Synthesis and Evaluation of Some New 4H-Pyran Derivatives as Antioxidant, Antibacterial and Anti-HCT-116 Cells of CRC, with Molecular Docking, Antiproliferative, Apoptotic and ADME Investigations
Abstract
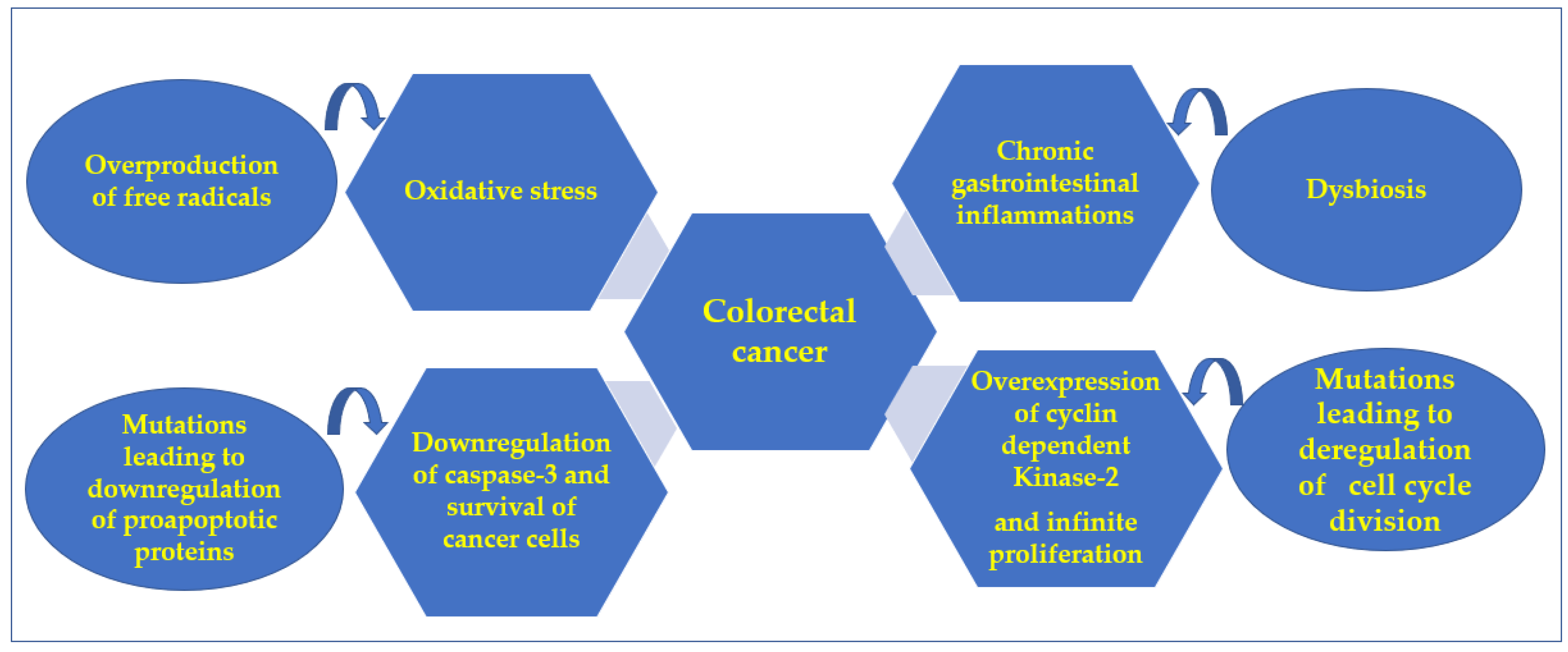
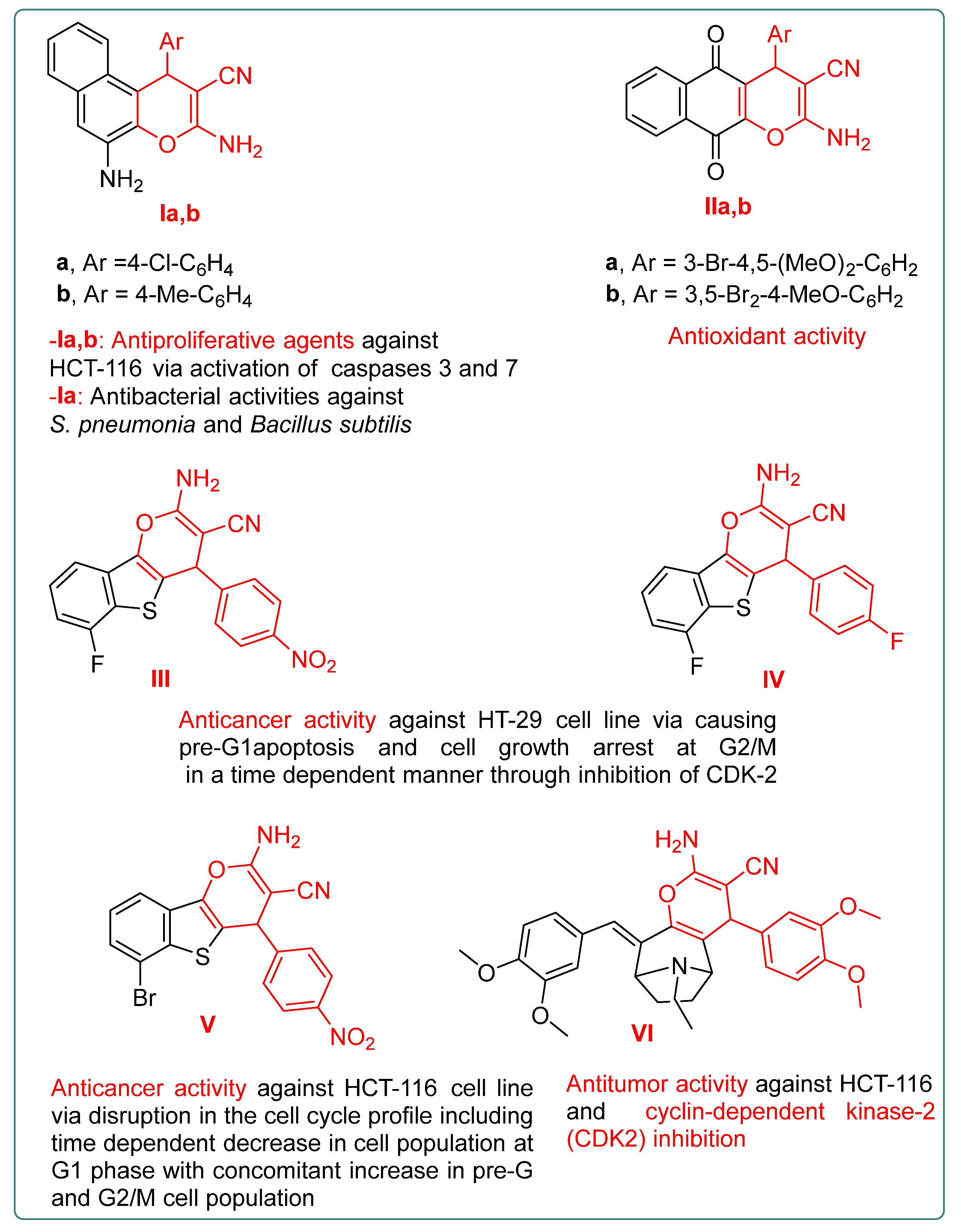
:1. Introduction
2. Results and Discussion
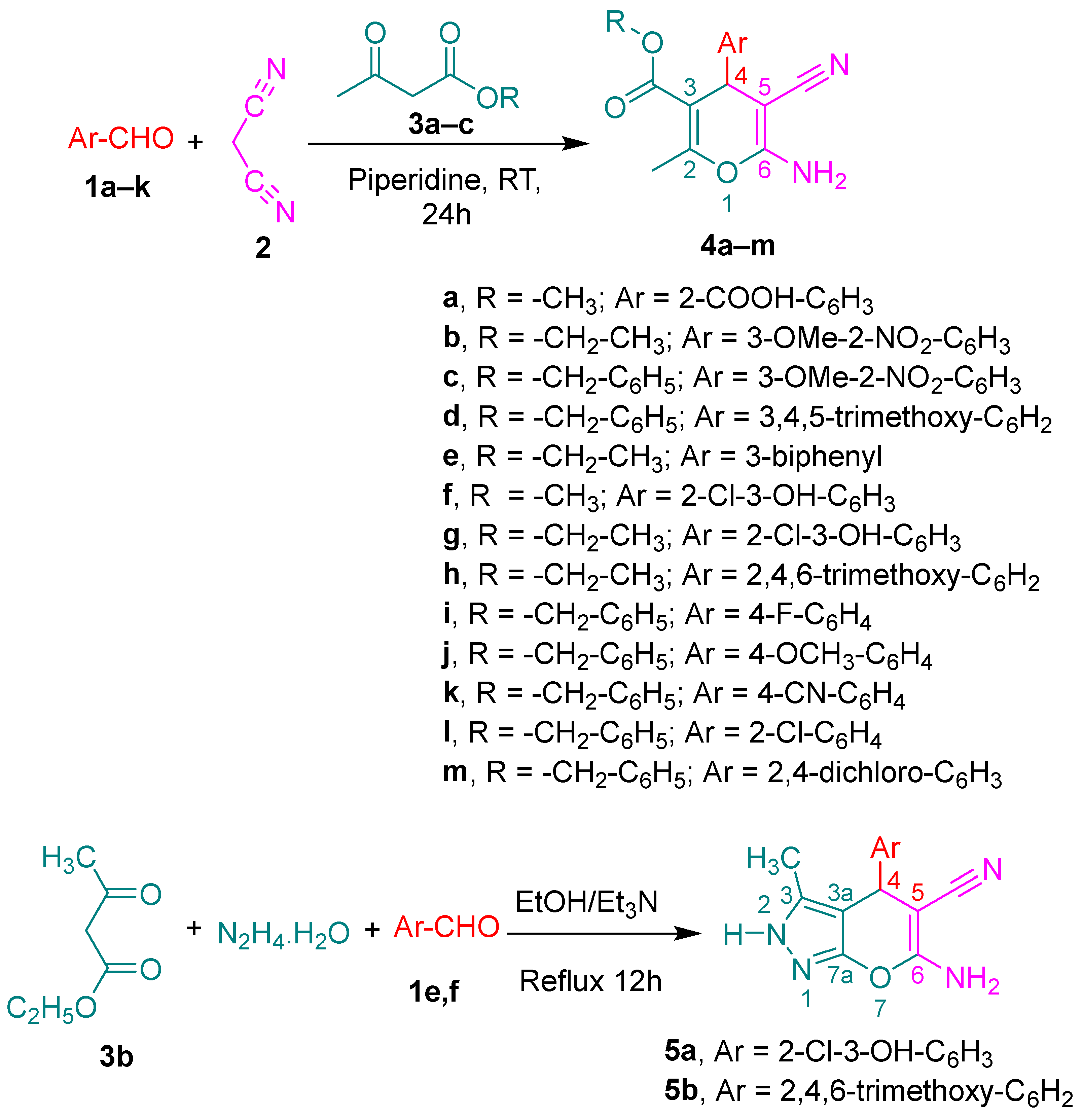
2.1. Chemistry
2.2. Biological Screening
2.2.1. Antioxidant Assays
2.2.2. Antibacterial Assays
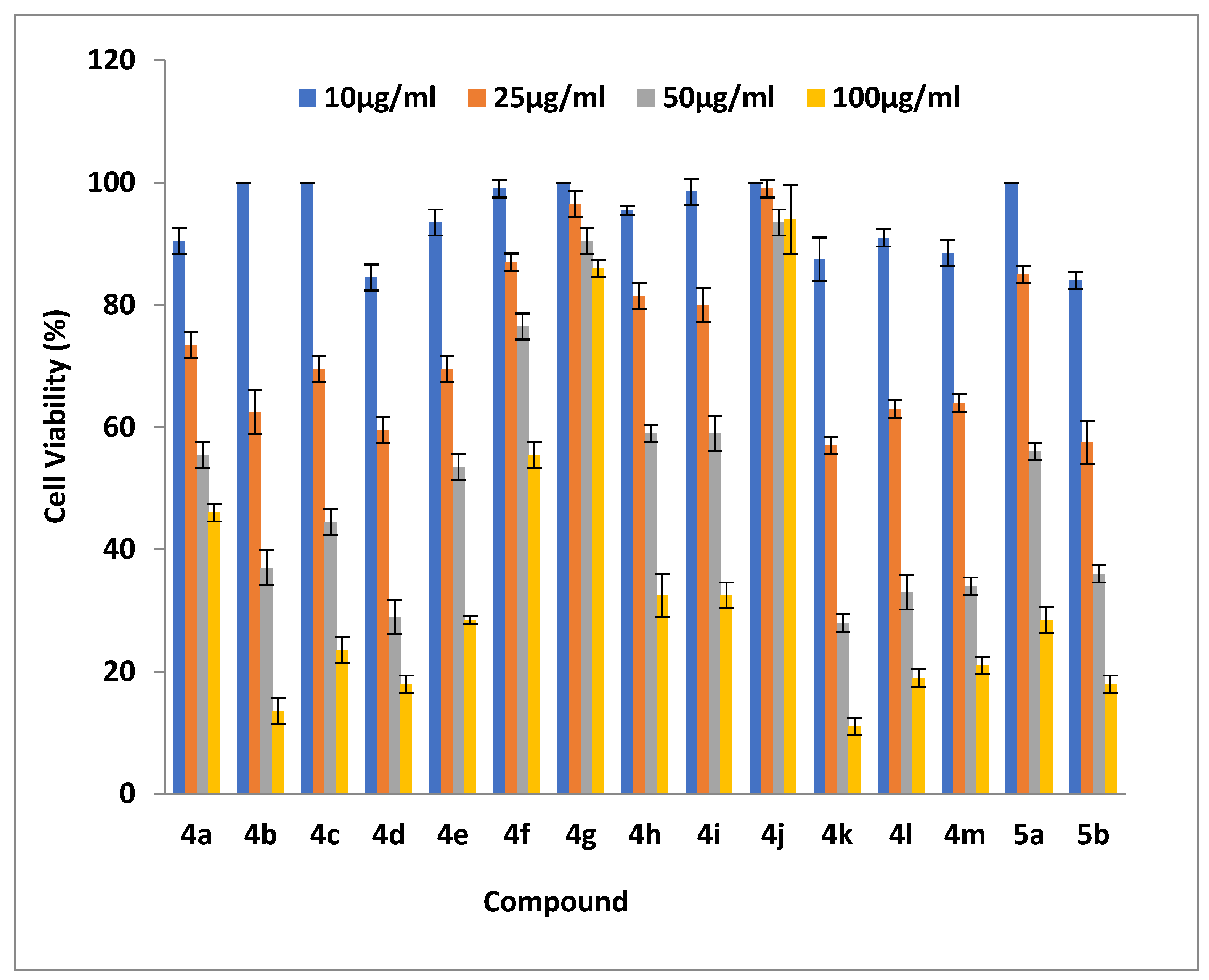
2.2.3. Cell Viability Assays
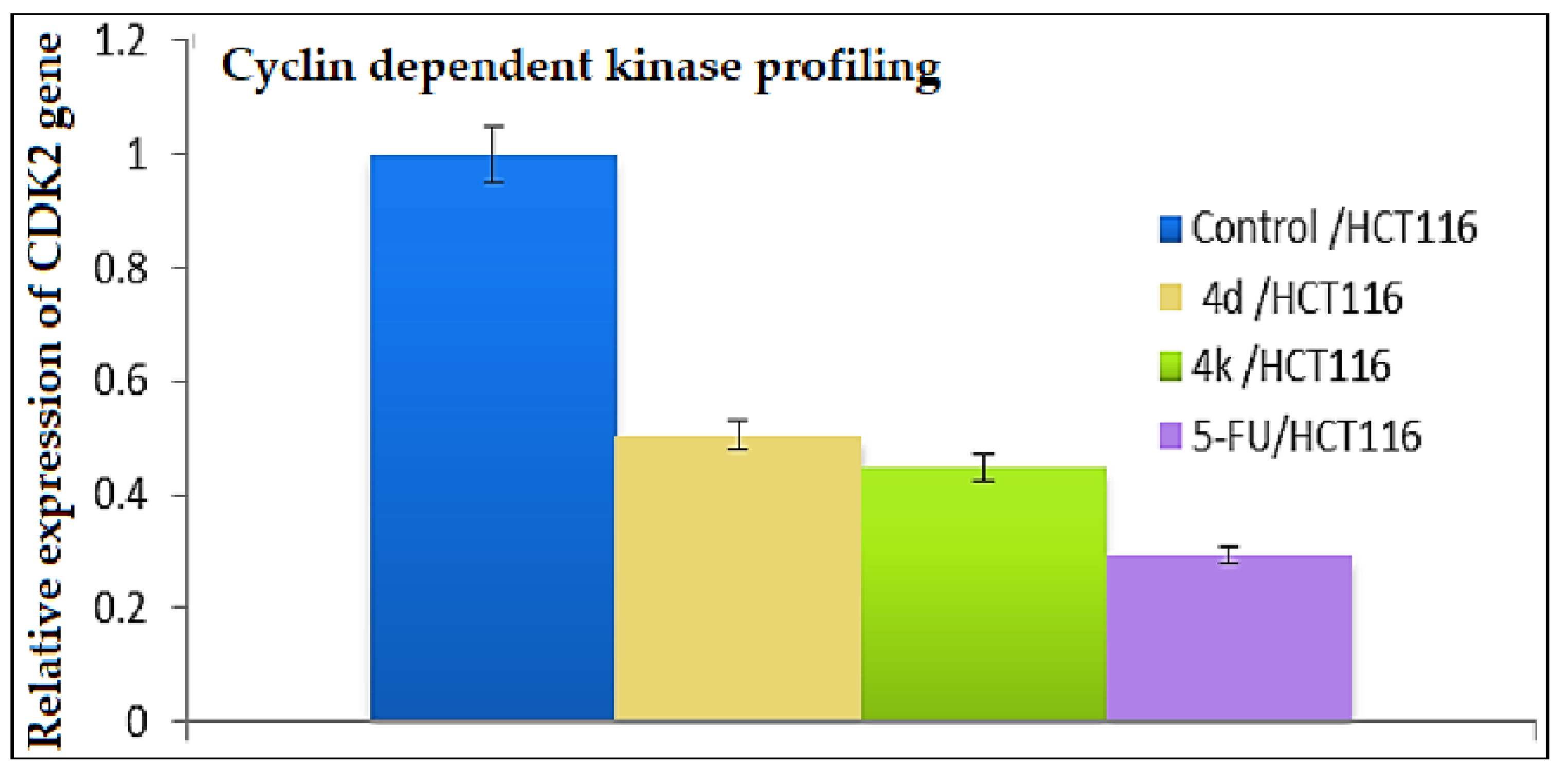
2.2.4. Investigation of the Underlying Mechanism of Action for the Antiproliferative Candidates
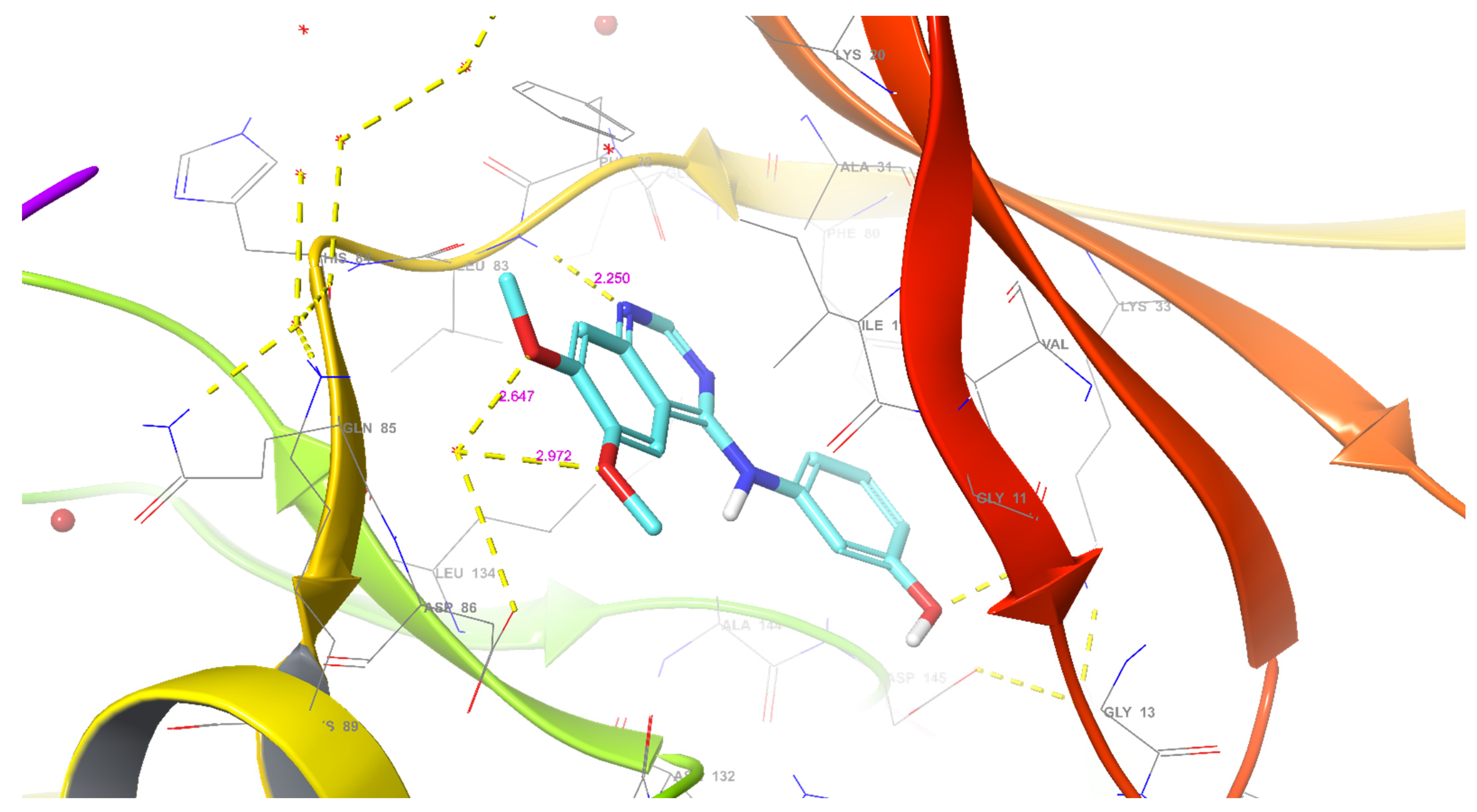
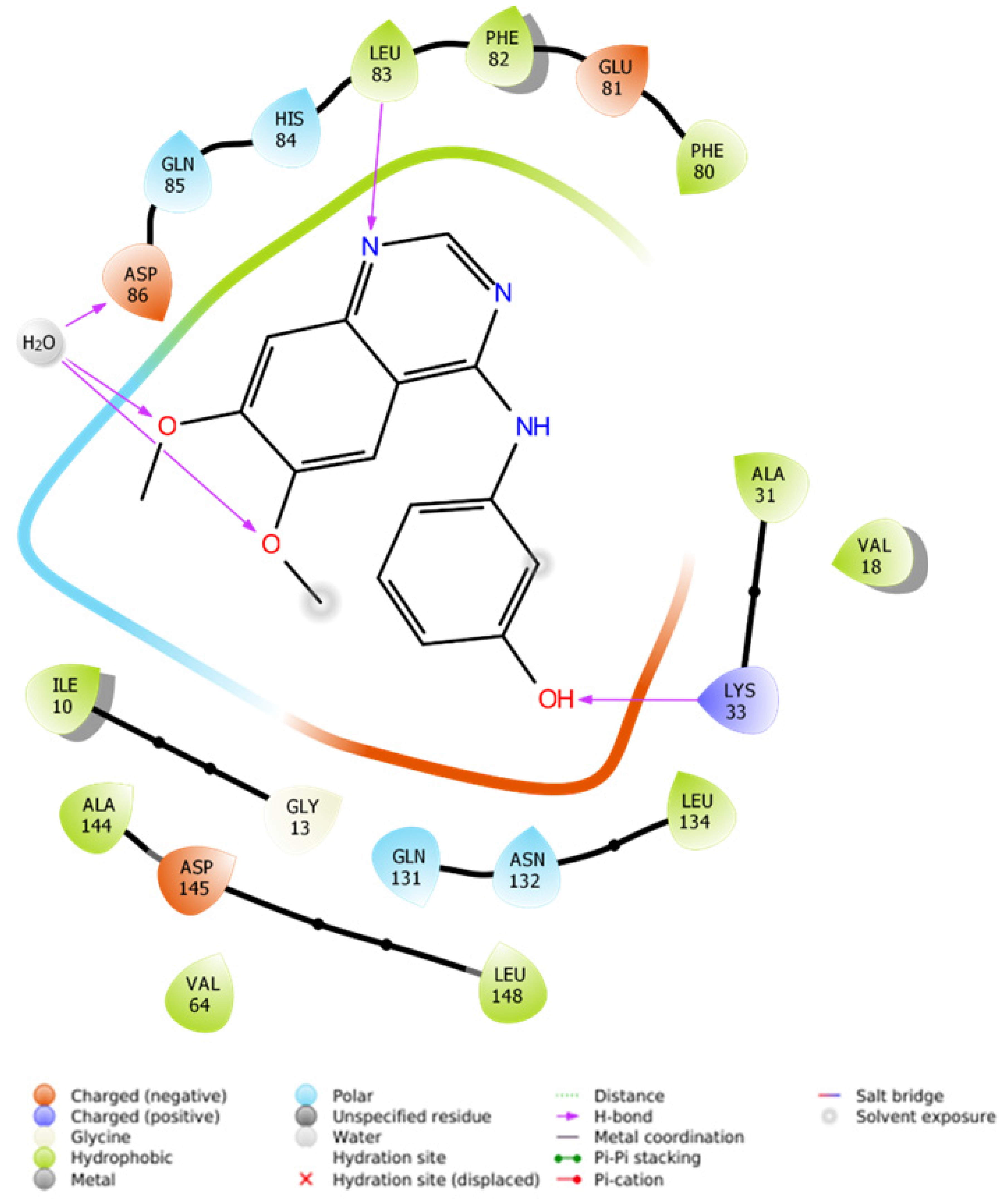
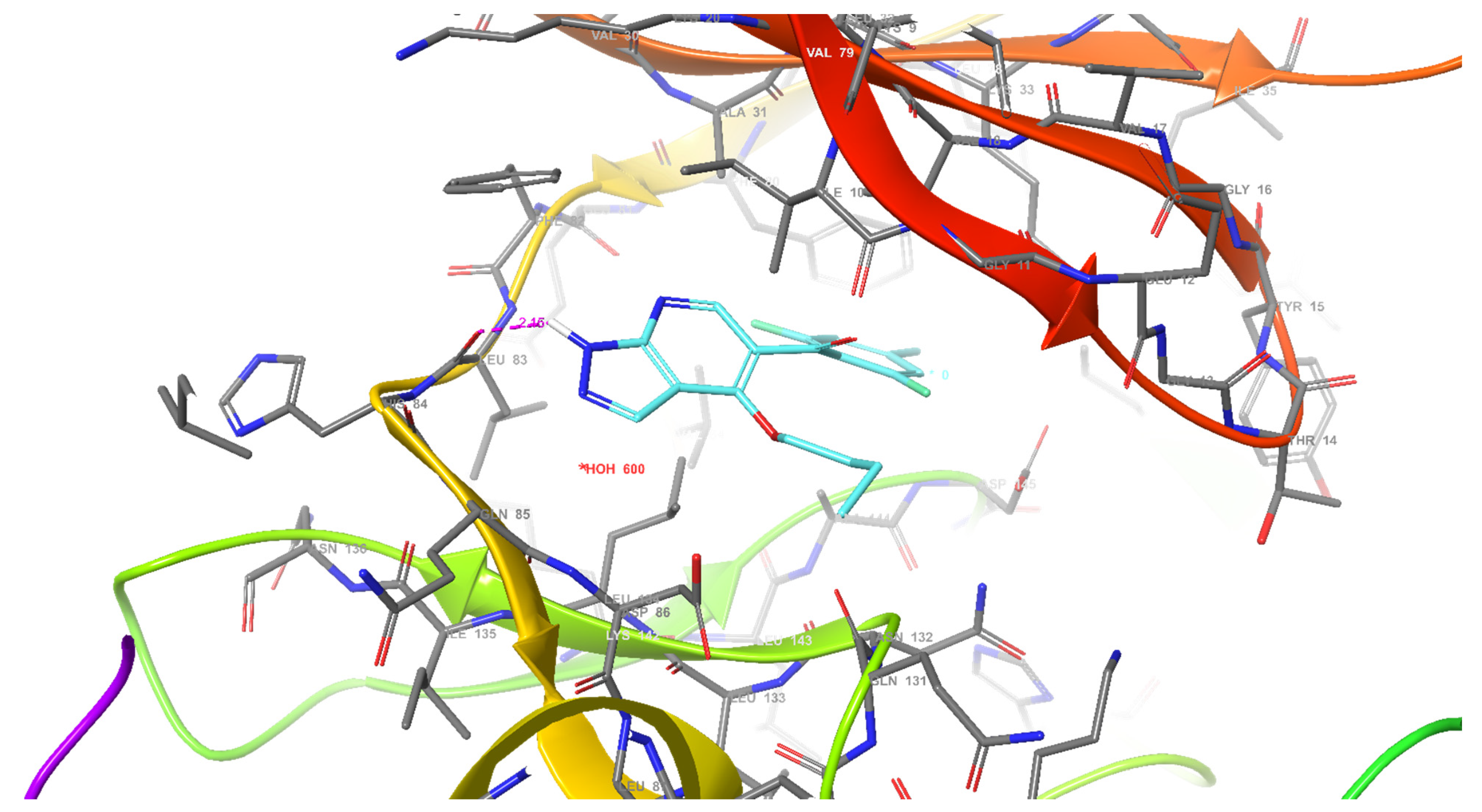
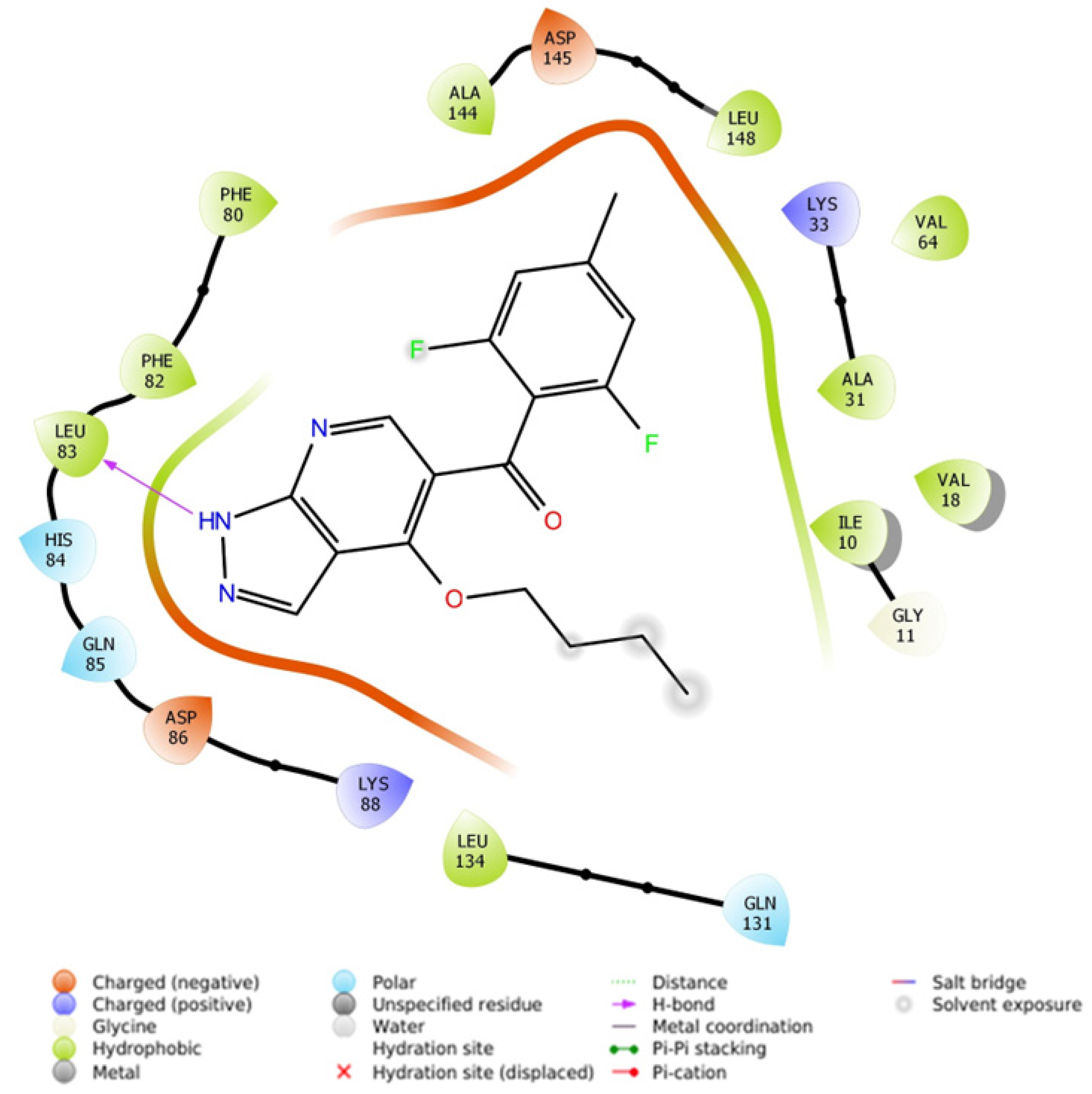
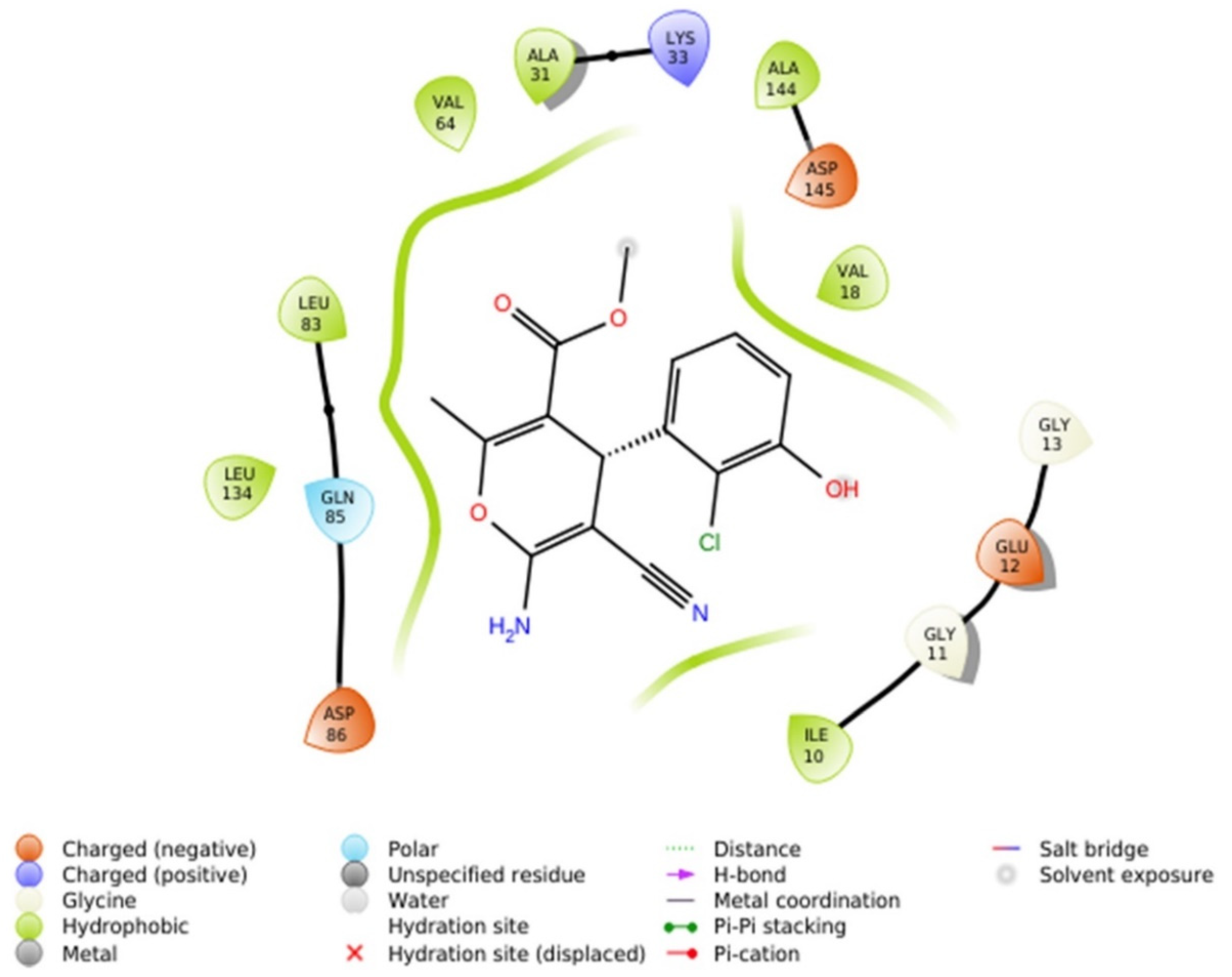
2.2.4.1. Molecular Docking Simulations against CDK2 as a Potential Molecular Target
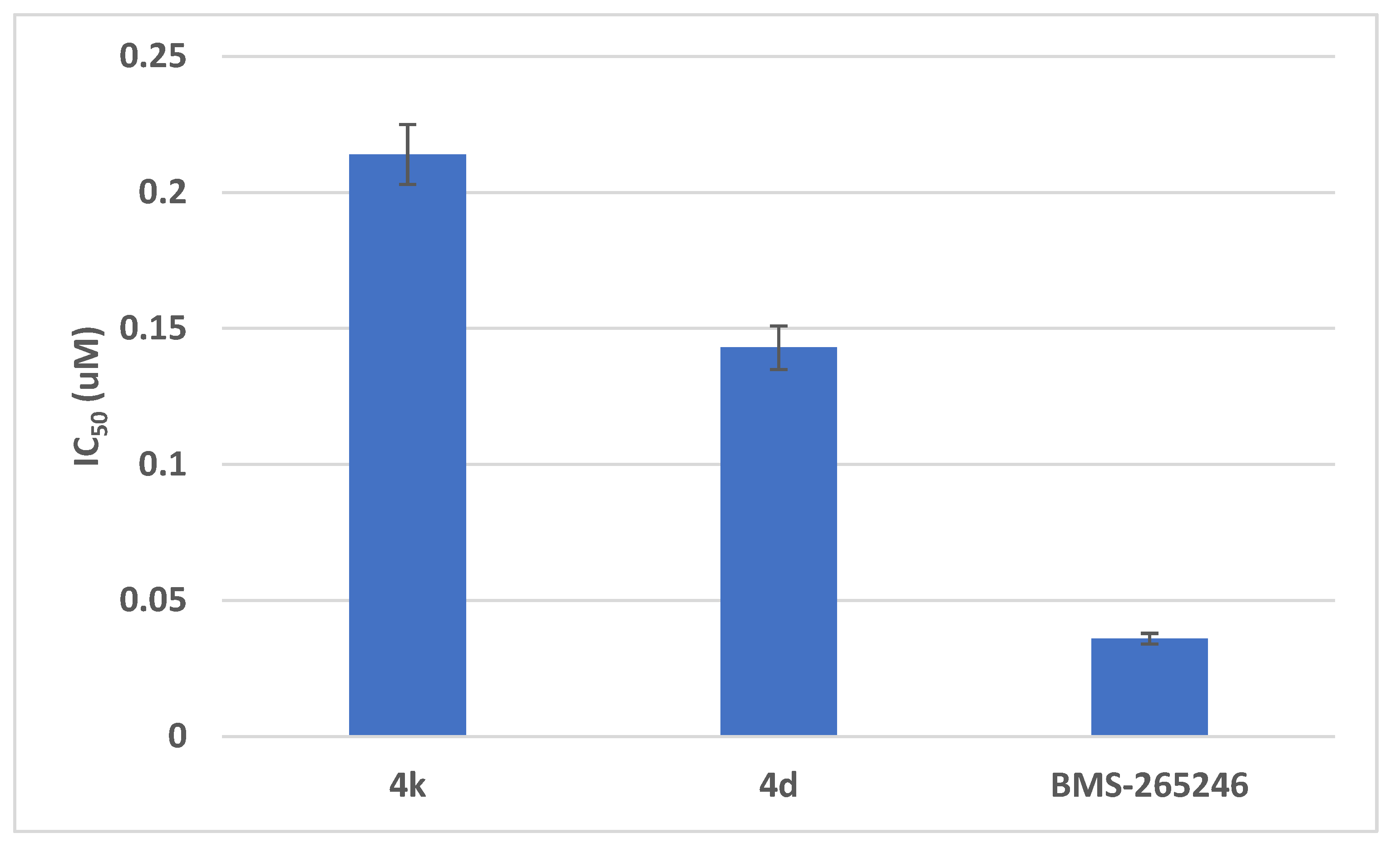
2.2.4.2. In Vitro CDK2 Inhibitory Assays
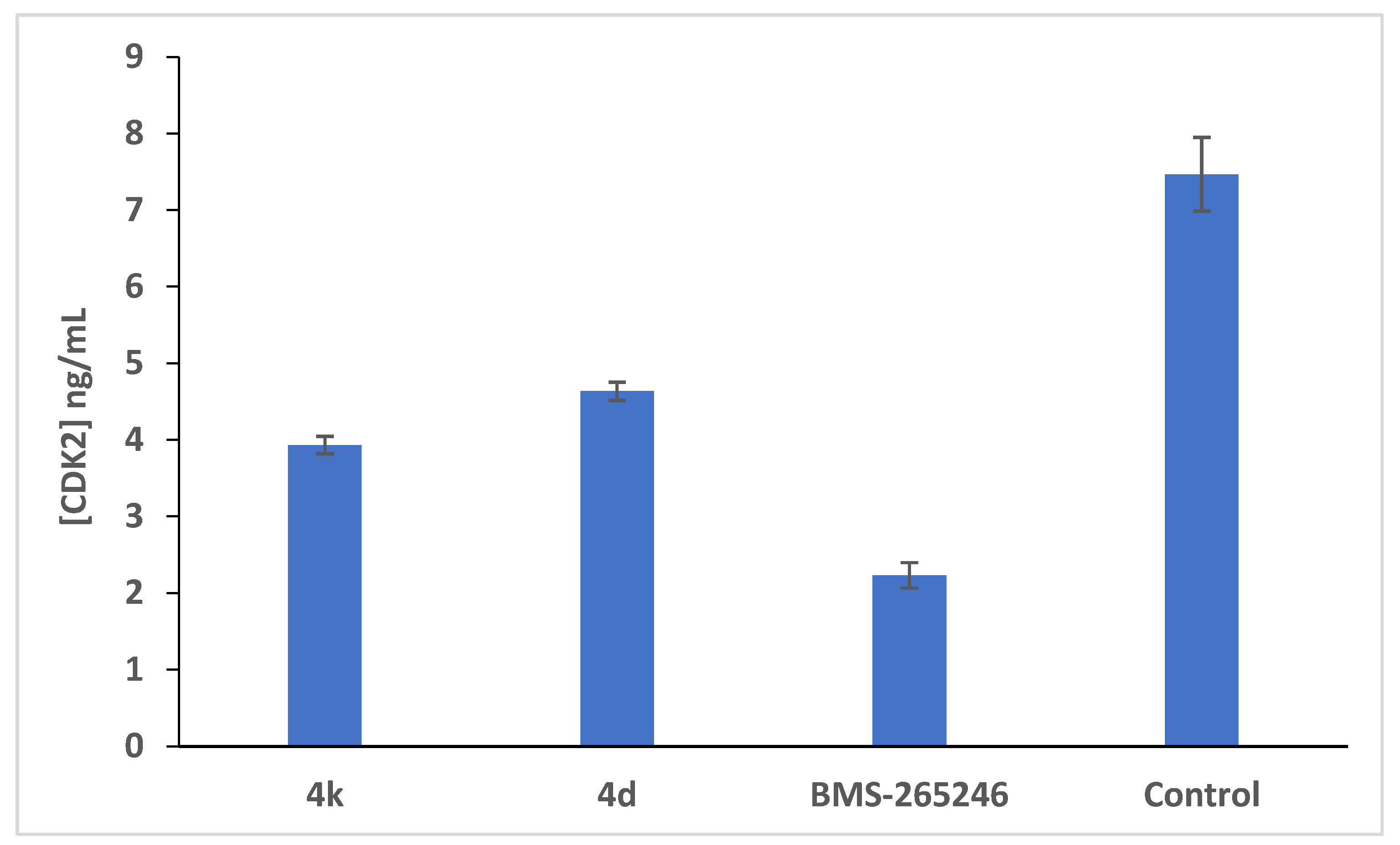
2.2.4.3. Further Mechanistic Studies via Quantitative Determination of the Concentration of CDK2 Protein and the Expression Profile of the CDK2 Gene in HCT-116 Cells Treated with the Cytotoxic Candidates
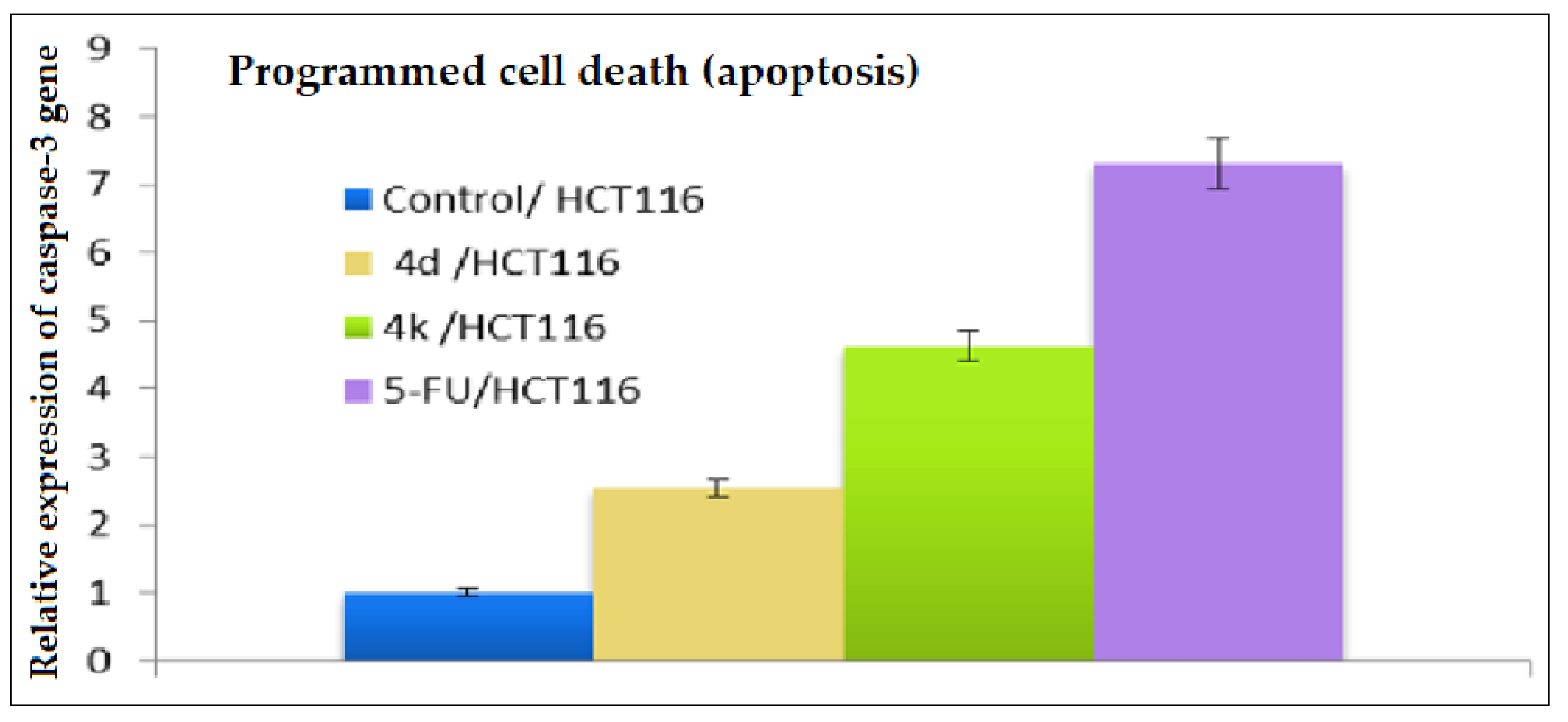
2.2.5. Investigation of the Apoptotic Potential via Real-Time PCR Determination of the Expression Profile of the Caspase-3 Gene in HCT-116 Cells Treated with Pyrans 4d and 4k
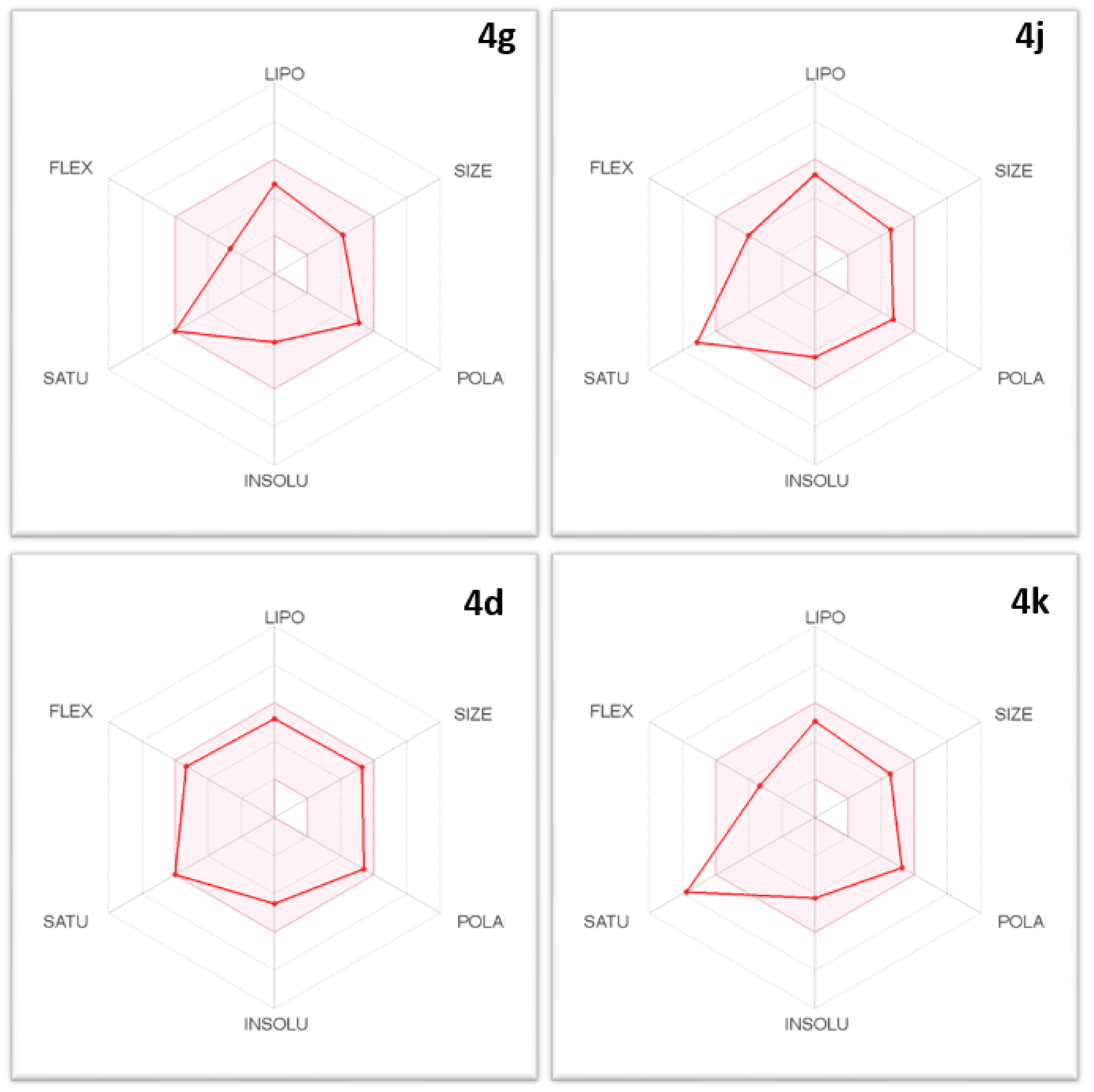
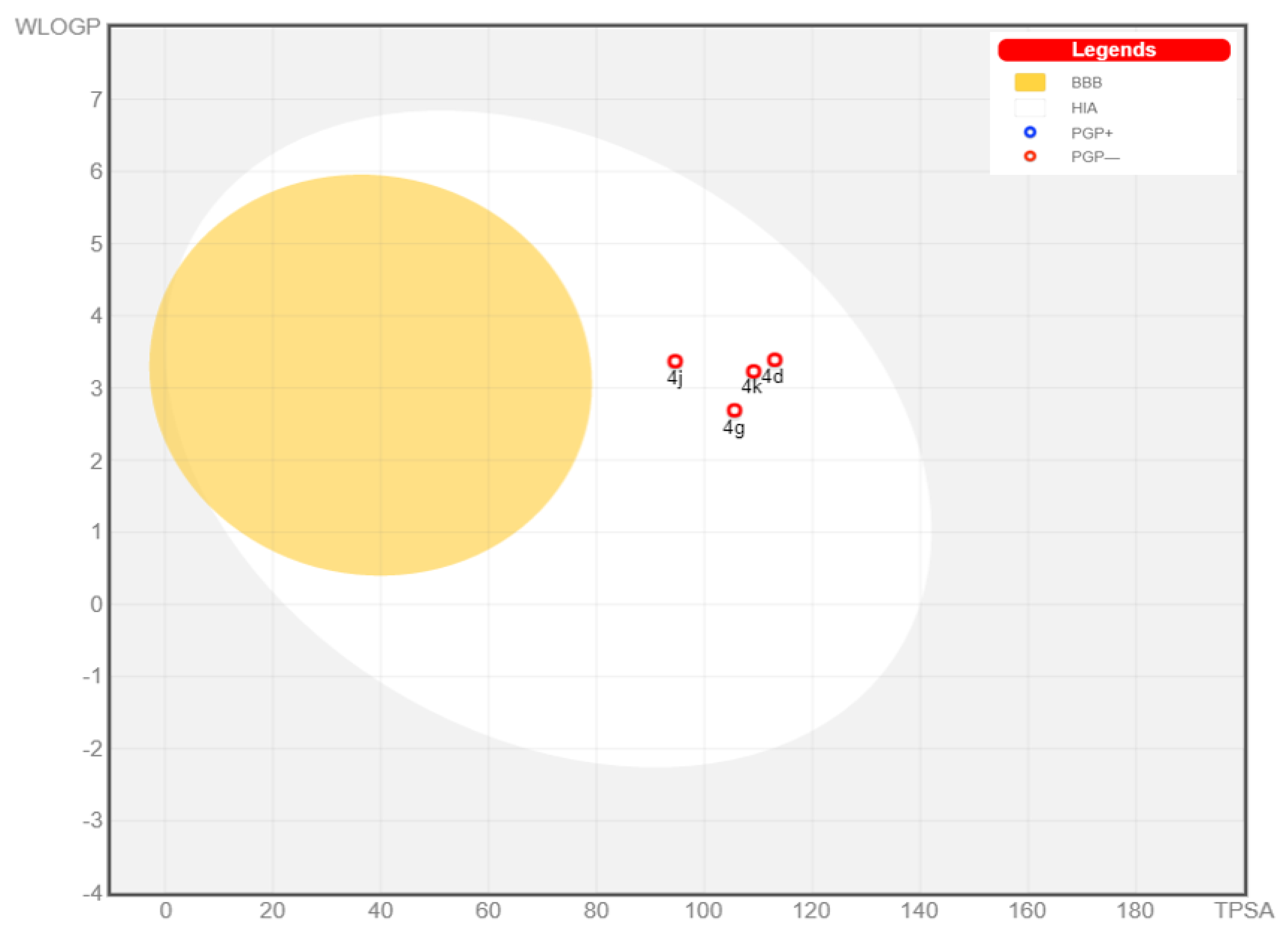
2.3. Physicochemical and Pharmacokinetic Properties and Lipinski’s Rule of Five
3. Materials and Methods
3.1. Chemistry
3.1.1. General
3.1.2. General Procedures for the Synthesis of 6-Amino-5-cyano-4-(aryl)-2-methyl-4H-pyran-3-carboxylic Acid Esters 4a–m
6-Amino-4-(2-carboxy-phenyl)-5-cyano-2-methyl-4H-pyran-3-carboxylic acid methyl ester 4a
6-Amino-5-cyano-4-(3-methoxy-2-nitro-phenyl)-2-methyl-4H-pyran-3-carboxylic acid ethyl ester 4b
6-Amino-5-cyano-4-(3-methoxy-2-nitro-phenyl)-2-methyl-4H-pyran-3-carboxylic acid benzyl ester 4c
6-Amino-5-cyano-2-methyl-4-(3,4,5-trimethoxy-phenyl)-4H-pyran-3-carboxylic acid benzyl ester 4d
6-Amino-4-biphenyl-3-yl-5-cyano-2-methyl-4H-pyran-3-carboxylic acid ethyl ester 4e
6-Amino-4-(2-chloro-3-hydroxy-phenyl)-5-cyano-2-methyl-4H-pyran-3-carboxylic acid methyl ester 4f
6-Amino-4-(2-chloro-3-hydroxy-phenyl)-5-cyano-2-methyl-4H-pyran-3-carboxylic acid ethyl ester 4g
6-Amino-5-cyano-2-methyl-4-(2,4,6-trimethoxy-phenyl)-4H-pyran-3-carboxylic acid ethyl ester 4h
6-Amino-5-cyano-4-(4-fluoro-phenyl)-2-methyl-4H-pyran-3-carboxylic acid benzyl ester 4i
6-Amino-5-cyano-4-(4-methoxy-phenyl)-2-methyl-4H-pyran-3-carboxylic acid benzyl ester 4j
6-Amino-5-cyano-4-(4-cyano-phenyl)-2-methyl-4H-pyran-3-carboxylic acid benzyl ester 4k
6-Amino-4-(2-chloro-phenyl)-5-cyano-2-methyl-4H-pyran-3-carboxylic acid benzyl ester 4l
6-Amino-5-cyano-4-(2,4-dichloro-phenyl)-2-methyl-4H-pyran-3-carboxylic acid benzyl ester 4m
3.1.3. General Procedures for Synthesis of 6-Amino-4-(aryl)-3-methyl-2,4-dihydro-pyrano[2,3-c]pyrazole-5-carbonitrile 5a,b
6-Amino-4-(2-chloro-3-hydroxy-phenyl)-3-methyl-2,4-dihydro-pyrano[2,3-c]pyrazole-5-carbonitrile 5a
6-Amino-3-methyl-4-(2,4,6-trimethoxy-phenyl)-2,4-dihydro-pyrano[2,3-c]pyrazole-5-carbonitrile 5b
3.2. Biology
3.2.1. Antioxidant Assays
DPPH Radical Scavenging Assay
Reducing Power Assay
3.2.2. Antibacterial Activity
3.2.3. Cell Culture
3.2.4. CDK2 Inhibitory Assay
3.2.5. In Vitro Quantitative Determination of CDK2 Concentration in HCT-116 Cells
3.2.6. Gene Expression Profiles
Design of the Primer
RNA Isolation and Reverse Transcription
Quantitative Real-Time PCR (qRT-PCR)
Data Analysis
3.3. In Silico Studies
ADME Evaluation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174–101181. [Google Scholar] [CrossRef]
- Currais, P.; Rosa, I.; Claro, I. Colorectal cancer carcinogenesis: From bench to bedside. World J. Gastrointest. Oncol. 2022, 14, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Carnero, A. Cell cycle deregulation: A common motif in cancer. Prog. Cell Cycle Res. 2003, 5, 5–18. [Google Scholar]
