Willardiine and Its Synthetic Analogues: Biological Aspects and Implications in Peptide Chemistry of This Nucleobase Amino Acid †
Abstract
1. Introduction
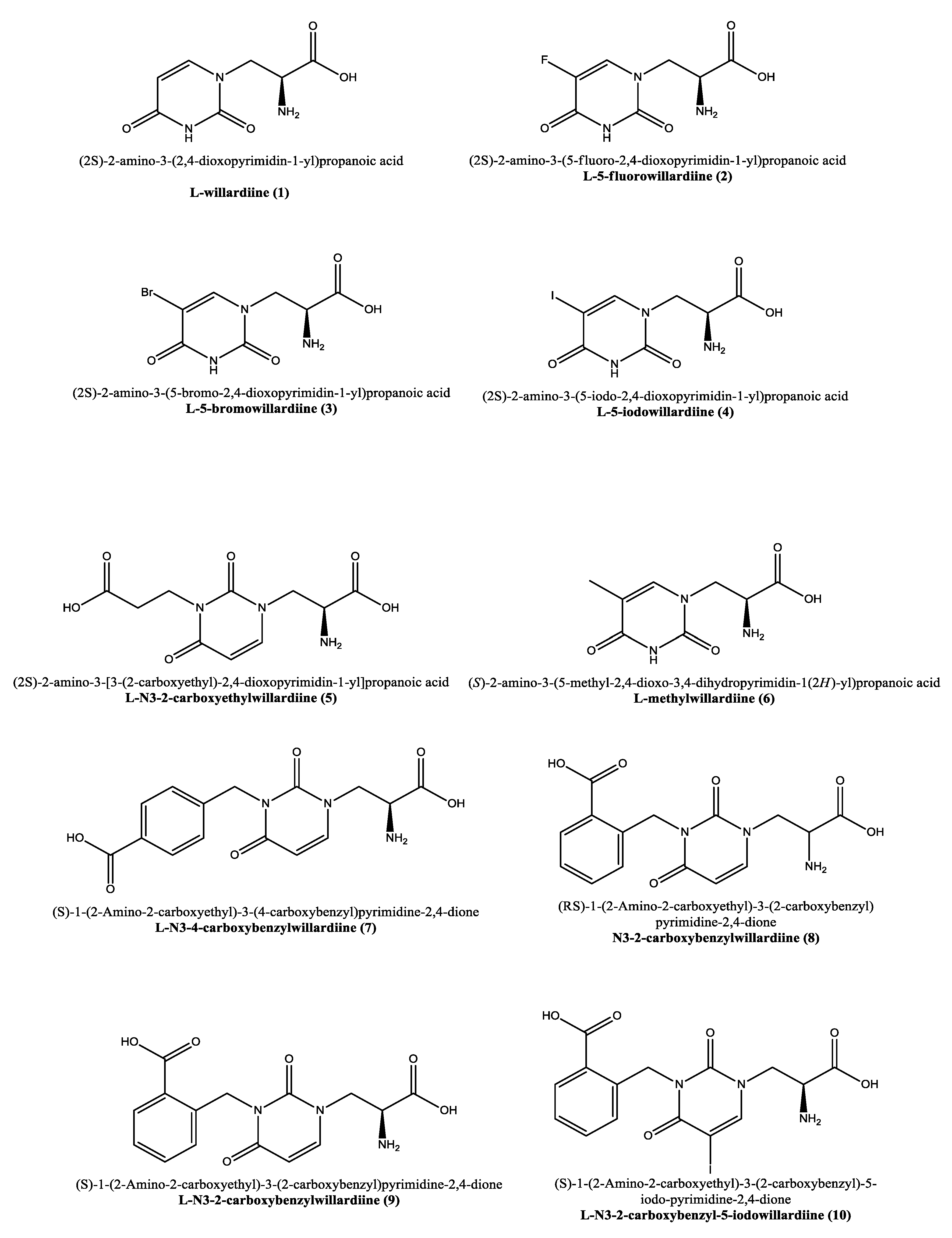
2. Willardiine and Its Analogues
2.1. Disease Relevance and Potential Pharmaceutical Role
2.1.1. AMPA Receptors in Neurological Disorders
2.1.2. Kainate Receptors in Neurological Disorders
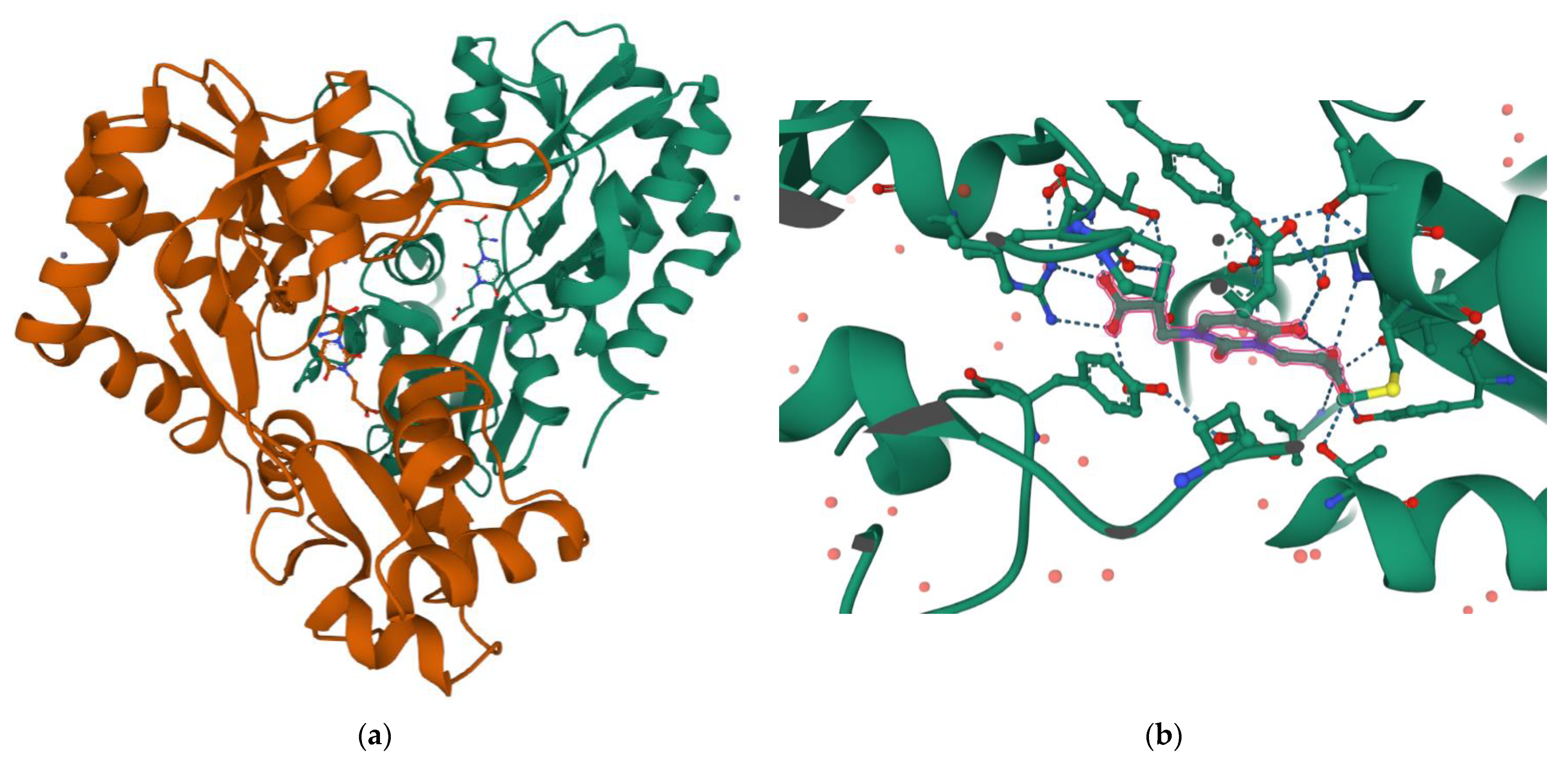
2.2. Molecular Insights on Bioactivity of Willardiine analogues
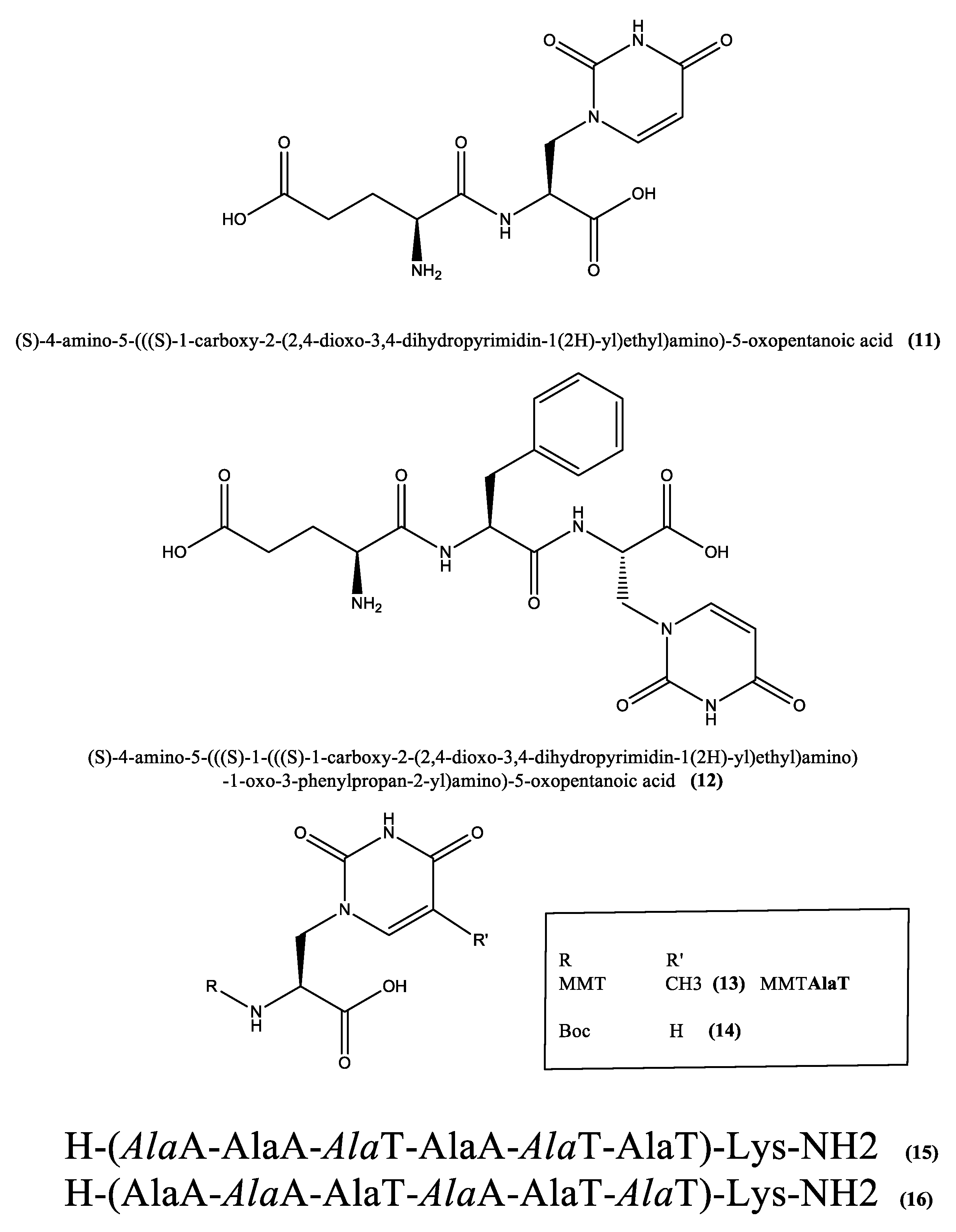
3. Willardiine in Peptide Structures
3.1. Charge Transfer and Transport in Nucleopeptides
3.2. Prebiotic Role of Nucleopeptides
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gmelin, R. Die freien Aminosäuren der Samen von Acacia Willardiana (Mimosaceae). Isolierung von Willardiin, Einer Neuen Pflanzlichen Aminosäure, Vermutlich L-Uracil-[β-(α-amino-propionsäure)]-(3). Hoppe-Seyler’s Zeitschrift für Physiol. Chem. 1959, 316, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, K.B.; Diederichsen, U. Nucleo amino acids as arginine mimetics in cyclic peptides. Lett. Pept. Sci. 2003, 10, 111–117. [Google Scholar] [CrossRef]
- Cheikh, A.B.; Orgel, L.E. Polymerization of amino acids containing nucleotide bases. J. Mol. Evol. 1990, 30, 315–321. [Google Scholar] [CrossRef]
- Musumeci, D.; Ullah, S.; Ikram, A.; Roviello, G.N. Novel insights on nucleopeptide binding: A spectroscopic and In Silico investigation on the interaction of a thymine-bearing tetrapeptide with a homoadenine DNA. J. Mol. Liq. 2022, 347, 117975. [Google Scholar] [CrossRef]
- Negi, V.S.; Pal, A.; Borthakur, D. Biochemistry of plants N–heterocyclic non-protein amino acids. Amino Acids 2021, 53, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Stensbol, T.; Madsen, U.; Krogsgaard-Larsen, P. The AMPA receptor binding site: Focus on agonists and competitive antagonists. Curr. Pharm. Des. 2002, 8, 857–872. [Google Scholar] [CrossRef]
- Kew, J.N.; Kemp, J.A. Ionotropic and metabotropic glutamate receptor structure and pharmacology. Psychopharmacology 2005, 179, 4–29. [Google Scholar] [CrossRef]
- Yelshanskaya, M.; Sobolevsky, A. Structural Insights into Function of Ionotropic Glutamate Receptors. Biochem. Suppl. Ser. A Membr. Cell Biol. 2022, 16, 190–206. [Google Scholar] [CrossRef]
- Bowie, D. The Many Faces of the AMPA-Type Ionotropic Glutamate Receptor; Elsevier: Amsterdam, The Netherlands, 2022; p. 108975. [Google Scholar]
- Pinzón-Parra, C.A.; Coatl-Cuaya, H.; Díaz, A.; Guevara, J.; Rodríguez-Moreno, A.; Flores, G. Long-term effect of neonatal antagonism of ionotropic glutamate receptors on dendritic spines and cognitive function in rats. J. Chem. Neuroanat. 2022, 119, 102054. [Google Scholar] [CrossRef]
- Hu, T.-M.; Wu, C.-L.; Hsu, S.-H.; Tsai, H.-Y.; Cheng, F.-Y.; Cheng, M.-C. Ultrarare Loss-of-Function Mutations in the Genes Encoding the Ionotropic Glutamate Receptors of Kainate Subtypes Associated with Schizophrenia Disrupt the Interaction with PSD95. J. Pers. Med. 2022, 12, 783. [Google Scholar] [CrossRef]
- Bowie, D. Ionotropic glutamate receptors & CNS disorders. CNS Neurol. Disord. Drug Targets 2008, 7, 129–143. [Google Scholar]
- Negrete-Díaz, J.V.; Falcón-Moya, R.; Rodríguez-Moreno, A. Kainate receptors: From synaptic activity to disease. FEBS J. 2021, 289, 5074–5088. [Google Scholar] [CrossRef]
