Novel Benzimidazole Derived Imine Ligand and Its Co(III) and Cu(II) Complexes as Anticancer Agents: Chemical Synthesis, DFT Studies, In Vitro and In Vivo Biological Investigations
Abstract
1. Introduction
2. Experimental
2.1. Materials and Methods
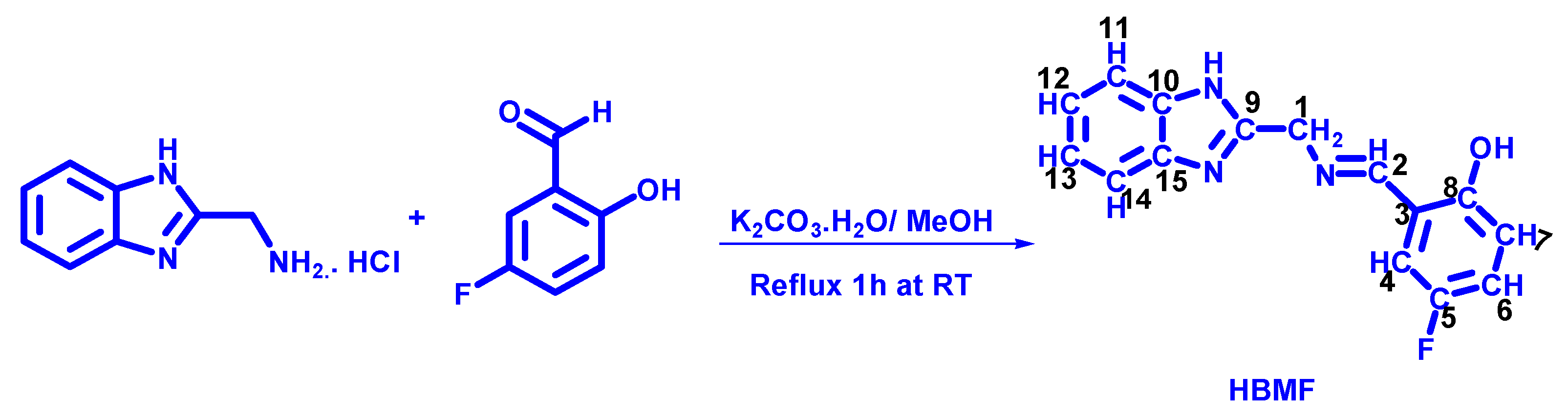
2.2. Preparation of 2-((E)-((1H-benzo[d]-2-yl)Methylimino)Methyl)-4-Fluoropphenol (HBMF)
2.3. Synthesis of Co(III) and Cu(II) Complexes in the Ratio 1:2 (C1 and C3)
2.4. Synthesis of Co(III) and Cu(II) Complexes in the Ratio 1:1:1 (C2 and C4)
2.5. Pharmacology
2.5.1. Cell Culture and In Vitro Treatment
2.5.2. MTT Assay
2.5.3. Animal Models and Ethics
2.5.4. Hematological Studies
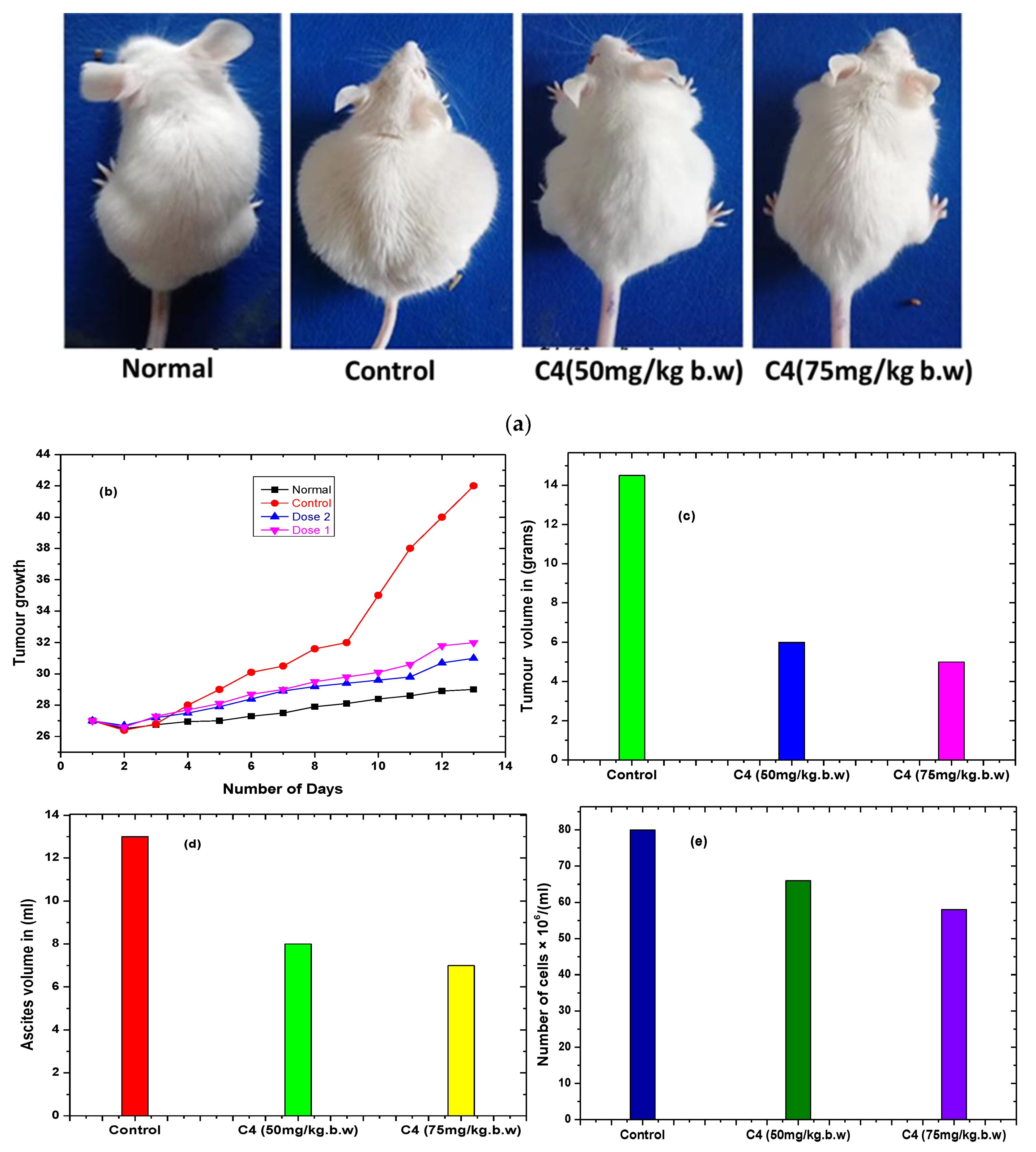
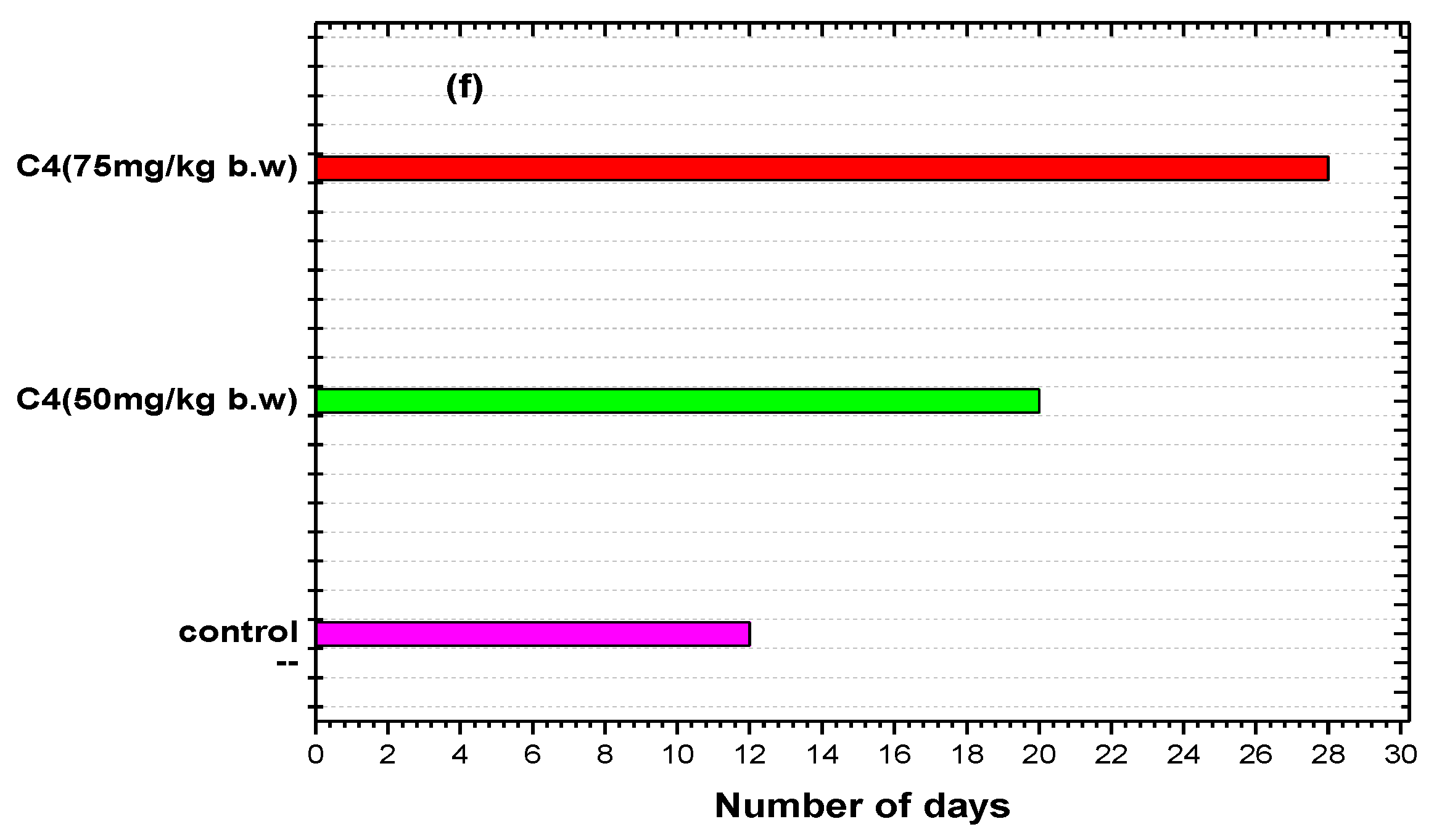
2.5.5. Animal Tumor Models and Treatment
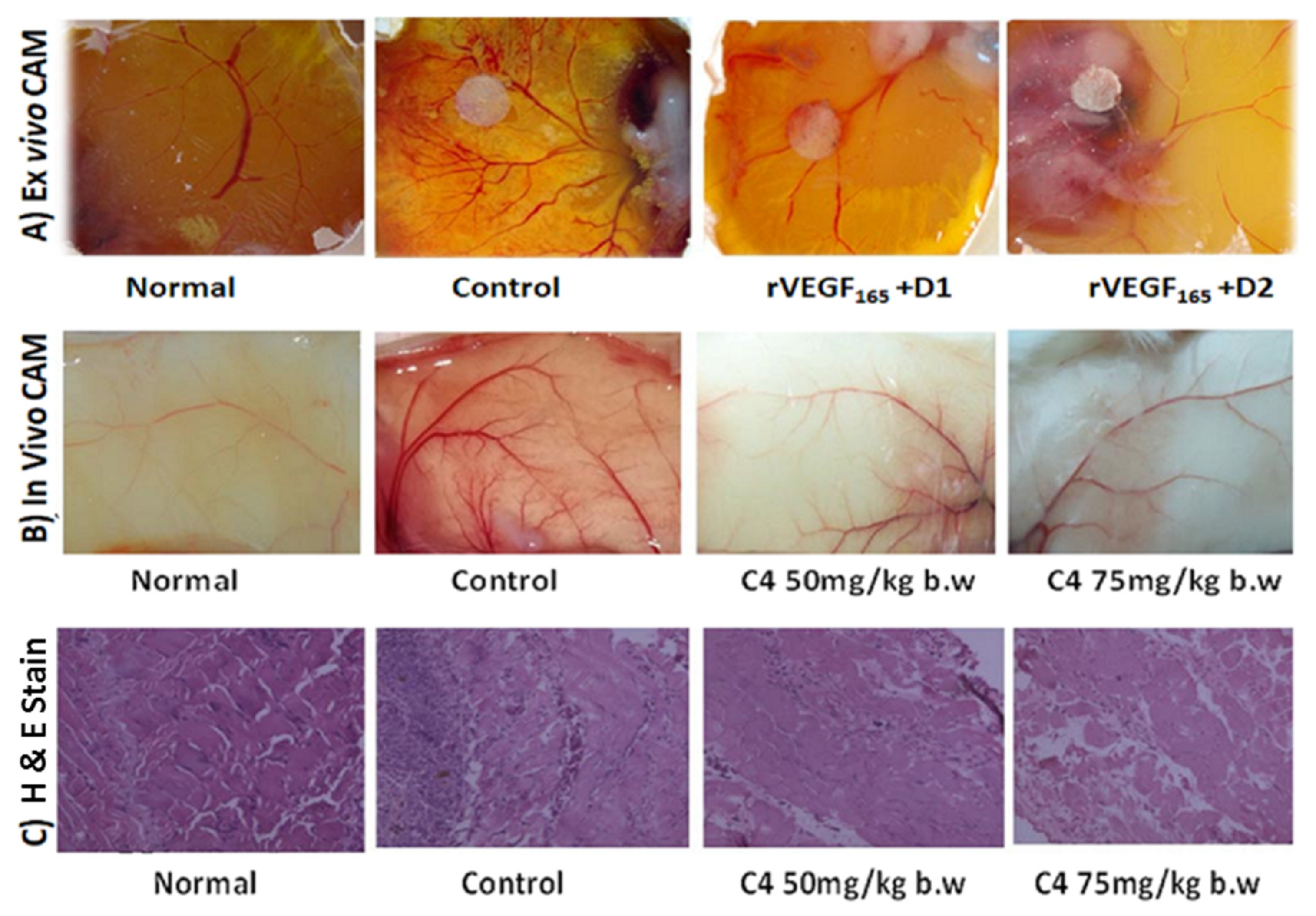
2.5.6. Chorioallontoic Membrane (CAM) Assay
2.5.7. Measure of Micro-Vessel Density (MVD)
2.6. Computational Details
3. Results and Discussion
3.1. Chemistry
3.1.1. Electronic Spectral Analysis
3.1.2. IR Spectral Studies
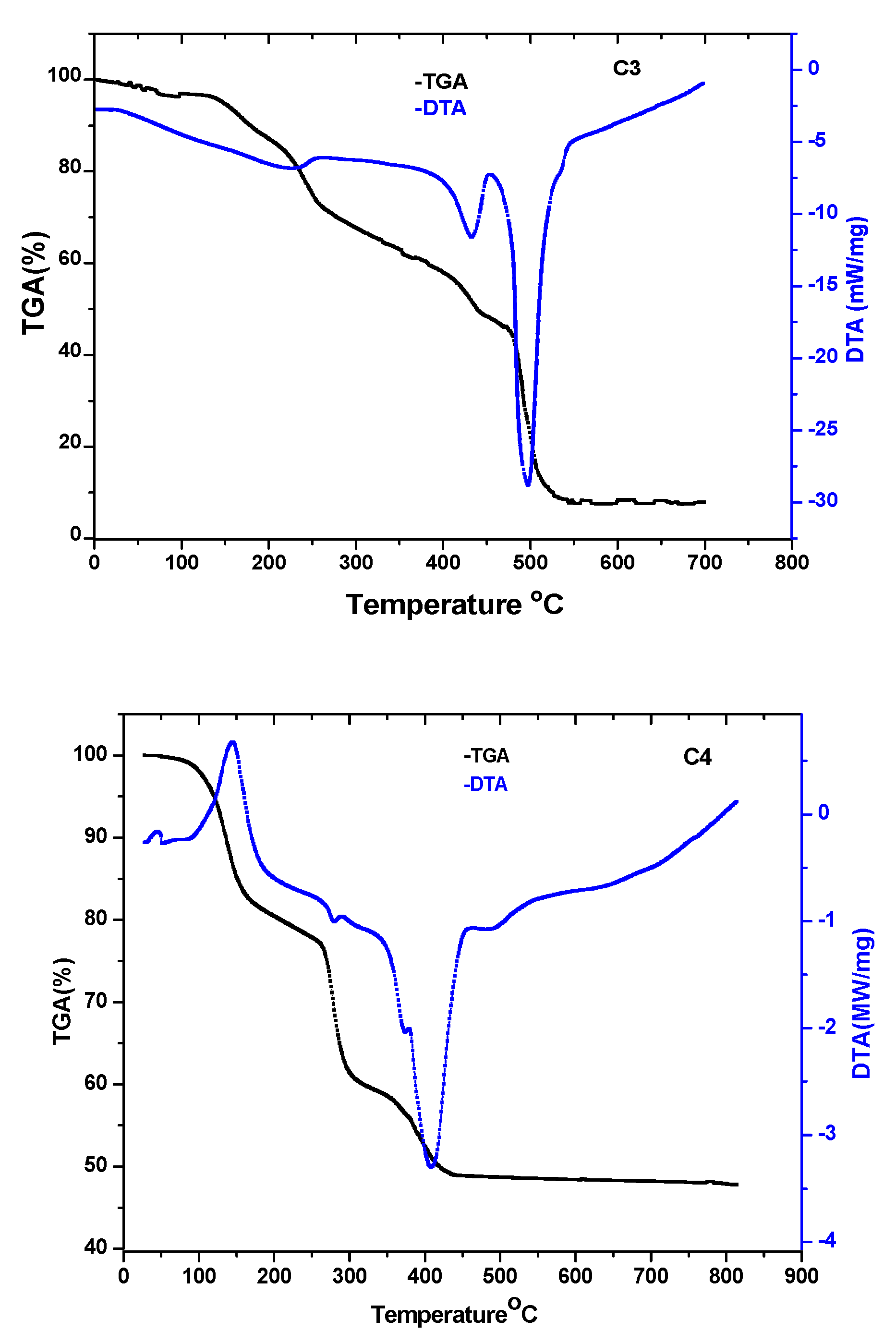
3.1.3. Thermogravimetric Analysis