- Pizzimenti, S.; Toaldo, C.; Pettazzoni, P.; Dianzani, M.U.; Barrera, G. The “two-faced” effects of reactive oxygen species and the lipid peroxidation product 4-hydroxynonenal in the hallmarks of cancer. Cancers 2010, 2, 338–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, T.; Wang, Z.; Zhang, J. Pathomechanisms of oxidative stress in inflammatory bowel disease and potential antioxidant therapies. Oxidative Med. Cell. Longev. 2017, 2017, 4535194. [Google Scholar] [CrossRef] [PubMed]
- Carini, F.; Mazzola, M.; Rappa, F.; Jurjus, A.; Geagea, A.G.; Al Kattar, S.; Bou-Assi, T.; Jurjus, R.; Damiani, P.; Leone, A.; et al. Colorectal carcinogenesis: Role of oxidative stress and antioxidants. Anticancer Res. 2017, 37, 4759–4766. [Google Scholar] [CrossRef] [Green Version]
- Aggarwal, V.; Tuli, H.S.; Varol, A.; Thakral, F.; Yerer, M.B.; Sak, K.; Varol, M.; Jain, A.; Khan, M.; Sethi, G. Role of reactive oxygen species in cancer progression: Molecular mechanisms and recent advancements. Biomolecules 2019, 9, 735. [Google Scholar] [CrossRef] [Green Version]
- Zahra, K.F.; Lefter, R.; Ali, A.; Abdellah, E.C.; Trus, C.; Ciobica, A.; Timofte, D. The involvement of the oxidative stress status in cancer pathology: A double view on the role of the antioxidants. Oxidative Med. Cell. Longev. 2021, 2021, 9965916. [Google Scholar] [CrossRef]
- Park, C.H.; Eun, C.S.; Han, D.S. Intestinal microbiota, chronic inflammation, and colorectal cancer. Intest. Res. 2018, 16, 338–345. [Google Scholar] [CrossRef] [Green Version]
- Zou, S.; Fang, L.; Lee, M.H. Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol. Rep. 2018, 6, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, A.; Tsoi, H.; Yu, J. Fusobacterium and escherichia: Models of colorectal cancer driven by microbiota and the utility of microbiota in colorectal cancer screening. Expert Rev. Gastroenterol. Hepatol. 2015, 9, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Antonic, V.; Stojadinovic, A.; Kester, K.E.; Weina, P.J.; Brücher, B.L.; Protic, M.; Avital, I.; Izadjoo, M. Significance of infectious agents in colorectal cancer development. J. Cancer 2013, 4, 227–240. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015, 6, 20. [Google Scholar] [CrossRef]
- Dai, Z.; Zhang, J.; Wu, Q.; Chen, J.; Liu, J.; Wang, L.; Chen, C.; Xu, J.; Zhang, H.; Shi, C.; et al. The role of microbiota in the development of colorectal cancer. Int. J. Cancer 2019, 145, 2032–2041. [Google Scholar] [CrossRef]
- Geravand, M.; Fallah, P.; Yaghoobi, M.H.; Soleimanifar, F.; Farid, M.; Zinatizadeh, N.; Yaslianifard, S. Investigation of enterococcus faecalis population in patients with polyp and colorectal cancer in comparison of healthy individuals. Arq. Gastroenterol. 2019, 56, 141–145. [Google Scholar] [CrossRef]
- Ting, N.L.N.; Lau, H.C.H.; Yu, J. Cancer pharmacomicrobiomics: Targeting microbiota to optimise cancer therapy outcomes. Gut 2022, 71, 1412–1425. [Google Scholar] [CrossRef]
- Bhat, S.; Muthunatarajan, S.; Mulki, S.S.; Archana Bhat, K.; Kotian, K.H. Bacterial infection among cancer patients: Analysis of isolates and antibiotic sensitivity pattern. Int. J. Microbiol. 2021, 2021, 8883700. [Google Scholar] [CrossRef]
- Benítez-Chao, D.F.; León-Buitimea, A.; Lerma-Escalera, J.A.; Morones-Ramírez, J.R. Bacteriocins: An overview of antimicrobial, toxicity, and biosafety assessment by in vivo models. Front. Microbiol. 2021, 12, 677–695. [Google Scholar] [CrossRef] [PubMed]
- Martelli, G.; Giacomini, D. Antibacterial and antioxidant activities for natural and synthetic dual-active compounds. Eur. J. Med. Chem. 2018, 158, 91–105. [Google Scholar] [CrossRef]
