Cationic Liposomes with Different Lipid Ratios: Antibacterial Activity, Antibacterial Mechanism, and Cytotoxicity Evaluations
Abstract
1. Introduction
2. Results and Discussion
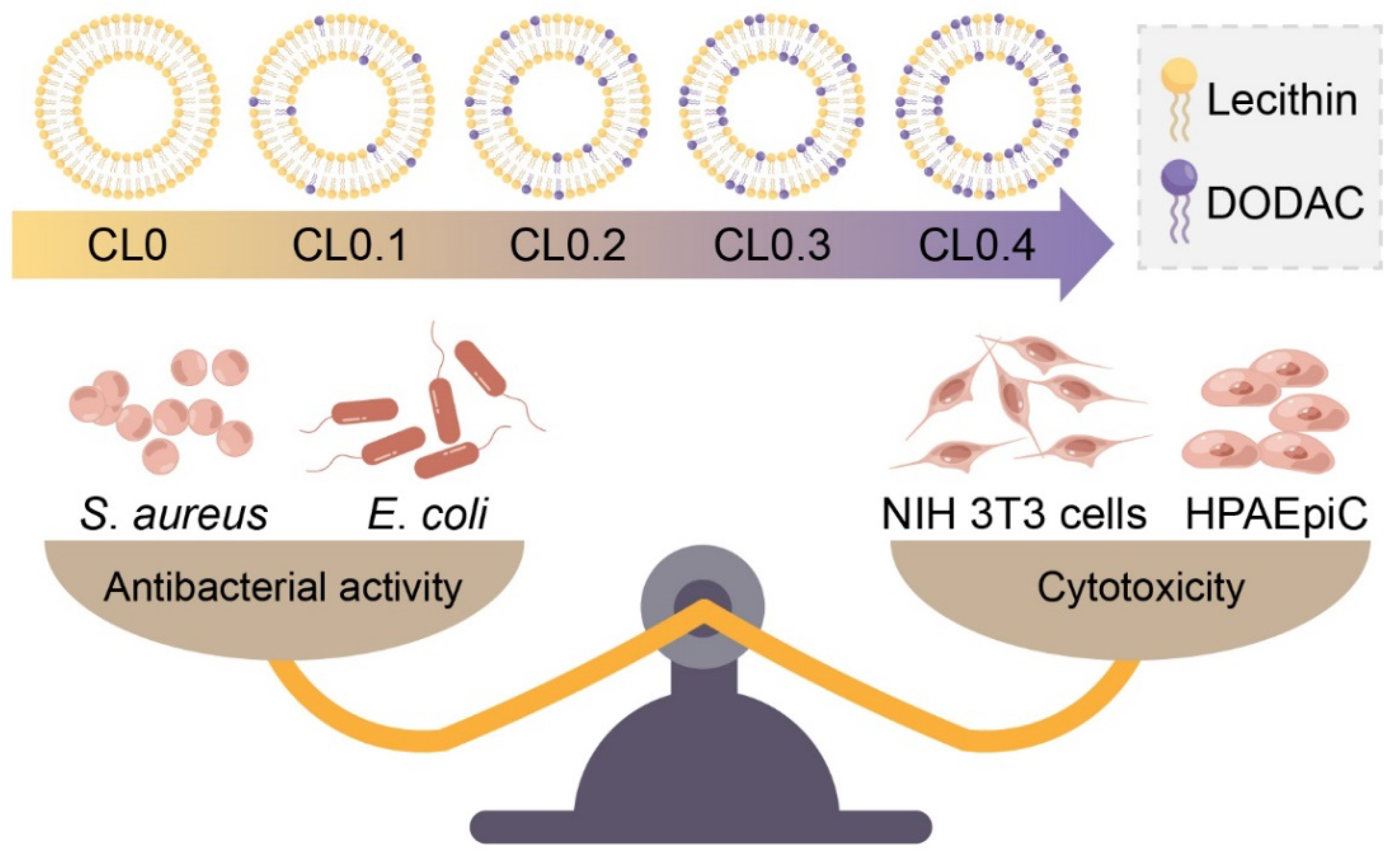
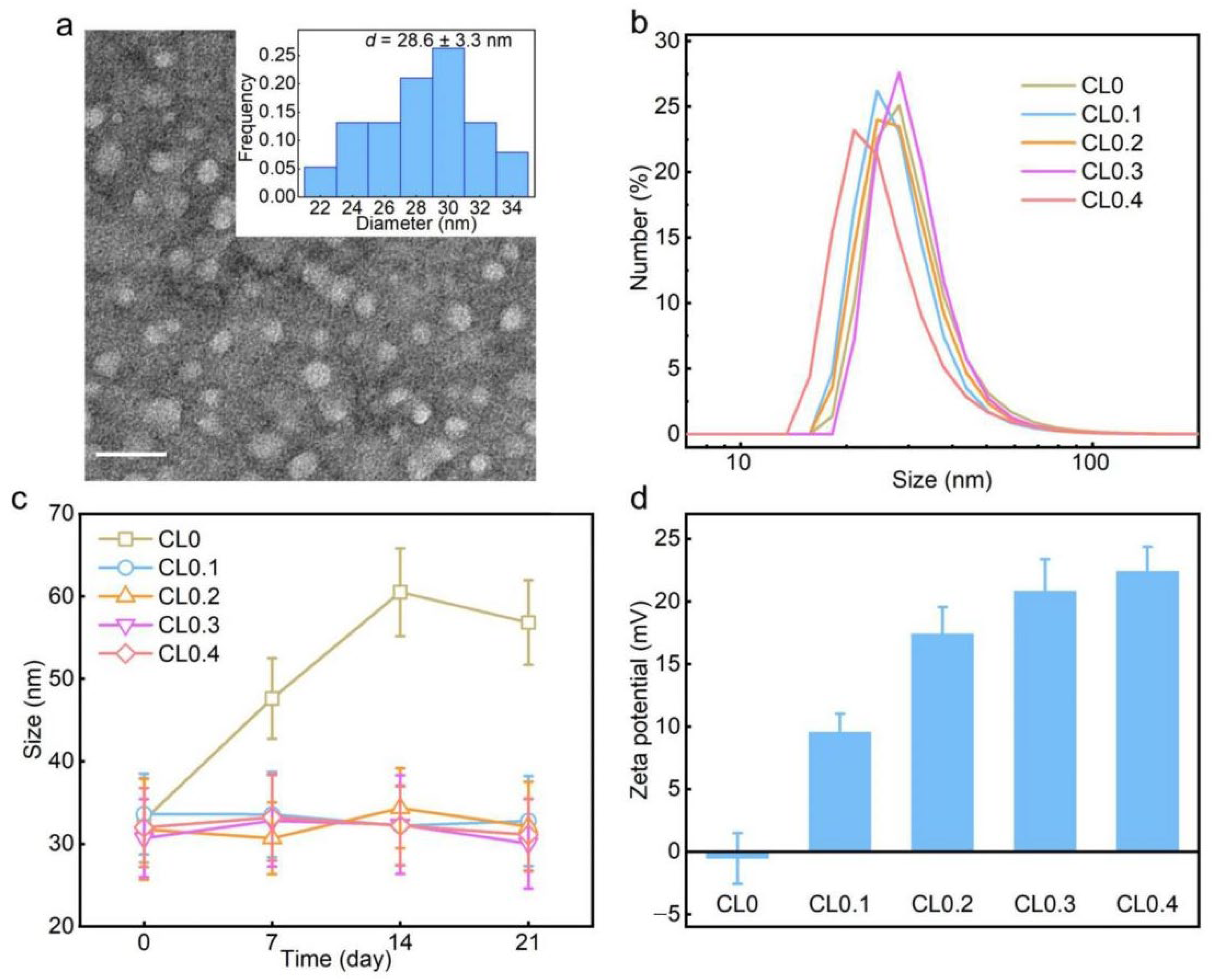
2.1. Characterization of CLs
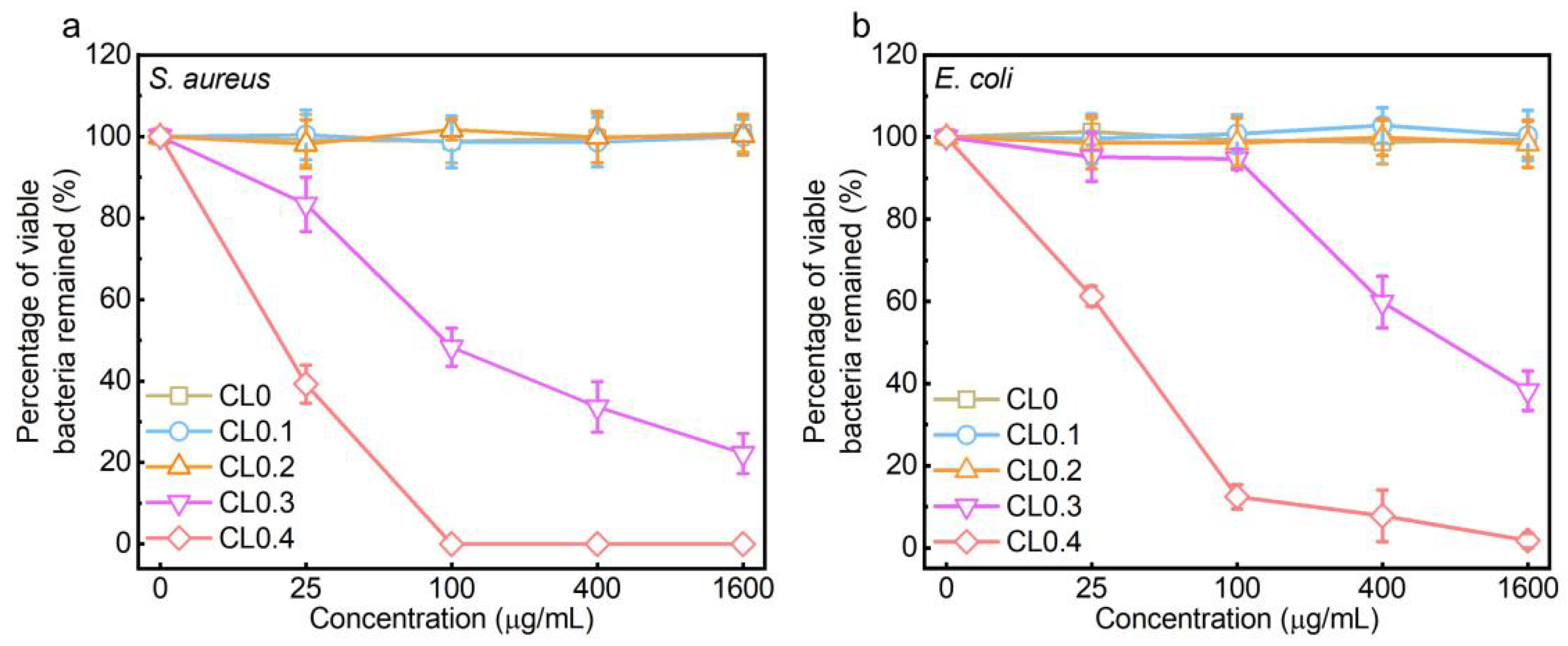
2.2. Antibacterial Activity of CLs
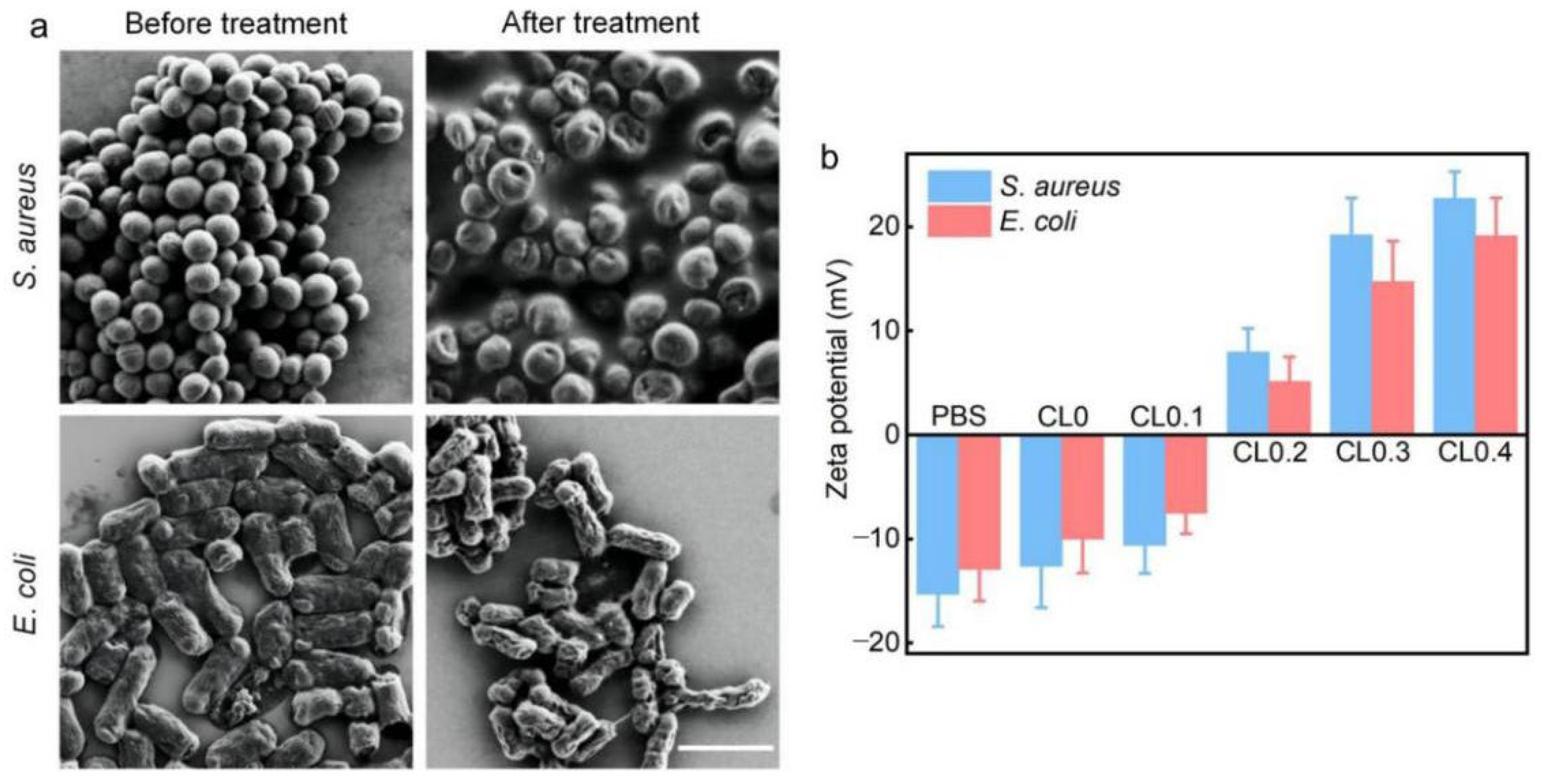
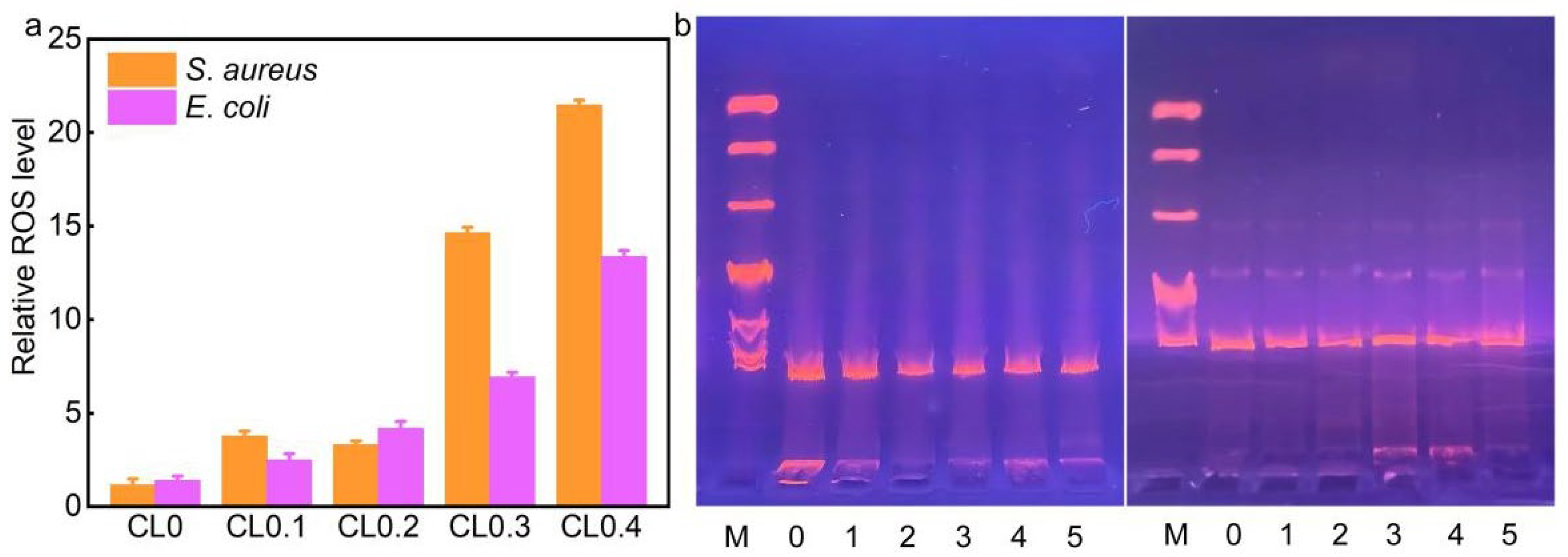
2.3. Antibacterial Mechanisms of CLs
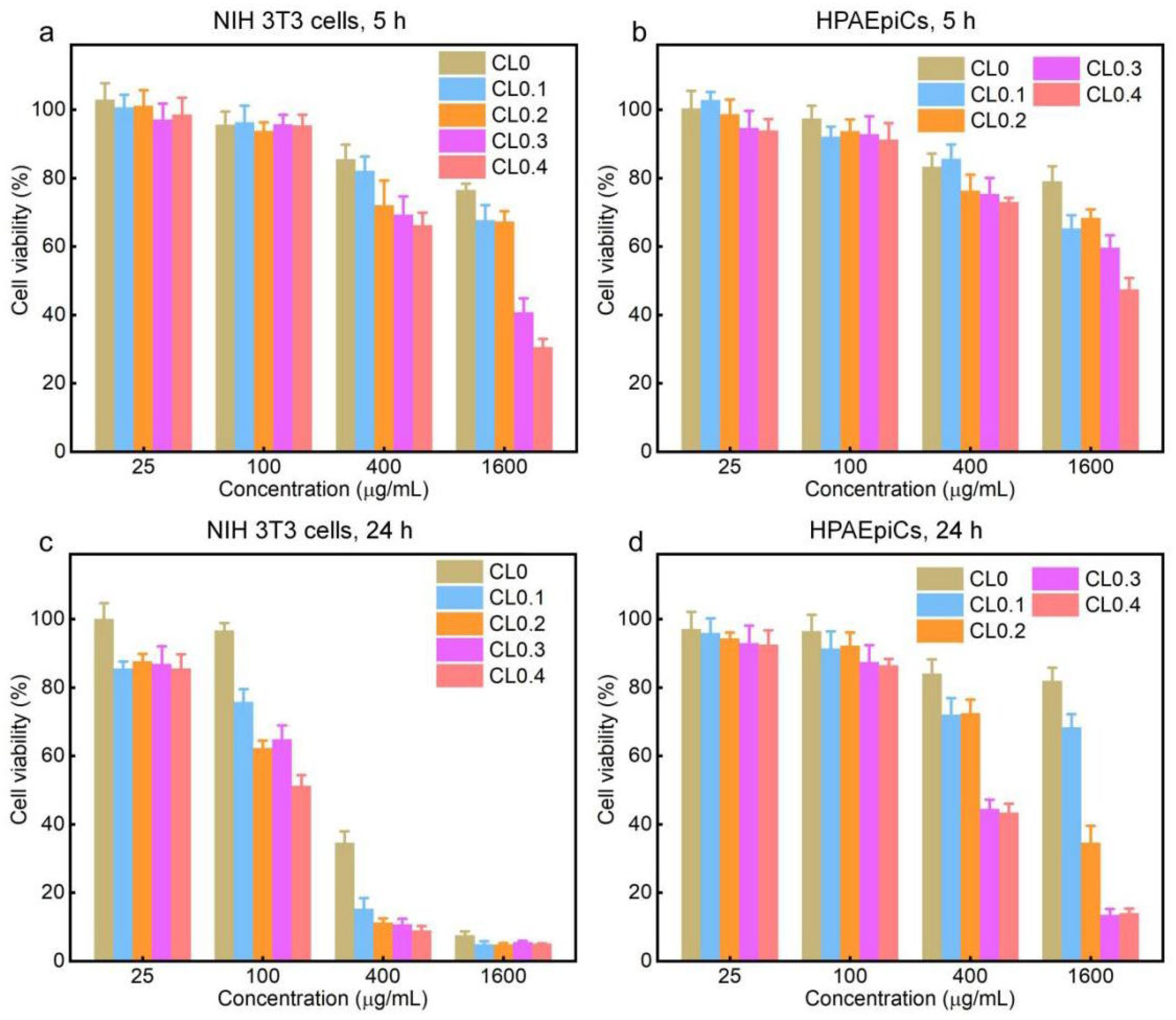
2.4. Cytotoxicity of CLs
3. Materials and Methods
3.1. Materials
3.2. Preparation of CLs
3.3. Characterization
3.4. Culture of Bacterial Cells
3.5. Agar Plate Count Assay
3.6. SEM Observation of Bacteria
3.7. Zeta Potential Measurements of Bacterial Suspensions
3.8. Analysis of Intracellular ROS Generation
3.9. DNA Extraction and Agarose Gel Electrophoresis Assay
3.10. MTT Assay
3.11. Apoptosis Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Penesyan, A.; Gillings, M.; Paulsen, I.T. Antibiotic discovery: Combatting bacterial resistance in cells and in biofilm communities. Molecules 2015, 20, 5286–5298. [Google Scholar] [CrossRef] [PubMed]
- Hamida, R.S.; Ali, M.A.; Goda, D.A.; Khalil, M.I.; Al-Zaban, M.I. Novel biogenic silver nanoparticle-induced reactive oxygen species inhibit the biofilm formation and virulence activities of methicillin-resistant Staphylococcus aureus (MRSA) strain. Front. Bioeng. Biotechnol. 2020, 8, 433. [Google Scholar] [CrossRef]
- Shah, B.A.; Yuan, B.; Yan, Y.; Din, S.T.U.; Sardar, A. Boost antimicrobial effect of CTAB-capped NixCu1−xO (0.0 ≤ x ≤ 0.05) nanoparticles by reformed optical and dielectric characters. J. Mater. Sci. 2021, 56, 13291–13312. [Google Scholar] [CrossRef]
- Goswami, S.R.; Singh, M. Microwave-mediated synthesis of zinc oxide nanoparticles: A therapeutic approach against Malassezia species. IET Nanobiotechnol. 2018, 12, 903–908. [Google Scholar] [CrossRef]
