Novel 2,6,9-Trisubstituted Purines as Potent CDK Inhibitors Alleviating Trastuzumab-Resistance of HER2-Positive Breast Cancers
Abstract
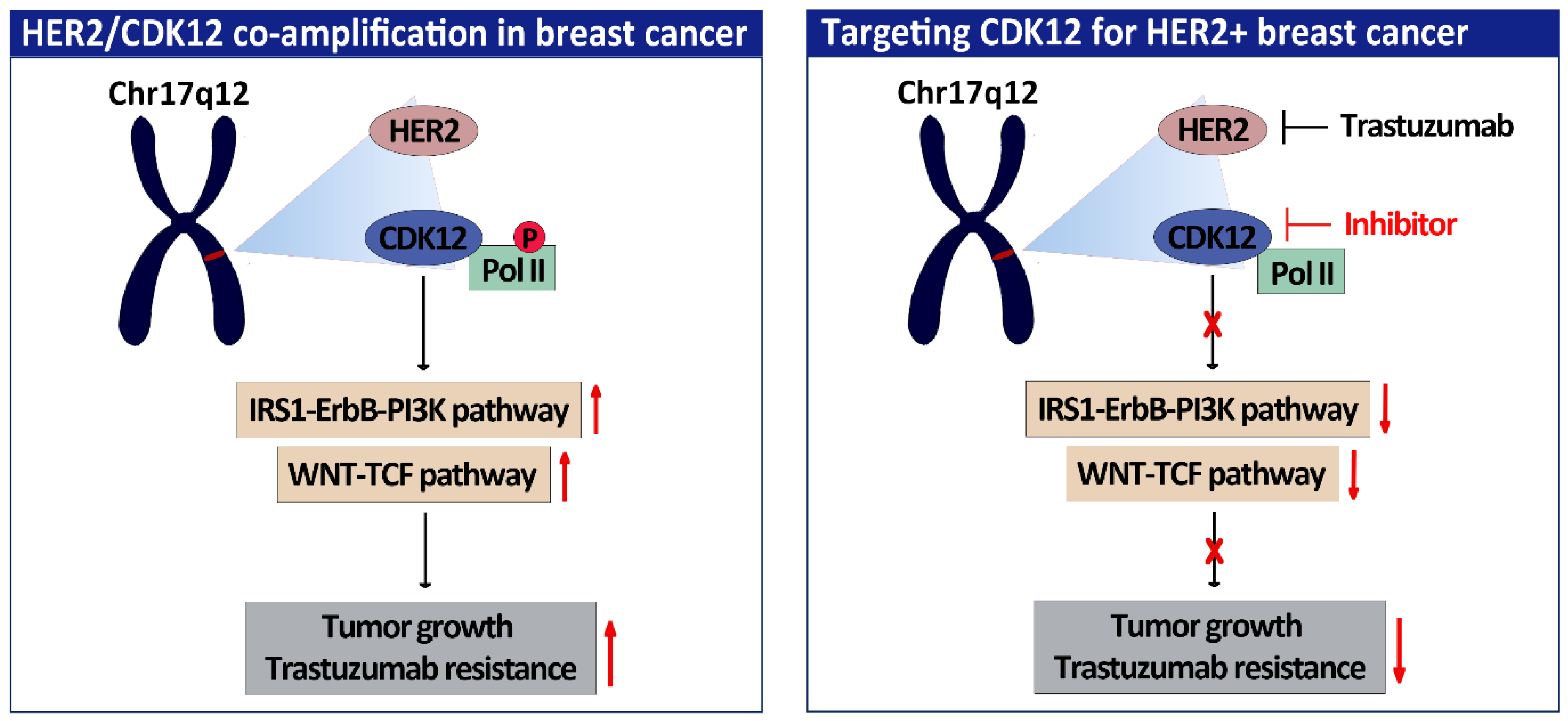
:1. Introduction
2. Results and Discussion
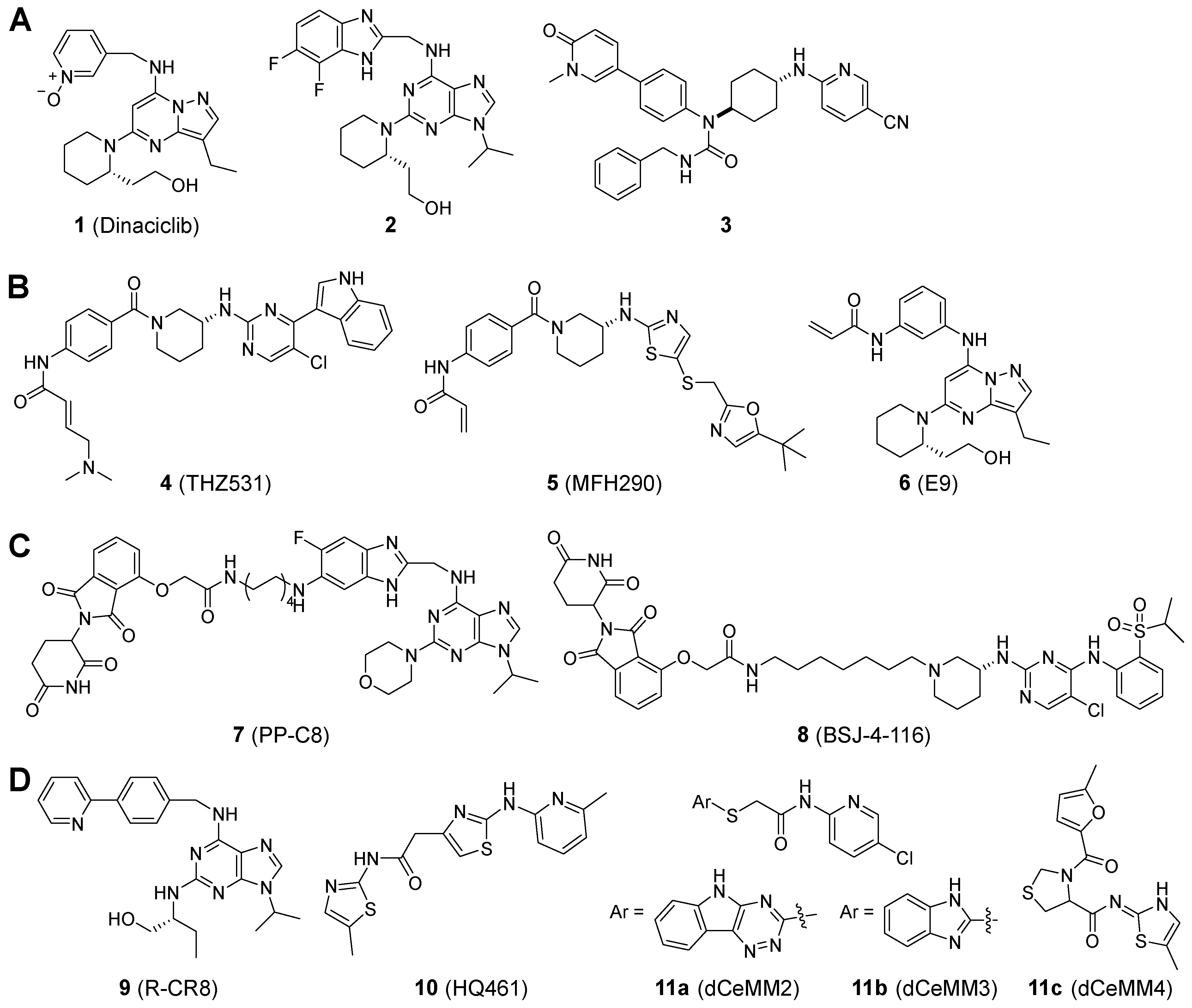
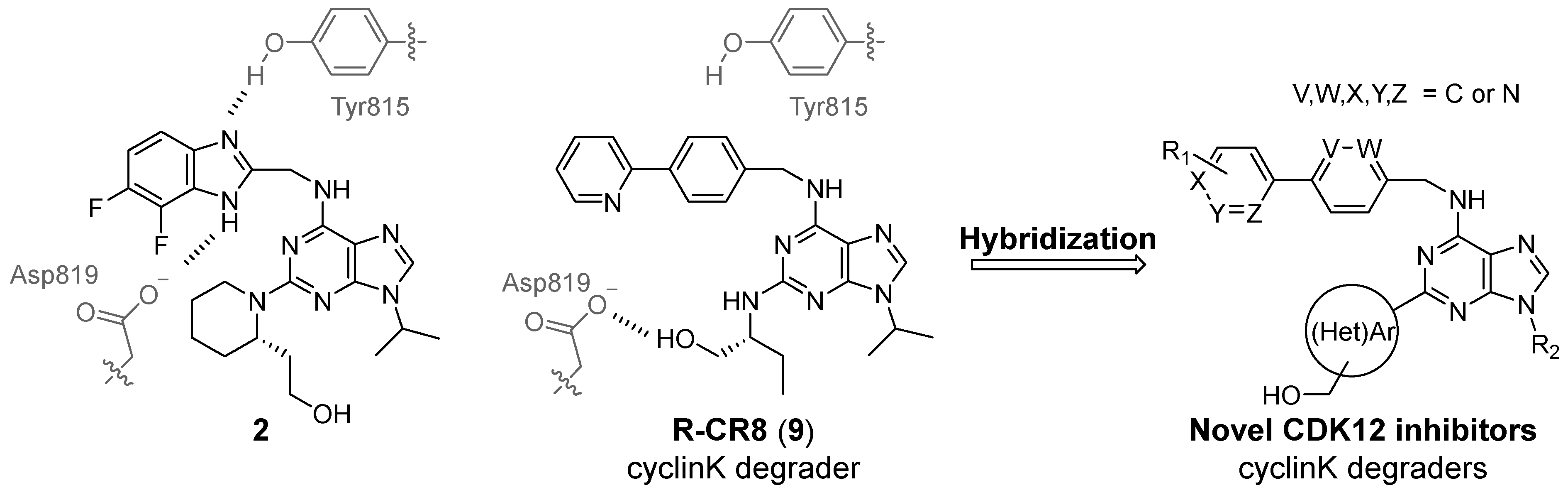
2.1. Inhibitor Design
2.2. Structure and Activity Relationship
2.3. Docking Analysis
2.4. In Vitro Kinome-Wide Inhibition Profiling of 30d
2.5. Liver Microsomal Stability and CYP Inhibition
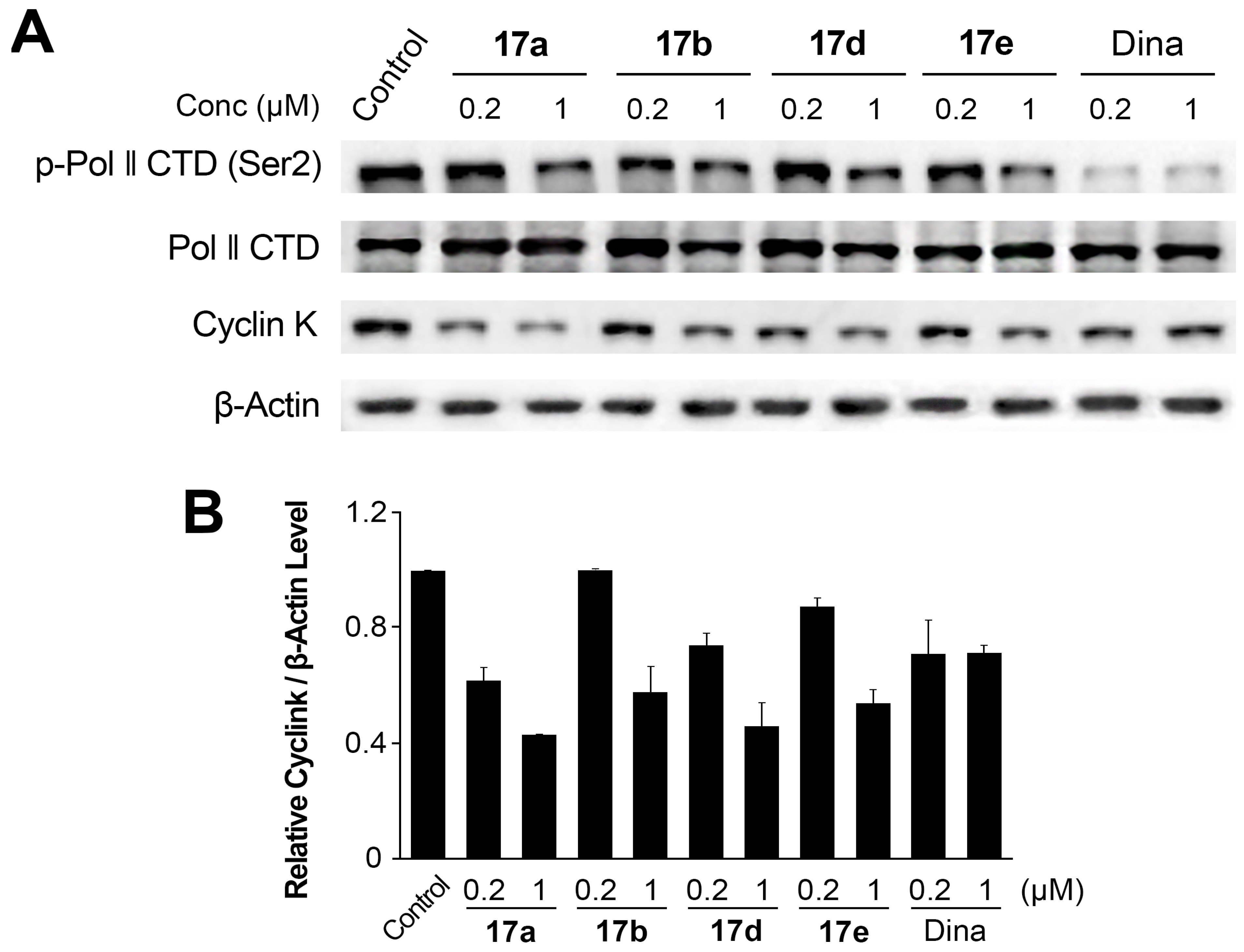
2.6. Suppression of Cyclink and PolII CTD Phosphorylation
2.7. Synergism between 30d and Trastuzumab
3. Materials and Methods
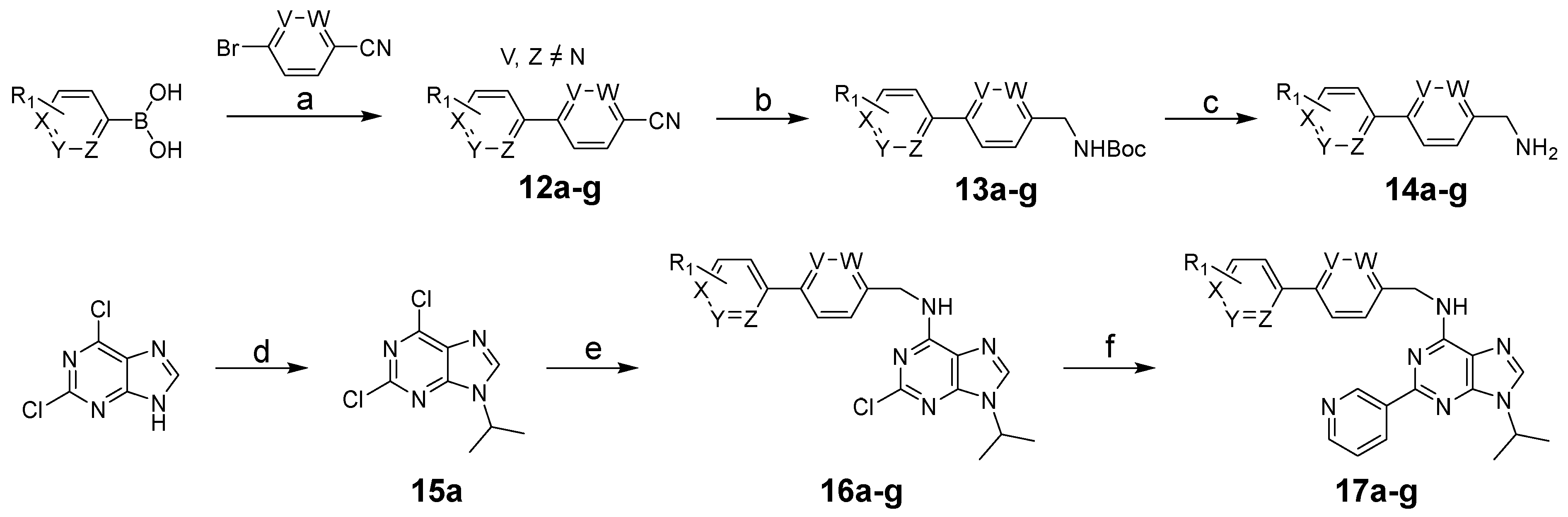
3.1. Chemistry
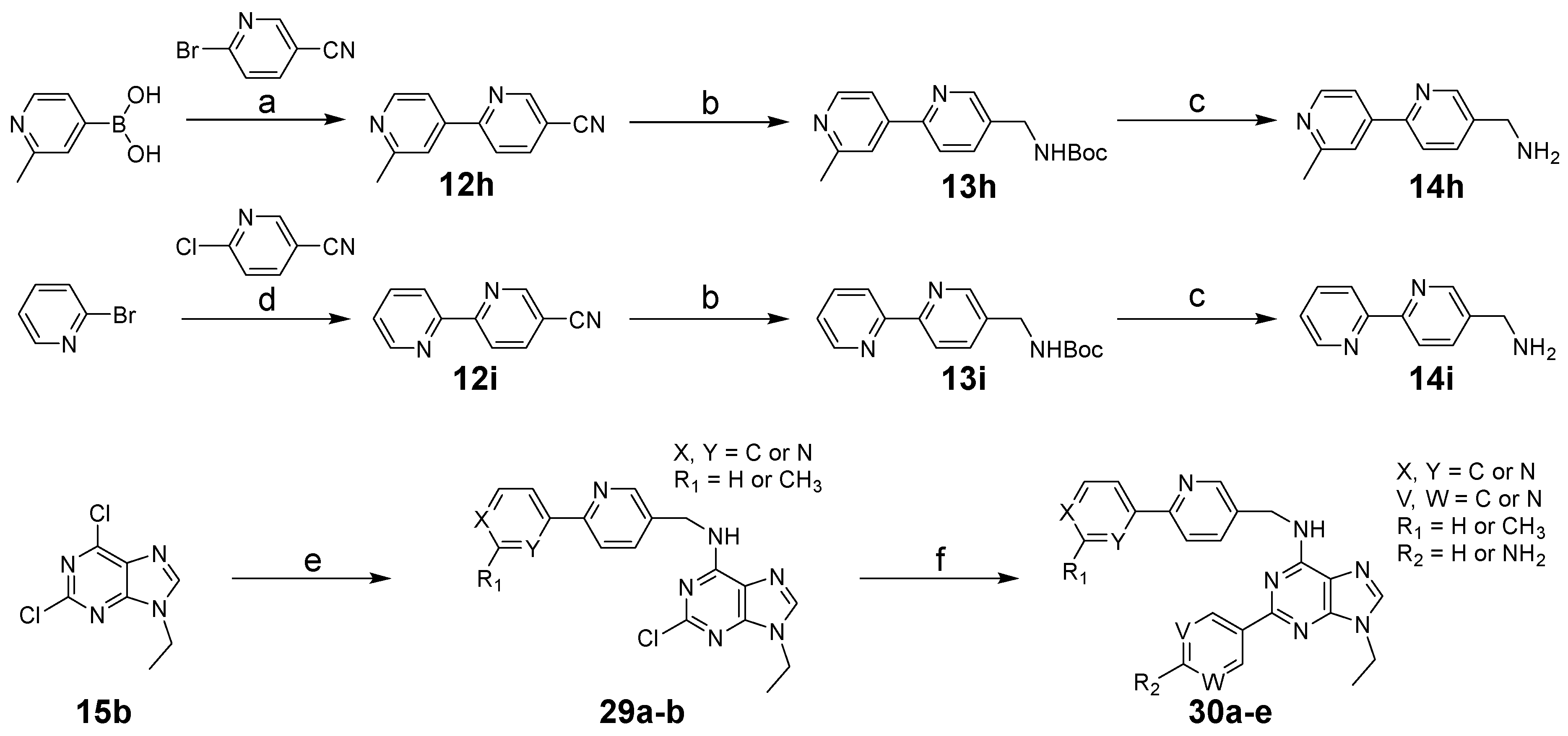
3.1.1. General Procedure for Synthesis of 12a–h
[2,3′-Bipyridine]-6′-carbonitrile (12a)
[3,3′-Bipyridine]-6-carbonitrile (12b)
6′-Methyl-[3,3′-bipyridine]-6-carbonitrile (12c)
[3,4′-Bipyridine]-6-carbonitrile (12d)
2′-Methyl-[3,4′-bipyridine]-6-carbonitrile (12e)
[2,3′-Bipyridine]-5-carbonitrile (12f)
[2,4′-Bipyridine]-6-carbonitrile (12g)
2′-Methyl-[2,4′-bipyridine]-5-carbonitrile (12h)
[2,2′-Bipyridine]-5-carbonitrile (12i)
3.1.2. General Procedure for Synthesis of 14a–i
[2,3′-Bipyridin]-6′-ylmethanamium (14a)
[3,3′-Bipyridine]-6-ylmethanamium (14b)
(6′-Methyl-[3,3′-bipyridin]-6-yl)methanamine (14c)
[3,4′-Bipyridine]-6-ylmethanamium (14d)
(2′-Methyl-[3,4′-bipyridin]-6-yl)methanamine (14e)
[2,3′-Bipyridine]-5-ylmethanamium (14f)
[2,4′-Bipyridine]-6-ylmethanamium (14g)
(2′-Methyl-[2,4′-bipyridin]-5-yl)methanamine (14h)
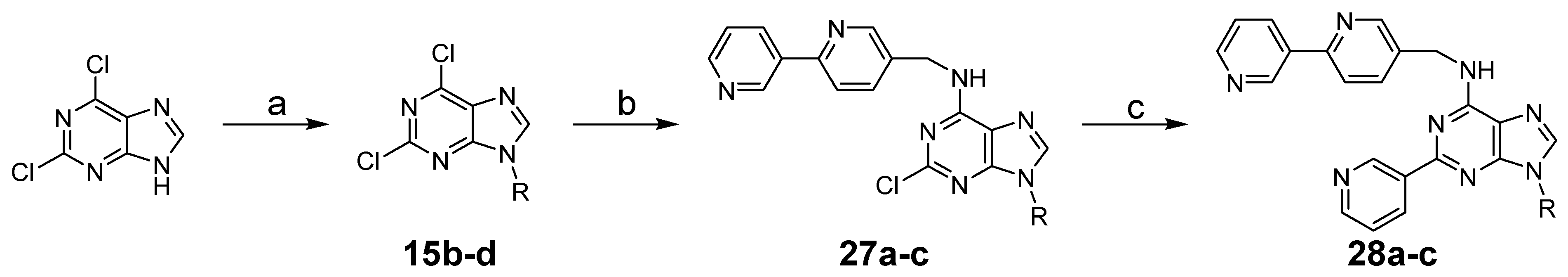
3.1.3. General Procedure for Synthesis of 15a–c
Procedure for Synthesis of 2,6-Dichloro-9-(tetrahydro-2H-pyran-4-yl)-9H-purine (15d)
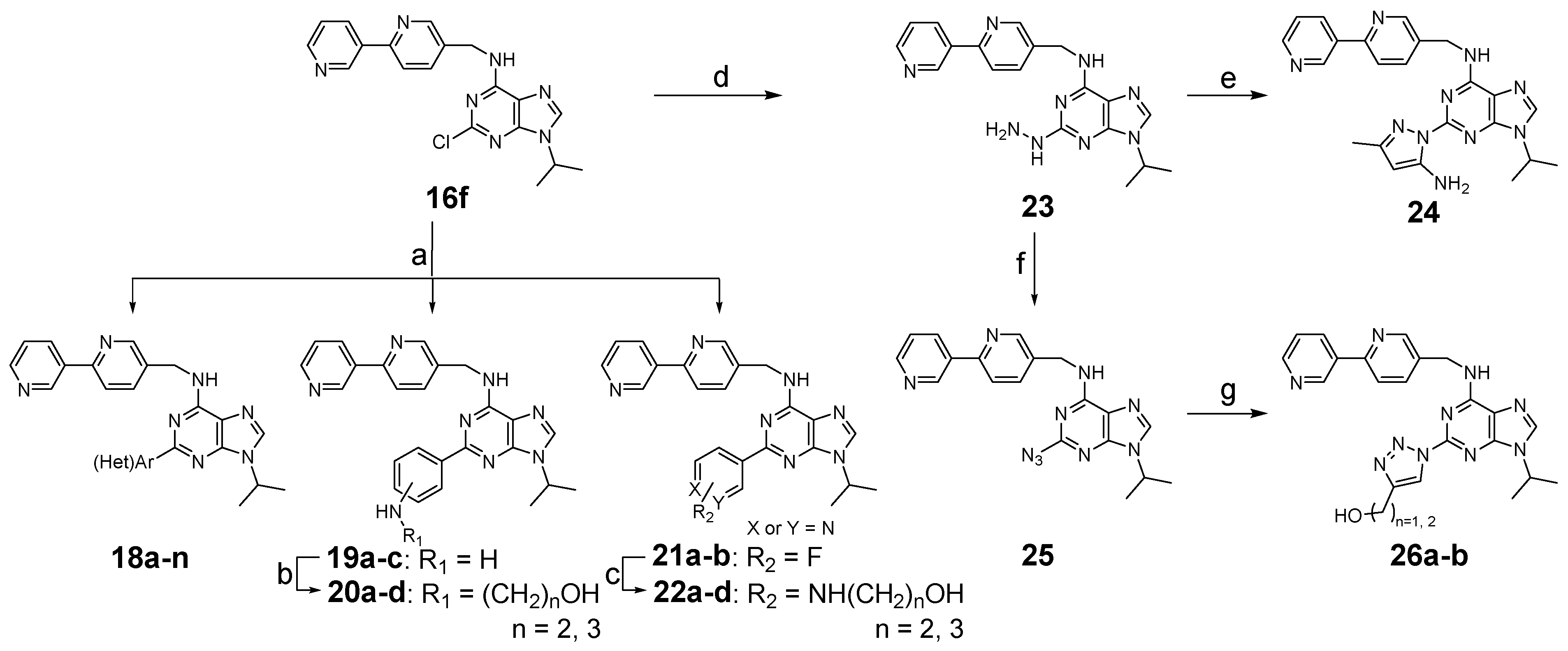
3.1.4. General Procedure for Synthesis of 16a–g, 27a–c, 29a–b
N-([2,3′-Bipyridin]-6′-ylmethyl)-2-chloro-9-isopropyl-9H-purin-6-amine (16a)
N-([3,3′-Bipyridin]-6-ylmethyl)-2-chloro-9-isopropyl-9H-purin-6-amine (16b)
2-Chloro-9-isopropyl-N-((6′-methyl-[3,3′-bipyridin]-6-yl)methyl)-9H-purin-6-amine (16c)
N-([3,4′-Bipyridin]-6-ylmethyl)-2-chloro-9-isopropyl-9H-purin-6-amine (16d)
2-Chloro-9-isopropyl-N-((2′-methyl-[3,4′-bipyridin]-6-yl)methyl)-9H-purin-6-amine (16e)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-chloro-9-isopropyl-9H-purin-6-amine (16f)
N-([2,4′-Bipyridin]-5-ylmethyl)-2-chloro-9-isopropyl-9H-purin-6-amine (16g)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-chloro-9-ethyl-9H-purin-6-amine (27a)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-chloro-9-cyclopentyl-9H-purin-6-amine (27b)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-chloro-9-(tetrahydro-2H-pyran-4-yl)-9H-purin-6-amine (27c)
