Antibiotic Resistance in the Drinking Water: Old and New Strategies to Remove Antibiotics, Resistant Bacteria, and Resistance Genes
Abstract
:1. Introduction
2. Antibiotic Resistance
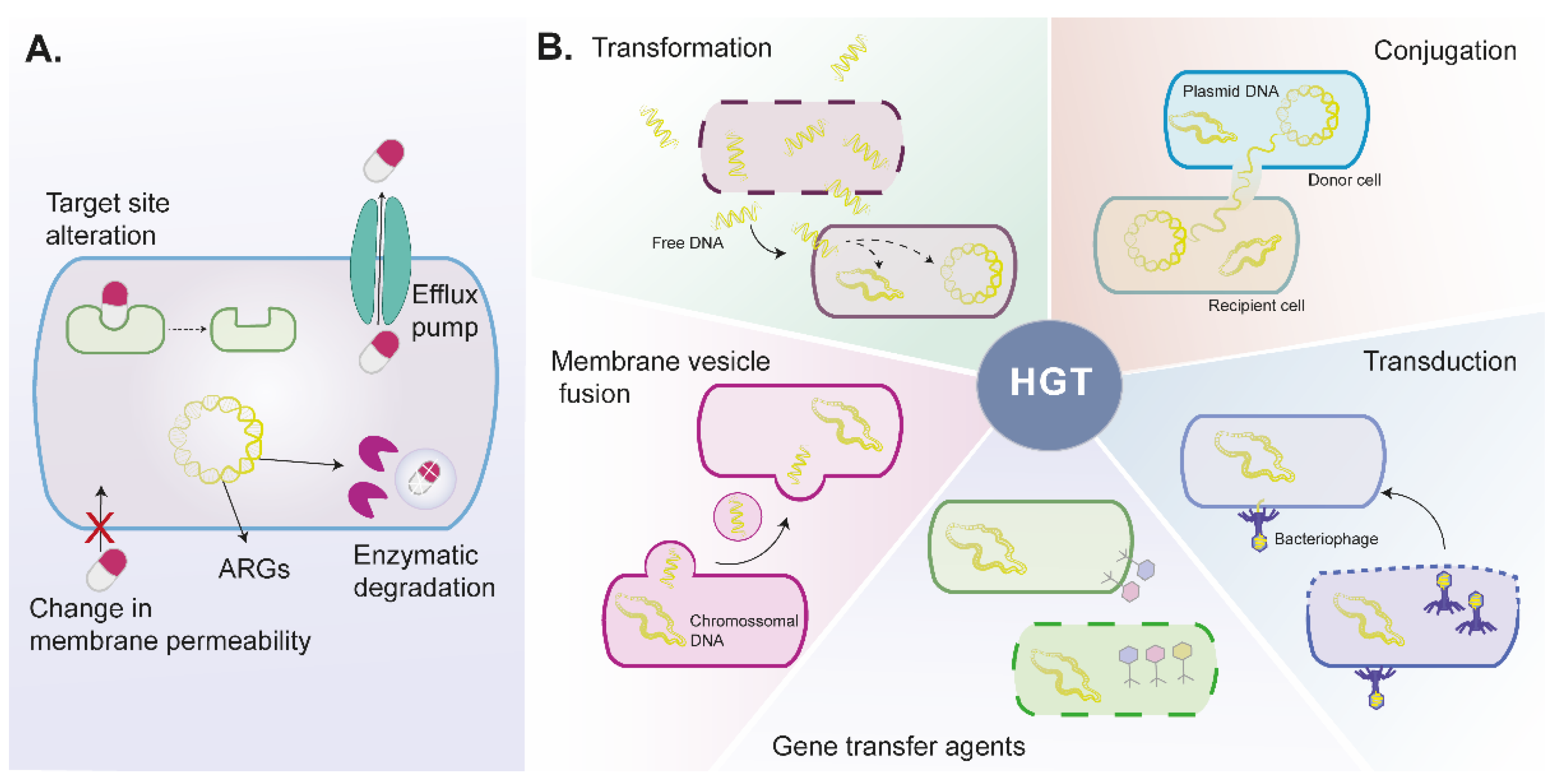
2.1. Bacteria Mechanisms to Antibiotic Resistance
2.1.1. Intrinsic Antibiotic Resistance
2.1.2. Acquired Antibiotic Resistance
Horizontal Gene Transfer
2.1.3. Biofilm Antimicrobial Resistance
3. Critical Factors for Antibiotic Resistance Widespread in Environment and Water Sources
3.1. Human and Veterinary Consumption
3.2. Health Care Facilities and the Pharmaceutical Industry
3.3. Biofilm Formation in Drinking Water Distribution Systems
3.4. Inefficient Antibiotic, ARB, and ARB Removal by Water Treatment Processes
4. Promising Strategies to Reduce Antibiotic Resistance
4.1. New Approaches for Antibiotics Elimination
4.1.1. Antibiotics Adsorption and Degradation Using Nanotechnology
4.1.2. Bioadsorption, Bioaccumulation, and Biodegradation of Antibiotics by Microalgae-Based Technologies
Microalgae-Bacterial Consortium
4.2. New Approaches for ARBs and ARGs Elimination
4.2.1. ARBs and ARGs Removal by Nanotechnology
4.2.2. ARBs and ARGs Removal by Microalgae-Based Technologies
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. Não há Tempo a Perder: Acautelar o Futuro Contra Infecções Resistentes aos Medicamentos; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Zhang, T.; Lv, K.; Lu, Q.; Wang, L.; Liu, X. Removal of antibiotic-resistant genes during drinking water treatment: A review. J. Environ. Sci. 2021, 104, 415–429. [Google Scholar] [CrossRef]
- Bortoloti, K.D.C.S.; Melloni, R.; Marques, P.S.; De Carvalho, B.M.F.; Andrade, M.C. Qualidade microbiológica de águas naturais quanto ao perfil de resistência de bactérias heterotróficas a antimicrobianos. Eng. Sanit. Ambient. 2018, 23, 717–725. [Google Scholar] [CrossRef]
- Iria, A. Efeitos da Presença de Antibióticos nas Origens de Agua. Contribuição para o Estudo da sua Remoção Através de Sistemas de Tratamento de Aguas. Ph.D. Thesis, Faculdade de Ciências e Tecnologia da Universidade Nova de Lisboa, Lisboa, Portugal, 2018. [Google Scholar]
- Collignon, P.; Beggs, J.J.; Walsh, T.; Gandra, S.; Laxminarayan, R. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: A univariate and multivariable analysis. Lancet Planet. Health 2018, 2, e398–e405. [Google Scholar] [CrossRef] [PubMed]
- Amarasiri, M.; Sano, D.; Suzuki, S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 2020, 50, 2016–2059. [Google Scholar] [CrossRef]
- Tan, Q.; Li, W.; Zhang, J.; Zhou, W.; Chen, J.; Li, Y.; Ma, J. Presence, dissemination and removal of antibiotic resistant bacteria and antibiotic resistance genes in urban drinking water system: A review. Front. Environ. Sci. Eng. 2019, 13, 36. [Google Scholar] [CrossRef]
- Homem, V.M.F.C. Tecnologias Alternativas de Remoção de Antibióticos de Águas Contaminadas; Faculdade de Engenharia da Universidade do Porto: Porto, Portugal, 2011; pp. 1–305. [Google Scholar]
- Sanganyado, E.; Gwenzi, W. Antibiotic resistance in drinking water systems: Occurrence, removal, and human health risks. Sci. Total Environ. 2019, 669, 785–797. [Google Scholar] [CrossRef]
- Xu, L.; Ouyang, W.; Qian, Y.; Su, C.; Su, J.; Chen, H. High-throughput profiling of antibiotic resistance genes in drinking water treatment plants and distribution systems. Environ. Pollut. 2016, 213, 119–126. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, T.; Chen, H. Antibiotic resistome promotion in drinking water during biological activated carbon treatment: Is it influenced by quorum sensing? Sci. Total Environ. 2018, 612, 1–8. [Google Scholar] [CrossRef]
- Hutchings, M.; Truman, A.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Felis, E.; Kalka, J.; Sochacki, A.; Kowalska, K.; Bajkacz, S.; Harnisz, M.; Korzeniewska, E. Antimicrobial pharmaceuticals in the aquatic environment—occurrence and environmental implications. Eur. J. Pharmacol. 2020, 866, 172813. [Google Scholar] [CrossRef]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Savin, M.; Bierbaum, G.; Hammerl, A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. ESKAPE Bacteria and Extended-Spectrum-B-Lactamase- Producing Escherichia coli Isolated from Wastewater and Process Water from German Poultry Slaughterhouses. Appl. Environ. Microbiol. 2020, 86, e02748-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrivastava, S.R.; Shrivastava, P.S.; Ramasamy, J. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med. Soc. 2018, 32, 76–77. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Beatson, S.A.; Paterson, D.L.; Walker, J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Nadeem, S.F.; Gohar, U.F.; Tahir, S.F.; Mukhtar, H.; Pornpukdeewattana, S.; Nukthamna, P.; Moula Ali, A.M.; Bavisetty, S.C.B.; Massa, S. Antimicrobial resistance: More than 70 years of war between humans and bacteria. Crit. Rev. Microbiol. 2020, 46, 578–599. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.S.; Sobkowiak, B.; Parreira, R.; Edgeworth, J.D.; Viveiros, M.; Clark, T.G.; Couto, I. Genetic diversity of norA, coding for a main efflux pump of Staphylococcus aureus. Front. Genet. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Uluseker, C.; Kaster, K.M.; Thorsen, K.; Basiry, D.; Shobana, S.; Jain, M.; Kumar, G.; Kommedal, R.; Pala-Ozkok, I. A Review on Occurrence and Spread of Antibiotic Resistance in Wastewaters and in Wastewater Treatment Plants: Mechanisms and Perspectives. Front. Microbiol. 2021, 12, 3003. [Google Scholar] [CrossRef]
- Papkou, A.; Hedge, J.; Kapel, N.; Young, B.; MacLean, R.C. Efflux pump activity potentiates the evolution of antibiotic resistance across S. aureus isolates. Nat. Commun. 2020, 11, 3970. [Google Scholar] [CrossRef]
- Fontaine, F.; Hequet, A.; Voisin-Chiret, A.-S.; Bouillon, A.; Lesnard, A.; Cresteil, T.; Jolivalt, C.; Rault, S. First Identification of Boronic Species as Novel Potential Inhibitors of the Staphylococcus aureus NorA Efflux Pump. J. Med. Chem. 2014, 57, 2536–2548. [Google Scholar] [CrossRef]
- Malmir, S.; Bahreinian, M.; Yeganeh, S.Z.; Mirnejad, R.; Moghaddam, M.M.; Saberi, F. Molecular Mechanisms of Resistance to Conventional Antibiotics in Bacteria. Int. J. Med. Rev. 2018, 5, 118–129. [Google Scholar] [CrossRef]
