Abstract
Hybrid natural products produced via mixed biosynthetic pathways are unique and often surprise one with unexpected medicinal properties in addition to their fascinating structural complexity/diversity. In view of chemical structures, hybridization is a way of diversifying natural products usually through dimerization of two similar or dissimilar subcomponents through a C–C or N–C covalent linkage. Here, we report four structurally attractive diterpene–alkaloid conjugates polyalongarins A–D (1–4), clerodane-containing aporphine and proaporphine alkaloids, the first of its kind from the barks of Taiwanese Polyalthia longifolia (Sonn.) Thwaites var. pendula. In addition to conventional spectroscopic analysis, single crystal X-ray crystallography was employed to determine the chemical structures and stereo-configurations of 1. Compounds 1–4 were subsequently subjected to in vitro antiviral examination against DENV2 by evaluating the expression level of the NS2B protein in DENV2-infected Huh-7 cells. These compounds display encouraging anti-DENV2 activity with superb EC50 (2.8–6.4 μM) and CC50 values (50.4–200 μM). The inhibitory mechanism of 1–4 on NS2B was further explored drawing on in-silico molecular docking analysis. Based on calculated binding affinities and predicted interactions between the functional groups of 1–4 and the allosteric-site residues of the DENV2 NS2B-NS3 protease, our analysis concludes that the clerodane–aporphine/proaporphine-type hybrids are novel and effective DENV NS2B-NS3 protease inhibitors.
1. Introduction
Dengue fever is a mosquito-transmitted infectious disease caused by dengue viruses (DENVs). DENVs belong to the genus Flavivirus in the Flaviviridae family of RNA viruses, of which a single positive-strand RNA genome can immediately translate into a single polyprotein chain by utilizing the host-cell translation machinery upon infection. This single polyprotein precursor is organized in a functionally inactive linear polyprotein NH2-C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5-COOH in the first place [1]. Each independent and functional unit becomes mature after the polyprotein being subject to cleavage by host or self-encoded proteases. The dengue-specific vaccine, Dengvaxia, has recently been approved as a prophylactic measure, while its efficacy and the risk of developing severe syndromes in seronegative patients raise considerable concerns [2]. Dengue fever remains an incurable disease with only supportive treatment. It has been well documented that the NS2B-NS3pro protease plays a role in post-translation maturation in the viral replication. The NS3pro is an intrinsic serine protease containing a characteristic catalytic tri-ad (His51, Asp75, and Ser135) at its N-terminal domain; the NS2B protein is crucial in stabilizing NS3pro, thus highlighting NS2B-NS3pro as an ideal drug target concerning development of flavivirus therapeutics [3,4,5].
Natural products are born as bioactive molecules with immense structural complexity/diversity, on which hybridization can further multiply the level of complexity/diversity via permuting/recombining scaffold units at multiplex spatial dimensions. The naturally occurring hybrids are relatively rare, while their uniqueness is otherwise warranted [6,7]. Most hybrids reported thus far are microbial secondary metabolites produced via mixed biosynthetic pathways, such as NRPS and PKS pathways [6]. They are often manifested by the manner of two identical or unidentical entities coupled simply with a C–C, O–C or N–C linkage(s) by means of radical addition (diradical coupling) or carbanionic/carbocationic chemistry [8]. Their origins have now been expanded to the kingdom of plants as exemplified by identification of dimeric diterpenes longimides A and B (two bisclerodane imides) [9] and alkaloids bidebilines A–D (four bisdehydroaporphines) in Polyalithia [10]. Polyalithia spp. belongs to Annonaceae, the largest family in Magnoliales, which are deciduous or evergreen flowering trees and shrubs in tropical, mid-latitude, or temperate region [11].
In this study, four novel diterpene–alkaloid hybrids named polyalongarins A–D (1–4) each assembled by one clerodane diterpene and one aporphine/proaporphine alkaloid, were isolated and identified for the first time from the barks of Taiwanese P. longifolia (Sonn.) Thwaites var. pendula (Figure 1). Both clerodane-type diterpenoids and aporphine/proaporphine-type alkaloids, individually, are abundant in Polyalthia (usually as ornamental trees in Southeast Asia and as a folk medicine in India) [12], which also exist in other plants, with a plethora of biological activities, such as anti-inflammation [13,14], anti-cancer [10], anti-plasmodium [15], anti-malaria [16], anti-microbes [17,18], etc. In this study, the isolated polyalongarins A–D (1–4) were examined for their biological evaluation, revealing a promising antiviral activity specific to the NS2B-NS3 protease of DENV2. The mode of action was in parallel explored with in-silico molecular docking simulation, whereby the target site of polyalongarins A–D is pinpointed at the allosteric regulation site rather than the catalytic site that does not come with the territory in this case.