2-Chloro-9-ethyl-N-((2′-methyl-[2,4′-bipyridin]-5-yl)methyl)-9H-purin-6-amine (29a)
N-([2,2′-Bipyridin]-5-ylmethyl)-2-chloro-9-ethyl-9H-purin-6-amine (29b)
3.1.5. General Procedure for Synthesis of 17a–g, 18a–n, 19a–c, 21a–b, 28a–c, 30a–e
N-([2,3′-Bipyridin]-6′-ylmethyl)-9-isopropyl-2-(pyridin-3-yl)-9H-purin-6-amine] (17a)
N-([3,3′-Bipyridin]-6-ylmethyl)-9-isopropyl-2-(pyridin-3-yl)-9H-purin-6-amine] (17b)
9-Isopropyl-N-((6′-methyl-[3,3′-bipyridin]-6-yl)methyl)-2-(pyridin-3-yl)-9H-purin-6-amine (17c)
N-([3,4′-Bipyridin]-6-ylmethyl)-9-isopropyl-2-(pyridin-3-yl)-9H-purin-6-amine] (17d)
9-Isopropyl-N-((2′-methyl-[3,4′-bipyridin]-6-yl)methyl)-2-(pyridin-3-yl)-9H-purin-6-amine (17e)
N-([2,3′-Bipyridin]-5-ylmethyl)-9-isopropyl-2-(pyridin-3-yl)-9H-purin-6-amine (17f)
N-([2,4′-Bipyridin]-5-ylmethyl)-9-isopropyl-2-(pyridin-3-yl)-9H-purin-6-amine] (17g)
N-([2,3′-Bipyridin]-5-ylmethyl)-9-isopropyl-2-phenyl-9H-purin-6-amine (18a)
3-(6-(([2,3′-Bipyridin]-5-ylmethyl)amino)-9-isopropyl-9H-purin-2-yl)benzamide (18b)
4-(6-(([2,3′-Bipyridin]-5-ylmethyl)amino)-9-isopropyl-9H-purin-2-yl)benzamide (18c)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-(1H-indol-5-yl)-9-isopropyl-9H-purin-6-amine (18d)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-(1H-indazol-6-yl)-9-isopropyl-9H-purin-6-amine (18e)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-(1H-indazol-5-yl)-9-isopropyl-9H-purin-6-amine (18f)
N-([2,3′-Bipyridin]-5-ylmethyl)-9-isopropyl-2-(quinolin-3-yl)-9H-purin-6-amine (18g)
N-([2,3′-Bipyridin]-5-ylmethyl)-9-isopropyl-2-(6-methylpyridin-3-yl)-9H-purin-6-amine (18h)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-(6-aminopyridin-3-yl)-9-isopropyl-9H-purin-6-amine] (18i)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-(2-aminopyridin-4-yl)-9-isopropyl-9H-purin-6-amine] (18j)
N-([2,3′-Bipyridin]-5-ylmethyl)-9-isopropyl-2-(pyrimidin-5-yl)-9H-purin-6-amine (18k)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-(2-aminopyrimidin-5-yl)-9-isopropyl-9H-purin-6-amine] (18l)
N-([2,3′-Bipyridin]-5-ylmethyl)-9-isopropyl-2-(1H-pyrazol-5-yl)-9H-purin-6-amine (18m)
N-([2,3′-Bipyridin]-5-ylmethyl)-9-isopropyl-2-(1H-pyrazol-4-yl)-9H-purin-6-amine (18n)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-(2-aminophenyl)-9-isopropyl-9H-purin-6-amine (19a)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-(3-aminophenyl)-9-isopropyl-9H-purin-6-amine (19b)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-(4-aminophenyl)-9-isopropyl-9H-purin-6-amine (19c)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-(6-fluoropyridin-3-yl)-9-isopropyl-9H-purin-6-amine (21a)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-(2-fluoropyridin-4-yl)-9-isopropyl-9H-purin-6-amine] (21b)
