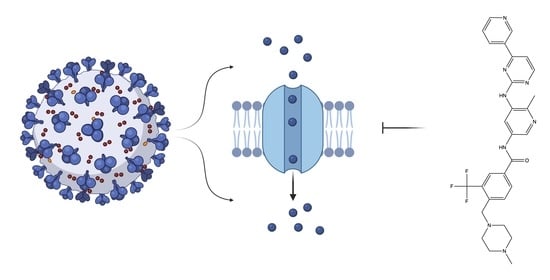
Targeting Viral Ion Channels: A Promising Strategy to Curb SARS-CoV-2
Abstract
:1. Introduction
2. Results
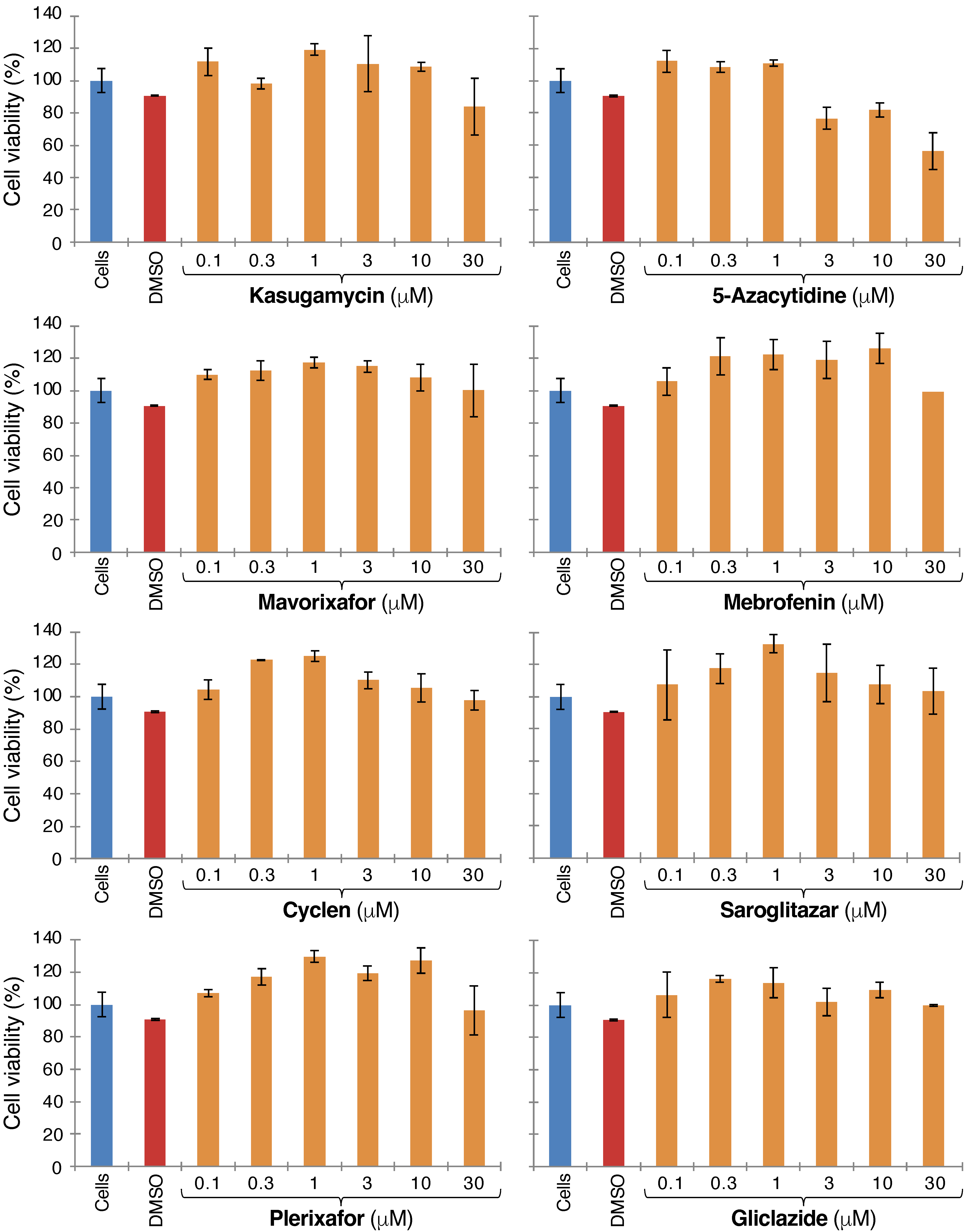
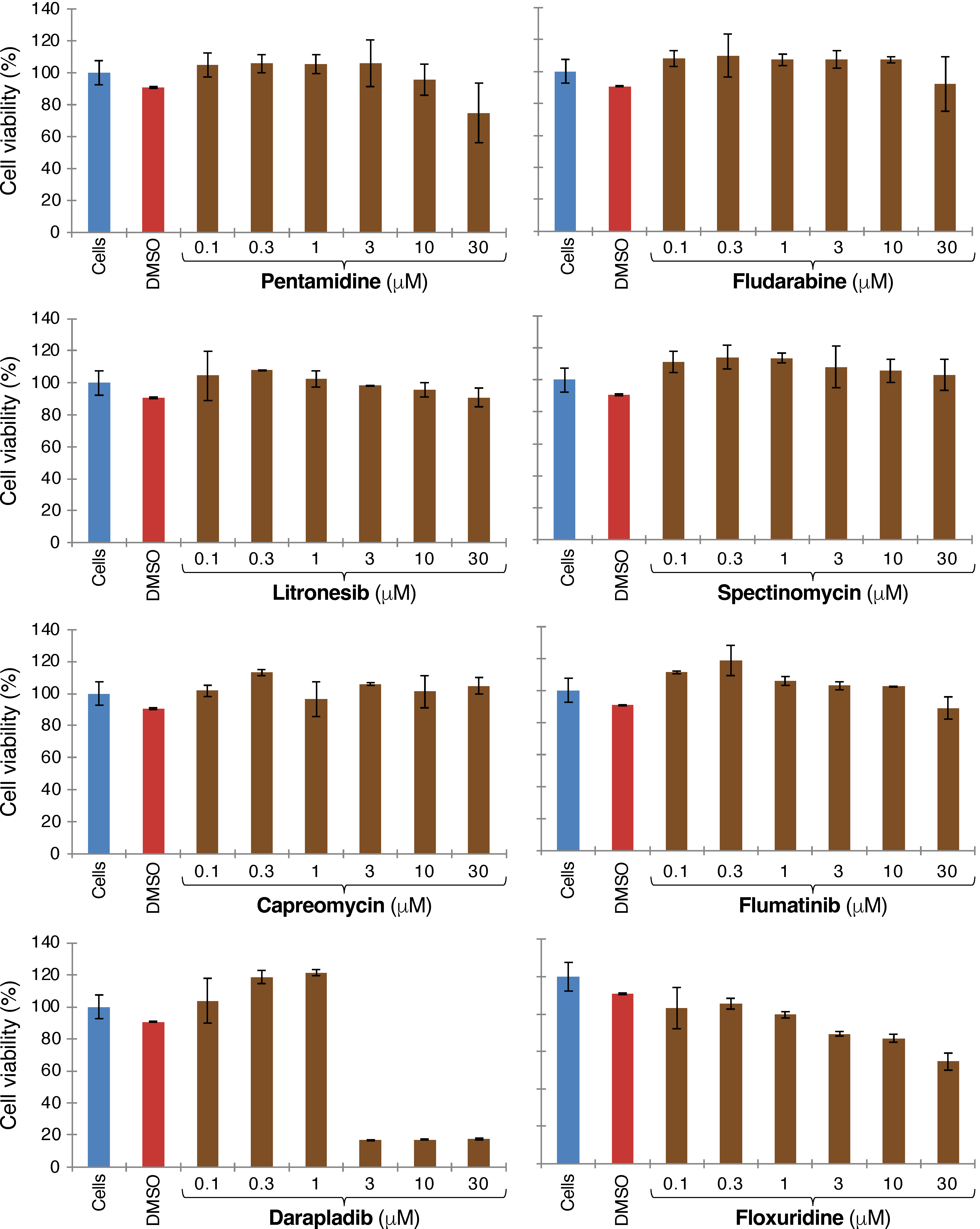
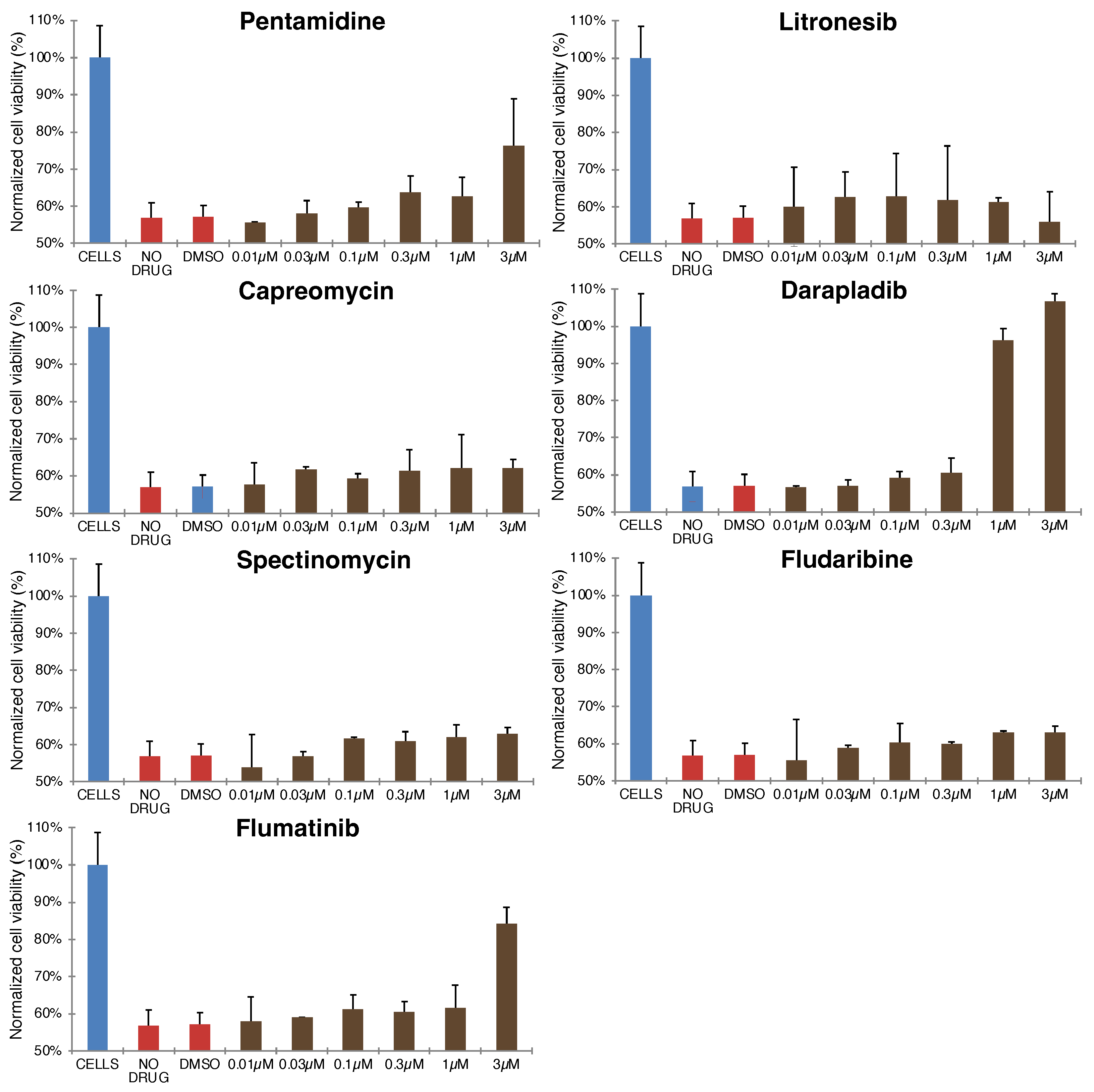
2.1. Blocker Toxicity
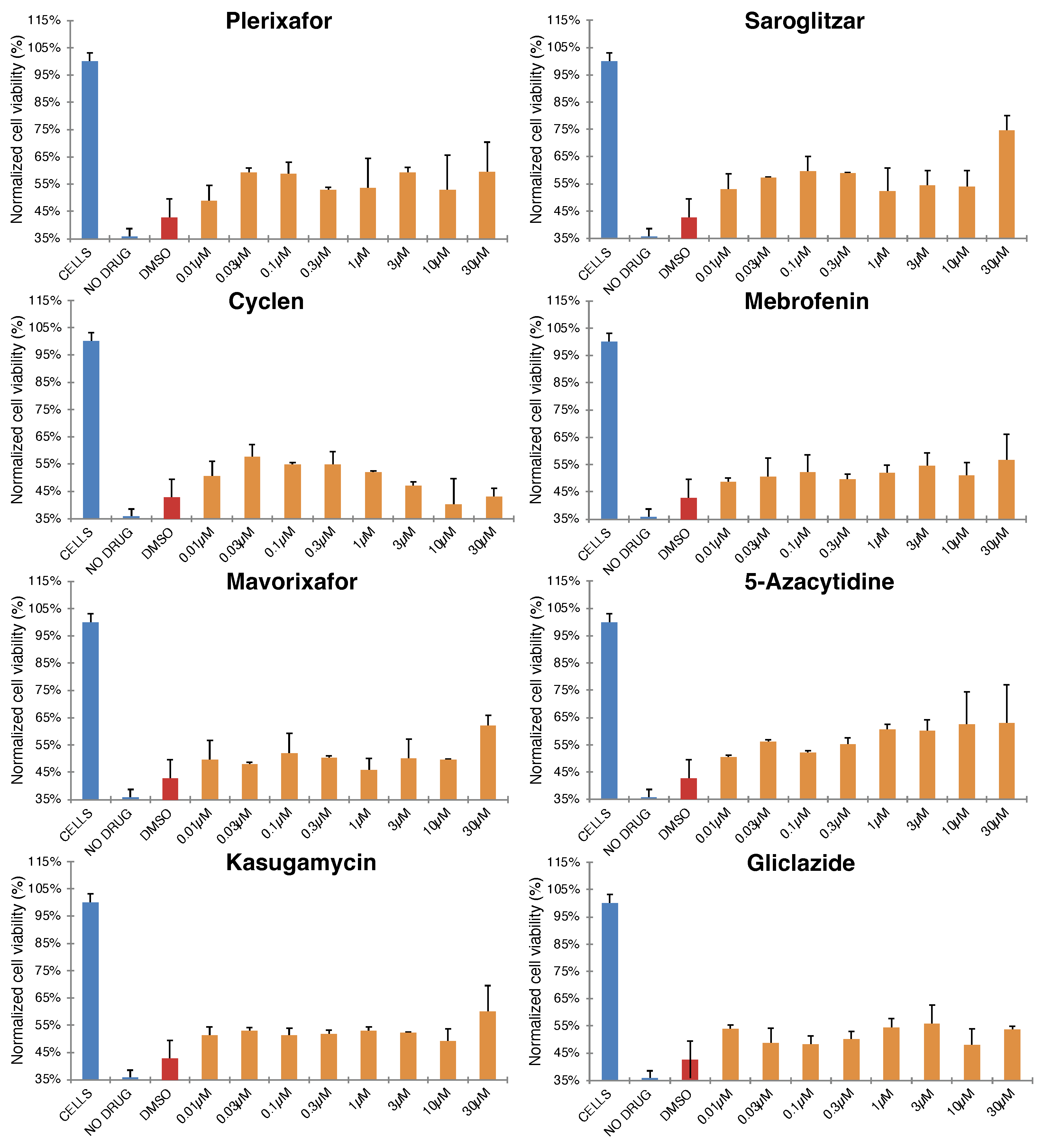
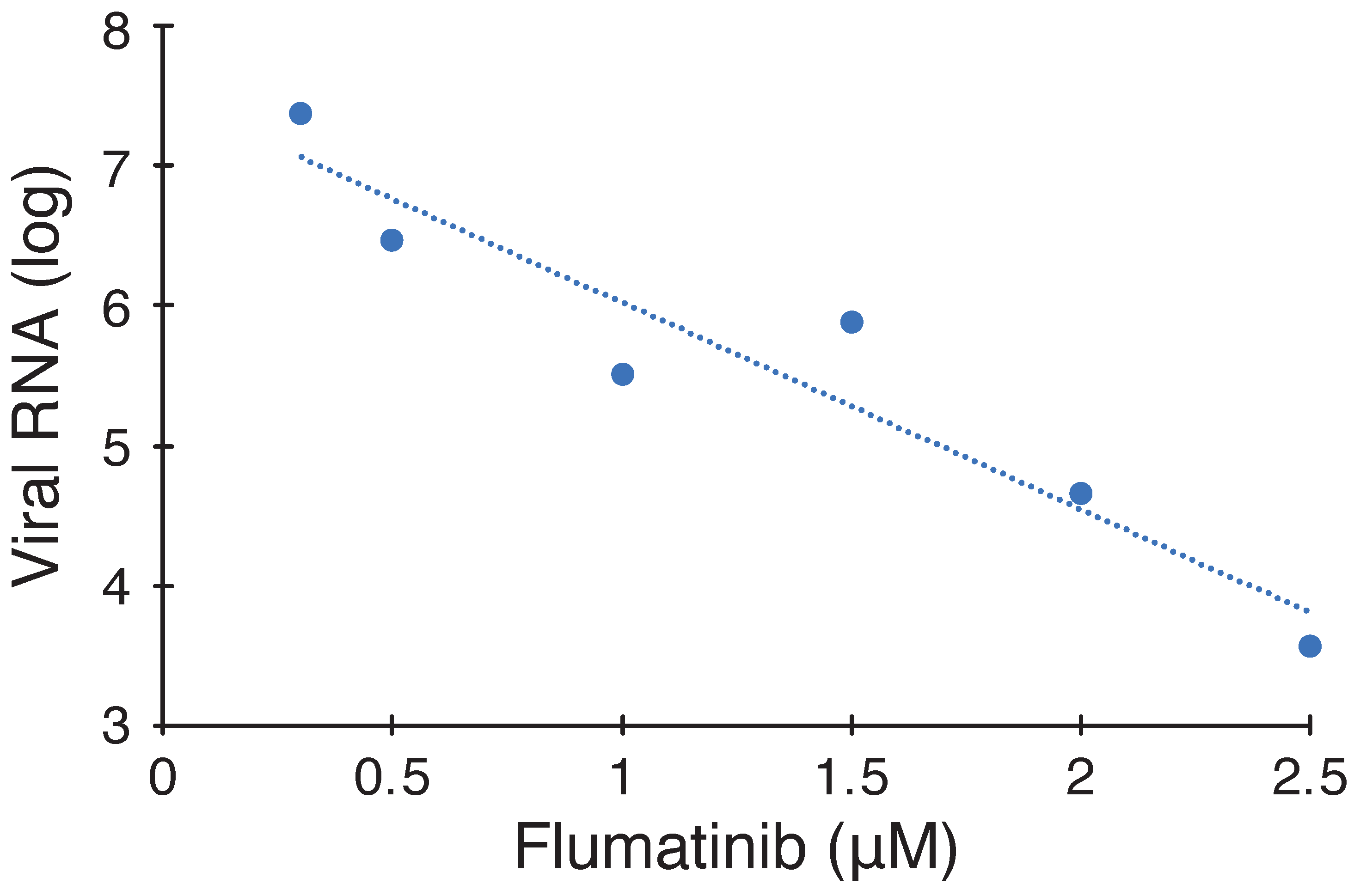
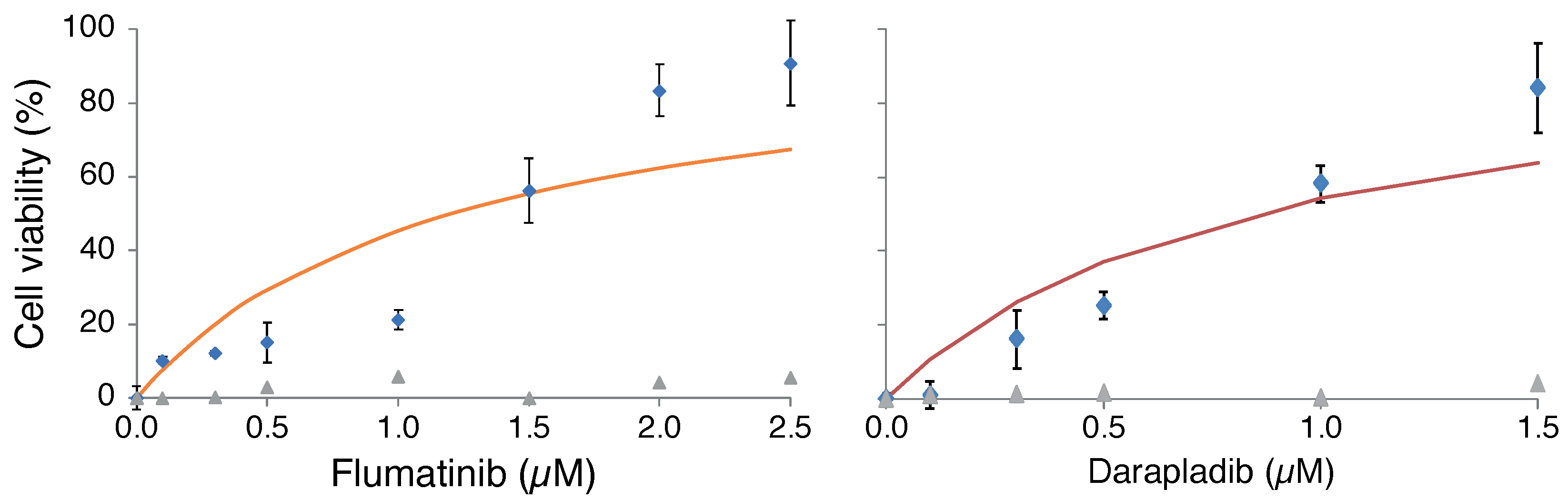
2.2. Antiviral Activity of Selected Blockers
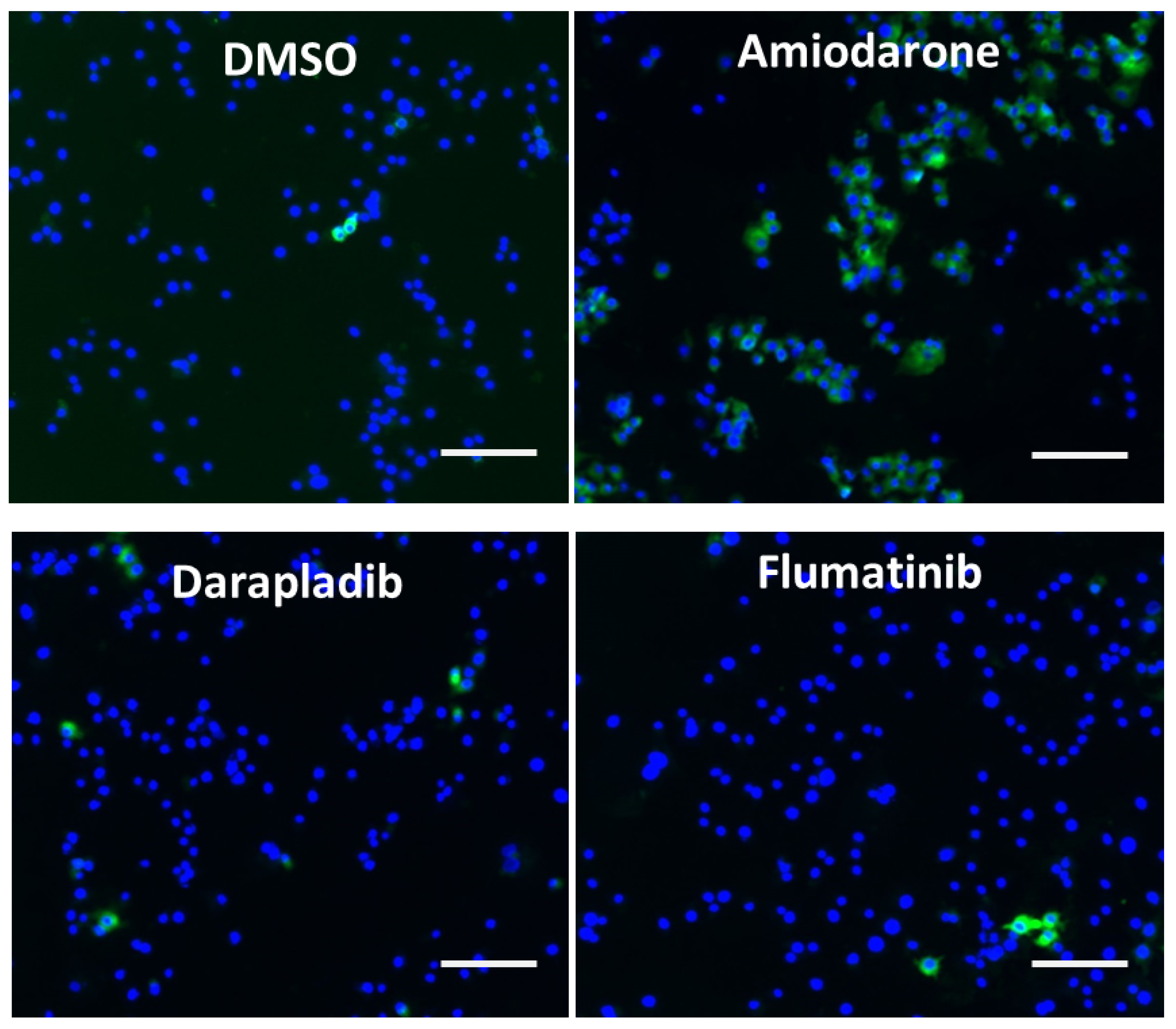
2.3. Drug Induced Phospholipidosis
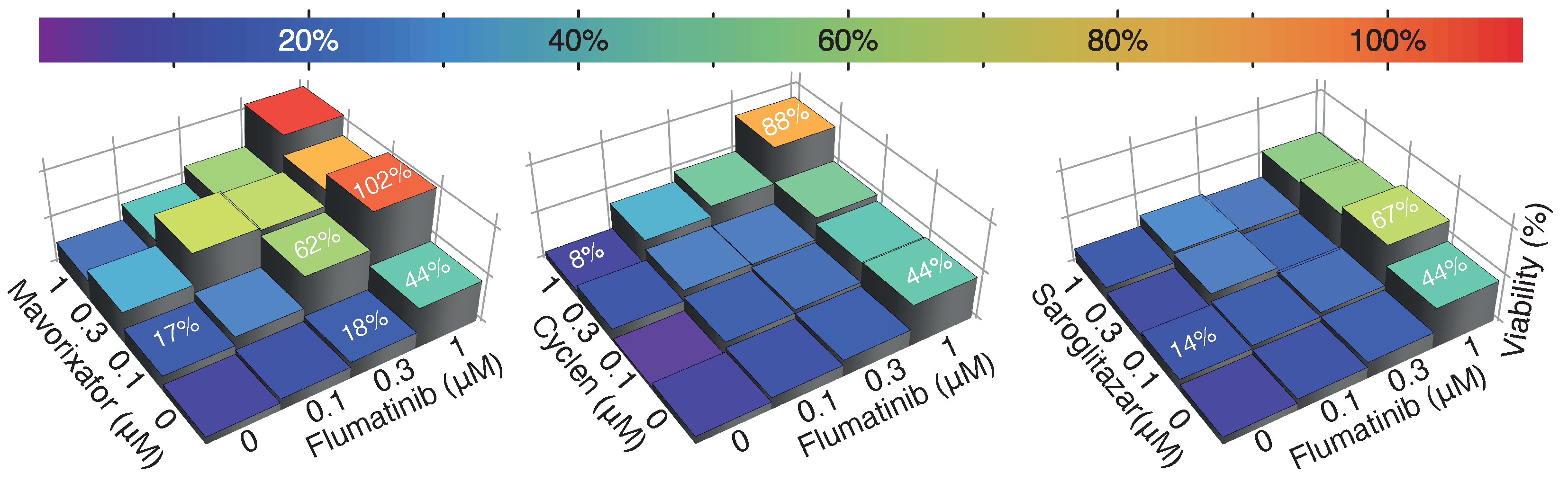
2.4. Drug Synergism
3. Discussion
4. Materials and Methods
4.1. Inhibitors
4.2. Cell Culture
4.3. Virus Culture and Infection
4.4. Compounds and Antiviral Screening Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Materials and Methods
Appendix A.1. RNA Extraction and Quantitative SARS-CoV2 RT-PCR
Appendix A.2. Drug-Induced Phospholipidosis
Appendix B. Figures