- Fishovitz, J.; Hermoso, J.; Chang, M.; Mobashery, S. Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life 2014, 66, 572–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zainab, S.M.; Junaidb, M.; Xub, N.; Malik, R.N. Antibiotics and antibiotic resistant genes (ARGs) in groundwater: A global review on dissemination, sources, interactions, environmental and human health risks. Water Res. 2020, 187, 116455. [Google Scholar] [CrossRef] [PubMed]
- Luo, A.; Wang, F.; Sun, D.; Liu, X.; Xin, B. Formation, Development, and Cross-Species Interactions in Biofilms. Front. Microbiol. 2022, 12, 757327. [Google Scholar] [CrossRef]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic resistance genes in bacteria: Occurrence, spread, and control. J. Basic Microbiol. 2021, 61, 1049–1070. [Google Scholar] [CrossRef]
- Uddin, J.; Dawan, J.; Jeon, G.; Yu, T.; He, X.; Ahn, J. The Role of Bacterial Membrane Vesicles in the Dissemination of Antibiotic Resistance and as Promising Carriers for Therapeutic Agent Delivery. Microorganisms 2020, 8, 670. [Google Scholar] [CrossRef]
- Lee, A.R.; Park, S.B.; Kim, S.W.; Jung, J.W.; Chun, J.H.; Kim, J.; Kim, Y.R.; Lazarte, J.M.S.; Jang, H.B.; Thompson, K.D.; et al. Membrane vesicles (MVs) from antibiotic-resistant Staphylococcus aureus transfer antibiotic-resistance to antibiotic-susceptible Escherichia coli. J. Appl. Microbiol. 2022, 1–14. [Google Scholar] [CrossRef]
- Von Wintersdorff, C.J.H.; Penders, J.; Van Niekerk, J.M.; Mills, N.D.; Majumder, S.; Van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Anderson, G.G.; O’Toole, G.A. Innate and induced resistance mechanisms of bacterial biofilms. In Bacterial Biofilms; Romeo, T., Ed.; Springer: Berlin, Germany, 2008; pp. 85–105. ISBN 978-3-540-75418-3. [Google Scholar]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Monzón, M.; Oteiza, C.; Leiva, J.; Lamata, M.; Amorena, B. Biofilm testing of Staphylococcus epidermidis clinical isolates: Low performance of vancomycin in relation to other antibiotics. Diagn. Microbiol. Infect. Dis. 2002, 44, 319–324. [Google Scholar] [CrossRef]
- Savage, V.J.; Chopra, I.; O’Neill, A.J. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob. Agents Chemother. 2013, 57, 1968–1970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Król, J.E.; Wojtowicz, A.J.; Rogers, L.M.; Heuer, H.; Smalla, K.; Krone, S.M.; Top, E.M. Invasion of E. coli biofilms by antibiotic resistance plasmids. Plasmid 2013, 70, 110–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemdan, B.A.; El-Taweel, G.E.; Goswami, P.; Pant, D.; Sevda, S. The role of biofilm in the development and dissemination of ubiquitous pathogens in drinking water distribution systems: An overview of surveillance, outbreaks, and prevention. World J. Microbiol. Biotechnol. 2021, 37, 36. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Jin, G.; Zhao, Y.; Shao, X. Prevalence and distribution analysis of antibiotic resistance genes in a large-scale aquaculture environment. Sci. Total Environ. 2020, 711, 134626. [Google Scholar] [CrossRef]
- Santos, L.; Ramos, F. Antimicrobial resistance in aquaculture: Current knowledge and alternatives to tackle the problem. Int. J. Antimicrob. Agents 2018, 52, 135–143. [Google Scholar] [CrossRef]
- Zhao, R.; Feng, J.; Liu, J.; Fu, W.; Li, X.; Li, B. Deciphering of microbial community and antibiotic resistance genes in activated sludge reactors under high selective pressure of different antibiotics. Water Res. 2019, 151, 388–402. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [Green Version]
- Bruyndonckx, R.; Adriaenssens, N.; Versporten, A.; Hens, N.; Monnet, D.L.; Molenberghs, G.; Goossens, H.; Weist, K.; Coenen, S. Consumption of antibiotics in the community, European Union/European Economic Area, 1997–2017. J. Antimicrob. Chemother. 2021, 76, 7–13. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Antimicrobial Consumption in the EU/EEA—Annual Epidemiological Report 2019; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2020. [Google Scholar]
- Xiang, Y.; Yang, Z.; Zhang, Y.; Xu, R.; Zheng, Y.; Hu, J.; Li, X.; Jia, M.; Xiong, W.; Cao, J. Influence of nanoscale zero-valent iron and magnetite nanoparticles on anaerobic digestion performance and macrolide, aminoglycoside, β-lactam resistance genes reduction. Bioresour. Technol. 2019, 294, 122139. [Google Scholar] [CrossRef]
- Kovalakova, P.; Cizmas, L.; McDonald, T.J.; Marsalek, B.; Feng, M.; Sharma, V.K. Occurrence and toxicity of antibiotics in the aquatic environment: A review. Chemosphere 2020, 251, 126351. [Google Scholar] [CrossRef] [PubMed]
- Obimakinde, S.; Fatoki, O.; Opeolu, B.; Olatunji, O. Veterinary pharmaceuticals in aqueous systems and associated effects: An update. Environ. Sci. Pollut. Res. 2017, 24, 3274–3297. [Google Scholar] [CrossRef]
- Watts, J.E.M.; Schreier, H.J.; Lanska, L.; Hale, M.S. The rising tide of antimicrobial resistance in aquaculture: Sources, sinks and solutions. Mar. Drugs 2017, 15, 158. [Google Scholar] [CrossRef] [Green Version]
- Tran, N.H.; Reinhard, M.; Gin, K.Y.H. Occurrence and fate of emerging contaminants in municipal wastewater treatment plants from different geographical regions-a review. Water Res. 2018, 133, 182–207. [Google Scholar] [CrossRef]
- Mackull’ak, T.; Cverenkárová, K.; Stanová, A.; Fehér, M.; Tamás, M.; Škulcová, A.; Gál, M.; Naumowicz, M.; Špalkov, V.; Bírošová, L. Hospital Wastewater—Source of Specific Micropollutants. Antibiotics 2021, 10, 1070. [Google Scholar] [CrossRef]
- Proia, L.; Adriana, A.; Jessica, S.; Carles, B.; Marinella, F.; Marta, L.; Luis, B.J.; Servais, P. Antibiotic resistance in urban and hospital wastewaters and their impact on a receiving freshwater ecosystem. Chemosphere 2018, 206, 70–82. [Google Scholar] [CrossRef]
- Marathe, N.P.; Berglund, F.; Razavi, M.; Pal, C.; Dröge, J.; Samant, S.; Kristiansson, E.; Joakim Larsson, D.G. Sewage effluent from an Indian hospital harbors novel carbapenemases and integron-borne antibiotic resistance genes. Microbiome 2019, 7, 97. [Google Scholar] [CrossRef] [Green Version]
- Laquaz, M.; Dagot, C.; Bazin, C.; Bastide, T.; Gaschet, M.; Ploy, M.C.; Perrodin, Y. Ecotoxicity and antibiotic resistance of a mixture of hospital and urban sewage in a wastewater treatment plant. Environ. Sci. Pollut. Res. 2018, 25, 9243–9253. [Google Scholar] [CrossRef]
- Petrovich, M.L.; Zilberman, A.; Kaplan, A.; Eliraz, G.R.; Wang, Y.; Langenfeld, K.; Duhaime, M.; Wigginton, K.; Poretsky, R.; Avisar, D.; et al. Microbial and Viral Communities and Their Antibiotic Resistance Genes Throughout a Hospital Wastewater Treatment System. Front. Microbiol. 2020, 11, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Rowe, W.P.M.; Baker-Austin, C.; Verner-Jeffreys, D.W.; Ryan, J.J.; Micallef, C.; Maskell, D.J.; Pearce, G.P. Overexpression of antibiotic resistance genes in hospital effluents over time. J. Antimicrob. Chemother. 2017, 72, 1617–1623. [Google Scholar] [CrossRef]