N-([2,3′-Bipyridin]-5-ylmethyl)-9-ethyl-2-(pyridin-3-yl)-9H-purin-6-amine (28a)
N-([2,3′-Bipyridin]-5-ylmethyl)-9-cyclopentyl-2-(pyridin-3-yl)-9H-purin-6-amine (28b)
N-([2,3′-Bipyridin]-5-ylmethyl)-2-(pyridin-3-yl)-9-(tetrahydro-2H-pyran-4-yl)-9H-purin-6-amine (28c)
2-(6-Aminopyridin-3-yl)-9-ethyl-N-((2′-methyl-[2,4′-bipyridin]-5-yl)methyl)-9H-purin-6-amine (30a)
2-(2-Aminopyrimidin-5-yl)-9-ethyl-N-((2′-methyl-[2,4′-bipyridin]-5-yl)methyl)-9H-purin-6-amine (30b)
9-Ethyl-N-((2′-methyl-[2,4′-bipyridin]-5-yl)methyl)-2-(pyrimidin-5-yl)-9H-purin-6-amine (30c)
N-([2,2′-Bipyridin]-5-ylmethyl)-2-(6-aminopyridin-3-yl)-9-ethyl-9H-purin-6-amine (30d)
N-([2,2′-Bipyridin]-5-ylmethyl)-2-(2-aminopyrimidin-5-yl)-9-ethyl-9H-purin-6-amine (30e)
3.1.6. General Procedure for Synthesis of 20a–d
2-((2-(6-(([2,3′-Bipyridin]-5-ylmethyl)amino)-9-isopropyl-9H-purin-2-yl)phenyl)amino)ethan-1-ol (20a)
3-((2-(6-(([2,3′-Bipyridin]-5-ylmethyl)amino)-9-isopropyl-9H-purin-2-yl)phenyl)amino)propan-1-ol (20b)
3-((3-(6-(([2,3′-Bipyridin]-5-ylmethyl)amino)-9-isopropyl-9H-purin-2-yl)phenyl)amino)propan-1-ol (20c)
3-((4-(6-(([2,3′-Bipyridin]-5-ylmethyl)amino)-9-isopropyl-9H-purin-2-yl)phenyl)amino)propan-1-ol (20d)
3.1.7. General Procedure for Synthesis of 22a–d
2-((5-(6-(([2,3′-Bipyridin]-5-ylmethyl)amino)-9-isopropyl-9H-purin-2-yl)pyridin-2-yl)amino)ethan-1-ol (22a)
3-((5-(6-(([2,3′-Bipyridin]-5-ylmethyl)amino)-9-isopropyl-9H-purin-2-yl)pyridin-2-yl)amino)propan-1-ol (22b)
2-((4-(6-(([2,3′-Bipyridin]-5-ylmethyl)amino)-9-isopropyl-9H-purin-2-yl)pyridin-2-yl)amino)ethan-1-ol] (22c)
3-((4-(6-(([2,3′-Bipyridin]-5-ylmethyl)amino)-9-isopropyl-9H-purin-2-yl)pyridin-2-yl)amino)propan-1-ol] (22d)
3.1.8. N-([2,3′-Bipyridin]-5-ylmethyl)-2-hydrazineyl-9-isopropyl-9H-purin-6-amine (23)
3.1.9. N-([2,3′-Bipyridin]-5-ylmethyl)-2-(5-amino-3-methyl-1H-pyrazol-1-yl)-9-isopropyl-9H-purin-6-amine (24)
3.1.10. N-([2,3′-Bipyridin]-5-ylmethyl)-2-azido-9-isopropyl-9H-purin-6-amine (25)
3.1.11. General Procedure for Synthesis of 26a–b
(1-(6-(([2,3′-Bipyridin]-5-ylmethyl)amino)-9-isopropyl-9H-purin-2-yl)-1H-1,2,3-triazol-4-yl)methanol (26a)
2-(1-(6-(([2,3′-Bipyridin]-5-ylmethyl)amino)-9-isopropyl-9H-purin-2-yl)-1H-1,2,3-triazol-4-yl)ethan-1-ol (26b)
3.2. Docking Analysis
3.3. Bioassays
3.3.1. Antibodies