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; Yu, T.; et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Pe naranda, S.; Bankamp, B.; Maher, K.; Chen, M.H.; et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003, 300, 1394–1399. [Google Scholar] [CrossRef] [Green Version]
- Skowronski, D.M.; De Serres, G. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2021, 384, 1576–1577. [Google Scholar] [CrossRef]
- El Sahly, H.M.; Baden, L.R.; Essink, B.; Doblecki-Lewis, S.; Martin, J.M.; Anderson, E.J.; Campbell, T.B.; Clark, J.; Jackson, L.A.; Fichtenbaum, C.J.; et al. Efficacy of the mRNA-1273 SARS-CoV-2 Vaccine at Completion of Blinded Phase. N. Engl. J. Med. 2021, 385, 1774–1785. [Google Scholar] [CrossRef]
- Edara, V.V.; Manning, K.E.; Ellis, M.; Lai, L.; Moore, K.M.; Foster, S.L.; Floyd, K.; Davis-Gardner, M.E.; Mantus, G.; Nyhoff, L.E.; et al. mRNA-1273 and BNT162b2 mRNA vaccines have reduced neutralizing activity against the SARS-CoV-2 omicron variant. Cell Rep. Med. 2022, 24, 100529. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, D.; Du, G.; Du, R.; Zhao, J.; Jin, Y.; Fu, S.; Gao, L.; Cheng, Z.; Lu, Q.; et al. Remdesivir in adults with severe COVID-19: A randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2020, 395, 1569–1578. [Google Scholar] [CrossRef] [PubMed]