- Stone, W.L.; Krishnan, K.; Campbell, S.E.; Palau, V.E. The role of antioxidants and pro-oxidants in colon cancer. World J. Gastrointest. Oncol. 2014, 6, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.; Ozsvari, B.; Lisanti, C.L.; Tanowitz, H.B.; Howell, A.; Martinez-Outschoorn, U.E.; Sotgia, F.; Michael, P.; Lisanti, M.P. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: Treating cancer like an infectious disease. Oncotarget 2015, 6, 4569–4584. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.T.; Duong, H.Q. The molecular characteristics of colorectal cancer: Implications for diagnosis and therapy. Oncol. Lett. 2018, 16, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, E.M.; Ravicz, J.R.; Liu, S.; Chawla, S.P.; Hall, F.L. Cell cycle checkpoint control: The cyclin G1/Mdm2/p53 axis emerges as a strategic target for broad-spectrum cancer gene therapy—A review of molecular mechanisms for oncologists. Mol. Clin. Oncol. 2018, 9, 115–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mani, S.; Wang, C.; Wu, K.; Francis, R.; Pestell, R. Cyclin-dependent kinase inhibitors: Novel anticancer agents. Expert Opin. Investig. Drugs 2000, 9, 1849–1870. [Google Scholar] [CrossRef]
- Ding, L.; Cao, J.; Lin, W.; Chen, H.; Xiong, X.; Ao, H.; Yu, M.; Lin, J.; Cui, Q. The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int. J. Mol. Sci. 2020, 21, 1960. [Google Scholar] [CrossRef] [Green Version]
- Coxon, C.R.; Anscombe, E.; Harnor, S.J.; Martin, M.P.; Carbain, B.; Golding, B.T.; Hardcastle, I.R.; Harlow, L.K.; Korolchuk, S.; Matheson, C.J.; et al. Cyclin-dependent kinase (CDK) inhibitors: Structure-activity relationships and insights into the CDK-2 selectivity of 6-substituted 2-arylaminopurines. J. Med. Chem. 2017, 60, 1746–1767. [Google Scholar] [CrossRef]
- Cam, W.R.; Masaki, T.; Shiratori, T.Y.; Kato, N.; Okamoto, M.; Yamaji, Y.; Igarashi, K.; Sano, T.; Omata, M. Activation of cyclin e-dependent kinase activity in colorectal cancer. Dig. Dis. Sci. 2001, 46, 2187–2198. [Google Scholar] [CrossRef]
- Lim, T.G.; Lee, S.Y.; Huang, Z.; Lim, D.Y.; Chen, H.; Jung, S.K.; Bode, A.M.; Lee, K.W.; Dong, Z. Curcumin suppresses proliferation of colon cancer cells by targeting CDK2. Cancer Prev. Res. 2014, 7, 466–474. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.N.; Li, H.; Yao, H.; Liu, X.; Li, L.; Leung, K.S.; Kung, H.F.; Lin, M.C. Adapalene inhibits the activity of cyclin-dependent kinase 2 in colorectal carcinoma. Mol. Med. Rep. 2015, 12, 6501–6508. [Google Scholar] [CrossRef] [Green Version]
- Ikwu, F.A.; Isyaku, Y.; Obadawo, B.S.; Lawal, H.A.; Ajibowu, S.A. In silico design and molecular docking study of CDK2 inhibitors with potent cytotoxic activity against HCT116 colorectal cancer cell line. J. Genet. Eng. Biotechnol. 2020, 18, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; You, J.; Wu, Q.; Meng, W.; He, Q.; Yang, B.; Zhu, C.; Cao, J. Cyclin-dependent kinases-based synthetic lethality: Evidence, concept, and strategy. Acta Pharm. Sin. B 2021, 11, 2738–2748. [Google Scholar] [CrossRef]
- Afifi, T.H.; Okasha, R.M.; Ahmed, H.E.; Ilaš, J.; Saleh, T.; Abd-El-Aziz, A.S. Structure-activity relationships and molecular docking studies of chromene and chromene based azo chromophores: A novel series of potent antimicrobial and anticancer agents. EXCLI J. 2017, 16, 868–902. [Google Scholar] [CrossRef] [PubMed]
- Al Nasr, I.S.; Jentzsch, J.; Shaikh, A.; Singh Shuveksh, P.; Koko, W.S.; Khan, T.A.; Ahmed, K.; Schobert, R.; Ersfeld, K.; Biersack, B. New pyrano-4H-benzo[g]chromene-5, 10-diones with antiparasitic and antioxidant activities. Chem. Biodivers. 2021, 18, e2000839–e2000851. [Google Scholar] [CrossRef] [PubMed]
- Mouineer, A.; Zaher, A.; El-Malah, A.; Sobh, E.A. Design, synthesis, antitumor activity, cell cycle analysis and ELISA assay for cyclin dependant kinase-2 of a new (4-aryl-6-flouro-4H-benzo [4,5] thieno [3,2-b] pyran) derivatives. Mediterr. J. Chem. 2017, 6, 165–179. [Google Scholar] [CrossRef]
- Zaher, A.F.; Abuel-Maaty, S.M.; El-Nassan, H.B.; Amer, S.A.; Abdelghany, T.M. Synthesis, antitumor screening and cell cycle analysis of novel benzothieno [3,2-b] pyran derivatives. J. Enzym. Inhib. Med. Chem. 2016, 31, 145–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samir, N.; George, R.F.; Elrazaz, E.Z.; Ayoub, I.M.; Shalaby, E.M.; Plaisier, J.R.; Demitri, N.; Wink, M. Synthesis of some tropane-based compounds targeting colon cancer. Future Med. Chem. 2020, 12, 2123–2140. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.J.; Gong, Q.T.; Wang, Y.; Yu, Y.; Liu, Y.H.; Wang, N.; Yu, X.Q. Biocatalytic tandem multicomponent reactions for one-pot synthesis of 2-Amino-4H-Pyran library and in vitro biological evaluation. Mol. Catal. 2020, 491, 110983–110992. [Google Scholar] [CrossRef]
