Copper (I)-Chloroquine Complexes: Interactions with DNA and Ferriprotoporphyrin, Inhibition of β-Hematin Formation and Relation to Antimalarial Activity
Abstract
1. Introduction
2. Results and Discussion
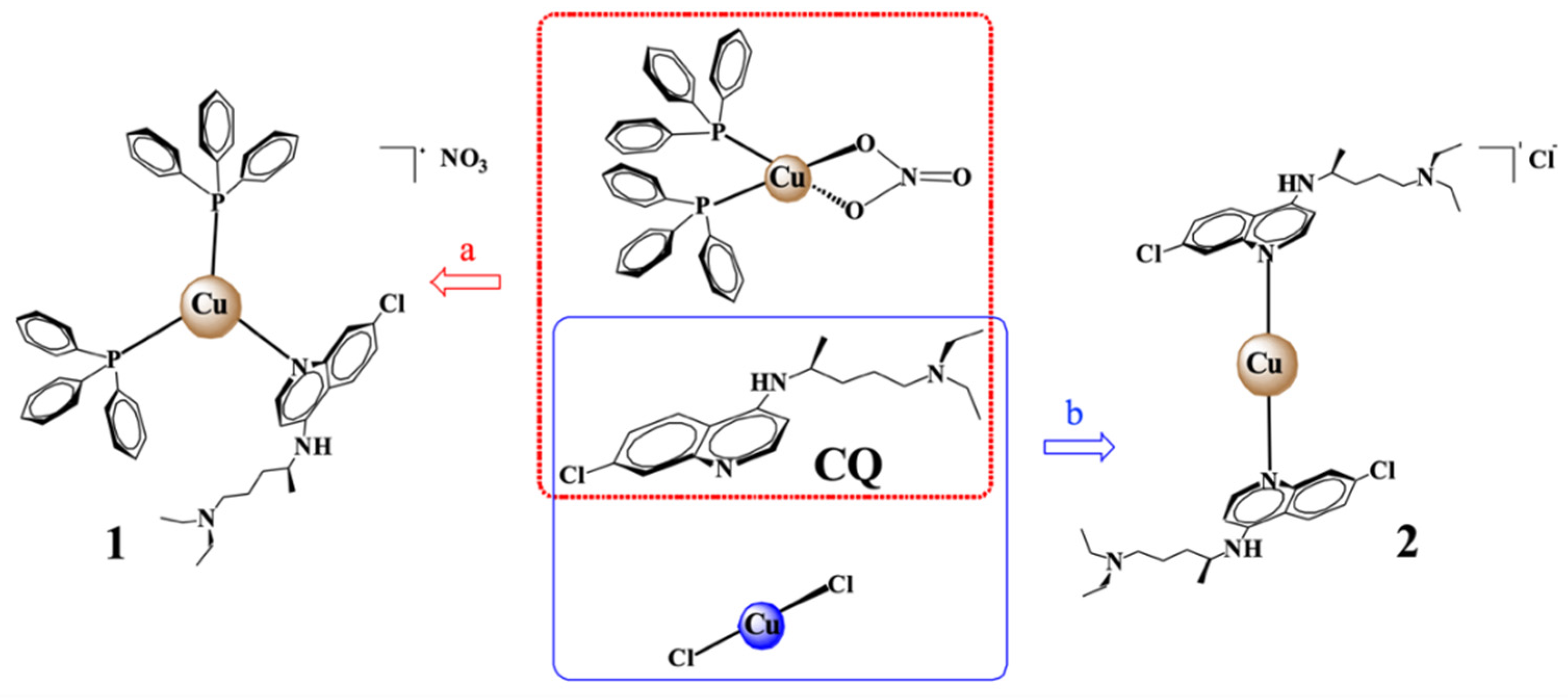
2.1. Synthesis of Copper Complexes
2.2. DNA as a Target
2.3. Interaction with Hemin and Inhibition of β-Hematin Formation
2.4. Antiplasmodial Activity
3. Materials and Methods
3.1. Synthesis of Copper Complexes
3.2. DNA as a Target
3.2.1. Spectrophotometric Titrations
3.2.2. Fluorimetric Titrations
3.2.3. Viscosity Study
3.2.4. Melting Temperature Study
3.3. Interaction with Hemin and Inhibition of β-Hematin Formation
3.3.1. Interaction with Ferriprotoporphyrin (Fe(III)PPIX) by UV-Vis Spectroscopic Titration
3.3.2. Determination of Inhibition of β-Haematin Formation by Infrared Spectroscopy
3.3.3. Determination of Inhibition of β-Haematin Formation by UV-Vis Spectroscopy
3.4. Antiplasmodial Activity Measurements
3.5. Cell Toxicity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. World Malaria Report; World Health Organization: Geneva, Switzerland, 2021; Available online: https://www.who.int/publications/i/item/9789240040496 (accessed on 3 June 2022).
