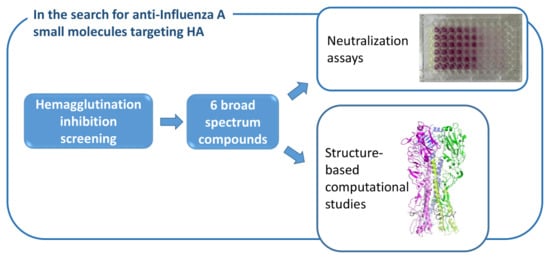
Broad-Spectrum Activity of Small Molecules Acting against Influenza a Virus: Biological and Computational Studies
Abstract
:1. Introduction
2. Results
2.1. Hemagglutination Inhibition (HI) Test against the A/H3N2 Strain
2.2. Compound Scaffold Validation
2.3. Neutralization Assays
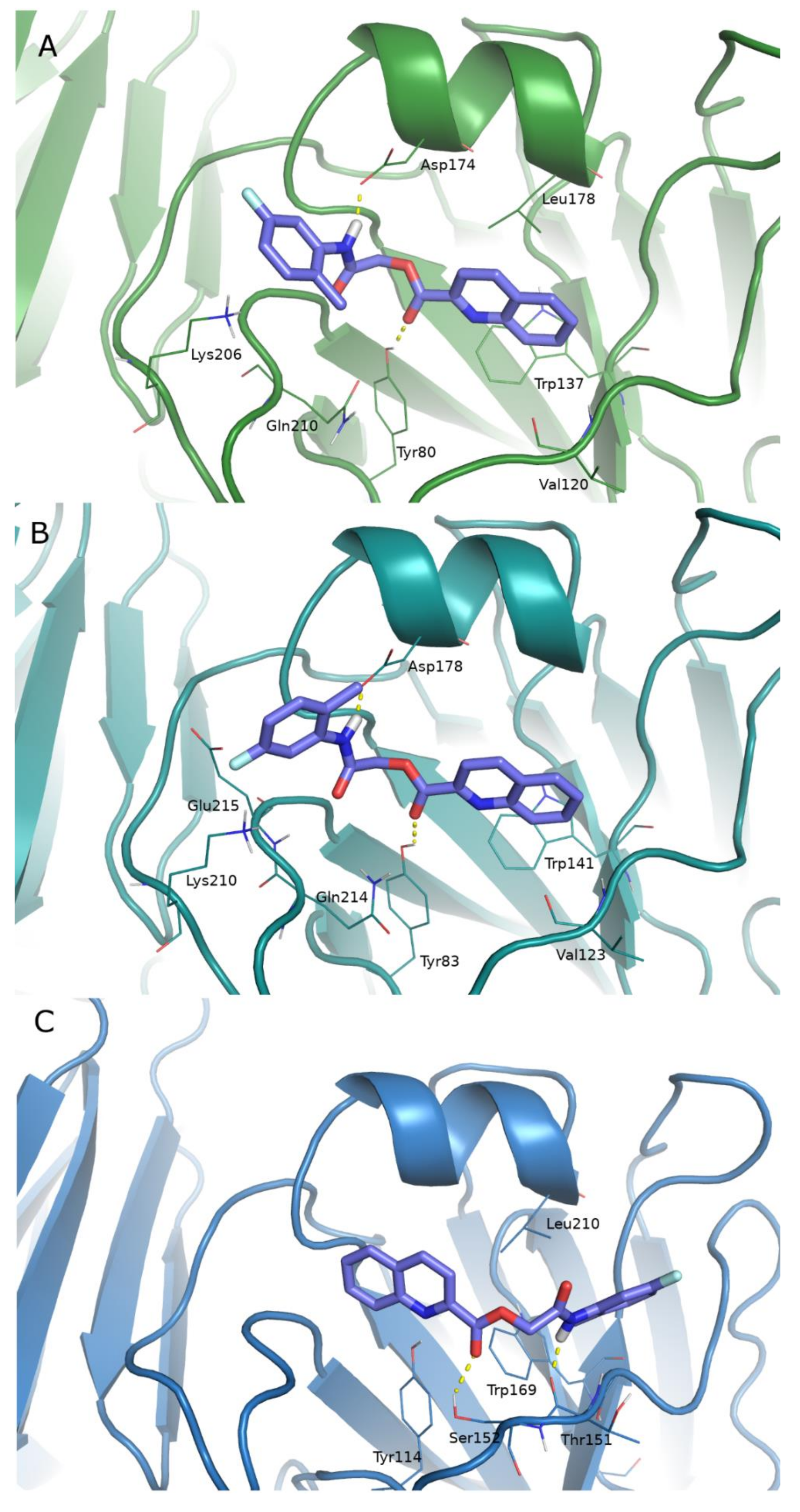
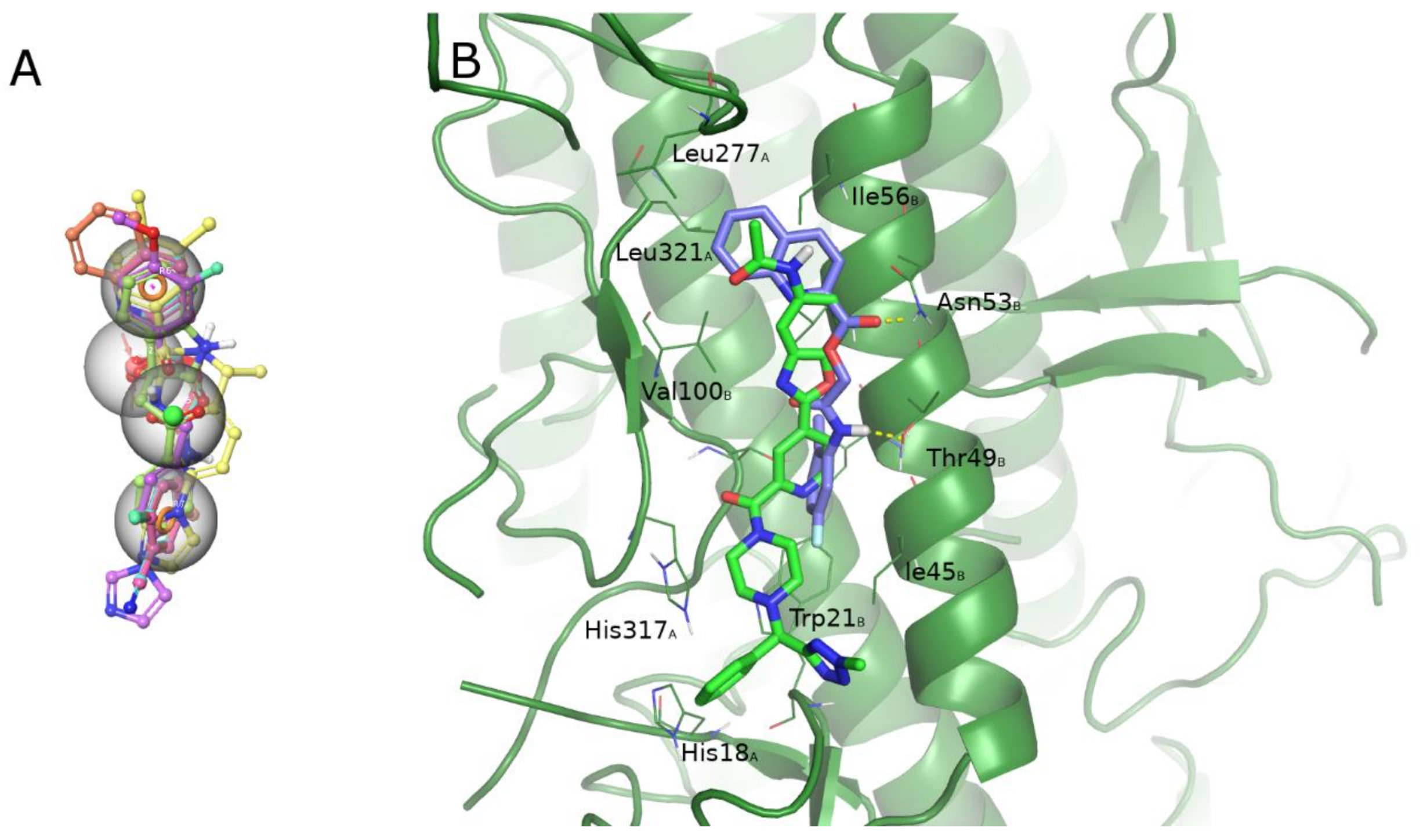
2.4. Computational Studies
2.5. Pharmacokinetic Profile
3. Discussion
4. Materials and Methods
4.1. Compounds
4.2. Biological Assay
4.2.1. Cells and Viral Strains
4.2.2. Cytotoxicity Assay
4.2.3. Hemagglutination Inhibition (HI) Assay
4.2.4. Neutralization Assay
4.3. Computational Studies
4.3.1. Homology Modeling
4.3.2. Protein Preparation
4.3.3. Binding Site Identification and Analysis
4.3.4. Receptor Grid Generation
4.3.5. Ligand Preparation
4.3.6. Docking Calculations
4.3.7. Induced Fit
4.3.8. Molecular Dynamics
4.3.9. Pharmacophore Alignment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Highly pathogenic avian influenza. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar] [CrossRef]
- Taubenberger, J.K.; Morens, D.M. 1918 Influenza: The Mother of All Pandemics. Emerg. Infect. Dis. 2006, 12, 15–22. [Google Scholar] [CrossRef]
- Garten, R.J.; Davis, C.T.; Russell, C.A.; Shu, B.; Lindstrom, S.; Balish, A.; Sessions, W.M.; Xu, X.; Skepner, E.; Deyde, V.; et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009, 325, 197–201. [Google Scholar] [CrossRef] [Green Version]
- Cheng, V.C.C.; To, K.K.W.; Tse, H.; Hung, I.F.N.; Yuen, K.Y. Two years after pandemic influenza A/2009/H1N1: What have we learned? Clin. Microbiol. Rev. 2012, 25, 223–263. [Google Scholar] [CrossRef] [Green Version]
- Rosano, A.; Bella, A.; Gesualdo, F.; Acampora, A.; Pezzotti, P.; Marchetti, S.; Ricciardi, W.; Rizzo, C. Investigating the impact of influenza on excess mortality in all ages in Italy during recent seasons (2013/14–2016/17 seasons). Int. J. Infect. Dis. 2019, 88, 127–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Up to 650,000 People Die of Respiratory Diseases Linked to Seasonal Flu Each Year. Available online: http://www.who.int/mediacentre/news/releases/2017/seasonal-flu/en/ (accessed on 1 March 2018).
- Green, H.K.; Andrews, N.; Fleming, D.; Zambon, M.; Pebody, R. Mortality attributable to Influenza in England and Wales prior to, during and after the 2009 pandemic. PLoS ONE 2013, 8, e79360. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, L.; Spreeuwenberg, P.; Lustig, R.; Taylor, R.J.; Fleming, D.M.; Kroneman, M.; Van Kerkhove, M.D.; Mounts, A.W.; Paget, W.J.; Echenique, H.; et al. Global Mortality Estimates for the 2009 Influenza Pandemic from the GLaMOR Project: A Modeling Study. PLoS Med. 2013, 10, e1001558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Wong, J.Y.; Wu, P.; Bond, H.S.; Lau, E.H.Y.; Sullivan, S.G.; Cowling, B.J. Heterogeneity in Estimates of the Impact of Influenza on Population Mortality: A Systematic Review. Am. J. Epidemiol. 2018, 187, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Paget, J.; Spreeuwenberg, P.; Charu, V.; Taylor, R.J.; Iuliano, A.D.; Bresee, J.; Simonsen, L.; Viboud, C. Global mortality associated with seasonal influenza epidemics: New burden estimates and predictors from the GLaMOR Project. J. Glob. Health 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Lee, I.; Park, S.; Hwang, M.-W.; Bae, J.-Y.; Lee, S.; Heo, J.; Park, M.S.; Garcia-Sastre, A.; Park, M.-S. Genetic Requirement for Hemagglutinin Glycosylation and Its Implications for Influenza A H1N1 Virus Evolution. J. Virol. 2013, 87, 7539–7549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, P.F.; Neumann, G.; Kawaoka, Y. Orthomyxoviruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Cohen, J.I., Griffin, D.E., Lamb, R.A., Martin, M.A., Racaniello, V.R., Roizman, B., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 1186–1243. [Google Scholar]
- Krammer, F.; Palese, P. Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov. 2015, 14, 167–182. [Google Scholar] [CrossRef]
- Krammer, F.; Palese, P.; Steel, J. Advances in Universal Influenza Virus Vaccine Design and Antibody Mediated Therapies Based on Conserved Regions of the Hemagglutinin. In Influenza Pathogenesis and Control; Oldstone, M.B.A., Compans, R.W., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume II, pp. 301–321. ISBN 978-3-319-11158-2. [Google Scholar]
- Coughlan, L.; Palese, P. Overcoming Barriers in the Path to a Universal Influenza Virus Vaccine. Cell Host Microbe 2018, 24, 18–24. [Google Scholar] [CrossRef] [Green Version]
- Colombo, R.E.; Fiorentino, C.; Dodd, L.E.; Hunsberger, S.; Haney, C.; Barrett, K.; Nabha, L.; Davey, R.T.; Olivier, K.N. A phase 1 randomized, double-blind, placebo-controlled, crossover trial of DAS181 (Fludase®) in adult subjects with well-controlled asthma. BMC Infect. Dis. 2016, 16, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikuchi, T.; Watanabe, A. Baloxavir heralds a new era in influenza virus biology. Respir. Investig. 2019, 57, 1–2. [Google Scholar] [CrossRef]