- Postila, P.A.; Ylilauri, M.; Pentikäinen, O.T. Full and partial agonism of ionotropic glutamate receptors indicated by molecular dynamics simulations. J. Chem. Inf. Model. 2011, 51, 1037–1047. [Google Scholar] [CrossRef]
- Henley, J.M.; Wilkinson, K.A. Synaptic AMPA receptor composition in development, plasticity and disease. Nat. Rev. Neurosci. 2016, 17, 337–350. [Google Scholar] [CrossRef]
- Sihra, T.S.; Rodríguez-Moreno, A. Presynaptic kainate receptor-mediated bidirectional modulatory actions: Mechanisms. Neurochem. Int. 2013, 62, 982–987. [Google Scholar] [CrossRef]
- Rodríguez-Moreno, A.; Sihra, T.S. Metabotropic actions of kainate receptors in the control of glutamate release in the hippocampus. Adv. Exp. Med. Biol. 2011, 717, 39–48. [Google Scholar] [CrossRef]
- Sihra, T.S.; Rodríguez-Moreno, A. Metabotropic actions of kainate receptors in the control of GABA release. Adv. Exp. Med. Biol. 2011, 717, 1–10. [Google Scholar] [CrossRef]
- Neto, J.X.L.; Fulco, U.L.; Albuquerque, E.L.; Corso, G.; Bezerra, E.M.; Caetano, E.W.; Da Costa, R.F.; Freire, V.N. A quantum biochemistry investigation of willardiine partial agonism in AMPA receptors. Phys. Chem. Chem. Phys. 2015, 17, 13092–13103. [Google Scholar] [CrossRef]
- Pasternack, A.; Coleman, S.K.; Jouppila, A.; Mottershead, D.G.; Lindfors, M.; Pasternack, M.; Keinänen, K. α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor channels lacking the N-terminal domain. J. Biol. Chem. 2002, 277, 49662–49667. [Google Scholar] [CrossRef]
- Kato, A.S.; Gill, M.B.; Yu, H.; Nisenbaum, E.S.; Bredt, D.S. TARPs differentially decorate AMPA receptors to specify neuropharmacology. Trends Neurosci. 2010, 33, 241–248. [Google Scholar] [CrossRef]
- Traynelis, S.F.; Wollmuth, L.P.; McBain, C.J.; Menniti, F.S.; Vance, K.M.; Ogden, K.K.; Hansen, K.B.; Yuan, H.; Myers, S.J.; Dingledine, R. Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol. Rev. 2010, 62, 405–496. [Google Scholar] [CrossRef]
- More, J.C.; Troop, H.M.; Dolman, N.P.; Jane, D.E. Structural requirements for novel willardiine derivatives acting as AMPA and kainate receptor antagonists. Br. J. Pharmacol. 2003, 138, 1093–1100. [Google Scholar] [CrossRef]
- Immel, J.R.; Bloom, S. carba-Nucleopeptides (cNPs): A Biopharmaceutical Modality Formed through Aqueous Rhodamine B Photoredox Catalysis. Angew. Chem. Int. Ed. 2022, 61, e202205606. [Google Scholar] [CrossRef]
- Giraud, T.; Hoschtettler, P.; Pickaert, G.; Averlant-Petit, M.-C.; Stefan, L. Emerging low-molecular weight nucleopeptide-based hydrogels: State of the art, applications, challenges and perspectives. Nanoscale 2022, 14, 4908–4921. [Google Scholar] [CrossRef]
- Huganir, R.L.; Nicoll, R.A. AMPARs and synaptic plasticity: The last 25 years. Neuron 2013, 80, 704–717. [Google Scholar] [CrossRef]
- Kessels, H.W.; Malinow, R. Synaptic AMPA receptor plasticity and behavior. Neuron 2009, 61, 340–350. [Google Scholar] [CrossRef]
- Cheng, G.-R.; Li, X.-Y.; Xiang, Y.-D.; Liu, D.; McClintock, S.M.; Zeng, Y. The implication of AMPA receptor in synaptic plasticity impairment and intellectual disability in fragile X syndrome. Physiol. Res. 2017, 66, 715–727. [Google Scholar] [CrossRef]
- Lee, K.; Goodman, L.; Fourie, C.; Schenk, S.; Leitch, B.; Montgomery, J.M. AMPA receptors as therapeutic targets for neurological disorders. Adv. Protein Chem. Struct. Biol. 2016, 103, 203–261. [Google Scholar]