3.3.2. Cell Culture
3.3.3. In Vitro Kinase Assay
3.3.4. Antiproliferation Assay
3.3.5. Western Blot
3.3.6. In Vitro Liver Microsomal Stability Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zubair, M.; Wang, S.; Ali, N. Advanced Approaches to Breast Cancer Classification and Diagnosis. Front. Pharmacol. 2020, 11, 632079. [Google Scholar] [CrossRef] [PubMed]
- Freudenberg, J.A.; Wang, Q.; Katsumata, M.; Drebin, J.; Nagatomo, I.; Greene, M.I. The role of HER2 in early breast cancer metastasis and the origins of resistance to HER2-targeted therapies. Exp. Mol. Pathol. 2009, 87, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ménard, S.; Pupa, S.M.; Campiglio, M.; Tagliabue, E. Biologic and therapeutic role of HER2 in cancer. Oncogene 2003, 22, 6570–6578. [Google Scholar] [CrossRef]
- Slamon, D.J.; Leyland-Jones, B.; Shak, S.; Fuchs, H.; Paton, V.; Bajamonde, A.; Fleming, T.; Eiermann, W.; Wolter, J.; Pegram, M.; et al. Use of Chemotherapy plus a Monoclonal Antibody against HER2 for Metastatic Breast Cancer That Overexpresses HER2. N. Engl. J. Med. 2001, 344, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Piccart-Gebhart, M.J.; Procter, M.; Leyland-Jones, B.; Goldhirsch, A.; Untch, M.; Smith, I.; Gianni, L.; Baselga, J.; Bell, R.H.; Jackisch, C.; et al. Trastuzumab after Adjuvant Chemotherapy in HER2-Positive Breast Cancer. N. Engl. J. Med. 2005, 353, 1659–1672. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.L.; Cobleigh, M.A.; Tripathy, D.; Gutheil, J.C.; Harris, L.N.; Fehrenbacher, L.; Slamon, D.J.; Murphy, M.; Novotny, W.F.; Burchmore, M.; et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J. Clin. Oncol. 2002, 20, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Pohlmann, P.R.; Mayer, I.A.; Mernaugh, R. Resistance to Trastuzumab in Breast Cancer. Clin. Cancer Res. 2009, 15, 7479–7491. [Google Scholar] [CrossRef]
- Nahta, R.; Yu, D.; Hung, M.-C.; Hortobagyi, G.N.; Esteva, F.J. Mechanisms of Disease: Understanding resistance to HER2-targeted therapy in human breast cancer. Nat. Clin. Pract. Oncol. 2006, 3, 269–280. [Google Scholar] [CrossRef]
- Sahlberg, K.K.; Hongisto, V.; Edgren, H.; Mäkelä, R.; Hellström, K.; Due, E.U.; Vollan, H.K.M.; Sahlberg, N.; Wolf, M.; Børresen-Dale, A.-L.; et al. The HER2 amplicon includes several genes required for the growth and survival of HER2 positive breast cancer cells. Mol. Oncol. 2013, 7, 392–401. [Google Scholar] [CrossRef]
- Choi, H.-J.; Jin, S.; Cho, H.; Won, H.Y.; An, H.W.; Jeong, G.Y.; Park, Y.U.; Kim, H.Y.; Park, M.K.; Son, T.; et al. CDK12 drives breast tumor initiation and trastuzumab resistance via WNT and IRS1-ErbB-PI3K signaling. EMBO Rep. 2019, 20, e48058. [Google Scholar] [CrossRef]