- Koshimichi, H.; Ishibashi, T.; Kawaguchi, N.; Sato, C.; Kawasaki, A.; Wajima, T. Safety, Tolerability, and Pharmacokinetics of the Novel Anti-influenza Agent Baloxavir Marboxil in Healthy Adults: Phase I Study Findings. Clin. Drug Investig. 2018, 38, 1189–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayden, F.G.; Sugaya, N.; Hirotsu, N.; Lee, N.; de Jong, M.D.; Hurt, A.C.; Ishida, T.; Sekino, H.; Yamada, K.; Portsmouth, S.; et al. Baloxavir Marboxil for Uncomplicated Influenza in Adults and Adolescents. N. Engl. J. Med. 2018, 379, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Beigel, J.H.; Voell, J.; Muñoz, P.; Kumar, P.; Brooks, K.M.; Zhang, J.; Iversen, P.; Heald, A.; Wong, M.; Davey, R.T. Safety, tolerability, and pharmacokinetics of radavirsen (AVI-7100), an antisense oligonucleotide targeting Influenza a M1/M2 translation. Br. J. Clin. Pharmacol. 2018, 84, 25–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tharakaraman, K.; Subramanian, V.; Viswanathan, K.; Sloan, S.; Yen, H.L.; Barnard, D.L.; Leung, Y.H.C.; Szretter, K.J.; Koch, T.J.; Delaney, J.C.; et al. A broadly neutralizing human monoclonal antibody is effective against H7N9. Proc. Natl. Acad. Sci. USA 2015, 112, 10890–10895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kallewaard, N.L.; Corti, D.; Collins, P.J.; Zhu, Q.; Gamblin, S.J.; Correspondence, J.J.S. Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes. Cell 2016, 166, 596–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilyushina, N.A.; Bovin, N.V.; Webster, R.G.; Govorkova, E.A. Combination chemotherapy, a potential strategy for reducing the emergence of drug-resistant influenza A variants. Antiviral Res. 2006, 70, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Yanmei, H.; Rami, M.; Chunlong, M.; Jiantao, Z.; Smee, D.F.; DeGrado, W.F.; Wang, J. An M2-V27A channel blocker demonstrates potent in vitro and in vivo antiviral activities against amantadine-sensitive and—Resistant influenza A viruses. Antiviral Res. 2017, 140, 45–54. [Google Scholar] [CrossRef]
- Deleu, S.; Kakuda, T.N.; Spittaels, K.; Vercauteren, J.J.; Hillewaert, V.; Lwin, A.; Leopold, L.; Hoetelmans, R.M.W. Single- and multiple-dose pharmacokinetics and safety of pimodivir, a novel, non-nucleoside polymerase basic protein 2 subunit inhibitor of the influenza A virus polymerase complex, and interaction with oseltamivir: A Phase 1 open-label study in healthy volunteers. Br. J. Clin. Pharmacol. 2018, 84, 2663–2672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gubareva, L.; Mohan, T. Antivirals Targeting the Neuraminidase. Cold Spring Harb. Perspect. Med. 2022, 12, a038455. [Google Scholar] [CrossRef] [PubMed]
- Massari, S.; Desantis, J.; Nizi, M.G.; Cecchetti, V.; Tabarrini, O. Inhibition of Influenza Virus Polymerase by Interfering with Its Protein-Protein Interactions. ACS Infect. Dis. 2021, 7, 1332–1350. [Google Scholar] [CrossRef]