- Grein, J.; Ohmagari, N.; Shin, D.; Diaz, G.; Asperges, E.; Castagna, A.; Feldt, T.; Green, G.; Green, M.L.; Lescure, F.X.; et al. Compassionate Use of Remdesivir for Patients with Severe Covid-19. N. Engl. J. Med. 2020, 382, 2327–2336. [Google Scholar] [CrossRef] [PubMed]
- WHO Solidarity Trial Consortium; Pan, H.; Peto, R.; Henao-Restrepo, A.M.; Preziosi, M.P.; Sathiyamoorthy, V.; Abdool Karim, Q.; Alejandria, M.M.; Hernández García, C.; Kieny, M.P.; et al. Repurposed Antiviral Drugs for COVID-19—Interim WHO Solidarity Trial Results. N. Engl. J. Med. 2021, 384, 497–511. [Google Scholar] [CrossRef]
- Jayk Bernal, A.; Gomes da Silva, M.M.; Musungaie, D.B.; Kovalchuk, E.; Gonzalez, A.; Delos Reyes, V.; Martín-Quirós, A.; Caraco, Y.; Williams-Diaz, A.; Brown, M.L.; et al. Molnupiravir for Oral Treatment of Covid-19 in Nonhospitalized Patients. N. Engl. J. Med. 2021, 386, 509–520. [Google Scholar] [CrossRef]