- Massoumi, H.; Kumar, R.; Chug, M.K.; Qian, Y.; Brisbois, E.J. Nitric oxide release and antibacterial efficacy analyses of S-Nitroso-N-Acetyl-penicillamine conjugated to titanium dioxide nanoparticles. ACS Appl. Bio Mater. 2022, 5, 2285–2295. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Wu, F.G. Two-dimensional materials for antimicrobial applications: Graphene materials and beyond. Chem. Asian J. 2018, 13, 3378–3410. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Yang, F.; Nie, C.; Yang, Y.; Ji, H.; He, C.; Cheng, C.; Zhao, C. Mussel-inspired synthesis of NIR-responsive and biocompatible Ag-graphene 2D nanoagents for versatile bacterial disinfections. ACS Appl. Mater. Interfaces 2018, 10, 296–307. [Google Scholar] [CrossRef]
- Zhu, J.; Hou, J.; Zhang, Y.; Tian, M.; He, T.; Liu, J.; Chen, V. Polymeric antimicrobial membranes enabled by nanomaterials for water treatment. J. Membr. Sci. 2018, 550, 173–197. [Google Scholar] [CrossRef]
- Farhangi, M.; Kobarfard, F.; Mahboubi, A.; Vatanara, A.; Mortazavi, S.A. Preparation of an optimized ciprofloxacin-loaded chitosan nanomicelle with enhanced antibacterial activity. Drug Dev. Ind. Pharm. 2018, 44, 1273–1284. [Google Scholar] [CrossRef]
- Zhang, M.; Yu, Z.; Lo, E.C.M. A new pH-responsive nano micelle for enhancing the effect of a hydrophobic bactericidal agent on mature Streptococcus mutans biofilm. Front. Microbiol. 2021, 12, 761583. [Google Scholar] [CrossRef]
- Gao, J.; Wang, S.; Dong, X.; Leanse, L.G.; Dai, T.; Wang, Z. Co-delivery of resolvin D1 and antibiotics with nanovesicles to lungs resolves inflammation and clears bacteria in mice. Commun. Biol. 2020, 3, 680. [Google Scholar] [CrossRef] [PubMed]
- Sarcina, L.; García-Manrique, P.; Gutiérrez, G.; Ditaranto, N.; Cioffi, N.; Matos, M.; Blanco-López, M.D.C. Cu nanoparticle-loaded nanovesicles with antibiofilm properties. Part I: Synthesis of new hybrid nanostructures. Nanostructures. 2020, 10, 1542. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Bao, Y.W.; Wu, F.G. Carbon dots for sensing and killing microorganisms. C 2019, 5, 33. [Google Scholar] [CrossRef]