- Lieve, N.; Stevaert, A.; Vanderlinden, E. Antiviral therapies on the horizon for influenza. Curr. Opin. Pharmacol. 2016, 30, 106–115. [Google Scholar] [CrossRef] [Green Version]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef]
- Ekiert, D.C.; Bhabha, G.; Elsliger, M.-A.; Friesen, R.H.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody recognition of a highly conserved influenza virus epitope: Implications for universal prevention and therapy. Science 2009, 324, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.T.; Palese, P. Universal epitopes of influenza virus hemagglutinins? Nat. Struct. Mol. Biol. 2009, 16, 233–234. [Google Scholar] [CrossRef]
- Hashem, A.M.; Van Domselaar, G.; Li, C.; Wang, J.; She, Y.M.; Cyr, T.D.; Sui, J.; He, R.; Marasco, W.A.; Li, X. Universal antibodies against the highly conserved influenza fusion peptide cross-neutralize several subtypes of influenza A virus. Biochem. Biophys. Res. Commun. 2010, 403, 247–251. [Google Scholar] [CrossRef]
- Chen, Z.; Cui, Q.; Caffrey, M.; Rong, L.; Du, R. Small molecule inhibitors of influenza virus entry. Pharmaceuticals 2021, 14, 587. [Google Scholar] [CrossRef]
- Pécheur, E.-I.; Borisevich, V.; Halfmann, P.; Morrey, J.D.; Smee, D.F.; Prichard, M.; Mire, C.E.; Kawaoka, Y.; Geisbert, T.W.; Polyak, S.J. The Synthetic Antiviral Drug Arbidol Inhibits Globally Prevalent Pathogenic Viruses. J. Virol. 2016, 90, 3086–3092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boriskin, Y.S.; Leneva, I.A.; Pécheur, E.-I.; Polyak, S.J. Arbidol: A broad-spectrum antiviral compound that blocks viral fusion. Curr. Med. Chem. 2008, 15, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Kadam, R.U.; Juraszek, J.; Brandenburg, B.; Buyck, C.; Schepens, W.B.G.; Kesteleyn, B.; Stoops, B.; Vreeken, R.J.; Vermond, J.; Goutier, W.; et al. Potent peptidic fusion inhibitors of influenza virus. Science 2017, 358, 496–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ammendolia, M.G.; Agamennone, M.; Pietrantoni, A.; Lannutti, F.; Siciliano, R.A.; de Giulio, B.; Amici, C.; Superti, F. Bovine lactoferrin-derived peptides as novel broad-spectrum inhibitors of influenza virus. Pathog. Glob. Health 2012, 106, 12–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scala, M.C.; Sala, M.; Pietrantoni, A.; Spensiero, A.; Di Micco, S.; Agamennone, M.; Bertamino, A.; Novellino, E.; Bifulco, G.; Gomez-Monterrey, I.M.; et al. Lactoferrin-derived Peptides Active towards Influenza: Identification of Three Potent Tetrapeptide Inhibitors. Sci. Rep. 2017, 7, 10593. [Google Scholar] [CrossRef] [Green Version]