- Martin, R.E.; Marchetti, R.V.; Cowan, A.I.; Howitt, S.M.; Bröer, S.; Kirk, K. Chloroquine Transport via the Malaria Parasite’s Chloroquine Resistance Transporter. Science 2009, 325, 1680–1682. [Google Scholar] [CrossRef] [PubMed]
- Chinappi, M.; Via, A.; Marcatili, P.; Tramontano, A. On the Mechanism of Chloroquine Resistance in Plasmodium falciparum. PLoS ONE 2010, 5, e14064. [Google Scholar] [CrossRef]
- Tilley, L.; Straimer, J.; Gnädig, N.F.; Ralph, S.A.; Fidock, D.A. Artemisinin Action and Resistance in Plasmodium falciparum. Trends Parasitol. 2016, 32, 682–696. [Google Scholar] [CrossRef]
- Biot, C.; Nosten, F.; Fraisse, L.; Ter-Minassian, D.; Khalife, J.; Dive, D. The antimalarial ferroquine: From bench to clinic. Parasite 2011, 18, 207–214. [Google Scholar] [CrossRef]
- Franz, K.J.; Metzler-Nolte, N. Introduction: Metals in Medicine. Chem. Rev. 2019, 119, 727–729. [Google Scholar] [CrossRef] [PubMed]
- Ong, Y.C.; Roy, S.; Andrews, P.C.; Gasser, G. Metal Compounds against Neglected Tropical Diseases. Chem. Rev. 2018, 119, 730–796. [Google Scholar] [CrossRef] [PubMed]
- Biot, C.; Castro, W.; Botté, C.Y.; Navarro, M. The therapeutic potential of metal-based antimalarial agents: Implications for the mechanism of action. Dalton Trans. 2012, 41, 6335–6349. [Google Scholar] [CrossRef]
- Sánchez-Delgado, R.A.; Navarro, M.; Pérez, H.; Urbina, J.A. Toward a Novel Metal-Based Chemotherapy against Tropical Diseases. 2. Synthesis and Antimalarial Activity In Vitro and In Vivo of New Ruthenium− and Rhodium−Chloroquine Complexes. J. Med. Chem. 1996, 39, 1095–1099. [Google Scholar] [CrossRef] [PubMed]
- Rajapakse, C.S.K.; Martínez, A.; Naoulou, B.; Jarzecki, A.A.; Suárez, L.; Deregnaucourt, C.; Sinou, V.; Schrével, J.; Musi, E.; Ambrosini, G.; et al. Synthesis, Characterization, and in vitro Antimalarial and Antitumor Activity of New Ruthenium(II) Complexes of Chloroquine. Inorg. Chem. 2009, 48, 1122–1131. [Google Scholar] [CrossRef]
- Navarro, M.; Castro, W.; Martínez, A.; Delgado, R.A.S. The mechanism of antimalarial action of [Au(CQ)(PPh3)]PF6: Structural effects and increased drug lipophilicity enhance heme aggregation inhibition at lipid/water interfaces. J. Inorg. Biochem. 2011, 105, 276–282. [Google Scholar] [CrossRef][Green Version]
- Gaál, A.; Orgován, G.; Mihucz, V.G.; Pape, I.; Ingerle, D.; Streli, C.; Szoboszlai, N. Metal transport capabilities of anticancer copper chelators. J. Trace Elem. Med. Biol. 2018, 47, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, X.; Chen, J.; Yang, Q.; Yang, L.; Xu, D.; Zhang, P.; Wang, X.; Liu, J. Hinokitiol copper complex inhibits proteasomal deubiquitination and induces paraptosis-like cell death in human cancer cells. Eur. J. Pharmacol. 2017, 815, 147–155. [Google Scholar] [CrossRef]