- Mahase, E. COVID-19: Pfizer’s paxlovid is 89% effective in patients at risk of serious illness, company reports. BMJ 2021, 375, n2713. [Google Scholar] [CrossRef]
- Kaczorowski, G.J.; McManus, O.B.; Priest, B.T.; Garcia, M.L. Ion channels as drug targets: The next GPCRs. J. Gen. Physiol. 2008, 131, 399–405. [Google Scholar] [CrossRef]
- Waszkielewicz, A.M.; Gunia, A.; Szkaradek, N.; Słoczyńska, K.; Krupińska, S.; Marona, H. Ion channels as drug targets in central nervous system disorders. Curr. Med. Chem. 2013, 20, 1241–1285. [Google Scholar] [CrossRef] [Green Version]
- McGivern, J.G.; Ding, M. Ion Channels and Relevant Drug Screening Approaches. SLAS Discov. 2020, 25, 413–419. [Google Scholar] [CrossRef]
- Garcia, M.L.; Kaczorowski, G.J. Ion channels find a pathway for therapeutic success. Proc. Natl. Acad. Sci. USA 2016, 113, 5472–5474. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeDiego, M.L.; Álvarez, E.; Almazán, F.; Rejas, M.T.; Lamirande, E.; Roberts, A.; Shieh, W.J.; Zaki, S.R.; Subbarao, K.; Enjuanes, L. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 2007, 81, 1701–1713. [Google Scholar] [PubMed] [Green Version]