- Scala, M.C.; Agamennone, M.; Pietrantoni, A.; Di Sarno, V.; Bertamino, A.; Superti, F.; Campiglia, P.; Sala, M. Discovery of a Novel Tetrapeptide against Influenza A Virus: Rational Design, Synthesis, Bioactivity Evaluation and Computational Studies. Pharmaceuticals 2021, 14, 959. [Google Scholar] [CrossRef]
- Superti, F.; Agamennone, M.; Pietrantoni, A.; Ammendolia, M.G. Bovine lactoferrin prevents influenza a virus infection by interfering with the fusogenic function of viral hemagglutinin. Viruses 2019, 11, 51. [Google Scholar] [CrossRef] [Green Version]
- Agamennone, M.; Pietrantoni, A.; Superti, F. Identification of small molecules acting against H1N1 Influenza A virus. Virology 2016, 488, 249–258. [Google Scholar] [CrossRef] [Green Version]
- Matsubara, T.; Onishi, A.; Yamaguchi, D.; Sato, T. Heptapeptide ligands against receptor-binding sites of influenza hemagglutinin toward anti-influenza therapy. Bioorganic Med. Chem. 2016, 24, 1106–1114. [Google Scholar] [CrossRef]
- Kim, J.I.; Lee, S.; Lee, G.Y.; Park, S.; Bae, J.-Y.; Heo, J.; Kim, H.-Y.; Woo, S.-H.; Lee, H.U.; Ahn, C.A.; et al. Novel Small Molecule Targeting the Hemagglutinin Stalk of Influenza Viruses. J. Virol. 2019, 93, e00878-19. [Google Scholar] [CrossRef] [Green Version]
- Leiva, R.; Barniol-Xicota, M.; Codony, S.; Ginex, T.; Vanderlinden, E.; Montes, M.; Caffrey, M.; Luque, F.J.; Naesens, L.; Vázquez, S. Aniline-Based Inhibitors of Influenza H1N1 Virus Acting on Hemagglutinin-Mediated Fusion. J. Med. Chem. 2018, 61, 98–118. [Google Scholar] [CrossRef]
- Vasile, F.; Panigada, M.; Siccardi, A.; Potenza, D.; Tiana, G. A combined nmr-computational study of the interaction between influenza virus hemagglutinin and sialic derivatives from human and avian receptors on the surface of transfected cells. Int. J. Mol. Sci. 2018, 19, 1267. [Google Scholar] [CrossRef] [Green Version]
- Le, K.P.; Do, P.-C.; Amaro, R.E.; Le, L. Molecular Docking of Broad-Spectrum Antibodies on Hemagglutinins of Influenza A Virus. Evol. Bioinform. Online 2019, 15, 1176934319876938. [Google Scholar] [CrossRef] [PubMed]
- Weis, W.; Brown, J.H.; Cusack, S.; Paulson, J.C.; Skehel, J.J.; Wiley, D.C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 1988, 333, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Kadam, R.U.; Wilson, I.A. A small-molecule fragment that emulates binding of receptor and broadly neutralizing antibodies to Influenza A hemagglutinin. Proc. Natl. Acad. Sci. USA 2018, 115, 4240–4245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrödinger Release 2021–2022: Maestro, Glide, LigPrep, Phase, Prime, MacroModel, SiteMap, Induced Fit Docking Protocol, Desmond Molecular Dynamics System, D.E. Shaw Research, New York, NY, 2021. Maestro-Desmond Interoperability Tools; Schrödinger, LLC: New York, NY, USA, 2021.