- Milaković, M.; Vestergaard, G.; González-Plaza, J.J.; Petrić, I.; Kosić-Vukšić, J.; Senta, I.; Kublik, S.; Schloter, M.; Udiković-Kolić, N. Effects of industrial effluents containing moderate levels of antibiotic mixtures on the abundance of antibiotic resistance genes and bacterial community composition in exposed creek sediments. Sci. Total Environ. 2020, 706, 136001. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Lu, X.T.; Zhang, J.Y.; Sui, Q.; Wang, R.; Chen, M.; Wei, Y. Occurrence of antibiotic resistance genes and mobile genetic elements in enterococci and genomic DNA during anaerobic digestion of pharmaceutical waste sludge with different pretreatments. Bioresour. Technol. 2017, 235, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Yan, Z.; Zhang, Y.; Xu, W.; Kong, D.; Shan, Z.; Wang, N. Behavior of antibiotic resistance genes under extremely high-level antibiotic selection pressures in pharmaceutical wastewater treatment plants. Sci. Total Environ. 2018, 612, 119–128. [Google Scholar] [CrossRef]
- Wang, K.; Zhuang, T.; Su, Z.; Chi, M.; Wang, H. Antibiotic residues in wastewaters from sewage treatment plants and pharmaceutical industries: Occurrence, removal and environmental impacts. Sci. Total Environ. 2021, 788, 147811. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Li, K.; Xin, R.; Han, Y.; Guo, Z.; Zou, W.; Wei, W.; Cui, X.; Zhang, Z.; Zhang, Y. Antibiotic resistomes in water supply reservoirs sediments of central China: Main biotic drivers and distribution pattern. Environ. Sci. Pollut. Res. 2022, 1–10. [Google Scholar] [CrossRef]
- Chan, S.; Pullerits, K.; Keucken, A.; Persson, K.M.; Paul, C.J.; Rådström, P. Bacterial release from pipe biofilm in a full-scale drinking water distribution system. NPJ Biofilms Microbiomes 2019, 5, 3–10. [Google Scholar] [CrossRef]
- Chen, J.; Li, W.; Tan, Q.; Sheng, D.; Li, Y.; Chen, S.; Zhou, W. Effect of disinfectant exposure and starvation treatment on the detachment of simulated drinking water biofilms. Sci. Total Environ. 2022, 807, 150896. [Google Scholar] [CrossRef]
- Zhu, N.J.; Ghosh, S.; Edwards, M.A.; Pruden, A. Interplay of Biologically Active Carbon Filtration and Chlorine-Based Disinfection in Mitigating the Dissemination of Antibiotic Resistance Genes in Water Reuse Distribution Systems. Environ. Sci. Technol. 2021, 55, 8329–8340. [Google Scholar] [CrossRef]
- Chen, J.; Li, W.; Zhang, J.; Qi, W.; Li, Y.; Chen, S.; Zhou, W. Prevalence of antibiotic resistance genes in drinking water and biofilms: The correlation with the microbial community and opportunistic pathogens. Chemosphere 2020, 259, 127483. [Google Scholar] [CrossRef]
- Liang, Y.; Li, H.; Chen, Z.; Yang, Y.; Shi, D.; Chen, T.; Yang, D.; Yin, J.; Zhou, S.; Cheng, C.; et al. Spatial behavior and source tracking of extracellular antibiotic resistance genes in a chlorinated drinking water distribution system. J. Hazard. Mater. 2022, 425, 127942. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, L.; Sun, X.; Wu, J.; Ma, L.; Zhou, Y.; Lin, K.; Luo, Y.; Cui, C. Risk assessment of antibiotic resistance genes in the drinking water system. Sci. Total Environ. 2021, 800, 149650. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; He, Y.; Li, X.; Nagarajan, D.; Chang, J.-S. Enhanced biodegradation of chlortetracycline via a microalgae-bacteria consortium. Bioresour. Technol. 2022, 343, 126149. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Bian, K.; Shi, P.; Ye, L.; Liu, C.H. Metagenomic profiling of antibiotic resistance genes and their associations with bacterial community during multiple disinfection regimes in a full-scale drinking water treatment plant. Water Res. 2020, 176, 115721. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Tian, Z.; Yu, J.; Yang, M.; Zhang, Y. Distribution and abundance of antibiotic resistance genes in sand settling reservoirs and drinking water treatment plants across the Yellow River, China. Water 2018, 10, 246. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Zhang, T.; Jiang, L.; Luo, Y.; Yao, S.; Zhang, D.; Lin, K.; Cui, C. Occurrence and reduction of antibiotic resistance genes in conventional and advanced drinking water treatment processes. Sci. Total Environ. 2019, 669, 777–784. [Google Scholar] [CrossRef]
- Xu, L.; Campos, L.C.; Canales, M.; Ciric, L. Drinking water biofiltration: Behaviour of antibiotic resistance genes and the association with bacterial community. Water Res. 2020, 182, 115954. [Google Scholar] [CrossRef]
- Destiani, R.; Templeton, M.R. Chlorination and ultraviolet disinfection of antibiotic-resistant bacteria and antibiotic resistance genes in drinking water. AIMS Environ. Sci. 2019, 6, 222–241. [Google Scholar] [CrossRef]
- Su, H.C.; Liu, Y.S.; Pan, C.G.; Chen, J.; He, L.Y.; Ying, G.G. Persistence of antibiotic resistance genes and bacterial community changes in drinking water treatment system: From drinking water source to tap water. Sci. Total Environ. 2018, 616, 453–461. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Y.; Wu, J.; Wang, J.; Cai, Y. Fate of antibiotic resistance genes in reclaimed water reuse system with integrated membrane process. J. Hazard. Mater. 2020, 382, 121025. [Google Scholar] [CrossRef]
- Liang, C.; Wei, D.; Zhang, S.; Ren, Q.; Shi, J.; Liu, L. Removal of antibiotic resistance genes from swine wastewater by membrane filtration treatment. Ecotoxicol. Environ. Saf. 2021, 210, 111885. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, L.; Zhang, T.; Jin, L.; Han, Q.; Zhang, D.; Lin, K.; Cui, C. Occurrence and removal of sulfonamide antibiotics and antibiotic resistance genes in conventional and advanced drinking water treatment processes. J. Hazard. Mater. 2018, 360, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, Y.; Jiang, L.; Yao, S.; Lin, K.; Zhou, Y.; Cui, C. Removal of antibiotic resistance genes and control of horizontal transfer risk by UV, chlorination and UV/chlorination treatments of drinking water. Chem. Eng. J. 2019, 358, 589–597. [Google Scholar] [CrossRef]
- Guo, X.; Li, J.; Yang, F.; Yang, J.; Yin, D. Prevalence of sulfonamide and tetracycline resistance genes in drinking water treatment plants in the Yangtze River Delta, China. Sci. Total Environ. 2014, 493, 626–631. [Google Scholar] [CrossRef]