- de Souza Siqueira, M.; da Silva-Filho, L.C. NbCl5-promoted the synthesis of 4H-pyrans through multicomponent reaction. Tetrahedron Lett. 2016, 57, 5050–5052. [Google Scholar] [CrossRef]
- Rahman, N.; Nongthombam, G.S.; Rani, J.W.; Nongrum, R.; Kharmawlong, G.K.; Nongkhlaw, R. An environment-friendly magnetic organo-nanomaterial as a potent catalyst in synthesis of pyranopyrazole derivatives. Curr. Organocatal. 2018, 5, 150–161. [Google Scholar] [CrossRef]
- Peng, Y.; Song, G. Amino-functionalized ionic liquid as catalytically active solvent for microwave-assisted synthesis of 4H-pyrans. Catal. Commun. 2007, 8, 111–114. [Google Scholar] [CrossRef]
- Al-Kadasi, M.A.; Osman, H.A.; Nazeruddin, G.M. Silica ammonium acetate as an efficient and recyclable heterogeneous catalyst for synthesis of 4H-pyran derivatives under ultrasound irradiation at ambient conditions. Am. Chem. Sci. J. 2014, 4, 587–599. [Google Scholar] [CrossRef]
- Shehab, W.S.; Ghoneim, A.A. Synthesis and biological activities of some fused pyran derivatives. Arab. J. Chem. 2016, 9, S966–S970. [Google Scholar] [CrossRef] [Green Version]
- Shahbazi, S.; Ghasemzadeh, M.A.; Shakib, P.; Zolfaghari, M.R.; Bahmani, M. Synthesis and antimicrobial study of 1, 4-dihydropyrano [2,3-c] pyrazole derivatives in the presence of amino-functionalized silica-coated cobalt oxide nanostructures as catalyst. Polyhedron 2019, 170, 172–179. [Google Scholar] [CrossRef]
- Kumar, D.; Reddy, V.B.; Sharad, S.; Dube, U.; Kapur, S. A facile one-pot green synthesis and antibacterial activity of 2-amino-4H-pyrans and 2-amino-5-oxo-5,6,7,8-tetrahydro-4H-chromenes. Eur. J. Med. Chem. 2009, 44, 3805–3809. [Google Scholar] [CrossRef]
- Shukla, P.; Sharma, A.; Fageria, L.; Chowdhury, R. Novel spiro/nonspiro pyranopyrazoles: Eco-friendly synthesis, in-vitro anticancer activity, DNA binding, and in-silico docking studies. Curr. Bioact. Compd. 2019, 15, 257–267. [Google Scholar] [CrossRef]
- Wang, D.C.; Xie, Y.M.; Fan, C.; Yao, S.; Song, H. Efficient and mild cyclization procedures for the synthesis of novel 2-amino-4H-pyran derivatives with potential antitumor activity. Chin. Chem. Lett. 2014, 25, 1011–1013. [Google Scholar] [CrossRef]
- Saleh, N.M.; El-Gazzar, M.G.; Aly, H.M.; Othman, R.A. Novel anticancer fused pyrazole derivatives as EGFR and VEGFR-2 dual TK inhibitors. Front. Chem. 2020, 7, 917–929. [Google Scholar] [CrossRef]
- El-Husseiny, W.M.; El-Sayed, M.A.A.; Abdel-Aziz, N.I.; El-Azab, A.S.; Ahmed, E.R.; Abdel-Aziz, A.A.M. Synthesis, antitumour and antioxidant activities of novel α, β-unsaturated ketones and related heterocyclic analogues: EGFR inhibition and molecular modelling study. J. Enzyme Inhib. Med. Chem. 2018, 33, 507–518. [Google Scholar] [CrossRef] [Green Version]
- El-Sayed, N.N.; Al-Otaibi, T.M.; Alonazi, M.; Masand, V.H.; Barakat, A.; Almarhoon, Z.M.; Ben Bacha, A. Synthesis and characterization of some new quinoxalin-2(1H) one and 2-methyl-3H-quinazolin-4-one derivatives targeting the onset and progression of CRC with SAR, molecular docking, and ADMET analyses. Molecules 2021, 26, 3121. [Google Scholar] [CrossRef]
- El-Sayed, N.N.; Almaneai, N.M.; Ben Bacha, A.; Al-Obeed, O.; Ahmad, R.; Abdulla, M.; Alafeefy, A.M. Synthesis and evaluation of anticancer, antiphospholipases, antiproteases, and antimetabolic syndrome activities of some 3H-quinazolin-4-one derivatives. J. Enzyme Inhib. Med. Chem. 2019, 34, 672–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahidi, F.; Zhong, Y. Measurement of antioxidant activity. J. Funct. Foods 2015, 18, 757–781. [Google Scholar] [CrossRef]
- Champlin, F.R.; Ellison, M.L.; Bullard, J.W.; Conrad, R.S. Effect of outer membrane permeabilisation on intrinsic resistance to low triclosan levels in pseudomonas aeruginosa. Int. J. Antimicrob. Agents 2005, 26, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.P.; Nagvekar, V.C.; Veeraraghavan, B.; Warrier, A.R.; Ts, D.; Ahdal, J.; Jain, R. Current perspectives on treatment of gram-positive infections in India: What is the way forward? Interdiscip. Perspect. Infect. Dis. 2019, 2019, 7601847. [Google Scholar] [CrossRef] [PubMed]