- Qin, Q.-P.; Meng, T.; Tan, M.-X.; Liu, Y.-C.; Luo, X.-J.; Zou, B.-Q.; Liang, H. Synthesis, crystal structure and biological evaluation of a new dasatinib copper(II) complex as telomerase inhibitor. Eur. J. Med. Chem. 2017, 143, 1597–1603. [Google Scholar] [CrossRef] [PubMed]
- Hernandes, M.S.; Britto, L.R. NADPH Oxidase and Neurodegeneration. Curr. Neuropharmacol. 2012, 10, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Kremer, M.L. Mechanism of the Fenton reaction. Evidence for a new intermediate. Phys. Chem. Chem. Phys. 1999, 1, 3595–3605. [Google Scholar] [CrossRef]
- Sangeetha, S.; Murali, M. Non-covalent DNA binding, protein interaction, DNA cleavage and cytotoxicity of [Cu(quamol)Cl]·H2O. Int. J. Biol. Macromol. 2018, 107, 2501–2511. [Google Scholar] [CrossRef] [PubMed]
- Kachadourian, R.; Brechbuhl, H.M.; Ruiz-Azuara, L.; Gracia-Mora, I.; Day, B.J. Casiopeína IIgly-induced oxidative stress and mitochondrial dysfunction in human lung cancer A549 and H157 cells. Toxicology 2010, 268, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, N.H.; Padhye, S.B.; Billington, D.C.; Rathbone, D.L.; Croft, S.L.; Kendrick, H.D.; Anson, C.E.; Powell, A.K. Synthesis and characterization of copper(II) complexes of pyridine-2-carboxamidrazones as potent antimalarial agents. Inorg. Chim. Acta 2003, 349, 23–29. [Google Scholar] [CrossRef]
- Tapanelli, S.; Habluetzel, A.; Pellei, M.; Marchiò, L.; Tombesi, A.; Capparè, A.; Santini, C. Novel metalloantimalarials: Transmission blocking effects of water soluble Cu(I), Ag(I) and Au(I) phosphane complexes on the murine malaria parasite Plasmodium berghei. J. Inorg. Biochem. 2017, 166, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Parra, Y.D.J.; Andueza, F.; Ferrer, R.; Colmenarez, J.B.; Acosta, M.E.; Charris, J.; Rosenthal, P.J.; Gut, J. [(7-chloroquinolin-4-yl)amino]acetophenones and their copper(II) derivatives: Synthesis, characterization, computational studies and antimalarial activity. EXCLI J. 2019, 18, 962–987. [Google Scholar] [CrossRef]
- Meshnick, S. Chloroquine as intercalator: A hypothesis revived. Parasitol. Today 1990, 6, 77–79. [Google Scholar] [CrossRef]
- Egan, T.J. Haemozoin Formation as a Target for the Rational Design of New Antimalarials. Drug Des. Rev.-Online 2004, 1, 93–110. [Google Scholar] [CrossRef]
- Cohen, S.N.; Yielding, K.L. Inhibition of DNA and RNA polymerase reactions by chloroquine. Proc. Natl. Acad. Sci. USA 1965, 54, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Bulloch, M.S.; Ralph, S.A. Is the AT-rich DNA of malaria parasites a drug target? Trends Pharmacol. Sci. 2022, 43, 266–268. [Google Scholar] [CrossRef] [PubMed]
- Schneider, L.N.; Krauel, E.T.; Deutsch, C.; Urbahns, K.; Bischof, T.; Maibom, K.A.M.; Landmann, J.; Keppner, F.; Kerpen, C.; Hailmann, M.; et al. Stable and Storable N(CF3)2 Transfer Reagents. Chem. Eur. J. 2021, 27, 10973–10978. [Google Scholar] [CrossRef] [PubMed]
