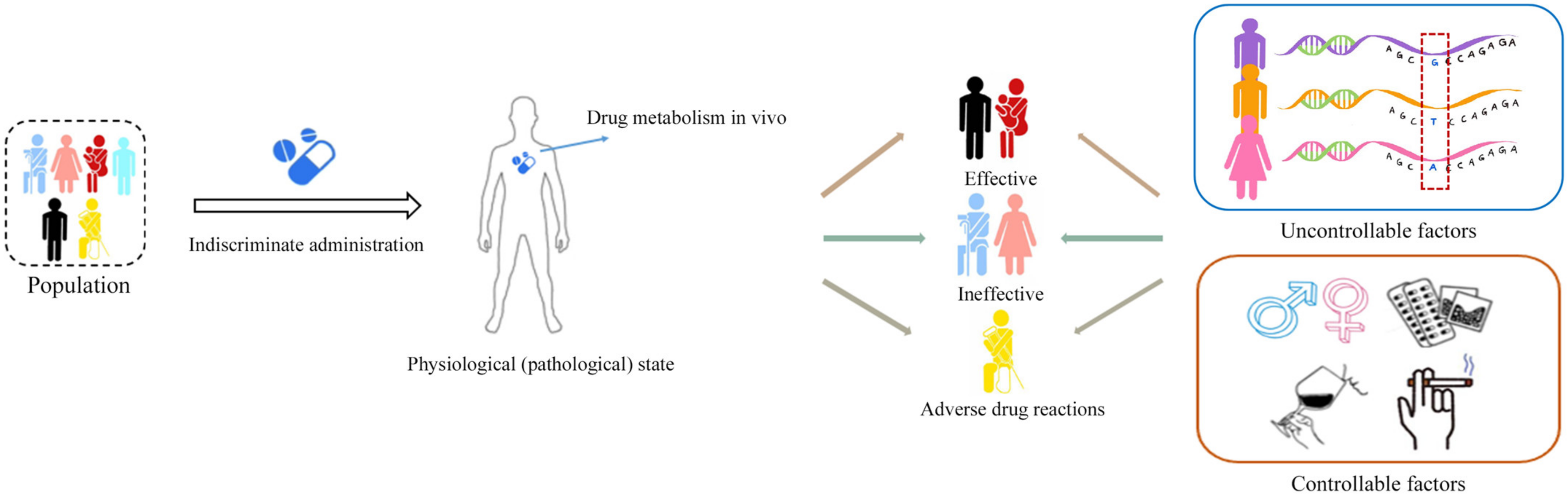
1. Introduction
2. Pharmacokinetics and Accurate Administration
3. Endogenous Metabolites and Pharmacometabolomics
4. Pharmacometabolomics Informs Pharmacokinetics
5. Pharmacokinetics-Related to Pharmacometabolomics for Predicting Drug Reactions
6. Challenges and Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Micaglio, E.; Locati, E.T.; Monasky, M.M.; Romani, F.; Heilbron, F.; Pappone, C. Role of Pharmacogenetics in Adverse Drug Reactions: An Update towards Personalized Medicine. Front. Pharmacol. 2021, 12, 651720. [Google Scholar] [CrossRef]
- Goetz, L.H.; Schork, N.J. Personalized medicine: Motivation, challenges, and progress. Fertil. Steril. 2018, 109, 952–963. [Google Scholar] [CrossRef]
- Sadee, W.; Wang, D.; Hartmann, K.; Toland, A.E. Pharmacogenomics: Driving Personalized Medicine. Pharmacol. Rev. 2023, 75, 789–814. [Google Scholar] [CrossRef]
- Zhou, K.; Bellenguez, C.; Spencer, C.C.A.; Bennett, A.J.; Coleman, R.L.; Tavendale, R.; Hawley, S.A.; Donnelly, L.A.; Schofield, C.; Groves, C.J.; et al. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat. Genet. 2011, 43, 117–120. [Google Scholar] [CrossRef]
- Henricks, L.M.; Lunenburg, C.A.T.C.; de Man, F.M.; Meulendijks, D.; Frederix, G.W.J.; Kienhuis, E.; Creemers, G.-J.; Baars, A.; Dezentjé, V.O.; Imholz, A.L.T.; et al. DPYD genotype-guided dose individualisation of fluoropyrimidine therapy in patients with cancer: A prospective safety analysis. Lancet Oncol. 2018, 19, 1459–1467. [Google Scholar] [CrossRef]
- Lauschke, V.M.; Milani, L.; Ingelman-Sundberg, M. Pharmacogenomic Biomarkers for Improved Drug Therapy-Recent Progress and Future Developments. AAPS J. 2017, 20, 4. [Google Scholar] [CrossRef]
- Betcher, H.K.; George, A.L. Pharmacogenomics in pregnancy. Semin. Perinatol. 2020, 44, 151222. [Google Scholar] [CrossRef]
- Lai, Y.; Varma, M.; Feng, B.; Stephens, J.C.; Kimoto, E.; El-Kattan, A.; Ichikawa, K.; Kikkawa, H.; Ono, C.; Suzuki, A.; et al. Impact of drug transporter pharmacogenomics on pharmacokinetic and pharmacodynamic variability—Considerations for drug development. Expert. Opin. Drug Metab. Toxicol. 2012, 8, 723–743. [Google Scholar] [CrossRef]
- Jones, E.M.; Lubock, N.B.; Venkatakrishnan, A.J.; Wang, J.; Tseng, A.M.; Paggi, J.M.; Latorraca, N.R.; Cancilla, D.; Satyadi, M.; Davis, J.E.; et al. Structural and functional characterization of G protein-coupled receptors with deep mutational scanning. Elife 2020, 9, e54895. [Google Scholar] [CrossRef]
- Mussap, M.; Loddo, C.; Fanni, C.; Fanos, V. Metabolomics in pharmacology—A delve into the novel field of pharmacometabolomics. Expert Rev. Clin. Pharmacol. 2020, 13, 115–134. [Google Scholar] [CrossRef]
- Liu, Z.; Xiang, Q.; Mu, G.; Xie, Q.; Zhou, S.; Wang, Z.; Chen, S.; Hu, K.; Gong, Y.; Jiang, J.; et al. The effect of smoking on residual platelet reactivity to clopidogrel: A systematic review and meta-analysis. Platelets 2020, 31, 3–14. [Google Scholar] [CrossRef]
- McClay, J.L. Epigenetic regulation of drug metabolism in aging. Aging 2021, 13, 16898–16899. [Google Scholar] [CrossRef]
- Pelliccia, F.; Rollini, F.; Marazzi, G.; Greco, C.; Gaudio, C.; Angiolillo, D.J. Drug-drug interactions between clopidogrel and novel cardiovascular drugs. Eur. J. Pharmacol. 2015, 765, 332–336. [Google Scholar] [CrossRef]
- Wishart, D.S. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef]
- Everett, J.R. Pharmacometabonomics in humans: A new tool for personalized medicine. Pharmacogenomics 2015, 16, 737–754. [Google Scholar] [CrossRef]
- Dong, D.; Yang, D.; Lin, L.; Wang, S.; Wu, B. Circadian rhythm in pharmacokinetics and its relevance to chronotherapy. Biochem. Pharmacol. 2020, 178, 114045. [Google Scholar] [CrossRef]
- Kantae, V.; Krekels, E.H.J.; Esdonk, M.J.V.; Lindenburg, P.; Harms, A.C.; Knibbe, C.A.J.; Van der Graaf, P.H.; Hankemeier, T. Integration of pharmacometabolomics with pharmacokinetics and pharmacodynamics: Towards personalized drug therapy. Metabolomics Off. J. Metabolomic Soc. 2017, 13, 9. [Google Scholar] [CrossRef]
- Cerny, M.A.; Spracklin, D.K.; Obach, R.S. Human Absorption, Distribution, Metabolism, and Excretion Studies: Origins, Innovations, and Importance. Drug Metab. Dispos. 2023, 51, 647–656. [Google Scholar] [CrossRef]
- Fang, Z.; Zhang, H.; Guo, J.; Guo, J. Overview of therapeutic drug monitoring and clinical practice. Talanta 2024, 266, 124996. [Google Scholar] [CrossRef]
- Ates, H.C.; Roberts, J.A.; Lipman, J.; Cass, A.E.G.; Urban, G.A.; Dincer, C. On-Site Therapeutic Drug Monitoring. Trends Biotechnol. 2020, 38, 1262–1277. [Google Scholar] [CrossRef]
- Decosterd, L.A.; Widmer, N.; André, P.; Aouri, M.; Buclin, T. The emerging role of multiplex tandem mass spectrometry analysis for therapeutic drug monitoring and personalized medicine. TrAC Trends Anal. Chem. 2016, 84, 5–13. [Google Scholar] [CrossRef]
- Lizza, B.D.; Raush, N.; Micek, S.T. Antibiotic Optimization in the Intensive Care Unit. Semin. Respir. Crit. Care Med. 2022, 43, 125–130. [Google Scholar] [CrossRef]
- Cusumano, J.A.; Klinker, K.P.; Huttner, A.; Luther, M.K.; Roberts, J.A.; LaPlante, K.L. Towards precision medicine: Therapeutic drug monitoring-guided dosing of vancomycin and β-lactam antibiotics to maximize effectiveness and minimize toxicity. Am. J. Health-Syst. Pharm. AJHP Off. J. Am. Soc. Health-Syst. Pharm. 2020, 77, 1104–1112. [Google Scholar] [CrossRef]
- Dhaese, S.; Van Vooren, S.; Boelens, J.; De Waele, J. Therapeutic drug monitoring of β-lactam antibiotics in the ICU. Expert Rev. Anti-Infect. Ther. 2020, 18, 1155–1164. [Google Scholar] [CrossRef]
- Hahn, R.Z.; Antunes, M.V.; Verza, S.G.; Perassolo, M.S.; Suyenaga, E.S.; Schwartsmann, G.; Linden, R. Pharmacokinetic and Pharmacogenetic Markers of Irinotecan Toxicity. Curr. Med. Chem. 2019, 26, 2085–2107. [Google Scholar] [CrossRef]
- de Man, F.M.; Goey, A.K.L.; van Schaik, R.H.N.; Mathijssen, R.H.J.; Bins, S. Individualization of Irinotecan Treatment: A Review of Pharmacokinetics, Pharmacodynamics, and Pharmacogenetics. Clin. Pharmacokinet. 2018, 57, 1229–1254. [Google Scholar] [CrossRef]
- Joerger, M. Metabolism of the taxanes including nab-paclitaxel. Expert Opin. Drug Metab. Toxicol. 2015, 11, 691–702. [Google Scholar] [CrossRef]
- Engels, F.K.; Loos, W.J.; van der Bol, J.M.; de Bruijn, P.; Mathijssen, R.H.J.; Verweij, J.; Mathot, R.A.A. Therapeutic drug monitoring for the individualization of docetaxel dosing: A randomized pharmacokinetic study. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 353–362. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Locati, M.; Curtale, G.; Mantovani, A. Diversity, Mechanisms, and Significance of Macrophage Plasticity. Annu. Rev. Pathol. 2020, 15, 123–147. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Shalova, I.N.; Biswas, S.K. Metabolic regulation of macrophage phenotype and function. Immunol. Rev. 2017, 280, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Burt, T.; Nandal, S. Pharmacometabolomics in Early-Phase Clinical Development. Clin. Transl. Sci. 2016, 9, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Clayton, T.A.; Lindon, J.C.; Cloarec, O.; Antti, H.; Charuel, C.; Hanton, G.; Provost, J.-P.; Le Net, J.-L.; Baker, D.; Walley, R.J.; et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature 2006, 440, 1073–1077. [Google Scholar] [CrossRef] [PubMed]