- Arnaout, M.K.; Tamburro, R.F.; Bodner, S.M.; Sandlund, J.T.; Rivera, G.K.; Pui, C.H.; Ribeiro, R.C. Bacillus cereus causing fulminant sepsis and hemolysis in two patients with acute leukemia. J. Pediatr. Hematol. Oncol. 1999, 21, 431–435. [Google Scholar] [CrossRef]
- Olmos, C.; Vilacosta, I.; López, J.; Sáez, C.; Anguita, M.; García-Granja, P.E.; Sarriá, C.; Silva, J.; Álvarez-Álvarez, B.; Martínez-Monzonis, M.A.; et al. Short-course antibiotic regimen compared to conventional antibiotic treatment for gram-positive cocci infective endocarditis: Randomized clinical trial (SATIE). BMC Infect. Dis. 2020, 20, 417–424. [Google Scholar] [CrossRef]
- Cascioferro, S.; Parrino, B.; Carbone, D.; Pecoraro, C.; Diana, P. Novel strategies in the war against antibiotic resistance. Future Med. Chem. 2021, 13, 529–531. [Google Scholar] [CrossRef]
- Fathima, A.; Rao, J.R. Selective toxicity of Catechin—A natural flavonoid towards bacteria. Appl. Microbiol. Biotechnol. 2016, 100, 6395–6402. [Google Scholar] [CrossRef]
- Damo, S.; Chazin, W.J.; Skaar, E.P.; Kehl-Fie, T.E. Inhibition of bacterial superoxide defense: A new front in the struggle between host and pathogen. Virulence 2012, 3, 325–328. [Google Scholar] [CrossRef]
- Donadio, G.; Mensitieri, F.; Santoro, V.; Parisi, V.; Bellone, M.L.; De Tommasi, N.; Izzo, V.; Dal Piaz, F. Interactions with microbial proteins driving the antibacterial activity of flavonoids. Pharmaceutics 2021, 13, 660. [Google Scholar] [CrossRef]
- Arakawa, H.; Maeda, M.; Okubo, S.; Shimamura, T. Role of hydrogen peroxide in bactericidal action of catechin. Biol. Pharm. Bull. 2004, 27, 277–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grzesik, M.; Bartosz, G.; Stefaniuk, I.; Pichla, M.; Namieśnik, J.; Sadowska-Bartosz, I. Dietary antioxidants as a source of hydrogen peroxide. Food Chem. 2019, 278, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Zubair, M.S.; Anam, S.; Khumaidi, A.; Susanto, Y.; Hidayat, M.; Ridhay, A. Molecular docking approach to identify potential anticancer compounds from Begonia (Begonia sp.). In Proceedings of the 1st International Conference on Science and Technology 2015 (ICST-2015), Universitas Gadjah Mada, Indonesia, 21 July 2016; Volume 1755, pp. 080005–080007. [Google Scholar]
- Chohan, T.A.; Qian, H.; Pan, Y.; Chen, J.Z. Cyclin-dependent kinase-2 as a target for cancer therapy: Progress in the development of CDK2 inhibitors as anti-cancer agents. Curr. Med. Chem. 2015, 22, 237–263. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Gao, W.; Zhang, L.; Pan, Y.; Zhang, S.; Wang, Y. Insights on structural characteristics and ligand binding mechanisms of CDK2. Int. J. Mol. Sci. 2015, 16, 9314–9340. [Google Scholar] [CrossRef] [Green Version]
- García-Sosa, A.T.; Mancera, R.L. The effect of a tightly bound water molecule on scaffold diversity in the computer-aided de novo ligand design of CDK2 inhibitors. J. Mol. Model. 2006, 12, 422–431. [Google Scholar] [CrossRef]
- Bramson, H.N.; Corona, J.; Davis, S.T.; Dickerson, S.H.; Edelstein, M.; Frye, S.V.; Gampe, R.T.; Harris, P.A.; Hassell, A.; Holmes, W.D.; et al. Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): Design, synthesis, enzymatic activities, and X-ray crystallographic analysis. J. Med. Chem. 2001, 44, 4339–4358. [Google Scholar] [CrossRef]
- Misra, R.N.; Xiao, H.Y.; Rawlins, D.B.; Shan, W.; Kellar, K.A.; Mulheron, J.G.; Sack, J.S.; Tokarski, J.S.; Kimball, S.D.; Webster, K.R. 1H-pyrazolo [3,4-b] pyridine inhibitors of cyclin-dependent kinases: Highly potent 2,6-difluorophenacyl analogues. Bioorg. Med. Chem. Lett. 2003, 13, 2405–2408. [Google Scholar] [CrossRef]
- Łukasik, P.; Baranowska-Bosiacka, I.; Kulczycka, K.; Gutowska, I. Inhibitors of cyclin-dependent kinases: Types and their mechanism of action. Int. J. Mol. Sci. 2021, 22, 2806. [Google Scholar] [CrossRef]
- Davis, S.T.; Benson, B.G.; Bramson, H.N.; Chapman, D.E.; Dickerson, S.H.; Dold, K.M.; Eberwein, D.J.; Edelstein, M.; Frye, S.V.; Gampe, R.T.J.; et al. Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors. Science 2001, 291, 134–137. [Google Scholar] [CrossRef] [Green Version]