- Chu, X.; Wu, F.; Sun, B.; Zhang, M.; Song, S.; Zhang, P.; Wang, Y.; Zhang, Q.; Zhou, N.; Shen, J. Genipin cross-linked carbon dots for antimicrobial, bioimaging and bacterial discrimination. Colloid Surf. B Biointerfaces 2020, 190, 110930. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; Yang, J.; Jia, H.R.; Li, Y.H.; Chen, Z.; Wu, F.G. Quaternized silicon nanoparticles with polarity-sensitive fluorescence for selectively imaging and killing Gram-positive bacteria. Adv. Funct. Mater. 2016, 26, 5958–5970. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, X.; Lin, F.; Guo, Y.; Wu, F.G. One-step synthesis of epoxy group-terminated organosilica nanodots: A versatile nanoplatform for imaging and eliminating multidrug-resistant bacteria and their biofilms. Small 2019, 15, 1901647. [Google Scholar] [CrossRef]
- He, X.; Xiong, L.H.; Zhao, Z.; Wang, Z.; Luo, L.; Lam, J.W.Y.; Kwok, R.T.K.; Tang, B.Z. AIE-based theranostic systems for detection and killing of pathogens. Theranostics 2019, 9, 3223–3248. [Google Scholar] [CrossRef]
- Panigrahi, A.; Are, V.N.; Jain, S.; Nayak, D.; Giri, S.; Sarma, T.K. Cationic organic nanoaggregates as AIE luminogens for wash-free imaging of bacteria and broad-spectrum antimicrobial application. ACS Appl. Mater. Interfaces 2020, 12, 5389–5402. [Google Scholar] [CrossRef]
- Majumdar, A.; Butola, B.S.; Thakur, S. Development and performance optimization of knitted antibacterial materials using polyester-silver nanocomposite fibres. Mater. Sci. Eng. C Mater. Biol. Appl. 2015, 54, 26–31. [Google Scholar] [CrossRef]
- Wang, Y.; Long, Y.; Zhang, D. Novel bifunctional V2O5/BiVO4 nanocomposite materials with enhanced antibacterial activity. J. Taiwan Inst. Chem. Eng. 2016, 68, 387–395. [Google Scholar] [CrossRef]
- Munir, M.U.; Ahmad, M.M. Nanomaterials aiming to tackle antibiotic-resistant bacteria. Pharmaceutics 2022, 14, 582. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Shen, H.; Liu, G.; Zhang, X.; Liu, X.; Liu, H.; Xu, P.; Chen, W.; Tian, Y.; Deng, H.; et al. CuO/TiO2 nanobelt with oxygen vacancies for visible-light-driven photocatalytic bacterial inactivation. ACS Appl. Nano Mater. 2022, 5, 10980–10990. [Google Scholar] [CrossRef]