- Clayton, T.A.; Baker, D.; Lindon, J.C.; Everett, J.R.; Nicholson, J.K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 14728–14733. [Google Scholar] [CrossRef]
- Saigusa, D.; Matsukawa, N.; Hishinuma, E.; Koshiba, S. Identification of biomarkers to diagnose diseases and find adverse drug reactions by metabolomics. Drug Metab. Pharmacokinet. 2021, 37, 100373. [Google Scholar] [CrossRef]
- Emwas, A.-H.; Luchinat, C.; Turano, P.; Tenori, L.; Roy, R.; Salek, R.M.; Ryan, D.; Merzaban, J.S.; Kaddurah-Daouk, R.; Zeri, A.C.; et al. Standardizing the experimental conditions for using urine in NMR-based metabolomic studies with a particular focus on diagnostic studies: A review. Metabolomics Off. J. Metabolomic Soc. 2015, 11, 872–894. [Google Scholar] [CrossRef]
- McCann, M.R.; George De la Rosa, M.V.; Rosania, G.R.; Stringer, K.A. L-Carnitine and Acylcarnitines: Mitochondrial Biomarkers for Precision Medicine. Metabolites 2021, 11, 51. [Google Scholar] [CrossRef]
- Sun, Y.; Kim, J.H.; Vangipuram, K.; Hayes, D.F.; Smith, E.M.L.; Yeomans, L.; Henry, N.L.; Stringer, K.A.; Hertz, D.L. Pharmacometabolomics reveals a role for histidine, phenylalanine, and threonine in the development of paclitaxel-induced peripheral neuropathy. Breast Cancer Res. Treat. 2018, 171, 657–666. [Google Scholar] [CrossRef]
- He, C.; Liu, Y.; Wang, Y.; Tang, J.; Tan, Z.; Li, X.; Chen, Y.; Huang, Y.; Chen, X.; Ouyang, D.; et al. 1H NMR based pharmacometabolomics analysis of metabolic phenotype on predicting metabolism characteristics of losartan in healthy volunteers. J. Chromatography. B Anal. Technol. Biomed. Life Sci. 2018, 1095, 15–23. [Google Scholar] [CrossRef]
- Chen, L.; Chen, C.-S.; Sun, Y.; Henry, N.L.; Stringer, K.A.; Hertz, D.L. Feasibility of pharmacometabolomics to identify potential predictors of paclitaxel pharmacokinetic variability. Cancer Chemother. Pharmacol. 2021, 88, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.M.; Sheau Chin, L.; Teh, C.-H.; Mostafa, H.; Mohamed Noor, D.A.; Sk Abdul Kader, M.A.; Kah Hay, Y.; Ibrahim, B. 1H NMR based pharmacometabolomics analysis of urine identifies metabolic phenotype of clopidogrel high on treatment platelets reactivity in coronary artery disease patients. J. Pharm. Biomed. Anal. 2017, 146, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.M. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol. Biol. 2015, 1277, 161–193. [Google Scholar] [CrossRef]
- Li, B.; He, X.; Jia, W.; Li, H. Novel Applications of Metabolomics in Personalized Medicine: A Mini-Review. Molecules 2017, 22, 1173. [Google Scholar] [CrossRef] [PubMed]
- Katsila, T.; Konstantinou, E.; Lavda, I.; Malakis, H.; Papantoni, I.; Skondra, L.; Patrinos, G.P. Pharmacometabolomics-aided Pharmacogenomics in Autoimmune Disease. eBioMedicine 2016, 5, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.M.; Sheau Chin, L.; Azri Mohamed Noor, D.; Sk Abdul Kader, M.A.; Kah Hay, Y.; Ibrahim, B. The Personalization of Clopidogrel Antiplatelet Therapy: The Role of Integrative Pharmacogenetics and Pharmacometabolomics. Cardiol. Res. Pract. 2017, 2017, 8062796. [Google Scholar] [CrossRef]
- Oh, J.; Yi, S.; Gu, N.; Shin, D.; Yu, K.-S.; Yoon, S.H.; Cho, J.-Y.; Jang, I.-J. Utility of Integrated Analysis of Pharmacogenomics and Pharmacometabolomics in Early Phase Clinical Trial: A Case Study of a New Molecular Entity. Genom. Inform. 2018, 16, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Phapale, P.B.; Kim, S.D.; Lee, H.W.; Lim, M.; Kale, D.D.; Kim, Y.L.; Cho, J.H.; Hwang, D.; Yoon, Y.R. An integrative approach for identifying a metabolic phenotype predictive of individualized pharmacokinetics of tacrolimus. Clin. Pharmacol. Ther. 2010, 87, 426–436. [Google Scholar] [CrossRef]
- Liu, L.; Cao, B.; Aa, J.; Zheng, T.; Shi, J.; Li, M.; Wang, X.; Zhao, C.; Xiao, W.; Yu, X.; et al. Prediction of the pharmacokinetic parameters of triptolide in rats based on endogenous molecules in pre-dose baseline serum. PLoS ONE 2012, 7, e43389. [Google Scholar] [CrossRef]
- Shin, K.H.; Choi, M.H.; Lim, K.S.; Yu, K.S.; Jang, I.J.; Cho, J.Y. Evaluation of endogenous metabolic markers of hepatic CYP3A activity using metabolic profiling and midazolam clearance. Clin. Pharmacol. Ther. 2013, 94, 601–609. [Google Scholar] [CrossRef]
- Huang, Q.; Aa, J.; Jia, H.; Xin, X.; Tao, C.; Liu, L.; Zou, B.; Song, Q.; Shi, J.; Cao, B.; et al. A Pharmacometabonomic Approach To Predicting Metabolic Phenotypes and Pharmacokinetic Parameters of Atorvastatin in Healthy Volunteers. J. Proteome Res. 2015, 14, 3970–3981. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.S.; Kerr, S.J.; Randolph, T.; Shireman, L.M.; Senn, T.; McCune, J.S. Prediction of Intravenous Busulfan Clearance by Endogenous Plasma Biomarkers Using Global Pharmacometabolomics. Metabolomics Off. J. Metabolomic Soc. 2016, 12, 161. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.-H.; Ahn, L.Y.; Choi, M.H.; Moon, J.-Y.; Lee, J.; Jang, I.-J.; Yu, K.-S.; Cho, J.-Y. Urinary 6β-Hydroxycortisol/Cortisol Ratio Most Highly Correlates With Midazolam Clearance Under Hepatic CYP3A Inhibition and Induction in Females: A Pharmacometabolomics Approach. AAPS J. 2016, 18, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Lee, J.W.; Hong, K.T.; Yu, K.-S.; Jang, I.-J.; Park, K.D.; Shin, H.Y.; Ahn, H.S.; Cho, J.-Y.; Kang, H.J. Pharmacometabolomics for predicting variable busulfan exposure in paediatric haematopoietic stem cell transplantation patients. Sci. Rep. 2017, 7, 1711. [Google Scholar] [CrossRef] [PubMed]
- Muhrez, K.; Benz-de Bretagne, I.; Nadal-Desbarats, L.; Blasco, H.; Gyan, E.; Choquet, S.; Montigny, F.; Emond, P.; Barin-Le Guellec, C. Endogenous metabolites that are substrates of organic anion transporter’s (OATs) predict methotrexate clearance. Pharmacol. Res. 2017, 118, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Gu, H.; Zhao, H.; Liu, Y.; Fu, S.; Wang, M.; Zhou, W.; Xie, Z.; Yu, H.; Huang, Z.; et al. Pharmacometabolomics in Endogenous Drugs: A New Approach for Predicting the Individualized Pharmacokinetics of Cholic Acid. J. Proteome Res. 2017, 16, 3529–3535. [Google Scholar] [CrossRef] [PubMed]
- Lesche, D.; Sigurdardottir, V.; Leichtle, A.B.; Nakas, C.T.; Christians, U.; Englberger, L.; Fiedler, M.; Largiadèr, C.R.; Mohacsi, P.; Sistonen, J. Targeted and global pharmacometabolomics in everolimus-based immunosuppression: Association of co-medication and lysophosphatidylcholines with dose requirement. Metabolomics Off. J. Metabolomic Soc. 2017, 14, 3. [Google Scholar] [CrossRef]
- Martínez-Ávila, J.C.; García Bartolomé, A.; García, I.; Dapía, I.; Tong, H.Y.; Díaz, L.; Guerra, P.; Frías, J.; Carcás Sansuan, A.J.; Borobia, A.M. Pharmacometabolomics applied to zonisamide pharmacokinetic parameter prediction. Metabolomics Off. J. Metabolomic Soc. 2018, 14, 70. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, S.H.; Yi, S.; Kim, A.H.; Kim, B.; Lee, S.; Yu, K.-S.; Jang, I.-J.; Cho, J.-Y. Quantitative prediction of hepatic CYP3A activity using endogenous markers in healthy subjects after administration of CYP3A inhibitors or inducers. Drug Metab. Pharmacokinet. 2019, 34, 247–252. [Google Scholar] [CrossRef]
- Xing, X.; Ma, P.; Huang, Q.; Qi, X.; Zou, B.; Wei, J.; Tao, L.; Li, L.; Zhou, G.; Song, Q. Predicting Pharmacokinetics Variation of Faropenem Using a Pharmacometabonomic Approach. J. Proteome Res. 2020, 19, 119–128. [Google Scholar] [CrossRef]