- Deshpande, A.; Sicinski, P.; Hinds, P.W. Cyclins and cdks in development and cancer: A perspective. Oncogene 2005, 24, 2909–2915. [Google Scholar] [CrossRef]
- Li, J.-Q.; Miki, H.; Ohmori, M.; Wu, F.; Funamoto, Y. Expression of cyclin E and cyclin-dependent kinase 2 correlates with metastasis and prognosis in colorectal carcinoma. Hum. Pathol. 2001, 32, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Thoma, O.M.; Neurath, M.F.; Waldner, M.J. Cyclin-dependent kinase inhibitors and their therapeutic potential in colorectal cancer treatment. Front. Pharmacol. 2021, 12, 757120–757129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, J. Role of apoptosis in colon cancer biology, therapy, and prevention. Curr. Colorectal. Cancer Rep. 2013, 9, 331–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdullah, M.; Syam, A.F.; Meilany, S.; Laksono, B.; Prabu, O.G.; Bekti, H.S.; Indrawati, L.; Makmun, D. The value of caspase-3 after the application of annona muricata leaf extract in COLO-205 colorectal cancer cell line. Gastro. Res. Prac. 2017, 2017, 4357165. [Google Scholar] [CrossRef] [Green Version]
- Hosseini, S.S.; Hajikhani, B.; Faghihloo, E.; Goudarzi, H. Increased expression of caspase genes in colorectal cancer cell line by nisin. Arch. Clin. Infect. Dis. 2020, 15, e97734. [Google Scholar] [CrossRef] [Green Version]
- Yadav, P.; Yadav, R.; Jain, S.; Vaidya, A. Caspase-3: A primary target for natural and synthetic compounds for cancer therapy. Chem. Biol. Drug Des. 2021, 98, 144–165. [Google Scholar] [CrossRef]
- Ahagh, M.H.; Dehghan, G.; Mehdipour, M.; Teimuri-Mofrad, R.; Payami, E.; Sheibani, N.; Ghaffari, M.; Asadi, M. Synthesis, characterization, anti-proliferative properties and DNA binding of benzochromene derivatives: Increased Bax/Bcl-2 ratio and caspase-dependent apoptosis in colorectal cancer cell line. Bioorg. Chem. 2019, 93, 103329–1033344. [Google Scholar] [CrossRef]
- Elgaafary, M.; Fouda, A.M.; Mohamed, H.M.; Hamed, A.; El-Mawgoud, H.K.; Jin, L.; Ulrich, J.; Simmet, T.; Syrovets, T.; El-Agrody, A.M. Synthesis of β-enaminonitrile-linked 8-methoxy-1H-benzo [f] chromene moieties and analysis of their antitumor mechanisms. Front. Chem. 2021, 9, 759148. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Sharom, F.J. The P-glycoprotein efflux pump: How does it transport drugs? J. Memb. Bio. 1997, 160, 161–175. [Google Scholar] [CrossRef]
- Giménez, B.G.; Santos, M.S.; Ferrarini, M.; Fernandes, J.P.S. Evaluation of blockbuster drugs under the rule-of-five. Die Pharm. 2010, 65, 148–152. [Google Scholar] [CrossRef]
- Bersuder, P.; Hole, M.; Smith, G. Antioxidants from a heated histidine-glucose model system. I: Investigation of the antioxidant role of histidine and isolation of antioxidants by high-performance liquid chromatography. J. Am. Oil Chem. Soc. 1998, 75, 181–187. [Google Scholar] [CrossRef]
- Oyaizu, M. Studies on products of browning reaction antioxidative activities of products of browning reaction prepared from glucosamine. Jpn. J. Nutr. Diet. 1986, 44, 307–315. [Google Scholar] [CrossRef] [Green Version]
- Vanden Berghe, D.A.; Vlietinck., A.J. Screening methods for antibacterial agents from higher plants. In Methods in Plant Biochemistry: Assays for Bioactivity; Dey, P.M., Harborne, J.B., Hostettman, K., Eds.; Academic Press: London, England, 1991; Volume 6, pp. 47–69. [Google Scholar]
- Gomez, K.A.; Gomez, A.A. Statistical Procedures for Agricultural Research, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 1984; p. 680. [Google Scholar]
- Schrödinger, LLC. Release 2015-2 (2015) LigPrep; Version 3.4; Schrödinger, LLC: New York, NY, USA, 2015; p. 26400175. [Google Scholar]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein—Ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [Green Version]
- Sumirtanurdin, R.; Sungkar, S.; Hisprastin, Y.; Sidharta, K.D.; Nurhikmah, D.D. Molecular docking simulation studies of curcumin and its derivatives as cyclin-dependent kinase 2 inhibitors. Turk. J. Pharm. Sci. 2020, 17, 417–423. [Google Scholar] [CrossRef]
- Available online: http://www.swissadme.ch/index.php (accessed on 10 January 2022).