- Shen, L.; Griffith, T.M.; Nyangaresi, P.O.; Qin, Y.; Pang, X.; Chen, G.; Li, M.; Lu, Y.; Zhang, B. Efficacy of UVC-LED in water disinfection on Bacillus species with consideration of antibiotic resistance issue. J. Hazard. Mater. 2020, 386, 121968. [Google Scholar] [CrossRef] [PubMed]
- Manaia, C.M.; Rocha, J.; Scaccia, N.; Marano, R.; Radu, E.; Biancullo, F.; Cerqueira, F.; Fortunato, G.; Iakovides, I.C.; Zammit, I.; et al. Antibiotic resistance in wastewater treatment plants: Tackling the black box. Environ. Int. 2018, 115, 312–324. [Google Scholar] [CrossRef]
- O’Flaherty, E.; Borrego, C.M.; Balcázar, J.L.; Cummins, E. Human exposure assessment to antibiotic-resistant Escherichia coli through drinking water. Sci. Total Environ. 2018, 616, 1356–1364. [Google Scholar] [CrossRef] [PubMed]
- Shannon, M.A.; Bohn, P.W.; Elimelech, M.; Georgiadis, J.G.; Mariñas, B.J.; Mayes, A.M. Science and technology for water purification in the coming decades. Nanosci. Technol. A Collect. Rev. Nat. J. 2009, 452, 337–346. [Google Scholar] [CrossRef]
- Wan, K.; Guo, L.; Ye, C.; Zhu, J.; Zhang, M.; Yu, X. Accumulation of antibiotic resistance genes in full-scale drinking water biological activated carbon (BAC) filters during backwash cycles. Water Res. 2021, 190, 116744. [Google Scholar] [CrossRef]
- Zhang, S.; Lin, W.; Yu, X. Effects of full-scale advanced water treatment on antibiotic resistance genes in the Yangtze Delta area in China. FEMS Microbiol. Ecol. 2016, 92, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Bai, X.; Ma, X.; Xu, F.; Li, J.; Zhang, H.; Xiao, X. The drinking water treatment process as a potential source of affecting the bacterial antibiotic resistance. Sci. Total Environ. 2015, 533, 24–31. [Google Scholar] [CrossRef]
- Le, T.H.; Ng, C.; Tran, N.H.; Chen, H.; Gin, K.Y.H. Removal of antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in municipal wastewater by membrane bioreactor systems. Water Res. 2018, 145, 498–508. [Google Scholar] [CrossRef] [PubMed]
- Benladghem, Z.; Seddiki, S.M.L.; Mahdad, Y.M. Identification of bacterial biofilms on desalination reverse osmosis membranes from the mediterranean sea. Biofouling 2020, 36, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.H.; Wang, Y.H.; Xue, S.; Chen, Z.; Luo, L.W.; Bai, Y.; Tong, X.; Hu, H.Y. Increased risks of antibiotic resistant genes (ARGs) induced by chlorine disinfection in the reverse osmosis system for potable reuse of reclaimed water. Sci. Total Environ. 2022, 815, 152860. [Google Scholar] [CrossRef] [PubMed]
- Slipko, K.; Reif, D.; Wögerbauer, M.; Hufnagl, P.; Krampe, J.; Kreuzinger, N. Removal of extracellular free DNA and antibiotic resistance genes from water and wastewater by membranes ranging from microfiltration to reverse osmosis. Water Res. 2019, 164, 114916. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, H.; Xiong, P.; Zhu, Q.; Liao, C.; Jiang, G. Occurrence, fate, and risk assessment of typical tetracycline antibiotics in the aquatic environment: A review. Sci. Total Environ. 2021, 753, 141975. [Google Scholar] [CrossRef] [PubMed]
- Ben, Y.; Hu, M.; Zhang, X.; Wu, S.; Wong, M.H.; Wang, M.; Andrews, C.B.; Zheng, C. Efficient detection and assessment of human exposure to trace antibiotic residues in drinking water. Water Res. 2020, 175, 115699. [Google Scholar] [CrossRef]
- Karkman, A.; Do, T.T.; Walsh, F.; Virta, M.P.J. Antibiotic-Resistance Genes in Waste Water. Trends Microbiol. 2018, 26, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Hu, C.; Shen, Y.; Shi, B.; Zhao, D.; Xing, X. Response of microorganisms in biofilm to sulfadiazine and ciprofloxacin in drinking water distribution systems. Chemosphere 2019, 218, 197–204. [Google Scholar] [CrossRef]
- Malakootian, M.; Olama, N.; Malakootian, M.; Nasiri, A. Photocatalytic degradation of metronidazole from aquatic solution by TiO2-doped Fe3+ nano-photocatalyst. Int. J. Environ. Sci. Technol. 2019, 16, 4275–4284. [Google Scholar] [CrossRef]
- Zheng, X.; Xu, S.; Wang, Y.; Sun, X.; Gao, Y.; Gao, B. Enhanced degradation of ciprofloxacin by graphitized mesoporous carbon (GMC)-TiO2 nanocomposite: Strong synergy of adsorption-photocatalysis and antibiotics degradation mechanism. J. Colloid Interface Sci. 2018, 527, 202–213. [Google Scholar] [CrossRef]
- Chaba, J.M.; Nomngongo, P.N. Preparation of V2O5-ZnO coated carbon nanofibers: Application for removal of selected antibiotics in environmental matrices. J. Water Process. Eng. 2018, 23, 50–60. [Google Scholar] [CrossRef]
- Jiang, Y.; Jing, X.; Zhu, K.; Peng, Z.; Zhang, J.; Liu, Y.; Zhang, W.; Ni, L.; Liu, Z. Ta3N5 nanoparticles/TiO2 hollow sphere (0D/3D) heterojunction: Facile synthesis and enhanced photocatalytic activities of levofloxacin degradation and H2 evolution. Dalt. Trans. 2018, 47, 13113–13125. [Google Scholar] [CrossRef]
- Kaur, A.; Gupta, G.; Ibhadon, A.O.; Salunke, D.B.; Sinha, A.S.K.; Kansal, S.K. A Facile synthesis of silver modified ZnO nanoplates for efficient removal of ofloxacin drug in aqueous phase under solar irradiation. J. Environ. Chem. Eng. 2018, 6, 3621–3630. [Google Scholar] [CrossRef]
- Song, Z.; Ma, Y.L.; Li, C.E. The residual tetracycline in pharmaceutical wastewater was effectively removed by using MnO2/graphene nanocomposite. Sci. Total Environ. 2019, 651, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Duan, P.; Chen, D.; Hu, X. Tin dioxide decorated on Ni-encapsulated nitrogen-doped carbon nanotubes for anodic electrolysis and persulfate activation to degrade cephalexin: Mineralization and degradation pathway. Chemosphere 2021, 269, 128740. [Google Scholar] [CrossRef]
- Bangari, R.S.; Sinha, N. Adsorption of tetracycline, ofloxacin and cephalexin antibiotics on boron nitride nanosheets from aqueous solution. J. Mol. Liq. 2019, 293, 111376. [Google Scholar] [CrossRef]
- Ravikumar, K.V.G.; Kubendiran, H.; Ramesh, K.; Rani, S.; Mandal, T.K.; Pulimi, M.; Natarajan, C.; Mukherjee, A. Batch and column study on tetracycline removal using green synthesized NiFe nanoparticles immobilized alginate beads. Environ. Technol. Innov. 2020, 17, 100520. [Google Scholar] [CrossRef]
- Gopal, G.; Sankar, H.; Natarajan, C.; Mukherjee, A. Tetracycline removal using green synthesized bimetallic nZVI-Cu and bentonite supported green nZVI-Cu nanocomposite: A comparative study. J. Environ. Manag. 2020, 254, 109812. [Google Scholar] [CrossRef]