- Liu, H.; Liu, K.; Dong, Z. Targeting CDK12 for Cancer Therapy: Function, Mechanism, and Drug Discovery. Cancer Res. 2021, 81, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, S.; Duckett, D.R.; Monastyrskyi, A. The promise and current status of CDK12/13 inhibition for the treatment of cancer. Future Med. Chem. 2021, 13, 117–141. [Google Scholar] [CrossRef] [PubMed]
- Paruch, K.; Dwyer, M.P.; Alvarez, C.; Brown, C.; Chan, T.-Y.; Doll, R.J.; Keertikar, K.; Knutson, C.; McKittrick, B.; Rivera, J.; et al. Discovery of Dinaciclib (SCH 727965): A Potent and Selective Inhibitor of Cyclin-Dependent Kinases. ACS Med. Chem. Lett. 2010, 1, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.F.; Cruz, C.; Greifenberg, A.K.; Dust, S.; Stover, D.; Chi, D.; Primack, B.; Cao, S.; Bernhardy, A.J.; Coulson, R.; et al. CDK12 Inhibition Reverses De Novo and Acquired PARP Inhibitor Resistance in BRCA Wild-Type and Mutated Models of Triple-Negative Breast Cancer. Cell Rep. 2016, 17, 2367–2381. [Google Scholar] [CrossRef] [PubMed]
- Mita, M.M.; Joy, A.A.; Mita, A.; Sankhala, K.; Jou, Y.-M.; Zhang, D.; Statkevich, P.; Zhu, Y.; Yao, S.-L.; Small, K.; et al. Randomized Phase II Trial of the Cyclin-Dependent Kinase Inhibitor Dinaciclib (MK-7965) Versus Capecitabine in Patients With Advanced Breast Cancer. Clin. Breast Cancer 2014, 14, 169–176. [Google Scholar] [CrossRef]
- Ito, M.; Tanaka, T.; Toita, A.; Uchiyama, N.; Kokubo, H.; Morishita, N.; Klein, M.G.; Zou, H.; Murakami, M.; Kondo, M.; et al. Discovery of 3-Benzyl-1-(trans-4-((5-cyanopyridin-2-yl)amino)cyclohexyl)-1-arylurea Derivatives as Novel and Selective Cyclin-Dependent Kinase 12 (CDK12) Inhibitors. J. Med. Chem. 2018, 61, 7710–7728. [Google Scholar] [CrossRef] [PubMed]
- Johannes, J.W.; Denz, C.R.; Su, N.; Wu, A.; Impastato, A.C.; Mlynarski, S.; Varnes, J.G.; Prince, D.B.; Cidado, J.; Gao, N.; et al. Structure-Based Design of Selective Noncovalent CDK12 Inhibitors. Chem. Med. Chem. 2018, 13, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Kwiatkowski, N.; Olson, C.M.; Dixon-Clarke, S.E.; Abraham, B.J.; Greifenberg, A.K.; Ficarro, S.B.; Elkins, J.M.; Liang, Y.; Hannett, N.M.; et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat. Chem. Biol. 2016, 12, 876–884. [Google Scholar] [CrossRef]