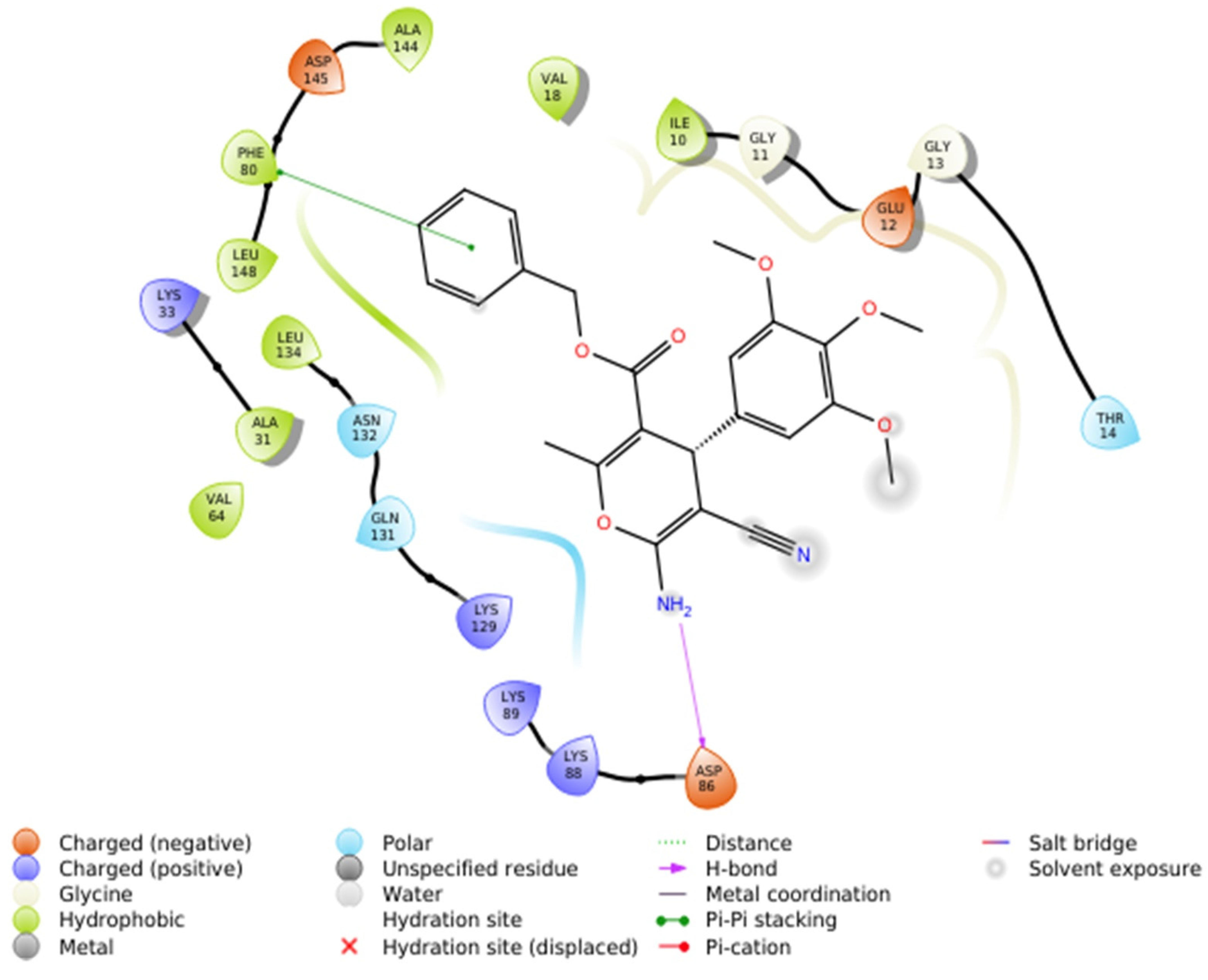
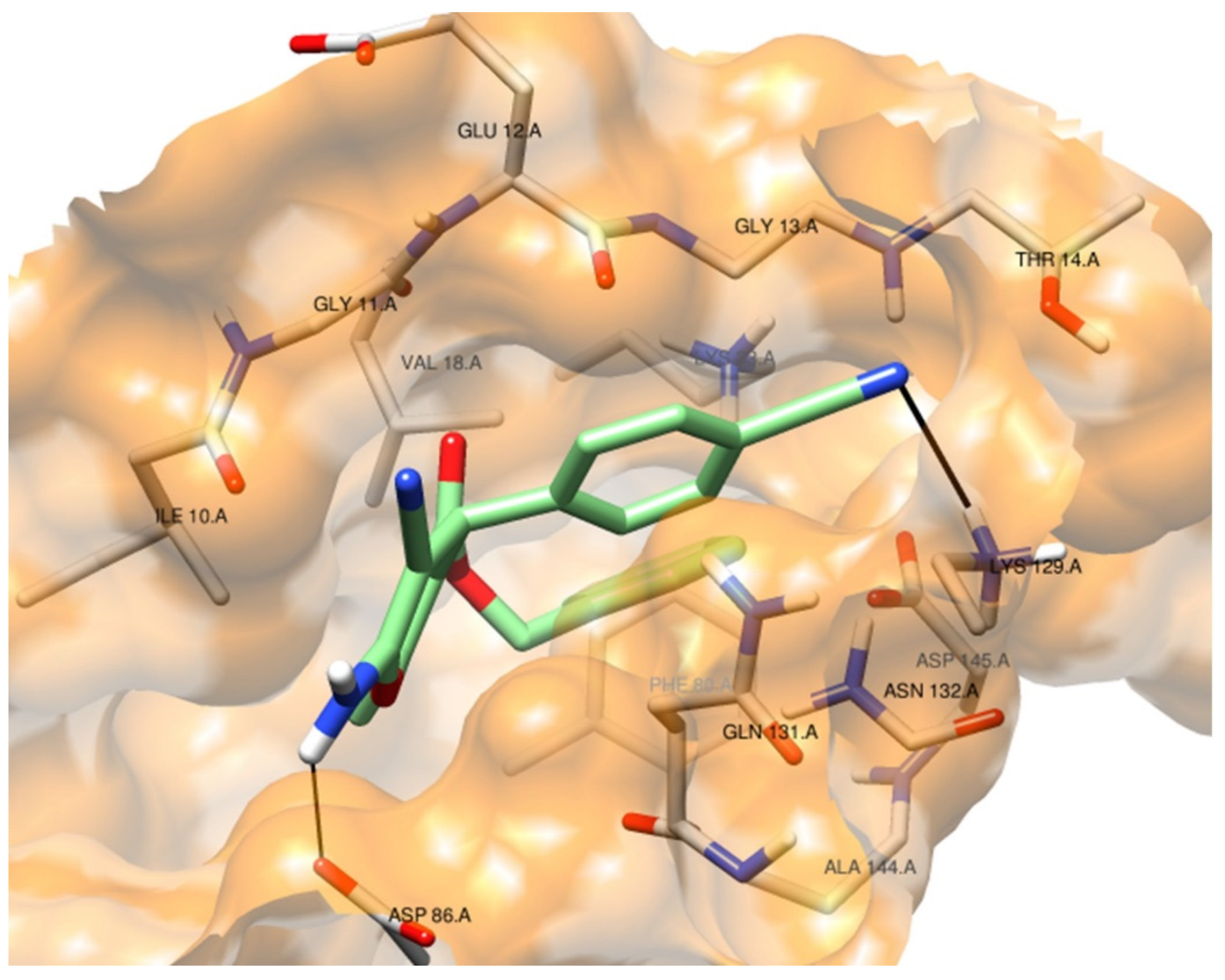
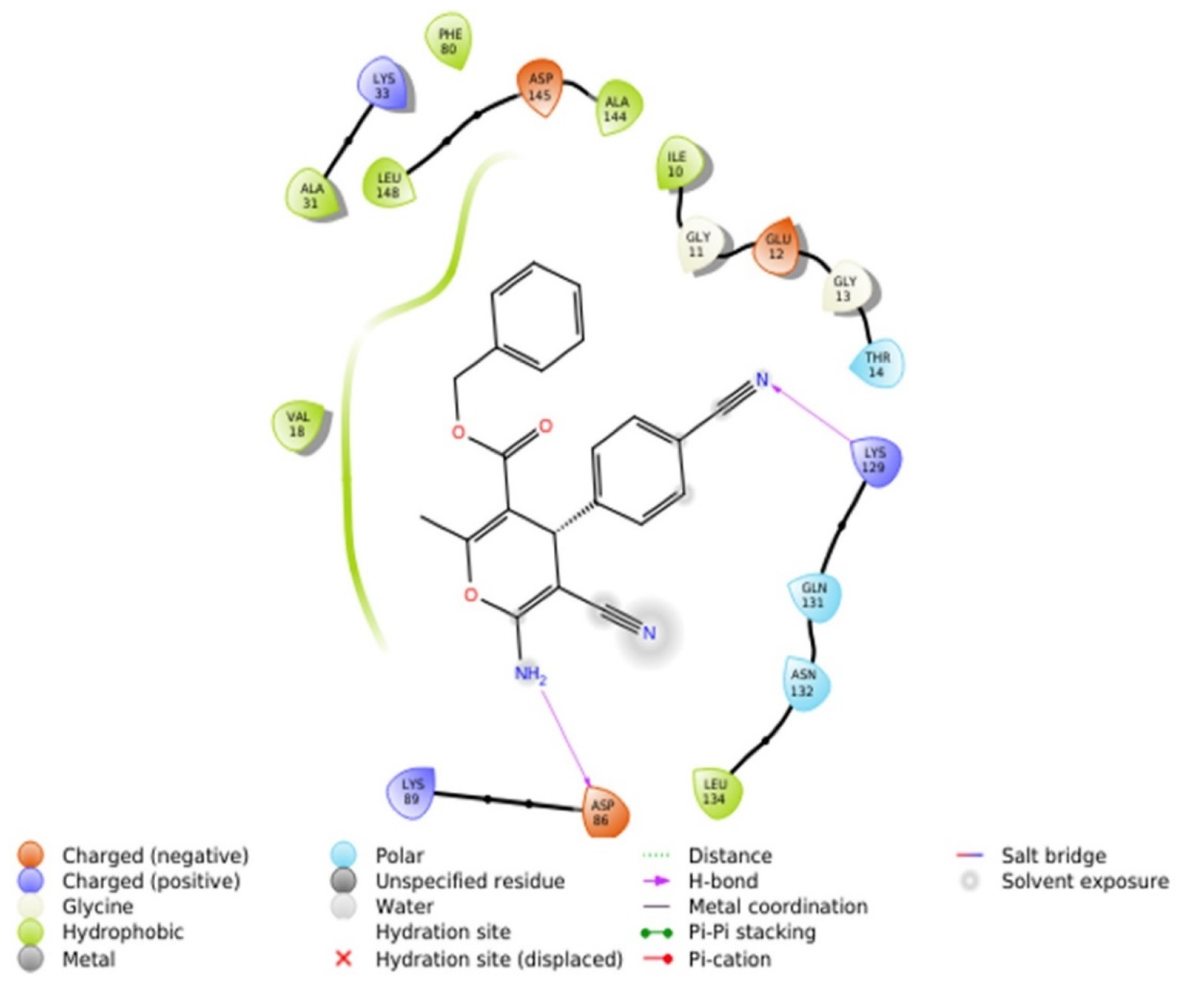
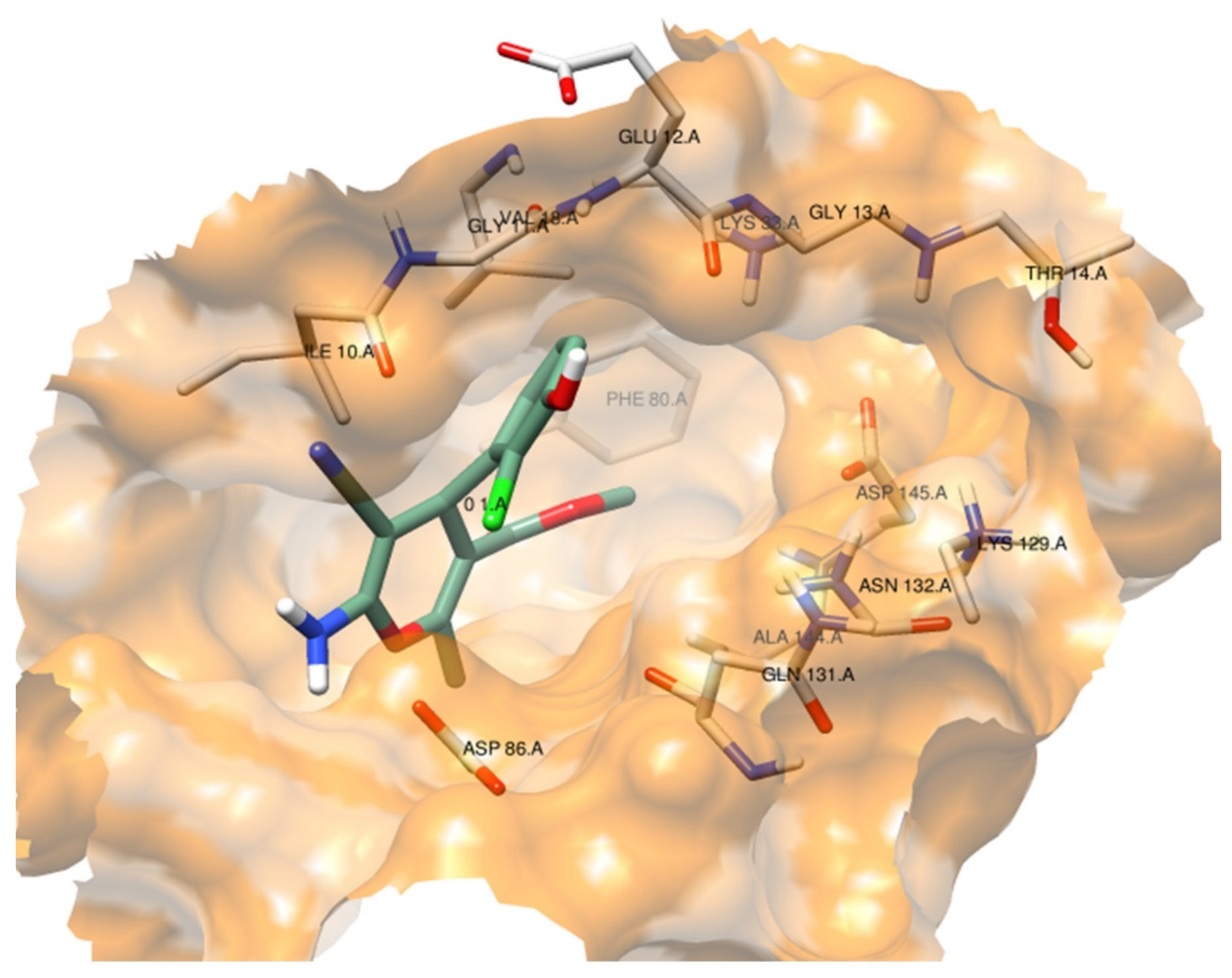

| Comp. No. | IC50 (mM) for DPPH Scavenging Potency | DPPH Radical Scavenging Effenciency (%) at 1 mg/mL | Reducing Power EC50 (mM) |
|---|---|---|---|
| 4a | 2.291 ± 0.134 | 24.15 ± 1.62 | 0.827 ± 0.045 |
| 4b | 2.560 ± 0.117 | 23.25 ± 1.06 | 1.197 ± 0.079 |
| 4c | 2.243 ± 0.050 | 57.10 ± 1.56 | 1.234 ± 0.067 |
| 4d | 0.504 ± 0.032 | 65.25 ± 1.77 | 0.149 ± 0.016 |
| 4e | 1.651 ± 0.097 | 39.25 ± 1.06 | 0.305 ± 0.039 |
| 4f | 3.336 ± 0.131 | 43.75 ± 1.77 | 0.398 ± 0.045 |
| 4g | 0.329 ± 0.042 | 90.50 ± 2.12 | 0.072 ± 0.004 |
| 4h | 1.309 ± 0.075 | 33.50 ± 2.12 | 0.221 ± 0.011 |
| 4i | 1.784 ± 0.115 | 26.05 ± 1.06 | 0.274 ± 0.039 |
| 4j | 0.194 ± 0.011 | 88.00 ± 1.41 | 0.074 ± 0.004 |
| 4k | 1.050 ± 0.075 | 40.55 ± 2.19 | 1.562 ± 0.102 |
| 4l | 0.919 ± 0.026 | 70.20 ± 1.70 | 0.801 ± 0.019 |
| 4m | 0.421 ± 0.017 | 69.0 ± 2.83 | 0.506 ± 0.034 |
| 5a | 0.826 ± 0.092 | 45.50 ± 2.12 | 0.363 ± 0.047 |
| 5b | 1.781 ± 0.082 | 37.50 ± 2.12 | 0.467 ± 0.041 |
| BHT | 0.245 ± 0.027 | 95.30 ± 0.42 | 0.089 ± 0.003 |
| B. cereus (ATCC 14579) | B. subtilis (ATCC 6633) | E. faecalis (ATCC 29122) | S. aureus (ATCC 25923) | S. epidermidis (ATCC 14990) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Comp. No. | IZ (mm) | IC50 (µM) | IZ (mm) | IC50 (µM) | IZ (mm) | IC50 (µM) | IZ (mm) | IC50 (µM) | IZ (mm) | IC50 (µM) |
| 4a | 7.50 ± 0.71 | 71.75 ± 2.04 | 5.20 ± 0.28 | 64.43 ± 3.37 | 9.00 ± 1.41 | 49.64 ± 1.81 | 8.00 ± 1.41 | 41.20 ± 2.48 | 9.50 ± 0.71 | 42.32 ± 1.33 |
| 4b | 16.50 ± 0.71 | 43.14 ± 1.98 | 19.50 ± 0.71 | 49.12 ± 1.39 | 15.50 ± 0.71 | 56.77 ± 1.59 | 20.50 ± 0.71 | 50.23 ± 1.39 | 23.50 ± 0.71 | 32.42 ± 1.39 |
| 4c | 14.00 ± 1.41 | 44.50 ± 0.83 | 11.00 ± 1.41 | 46.27 ± 1.68 | 10.00 ± 1.41 | 41.05 ± 2.18 | 15.50 ± 0.71 | 37.01 ± 2.02 | 16.65 ± 0.50 | 30.37 ± 1.00 |
| 4d | 9.50 ± 0.71 | 39.52 ± 0.80 | 12.00 ± 1.41 | 40.32 ± 1.95 | 14.50 ± 0.71 | 29.10 ± 2.27 | 9.00 ± 1.41 | 24.40 ± 1.15 | 8.50 ± 0.71 | 26.69 ± 1.15 |
| 4e | 23.00 ± 1.41 | 33.57 ± 1.58 | 20.50 ± 0.71 | 37.73 ± 1.58 | 15.50 ± 0.71 | 37.32 ± 2.16 | 16.50 ± 2.10 | 34.82 ± 2.16 | 20.50 ± 2.12 | 30.80 ± 1.58 |
| 4f | 22.50 ± 0.71 | 42.40 ± 2.65 | 19.50 ± 0.71 | 36.79 ± 0.87 | 19.00 ± 1.41 | 34.61 ± 0.59 | 16.50 ± 0.71 | 38.35 ± 1.31 | 14.50 ± 0.71 | 33.05 ± 2.65 |
| 4g | 29.00 ± 1.41 | 29.42 ± 1.49 | 28.00 ± 1.41 | 25.69 ± 1.70 | 24.50 ± 0.71 | 31.82 ± 1.49 | 27.50 ± 0.71 | 27.78 ± 1.25 | 29.75 ± 0.35 | 30.32 ± 1.49 |
| 4h | 4.50 ± 0.71 | 46.48 ± 1.52 | 6.00 ± 1.41 | 50.08 ± 2.83 | 8.00 ± 1.41 | 45.94 ± 0.75 | 17.50 ± 0.71 | 32.72 ± 0.93 | 16.50 ± 0.71 | 35.79 ± 1.52 |
| 4i | 14.50 ± 0.71 | 38.01 ± 1.37 | 9.50 ± 0.71 | 31.56 ± 1.95 | 6.50 ± 0.71 | 37.46 ± 1.37 | 9.50 ± 2.12 | 40.21 ± 1.37 | 11.50 ± 0.71 | 45.97 ± 0.96 |
| 4j | 24.50 ± 0.71 | 27.63 ± 1.51 | 21.50 ± 0.71 | 23.64 ± 0.37 | 23.20 ± 0.57 | 28.16 ± 1.51 | 25.25 ± 1.06 | 33.34 ± 1.70 | 26.65 ± 0.92 | 33.34 ± 1.70 |
| 4k | 7.00 ± 1.41 | 48.06 ± 1.35 | 4.50 ± 0.71 | 42.14 ± 2.48 | 5.50 ± 0.71 | 45.37 ± 0.57 | 5.75 ± 0.35 | 43.89 ± 0.57 | 4.50 ± 0.71 | 50.22 ± 2.48 |
| 4l | 10.00 ± 1.41 | 33.22 ± 2.42 | 11.50 ± 0.71 | 32.56 ± 1.50 | 6.00 ± 1.41 | 37.81 ± 1.50 | 9.00 ± 1.41 | 31.12 ± 0.55 | 10.50 ± 0.71 | 33.35 ± 1.10 |
| 4m | 18.50 ± 0.71 | 35.76 ± 1.20 | 21.50 ± 0.71 | 32.75 ± 2.05 | 20.50 ± 0.71 | 35.28 ± 2.22 | 17.50 ± 0.71 | 33.23 ± 1.01 | 18.50 ± 0.71 | 35.04 ± 1.88 |
| 5a | 12.50 ± 0.71 | 62.43 ± 0.46 | 12.50 ± 0.71 | 62.43 ± 0.46 | 10.50 ± 0.71 | 51.86 ± 1.39 | 14.00 ± 1.40 | 43.93 ± 1.39 | 13.50 ± 0.71 | 54.84 ± 1.88 |
| 5b | 8.00 ± 1.41 | 42.65 ± 1.66 | 8.50 ± 0.71 | 41.77 ± 1.23 | 9.75 ± 1.06 | 45.13 ± 1.87 | 5.50 ± 0.71 | 39.14 ± 1.66 | 6.60 ± 0.57 | 50.05 ± 0.61 |
| Ampicillin | 23.50 ± 0.71 | 25.76 ± 0.57 | 25.50 ± 0.71 | 37.20 ± 1.43 | 24.50 ± 0.71 | 36.49 ± 3.03 | 21.50 ± 0.71 | 38.64 ± 2.00 | 22.50 ± 0.71 | 50.09 ± 3.43 |
| E. coil (ATCC 25966) | K. pneumonia (ATCC 700603) | P. aeruginosa (ATCC 27853) | S. enteric (ATCC 43972) | |||||
|---|---|---|---|---|---|---|---|---|
| Comp. No. | IZ (mm) | IC50 (µM) | IZ (mm) | IC50 (µM) | IZ (mm) | IC50 (µM) | IZ (mm) | IC50 (µM) |
| 4a | 4.5 ± 0.71 | 138.41 ± 6.68 | 2.5 ± 0.7 | 131.09 ± 3.50 | 5.5 ± 0.7 | 37.6 ± 0.5 | 4.5 ± 0.7 | 119.63 ± 1.91 |
| 4b | 2.5 ± 0.71 | 100.19 ± 3.90 | 1.5 ± 0.7 | 99.07 ± 2.50 | 2.5 ± 0.7 | 84.05 ± 5.01 | 1.5 ± 0.7 | 118.14 ± 3.34 |