- Ortego, J.; Ceriani, J.E.; Pati no, C.; Plana, J.; Enjuanes, L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology 2007, 368, 296–308. [Google Scholar] [PubMed] [Green Version]
- Hyser, J.M. Viroporins. In Electrophysiology of Unconventional Channels and Pores; Springer: Berlin/Heidelberg, Germany, 2015; pp. 153–181. [Google Scholar]
- Issa, E.; Merhi, G.; Panossian, B.; Salloum, T.; Tokajian, S. SARS-CoV-2 and ORF3a: Nonsynonymous mutations, functional domains, and viral pathogenesis. Msystems 2020, 5, e00266-20. [Google Scholar] [PubMed]
- Ren, Y.; Shu, T.; Wu, D.; Mu, J.; Wang, C.; Huang, M.; Han, Y.; Zhang, X.Y.; Zhou, W.; Qiu, Y.; et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020, 17, 881–883. [Google Scholar]
- Mandala, V.S.; McKay, M.J.; Shcherbakov, A.A.; Dregni, A.J.; Kolocouris, A.; Hong, M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020, 27, 1202–1208. [Google Scholar] [CrossRef]
- Kern, D.M.; Sorum, B.; Mali, S.S.; Hoel, C.M.; Sridharan, S.; Remis, J.P.; Toso, D.B.; Kotecha, A.; Bautista, D.M.; Brohawn, S.G. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat. Struct. Mol. Biol. 2021, 28, 573–582. [Google Scholar] [CrossRef]
- Tomar, P.P.S.; Krugliak, M.; Arkin, I.T. Blockers of the SARS-CoV-2 3a Channel Identified by Targeted Drug Repurposing. Viruses 2021, 13, 532. [Google Scholar]
- Tomar, P.P.S.; Krugliak, M.; Arkin, I.T. Identification of SARS-CoV-2 E Channel Blockers from a Repurposed Drug Library. Pharmaceuticals 2021, 14, 604. [Google Scholar]
- Ogando, N.S.; Dalebout, T.J.; Zevenhoven-Dobbe, J.C.; Limpens, R.W.A.L.; van der Meer, Y.; Caly, L.; Druce, J.; de Vries, J.J.C.; Kikkert, M.; Bárcena, M.; et al. SARS-coronavirus-2 replication in Vero E6 cells: Replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020, 101, 925–940. [Google Scholar] [CrossRef]
- Tummino, T.A.; Rezelj, V.V.; Fischer, B.; Fischer, A.; O’Meara, M.J.; Monel, B.; Vallet, T.; White, K.M.; Zhang, Z.; Alon, A.; et al. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science 2021, 373, 541–547. [Google Scholar] [PubMed]