- Liu, Y.; Hao, M.; Leggett, A.L.; Gao, Y.; Ficarro, S.B.; Che, J.; He, Z.; Olson, C.M.; Marto, J.A.; Kwiatkowski, N.P.; et al. Discovery of MFH290: A Potent and Highly Selective Covalent Inhibitor for Cyclin-Dependent Kinase 12/13. J. Med. Chem. 2020, 63, 6708–6726. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, T.; Terai, H.; Ficarro, S.B.; Kwiatkowski, N.; Hao, M.-F.; Sharma, B.; Christensen, C.L.; Chipumuro, E.; Wong, K.-K.; et al. Overcoming Resistance to the THZ Series of Covalent Transcriptional CDK Inhibitors. Cell Chem. Biol. 2018, 25, 135–142.e5. [Google Scholar] [CrossRef]
- Jiang, B.; Gao, Y.; Che, J.; Lu, W.; Kaltheuner, I.H.; Dries, R.; Kalocsay, M.; Berberich, M.J.; Jiang, J.; You, I.; et al. Discovery and resistance mechanism of a selective CDK12 degrader. Nat. Chem. Biol. 2021, 17, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Niu, T.; Li, K.; Jiang, L.; Zhou, Z.; Hong, J.; Chen, X.; Dong, X.; He, Q.; Cao, J.; Yang, B.; et al. Noncovalent CDK12/13 dual inhibitors-based PROTACs degrade CDK12-Cyclin K complex and induce synthetic lethality with PARP inhibitor. Eur. J. Med. Chem. 2022, 228, 114012. [Google Scholar] [CrossRef] [PubMed]
- Słabicki, M.; Kozicka, Z.; Petzold, G.; Li, Y.-D.; Manojkumar, M.; Bunker, R.D.; Donovan, K.A.; Sievers, Q.L.; Koeppel, J.; Suchyta, D.; et al. The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K. Nature 2020, 585, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Chen, P.; Cao, L.; Li, Y.; Zeng, Z.; Cui, Y.; Wu, Q.; Li, J.; Wang, J.-H.; Dong, M.-Q.; et al. Discovery of a molecular glue promoting CDK12-DDB1 interaction to trigger cyclin K degradation. eLife 2020, 9, e59994. [Google Scholar] [CrossRef] [PubMed]
- Mayor-Ruiz, C.; Bauer, S.; Brand, M.; Kozicka, Z.; Siklos, M.; Imrichova, H.; Kaltheuner, I.H.; Hahn, E.; Seiler, K.; Koren, A.; et al. Rational discovery of molecular glue degraders via scalable chemical profiling. Nat. Chem. Biol. 2020, 16, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Caddick, S.; Judd, D.B.; Lewis, A.K.K.; Reich, M.T.; Williams, M.R. A generic approach for the catalytic reduction of nitriles. Tetrahedron 2003, 59, 5417–5423. [Google Scholar] [CrossRef]
- Luzung, M.R.; Patel, J.S.; Yin, J. A Mild Negishi Cross-Coupling of 2-Heterocyclic Organozinc Reagents and Aryl Chlorides. J. Org. Chem. 2010, 75, 8330–8332. [Google Scholar] [CrossRef]
- Lutzen, A.; Hapke, M. Synthesis of 5-substituted 2,2′-bipyridines from substituted 2-chloropyridines by a modified Negishi cross-coupling reaction. Eur. J. Org. Chem. 2002, 2002, 2292–2297. [Google Scholar] [CrossRef]
- Library of Integrated Network-Based Cellular Signatures. Available online: https://lincs.hms.harvard.edu/db/datasets/20128/results (accessed on 1 March 2022).