- Navarro, M.; Goitia, H.; Silva, P.; Velásquez, M.; Ojeda, L.; Fraile, G. Synthesis and characterization of new copper– and zinc–chloroquine complexes and their activities on respiratory burst of polymorphonuclear leukocytes. J. Inorg. Biochem. 2005, 99, 1630–1636. [Google Scholar] [CrossRef] [PubMed]
- Wasi, N.; Singh, H.; Gajanana, A.; Raichowdhary, A. Synthesis of metal complexes of antimalarial drugs and in vitro evaluation of their activity against Plasmodium falciparum. Inorg. Chim. Acta 1987, 135, 133–137. [Google Scholar] [CrossRef]
- Navarro, M.; Castro, W.; Biot, C. Bioorganometallic Compounds with Antimalarial Targets: Inhibiting Hemozoin Formation. Organometallics 2012, 31, 5715–5727. [Google Scholar] [CrossRef]
- Biot, C.; Delhaes, L.; N’Diaye, C.; Maciejewski, L.; Camus, D.; Dive, D.; Brocard, J. Synthesis and antimalarial activity in vitro of potential metabolites of ferrochloroquine and related compounds. Bioorg. Med. Chem. 1999, 7, 2843–2847. [Google Scholar] [CrossRef]
- Foley, M.; Tilley, L. Quinoline Antimalarials Mechanisms of Action and Resistance and Prospects for New Agents. Pharmacol. Ther. 1998, 79, 55–87. [Google Scholar] [CrossRef]
- Ziegler, J.; Linck, R.; Wright, D. Heme Aggregation Inhibitors: Antimalarial Drugs Targeting an Essential Biomineralization Process. Curr. Med. Chem. 2001, 8, 171–189. [Google Scholar] [CrossRef] [PubMed]
- Hyde, J.E. Exploring the folate pathway in Plasmodium falciparum. Acta Trop. 2005, 94, 191–206. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, A.M.; Kumar, N. Antimalarial Action of Artesunate Involves DNA Damage Mediated by Reactive Oxygen Species. Antimicrob. Agents Chemother. 2015, 59, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Long, E.C.; Barton, J.K. On demonstrating DNA intercalation. Acc. Chem. Res. 1990, 23, 271–273. [Google Scholar] [CrossRef]
- Graves, D.E.; Watkins, C.L.; Yielding, L.W. Ethidium bromide and its photoreactive analogs: Spectroscopic anlysis of deoxyribonucleic acid binding properties. Biochemistry 1981, 20, 1887–1892. [Google Scholar] [CrossRef]
- Martínez, A.; Rajapakse, C.S.; Sánchez-Delgado, R.A.; Varela-Ramirez, A.; Lema, C.; Aguilera, R.J. Arene–Ru(II)–chloroquine complexes interact with DNA, induce apoptosis on human lymphoid cell lines and display low toxicity to normal mammalian cells. J. Inorg. Biochem. 2010, 104, 967–977. [Google Scholar] [CrossRef]
- Haq, I.; Lincoln, P.; Suh, D.; Norden, B.; Chowdhry, B.Z.; Chaires, J.B. Interaction of.DELTA.- and.LAMBDA.-[Ru(phen)2DPPZ]2+ with DNA: A Calorimetric and Equilibrium Binding Study. J. Am. Chem. Soc. 1995, 117, 4788–4796. [Google Scholar] [CrossRef]
- Ganguli, P.K.; Theophanides, T. Premelting phenomenon in DNA caused by the antitumor drug cis-dichlorodiammineplatinum. Inorg. Chim. Acta 1981, 55, L43–L45. [Google Scholar] [CrossRef]
- Rosenthal, P.J. (Ed.) Antimalarial Chemotherapy: Mechanisms of Action, Resistance, and New Directions in Drug Discovery; Humana Press: New York, NY, USA, 2001; ISBN 0896036707. [Google Scholar]