- Lees, A.; Fahn, S.; Eggert, K.M.; Jankovic, J.; Lang, A.; Micheli, F.; Maral Mouradian, M.; Oertel, W.H.; Olanow, C.W.; Poewe, W. Perampanel, an AMPA antagonist, found to have no benefit in reducing “off” time in Parkinson’s disease. Mov. Disord. 2012, 27, 284–288. [Google Scholar] [CrossRef]
- Walker, F.O. Huntington’s disease. Lancet 2007, 369, 218–228. [Google Scholar] [CrossRef]
- Salpietro, V.; Dixon, C.L.; Guo, H.; Bello, O.D.; Vandrovcova, J.; Efthymiou, S.; Maroofian, R.; Heimer, G.; Burglen, L.; Valence, S. AMPA receptor GluA2 subunit defects are a cause of neurodevelopmental disorders. Nat. Commun. 2019, 10, 3094. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-W.; Park, K.; Kang, R.J.; Gonzales, E.L.T.; Kim, D.G.; Oh, H.A.; Seung, H.; Ko, M.J.; Kwon, K.J.; Kim, K.C. Pharmacological modulation of AMPA receptor rescues social impairments in animal models of autism. Neuropsychopharmacology 2019, 44, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Jaso, B.A.; Niciu, M.J.; Iadarola, N.D.; Lally, N.; Richards, E.M.; Park, M.; Ballard, E.D.; Nugent, A.C.; Machado-Vieira, R.; Zarate, C.A. Therapeutic modulation of glutamate receptors in major depressive disorder. Curr. Neuropharmacol. 2017, 15, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, J.D. Memory, plasticity and sleep-A role for calcium permeable AMPA receptors? Front. Mol. Neurosci. 2012, 5, 49. [Google Scholar] [CrossRef] [PubMed]
- Shaltiel, G.; Maeng, S.; Malkesman, O.; Pearson, B.; Schloesser, R.; Tragon, T.; Rogawski, M.; Gasior, M.; Luckenbaugh, D.; Chen, G. Evidence for the involvement of the kainate receptor subunit GluR6 (GRIK2) in mediating behavioral displays related to behavioral symptoms of mania. Mol. Psychiatry 2008, 13, 858–872. [Google Scholar] [CrossRef]
- Schiffer, H.; Heinemann, S. Association of the human kainate receptor GluR7 gene (GRIK3) with recurrent major depressive disorder. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2007, 144, 20–26. [Google Scholar] [CrossRef]
- Nair, J.D.; Wilkinson, K.A.; Henley, J.M.; Mellor, J.R. Kainate receptors and synaptic plasticity. Neuropharmacology 2021, 196, 108540. [Google Scholar] [CrossRef]
- Li, H.; Li, J.; Guan, Y.; Wang, Y. The emerging role of kainate receptor functional dysregulation in pain. Mol. Pain 2021, 17, 1744806921990944. [Google Scholar] [CrossRef]
- Henley, J.M.; Nair, J.D.; Seager, R.; Yucel, B.P.; Woodhall, G.; Henley, B.S.; Talandyte, K.; Needs, H.I.; Wilkinson, K.A. Kainate and AMPA receptors in epilepsy: Cell biology, signalling pathways and possible crosstalk. Neuropharmacology 2021, 195, 108569. [Google Scholar] [CrossRef]
- Crépel, V.; Mulle, C. Physiopathology of kainate receptors in epilepsy. Curr. Opin. Pharmacol. 2015, 20, 83–88. [Google Scholar] [CrossRef]
- Patneau, D.K.; Mayer, M.L.; Jane, D.E.; Watkins, J.C. Activation and desensitization of AMPA/kainate receptors by novel derivatives of willardiine. J. Neurosci. 1992, 12, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, L.; Beaver, K.; Jane, D.; Taylor, P.; Sunter, D.; Roberts, P. Characterization of the pharmacology and regional distribution of (S)-[3H]-5-fluorowillardiine binding in rat brain. Br. J. Pharmacol. 1995, 116, 2033–2039. [Google Scholar] [CrossRef] [PubMed]