- Mi, X.; Li, Y.; Ning, X.; Jia, J.; Wang, H.; Xia, Y.; Sun, Y.; Zhan, S. Electro-Fenton degradation of ciprofloxacin with highly ordered mesoporous MnCo2O4-CF cathode: Enhanced redox capacity and accelerated electron transfer. Chem. Eng. J. 2019, 358, 299–309. [Google Scholar] [CrossRef]
- Kaur, M.; Mehta, S.K.; Kumar, S. Visible light driven photocatalytic degradation of o floxacin and malachite green dye using cadmium sulphide nanoparticles. J. Environ. Chem. Eng. 2018, 6, 3631–3639. [Google Scholar] [CrossRef]
- Sudhaik, A.; Raizada, P.; Shandilya, P.; Singh, P. Magnetically recoverable graphitic carbon nitride and NiFe2O4 based magnetic photocatalyst for degradation of oxytetracycline antibiotic in simulated wastewater under solar light. J. Environ. Chem. Eng. 2018, 6, 3874–3883. [Google Scholar] [CrossRef]
- Pham, V.L.; Kim, D.; Ko, S. Oxidative degradation of the antibiotic oxytetracycline by Cu@Fe3O4 core-shell nanoparticles. Sci. Total Environ. 2018, 631–632, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Moussavi, G.; Mashayekh-salehi, A.; Kamyar, Y.; Anoshiravan, M. The catalytic destruction of antibiotic tetracycline by sulfur-doped manganese oxide (SeMgO) nanoparticles. J. Environ. Manag. 2018, 210, 131–138. [Google Scholar] [CrossRef]
- Dong, G.; Huang, L.; Wu, X.; Wang, C.; Liu, Y.; Liu, G.; Wang, L.; Liu, X.; Xia, H. Effect and mechanism analysis of MnO2 on permeable reactive barrier (PRB) system for the removal of tetracycline. Chemosphere 2018, 193, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Jiang, Z.; Zhang, C.; Deng, J.; Hou, K.; Cheng, Y.; Zhang, L.; Zeng, G. Removal of tetracycline by Fe/Ni bimetallic nanoparticles in aqueous solution. J. Colloid Interface Sci. 2018, 513, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Wang, J.; Li, H.; Mao, S.; Wang, D.; Xiang, Z.; Guo, R.; Chen, J. The dual function of the algal treatment: Antibiotic elimination combined with CO2 fixation. Chemosphere 2018, 211, 192–201. [Google Scholar] [CrossRef]
- Kiki, C.; Rashid, A.; Wang, Y.; Li, Y.; Zeng, Q.; Yu, C.P.; Sun, Q. Dissipation of antibiotics by microalgae: Kinetics, identification of transformation products and pathways. J. Hazard. Mater. 2020, 387, 121985. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, W.; Li, J.; Yuan, M.; Zhang, J.; Xu, F.; Xu, H.; Zheng, X.; Wang, L. Ecotoxicological effects of sulfonamides and fluoroquinolones and their removal by a green alga (Chlorella vulgaris) and a cyanobacterium (Chrysosporum ovalisporum). Environ. Pollut. 2020, 263, 114554. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, R.; Li, H.; Du, Q.; Lu, J.; Huang, Y.; Yan, Z.; Chen, J. Mechanism analysis for the process-dependent driven mode of NaHCO3 in algal antibiotic removal: Efficiency, degradation pathway and metabolic response. J. Hazard. Mater. 2020, 394, 122531. [Google Scholar] [CrossRef]
- Yang, L.; Ren, L.; Tan, X.; Chu, H.; Chen, J.; Zhang, Y.; Zhou, X. Removal of ofloxacin with biofuel production by oleaginous microalgae Scenedesmus obliquus. Bioresour. Technol. 2020, 315, 123738. [Google Scholar] [CrossRef]
- Grimes, K.L.; Dunphy, L.J.; Loudermilk, E.M.; Melara, A.J.; Kolling, G.L.; Papin, J.A.; Colosi, L.M. Evaluating the efficacy of an algae-based treatment to mitigate elicitation of antibiotic resistance. Chemosphere 2019, 237, 124421. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.K.; Choi, T.R.; Gurav, R.; Bhatia, S.K.; Park, Y.L.; Kim, H.J.; Kan, E.; Yang, Y.H. Adsorption behavior of tetracycline onto Spirulina sp. (microalgae)-derived biochars produced at different temperatures. Sci. Total Environ. 2020, 710, 136282. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.Q.; Govindwar, S.; Kurade, M.B.; Paeng, K.J.; Roh, H.S.; Khan, M.A.; Jeon, B.H. Toxicity of sulfamethazine and sulfamethoxazole and their removal by a green microalga, Scenedesmus obliquus. Chemosphere 2019, 218, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yao, Y.; Ye, W.; Qian, R.; Chen, H.; Liang, J.; Ye, J. Enhanced removal of antibiotics and decreased antibiotic resistance genes in the photo-sequencing batch reactor during the aquaculture wastewater treatment. Environ. Technol. 2021, 1–12. [Google Scholar] [CrossRef]
- Xiong, Q.; Liu, Y.S.; Hu, L.X.; Shi, Z.Q.; Cai, W.W.; He, L.Y.; Ying, G.G. Co-metabolism of sulfamethoxazole by a freshwater microalga Chlorella pyrenoidosa. Water Res. 2020, 175, 115656. [Google Scholar] [CrossRef]
- Peng, Y.Y.; Gao, F.; Yang, H.L.; Wu, H.W.J.; Li, C.; Lu, M.M.; Yang, Z.Y. Simultaneous removal of nutrient and sulfonamides from marine aquaculture wastewater by concentrated and attached cultivation of Chlorella vulgaris in an algal biofilm membrane photobioreactor (BF-MPBR). Sci. Total Environ. 2020, 725, 138524. [Google Scholar] [CrossRef]
- Pan, M.; Lyu, T.; Zhan, L.; Matamoros, V.; Angelidaki, I.; Cooper, M.; Pan, G. Mitigating antibiotic pollution using cyanobacteria: Removal efficiency, pathways and metabolism. Water Res. 2021, 190, 116735. [Google Scholar] [CrossRef]
- Xie, P.; Chen, C.; Zhang, C.; Su, G.; Ren, N.; Ho, S.H. Revealing the role of adsorption in ciprofloxacin and sulfadiazine elimination routes in microalgae. Water Res. 2020, 172, 115475. [Google Scholar] [CrossRef]
- da Silva Rodrigues, D.A.; da Cunha, C.C.R.F.; do Espirito Santo, D.R.; de Barros, A.L.C.; Pereira, A.R.; de Queiroz Silva, S.; da Fonseca Santiago, A.; de Cássia Franco Afonso, R.J. Removal of cephalexin and erythromycin antibiotics, and their resistance genes, by microalgae-bacteria consortium from wastewater treatment plant secondary effluents. Environ. Sci. Pollut. Res. 2021, 28, 67822–67832. [Google Scholar] [CrossRef]
- da Silva Rodrigues, D.A.; da Cunha, C.C.R.F.; Freitas, M.G.; de Barros, A.L.C.; e Castro, P.B.N.; Pereira, A.R.; de Queiroz Silva, S.; da Fonseca Santiago, A.; de Cássia Franco Afonso, R.J. Biodegradation of sulfamethoxazole by microalgae-bacteria consortium in wastewater treatment plant effluents. Sci. Total Environ. 2020, 749, 141441. [Google Scholar] [CrossRef]
- Gautam, P.K.; Singh, A.; Misra, K.; Sahoo, A.K.; Samanta, S.K. Synthesis and applications of biogenic nanomaterials in drinking and wastewater treatment. J. Environ. Manag. 2019, 231, 734–748. [Google Scholar] [CrossRef]
- Ali, I.; Peng, C.; Khan, Z.M.; Naz, I.; Sultan, M.; Ali, M.; Abbasi, I.A.; Islam, T.; Ye, T. Overview of microbes based fabricated biogenic nanoparticles for water and wastewater treatment. J. Environ. Manag. 2019, 230, 128–150. [Google Scholar] [CrossRef]