- Willyard, C. Flu on the farm. Nature 2019, 573, S62–S63. [Google Scholar] [CrossRef] [Green Version]
- Shi, W.; Gao, G.F. Emerging H5N8 avian influenza viruses. Science 2021, 372, 784–786. [Google Scholar] [CrossRef]
- Hussein, A.F.A.; Cheng, H.; Tundup, S.; Antanasijevic, A.; Varhegyi, E.; Perez, J.; AbdulRahman, E.M.; Elenany, M.G.; Helal, S.; Caffrey, M.; et al. Identification of entry inhibitors with 4-aminopiperidine scaffold targeting group 1 influenza A virus. Antiviral Res. 2020, 177, 104782. [Google Scholar] [CrossRef]
- Basu, A.; Antanasijevic, A.; Wang, M.; Li, B.; Mills, D.M.; Ames, J.A.; Nash, P.J.; Williams, J.D.; Peet, N.P.; Moir, D.T.; et al. New Small Molecule Entry Inhibitors Targeting Hemagglutinin-Mediated Influenza A Virus Fusion. J. Virol. 2014, 88, 1447–1460. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.; Kadam, R.U.; Lee, C.C.D.; Woehl, J.L.; Wu, N.C.; Zhu, X.; Kitamura, S.; Wilson, I.A.; Wolan, D.W. An influenza A hemagglutinin small-molecule fusion inhibitor identified by a new high-throughput fluorescence polarization screen. Proc. Natl. Acad. Sci. USA 2020, 117, 18431–18438. [Google Scholar] [CrossRef]
- Van Dongen, M.J.P.; Kadam, R.U.; Juraszek, J.; Lawson, E.; Brandenburg, B.; Schmitz, F.; Schepens, W.B.G.; Stoops, B.; Van Diepen, H.A.; Jongeneelen, M.; et al. A small-molecule fusion inhibitor of influenza virus is orally active in mice. Science 2019, 363, eaar6221. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Lin, X.; Qiu, Z.; Li, W.; Zhu, L.; Wang, L.; Li, S.; Li, H.; Lin, W.; Yang, M.; et al. Design and Synthesis of Benzenesulfonamide Derivatives as Potent Anti-Influenza Hemagglutinin Inhibitors. ACS Med. Chem. Lett. 2011, 2, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Si, L.; Wang, Y.; Wu, Y.; Yu, F.; Jiao, P.; Shi, Y.; Wang, H.; Xiao, S.; Fu, G.; et al. Discovery of pentacyclic triterpenoids as potential entry inhibitors of influenza viruses. J. Med. Chem. 2014, 57, 10058–10071. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, F.; Meng, L.; Sun, J.; Su, Y.; Shao, L.; Zhou, D.; Yu, F. Synthesis, Structure Activity Relationship and Anti-influenza A Virus Evaluation of Oleanolic Acid-Linear Amino Derivatives. Chem. Pharm. Bull. 2019, 67, 1201–1207. [Google Scholar] [CrossRef] [Green Version]
- Yusuf, M.; Konc, J.; Sy Bing, C.; Trykowska Konc, J.; Ahmad Khairudin, N.B.; Janezic, D.; Wahab, H.A. Structurally conserved binding sites of hemagglutinin as targets for influenza drug and vaccine development. J. Chem. Inf. Model. 2013, 53, 2423–2436. [Google Scholar] [CrossRef]
- Lao, J.; Vanet, A. A New Strategy to Reduce Influenza Escape: Detecting Therapeutic Targets Constituted of Invariance Groups. Viruses 2017, 9, 38. [Google Scholar] [CrossRef] [Green Version]