3.1.4. Measurements of Molar Conductance and Magnetic Moment
3.1.5. Mass Spectra
3.1.6. ESR Spectra
3.2. Biology
3.2.1. Pharmacology
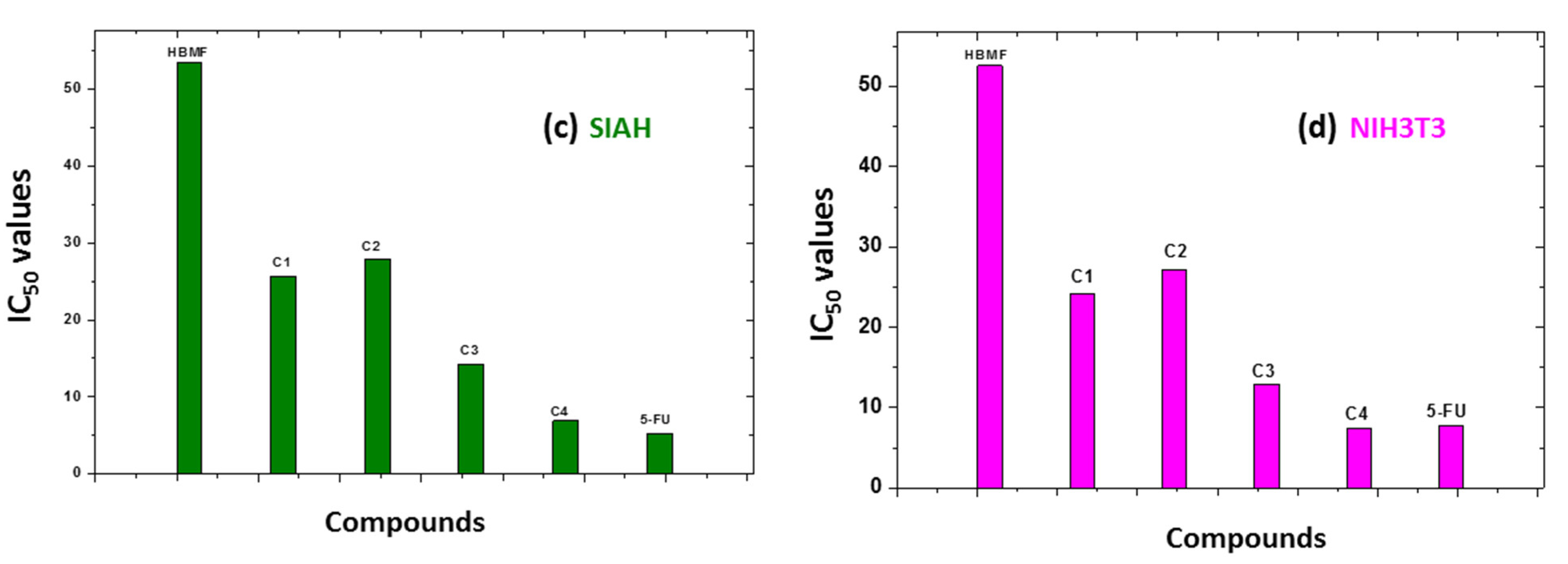
Cytotoxicity and Structural-Activity Relationship
Hematological and Serum Profile Parameters of Cobalt Complex
Complex C4 Inhibits EAC Tumor Cell Growth
Angiogenesis
3.3. Computational Results
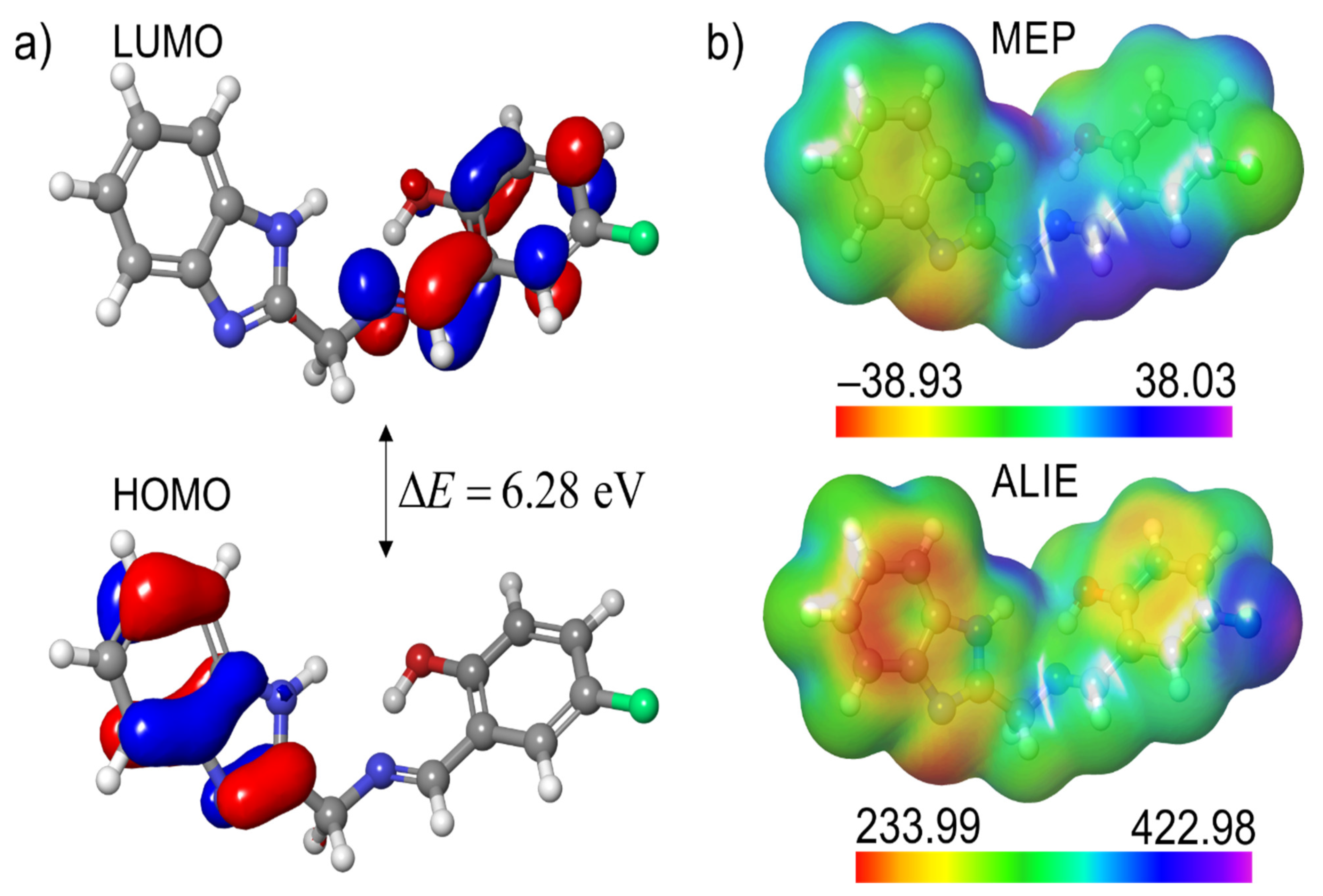
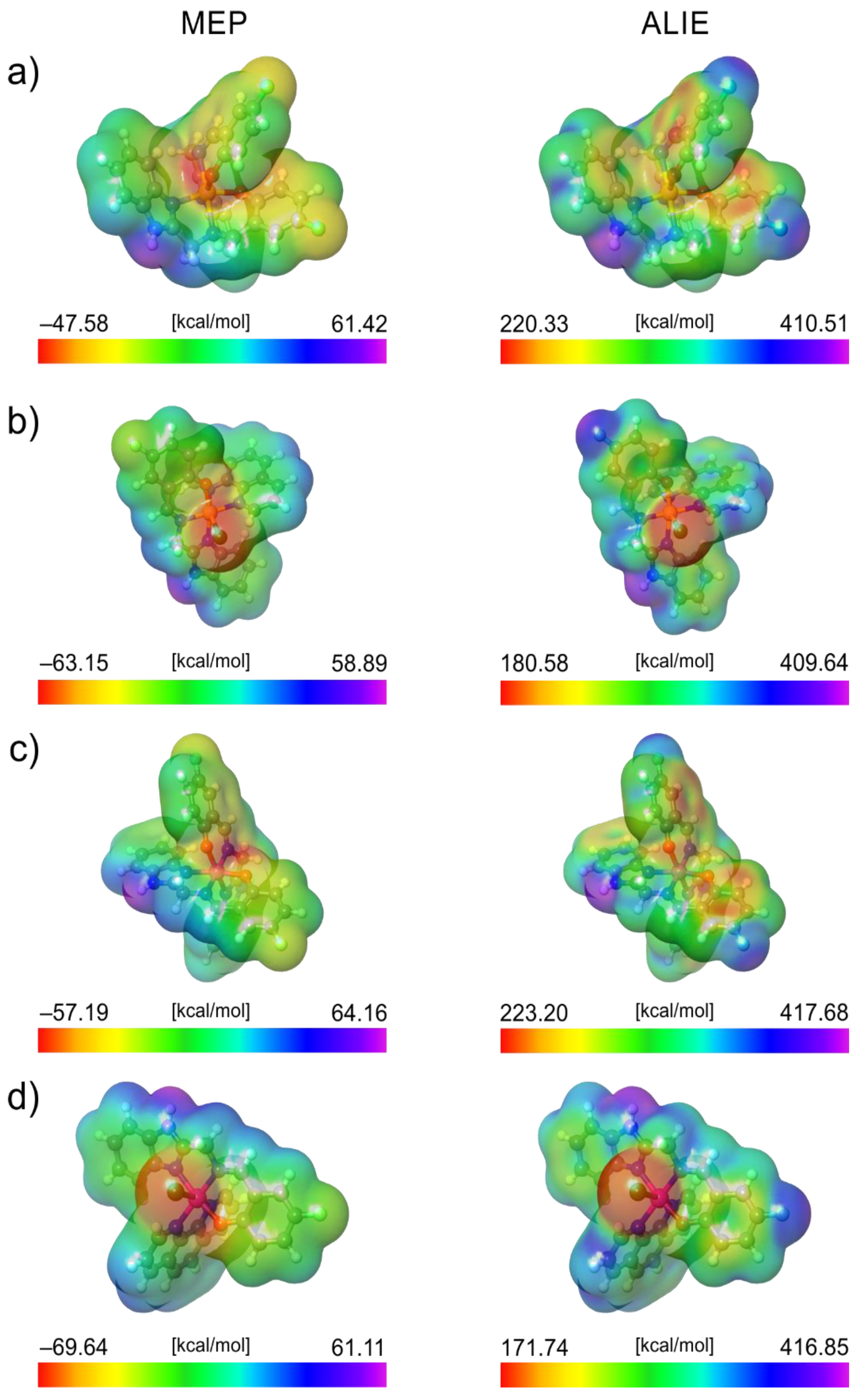
3.3.1. Local Reactivity of Ligand
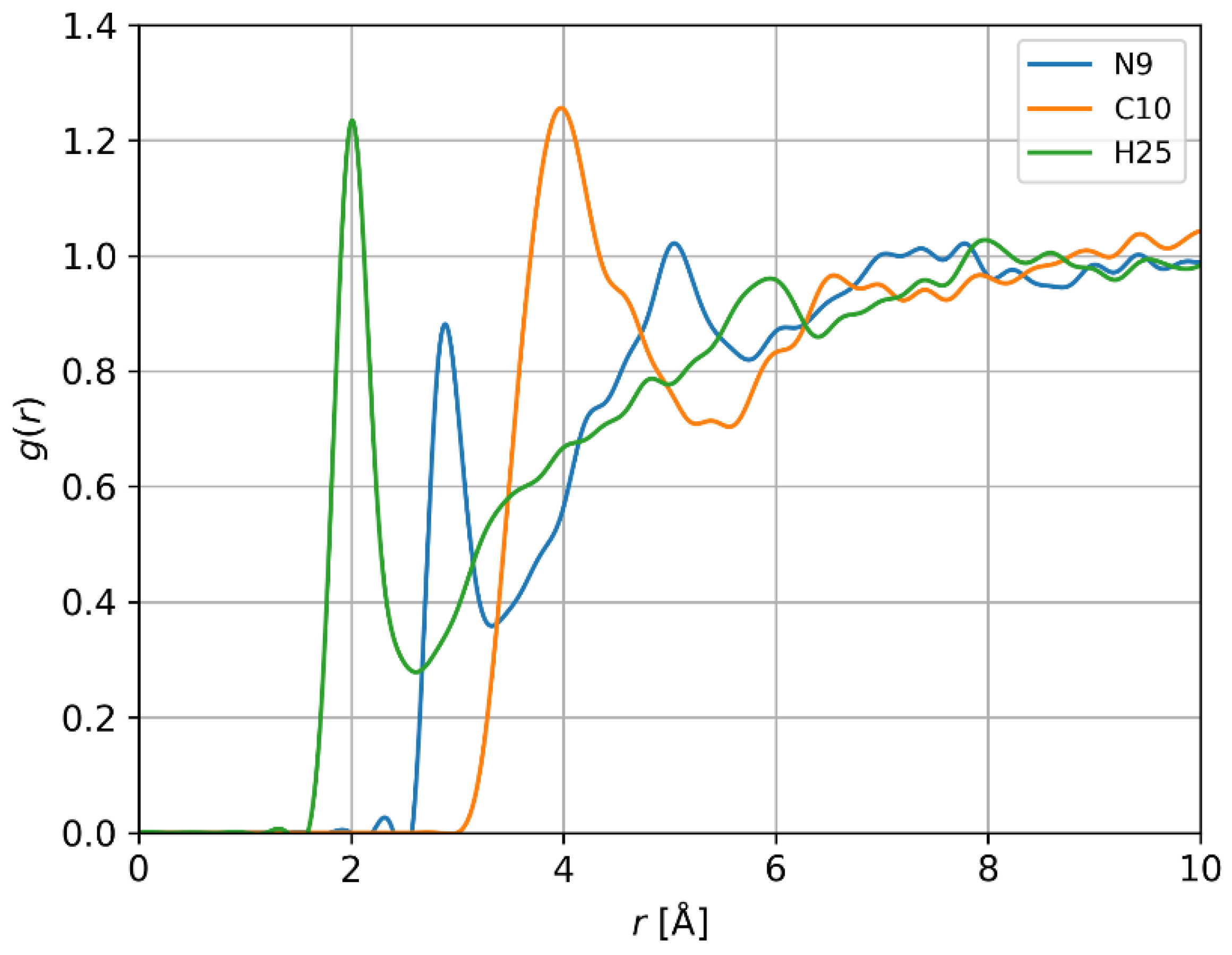
3.3.2. Interactions of HBMF with Water
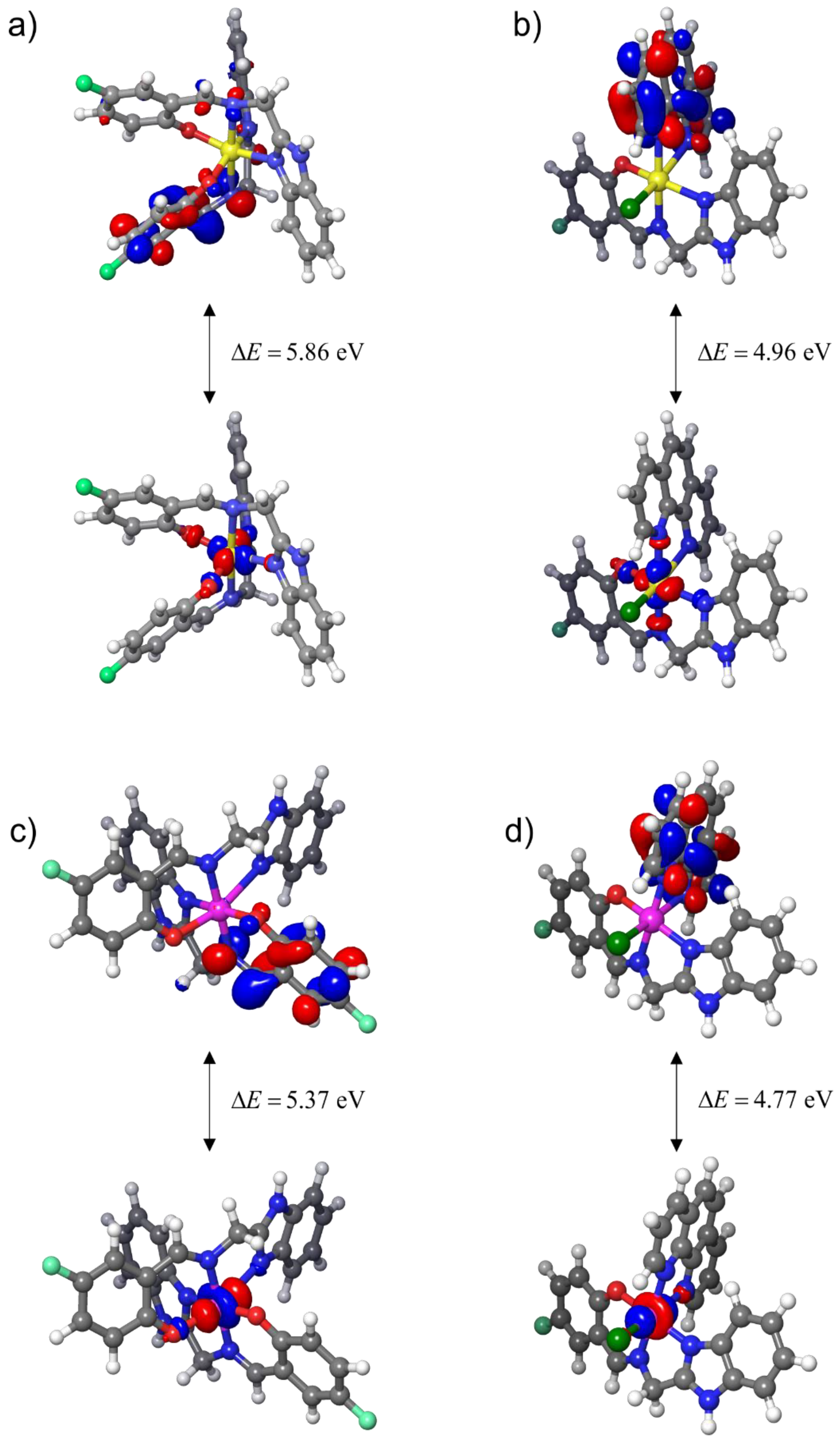
3.3.3. Local Reactivity of Complexes
3.3.4. Simulation of UV Spectra
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ndagi, U.; Mhlongo, N.; Soliman, E. Metal complexes in cancer therapy an update from drug design perspective. Drug Des. Dev. Ther. 2017, 11, 599–616. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Tchounwon, P. Cisplatin in cancer therapy: Molecular mechanism of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Alam, R.; Wahi, D.; Singh, R.; Sinha, D.; Tandon, V.; Grover, A.; Rahisuddin. Design, synthesis, cytotoxicity, HuTopoIIαinbitory