- Lunn, M.; Ganakas, A.; Mercer, L.; Lawrence, A.; Beart, P. Localisation and properties of AMPA-insensitive kainate sites: Receptor autoradiography and gene expression in rat brain. Neurosci. Lett. 1996, 204, 121–124. [Google Scholar] [CrossRef]
- Larm, J.A.; Cheung, N.S.; Beart, P.M. (S)-5-fluorowillardiine-mediated neurotoxicity in cultured murine cortical neurones occurs via AMPA and kainate receptors. Eur. J. Pharmacol. 1996, 314, 249–254. [Google Scholar] [CrossRef]
- JENSEN, R.J. Responses of directionally selective retinal ganglion cells to activation of AMPA glutamate receptors. Vis. Neurosci. 1999, 16, 205–219. [Google Scholar] [CrossRef]
- Olivera, S.; Rodriguez-Ithurralde, D.; Henley, J.M. Regional localization and developmental profile of acetylcholinesterase-evoked increases in [3H]-5-fluorowillardiine binding to AMPA receptors in rat brain. Br. J. Pharmacol. 2001, 133, 1055–1062. [Google Scholar] [CrossRef]
- Kessler, M.; Arai, A.C. Use of [3H] fluorowillardiine to study properties of AMPA receptor allosteric modulators. Brain Res. 2006, 1076, 25–41. [Google Scholar] [CrossRef]
- Dolman, N.P.; Troop, H.M.; More, J.C.; Alt, A.; Knauss, J.L.; Nistico, R.; Jack, S.; Morley, R.M.; Bortolotto, Z.A.; Roberts, P.J. Synthesis and pharmacology of willardiine derivatives acting as antagonists of kainate receptors. J. Med. Chem. 2005, 48, 7867–7881. [Google Scholar] [CrossRef]
- Martinez, M.; Ahmed, A.H.; Loh, A.P.; Oswald, R.E. Thermodynamics and Mechanism of the Interaction of Willardiine Partial Agonists with a Glutamate Receptor: Implications for Drug Development. Biochemistry 2014, 53, 3790–3795. [Google Scholar] [CrossRef]
- Ahmed, A.H.; Thompson, M.D.; Fenwick, M.K.; Romero, B.; Loh, A.P.; Jane, D.E.; Sondermann, H.; Oswald, R.E. Mechanisms of antagonism of the GluR2 AMPA receptor: Structure and dynamics of the complex of two willardiine antagonists with the glutamate binding domain. Biochemistry 2009, 48, 3894–3903. [Google Scholar] [CrossRef] [PubMed]
- GN Roviello, G Oliviero, A Di Napoli, N Borbone Synthesis, self-assembly-behavior and biomolecular recognition properties of thyminyl dipeptides. Arab. J. Chem. 2020, 13, 1966–1974. [CrossRef]
- Roviello, G.N.; Vicidomini, C.; Costanzo, V.; Roviello, V. Nucleic acid binding and other biomedical properties of artificial oligolysines. Int. J. Nanomed. 2016, 11, 5897–5904. [Google Scholar] [CrossRef]
- Roviello, G.N. Novel insights into nucleoamino acids: Biomolecular recognition and aggregation studies of a thymine-conjugated l-phenyl alanine. Amino Acids 2018, 50, 933–941. [Google Scholar] [CrossRef]
- Musumeci, D.; Mokhir, A.; Roviello, G.N. Synthesis and nucleic acid binding evaluation of a thyminyl L-diaminobutanoic acid-based nucleopeptide. Bioorg. Chem. 2020, 100, 103862. [Google Scholar] [CrossRef]
- Roviello, G.N.; Benedetti, E.; Pedone, C.; Bucci, E.M. Nucleobase-containing peptides: An overview of their characteristic features and applications. Amino Acids 2010, 39, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Roviello, G.N.; Musumeci, D. Synthetic approaches to nucleopeptides containing all four nucleobases, and nucleic acid-binding studies on a mixed-sequence nucleo-oligolysine. RSC Adv. 2016, 6, 63578–63585. [Google Scholar] [CrossRef]