- Nasrollahzadeh, M.; Sajjadi, M.; Iravani, S.; Varma, R.S. Green-synthesized nanocatalysts and nanomaterials for water treatment: Current challenges and future perspectives. J. Hazard. Mater. 2021, 401, 123401. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Mourato, C.; Sanches, S.; Noronha, J.P.; Crespo, M.T.B.; Pereira, I.A.C. Biogenic platinum and palladium nanoparticles as new catalysts for the removal of pharmaceutical compounds. Water Res. 2017, 108, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Mu, T.; Zhang, Y.; Shi, W.; Chen, G.; Liu, Y.; Huang, M. A novel UiO-66/PSF-composite membrane for the rejection of multiple antibiotics: Numerical simulation and experiment verification. Chemosphere 2021, 269, 128686. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, Z.; Wang, W.; Wang, Y.; Gao, B.; Wang, Z. Enhanced antifouling and antimicrobial thin film nanocomposite membranes with incorporation of Palygorskite/titanium dioxide hybrid material. J. Colloid Interface Sci. 2019, 537, 1–10. [Google Scholar] [CrossRef]
- Gokulakrishnan, S.A.; Arthanareeswaran, G.; László, Z.; Veréb, G.; Kertész, S.; Kweon, J. Recent development of photocatalytic nanomaterials in mixed matrix membrane for emerging pollutants and fouling control, membrane cleaning process. Chemosphere 2021, 281, 130891. [Google Scholar] [CrossRef]
- Xiong, Q.; Hu, L.X.; Liu, Y.S.; Zhao, J.L.; He, L.Y.; Ying, G.G. Microalgae-based technology for antibiotics removal: From mechanisms to application of innovational hybrid systems. Environ. Int. 2021, 155, 106594. [Google Scholar] [CrossRef]
- Leng, L.; Wei, L.; Xiong, Q.; Xu, S.; Li, W.; Lv, S.; Lu, Q.; Wan, L.; Wen, Z.; Zhou, W. Use of microalgae based technology for the removal of antibiotics from wastewater: A review. Chemosphere 2020, 238, 124680. [Google Scholar] [CrossRef]
- Hena, S.; Gutierrez, L.; Croué, J.P. Removal of pharmaceutical and personal care products (PPCPs) from wastewater using microalgae: A review. J. Hazard. Mater. 2021, 403, 124041. [Google Scholar] [CrossRef]
- Daneshvar, E.; Zarrinmehr, M.J.; Hashtjin, A.M.; Farhadian, O.; Bhatnagar, A. Versatile applications of freshwater and marine water microalgae in dairy wastewater treatment, lipid extraction and tetracycline biosorption. Bioresour. Technol. 2018, 268, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Norvill, Z.N.; Toledo-Cervantes, A.; Blanco, S.; Shilton, A.; Guieysse, B.; Muñoz, R. Photodegradation and sorption govern tetracycline removal during wastewater treatment in algal ponds. Bioresour. Technol. 2017, 232, 35–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Zou, J.; Feng, L.; Zhang, L.; Liu, Y. Chlorella vulgaris enhance the photodegradation of chlortetracycline in aqueous solution via extracellular organic matters (EOMs): Role of triplet state EOMs. Water Res. 2019, 149, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Franklin, A.M.; Brinkman, N.E.; Jahne, M.A.; Keely, S.P. Twenty-first century molecular methods for analyzing antimicrobial resistance in surface waters to support One Health assessments. J. Microbiol. Methods 2021, 184, 106174. [Google Scholar] [CrossRef]
- Hao, H.; Shi, D.-Y.; Yang, D.; Yang, Z.-W.; Qiu, Z.-G.; Liu, W.-L.; Shen, Z.-Q.; Yin, J.; Wang, H.-R.; Li, J.-W.; et al. Profiling of intracellular and extracellular antibiotic resistance genes in tap water. J. Hazard. Mater. 2019, 365, 340–345. [Google Scholar] [CrossRef]
- Han, Z.; Zhang, Y.; An, W.; Lu, J.; Hu, J.; Yang, M. Antibiotic resistomes in drinking water sources across a large geographical scale: Multiple drivers and co-occurrence with opportunistic bacterial pathogens. Water Res. 2020, 183, 116088. [Google Scholar] [CrossRef]
- Ji, X.; Tang, Y.; Ye, J.; Wu, S.; Hou, M.; Huang, S.; Wang, R. The effect of carbon-based copper nanocomposites on Microcystis aeruginosa and the movability of antibiotic resistance genes in urban water. Chemosphere 2022, 286, 131744. [Google Scholar] [CrossRef]
- Su, Y.; Wu, D.; Xia, H.; Zhang, C.; Shi, J.; Wilkinson, K.J.; Xie, B. Metallic nanoparticles induced antibiotic resistance genes attenuation of leachate culturable microbiota: The combined roles of growth inhibition, ion dissolution and oxidative stress. Environ. Int. 2019, 128, 407–416. [Google Scholar] [CrossRef]
- Lu, X.; Hou, J.; Yang, K.; Zhu, L.; Xing, B.; Lin, D. Binding Force and Site-Determined Desorption and Fragmentation of Antibiotic Resistance Genes from Metallic Nanomaterials. Environ. Sci. Technol. 2021, 55, 9305–9316. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Li, B.; Geng, M.; Wang, Y.; Zhao, J.; Jin, B.; Li, Y. Response of extracellular polymeric substances and microbial community structures on resistance genes expression in wastewater treatment containing copper oxide nanoparticles and humic acid. Bioresour. Technol. 2021, 340, 125741. [Google Scholar] [CrossRef]
- Abu-Saied, M.A.; Elnouby, M.; Taha, T.; El-Shafeey, M.; Alshehri, A.G.; Alamri, S.; Alghamdi, H.; Shati, A.; Alrumman, S.; Al-Kahtani, M.; et al. Potential decontamination of drinking water pathogens through k-carrageenan integrated green bottle fly bio-synthesized silver nanoparticles. Molecules 2020, 25, 1936. [Google Scholar] [CrossRef] [Green Version]
- Duan, W.; Gao, J.; Wu, Z.; Dai, H.; Wang, Z.; Li, D.; Wang, Y.; Liu, J. Enhanced removal of antibiotic resistance genes by nanoscale iron-cobalt particles modified with Ginkgo biloba L. leaf: Combining Illumina MiSeq sequencing and oligotyping analysis. Bioresour. Technol. 2021, 321, 124453. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Bhanjana, G.; Heydarifard, S.; Dilbaghi, N.; Nazhad, M.M.; Kumar, V.; Kim, K.H.; Kumar, S. Enhanced antibacterial profile of nanoparticle impregnated cellulose foam filter paper for drinking water filtration. Carbohydr. Polym. 2018, 202, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Pinto, J.; Magrì, D.; Valentini, P.; Palazon, F.; Heredia-Guerrero, J.A.; Lauciello, S.; Barroso-Solares, S.; Ceseracciu, L.; Pompa, P.P.; Athanassiou, A.; et al. Antibacterial Melamine Foams Decorated with in Situ Synthesized Silver Nanoparticles. ACS Appl. Mater. Interfaces 2018, 10, 16095–16104. [Google Scholar] [CrossRef]
- Ren, S.; Boo, C.; Guo, N.; Wang, S.; Elimelech, M.; Wang, Y. Photocatalytic Reactive Ultrafiltration Membrane for Removal of Antibiotic Resistant Bacteria and Antibiotic Resistance Genes from Wastewater Effluent. Environ. Sci. Technol. 2018, 52, 8666–8673. [Google Scholar] [CrossRef]