- Parry, D.; Guzi, T.; Shanahan, F.; Davis, N.; Prabhavalkar, D.; Wiswell, D.; Seghezzi, W.; Paruch, K.; Dwyer, M.P.; Doll, R.; et al. Dinaciclib (SCH 727965), a Novel and Potent Cyclin-Dependent Kinase Inhibitor. Mol. Cancer Ther. 2010, 9, 2344–2353. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera? A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef] [PubMed]

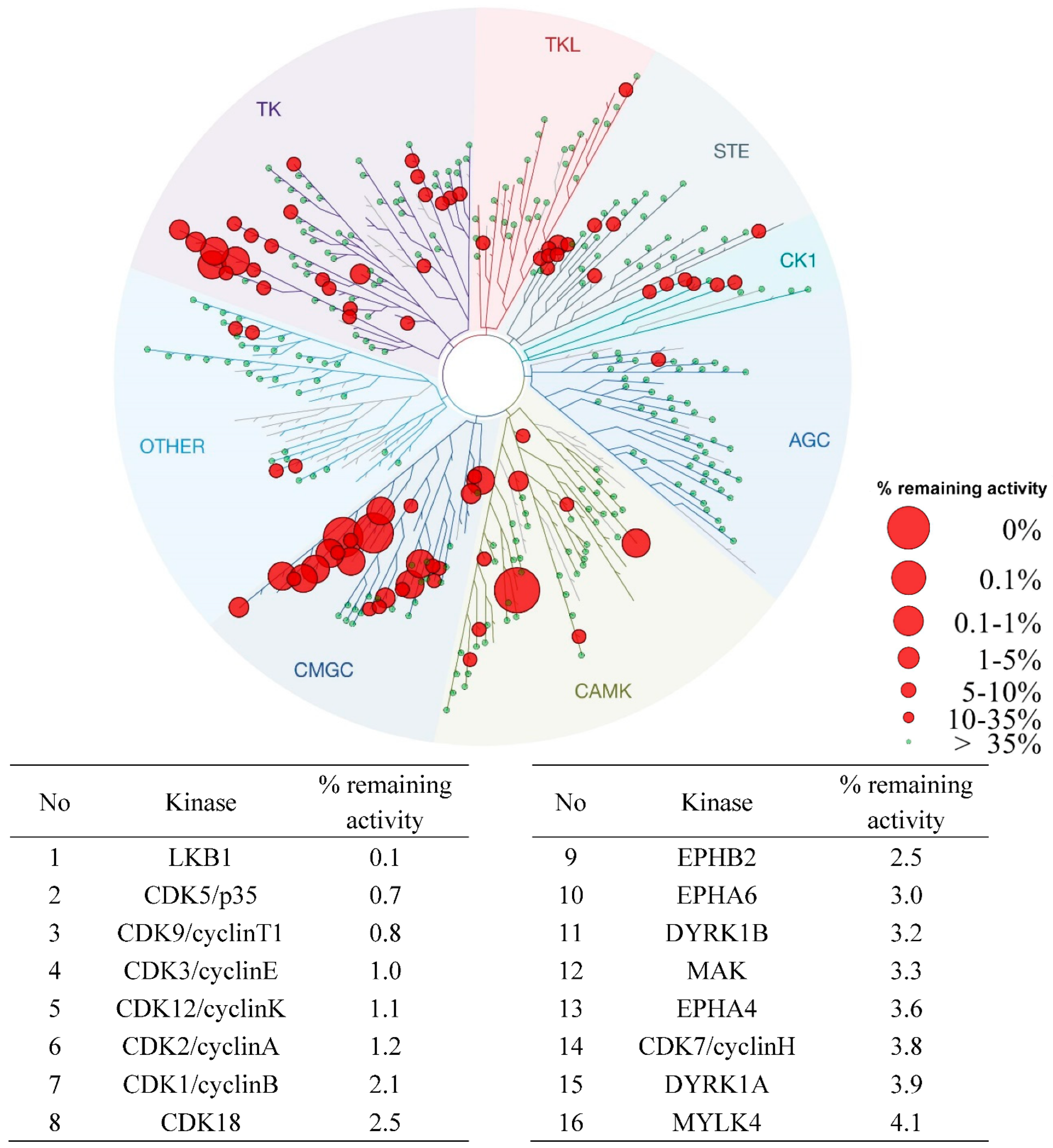

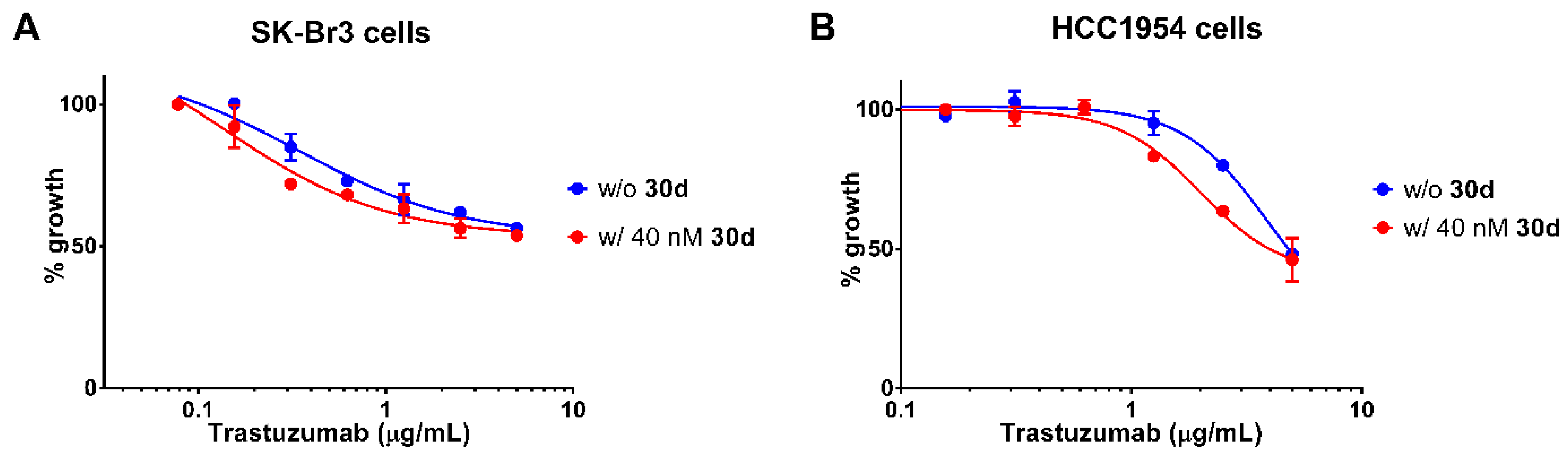
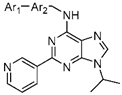 | ||||
|---|---|---|---|---|
| Compound | Ar1-Ar2 | CDK12/CyclinK (IC50/μM) | SK-Br3 (GI50/μM) * | HCC1954 (GI50/μM) * |
| 17a | 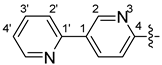 | 0.433 | 0.151 ± 0.025 | 0.106 ± 0.011 |
| 17b | 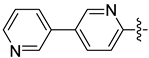 | 0.582 | 0.862 ± 0.178 | 0.627 ± 0.128 |
| 17c | 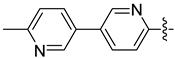 | 1.109 | 24.560 ± 4.992 | 13.580 ± 1.160 |
| 17d | 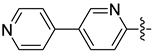 | 0.627 | 0.741 ± 0.034 | 0.887 ± 0.040 |
| 17e | 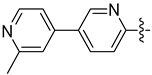 | 0.481 | 0.277 ± 0.038 | 0.230 ± 0.044 |
| 17f | 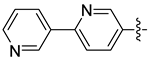 | 0.221 | 0.410 ± 0.034 | 0.247 ± 0.030 |
| 17g |  | 0.153 | 0.804 ± 0.095 | 0.494 ± 0.048 |
 | ||||
|---|---|---|---|---|
| Compound | R | CDK12/CyclinK (IC50/μM) | SK-Br3 (GI50/μM) * | HCC1954 (GI50/μM) * |
| 18a | 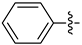 | 0.623 | 1.692 ± 0.243 | 1.116 ± 0.260 |
| 19a | 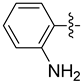 | 0.619 | 2.607 ± 0.899 | 2.634 ± 0.448 |
| 20a | 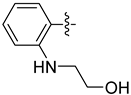 | 0.799 | 2.304 ± 0.318 | 1.557 ± 0.116 |
| 20b | 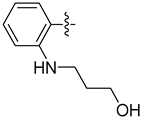 | 3.012 | 11.240 ± 1.117 | 6.051 ± 0.351 |
| 19b | 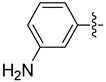 | 0.170 | 0.614 ± 0.055 | 0.341 ± 0.059 |
| 20c | 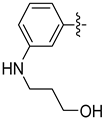 | 0.257 | 0.732 ± 0.177 | 0.502 ± 0.025 |
| 19c | 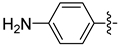 | 0.150 | 0.795 ± 0.063 | 0.497 ± 0.088 |
| 20d | 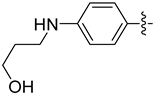 | 0.499 | 1.093 ± 0.127 | 0.907 ± 0.068 |
| 18b |  | 0.504 | 1.625 ± 0.248 | 1.137 ± 0.304 |
| 18c | 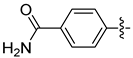 | 0.484 | 3.711 ± 0.450 | 3.141 ± 0.201 |
| 18d | 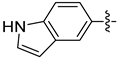 | 0.155 | 1.496 ± 0.112 | 1.351 ± 0.243 |
| 18e | 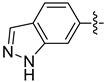 | 0.179 | 0.608 ± 0.115 | 0.399 ± 0.051 |
| 18f | 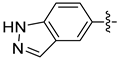 | 0.104 | 2.580 ± 0.167 | 1.865 ± 0.148 |
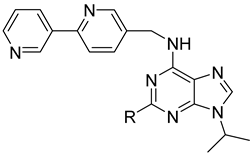 | ||||
|---|---|---|---|---|
| Compound | R | CDK12/CyclinK (IC50/μM) | SK-Br3 (GI50/μM) * | HCC1954 (GI50/μM) * |
| 18g |  | 0.486 | 3.715 ± 0.557 | 4.230 ± 0.214 |
| 17f | 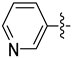 | 0.221 | 0.410 ± 0.034 | 0.247 ± 0.030 |
| 21a | 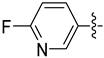 | 0.147 | 1.696 ± 0.165 | 1.335 ± 0.393 |
| 18h | 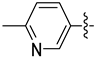 | 0.090 | 1.270 ± 0.330 | 0.942 ± 0.125 |
| 18i | 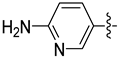 | 0.065 | 0.376 ± 0.067 | 0.278 ± 0.007 |
| 22a | 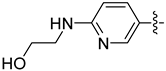 | 0.053 | 0.262 ± 0.021 | 0.174 ± 0.019 |
| 22b |  | 0.304 | 1.183 ± 0.118 | 0.977 ± 0.070 |
| 21b | 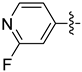 | 0.164 | 1.679 ± 0.267 | 1.274 ± 0.021 |
| 18j |  | 0.094 | 0.710 ± 0.024 | 0.354 ± 0.060 |
| 22c |  | 0.086 | 0.473 ± 0.087 | 0.433 ± 0.052 |
| 22d | 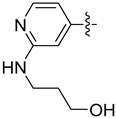 | 0.113 | 0.480 ± 0.026 | 0.387 ± 0.037 |
| 18k |  | 0.051 | 0.318 ± 0.149 | 0.310 ± 0.041 |
| 18l |  | 0.087 | 0.242 ± 0.042 | 0.162 ± 0.011 |
| 18m | 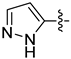 | 0.501 | 1.717 ± 0.336 | 1.132 ± 0.050 |