- Gaush, C.R.; Smith, T.F. Replication and plaque assay of influenza virus in an established line of canine kidney cells. Appl. Microbiol. 1968, 16, 588–594. [Google Scholar] [CrossRef]
- Rimmelzwaan, G.F.; Baars, M.; Claas, E.C.J.; Osterhaus, A.D.M.E. Comparison of RNA hybridization, hemagglutination assay, titration of infectious virus and immunofluorescence as methods for monitoring influenza virus replication in vitro. J. Virol. Methods 1998, 74, 57–66. [Google Scholar] [CrossRef]
- Marchetti, M.; Trybala, E.; Superti, F.; Johansson, M.; Bergström, T. Inhibition of herpes simplex virus infection by lactoferrin is dependent on interference with the virus binding to glycosaminoglycans. Virology 2004, 318, 405–413. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; McBride, R.; Nycholat, C.M.; Paulson, J.C.; Wilson, I.A. Structural Characterization of the Hemagglutinin Receptor Specificity from the 2009 H1N1 Influenza Pandemic. J. Virol. 2012, 86, 982–990. [Google Scholar] [CrossRef] [Green Version]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.L.; Smondyrev, A.M.; Rao, S.N. PHASE: A novel approach to pharmacophore modeling and 3D database searching. Chem. Biol. Drug Des. 2006, 67, 370–372. [Google Scholar] [CrossRef] [PubMed]




| ID | Structure | HI Titer (μM) | ||
|---|---|---|---|---|
| A/Parma H1N1 a | A/Roma H1N1 a | A/Parma H3N2 | ||
| 1 | 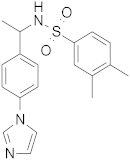 | 0.48 | 0.24 | 0.48 |
| 2 | 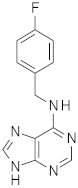 | 0.48 | 0.48 | - |
| 3 |  | 0.48 | 0.48 | - |
| 4 | 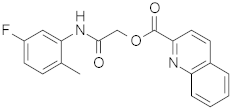 | 0.48 | 0.48 | 0.48 |
| 5 | 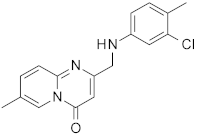 | 0.48 | 0.48 | - |
| ID | Structure | HI Titer (pM) | |||
|---|---|---|---|---|---|
| A/Roma H1N1 | A/Parma H1N1 | A/Parma H3N2 | Ref Cmpd | ||
| 6 | 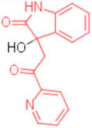 | 0.012 | 0.012 | 480 | 4 |
| 7 | 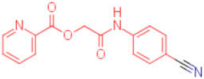 | 0.047 | 0.012 | 12,500 | 4 |
| 8 | 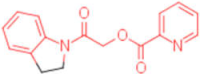 | 0.012 | 0.012 | - | 4 |
| 9 | 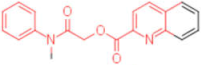 | 0.012 | 0.012 | - | 4 |
| 10 | 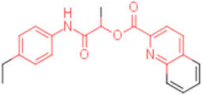 | 0.012 | 0.012 | - | 4 |
| 11 |  | 0.012 | 0.012 | - | 4 |
| 12 | 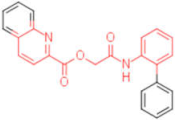 | 0.012 | 0.012 | - | 4 |
| 13 |  | 24 | 0.012 | 6 | 4 |
| 14 | 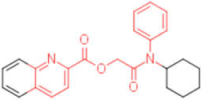 | - | 0.012 | - | 4 |
| 15 | 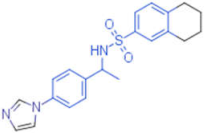 | 0.012 | - | - | 1 |
| 16 |  | 0.012 | 0.012 | - | 1 |
| 17 | 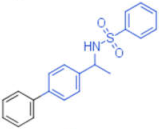 | - | 0.012 | - | 1 |
| 18 | 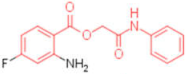 | 0.012 | 0.012 | - | 4 |
| 19 | 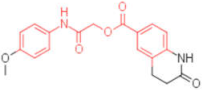 | - | 0.012 | - | 4 |
| 20 | 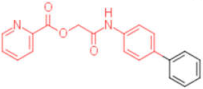 | 0.012 | 0.012 | - | 4 |
| 21 | 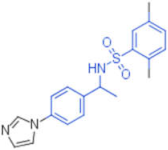 | 0.012 | - | - | 1 |
| 22 | 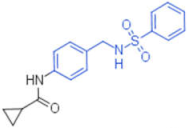 | 0.012 | - | - | 1 |