- Liu, H.; Hua, X.; Zhang, Y.N.; Zhang, T.; Qu, J.; Nolte, T.M.; Chen, G.; Dong, D. Electrocatalytic inactivation of antibiotic resistant bacteria and control of antibiotic resistance dissemination risk. Environ. Pollut. 2021, 291, 118189. [Google Scholar] [CrossRef]
- Pu, Q.; Fan, X.T.; Sun, A.Q.; Pan, T.; Li, H.; Bo Lassen, S.; An, X.L.; Su, J.Q. Co-effect of cadmium and iron oxide nanoparticles on plasmid-mediated conjugative transfer of antibiotic resistance genes. Environ. Int. 2021, 152, 106453. [Google Scholar] [CrossRef]
- Hu, Z.; Zhang, L.; Zhong, L.; Zhou, Y.; Xue, J.; Li, Y. Preparation of an antibacterial chitosan-coated biochar-nanosilver composite for drinking water purification. Carbohydr. Polym. 2019, 219, 290–297. [Google Scholar] [CrossRef]
- Xie, L.; Shu, Y.; Hu, Y.; Cheng, J.; Chen, Y. SWNTs-PAN/TPU/PANI composite electrospun nanofiber membrane for point-of-use efficient electrochemical disinfection: New strategy of CNT disinfection. Chemosphere 2020, 251, 126286. [Google Scholar] [CrossRef]
- Peñalva, G.; Högberg, L.D.; Weist, K.; Vlahović-Palčevski, V.; Heuer, O.; Monnet, D.L.; Vandael, E.; Ivanov, I.N.; Payerl-Pal, M.; Skjold Selle Pedersen, K.; et al. Decreasing and stabilising trends of antimicrobial consumption and resistance in Escherichia coli and Klebsiella pneumoniae in segmented regression analysis, European Union/European Economic Area, 2001 to 2018. Eurosurveillance 2019, 24, 1900656. [Google Scholar] [CrossRef]
- Commision, E. COMMISSION IMPLEMENTING DECISION (EU) 2015/495 of 20 March 2015 Establishing a Watch List of SubStances for Union-Wide Monitoring in the Field of Water Policy Pursuant to Directive 2008/105/EC of the European Parliament and of the Council; European Commission: Brussels, Belgium, 2015; Volume L78/40. [Google Scholar]
- Commision, E. COMMISSION IMPLEMENTING DECISION (EU) 2018/840 of 5 June 2018 Establishing a Watch List of SubStances for Union-Wide Monitoring in the Field of Water Policy Pursuant to Directive 2008/105/EC of the European Parliament and of the Council and Repealing Comm; European Commission: Brussels, Belgium, 2018; Volume 61. [Google Scholar]
- Cortes, L.G.; Marinov, D.; Sanseverino, I.; Cuenca, A.N.; Niegowska, M.; Rodriguez, E.P.; Lettieri, T. JRC Technical Report: Selection of Substances for the 3rd Watch List under WFD; Office of the European Union: Luxembourg, 2020. [Google Scholar]
| Antibiotic Class | ARG | Water Treatment Process | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CO + SE + SF + CL | SF | CL | OZ + CL | OZ | UV | BAC | GAC | MF | Ref | ||
| Aminoglycosides | aadA | ↓ | ↓ | ↓ | ↑ | [68] | |||||
| aadB | ND/↓ | ↑ | [69] | ||||||||
| aadE | ↓ | ↑ | |||||||||
| aphA1 | ND/↑/↓ | ||||||||||
| strA | ↑/↓ | [69,70] | |||||||||
| strB | ↑/↓ | ||||||||||
| B-lactams | blaOXA-1 | ↓ | ↑ | [70] | |||||||
| blaCTX-M | ↑ | [71] | |||||||||
| blaTEM-1 | ↓ | ↓ | ↓ | ↑ | [70,71,72] | ||||||
| Chloramphenicol | cmlA | ↓ | ↑ | ↑ | [73] | ||||||
| dfrA1 | ↑ | [71] | |||||||||
| dfrA12 | ↑ | ||||||||||
| Efflux pump | opxB | ↓ | ↓ | ↑ | [73] | ||||||
| mexF | ↑ | ↑ | ↑ | ↑ | [68] | ||||||
| mexT | ↑ | ↑ | ↑ | ↑ | |||||||
| mexW | ↑ | ↑ | ↑ | ↑ | |||||||
| Florfenicol | floR | ↓ | ↓ | ↑ | [73] | ||||||
| Lincosamides | cfr | ↓ | ↑ | ↑ | |||||||
| Macrolides | ereA | ND | [69] | ||||||||
| ermB | ND/↓ ↑ | ↓ | ↓ | ↑ | [69] [70] | ||||||
| ermC | ↑ | [70] | |||||||||
| ermF | ND | ↓ | [69,74,75] | ||||||||
| ermG | ND | ||||||||||
| ermX | ND | ||||||||||
| mefA | ND | ||||||||||
| mph(A) | ↓ | ↓ | [72] | ||||||||
| Polypeptides | bacA | ↓ | ↓ | ↓ | ↑ | [68] | |||||
| Quinolones | qepA | ↓ | ND | ↑ | [70,73] | ||||||
| qnrA | ↓ | ND | ↑ | ||||||||
| qnrB | ↑ | ND | ↑ | ↑ | |||||||
| qnrD | ND | ↑ | [73] | ||||||||
| qnrS | ND | ↑ | ↑ | ||||||||
| Sulfonamides | sul1 | ↓ | ↓ | ↓ | ↓ | ↓ | ↑/↓ | ↑ | ↓/↑ | [68,70,71,72,73,74,75,76,77,78] | |
| sul2 | ↓ | ↓ | ↓ | ↑ | ↓/↑ | [69,70,71,73,74,75,76,78] | |||||
| sul3 | ↓ | [70] | |||||||||
| Tetracyclines | tetA | ↓ | ↓ | ↓ | ↑ | ↓ | ↑ | ↑ | ↑ | [69,70,71,72,73,78] | |
| tetB | ↑ | ↓ | [70,74] | ||||||||
| tetC | ↓ | ||||||||||
| tetM | ↑ | ↓ | ↑ | ↑ | ↓/↑ | [70,73,75,78] | |||||
| tetG | ND | ↑ | ↓ | [69,71,74] | |||||||
| tetL | ↓ | [79] | |||||||||
| tetO | ND/↓ | ↓ | ↓ | ↑ | ↑ | [69,73,78] | |||||
| tetQ | ND | ↓ | ↓ | ↑ | ↓ | [69,71,73,75] | |||||
| tetS | ↓ | ↑ | [73] | ||||||||
| tetW | ND/↑ | ↓ | ↑ | ↑ | ↓/↑ | [69,70,71,73,75,78] | |||||
| tetX | ND/↓ | ↓ | ↓ | ↑ | ↓/↑ | [74,78] | |||||
| Strategy | Target | Removal Efficiency | Ref |
|---|---|---|---|
| Nanomaterials (nanocomposites, nanofibers, NPs) | |||
| TiO2-doped Fe3+ nano-photocatalyst | Metronidazole | 97% (ci = 80 mg/mL, pH 11, 2 h) 69.85% (pH 6, 2 h) | [94] |
| Graphitized mesoporous carbon TiO2 nanocomposites | Ciprofloxacin | 100% (ci = 1.5 mg/L, 1.5 h) | [95] |
| V2O5-ZnO NPs coated carbon nanofibers | Ciprofloxacin Cinoxacin | Adsorption of 87.70 mg/g (ci = 10–200 mg/L, pH 6.5, 20 min) Adsorption of 71.4 mg/g (ci = 10–200 mg/L, pH 6.5, 20 min) | [96] |
| Ta3N5 NPs/TiO2 hollow nanosphere composite | Levofloxacin Ciprofloxacin Tetracycline hydrochloride | 93% (2 h); 89.76% after 4 cycles 93.2% (3 h) 92.2% (3 h) | [97] |
| Silver modified ZnO nanoplates | Ofloxacin | 98% (ci = 10 mg/mL, pH 7, 2,30 h) | [98] |
| MnO2/graphene nanocomposite | Tetracycline | 99.4% | [99] |
| SnO2/Ni@N carbon nanotubes | Cephalexin | >70% (electropersulfate oxidation) | [100] |
| Boron Nitride Nanosheets | Tetracycline Ofloxacin Cephalexin | Adsorption of 346.66 mg/g, pH 8 Adsorption of 72.50 mg/g (pH 8) Adsorption 225.0 mg/g (pH 12) | [101] |
| Green GS-NiFe beads nanocomposite | Tetracycline (ci = 20 mg/L) | Adsorption/degradation of 487 ± 6.85 mg/g Adsorption/degradation of 420 ± 10.21 mg/g Adsorption/degradation of 408 ± 12.35 mg/g | [102] |
| Green bimetallic nZVI-Cu NPs (pomegranate ring extract) | Tetracycline | 72% (ci = 10 mg/L, pH 7) | [103] |
| Bentonite supported green nZVI-Cu nanocomposite | 95% (pH 7) | ||
| MnCo2O4 NPs | Ciprofloxacin | 100% (pH 3, 5 h) | [104] |
| CdS NPs | 79.50% (ci = 10 mg/mL, pH 9, 80 min) | [105] | |
| NiFe2O4 NPs loaded graphitic carbon nitride | Oxytetracycline | 100% (pH 5, 8 h) | [106] |
| ZV Cu (core) and Fe3O4 (shell) NPs | Oxytetracycline | >99% (ci = 20 mg/mL, pH3, 10 min) | [107] |
| S-doped MgO NPs | Tetracycline | 90% (pH neutral, 10 min) | [108] |
| PRB columns packed with ZVI | Tetracycline (ci = 20 mg/L, pH 6.5, 30 days) | 65% | [109] |
| PRB columns packed with MnO2 | 50% | ||