- Lin, F.; Duan, Q.Y.; Wu, F.G. Conjugated polymer-based photothermal therapy for killing microorganisms. ACS Appl. Polym. Mater. 2020, 2, 4331–4344. [Google Scholar] [CrossRef]
- Shao, M.; Fan, Y.; Zhang, K.; Hu, Y.; Xu, F.J. One nanosystem with potent antibacterial and gene-delivery performances accelerates infected wound healing. Nano Today 2021, 39, 101224. [Google Scholar] [CrossRef]
- Lin, J.; Hu, J.; Wang, W.; Liu, K.; Zhou, C.; Liu, Z.; Kong, S.; Lin, S.; Deng, Y.; Guo, Z. Thermo and light-responsive strategies of smart titanium-containing composite material surface for enhancing bacterially anti-adhesive property. Chem. Eng. J. 2021, 407, 125783. [Google Scholar] [CrossRef]
- Ge, J.; Neofytou, E.; Cahill III, T.J.; Beygui, R.E.; Zare, R.N. Drug release from electric-field-responsive nanoparticles. ACS Nano 2012, 6, 227–233. [Google Scholar] [CrossRef]
- Elbourne, A.; Cheeseman, S.; Atkin, P.; Truong, N.P.; Syed, N.; Zavabet, A.; Mohiuddin, M.; Esrafilzadeh, D.; Cozzolino, D.; McConville, C.F.; et al. Antibacterial liquid metals: Biofilm treatment via magnetic activation. ACS Nano 2020, 14, 802–817. [Google Scholar] [CrossRef]
- Duan, B.; Shao, X.; Han, Y.; Li, Y.; Zhao, Y. Mechanism and application of ultrasound-enhanced bacteriostasis. J. Clean Prod. 2021, 290, 125750. [Google Scholar] [CrossRef]
- Jebel, F.S.; Almasi, H. Morphological, physical, antimicrobial and release properties of ZnO nanoparticles-loaded bacterial cellulose films. Carbohydr. Polym. 2016, 149, 8–19. [Google Scholar] [CrossRef]
- Gomez, A.G.; Hosseinidoust, Z. Liposomes for antibiotic encapsulation and delivery. ACS Infect. Dis. 2020, 6, 896–908. [Google Scholar] [CrossRef]
- Bangham, A.D.; Horne, R.W. Negative staining of phospholipids and their structural modification by surface-active agents as observed in the electron microscope. J. Mol. Biol. 1964, 8, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Horne, R.W.; Bangham, A.D.; Whittaker, V.P. Negatively stained lipoprotein membranes. Nature 1963, 200, 1340. [Google Scholar] [CrossRef] [PubMed]