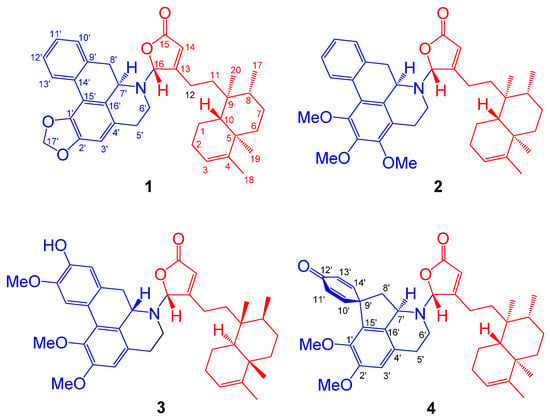
Figure 1.
The chemical structures of polyalongarins A–D (1–4).
2. Results
2.1. Structure Elucidation of Polyalongarins A–D (1–4)
Polyalongarin A (1) was obtained as colorless crystals with specific rotation [α] = –181 (c 0.05, CHCl3). On the basis of the HRESIMS spectrum, polyalongarin A (1) [m/z 566.32658 [M + H]+ (calcd. for C37H44NO4, 566.32649)] complies with the “nitrogen rule”, revealing that 1 is an alkaloid possessing a molecular formula of C37H43NO4 with 17 degrees of unsaturation (DOU). According to 13C NMR and DEPTs spectra, the 37 carbons can be grouped into four methyls, ten methylenes (one nitrogen-containing methylene at δC 41.4 and one dioxymethylene at δC 100.9), four methines (one nitrogen-containing methine at δC 56.3 and one N,O-mixed acetal at δC 93.9), seven olefinic methines, two quaternary carbons, nine sp2 quaternary carbons, and one carboxyl group at δC 171.5. Having determined proton-carbon single bond correlations by HMQC, the planar moiety of 1 was concluded by 2D NMR (1H-1H COSY and HMBC) (Figure 2); thereby, the 1H- and 13C-NMR signals were unequivocally assigned as summarized in Table 1.
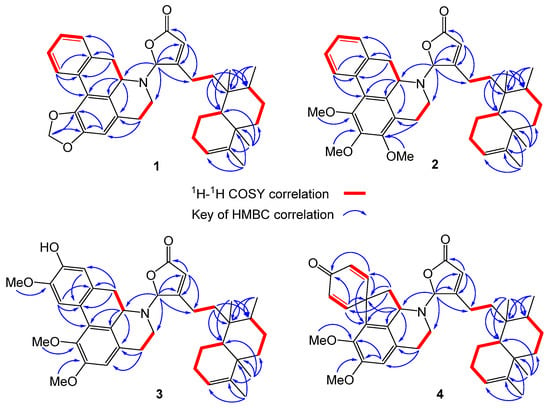
Figure 2.
1H-1H COSY (red bold) and key HMBC (blue arrow) correlations of compounds 1–4.

Table 1.
1H- and 13C- NMR spectroscopic data of Polyalongarin A-D (1–4 in CDCl3).
The α,β-unsaturated γ-lactone unit was confirmed based on the HMBC correlations for H-14 (δH 5.98 s) and H-16 (δH 6.19 s) extended to C-13 (δC 169.8), C-15 (δC 171.5), and C-16 (δC 93.9), where the characteristic chemical shift value at δC 93.9 (C-16) stands for a strong electron-withdrawing group adjacent to the γ-position of the five-membered ring. Next, a tetramethyl-octahydronaphthalene bicyclic ring (a decalin-derivative) was established by two 1H-1H COSY correlations of H-10/H2-1/H2-2/H-3 and H2-6/H2-7/H-8/CH3-17 alongside the HMBC correlations for the four methyls to their neighbor carbons (Figure 2): CH3-17 (δH 0.79 d, J = 4.5) to C-7, C-8, and C-9; CH3-18 (δH 1.58 s) to C-3, C-4, and C-5; CH3-19 (δH 1.01 s) to C-4, C-5, C-6, and C-10; and CH3-20 (δH 0.78 s) to C-8, C-9, C-10. Given the HMBC cross peaks of H-14/C-12, H-16/C-12, and CH3-20/C-11 and the 1H-1H COSY correlations of H2-11/H2-12, the α,β-unsaturated γ-lactone ring is determined in connection with the tetramethyl-octahydronaphthalene bicyclic ring through a short C2 chain known as a type III clerodane diterpenoid [19]. On the other hand, the remaining twelve olefinic carbons and eleven DOU suggest that the alkaloid portion is made of a five-membered ring structure and two benzene rings. Three correlations of H-5′/H-6′, H-7′/H-8′, and H-10′/H-11′/H-12′/H-13′ were observed in the COSY spectrum of 1, as well as the long range 1H-13C correlations of H-3′/C-1′, C-2′, C-4′, C-5′, C-16′; H-5′/C-3′, C-4′; H-7′/C-4′, C-6′, C-15′, C-16′; H-8′/C-9′, C-10′, C-14′, C-16′; H-10′/C-8′, C-9′, C-14′; H-13′/C-9′, C-14′, C-15′; and H2-17/C-1′ and C-2′ were deduced from the HMBC spectrum. Taken together, a benzylisoquinoline named anonaine [17,18] was determined. Moreover, an unexpected but crucial HMBC correlation of H-16/C-6′, C-7′ was observed, where the strong electron-pulled phenomenon at C-16 resulted from the N atom of anonaine and the carboxyl group. The C-N bond that connects the clerodane and the anonaine thereby brings about the unprecedented hybrid of clerodane–aprophine alkaloid, named polyalongarin A (1) here.