| PRB columns packed with ZVI and MnO2 | 85% (pH 6.5, 30 days) | ||
| Fe/Ni bimetallic NPs | Tetracycline | 97.4% (ci = 100 mg/mL, pH 5, 3 h) | [110] |
| Microalgae | |||
| Microcystis aeruginosa | Cefradine Amoxicillin | 37.08% 60.89% | [111] |
| Chlorella pyrenoidosa | Cefradine Amoxicillin | 42.63% 71.25% | |
| Haematococcus pluvialis | Sulfonamides | 42–100% (mean 93%) of sulfamerazine, sulfamethoxazole, sulfamonomethoxine | [112] |
| Selenastrum capricornutum | Macrolides Fluoroquinolones | 9–99% (mean 82%) of trimethoprim, clarithromycin azithromycin, roxithromycin 9–99% (mean 82%) of lomefloxacin, levofloxacin, flumequine | |
| Scenedesmus quadricauda | Sulfonamides | 23–98% (mean 78%) | |
| Chlorella vulgaris | Macrolides Fluoroquinolones | 10–100% (mean 47%) 10–100% (mean 47%) of fluoroquinolones (lomefloxacin, levofloxacin, flumequine) | |
| Chlorella vulgaris | Enrofloxacin Sulfadiazine Sulfamethazine Norfloxacin | 53–73% 11–24% 16–33% Inefficient removal | [113] |
| Chrysosporum ovalisporum | Enrofloxacin Sulfadiazine Sulfamethazine Norfloxacin | 58–79% 10–20% 14–27% Inefficient removal | |
| Chlorella pyrenoidosa | cefuroxime sodium | 60% (within 48 h) 92.9% (with NaHCO3 addition) | [114] |
| Scenedesmus obliquus | Ofloxacin | 9.95–39.24% | [115] |
| Scenedesmus dimorphus | Ofloxacin | 93% | [116] |
| Spirulina sp.-derived biochar | Tetracycline | Adsorption of 61% (120 h; ↓ adsorption along with cycles) | [117] |
| Scenedesmus obliquus | Sulfamethazine Sulfamethoxazole | 31.4–62.3% (12 days) 27.7–46.8% (12 days) | [118] |
| Chlorella micrococcus (photo-sequencing batch reactor) | Trimethoprim Sulfamethoxazole Sulfamethazine Sulfamerazine Norfloxacin Enrofloxacin | 91.8% 85.5% 85.5% 85.5% 98% 100% | [119] |
| Chlorella pyrenoidosa | Sulfamethoxazole | Biodegradation of 14.9% (11 days) 99.3% (with sodium acetate addition, 5 days) | [120] |
| Chlorella vulgaris (batch culture) | Sulfadiazine Sulfamethazine Sulfamethoxazole | 32.06% (12 days) 31.17% (12 days) 34.07% (12 days) | [121] |
| Chlorella vulgaris biofilm membrane photobioreactor | Sulfadiazine Sulfamethazine Sulfamethoxazole | 79.2% (1 day) 76.7% (1 day) 82.1% (1 day) | |
| M. aeruginosa | Tetracycline | 98% (2 days) | [122] |
| Chlamydomonas sp. Tai-03 | Ciprofloxacin Sulfadiazine | 100% (65.05% by biodegradation) 54.53% (35.60% by photolysis) | [123] |
| Microalgae-bacteria consortium | Cephalexin Erythromycin | 96.54% (7 days) 92.38% (7 days) | [124] |
| Sulfamethoxazole | 54.34% (42.86% by biodegradation) | [125] | |
| Strategy | Result | Ref |
|---|---|---|
| Nanotechnology (nanoparticles, nanocomposites, nanofibers) | ||
| GNICPs (Ginkgo biloba L. modified iron-cobalt NPs) | ↓ bacterial abundance (↓16S rRNA) ↓ ARGs: blaTEM, sul1, qnrA, acrA-02, mexB, tetM-01, ermB, mefA, ereA ↓MGEs: intI1, intI3, tnp-04, and TP614 Altered microbial community composition | [147] |
| Cd2+ and Fe2O3 NPs | ↑ conjugative transfer frequencies ↑ cell membrane permeability ↑ antioxidant enzymes (SOD, CAT) ↑ mRNA expression of trfAp and trfBp | [152] |
| Metallic (Cu, Zn, CuO, ZnO) NPs | ↓ bacterial growth ↓ ARGs: sul1, aadA1 and MGE: intl1 ↑ ROS production ↑ bacterial cell membrane permeability | [143] |
| nTiO2 NPs | Adsorption of tetM-plasmid (0.06/min and 4.29 mg/g) | [144] |
| nZVI NPs | Adsorption of tetM-plasmid (0.05/min and 2.15 mg/g) ARGs fragmentation | |
| CuO NPs (with humic acid) | ↓ absolute ARGs: macB, mexF and MGE: intl1 ↓ absolute metallic-resistance genes: copA, cusA Modulation of EPS production | [145] |
| CNTs/AG/Ti electrode (Carbon nanotubes/agarose/titanium) | ↑ ROS production bacterial cell membrane damage ↓ ampicilin-resistant E. coli (100%, 1.8.V, 30 min) blaTEM-1 degradation (100%, 2 V (PBS), 30 min) | [151] |
| Water-resistant cellulose foam paper coated with CuO, ZnO, or Ag2O NPs | Enhanced cellulose filter paper antibacterial activity against E. coli, P. aeruginosa, Bacillus Subtilis, and Bacillus cereus Ag2O NPs produced the highest antibacterial activity | [148] |
| Melamine foams with Ag NPs | Antibacterial activity against E. coli | [149] |
| PVDF membrane functionalized with TiO2 NPs | 99.9% retention of tetracycline, chloramphenicol, and sulfadiazine-resistant bacteria ↓ ARGs: floR (97.8%), sul1 (99.5%), sul2 (98.8%), intI1 (93.7%), tetC (20.6%), tetW (27.2%), tetQ (2.0%) Inhibition of HGT | [150] |
| Chitosan/biochar-nanosilver (C-Ag) composite | Sustainable antibacterial activity against E. coli (>50 days) Good reusability | [153] |
| Carbon-based copper nanocomposites | ↓ absolute ARGs and MGEs ↓ HGT mediated by plasmids and MGEs | [142] |
| SWNTs-PAN/TPU/PANI composite electrospun nanofiber membrane | Complete removal of S. aureus and E. coli Good durability and stability over various cycles | [154] |
| k-carrageenan/Ag NPs film | Antimicrobial activity against Vibrio cholerae, Candida albicans, P. aeruginosa, E. coli, K. pneumoniae, and Bacillus cereus Inhibition of bacteria growth | [146] |
| Microalgae | ||
| Chlorella micrococcus (photo-sequencing batch reactor) | ↓ 78% ARG absolute abundance ↓ Rhodocyclaceae and Burkholderiaceae bacteria families | [119] |
| Microalgae-bacteria consortium | ↓ ARGs: blaTEM (72%) and ermB (97%) absolute abundance | [124] |
| ↑ ARG: sul1 | [125] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duarte, A.C.; Rodrigues, S.; Afonso, A.; Nogueira, A.; Coutinho, P. Antibiotic Resistance in the Drinking Water: Old and New Strategies to Remove Antibiotics, Resistant Bacteria, and Resistance Genes. Pharmaceuticals 2022, 15, 393. https://doi.org/10.3390/ph15040393
Duarte AC, Rodrigues S, Afonso A, Nogueira A, Coutinho P. Antibiotic Resistance in the Drinking Water: Old and New Strategies to Remove Antibiotics, Resistant Bacteria, and Resistance Genes. Pharmaceuticals. 2022; 15(4):393. https://doi.org/10.3390/ph15040393
Chicago/Turabian StyleDuarte, Ana Catarina, Sílvia Rodrigues, Andrea Afonso, António Nogueira, and Paula Coutinho. 2022. "Antibiotic Resistance in the Drinking Water: Old and New Strategies to Remove Antibiotics, Resistant Bacteria, and Resistance Genes" Pharmaceuticals 15, no. 4: 393. https://doi.org/10.3390/ph15040393
APA StyleDuarte, A. C., Rodrigues, S., Afonso, A., Nogueira, A., & Coutinho, P. (2022). Antibiotic Resistance in the Drinking Water: Old and New Strategies to Remove Antibiotics, Resistant Bacteria, and Resistance Genes. Pharmaceuticals, 15(4), 393. https://doi.org/10.3390/ph15040393