- Xing, X.; Ma, P.; Huang, Q.; Qi, X.; Zou, B.; Wei, J.; Tao, L.; Li, L.; Zhou, G.; Song, Q. Integration analysis of metabolites and single nucleotide polymorphisms improves the prediction of drug response of celecoxib. Metabolomics Off. J. Metabolomic Soc. 2020, 16, 41. [Google Scholar] [CrossRef] [PubMed]
- An, Z.; Wang, X.; Li, P.; He, J.; Liu, L. Exploring the metabolic characteristics and pharmacokinetic variation of paroxetine in healthy volunteers using a pharmacometabonomic approach. J. Pharm. Biomed. Anal. 2021, 204, 114224. [Google Scholar] [CrossRef] [PubMed]
- Silveira, A.M.R.; Duarte, G.H.B.; Fernandes, A.M.A.d.P.; Garcia, P.H.D.; Vieira, N.R.; Antonio, M.A.; Carvalho, P.d.O. Serum Predose Metabolic Profiling for Prediction of Rosuvastatin Pharmacokinetic Parameters in Healthy Volunteers. Front. Pharmacol. 2021, 12, 752960. [Google Scholar] [CrossRef] [PubMed]
- Molloy, B.; Mullin, L.; King, A.; Gethings, L.A.; Plumb, R.S.; Wilson, I.D. The Pharmacometabodynamics of Gefitinib after Intravenous Administration to Mice: A Preliminary UPLC-IM-MS Study. Metabolites 2021, 11, 379. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Wang, G.; Hu, T.; Li, H.; An, Z. Integration Analysis of Pharmacokinetics and Metabolomics to Predict Metabolic Phenotype and Drug Exposure of Remdesivir. Front. Pharmacol. 2021, 12, 779135. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Liu, L.; Hu, T.; An, Z. Integrative Analysis of Pharmacokinetic and Metabolomic Profiles for Predicting Metabolic Phenotype and Drug Exposure Caused by Sotorasib in Rats. Front. Oncol. 2022, 12, 778035. [Google Scholar] [CrossRef]
- Tee, K.B.; Ibrahim, L.; Hashim, N.M.; Saiman, M.Z.; Zakaria, Z.H.; Huri, H.Z. Pharmacokinetic-Pharmacometabolomic Approach in Early-Phase Clinical Trials: A Way Forward for Targeted Therapy in Type 2 Diabetes. Pharmaceutics 2022, 14, 1268. [Google Scholar] [CrossRef]
- McCune, J.S.; Navarro, S.L.; Baker, K.S.; Risler, L.J.; Phillips, B.R.; Randolph, T.W.; Shireman, L.; Schoch, H.G.; Deeg, H.J.; Zhang, Y.; et al. Prediction of Busulfan Clearance by Predose Plasma Metabolomic Profiling. Clin. Pharmacol. Ther. 2023, 113, 370–379. [Google Scholar] [CrossRef]
- Winnike, J.H.; Li, Z.; Wright, F.A.; Macdonald, J.M.; O’Connell, T.M.; Watkins, P.B. Use of pharmaco-metabonomics for early prediction of acetaminophen-induced hepatotoxicity in humans. Clin. Pharmacol. Ther. 2010, 88, 45–51. [Google Scholar] [CrossRef]
- Rahmioglu, N.; Le Gall, G.; Heaton, J.; Kay, K.L.; Smith, N.W.; Colquhoun, I.J.; Ahmadi, K.R.; Kemsley, E.K. Prediction of variability in CYP3A4 induction using a combined 1H NMR metabonomics and targeted UPLC-MS approach. J. Proteome Res. 2011, 10, 2807–2816. [Google Scholar] [CrossRef]
- Condray, R.; Dougherty, G.G.; Keshavan, M.S.; Reddy, R.D.; Haas, G.L.; Montrose, D.M.; Matson, W.R.; McEvoy, J.; Kaddurah-Daouk, R.; Yao, J.K. 3-Hydroxykynurenine and clinical symptoms in first-episode neuroleptic-naive patients with schizophrenia. Int. J. Neuropsychopharmacol. 2011, 14, 756–767. [Google Scholar] [CrossRef] [PubMed]
- Kaddurah-Daouk, R.; Boyle, S.H.; Matson, W.; Sharma, S.; Matson, S.; Zhu, H.; Bogdanov, M.B.; Churchill, E.; Krishnan, R.R.; Rush, A.J.; et al. Pretreatment metabotype as a predictor of response to sertraline or placebo in depressed outpatients: A proof of concept. Transl. Psychiatry 2011, 1, e26. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.N.; Kim, M.; Wen, H.; Kang, S.; Yang, H.-J.; Choi, M.-J.; Lee, H.S.; Choi, D.; Park, I.S.; Suh, Y.J.; et al. Predicting idiopathic toxicity of cisplatin by a pharmacometabonomic approach. Kidney Int. 2011, 79, 529–537. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Backshall, A.; Sharma, R.; Clarke, S.J.; Keun, H.C. Pharmacometabonomic profiling as a predictor of toxicity in patients with inoperable colorectal cancer treated with capecitabine. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 3019–3028. [Google Scholar] [CrossRef]
- Trupp, M.; Zhu, H.; Wikoff, W.R.; Baillie, R.A.; Zeng, Z.-B.; Karp, P.D.; Fiehn, O.; Krauss, R.M.; Kaddurah-Daouk, R. Metabolomics reveals amino acids contribute to variation in response to simvastatin treatment. PLoS ONE 2012, 7, e38386. [Google Scholar] [CrossRef]
- Coen, M.; Goldfain-Blanc, F.; Rolland-Valognes, G.; Walther, B.; Robertson, D.G.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Pharmacometabonomic investigation of dynamic metabolic phenotypes associated with variability in response to galactosamine hepatotoxicity. J. Proteome Res. 2012, 11, 2427–2440. [Google Scholar] [CrossRef]
- Kapoor, S.R.; Filer, A.; Fitzpatrick, M.A.; Fisher, B.A.; Taylor, P.C.; Buckley, C.D.; McInnes, I.B.; Raza, K.; Young, S.P. Metabolic profiling predicts response to anti-tumor necrosis factor α therapy in patients with rheumatoid arthritis. Arthritis Rheum. 2013, 65, 1448–1456. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Bogdanov, M.B.; Wikoff, W.R.; Zhu, H.; Boyle, S.H.; Churchill, E.; Wang, Z.; Rush, A.J.; Krishnan, R.R.; Pickering, E.; et al. Pharmacometabolomic mapping of early biochemical changes induced by sertraline and placebo. Transl. Psychiatry 2013, 3, e223. [Google Scholar] [CrossRef]
- Zhu, H.; Bogdanov, M.B.; Boyle, S.H.; Matson, W.; Sharma, S.; Matson, S.; Churchill, E.; Fiehn, O.; Rush, J.A.; Krishnan, R.R.; et al. Pharmacometabolomics of response to sertraline and to placebo in major depressive disorder—Possible role for methoxyindole pathway. PLoS ONE 2013, 8, e68283. [Google Scholar] [CrossRef]
- Ellero-Simatos, S.; Lewis, J.P.; Georgiades, A.; Yerges-Armstrong, L.M.; Beitelshees, A.L.; Horenstein, R.B.; Dane, A.; Harms, A.C.; Ramaker, R.; Vreeken, R.J.; et al. Pharmacometabolomics reveals that serotonin is implicated in aspirin response variability. CPT Pharmacomet. Syst. Pharmacol. 2014, 3, e125. [Google Scholar] [CrossRef]
- Zhao, Y.-Y.; Chen, H.; Tian, T.; Chen, D.-Q.; Bai, X.; Wei, F. A pharmaco-metabonomic study on chronic kidney disease and therapeutic effect of ergone by UPLC-QTOF/HDMS. PLoS ONE 2014, 9, e115467. [Google Scholar] [CrossRef] [PubMed]
- Puskarich, M.A.; Finkel, M.A.; Karnovsky, A.; Jones, A.E.; Trexel, J.; Harris, B.N.; Stringer, K.A. Pharmacometabolomics of l-carnitine treatment response phenotypes in patients with septic shock. Ann. Am. Thorac. Soc. 2015, 12, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Rotroff, D.M.; Shahin, M.H.; Gurley, S.B.; Zhu, H.; Motsinger-Reif, A.; Meisner, M.; Beitelshees, A.L.; Fiehn, O.; Johnson, J.A.; Elbadawi-Sidhu, M.; et al. Pharmacometabolomic Assessments of Atenolol and Hydrochlorothiazide Treatment Reveal Novel Drug Response Phenotypes. CPT Pharmacomet. Syst. Pharmacol. 2015, 4, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Ellero-Simatos, S.; Beitelshees, A.L.; Lewis, J.P.; Yerges-Armstrong, L.M.; Georgiades, A.; Dane, A.; Harms, A.C.; Strassburg, K.; Guled, F.; Hendriks, M.M.W.B.; et al. Oxylipid Profile of Low-Dose Aspirin Exposure: A Pharmacometabolomics Study. J. Am. Heart Assoc. 2015, 4, e002203. [Google Scholar] [CrossRef]
- Weng, L.; Gong, Y.; Culver, J.; Gardell, S.J.; Petucci, C.; Morse, A.M.; Frye, R.F.; Turner, S.T.; Chapman, A.; Boerwinkle, E.; et al. Presence of arachidonoyl-carnitine is associated with adverse cardiometabolic responses in hypertensive patients treated with atenolol. Metabolomics Off. J. Metabolomic Soc. 2016, 12, 160. [Google Scholar] [CrossRef]