- Egan, T.J.; Mavuso, W.W.; Ross, D.C.; Marques, H.M. Thermodynamic factors controlling the interaction of quinoline antimalarial drugs with ferriprotoporphyrin IX. J. Inorg. Biochem. 1997, 68, 137–145. [Google Scholar] [CrossRef]
- Egan, T.J.; Ross, D.C.; Adams, P.A. Quinoline anti-malarial drugs inhibit spontaneous formation of β-haematin (malaria pigment). FEBS Lett. 1994, 352, 54–57. [Google Scholar] [CrossRef]
- Domínguez, J.N.; León, C.; Rodrigues, J.; de Domínguez, N.G.; Gut, J.; Rosenthal, P.J. Synthesis and antimalarial activity of sulfonamide chalcone derivatives. Il Farm. 2005, 60, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Macedo, T.S.; Villarreal, W.; Couto, C.C.; Moreira, D.R.M.; Navarro, M.; Machado, M.; Prudêncio, M.; Batista, A.A.; Soares, M.B.P. Platinum(ii)–chloroquine complexes are antimalarial agents against blood and liver stages by impairing mitochondrial function. Metallomics 2017, 9, 1548–1561. [Google Scholar] [CrossRef] [PubMed]
- Macedo, T.S.; Colina-Vegas, L.; Da Paixão, M.; Navarro, M.; Barreto, B.C.; Oliveira, P.C.M.; Macambira, S.G.; Machado, M.; Prudêncio, M.; D’Alessandro, S.; et al. Chloroquine-containing organoruthenium complexes are fast-acting multistage antimalarial agents. Parasitology 2016, 143, 1543–1556. [Google Scholar] [CrossRef] [PubMed]
- Navarro, M.; Castro, W.; Madamet, M.; Amalvict, R.; Benoit, N.; Pradines, B. Metal-chloroquine derivatives as possible anti-malarial drugs: Evaluation of anti-malarial activity and mode of action. Malar. J. 2014, 13, 471. [Google Scholar] [CrossRef] [PubMed]
- Scatchard, G. The Attractions of Proteins for Small Molecules and Ions. Ann. N. Y. Acad. Sci. 1949, 51, 660–672. [Google Scholar] [CrossRef]
- Wei, C.; Jia, G.; Yuan, J.; Feng, Z.; Li, C. A Spectroscopic Study on the Interactions of Porphyrin with G-Quadruplex DNAs. Biochemistry 2006, 45, 6681–6691. [Google Scholar] [CrossRef]
- Higuera-Padilla, A.R.; Capote, J.; Ortega, D.; Castro, W.; Rodriguez-Cordero, M.; Coll, D.; Hernández-Medina, F.; Fernández-Mestre, M.; Urdanibia, I.; Taylor, P.; et al. Synthesis, characterization and biological activity of platinum(II) complexes with a tetrapyrazole ligand. Polyhedron 2015, 102, 321–328. [Google Scholar] [CrossRef]
- Lambros, C.; Vanderberg, J.P. Synchronization of Plasmodium falciparum Erythrocytic Stages in Culture. J. Parasitol. 1979, 65, 418–420. [Google Scholar] [CrossRef]
- Bogreau, H.; Renaud, F.; Bouchiba, H.; Durand, P.; Assi, S.-B.; Henry, M.-C.; Garnotel, E.; Pradines, B.; Fusai, T.; Wade, B.; et al. Genetic Diversity and Structure of African Plasmodium falciparum Populations in Urban and Rural areas. Am. J. Trop. Med. Hyg. 2006, 74, 953–959. [Google Scholar] [CrossRef]
- Henry, M.; Diallo, I.; Bordes, J.; Ka, S.; Pradines, B.; Diatta, B.; M’Baye, P.S.; Sane, M.; Thiam, M.; Gueye, P.M.; et al. Urban Malaria in Dakar, Senegal: Chemosusceptibility and Genetic Diversity of Plasmodium falciparum Isolates. Am. J. Trop. Med. Hyg. 2006, 75, 146–151. [Google Scholar] [CrossRef]