| 18n | 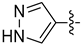 | 1.433 | 1.993 ± 0.120 | 1.172 ± 0.027 |
| 24 | 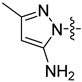 | 2.345 | 5.160 ± 0.365 | 3.436 ± 0.177 |
| 26a | 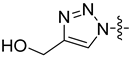 | 2.051 | 13.260 ± 3.154 | 9.967 ± 1.857 |
| 26b | 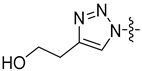 | 0.539 | 8.303 ± 0.882 | 6.158 ± 0.429 |
 | ||||
|---|---|---|---|---|
| Compound | R | CDK12/CyclinK (IC50/μM) | SK-Br3 (GI50/μM) * | HCC1954 (GI50/μM) * |
| 17f |  | 0.221 | 0.410 ± 0.034 | 0.247 ± 0.030 |
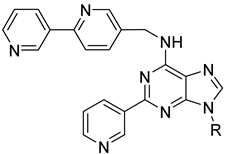
| 28a |  | 0.016 | 0.215 ± 0.053 | 0.142 ± 0.014 |
| 28b | 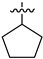 | 0.293 | 2.597 ± 0.310 | 2.140 ± 0.176 |
| 28c | 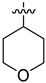 | >10 | >100 | 44.360 ± 2.461 |
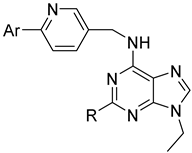 | |||||
|---|---|---|---|---|---|
| Compound | Ar | R | CDK12/CyclinK (IC50/μM) * | SK-Br3 (GI50/μM) * | HCC1954 (GI50/μM) * |
| 30a |  | 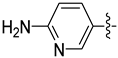 | 0.029 ± 0.015 | 0.136 ± 0.016 | 0.104 ± 0.003 |
| 30b |  |  | 0.028 ± 0.008 | 0.090 ± 0.008 | 0.080 ± 0.007 |
| 30c | 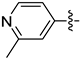 | 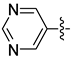 | 0.064 ± 0.012 | 0.189 ± 0.057 | 0.157 ± 0.022 |
| 30d | 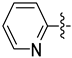 | 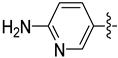 | 0.021 ± 0.007 | 0.046 ± 0.007 | 0.036 ± 0.005 |
| 30e |  | 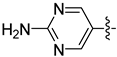 | 0.085 ± 0.011 | 0.052 ± 0.003 | 0.034 ± 0.002 |
| THZ531 | 0.051 ± 0.007 | 0.030 ± 0.006 | 0.195 ± 0.050 | ||
| Dinaciclib | 0.0066 ± 0.0002 | 0.012 ± 0.001 | 0.012 ± 0.001 | ||
| 2 | 0.0086 ± 0.0017 | 0.094 ± 0.012 | 0.069 ± 0.006 | ||
| R-CR8 (9) | 0.287 ± 0.036 | 0.101 ± 0.014 | 0.083 ± 0.011 | ||
| Compound | Liver Microsomal Stability (% Remaining) | CYP % Activity at 10 μM | ||||||
|---|---|---|---|---|---|---|---|---|
| Human | Dog | Mouse | 1A2 | 2C9 | 2C19 | 2D6 | 3A4 | |
| 30a | 32.2 | 45.1 | 35.3 | 59.3 | 73.0 | 76.8 | 89.6 | 38.9 |
| 30b | 40.6 | 53.6 | 45.0 | 74.9 | 82.9 | 75.4 | 85.7 | 72.4 |
| 30c | 27.3 | 79.5 | 34.0 | 68.6 | 67.5 | 73.9 | 88.8 | 31.8 |
| 30d | 64.8 | 46.2 | 49.9 | 74.7 | 57.9 | 68.1 | 81.0 | 43.0 |
| 30e | 100 | 67.6 | 85.0 | 87.6 | 81.6 | 72.6 | 92.1 | 92.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuchukulla, R.R.; Hwang, I.; Park, S.W.; Moon, S.; Kim, S.H.; Kim, S.; Chung, H.W.; Ji, M.-J.; Park, H.-M.; Kong, G.; et al. Novel 2,6,9-Trisubstituted Purines as Potent CDK Inhibitors Alleviating Trastuzumab-Resistance of HER2-Positive Breast Cancers. Pharmaceuticals 2022, 15, 1041. https://doi.org/10.3390/ph15091041
Kuchukulla RR, Hwang I, Park SW, Moon S, Kim SH, Kim S, Chung HW, Ji M-J, Park H-M, Kong G, et al. Novel 2,6,9-Trisubstituted Purines as Potent CDK Inhibitors Alleviating Trastuzumab-Resistance of HER2-Positive Breast Cancers. Pharmaceuticals. 2022; 15(9):1041. https://doi.org/10.3390/ph15091041
Chicago/Turabian StyleKuchukulla, Ratnakar Reddy, Injeoung Hwang, Sang Won Park, Sojeong Moon, Suhn Hyung Kim, Sumin Kim, Hwan Won Chung, Mi-Jung Ji, Hyun-Mee Park, Gu Kong, and et al. 2022. "Novel 2,6,9-Trisubstituted Purines as Potent CDK Inhibitors Alleviating Trastuzumab-Resistance of HER2-Positive Breast Cancers" Pharmaceuticals 15, no. 9: 1041. https://doi.org/10.3390/ph15091041
APA StyleKuchukulla, R. R., Hwang, I., Park, S. W., Moon, S., Kim, S. H., Kim, S., Chung, H. W., Ji, M.-J., Park, H.-M., Kong, G., & Hur, W. (2022). Novel 2,6,9-Trisubstituted Purines as Potent CDK Inhibitors Alleviating Trastuzumab-Resistance of HER2-Positive Breast Cancers. Pharmaceuticals, 15(9), 1041. https://doi.org/10.3390/ph15091041