- Rotroff, D.M.; Oki, N.O.; Liang, X.; Yee, S.W.; Stocker, S.L.; Corum, D.G.; Meisner, M.; Fiehn, O.; Motsinger-Reif, A.A.; Giacomini, K.M.; et al. Pharmacometabolomic Assessment of Metformin in Non-diabetic, African Americans. Front. Pharmacol. 2016, 7, 135. [Google Scholar] [CrossRef]
- Phua, L.C.; Goh, S.; Tai, D.W.M.; Leow, W.Q.; Alkaff, S.M.F.; Chan, C.Y.; Kam, J.H.; Lim, T.K.H.; Chan, E.C.Y. Metabolomic prediction of treatment outcome in pancreatic ductal adenocarcinoma patients receiving gemcitabine. Cancer Chemother. Pharmacol. 2018, 81, 277–289. [Google Scholar] [CrossRef]
- Elbadawi-Sidhu, M.; Baillie, R.A.; Zhu, H.; Chen, Y.-D.I.; Goodarzi, M.O.; Rotter, J.I.; Krauss, R.M.; Fiehn, O.; Kaddurah-Daouk, R. Pharmacometabolomic signature links simvastatin therapy and insulin resistance. Metabolomics Off. J. Metabolomic Soc. 2017, 13, 11. [Google Scholar] [CrossRef]
- Nguyen, T.V.; Reuter, J.M.; Gaikwad, N.W.; Rotroff, D.M.; Kucera, H.R.; Motsinger-Reif, A.; Smith, C.P.; Nieman, L.K.; Rubinow, D.R.; Kaddurah-Daouk, R.; et al. The steroid metabolome in women with premenstrual dysphoric disorder during GnRH agonist-induced ovarian suppression: Effects of estradiol and progesterone addback. Transl. Psychiatry 2017, 7, e1193. [Google Scholar] [CrossRef]
- Tan, G.; Zhao, B.; Li, Y.; Liu, X.; Zou, Z.; Wan, J.; Yao, Y.; Xiong, H.; Wang, Y. Pharmacometabolomics identifies dodecanamide and leukotriene B4 dimethylamide as a predictor of chemosensitivity for patients with acute myeloid leukemia treated with cytarabine and anthracycline. Oncotarget 2017, 8, 88697–88707. [Google Scholar] [CrossRef][Green Version]
- Lee, J.; Kim, A.H.; Yi, S.; Lee, S.; Yoon, S.H.; Yu, K.-S.; Jang, I.-J.; Cho, J.-Y. Distribution of Exogenous and Endogenous CYP3A Markers and Related Factors in Healthy Males and Females. AAPS J. 2017, 19, 1196–1204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, W.; Chen, J.; Li, R.; Zhang, Z.; Huang, Y.; Xu, F. Branched-Chain Amino Acids as Predictors for Individual Differences of Cisplatin Nephrotoxicity in Rats: A Pharmacometabonomics Study. J. Proteome Res. 2017, 16, 1753–1762. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Jeong, G.-H.; Lee, I.-K.; Yoon, Y.-R.; Liu, K.-H.; Gu, N.; Shin, K.-H. A Pharmacometabolomic Approach to Predict Response to Metformin in Early-Phase Type 2 Diabetes Mellitus Patients. Molecules 2018, 23, 1579. [Google Scholar] [CrossRef]
- Jiang, L.; Lee, S.C.; Ng, T.C. Pharmacometabonomics Analysis Reveals Serum Formate and Acetate Potentially Associated with Varying Response to Gemcitabine-Carboplatin Chemotherapy in Metastatic Breast Cancer Patients. J. Proteome Res. 2018, 17, 1248–1257. [Google Scholar] [CrossRef] [PubMed]
- Malkawi, A.K.; Alzoubi, K.H.; Jacob, M.; Matic, G.; Ali, A.; Al Faraj, A.; Almuhanna, F.; Dasouki, M.; Abdel Rahman, A.M. Metabolomics Based Profiling of Dexamethasone Side Effects in Rats. Front. Pharmacol. 2018, 9, 46. [Google Scholar] [CrossRef]
- Lewis, T.; Chalise, P.; Gauldin, C.; Truog, W. Pharmacometabolomics of Respiratory Phenotypic Response to Dexamethasone in Preterm Infants at Risk for Bronchopulmonary Dysplasia. Clin. Transl. Sci. 2019, 12, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.I.; Perry-Walker, K.; Finnell, R.H.; Pennell, K.D.; Tran, V.; May, R.C.; McElrath, T.F.; Meador, K.J.; Pennell, P.B.; Jones, D.P. Metabolome-wide association study of anti-epileptic drug treatment during pregnancy. Toxicol. Appl. Pharmacol. 2019, 363, 122–130. [Google Scholar] [CrossRef]
- Cao, Z.; Miller, M.S.; Lubet, R.A.; Grubbs, C.J.; Beger, R.D. Pharmacometabolomic Pathway Response of Effective Anticancer Agents on Different Diets in Rats with Induced Mammary Tumors. Metabolites 2019, 9, 149. [Google Scholar] [CrossRef]
- Evans, C.R.; Karnovsky, A.; Puskarich, M.A.; Michailidis, G.; Jones, A.E.; Stringer, K.A. Untargeted Metabolomics Differentiates l-Carnitine Treated Septic Shock 1-Year Survivors and Nonsurvivors. J. Proteome Res. 2019, 18, 2004–2011. [Google Scholar] [CrossRef]
- Gao, Y.; Li, W.; Chen, J.; Wang, X.; Lv, Y.; Huang, Y.; Zhang, Z.; Xu, F. Pharmacometabolomic prediction of individual differences of gastrointestinal toxicity complicating myelosuppression in rats induced by irinotecan. Acta Pharm. Sinica. B 2019, 9, 157–166. [Google Scholar] [CrossRef]
- Cunningham, K.; Claus, S.P.; Lindon, J.C.; Holmes, E.; Everett, J.R.; Nicholson, J.K.; Coen, M. Pharmacometabonomic characterization of xenobiotic and endogenous metabolic phenotypes that account for inter-individual variation in isoniazid-induced toxicological response. J. Proteome Res. 2012, 11, 4630–4642. [Google Scholar] [CrossRef]
- Hu, T.; An, Z.; Sun, Y.; Wang, X.; Du, P.; Li, P.; Chi, Y.; Liu, L. Longitudinal Pharmacometabonomics for Predicting Malignant Tumor Patient Responses to Anlotinib Therapy: Phenotype, Efficacy, and Toxicity. Front. Oncol. 2020, 10, 548300. [Google Scholar] [CrossRef] [PubMed]
- Broughton-Neiswanger, L.E.; Rivera-Velez, S.M.; Suarez, M.A.; Slovak, J.E.; Hwang, J.K.; Villarino, N.F. Pharmacometabolomics with a combination of PLS-DA and random forest algorithm analyses reveal meloxicam alters feline plasma metabolite profiles. J. Vet. Pharmacol. Ther. 2020, 43, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Puskarich, M.A.; Jennaro, T.S.; Gillies, C.E.; Evans, C.R.; Karnovsky, A.; McHugh, C.E.; Flott, T.L.; Jones, A.E.; Stringer, K.A. Pharmacometabolomics identifies candidate predictor metabolites of an L-carnitine treatment mortality benefit in septic shock. Clin. Transl. Sci. 2021, 14, 2288–2299. [Google Scholar] [CrossRef]
- Du, Z.; Lu, Y.; Sun, J.; Chang, K.; Lu, M.; Fang, M.; Zeng, X.; Zhang, W.; Song, J.; Guo, X.; et al. Pharmacokinetics/pharmacometabolomics-pharmacodynamics reveals the synergistic mechanism of a multicomponent herbal formula, Baoyuan decoction against cardiac hypertrophy. Biomed. Pharmacother. Biomed. Pharmacother. 2021, 139, 111665. [Google Scholar] [CrossRef] [PubMed]
- Sha’aban, A.; Zainal, H.; Khalil, N.A.; Abd Aziz, F.; Ch’ng, E.S.; Teh, C.-H.; Mohammed, M.; Ibrahim, B. Prediction of Low-Dose Aspirin-Induced Gastric Toxicity Using Nuclear Magnetic Resonance Spectroscopy-Based Pharmacometabolomics in Rats. Molecules 2022, 27, 2126. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.-J.; Chiang, K.-M.; Xiu, L.-L.; Chung, C.-M.; Lo, C.-J.; Shiao, M.-S.; Cheng, M.-L.; Kuo, C.-C.; Yang, H.-C.; Pan, W.-H. Pharmacometabolomic study of drug response to antihypertensive medications for hypertension marker identification in Han Chinese individuals in Taiwan. Comput. Struct. Biotechnol. J. 2022, 20, 6458–6466. [Google Scholar] [CrossRef]
- Guan, S.; Chen, X.; Chen, Y.; Wan, G.; Su, Q.; Liang, H.; Yang, Y.; Fang, W.; Huang, Y.; Zhao, H.; et al. FOXO3 mutation predicting gefitinib-induced hepatotoxicity in NSCLC patients through regulation of autophagy. Acta Pharm. Sinica B 2022, 12, 3639–3649. [Google Scholar] [CrossRef]
- Liu, D.; Huang, J.; Gao, S.; Jin, H.; He, J. A temporo-spatial pharmacometabolomics method to characterize pharmacokinetics and pharmacodynamics in the brain microregions by using ambient mass spectrometry imaging. Acta Pharm. Sinica. B 2022, 12, 3341–3353. [Google Scholar] [CrossRef]
- Chen, C.; Krausz, K.W.; Idle, J.R.; Gonzalez, F.J. Identification of novel toxicity-associated metabolites by metabolomics and mass isotopomer analysis of acetaminophen metabolism in wild-type and Cyp2e1-null mice. J. Biol. Chem. 2008, 283, 4543–4559. [Google Scholar] [CrossRef]