- Bangham, A.D.; Horne, R.W. Action of saponin on biological cell membranes. Nature 1962, 196, 952–953. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gong, J.; Wei, Z. Strategies for liposome drug delivery systems to improve tumor treatment efficacy. AAPS PharmSciTech 2022, 23, 27. [Google Scholar] [CrossRef] [PubMed]
- Passi, M.; Shahid, S.; Chockalingam, S.; Sundar, I.K.; Packirisamy, G. Conventional and nanotechnology based approaches to combat chronic obstructive pulmonary disease: Implications for chronic airway diseases. Int. J. Nanomed. 2020, 15, 3803–3826. [Google Scholar] [CrossRef] [PubMed]
- Hallaj-Nezhadi, S.; Hassan, M. Nanoliposome-based antibacterial drug delivery. Drug Deliv. 2015, 22, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Teng, W.; Cao, J.; Wang, J. Liposomes as delivery system for applications in meat products. Foods 2022, 11, 3017. [Google Scholar] [CrossRef]
- Filipczak, N.; Pan, J.; Yalamarty, S.S.K.; Torchilin, V.P. Recent advancements in liposome technology. Adv. Drug Deliv. Rev. 2020, 156, 4–22. [Google Scholar] [CrossRef]
- Ferreira, M.; Ogren, M.; Dias, J.N.R.; Silva, M.; Gil, S.; Tavares, L.; Aires-da-Silva, F.; Gaspar, M.M.; Aguiar, S.I. Liposomes as antibiotic delivery systems: A promising nanotechnological strategy against antimicrobial resistance. Molecules 2021, 26, 2047. [Google Scholar] [CrossRef]
- Natsaridis, E.; Gkartziou, F.; Mourtas, S.; Stuart, M.C.A.; Kolonitsiou, F.; Klepetsanis, P.; Spiliopoulou, I.; Antimisiaris, S.G. Moxifloxacin liposomes: Effect of liposome preparation method on physicochemical properties and antimicrobial activity against Staphylococcus epidermidis. Pharmaceutics 2022, 14, 370. [Google Scholar] [CrossRef]