- Roviello, G.N.; Musumeci, D.; Moccia, M.; Castiglione, M.; Sapio, R.; Valente, M.; Bucci, E.M.; Perretta, G.; Pedone, C. dabPNA: Design, synthesis, and DNA binding studies. Nucleosides Nucleotides Nucleic Acids 2007, 26, 1307–1310. [Google Scholar] [CrossRef]
- Terracciano, M.; De Stefano, L.; Borbone, N.; Politi, J.; Oliviero, G.; Nici, F.; Casalino, M.; Piccialli, G.; Dardano, P.; Varra, M.; et al. Solid phase synthesis of a thrombin binding aptamer on macroporous silica for label free optical quantification of thrombin. RSC Adv. 2016, 6, 86762–86769. [Google Scholar] [CrossRef]
- Oliviero, G.; Amato, J.; Borbone, N.; D’Errico, S.; Piccialli, G.; Bucci, E.; Piccialli, V.; Mayol, L. Synthesis of 4-N-alkyl and ribose-modified AICAR analogues on solid support. Tetrahedron 2008, 64, 6475–6481. [Google Scholar] [CrossRef]
- Oliviero, G.; Amato, J.; Borbone, N.; D’Errico, S.; Piccialli, G.; Mayol, L. Synthesis of N-1 and ribose modified inosine analogues on solid support. Tetrahedron Lett. 2007, 48, 397–400. [Google Scholar] [CrossRef]
- Oliviero, G.; D’Errico, S.; Borbone, N.; Amato, J.; Piccialli, V.; Varra, M.; Piccialli, G.; Mayol, L. A solid-phase approach to the synthesis of N-1-alkyl analogues of cyclic inosine-diphosphate-ribose (cIDPR). Tetrahedron 2010, 66, 1931–1936. [Google Scholar] [CrossRef]
- D’Errico, S.; Oliviero, G.; Amato, J.; Borbone, N.; Cerullo, V.; Hemminki, A.; Piccialli, V.; Zaccaria, S.; Mayol, L.; Piccialli, G. Synthesis and biological evaluation of unprecedented ring-expanded nucleosides (RENs) containing the imidazo[4,5-d][1,2,6]oxadiazepine ring system. Chem. Commun. 2012, 48, 9310. [Google Scholar] [CrossRef]
- Oliviero, G.; D’Errico, S.; Borbone, N.; Amato, J.; Piccialli, V.; Piccialli, G.; Mayol, L. Facile Solid-Phase Synthesis of AICAR 5′-Monophosphate (ZMP) and Its 4-N-Alkyl Derivatives. Eur. J. Org. Chem. 2010, 2010, 1517–1524. [Google Scholar] [CrossRef]
- D’Errico, S.; Oliviero, G.; Borbone, N.; Amato, J.; D’Alonzo, D.; Piccialli, V.; Mayol, L.; Piccialli, G. A Facile Synthesis of 5′-Fluoro-5′-deoxyacadesine (5′-F-AICAR): A Novel Non-phosphorylable AICAR Analogue. Molecules 2012, 17, 13036–13044. [Google Scholar] [CrossRef] [PubMed]
- Galeone, A.; Mayol, L.; Oliviero, G.; Piccialli, G.; Varra, M. Synthesis of a novel N-1 carbocyclic, N-9 butyl analogue of cyclic ADP ribose (cADPR). Tetrahedron 2002, 58, 363–368. [Google Scholar] [CrossRef]
- Roviello, G.N.; Gaetano, S.D.; Capasso, D.; Cesarani, A.; Bucci, E.M.; Pedone, C. Synthesis, spectroscopic studies and biological activity of a novel nucleopeptide with Moloney murine leukemia virus reverse transcriptase inhibitory activity. Amino Acids 2009, 38, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Roviello, G.N.; Di Gaetano, S.; Capasso, D.; Franco, S.; Crescenzo, C.; Bucci, E.M.; Pedone, C. RNA-Binding and Viral Reverse Transcriptase Inhibitory Activity of a Novel Cationic Diamino Acid-Based Peptide. J. Med. Chem. 2011, 54, 2095–2101. [Google Scholar] [CrossRef] [PubMed]
- Seley-Radtke, K.L.; Yates, M.K. The evolution of nucleoside analogue antivirals: A review for chemists and non-chemists. Part 1: Early structural modifications to the nucleoside scaffold. Antivir. Res. 2018, 154, 66–86. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Tomizaki, K.-y.; Takahashi, T.; Usui, K.; Kajikawa, K.; Mihara, H. Interactions between peptides containing nucleobase amino acids and T7 phages displayingS. cerevisiae proteins. Biopolymers 2007, 88, 131–140. [Google Scholar] [CrossRef]