- Pyo, J.S.; Ju, H.K.; Park, J.H.; Kwon, S.W. Determination of volatile biomarkers for apoptosis and necrosis by solid-phase microextraction-gas chromatography/mass spectrometry: A pharmacometabolomic approach to cisplatin’s cytotoxicity to human lung cancer cell lines. J. Chromatography. B Anal. Technol. Biomed. Life Sci. 2008, 876, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Fitzgerald, E.F.; Gomez, M.I.; Lambert, G.H.; Longnecker, M.P. The ratio of specific polychlorinated biphenyls as a surrogate biomarker of cytochrome P4501A2 activity: A pharmaco-metabonomic study in humans. Cancer Epidemiol. Biomark. Prev. A Publ. Am. Assoc. Cancer Res. Cosponsored By Am. Soc. Prev. Oncol. 2008, 17, 1013–1015. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Hebbring, S.; Zhu, H.; Jenkins, G.D.; Biernacka, J.; Snyder, K.; Drews, M.; Fiehn, O.; Zeng, Z.; Schaid, D.; et al. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: Pharmacometabolomics-informed pharmacogenomics. Clin. Pharmacol. Ther. 2011, 89, 97–104. [Google Scholar] [CrossRef] [PubMed]
- du Preez, I.; Loots, D.T. Altered fatty acid metabolism due to rifampicin-resistance conferring mutations in the rpoB Gene of Mycobacterium tuberculosis: Mapping the potential of pharmaco-metabolomics for global health and personalized medicine. Omics A J. Integr. Biol. 2012, 16, 596–603. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Zhu, H.; Sharma, S.; Bogdanov, M.; Rozen, S.G.; Matson, W.; Oki, N.O.; Motsinger-Reif, A.A.; Churchill, E.; Lei, Z.; et al. Alterations in metabolic pathways and networks in Alzheimer’s disease. Transl. Psychiatry 2013, 3, e244. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Han, Y.; Yan, G.; Wang, X. Urinary metabolic biomarker and pathway study of hepatitis B virus infected patients based on UPLC-MS system. PLoS ONE 2013, 8, e64381. [Google Scholar] [CrossRef]
- Park, J.; Noh, K.; Lee, H.W.; Lim, M.-s.; Seong, S.J.; Seo, J.J.; Kim, E.-J.; Kang, W.; Yoon, Y.-R. Pharmacometabolomic approach to predict QT prolongation in guinea pigs. PLoS ONE 2013, 8, e60556. [Google Scholar] [CrossRef]
- Yerges-Armstrong, L.M.; Ellero-Simatos, S.; Georgiades, A.; Zhu, H.; Lewis, J.P.; Horenstein, R.B.; Beitelshees, A.L.; Dane, A.; Reijmers, T.; Hankemeier, T.; et al. Purine pathway implicated in mechanism of resistance to aspirin therapy: Pharmacometabolomics-informed pharmacogenomics. Clin. Pharmacol. Ther. 2013, 94, 525–532. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Yan, K.; Pence, L.; Simpson, P.M.; Gill, P.; Letzig, L.G.; Beger, R.D.; Sullivan, J.E.; Kearns, G.L.; Reed, M.D.; et al. Targeted liquid chromatography-mass spectrometry analysis of serum acylcarnitines in acetaminophen toxicity in children. Biomark. Med. 2014, 8, 147–159. [Google Scholar] [CrossRef]
- Villaseñor, A.; Ramamoorthy, A.; Silva dos Santos, M.; Lorenzo, M.P.; Laje, G.; Zarate, C.; Barbas, C.; Wainer, I.W. A pilot study of plasma metabolomic patterns from patients treated with ketamine for bipolar depression: Evidence for a response-related difference in mitochondrial networks. Br. J. Pharmacol. 2014, 171, 2230–2242. [Google Scholar] [CrossRef]
- Nam, H.W.; Karpyak, V.M.; Hinton, D.J.; Geske, J.R.; Ho, A.M.C.; Prieto, M.L.; Biernacka, J.M.; Frye, M.A.; Weinshilboum, R.M.; Choi, D.S. Elevated baseline serum glutamate as a pharmacometabolomic biomarker for acamprosate treatment outcome in alcohol-dependent subjects. Transl. Psychiatry 2015, 5, e621. [Google Scholar] [CrossRef] [PubMed]
- Blasco, H.; Patin, F.; Descat, A.; Garçon, G.; Corcia, P.; Gelé, P.; Lenglet, T.; Bede, P.; Meininger, V.; Devos, D.; et al. A pharmaco-metabolomics approach in a clinical trial of ALS: Identification of predictive markers of progression. PLoS ONE 2018, 13, e0198116. [Google Scholar] [CrossRef] [PubMed]
- Navarro, S.L.; Randolph, T.W.; Shireman, L.M.; Raftery, D.; McCune, J.S. Pharmacometabonomic Prediction of Busulfan Clearance in Hematopoetic Cell Transplant Recipients. J. Proteome Res. 2016, 15, 2802–2811. [Google Scholar] [CrossRef] [PubMed]
- Miolo, G.; Muraro, E.; Caruso, D.; Crivellari, D.; Ash, A.; Scalone, S.; Lombardi, D.; Rizzolio, F.; Giordano, A.; Corona, G. Pharmacometabolomics study identifies circulating spermidine and tryptophan as potential biomarkers associated with the complete pathological response to trastuzumab-paclitaxel neoadjuvant therapy in HER-2 positive breast cancer. Oncotarget 2016, 7, 39809–39822. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, F.A.; Shahin, M.H.; Gong, Y.; McDonough, C.W.; Beitelshees, A.L.; Gums, J.G.; Chapman, A.B.; Boerwinkle, E.; Turner, S.T.; Frye, R.F.; et al. Novel plasma biomarker of atenolol-induced hyperglycemia identified through a metabolomics-genomics integrative approach. Metabolomics Off. J. Metabolomic Soc. 2016, 12, 129. [Google Scholar] [CrossRef]
- Hao, D.; Sarfaraz, M.O.; Farshidfar, F.; Bebb, D.G.; Lee, C.Y.; Card, C.M.; David, M.; Weljie, A.M. Temporal characterization of serum metabolite signatures in lung cancer patients undergoing treatment. Metabolomics Off. J. Metabolomic Soc. 2016, 12, 58. [Google Scholar] [CrossRef]
- Do, E.Y.; Gwon, M.-R.; Kim, B.K.; Ohk, B.; Lee, H.W.; Kang, W.Y.; Seong, S.J.; Kim, H.-J.; Yoon, Y.-R. Metabolomic analysis of healthy human urine following administration of glimepiride using a liquid chromatography-tandem mass spectrometry. Transl. Clin. Pharmacol. 2017, 25, 67–73. [Google Scholar] [CrossRef]
- Amin, A.M.; Sheau Chin, L.; Teh, C.-H.; Mostafa, H.; Mohamed Noor, D.A.; Abdul Kader, M.A.S.K.; Kah Hay, Y.; Ibrahim, B. Pharmacometabolomics analysis of plasma to phenotype clopidogrel high on treatment platelets reactivity in coronary artery disease patients. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2018, 117, 351–361. [Google Scholar] [CrossRef]
- Combrink, M.; du Preez, I.; Ronacher, K.; Walzl, G.; Loots, D.T. Time-Dependent Changes in Urinary Metabolome Before and After Intensive Phase Tuberculosis Therapy: A Pharmacometabolomics Study. Omics A J. Integr. Biol. 2019, 23, 560–572. [Google Scholar] [CrossRef]
- Vargas, D.A.; Prieto, M.D.; Martínez-Valencia, A.J.; Cossio, A.; Burgess, K.E.V.; Burchmore, R.J.S.; Gómez, M.A. Pharmacometabolomics of Meglumine Antimoniate in Patients With Cutaneous Leishmaniasis. Front. Pharmacol. 2019, 10, 657. [Google Scholar] [CrossRef]
- Zapata, R.C.; Rosenthal, S.B.; Fisch, K.; Dao, K.; Jain, M.; Osborn, O. Metabolomic profiles associated with a mouse model of antipsychotic-induced food intake and weight gain. Sci. Rep. 2020, 10, 18581. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Li, X.; Zhang, X.; Han, F.; Lu, X.; Liu, L.; Zhang, J.; Dong, M.; Yang, H.; Li, H. Pharmacometabolomics Identifies 3-Hydroxyadipic Acid, d-Galactose, Lysophosphatidylcholine (P-16:0), and Tetradecenoyl-l-Carnitine as Potential Predictive Indicators of Gemcitabine Efficacy in Pancreatic Cancer Patients. Front. Oncol. 2019, 9, 1524. [Google Scholar] [CrossRef] [PubMed]
- Bawadikji, A.A.; Teh, C.-H.; Sheikh Abdul Kader, M.A.B.; Abdul Wahab, M.J.B.; Syed Sulaiman, S.A.; Ibrahim, B. Plasma Metabolites as Predictors of Warfarin Outcome in Atrial Fibrillation. Am. J. Cardiovasc. Drugs Drugs Devices Other Interv. 2020, 20, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Wu, J.; Kim, S.; LoRusso, P.; Li, J. Pharmacometabolomics Reveals Irinotecan Mechanism of Action in Cancer Patients. J. Clin. Pharmacol. 2019, 59, 20–34. [Google Scholar] [CrossRef] [PubMed]