activity and molecular docking studies of pyrazolederivatives as potential anticancer agents. Bioorg. Chem. 2016, 69, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Vasamthakumar, B.C.; Lohith, G.K.; Revanasiddappa, H.D. Co(III) and Cu(II) Benzimidazole base complexes: Synthesis, Characterization and DNA binding and cleavage activity. J. Chem. Biol. Phys. Sci. 2017, 7, 882–895. [Google Scholar]
- Song, W.-J.; Cheng, J.-P.; Jiang, D.-H.; Guo, L.; Cai, M.-F.; Yang, H.-B.; Lin, Q.-Y. Synthesis, interaction with DNAand antiproliferative activities of two novel Cu(II) complexes with Schiff base of benzimidazole. Spectrochim. A Mol. Biomol. Spectrosc. 2014, 121, 70–76. [Google Scholar] [CrossRef]
- Sunitha, M.; Jogi, P.; Ushaiah, B.; Gayanakumari, C. Synthesis, Characterization and Antimicrobial activity of transition metal complexes of Schiff base ligand derived from 3- ethoxy salicylaldehyde and 2-(2-aminophenyl) 1-H-benzimidazole. E-J. Chem. 2012, 9, 2516–2523. [Google Scholar] [CrossRef]
- Savithri, K.; Vasantha kumar, B.V.; Vivek, H.K.; Revanasiddappa, H.D. Synthesis and Characterization of Cobalt(III) and Copper(II) Complexes of 2-((E)-(6-Fluorobenzo[d]thiazol-2-ylimino) methyl)-4-chlorophenol: DNA Binding and Nuclease Studies—SOD and Antimicrobial Activities. Int. J. Spectrosc. 2018, 2018, 8759372. [Google Scholar] [CrossRef]
- Esteghamat-Panah, R.; Farrokhpour, H.; Hadadzadeh, H.; Abyar, F.; Rudbari, H.A. An experimental and quantum chemical study on the non-covalent interactions of a cyclometallated Rh(III) complex with DNA and BSA. RSC Adv. 2016, 6, 23913–23929. [Google Scholar] [CrossRef]
- Rajarajeswari, C.; Loganathan, R.; Palaniandavar, M.; Suresh, E.; Riyasdeend, A.; Abdulkadhar, M. Copper(II) complexes with 2NO and 3N donor ligands: Synthesis, structures and chemical nuclease and anticancer activities. Dalton Trans. 2013, 42, 8347–8363. [Google Scholar] [CrossRef]
- Wright, J.B. Structure and antioxidant superoxide dismutase. Chem. Rev. 1951, 48, 397–541. [Google Scholar] [CrossRef]
- Paularokiadoss, F.T.; Christopher Jeyakumar, R.; Thomas, A.; Sekar, D.; Bhakiaraj. Group 13 monohalides [AX (A = B, Al, Ga and In; X = Halogens)] as alternative ligands for carbonyl in organometallics: Electronic structure and bonding analysis. Comput. Theor. Chem. 2022, 1209, 113587. [Google Scholar] [CrossRef]