| Compounds | Absorption Titration | Emission Titration | DNA Thermal Denaturation | |||
|---|---|---|---|---|---|---|
| Kb1 (×107 M−1) a | Kb2 (×105 M−1) a | Kb1 (×107 M−1) b | Kb2 (×105 M−1) b | Tm °C * | ΔTm °C ** | |
| [Cu(CQ)(PPh3)2]NO3 (1) | 1.20 ± 0.35 | 3.15 ± 0.51 | 4.66 ± 0.03 | 13.8 ± 0.47 | 80.4 ± 0.1 | −0.3 |
| [Cu(CQ)2]Cl (2) | 2.28 ± 0.01 | 4.58 ± 0.50 | 1.58 ± 0.56 | 12.8 ± 4.16 | 80.4 ± 0.1 | −0.3 |
| CQDP | 1.07 ± 0.05 | 1.94 ± 0.19 | 5.61 ± 1.46 | 3.14 ± 1.53 | 80.7 ± 0.1 | 0.0 |
| Compound | Log K | Inhibition of β-Hematin Formation a | HAI50 (mM) in Buffer b |
|---|---|---|---|
| 1 | 5.33 ± 0.10 | + | 0.51 ± 0.01 (1.3) |
| 2 | 3.55 ± 0.06 | + | 0.66 ± 0.03 (1.0) |
| CQDP | 5.35 ± 0.03 | + | 0.64 ± 0.01 (1.0) |
| CQ-Susceptible Strain (3D7) | CQ-Resistant Strain (W2) | Susceptible Index | Resistance Index | Cell J744 | Selectivity Index (SI) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Compound | IC50 (nM) | Relative activity | IC50 (nM) | Relative activity | 3D7 IC50/ W2 IC50 | W2 IC50/ 3D7 IC50 | CC50 (µM) | J744 CC50/ 3D7 IC50 | J744 CC50/ W2 IC50 |
| Chloroquine | 8 | -- | 404 | -- | 0.02 | 50.5 | 35.1 ± 11.0 | 4387 | 86.9 |
| [Cu(CQ)(PPh3)2]NO3 | 6 | 1.33 | 231 | 1.75 | 0.03 | 38.5 | 2.9 ± 0.8 | 483 | 12.6 |
| Cu(CQ)2Cl | 13 | 0.62 | 279 | 1.45 | 0.05 | 21.5 | 29.7 ± 7.0 | 2284 | 106.4 |
| Gentian Violet | -- | -- | -- | -- | -- | -- | 1.0 ± 0.03 | -- | -- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villarreal, W.; Castro, W.; González, S.; Madamet, M.; Amalvict, R.; Pradines, B.; Navarro, M. Copper (I)-Chloroquine Complexes: Interactions with DNA and Ferriprotoporphyrin, Inhibition of β-Hematin Formation and Relation to Antimalarial Activity. Pharmaceuticals 2022, 15, 921. https://doi.org/10.3390/ph15080921
Villarreal W, Castro W, González S, Madamet M, Amalvict R, Pradines B, Navarro M. Copper (I)-Chloroquine Complexes: Interactions with DNA and Ferriprotoporphyrin, Inhibition of β-Hematin Formation and Relation to Antimalarial Activity. Pharmaceuticals. 2022; 15(8):921. https://doi.org/10.3390/ph15080921
Chicago/Turabian StyleVillarreal, Wilmer, William Castro, Sorenlis González, Marylin Madamet, Rémy Amalvict, Bruno Pradines, and Maribel Navarro. 2022. "Copper (I)-Chloroquine Complexes: Interactions with DNA and Ferriprotoporphyrin, Inhibition of β-Hematin Formation and Relation to Antimalarial Activity" Pharmaceuticals 15, no. 8: 921. https://doi.org/10.3390/ph15080921
APA StyleVillarreal, W., Castro, W., González, S., Madamet, M., Amalvict, R., Pradines, B., & Navarro, M. (2022). Copper (I)-Chloroquine Complexes: Interactions with DNA and Ferriprotoporphyrin, Inhibition of β-Hematin Formation and Relation to Antimalarial Activity. Pharmaceuticals, 15(8), 921. https://doi.org/10.3390/ph15080921