- Patel, A.; Dey, S.; Shokeen, K.; Karpińsk, T.M.; Sivaprakasam, S.; Kumar, S.; Manna, D. Sulfonium-based liposome-encapsulated antibiotics deliver a synergistic antibacterial activity. RSC Med. Chem. 2021, 12, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Vandera, K.K.A.; Picconi, P.; Valero, M.; González-Gaitano, G.; Woods, A.; Zain, N.M.M.; Bruce, K.D.; Clifton, L.A.; Skoda, M.W.A.; Rahman, K.M.; et al. Antibiotic-in-cyclodextrin-in-liposomes: Formulation development and interactions with model bacterial membranes. Mol. Pharm. 2020, 17, 2354–2369. [Google Scholar] [CrossRef] [PubMed]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. Fems Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef]
- Pham, T.D.M.; Ziora, Z.M.; Blaskovich, M.A.T. Quinolone antibiotics. MedChemComm 2019, 10, 1719–1739. [Google Scholar] [CrossRef] [PubMed]
- Mugabe, C.; Halwani, M.; Azghani, A.O.; Lafrenie, R.M.; Omri, A. Mechanism of enhanced activity of liposome-entrapped aminoglycosides against resistant strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2006, 50, 2016–2022. [Google Scholar] [CrossRef] [PubMed]
- Alhajlan, M.; Alhariri, M.; Omri, A. Efficacy and safety of liposomal clarithromycin and its effect on Pseudomonas aeruginosa virulence factors. Antimicrob. Agents Chemother. 2013, 57, 2694–2704. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.; Pinto, S.N.; Aires-da-Silva, F.; Bettencourt, A.; Aguiar, S.I.; Gaspar, M.M. Liposomes as a nanoplatform to improve the delivery of antibiotics into Staphylococcus aureus biofilms. Pharmaceutics 2021, 13, 321. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, Z.; Zhao, W.; Lu, T.; Wang, R.; Mei, Q.; Chen, T. Enhanced bactericidal potency of nanoliposomes by modification of the fusion activity between liposomes and bacterium. Int. J. Nanomed. 2013, 8, 2351–2360. [Google Scholar] [CrossRef]
- Tsai, T.; Yang, Y.T.; Wang, T.H.; Chien, H.F.; Chen, C.T. Improved photodynamic inactivation of Gram-positive bacteria using hematoporphyrin encapsulated in liposomes and micelles. Lasers Surg. Med. 2009, 41, 316–322. [Google Scholar] [CrossRef]
- Nisnevitch, M.; Nakonechny, F.; Nitzan, Y. Photodynamic antimicrobial chemotherapy by liposome-encapsulated water-soluble photosensitizers. Russ. J. Bioorg. Chem. 2010, 36, 363–369. [Google Scholar] [CrossRef]