- McCune, J.S.; McKiernan, J.S.; van Maarseveen, E.; Huitema, A.D.R.; Randolph, T.W.; Deeg, H.J.; Nakamura, R.; Baker, K.S. Prediction of Acute Graft versus Host Disease and Relapse by Endogenous Metabolomic Compounds in Patients Receiving Personalized Busulfan-Based Conditioning. J. Proteome Res. 2021, 20, 684–694. [Google Scholar] [CrossRef]
- Cristoni, S.; Bernardi, L.R.; Malvandi, A.M.; Larini, M.; Longhi, E.; Sortino, F.; Conti, M.; Pantano, N.; Puccio, G. A case of personalized and precision medicine: Pharmacometabolomic applications to rare cancer, microbiological investigation, and therapy. Rapid Commun. Mass. Spectrom. RCM 2021, 35, e8976. [Google Scholar] [CrossRef]
- Kachroo, P.; Sordillo, J.E.; Lutz, S.M.; Weiss, S.T.; Kelly, R.S.; McGeachie, M.J.; Wu, A.C.; Lasky-Su, J.A. Pharmaco-Metabolomics of Inhaled Corticosteroid Response in Individuals with Asthma. J. Pers. Med. 2021, 11, 1148. [Google Scholar] [CrossRef]
- Guan, S.; Chen, X.; Xin, S.; Liu, S.; Yang, Y.; Fang, W.; Huang, Y.; Zhao, H.; Zhu, X.; Zhuang, W.; et al. Establishment and application of a predictive model for gefitinib-induced severe rash based on pharmacometabolomic profiling and polymorphisms of transporters in non-small cell lung cancer. Transl. Oncol. 2021, 14, 100951. [Google Scholar] [CrossRef]
- Velenosi, T.J.; Krausz, K.W.; Hamada, K.; Dorsey, T.H.; Ambs, S.; Takahashi, S.; Gonzalez, F.J. Pharmacometabolomics reveals urinary diacetylspermine as a biomarker of doxorubicin effectiveness in triple negative breast cancer. NPJ Precis. Oncol. 2022, 6, 70. [Google Scholar] [CrossRef]
- He, X.; Yang, X.; Yan, X.; Huang, M.; Xiang, Z.; Lou, Y. Individualized Dosage of Tacrolimus for Renal Transplantation Patients Based on Pharmacometabonomics. Molecules 2022, 27, 3517. [Google Scholar] [CrossRef]
- Siani-Rose, M.; Cox, S.; Goldstein, B.; Abrams, D.; Taylor, M.; Kurek, I. Cannabis-Responsive Biomarkers: A Pharmacometabolomics-Based Application to Evaluate the Impact of Medical Cannabis Treatment on Children with Autism Spectrum Disorder. Cannabis Cannabinoid Res. 2023, 8, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.; Lee, H.-S.; Joo, E.Y.; Shon, Y.-M.; Hong, S.B.; Seo, D.-W.; Lee, S.-Y. Levetiracetam Therapeutic Drug Monitoring in a Large Cohort of Korean Epileptic Patients. Pharmaceuticals 2021, 14, 826. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Schmidt, M.A.; Kaddurah-Daouk, R. Current Concepts in Pharmacometabolomics, Biomarker Discovery, and Precision Medicine. Metabolites 2020, 10, 129. [Google Scholar] [CrossRef] [PubMed]
- Khamis, M.M.; Adamko, D.J.; El-Aneed, A. Mass spectrometric based approaches in urine metabolomics and biomarker discovery. Mass. Spectrom. Rev. 2017, 36, 115–134. [Google Scholar] [CrossRef]
- Smith, L.; Villaret-Cazadamont, J.; Claus, S.P.; Canlet, C.; Guillou, H.; Cabaton, N.J.; Ellero-Simatos, S. Important Considerations for Sample Collection in Metabolomics Studies with a Special Focus on Applications to Liver Functions. Metabolites 2020, 10, 104. [Google Scholar] [CrossRef]
- Hernandes, V.V.; Barbas, C.; Dudzik, D. A review of blood sample handling and pre-processing for metabolomics studies. Electrophoresis 2017, 38, 2232–2241. [Google Scholar] [CrossRef]
- Lepoittevin, M.; Blancart-Remaury, Q.; Kerforne, T.; Pellerin, L.; Hauet, T.; Thuillier, R. Comparison between 5 extractions methods in either plasma or serum to determine the optimal extraction and matrix combination for human metabolomics. Cell Mol. Biol. Lett. 2023, 28, 43. [Google Scholar] [CrossRef]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. A Conversation on Data Mining Strategies in LC-MS Untargeted Metabolomics: Pre-Processing and Pre-Treatment Steps. Metabolites 2016, 6, 40. [Google Scholar] [CrossRef]
- Everett, J.R. From Metabonomics to Pharmacometabonomics: The Role of Metabolic Profiling in Personalized Medicine. Front. Pharmacol. 2016, 7, 297. [Google Scholar] [CrossRef]
- Xi, B.; Gu, H.; Baniasadi, H.; Raftery, D. Statistical analysis and modeling of mass spectrometry-based metabolomics data. Methods Mol. Biol. 2014, 1198, 333–353. [Google Scholar] [CrossRef]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis--a marriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Dong, N.; Yun, Y.; Deng, B.; Ren, D.; Liu, S.; Liang, Y. Chemometric methods in data processing of mass spectrometry-based metabolomics: A review. Anal. Chim. Acta 2016, 914, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Corona, G.; Di Gregorio, E.; Buonadonna, A.; Lombardi, D.; Scalone, S.; Steffan, A.; Miolo, G. Pharmacometabolomics of trabectedin in metastatic soft tissue sarcoma patients. Front. Pharmacol. 2023, 14, 1212634. [Google Scholar] [CrossRef] [PubMed]

| Pharmacokinetics-Related Pharmacometabolomics | |||||
|---|---|---|---|---|---|
| Number | Year | Drug | Object | Analytical Technique | Results with Pharmacometabolomics |
| 1 | 2010 | Tacrolimus | Healthy human volunteers | LC-MS | Predicting individualized PK of tacrolimus [48] |
| 2 | 2012 | Triptolide | Rats | GC-MS | Predicting the PK of Triptolide in in rats with different metabolic patterns [49] |
| 3 | 2013 | Midazolam | Healthy male volunteers | GC-MS | Establishing the CL equation for predicting midazolam [50] |
| 4 | 2015 | Atorvastatin | Healthy Volunteers | LC-MS | Predicting pharmacokinetic differences of atorvastatin in individuals [51] |
| 5 | 2016 | Busulfan | Allogeneic hematopoietic cell transplant recipients | LC-MS | Establishing a model for predicting the clearance rate of busulfan [52] |
| 6 | 2016 | Midazolam | Healthy female volunteers | LC-MS | Predicting the activity of liver CYP in women under different states [53] |
| 7 | 2017 | Busulfan | Paediatric haematopoietic stem cell transplantation patients | LC-MS | Potential biomarkers for predicting exposure to busulfan [54] |
| 8 | 2017 | Methotrexate | Patients treated with high-dose methotrexate | GC-MS | Predicting the clearance rate of methotrexate [55] |
| 9 | 2017 | Cholic acid | Rats | LC-MS | Using bile acid as an example to predict individualized PK [56] |
| 10 | 2017 | Everolimus | Heart transplant recipients | UPLC-MS/MS | Evaluating the factors affecting the metabolism of Everolimus and determine metabolic biomarkers [57] |
| 11 | 2018 | Zonisamide | Healthy human volunteers | LC-MS | Identification of endogenous metabolites that can predict the distribution of zonisamide [58] |
| 12 | 2018 | Losartan | Healthy male volunteers | NMR | Predicting individualized PK characteristics of losartan [40] |
| 13 | 2018 | New candidate drug | Human | LC-MS | Explaining the pharmacokinetic and pharmacodynamic characteristics of new candidate drugs [47] |
| 14 | 2019 | Midazolam | Healthy human volunteers | GC-MS LC-MS | Establishing an equation for predicting the clearance rate of midazolam [59] |
| 15 | 2020 | Faropenem | Healthy male volunteers | GC-MS LC-MS | Predicting individual PK parameters of Faropenem [60] |
| 16 | 2020 | Celecoxib | Healthy human volunteers | UPLC-MS/MS | Monitoring PK of celecoxib and establishing prediction models [61] |
| 17 | 2021 | Paroxetine | Healthy human volunteers | LC-MS | Screening and identification of endogenous markers that can predict paroxetine PK [62] |
| 18 | 2021 | Rosuvastatin | Healthy human volunteers | LC-MS | Predicting PK parameters of rosuvastatin [63] |