- Uozumi, R.; Takahashi, T.; Yamazaki, T.; Granholm, V.; Mihara, H. Design and conformational analysis of natively folded β-hairpin peptides stabilized by nucleobase interactions. Biopolymers 2010, 94, 830–842. [Google Scholar] [CrossRef] [PubMed]
- Roviello, V.; Musumeci, D.; Mokhir, A.; Roviello, G.N. Evidence of protein binding by a nucleopeptide based on a thymine-decorated L-diaminopropanoic acid through CD and in silico studies. Curr. Med. Chem. 2021, 28, 5004–5015. [Google Scholar] [CrossRef]
- Musumeci, D.; Roviello, V.; Roviello, G.N. DNA- and RNA-binding ability of oligoDapT, a nucleobase-decorated peptide, for biomedical applications. Int. J. Nanomed. 2018, 13, 2613–2629. [Google Scholar] [CrossRef]
- Roviello, G.N.; Vicidomini, C.; Di Gaetano, S.; Capasso, D.; Musumeci, D.; Roviello, V. Solid phase synthesis and RNA-binding activity of an arginine-containing nucleopeptide. RSC Adv. 2016, 6, 14140–14148. [Google Scholar] [CrossRef]
- Geotti-Bianchini, P.; Beyrath, J.; Chaloin, O.; Formaggio, F.; Bianco, A. Design and synthesis of intrinsically cell-penetrating nucleopeptides. Org. Biomol. Chem. 2008, 6, 3661. [Google Scholar] [CrossRef]
- Roviello, G.; Musumeci, D.; Castiglione, M.; Bucci, E.M.; Pedone, C.; Benedetti, E. Solid phase synthesis and RNA-binding studies of a serum-resistant nucleo-epsilon-peptide. J. Pept. Sci. 2009, 15, 155–160. [Google Scholar] [CrossRef]
- De Napoli, L.; Messere, A.; Montesarchio, D.; Piccialli, G.; Benedetti, E.; Bucci, E.; Rossi, F. A new solid-phase synthesis of oligonucleotides 3′-conjugated with peptides. Bioorg. Med. Chem. 1999, 7, 395–400. [Google Scholar] [CrossRef]
- Roviello, G.N.; Roviello, G.; Musumeci, D.; Bucci, E.M.; Pedone, C. Dakin–West reaction on 1-thyminyl acetic acid for the synthesis of 1,3-bis(1-thyminyl)-2-propanone, a heteroaromatic compound with nucleopeptide-binding properties. Amino Acids 2012, 43, 1615–1623. [Google Scholar] [CrossRef] [PubMed]
- Bucci, R.; Bossi, A.; Erba, E.; Vaghi, F.; Saha, A.; Yuran, S.; Maggioni, D.; Gelmi, M.L.; Reches, M.; Pellegrino, S. Nucleobase morpholino β amino acids as molecular chimeras for the preparation of photoluminescent materials from ribonucleosides. Sci. Rep. 2020, 10, 19331. [Google Scholar] [CrossRef]
- Wang, H.; Feng, Z.; Qin, Y.; Wang, J.; Xu, B. Nucleopeptide Assemblies Selectively Sequester ATP in Cancer Cells to Increase the Efficacy of Doxorubicin. Angew. Chem. Int. Ed. Engl. 2018, 57, 4931–4935. [Google Scholar] [CrossRef]
- Höger, G.A.; Wiegand, M.; Worbs, B.; Diederichsen, U. Membrane-Associated Nucleobase-Functionalized β-Peptides (β-PNAs) Affecting Membrane Support and Lipid Composition. ChemBioChem 2020, 21, 2599–2603. [Google Scholar] [CrossRef]
- Dolman, N.P.; More, J.C.; Alt, A.; Knauss, J.L.; Troop, H.M.; Bleakman, D.; Collingridge, G.L.; Jane, D.E. Structure-activity relationship studies on N3-substituted willardiine derivatives acting as AMPA or kainate receptor antagonists. J. Med. Chem. 2006, 49, 2579–2592. [Google Scholar] [CrossRef] [PubMed]
- Mik, V.; Mickova, Z.; Dolezal, K.; Frebort, I.; Pospisil, T. Activity of (+)-Discadenine as a Plant Cytokinin. J. Nat. Prod. 2017, 80, 2136–2140. [Google Scholar] [CrossRef] [PubMed]