- Kuang, Y.; Song, H.L.; Yang, G.P.; Pei, Q.; Yang, X.Y.; Ye, L.; Yang, S.; Wu, S.T.; Guo, C.; He, Q.N.; et al. Effect of high-fat diet on the pharmacokinetics and safety of flumatinib in healthy Chinese subjects. Cancer Chemother. Pharmacol. 2020, 86, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Stone, N.D.; Dunaway, S.B.; Flexner, C.; Tierney, C.; Calandra, G.B.; Becker, S.; Cao, Y.J.; Wiggins, I.P.; Conley, J.; MacFarland, R.T.; et al. Multiple-dose escalation study of the safety, pharmacokinetics, and biologic activity of oral AMD070, a selective CXCR4 receptor inhibitor, in human subjects. Antimicrob. Agents Chemother. 2007, 51, 2351–2358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toptan, T.; Hoehl, S.; Westhaus, S.; Bojkova, D.; Berger, A.; Rotter, B.; Hoffmeier, K.; Cinatl, J.; Ciesek, S.; Widera, M. Optimized qRT-PCR approach for the detection of intra-and extra-cellular SARS-CoV-2 RNAs. Int. J. Mol. Sci. 2020, 21, 4396. [Google Scholar]
- Bock, S.; Hoffmann, B.; Beer, M.; Wernike, K. Saving Resources: SARS-CoV-2 Diagnostics by Real-Time RT-PCR Using Reduced Reaction Volumes. Diseases 2021, 9, 84. [Google Scholar]
- Morelli, J.; Buehrle, M.; Pognan, F.; Barone, L.; Fieles, W.; Ciaccio, P. Validation of an in vitro screen for phospholipidosis using a high-content biology platform. Cell Biol. Toxicol. 2006, 22, 15–27. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, A.; Arkin, I.T. Targeting Viral Ion Channels: A Promising Strategy to Curb SARS-CoV-2. Pharmaceuticals 2022, 15, 396. https://doi.org/10.3390/ph15040396
Singh A, Arkin IT. Targeting Viral Ion Channels: A Promising Strategy to Curb SARS-CoV-2. Pharmaceuticals. 2022; 15(4):396. https://doi.org/10.3390/ph15040396
Chicago/Turabian StyleSingh, Anamika, and Isaiah T. Arkin. 2022. "Targeting Viral Ion Channels: A Promising Strategy to Curb SARS-CoV-2" Pharmaceuticals 15, no. 4: 396. https://doi.org/10.3390/ph15040396
APA StyleSingh, A., & Arkin, I. T. (2022). Targeting Viral Ion Channels: A Promising Strategy to Curb SARS-CoV-2. Pharmaceuticals, 15(4), 396. https://doi.org/10.3390/ph15040396