| 19 | 2021 | Paclitaxel | Female patients with oligometastatic breast cancer | LC-MS | Identification of pretherapeutic metabolites that may be associated with PK variability in paclitaxel [41] |
| 20 | 2021 | Gefitinib | Mice | UPLC-IM-MS | Analyzing the urine profile of gefitinib and analyzing PK [64] |
| 21 | 2022 | Remdesivir | Rats | LC-MS | Predicting AUC and Cmax of drugs [65] |
| 22 | 2022 | Sotorasib | Rats | LC-MS | Predicting drug exposure/toxicity biomarkers [66] |
| 23 | 2022 | Metformin | Healthy human volunteers | UPLC-QTOF-MS | Predicting the dose of drugs in clinical trials [67] |
| 24 | 2023 | Busulfan | Patients receiving HCT conditioning with Busulfan | LC-MS | Predicting the clearance rate of busulfan [68] |
| Pharmacometabolomics related to drug administration response (effective, ineffective and toxic) | |||||
| 1 | 2006 | Paracetamol | Rats | NMR | Predicting the degree of liver injury after paracetamol administration [34] |
| 2 | 2009 | Paracetamol | Healthy male volunteers | NMR | Determination of predictive factors for common metabolites based on urine metabolism profiles [35] |
| 3 | 2010 | Paracetamol | Healthy human volunteers | NMR | Identification of relevant metabolites to distinguish susceptibility to acetaminophen induced liver injury [69] |
| 4 | 2011 | CYP3A4 inducer | Healthy human volunteers | NMR | Predicting metabolic characteristics related to induced changes in CYP3A4 activity [70] |
| 5 | 2011 | 3-Hydroxykynurenine | Patients with schizophrenia in first episode | LCECA | Predicting the severity of clinical symptoms in the early stages of the disease and before exposure to antipsychotic drugs [71] |
| 6 | 2011 | Sertraline | Patients with major depression | LCECA | Predicting whether depression patients respond to sertraline [72] |
| 7 | 2011 | Cisplatinum | Rats | NMR | Idiopathic and pre administration prediction of cisplatin induced nephrotoxicity [73] |
| 8 | 2011 | Capecitabine | Patients with colorectal cancer | 1HNMR | Predicting the toxicity of capecitabine in patients with advanced colorectal cancer [74] |
| 9 | 2012 | Simvastatin | Healthy, first treatment drug patients | GC-MS | Identifying metabolites that can predict LDL-C responses [75] |
| 10 | 2012 | Galactosamine | Rats | 1HNMR | Analyzing the metabolic spectrum before and after administration to understand the variable response phenotype induced by galactosamine [76] |
| 11 | 2013 | anti-tumor necrosis factor (ANF) | Patients with two types of arthritis | NMR | Predicting the response of patients with rheumatoid arthritis and psoriatic arthritis to TNF antagonists [77] |
| 12 | 2013 | Sertraline | Patients with major depression | GC-TOF | Biomarkers found to separate responders and non-responders to sertraline treatment [78] |
| 13 | 2013 | Sertraline | Patients with major depression | LCECA, GCTOF-MS | Distinguishing between responders and no-responders of sertraline or placebo [79] |
| 14 | 2014 | Aspirin | Healthy human volunteers | LC-MS | Identification of serotonin associated with aspirin response variability [80] |
| 15 | 2014 | Ergone | Rats | UPLC-QTOF/HDMS | Metabolic analysis of adenine induced chronic kidney disease [81] |
| 16 | 2015 | L-Carnitine | Septic patient | NMR | Identification of endogenous biomarkers for distinguishing between response to L-carnitine treatment in sepsis [82] |
| 17 | 2015 | Atenolol and Hydrochlorothiazide | Hypertensive patient | GC-MS | Identifying the characteristics of metabolites related to the treatment of two drugs and establishing predictive models [83] |
| 18 | 2015 | Aspirin | Healthy human volunteers | LC-MS/MS | Studying the metabolic characteristics of aspirin exposure and evaluate changes in related reactions [84] |
| 19 | 2016 | Atenolol | Hypertensive patient | LC-MS | The relationship between baseline serum acylcarnitine levels and cardiometabolic responses after exposure to atenolol was studied [85] |
| 20 | 2016 | Metformin | Non-diabetic | GC-TOF | Identification of metabolic characteristics of metformin exposure and its pharmacological effects on oral glucose tolerance [86] |
| 21 | 2017 | Clopidogrel | CAD patient | NMR | Identification of endogenous metabolites associated with clopidogrel HTPR in urine reveals relevant pathways and conditions [42] |
| 22 | 2017 | Gemcitabine | Patients with pancreatic ductal adenocarcinoma receiving gemcitabine | GC-MS | Identification of relevant differential PDAC metabolites that can predict response to gemcitabine treatment [87] |
| 23 | 2017 | Simvastatin | Patients treated with simvastatin | GC-MS | Predicting the risk of developing hyperglycemia or insulin resistance during simvastatin treatment [88] |
| 24 | 2017 | Estradiol and/or progesterone | Patients with premenstrual anxiety disorder | UPLC/MS-MS | Determining steroid-specific metabolites resulting from treatment with estradiol and/or progesterone [89] |
| 25 | 2017 | Cytarabine and Anthracycline | Patients with acute myeloid leukemia | UHPLC-Q-TOF | Statistical modeling of chemotherapy response in de novo AML patients treated with cytarabine and anthracyclines [90] |
| 26 | 2017 | Midazolam | Healthy human volunteers | GC-MS, LC-MS/MS | Validation of endogenous versus exogenous markers to assess CYP3A activity and predict treatment effects [91] |
| 27 | 2017 | Cisplatinum | Rats | LC-MS/MS, GC-MS | Discovery of predicted metabolites in serum prior to cisplatin administration and construction and validation of predictive models [92] |
| 28 | 2018 | Paclitaxel | Female adult patients with oligometastatic breast cancer | NMR | Predicting metabolic changes induced by PN and paclitaxel [39] |
| 29 | 2018 | Metformin | Early-stage type 2 diabetic patients | GC-MS | Predicting the efficacy of metformin [93] |
| 30 | 2018 | Gemcitabine-carboplatin chemotherapy | Patients with metastatic breast cancer | 1H-NMR | Determining predictive metabolites for response to chemotherapy in patients with metastatic breast cancer [94] |
| 31 | 2018 | Dexamethasone | Rats with osteoporosis | LC-MS/MS | Predicting side effects associated with dexamethasone treatment [95] |
| 32 | 2019 | Dexamethasone | Preterm infants treated with dexamethasone | GC-MS | Identifying changes in metabolites before and after dexamethasone treatment can be used to distinguish between responders and no-responders [96] |
| 33 | 2019 | Lamotrigine and levetiracetam | Pregnant women with epilepsy | LC-HRMS | Assessing the risk of pregnant women receiving antiepileptic drug treatment [97] |
| 34 | 2019 | Tamoxifen | Rats | GC-MS LC-MS | Screening of potential pharmacodynamic biomarkers in rats treated with antitumor drugs under different metabolic patterns [98] |
| 35 | 2019 | L-Carnitine | Subjects with vasopressor-dependent septic shock treated with levocarnitine | LC-MS | Identifying differential metabolites in patients can be used to distinguish between 1-year survivors and non survivors [99] |
| 36 | 2019 | Irinotecan | Rats | GC-MS LC-MS | Establishing a model for predicting delayed diarrhea and CPT-11 bone marrow suppression toxicity [100] |
| 37 | 2019 | Isoniazide | Rats | 1HNMR | Determining the variability of isoniazid toxicity reactions can be used to distinguish whether adverse reactions have occurred [101] |
| 38 | 2020 | Anlotinib | Terminal cancer patients | LC-MS | Exploring the utility of longitudinal pharmacometabolomics in predicting response to erlotinib in patients with nasty tumors [102] |