- Stanojev, J.S.; Armakovic, B.; Bajac, J.; Matovic, V.; Srdic, V. PbSe sensitized with iodine and oxygen: A combined computational and experimental study. J. Alloys Compd. 2022, 896, 163119. [Google Scholar] [CrossRef]
- Al-Otaibi, J.S.; Mary, Y.S.; Ullah, Z.; Kwon, H.W. Adsorption behavior and solvent effects of an adamantane-triazole derivative on metal clusters—DFT simulation studies. J. Mol. Liq. 2022, 345, 118242. [Google Scholar] [CrossRef]
- Reichert, T.; Vučićević, M.; Hillman, P.; Bleicher, M.; Armaković, S.S.; Armaković, J. Sumanene as a delivery system for 5-fluorouracil drug—DFT, SAPT and MD study. J. Mol. Liq. 2021, 342, 117526. [Google Scholar] [CrossRef]
- Suntharalingam, K.; Mendoza, O.; Duarte, A.A.; Mann, D.J.; Vilar, R. A platinum complex that binds noncovalently to DNA and induces cell death via a different mechanism than cisplatin. Metallomics 2013, 5, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Kumaravel, G.; Ponya Utthra, P.; Raman, N. Exploiting the biological efficacy of benzimidazole based Schiff base complexes with l-Histidine as a co-ligand: Combined molecular docking, DNA interaction, antimicrobial and cytotoxic studies. Bioorg. Chem. 2018, 77, 269–279. [Google Scholar] [CrossRef]
- Schildkraut, C.L.; Marmur, J.; Doty, P. The formation of hybrid DNA molecules and their use in studies of DNA homologies. J. Mol. Biol. 1961, 3, 595–617. [Google Scholar] [CrossRef]
- Krishnamurthy, G.; Shashikal, J. Complexes of zinc(II) with 1,2-disubstituted benzimidazoles. J. Chem. Res. 2006, 14, 766–768. [Google Scholar] [CrossRef]
- Prabhakar, B.T.; Ara Khanum, S.; Jayashree, K.; Bharathi, P.; Shashikanthc, S. Anti-tumor and proapoptotic effect of novel synthetic benzophenone analogues in Ehrlich ascites tumor cells. Bioorg. Med. Chem. 2006, 14, 435–446. [Google Scholar] [CrossRef]
- Danihelová, M.; Veverka, M.; Sturdík, E.; Jantová, S. Antioxidant action and cytotoxicity on HeLa and NIH-3T3 cells of new quercetin derivatives. Toxicology 2013, 6, 209–216. [Google Scholar] [CrossRef]
- MacroModel Schrödinger Release 2022-1: MacroModel; Schrödinger LLC: New York, NY, USA, 2022.