- Rozan, P.; Kuo, Y.H.; Lambein, F. Amino acids in seeds and seedlings of the genus Lens. Phytochemistry 2001, 58, 281–289. [Google Scholar] [CrossRef]
- Ignatowska, J.; Mironiuk-Puchalska, E.; Grześkowiak, P.; Wińska, P.; Wielechowska, M.; Bretner, M.; Karatsai, O.; Jolanta Rędowicz, M.; Koszytkowska-Stawińska, M. New insight into nucleo α-amino acids—Synthesis and SAR studies on cytotoxic activity of β-pyrimidine alanines. Bioorg. Chem. 2020, 100, 103864. [Google Scholar] [CrossRef]
- Walsh, C.T.; Zhang, W. Chemical Logic and Enzymatic Machinery for Biological Assembly of Peptidyl Nucleoside Antibiotics. ACS Chem. Biol. 2011, 6, 1000–1007. [Google Scholar] [CrossRef]
- Filippov, D.; Kuyl-Yeheskiely, E.; Van der Marel, G.A.; Tesser, G.I.; Van Boom, J.H. Synthesis of a nucleopeptide fragment from poliovirus genome. Tetrahedron Lett. 1998, 39, 3597–3600. [Google Scholar] [CrossRef]
- Roviello, G.N.; Roviello, V.; Autiero, I.; Saviano, M. Solid phase synthesis of TyrT, a thymine-tyrosine conjugate with poly(A) RNA-binding ability. RSC Adv. 2016, 6, 27607–27613. [Google Scholar] [CrossRef]
- Kramer, R.A.; Bleicher, K.H.; Wennemers, H. Design and Synthesis of Nucleoproline Amino Acids for the Straightforward Preparation of Chiral and Conformationally Constrained Nucleopeptides. Helv. Chim. Acta 2012, 95, 2621–2634. [Google Scholar] [CrossRef]
- Roviello, G.N.; Crescenzo, C.; Capasso, D.; Di Gaetano, S.; Franco, S.; Bucci, E.M.; Pedone, C. Synthesis of a novel Fmoc-protected nucleoaminoacid for the solid phase assembly of 4-piperidyl glycine/L-arginine-containing nucleopeptides and preliminary RNA: Interaction studies. Amino Acids 2010, 39, 795–800. [Google Scholar] [CrossRef]
- Xie, J.; Noel, O. Synthesis of Nucleo Aminooxy Acid Derivatives. Synthesis 2012, 45, 134–140. [Google Scholar] [CrossRef][Green Version]
- Pomplun, S.; Gates, Z.P.; Zhang, G.; Quartararo, A.J.; Pentelute, B.L. Discovery of nucleic acid binding molecules from combinatorial biohybrid nucleobase peptide libraries. J. Am. Chem. Soc. 2020, 142, 19642–19651. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, I.; Larsen, P.O. γ-Glutamylwillardiine and γ-glutamylphenylalanylwillardiine from seeds of Fagus silvatica. Phytochemistry 1974, 13, 2799–2802. [Google Scholar] [CrossRef]
- Sisido, M.; Kuwahara, M. Novel Peptide Nucleic Acids with Improved Solubility and DNA-Binding Ability. In Self-Assembling Peptide Systems in Biology, Medicine and Engineering; Springer: Berlin/Heidelberg, Germany, 2002; pp. 295–309. [Google Scholar]
- Roviello, G.N.; Gröschel, S.; Pedone, C.; Diederichsen, U. Synthesis of novel MMT/acyl-protected nucleo alanine monomers for the preparation of DNA/alanyl-PNA chimeras. Amino Acids 2010, 38, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Talukder, P.; Dedkova, L.M.; Ellington, A.D.; Yakovchuk, P.; Lim, J.; Anslyn, E.V.; Hecht, S.M. Synthesis of alanyl nucleobase amino acids and their incorporation into proteins. Bioorg. Med. Chem. 2016, 24, 4177–4187. [Google Scholar] [CrossRef]
- Diederichsen, U.; Weicherding, D.; Diezemann, N. Side chain homologation of alanyl peptide nucleic acids: Pairingselectivity and stacking. Org. Biomol. Chem. 2005, 3, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Geotti-Bianchini, P.; Crisma, M.; Peggion, C.; Bianco, A.; Formaggio, F. Conformationally controlled, thymine-based α-nucleopeptides. Chem. Commun. 2009, 22, 3178–3180. [Google Scholar] [CrossRef]