| 4c | 2.5 ± 0.71 | 93.74 ± 4.98 | 3.5 ± 0.7 | 112.25 ± 1.42 | 7.5 ± 0.7 | 85.90 ± 2.61 | 8.5 ± 0.7 | 100.26 ± 2.61 |
| 4d | 3.5 ± 0.71 | 88.67 ± 2.52 | 4.5 ± 0.7 | 72.63 ± 0.69 | 6.5 ± 0.7 | 67.36 ± 1.37 | 7.0 ± 0.0 | 65.77 ± 2.29 |
| 4e | 2.7 ± 0.35 | 103.22 ± 3.05 | 2.5 ± 0.7 | 90.73 ± 2.78 | 4.5 ± 0.7 | 87.96 ± 2.78 | 3.5 ± 0.7 | 96.14 ± 2.50 |
| 4f | 7.5 ± 0.71 | 95.72 ± 5.61 | 4.5 ± 0.7 | 82.00 ± 3.43 | 6.0 ± 1.4 | 104.76 ± 2.81 | 6.5 ± 0.7 | 94.16 ± 3.43 |
| 4g | 11.0 ± 1.41 | 61.84 ± 5.38 | 11.5 ± 0.7 | 61.84 ± 2.99 | 13.5 ± 0.7 | 54.07 ± 4.48 | 11.5 ± 0.7 | 53.02 ± 3.29 |
| 4h | 1.5 ± 0.71 | 114.05 ± 0.80 | 3.5 ± 0.7 | 118.33 ± 2.67 | 4.5 ± 0.7 | 89.48 ± 2.94 | 5.5 ± 0.7 | 111.65 ± 2.94 |
| 4i | 5.5 ± 0.71 | 82.57 ± 1.65 | 6.5 ± 0.7 | 79.86 ± 3.02 | 8.5 ± 0.7 | 62.02 ± 2.20 | 8.0 ± 1.4 | 91.52 ± 2.74 |
| 4j | 10.5 ± 0.71 | 57.12 ± 0.80 | 12.0 ± 1.4 | 48.35 ± 3.99 | 14.5 ± 0.7 | 49.68 ± 4.78 | 14.5 ± 0.7 | 51.81 ± 2.92 |
| 4k | 4.5 ± 0.71 | 80.24 ± 4.31 | 2.5 ± 0.7 | 77.82 ± 2.96 | 3.5 ± 0.7 | 73.24 ± 3.50 | 2.5 ± 0.7 | 75.53 ± 2.42 |
| 4l | 3.5 ± 0.7 | 90.07 ± 2.63 | 2.5 ± 0.7 | 77.46 ± 7.08 | 4.5 ± 0.7 | 72.74 ± 2.63 | 4.0 ± 0.0 | 77.60 ± 1.58 |
| 4m | 5.5 ± 0.71 | 69.35 ± 2.89 | 6.5 ± 0.7 | 66.70 ± 2.41 | 9.5 ± 0.7 | 60.92 ± 2.17 | 8.5 ± 0.7 | 61.77 ± 2.17 |
| 5a | 2.8 ± 0.21 | 109.67 ± 5.95 | 6.0 ± 1.4 | 96.46 ± 4.96 | 5.5 ± 0.7 | 85.89 ± 3.30 | 4.5 ± 0.7 | 104.72 ± 1.32 |
| 5b | 3.5 ± 0.71 | 116.84 ± 4.09 | 4.5 ± 0.7 | 109.54 ± 1.75 | 6.5 ± 0.7 | 92.30 ± 1.46 | 7.5 ± 0.7 | 106.03 ± 2.92 |
| Ampicillin | 23.5 ± 0.71 | 57.96 ± 4.87 | 21.0 ± 1.4 | 32.91 ± 2.29 | 21.5 ± 0.7 | 41.50 ± 1.72 | 19.5 ± 0.7 | 14.88 ± 0.34 |
| Comp. No. | Mean IC50 (µg/mL) | Mean IC50 (µM) |
|---|---|---|
| 4a | 51.99 ± 4.54 | 165.43 ± 14.44 |
| 4b | 36.51 ± 3.18 | 101.60 ± 8.83 |
| 4c | 43.01 ± 2.03 | 102.07 ± 4.82 |
| 4d | 32.78 ± 2.22 | 75.10 ± 5.09 |
| 4e | 49.71 ± 3.53 | 137.92 ± 9.78 |
| 4f | 106.67 ± 4.29 | 332.59 ± 13.38 |
| 4g | ND 1 | ND 1 |
| 4h | 69.21 ± 4.29 | 184.89 ± 11.46 |
| 4i | 65.95 ± 4.53 | 180.99 ± 12.42 |
| 4j | ND 1 | ND 1 |
| 4k | 31.90 ± 2.83 | 85.88 ± 7.61 |
| 4l | 35.55 ± 1.77 | 93.35 ± 4.63 |
| 4m | 37.10 ± 2.10 | 89.33 ± 5.05 |
| 5a | 60.00 ± 3.28 | 189.21 ± 10.84 |
| 5b | 33.52 ± 2.42 | 97.91 ± 7.06 |
| Ligands | Glide Score XP | Glide Energy Kcal/mol |
|---|---|---|
| 4k | −6.383 | −43.311 |
| 4d | −6.224 | −48.700 |
| 4f | −4.160 | −30.726 |
| Co-crystallized ligand (DTQ 1) | −9.649 | −49.122 |
| BMS-265246 | −7.187 | −47.340 |
| Comp. No. | Properties | Comment |
|---|---|---|
| 4g |
| No violation |
| ||
| ||
| 4j |
| No violation |
| ||
| 4d |
| No violation |
| ||
| ||
| ||
| 4k |
| No violation |
| ||
| Gene Name | Forward Primer | Reverse Primer |
|---|---|---|
| Caspase-3 | F 5′-GGAAGCGAATCAATGGACTCTGG-3′ | R 5′-GCATCGACATCTGTACCAGACC-3′ |
| Housekeeping gene (GAPDH) | F 5′-GCACCGTCAAGGCTGAGAAC-3′ | R 5′-ATGGTGGTGAAGACGCCAGT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Sayed, N.N.E.; Zaki, M.E.A.; Al-Hussain, S.A.; Ben Bacha, A.; Berredjem, M.; Masand, V.H.; Almarhoon, Z.M.; Omar, H.S. Synthesis and Evaluation of Some New 4H-Pyran Derivatives as Antioxidant, Antibacterial and Anti-HCT-116 Cells of CRC, with Molecular Docking, Antiproliferative, Apoptotic and ADME Investigations. Pharmaceuticals 2022, 15, 891. https://doi.org/10.3390/ph15070891
El-Sayed NNE, Zaki MEA, Al-Hussain SA, Ben Bacha A, Berredjem M, Masand VH, Almarhoon ZM, Omar HS. Synthesis and Evaluation of Some New 4H-Pyran Derivatives as Antioxidant, Antibacterial and Anti-HCT-116 Cells of CRC, with Molecular Docking, Antiproliferative, Apoptotic and ADME Investigations. Pharmaceuticals. 2022; 15(7):891. https://doi.org/10.3390/ph15070891
Chicago/Turabian StyleEl-Sayed, Nahed N. E., Magdi E. A. Zaki, Sami A. Al-Hussain, Abir Ben Bacha, Malika Berredjem, Vijay H. Masand, Zainab M. Almarhoon, and Hanaa S. Omar. 2022. "Synthesis and Evaluation of Some New 4H-Pyran Derivatives as Antioxidant, Antibacterial and Anti-HCT-116 Cells of CRC, with Molecular Docking, Antiproliferative, Apoptotic and ADME Investigations" Pharmaceuticals 15, no. 7: 891. https://doi.org/10.3390/ph15070891
APA StyleEl-Sayed, N. N. E., Zaki, M. E. A., Al-Hussain, S. A., Ben Bacha, A., Berredjem, M., Masand, V. H., Almarhoon, Z. M., & Omar, H. S. (2022). Synthesis and Evaluation of Some New 4H-Pyran Derivatives as Antioxidant, Antibacterial and Anti-HCT-116 Cells of CRC, with Molecular Docking, Antiproliferative, Apoptotic and ADME Investigations. Pharmaceuticals, 15(7), 891. https://doi.org/10.3390/ph15070891