- Yang, D.; Pornpattananangkul, D.; Nakatsuji, T.; Chan, M.; Carson, D.; Huang, C.M.; Zhang, L. The antimicrobial activity of liposomal lauric acids against Propionibacterium acnes. Biomaterials 2009, 30, 6035–6040. [Google Scholar] [CrossRef] [PubMed]
- Jackman, J.A.; Yoon, B.K.; Li, D.; Cho, N.J. Nanotechnology formulations for antibacterial free fatty acids and monoglycerides. Molecules 2016, 21, 305. [Google Scholar] [CrossRef] [PubMed]
- Eid, K.A.M.; Azzazy, H.M.E. Sustained broad-spectrum antibacterial effects of nanoliposomes loaded with silver nanoparticles. Nanomedicine 2014, 9, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Almeida, B.; Nag, O.K.; Rogers, K.E.; Delehanty, J.B. Recent progress in bioconjugation strategies for liposome-mediated drug delivery. Molecules 2020, 25, 5672. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.Y.; Ryu, J.Y.; Lee, H.A.R.; Hong, S.H.; Park, H.S.; Hong, K.S.; Park, S.G.; Kim, H.P.; Yoon, T.J. Lecithin nano-liposomal particle as a CRISPR/Cas9 complex delivery system for treating type 2 diabetes. J. Nanobiotechnol. 2019, 17, 19. [Google Scholar] [CrossRef]
- Allen, T.M.; Cullis, P.R. Liposomal drug delivery systems: From concept to clinical applications. Adv. Drug Deliv. Rev. 2013, 65, 36–48. [Google Scholar] [CrossRef]
- Large, D.E.; Abdelmessih, R.G.; Fink, E.A.; Auguste, D.T. Liposome composition in drug delivery design, synthesis, characterization, and clinical application. Adv. Drug Deliv. Rev. 2021, 176, 113851. [Google Scholar] [CrossRef]
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975–999. [Google Scholar] [CrossRef]
- Cui, S.; Wang, Y.; Gong, Y.; Lin, X.; Zhao, Y.; Zhi, D.; Zhou, Q.; Zhang, S. Correlation of the cytotoxic effects of cationic lipids with their headgroups. Toxicol. Res. 2018, 7, 473–479. [Google Scholar] [CrossRef]
- Inglut, C.T.; Sorrin, A.J.; Kuruppu, T.; Vig, S.; Cicalo, J.; Ahmad, H.; Huang, H.C. Immunological and toxicological considerations for the design of liposomes. Nanomaterials 2020, 10, 190. [Google Scholar] [CrossRef]