- Geotti-Bianchini, P.; Moretto, A.; Peggion, C.; Beyrath, J.; Bianco, A.; Formaggio, F. Replacement of Ala by Aib improves structuration and biological stability in thymine-based α-nucleopeptides. Org. Biomol. Chem. 2010, 8, 1315–1321. [Google Scholar] [CrossRef]
- Ranevski, R. Synthese und Untersuchung von Alanyl-PNA Oligomeren und deren Einfluß auf β-Faltblatt Strukturen. Ph.D. Thesis, Georg-August-Universitat, Göttingen, Germany, 2006. [Google Scholar]
- Sturm, C. Theoretical Investigation of the Geometrical Arrangements of Alpha-Alanyl-Peptide Nucleic Acid Hexamer Dimers and the Underlying Interstrand Binding Motifs. Ph.D. Thesis, Universität Würzburg, Würzburg, Germany, 2006. [Google Scholar]
- Lewis, F.D.; Liu, X.; Liu, J.; Miller, S.E.; Hayes, R.T.; Wasielewski, M.R. Direct measurement of hole transport dynamics in DNA. Nature 2000, 406, 51–53. [Google Scholar] [CrossRef]
- Weicherding, D.; Davis, W.; Hess, S.; Von Feilitzsch, T.; Michel-Beyerle, M.; Diederichsen, U. Femtosecond time-resolved guanine oxidation in acridine modified alanyl peptide nucleic acids. Bioorg. Med. Chem. Lett. 2004, 14, 1629–1632. [Google Scholar] [CrossRef]
- Cleaves, H.J. Prebiotic chemistry: What we know, what we don’t. Evol. Educ. Outreach 2012, 5, 342–360. [Google Scholar] [CrossRef]
- Leslie, E.O. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 99–123. [Google Scholar] [CrossRef] [PubMed]
- Scognamiglio, P.L.; Platella, C.; Napolitano, E.; Musumeci, D.; Roviello, G.N. From prebiotic chemistry to supramolecular biomedical materials: Exploring the properties of self-assembling nucleobase-containing peptides. Molecules 2021, 26, 3558. [Google Scholar] [CrossRef] [PubMed]
- Frenkel-Pinter, M.; Samanta, M.; Ashkenasy, G.; Leman, L.J. Prebiotic peptides: Molecular hubs in the origin of life. Chem. Rev. 2020, 120, 4707–4765. [Google Scholar] [CrossRef] [PubMed]
| Compound | EC50 (μM) | ±SD |
|---|---|---|
| 1 | 44.8 | 15.0 |
| 2 | 1.47 | 0.39 |
| 3 | 8.82 | 1.29 |
| 4 | 19.2 | 1.92 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palumbo, R.; Omodei, D.; Vicidomini, C.; Roviello, G.N. Willardiine and Its Synthetic Analogues: Biological Aspects and Implications in Peptide Chemistry of This Nucleobase Amino Acid. Pharmaceuticals 2022, 15, 1243. https://doi.org/10.3390/ph15101243
Palumbo R, Omodei D, Vicidomini C, Roviello GN. Willardiine and Its Synthetic Analogues: Biological Aspects and Implications in Peptide Chemistry of This Nucleobase Amino Acid. Pharmaceuticals. 2022; 15(10):1243. https://doi.org/10.3390/ph15101243
Chicago/Turabian StylePalumbo, Rosanna, Daniela Omodei, Caterina Vicidomini, and Giovanni N. Roviello. 2022. "Willardiine and Its Synthetic Analogues: Biological Aspects and Implications in Peptide Chemistry of This Nucleobase Amino Acid" Pharmaceuticals 15, no. 10: 1243. https://doi.org/10.3390/ph15101243
APA StylePalumbo, R., Omodei, D., Vicidomini, C., & Roviello, G. N. (2022). Willardiine and Its Synthetic Analogues: Biological Aspects and Implications in Peptide Chemistry of This Nucleobase Amino Acid. Pharmaceuticals, 15(10), 1243. https://doi.org/10.3390/ph15101243