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Rassolov, V.A.; Ratner, M.A.; Pople, J.A.; Redfern, P.C.; Curtiss, L.A. 6-31G* basis set for third-row atoms. J. Comput. Chem. 2001, 22, 976–984. [Google Scholar] [CrossRef]
- Bannwarth, C.E.; Caldeweyher, S.; Ehlert, A.; Hansen, P.; Pracht, J.; Seibert, S.; Spicher, S.; Grimme, E. Extended tight-binding quantum chemistry methods, Wiley Interdiscip. Rev. Comput. Mol. Sci. 2021, 11, e1493. [Google Scholar]
- Armaković, S.; Armaković, S.J. Atomistica.online—Web application for generating input files for ORCA molecular modelling package made with the Anvil platform. Mol. Simul. 2022, 49, 117–123. [Google Scholar] [CrossRef]
- Thompson, J.D.; Winget, P.; Truhlar, D.G. MIDIX basis set for the lithium atom: Accurate geometries and atomic partial charges for lithium compounds with minimal computational cost. Phys. Chem. Commun. 2001, 4, 72–77. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other function. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar] [CrossRef]
- Hay, P.J.; Wadt, W.R. Ab initio effective core potentials for molecular calculations. Potentials for K to Au including the outermost core orbitals. J. Chem. Phys. 1985, 82, 299. [Google Scholar] [CrossRef]
- Yanai, T.; Tew, D.P.; Handy, N.C. A new hybrid exchange–correlation functional using the Coulomb-attenuating method (CAM-B3LYP). Chem. Phys. Lett. 2004, 393, 51–57. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermolecular Forces; Springer: Berlin/Heidelberg, Germany, 1981; pp. 331–342. [Google Scholar]
- Bochevarov, A.D.; Harder, E.; Hughes, T.F.; Greenwood, J.R.; Braden, D.A.; Philipp, D.M.; Rinaldo, D.; Halls, M.D.; Zhang, J.; Friesner, R.A. Jaguar: A high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 2013, 113, 2110–2142. [Google Scholar] [CrossRef]
- Jacobson, L.D.; Bochevarov, A.D.; Watson, M.A.; Hughes, T.F.; Rinaldo, D.; Ehrlich, S.; Steinbrecher, T.B.; Vaitheeswaran, S.; Philipp, D.M.; Halls, M.D. Automated transition state search and its application to diverse types of organic reactions. J. Chem. Theory Comput. 2017, 13, 5780–5797. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2021-4: Desmond Molecular Dynamics System; D E. Shaw Research Maestro-Desmond Interoperability Tools; Schrödinger: New York, NY, USA, 2021.
- Prakasha, G.; Shiva Prasad, K.; Syed, I.; Revanasiddappa, H.D. Novel Benzimidazole Derived Imine-Based Ligand and its Co(III), Ni(II), Cu(II) and Pt(II) Complexes: Chemical Synthesis, Structure, Antimicrobial, DNA Interaction Studies and Nuclease Activity. Lett. Appl. Nanobiosci. 2020, 9, 1655–1672. [Google Scholar]
- Vijay Avin, B.R.; Thirusangu, P.; Lakshmi Ranganatha, V.; Firdouse, A.; Prabhakar, B.T.; Ara Khanum, S. Synthesis and tumor inhibitory activity of novel coumarin analogs targeting angiogenesis and apoptosis. Eur. J. Med. Chem. 2014, 75, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Weidner, N.; Semple, J.P.; Welch, W.R.; Folkman, J. Tumor Angiogenesis and Metastasis—Correlation in Invasive Breast Carcinoma. N. Engl. J. Med. 1991, 324, 1–8. [Google Scholar] [CrossRef]
- Singh, Y.P.; Patel, R.N.; Singh, Y.; Butcher, R.J.; Vishakarma, P.K. Structure and antioxidant superoxide dismutase activity of copper(II) hydrazone complex. Polyhedron 2017, 122, 1–15. [Google Scholar] [CrossRef]
- Ommenya, F.K.; Nyawade, E.A.; Andala, D.M.; Kinyua, J. Synthesis, Characterization and Antibacterial Activity of Schiff Base, 4-Chloro-2-{(E)-[(4-Fluorophenyl)imino]methyl}phenol Metal (II) Complexes. J. Chem. 2020, 2020, 1745236. [Google Scholar] [CrossRef]
- Hussain, A.; Al Ajmi, F.M.; Rehman, T.; Amir, S.; Husain, M.; Alsalme, A.; Siddiqui, A.; Khan, A. Copper(II) complexes as potential anticancer and Nonsteroidal antiinfammatory agents: In vitro and in vivo studies. Sci. Rep. 2019, 9, 5237. [Google Scholar]
- Chauhan, M.; Banerjee, K.; Arjmand, F. DNA Binding Studies of Novel Copper(II) Complexes Containing l-Tryptophan as Chiral Auxiliary: In Vitro Antitumor Activity of Cu−Sn2 Complex in Human Neuroblastoma Cells. Inorg. Chem. 2007, 46, 3072–3082. [Google Scholar] [CrossRef]
- Gurupadaswamy, H.D.; Girish, V.; Kavitha, C.V.; Raghavan, S.C.; Khanum, S.A. Synthesis and evaluation of 2,5-di(4-aryloylaryloxymethyl)- 1,3,4-oxadiazoles as anti-cancer agents. Eur. J. Med. Chem. 2013, 63, 536–543. [Google Scholar] [CrossRef]
- Fuh-An, Y.; Chih-Wei, G.; Yao-Jung, C.; Jyh-Horung, C.; Shin-Shin, W.; Jo-Yu, T.; Lian-Pin, H.; Shanmugam, E. ESR, zero-field splitting, and magnetic exchange of exchange-coupled copper(II)-copper(II) pairs in copper(II) tetraphenylporphyrin N-oxide. Inorg. Chem. 2007, 46, 578–585. [Google Scholar]