| 23 | 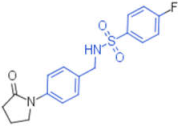 | 12 | 0.012 | - | 1 |
| 24 | 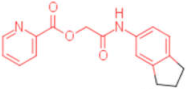 | 0.024 | 0.047 | - | 4 |
| 25 | 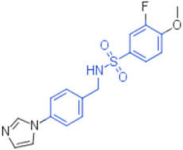 | 0.012 | 0.012 | 50,000 | 1 |
| 26 | 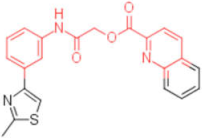 | 0.093 | 0.047 | - | 4 |
| 27 |  | - | - | 25,000 | 1 |
| 28 |  | 0.093 | 0.047 | - | 1 |
| ID | CC50 *(μM) | A/Roma H1N1 | A/Parma H1N1 | A/Parma H3N2 | |||
|---|---|---|---|---|---|---|---|
| EC50 (nM) ∞ | SI ^ | EC50 (nM) ∞ | SI ^ | EC50 (nM) ∞ | SI ^ | ||
| 1 | 300 | 500 +/−0.01 p < 0.01 a | 6 × 102 a | 125 +/−0.01 p < 0.01 a | 2.4 × 103 a | 98 +/−0.01 p < 0.01 | 3 × 103 |
| 4 | 250 | 62 +/−0.07 p < 0.01 a | 4 × 103 a | 125 +/−0.03 p < 0.01 a | 2 × 103 a | 125 +/−0.02 p < 0.01 | 2 × 103 |
| 6 | 31.25 | 200 +/−0.001 p < 0.02 | 1.56 × 102 | 900 +/−0.01 p < 0.01 | 0.34 × 102 | 300 +/−0.001 p < 0.01 | 1.04 × 102 |
| 7 | 31.25 | 800 +/−0.05 p < 0.01 | 0.39 × 102 | 900 +/−0.003 p < 0.01 | 0.34 × 102 | 700 +/−0.002 p < 0.01 | 0.44 × 102 |
| 13 | 31.25 | 700 +/−0.04 p < 0.01 | 0.47 × 102 | 700 +/−0.04 p < 0.01 | 0.44 × 102 | 500 +/0.05 p < 0.01 | 0.62 × 102 |
| 25 | 31.25 | 900 +/−0.02 p < 0.01 | 0.34 × 102 | 700 +/−0.002 p < 0.01 | 0.44 × 102 | 800 +/0.01 p < 0.01 | 0.39 × 102 |
| Cmpd | mol MW | QPlogPo/w a | QPlogS b | QPlogHERG c | QPPCaco d | #metab e | QPlogKhsg f | PercentHumanOralAbsorption | PSA g |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 355.454 | 3.941 | −5.442 | −6.374 | 1074.843 | 3 | 0.405 | 100.000 | 60.952 |
| 4 | 338.337 | 3.947 | −5.292 | −6.437 | 1607.601 | 3 | 0.321 | 100.000 | 72.475 |
| 6 | 268.271 | 1.351 | −2.292 | −4.267 | 255.939 | 3 | −0.307 | 77.956 | 97.449 |
| 7 | 288.278 | 2.779 | −4.527 | −6.168 | 670.015 | 3 | −0.007 | 93.798 | 84.973 |
| 13 | 361.390 | 2.138 | −2.551 | −4.094 | 634.323 | 2 | −0.215 | 89.620 | 72.606 |
| 25 | 281.270 | 1.431 | −4.575 | −6.339 | 150.454 | 2 | −0.357 | 74.293 | 110.030 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agamennone, M.; Superti, F. Broad-Spectrum Activity of Small Molecules Acting against Influenza a Virus: Biological and Computational Studies. Pharmaceuticals 2022, 15, 301. https://doi.org/10.3390/ph15030301
Agamennone M, Superti F. Broad-Spectrum Activity of Small Molecules Acting against Influenza a Virus: Biological and Computational Studies. Pharmaceuticals. 2022; 15(3):301. https://doi.org/10.3390/ph15030301
Chicago/Turabian StyleAgamennone, Mariangela, and Fabiana Superti. 2022. "Broad-Spectrum Activity of Small Molecules Acting against Influenza a Virus: Biological and Computational Studies" Pharmaceuticals 15, no. 3: 301. https://doi.org/10.3390/ph15030301
APA StyleAgamennone, M., & Superti, F. (2022). Broad-Spectrum Activity of Small Molecules Acting against Influenza a Virus: Biological and Computational Studies. Pharmaceuticals, 15(3), 301. https://doi.org/10.3390/ph15030301