- Cai, W.; Liu, J.; Zheng, L.; Xu, Z.; Chen, J.; Zhong, J.; Song, Z.; Xu, X.; Chen, S.; Jiao, C.; et al. Study on the anti-infection ability of vancomycin cationic liposome combined with polylactide fracture internal fixator. Int. J. Biol. Macromol. 2021, 167, 834–844. [Google Scholar] [CrossRef] [PubMed]
- Drulis-Kawa, Z.; Gubernator, J.; Dorotkiewicz-Jach, A.; Doroszkiewicz, W.; Kozubek, A. A comparison of the in vitro antimicrobial activity of liposomes containing meropenem and gentamicin. Cell. Mol. Biol. Lett. 2006, 11, 360–375. [Google Scholar] [CrossRef] [PubMed]
- Bombelli, C.; Bordi, F.; Ferro, S.; Giansanti, L.; Jori, G.; Mancini, G.; Mazzuca, C.; Monti, D.; Ricchelli, F.; Sennato, S.; et al. New cationic liposomes as vehicles of m-tetrahydroxyphenylchlorin in photodynamic therapy of infectious diseases. Mol. Pharm. 2008, 5, 672–679. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, P.; Zhang, X.; Li, F.; Xu, K.-F.; Li, Y.-H.; Liu, X.; Yang, J.; Zhu, B.; Wu, F.-G. Cationic Liposomes with Different Lipid Ratios: Antibacterial Activity, Antibacterial Mechanism, and Cytotoxicity Evaluations. Pharmaceuticals 2022, 15, 1556. https://doi.org/10.3390/ph15121556
Lu P, Zhang X, Li F, Xu K-F, Li Y-H, Liu X, Yang J, Zhu B, Wu F-G. Cationic Liposomes with Different Lipid Ratios: Antibacterial Activity, Antibacterial Mechanism, and Cytotoxicity Evaluations. Pharmaceuticals. 2022; 15(12):1556. https://doi.org/10.3390/ph15121556
Chicago/Turabian StyleLu, Pengpeng, Xinping Zhang, Feng Li, Ke-Fei Xu, Yan-Hong Li, Xiaoyang Liu, Jing Yang, Baofeng Zhu, and Fu-Gen Wu. 2022. "Cationic Liposomes with Different Lipid Ratios: Antibacterial Activity, Antibacterial Mechanism, and Cytotoxicity Evaluations" Pharmaceuticals 15, no. 12: 1556. https://doi.org/10.3390/ph15121556
APA StyleLu, P., Zhang, X., Li, F., Xu, K.-F., Li, Y.-H., Liu, X., Yang, J., Zhu, B., & Wu, F.-G. (2022). Cationic Liposomes with Different Lipid Ratios: Antibacterial Activity, Antibacterial Mechanism, and Cytotoxicity Evaluations. Pharmaceuticals, 15(12), 1556. https://doi.org/10.3390/ph15121556