- Thamilarasan, V.; Sengottuvelan, N.; Sudha, A.; Srinivasan, P.; Chakkaravarthi, G. Cobalt(III) complexes as potential anticancer agents: Physicochemical, structural, cytotoxic activity and DNA/protein interactions. Photochem. Photobiol. 2016, 162, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Mazumder, U.K.; Kumar, R.S.; Sivakumar, T.; Vamsi, M.L.M. Antitumor Activity and Antioxidant Status of Caesalpinia bonducella Against Ehrlich Ascites Carcinoma in Swiss Albino Mice. J. Pharmacol. Sci. 2004, 94, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Staton, C.A.; Reed, M.W.R.; Brown, N.J. A critical analysis of current in vitro and in vivo angiogenesis assays. Int. J. Exp. Pathol. 2008, 90, 195–221. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Folkman, J. Angiogenesis: An organizing principle for drug discovery. Nat. Rev. Drug Discov. 2007, 6, 273–286. [Google Scholar] [CrossRef]
- Kumar, S.N.; Krishnamurthy, G.; Yadav, D.; Malojirao, V.H.; Naik, R.T.R.; Kandagallae, S.; Prabhakar, B.T. Synthesis, characterization and tumor inhibitory activity of a novel Pd(II) complex derived from methanethiol-bridged (2-((1H-benzo[d]imidazol2-yl)methylthio)-1H-benzo[d]imidazol6-yl)(phenyl)methanon. New J. Chem. 2019, 43, 790–806. [Google Scholar]
- Politzer, P.; Murray, J.S.; Bulat, F.A. Average local ionization energy: A review. J. Mol. Model. 2010, 16, 1731–1742. [Google Scholar] [CrossRef]
- Sjoberg, P.; Politzer, P. Use of the electrostatic potential at the molecular surface to interpret and predict nucleophilic processes. J. Phys. Chem. 1990, 94, 3959–3961. [Google Scholar] [CrossRef]
- Murray, J.S.; Politzer, P. The electrostatic potential: An overview. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2011, 1, 153–163. [Google Scholar] [CrossRef]
- Politzer, P.; Murray, J.S. The average local ionization energy: Concepts and applications. Theor. Comput. Chem. 2007, 7, 119–137. [Google Scholar]
- Grimme, S.; Bannwarth, C.; Shushkov, P. A Robust and Accurate Tight-Binding Quantum Chemical Method for Structures, Vibrational Frequencies, and Non-covalent Interactions of Large Molecular Systems Parametrized for All spd-Block Elements (Z = 1–86). J. Chem. Theory Comput. 2017, 13, 1989–2009. [Google Scholar] [CrossRef] [PubMed]
- Bannwarth, C.; Caldeweyher, E.; Ehlert, S.; Hansen, A.; Pracht, P.; Seibert, J.; Spicher, S.; Grimme, S. Extended tight-binding quantum chemistry methods, WIREs. Comput. Mol. Sci. 2021, 11, e1493. [Google Scholar] [CrossRef]
- Bannwarth, C.; Ehlert, S.; Grimme, S. GFN2-xTB—An Accurate and Broadly Parametrized Self-Consistent Tight-Binding Quantum Chemical Method with Multipole Electrostatics and Density-Dependent Dispersion Contributions. J. Chem. Theory Comput. 2019, 15, 1652–1671. [Google Scholar] [CrossRef] [PubMed]

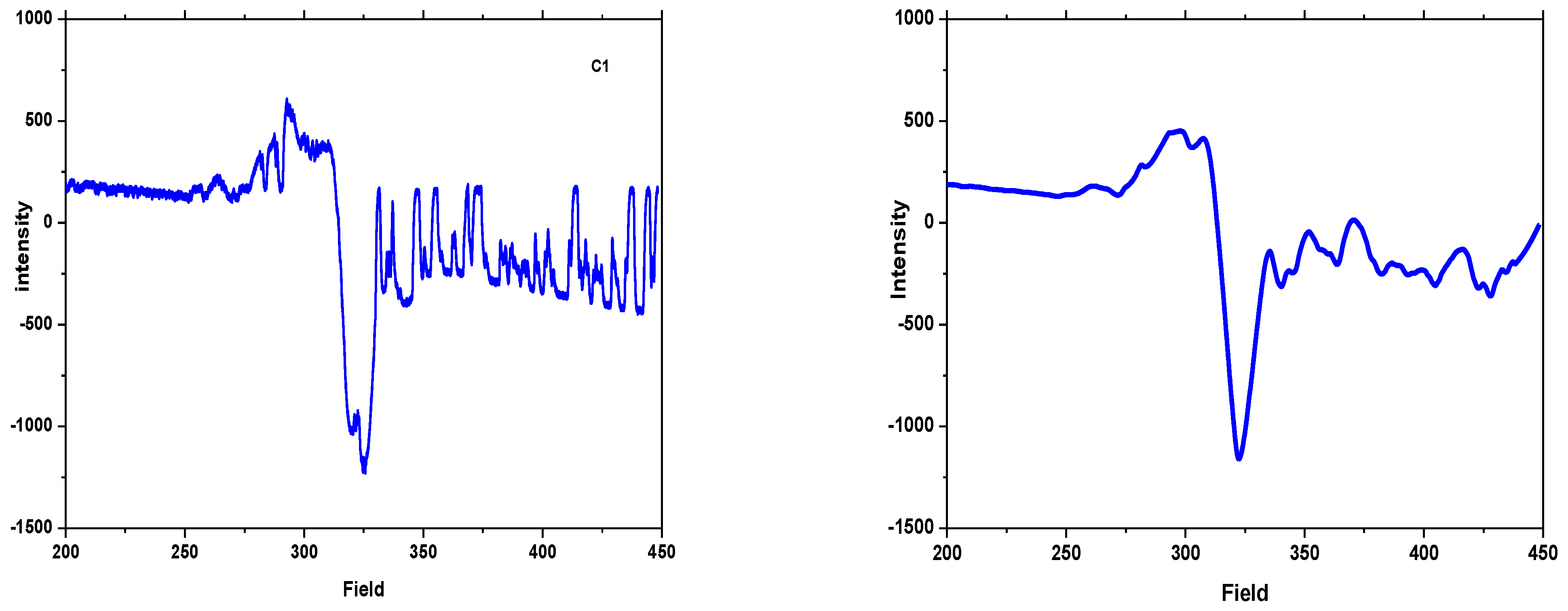


| Complexes | g⊥ | g|| | gavg | G | A‖ | α2 | f = g‖/A‖ |
|---|---|---|---|---|---|---|---|
| C1 | 2.0243 | 2.0708 | 2.0398 | 3.5822 | 154 | 0.72 | 134 |
| C2 | 2.0332 | 2.1591 | 2.0751 | 3.7510 | 171 | 0.67 | 126 |
| Hematological and Serum Profile Parameters | Normal Mice | Treated Mice |
|---|---|---|
| Alkaline Phosphatase (IU/L) | 136.78 ± 1.30 | 134.45 ± 2.1 |
| Creatinine (mg/dL) | 0.39 ± 1.3 | 0.43 ± 1.25 |
| Urea (mg/dL) | 48 ± 2.2 | 43 ± 2.3 |
| RBC (106/μL) | 5.7 ± 0.5 | 5.4 ± 0.6 |
| WBC (106/μL) | 3.56 ± 2.3 | 3.5 ± 2.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
G, P.; Revanasiddappa, H.D.; B, J.; T, P.B.; Shivamallu, C.; Viswanath, P.M.; Achar, R.R.; Silina, E.; Stupin, V.; Manturova, N.; et al. Novel Benzimidazole Derived Imine Ligand and Its Co(III) and Cu(II) Complexes as Anticancer Agents: Chemical Synthesis, DFT Studies, In Vitro and In Vivo Biological Investigations. Pharmaceuticals 2023, 16, 125. https://doi.org/10.3390/ph16010125
G P, Revanasiddappa HD, B J, T PB, Shivamallu C, Viswanath PM, Achar RR, Silina E, Stupin V, Manturova N, et al. Novel Benzimidazole Derived Imine Ligand and Its Co(III) and Cu(II) Complexes as Anticancer Agents: Chemical Synthesis, DFT Studies, In Vitro and In Vivo Biological Investigations. Pharmaceuticals. 2023; 16(1):125. https://doi.org/10.3390/ph16010125
Chicago/Turabian StyleG, Prakasha, H. D. Revanasiddappa, Jayalakshmi B, Prabhakar B. T, Chandan Shivamallu, Prashant M. Viswanath, Raghu Ram Achar, Ekaterina Silina, Victor Stupin, Natalia Manturova, and et al. 2023. "Novel Benzimidazole Derived Imine Ligand and Its Co(III) and Cu(II) Complexes as Anticancer Agents: Chemical Synthesis, DFT Studies, In Vitro and In Vivo Biological Investigations" Pharmaceuticals 16, no. 1: 125. https://doi.org/10.3390/ph16010125
APA StyleG, P., Revanasiddappa, H. D., B, J., T, P. B., Shivamallu, C., Viswanath, P. M., Achar, R. R., Silina, E., Stupin, V., Manturova, N., Shati, A. A., Alfaifi, M. Y., Elbehairi, S. E. I., Armaković, S. J., Armaković, S., & Kollur, S. P. (2023). Novel Benzimidazole Derived Imine Ligand and Its Co(III) and Cu(II) Complexes as Anticancer Agents: Chemical Synthesis, DFT Studies, In Vitro and In Vivo Biological Investigations. Pharmaceuticals, 16(1), 125. https://doi.org/10.3390/ph16010125