| 39 | 2020 | Meloxicam | Cats | GC-MS | Predicting adverse reactions of meloxicam [103] |
| 40 | 2021 | L-Carnitine | Septic patient | NMR | Different efficacy of L-carnitine found in patients with different metabolic profiles [104] |
| 41 | 2021 | Baoyuan decoction | Rats | UPLC-MS/MS | Analysis of endogenous metabolites associated with oral administration of Baoyuan decoction to predict PD metrics [105] |
| 42 | 2022 | Aspirin | Rats | NMR | Predicting gastric toxicity associated with LDA induced coronary artery disease [106] |
| 43 | 2022 | Angiotensin-converting enzyme inhibitors, angiotensin receptor blockers, calcium channel blockers, and diuretics | Hypertensive patient | LC-MS | Metabolic profiles based on metabolic profiles comparing metabolic profiles between four antihypertensive drug groups and non-drug groups [107] |
| 44 | 2022 | Gefitinib | Patients with non-small cell carcinoma | LC-MS/MS | Identification of biomarkers inducing liver toxicity [108] |
| 45 | 2022 | Olanzapine | Rats | AFADESI-MSI | Identification of metabolites and drug-related treatments and adverse reactions [109] |
| Pharmacometabolomics related to biomarkers | |||||
| 1 | 2008 | Paracetamol | Mice | LC-MS | Identification of biomarkers related to toxic reactions [110] |
| 2 | 2008 | Cisplatinum | Lung cancer patients | GC-MS | Discovering new biomarkers related to cisplatin therapy [111] |
| 3 | 2008 | Polychlorinated biphenyls | Mohawk men and women | LC | Validation of biomarkers based on serum metabolic profiles [112] |
| 4 | 2011 | Citalopram and escitalopram | Patient with major depression | GC-MS | Discovering biomarkers for citalopram/escitalopram treatment [113] |
| 5 | 2012 | Rifampicin | Bacterial strains | GC-MS | Comparing the fatty acid metabolites of two strains [114] |
| 6 | 2013 | AD subjects, mild cognitive impairment, and control | LCECA | Identify functionally relevant alterations in metabolic networks and pathways in AD [115] | |
| 7 | 2013 | Hepatitis B virus patients | UPLC-Q-TOF-HDMS | Identifying urine biomarkers for HBV [116] | |
| 8 | 2013 | Sparfloxacin | hamsters | LC-MS/MS | Predicting metabolic changes directly related to physiological or pathological functions and drug toxicity [117] |
| 9 | 2013 | Aspirin | Healthy human volunteers | GC-MS | Analyze serum samples from good and poor responders to aspirin for changes in metabolite levels [118] |
| 10 | 2014 | Acetaminophen | Children treated with Acetaminophen | LC-MS | Studying the association between APAP induced hepatotoxicity and long-chain acylcarnitine in children with APAP toxicity [119] |
| 11 | 2014 | Ketamine | People with bi-directional depression | LC-QTOF-MS | A metabolomic approach to identify potential markers of ketamine response and non-response [120] |
| 12 | 2015 | Acamprosate | Patients with alcohol use disorders | LC-MS | Discovering an increase in baseline serum glutamate levels as a potential biomarker associated with a positive reaction to akanic acid [121] |
| 13 | 2015 | Olesoxime | Patients with Amyotrophic Lateral Sclerosis | HPLC-MS/MS | Detection of metabolomic profiles of patients treated with Olesoxime and placebo and prediction modeling [122] |
| 14 | 2016 | Busulfan | Patients treated with Busulfan | LC-MS | Identification of Potential Other Metabolites Predicts Intravenous Leucovorin Clearance in HCT Subjects [123] |
| 15 | 2016 | Trastuzumab and Paclitaxel | Patients treated with trastuzumab-paclitaxel | LC-MS | Identification of biomarkers associated with trastuzumab paclitaxel therapy [124] |
| 16 | 2016 | Atenolol | Hypertensive patient | GC-MS | Identification of biomarkers related to glucose changes after atenolol treatment [125] |
| 17 | 2016 | Busulfan | Allogeneic hematopoietic cell transplant recipients | LC-MS | Identification of biomarkers predictive of leucovorin clearance by targeted drug metabolomics [52] |
| 18 | 2016 | Patients with liver cancer | GC–MS | Prognostic biomarkers for identifying clinical outcomes in lung cancer patients [126] | |
| 19 | 2017 | Glimepiride | Healthy human volunteers | LC-MS/MS | Identification of endogenous metabolites affected by glimepiride administration [127] |
| 20 | 2018 | Clopidogrel | Patients with coronary artery disease | 1H NMR | Identifying metabolic phenotypes associated with clopidogrel blood and identify relevant biomarkers [128] |
| 21 | 2019 | Tuberculosis patient | GCxGC-TOFMS | Determining the changes in human urine metabolome induced by TB treatment and the extent of treatment [129] | |
| 22 | 2019 | Glucosamine antimonate | Patients with cutaneous leishmaniasis | LC-MS | Prediction and prognostic candidate biomarkers for determining the treatment outcome of meglumine antimoniate [130] |
| 23 | 2020 | Olanzapine | Mice | LC-MS | Identifying metabolites biomarkers in plasma associated with AP induced overeating and weight gain [131] |
| 24 | 2020 | Gemcitabine | Mice | LC-MS | Metabolites of potential biomarkers to identify the efficacy of gemcitabine in patients with pancreatic cancer [132] |
| 25 | 2020 | Warfarin | Patients treated with warfarin | NMR | Predicting INR based reactions in patients receiving warfarin treatment [133] |
| 26 | 2020 | Irinotecan | Cancer patients | LC-MS/MS | Detection of related metabolic changes for predicting the efficacy or toxicity of irinotecan [134] |
| 27 | 2021 | Busulfan | Patients receiving HCT conditioning with Busulfan | LC-MS | Identification of biomarkers related to HCT patients [135] |
| 28 | 2021 | Patients with high-density diffuse peritoneal carcinomatosis | LC-MS | Detection of metabolites associated with the propensity of cancer patients to experience oxidative stress and develop infections [136] | |
| 29 | 2021 | Inhaled corticosteroids | Patient with asthma | UPLC-MS/MS | Evaluate plasma metabolomics indicators of inhaled corticosteroids to determine relevant metabolites [137] |
| 30 | 2021 | Gefitinib | Patient with rash victim | HPLC/MS-MS | Development of a predictive model for gefitinib-induced rash and validation of the model [138] |
| 31 | 2022 | Adriamycin | Mice | LC-MS | Identification of urinary biomarkers associated with sensitivity or resistance to doxorubicin [139] |
| 32 | 2022 | Tacrolimus | Kidney transplant patients | UPLC/Q-TOF-MS | Identifying relevant metabolites as biomarkers [140] |
| 33 | 2023 | Medical cannabis | Children with autism spectrum disorders | CE-TOF-MS, RRLC-TOF-MS | Determining corresponding cannabis reactivity biomarkers [141] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jian, J.; He, D.; Gao, S.; Tao, X.; Dong, X. Pharmacokinetics in Pharmacometabolomics: Towards Personalized Medication. Pharmaceuticals 2023, 16, 1568. https://doi.org/10.3390/ph16111568
Jian J, He D, Gao S, Tao X, Dong X. Pharmacokinetics in Pharmacometabolomics: Towards Personalized Medication. Pharmaceuticals. 2023; 16(11):1568. https://doi.org/10.3390/ph16111568
Chicago/Turabian StyleJian, Jingai, Donglin He, Songyan Gao, Xia Tao, and Xin Dong. 2023. "Pharmacokinetics in Pharmacometabolomics: Towards Personalized Medication" Pharmaceuticals 16, no. 11: 1568. https://doi.org/10.3390/ph16111568
APA StyleJian, J., He, D., Gao, S., Tao, X., & Dong, X. (2023). Pharmacokinetics in Pharmacometabolomics: Towards Personalized Medication. Pharmaceuticals, 16(11), 1568. https://doi.org/10.3390/ph16111568