Given that there are four intriguing but dizzy diastereomers trans–cis (TC), trans–trans (TT), cis–cis (CC), and cis–trans (CT) in the decalin system, the neo- or ent-neo-absolute configurations of clerodanes were determined on the basis of the stereo-configurations of C-5, C-8, C-9, and C-10 [20]. In brief, the absence of NOESY correlations for H-10/CH3-19 but the appearance of that for H-10/H-8 and CH3-17/CH3-19/CH3-20 supports that the clerodane skeleton in compound 1 is a type of TC. The C-7′ in anonaine is usually a R-configuration with a negative optical rotation as seen in previous reports, while the chirality of C-16 is uncertain merely based on the NOESY correlations (Figure 3A) of H-16/H-7′/H-8′α and H-16/H-8′β. To resolve this uncertainty, we resorted to X-ray crystallography. The crystals of 1 were grown in a MeOH/CH2Cl2 solvent system and subsequently subjected to X-ray diffraction, whereby the crystal structure undoubtedly depicts the absolute stereochemistry of 1. The ORTEP diagram for the crystal structure of 1 (Figure 3B) confirms the TC type configuration of clerodane (neo-TC), namely, the R-configuration of C-16 and the P helical conformation between the five-membered γ-lactone ring and aprophine.
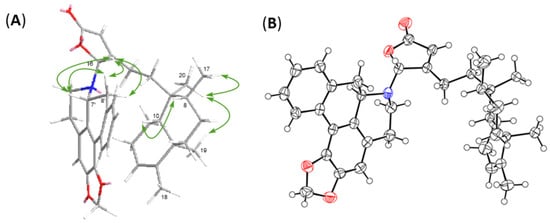
Figure 3.
Main NOESY correlations (A) and ORTEP diagram (B) of compound 1.
The framework of polyalongarin B (2) is akin to that of 1, in that both share similar UV, IR, NMR, optical rotation and circular dichroism (CD) spectra. Its molecular formula is C39H49NO5 with 16 DOU according to HRESIMS [m/z 612.36844 [M + H]+ (calcd. for C39H50NO5, 612.36835)]. There are three additional aliphatic methoxy groups with chemical shifts at δH 3.95 (3H, s), 3.70 (3H, s), and 3.92 (3H, s) in 1H-NMR spectrum (Table 1), whereas the absence of dioxymethylene is observed from 13C-NMR spectrum (Table 1). In conjunction with HMBC correlations of H2-5′/C-3′ and C-4′; 1′-OMe/C-1′; 2′-OMe/C-2′; and 3′-OMe/C-3′ (Figure 2), 2 was determined to be a clerodane-containing 1′,2′,3′-trimethoxy-aprophine alkaloid similar with 1.
Based on spectroscopic analysis, 3 is likely a clerodane–aprophine alkaloid. As a matter of fact, it is a hydroxylated analog of 2 with molecular formula of C39H49NO6 (16 DOU) derived from HRESIMS [m/z 628.36295 [M + H]+ (calcd. for C39H50NO6, 628.32326)]. Given the absence of 1H-1H COSY fragment of H-10′/H-11′/H-12′/H-13′ and atypical HMBC correlations (H-10′/C-9′, C-11′, C-12′, and C-14′; H-13′/C-9′, C-11′, C-12′, and C-14′; 1′-OMe/C-1′; 2′-OMe/C-2′; and 12′-OMe/C-12′), the chemical entity 3 is determined to be a 1′,2′,12′-trimethoxy-11′-hydroxy-aprophine alkaloid. Given the optical rotation [35.4 (c 0.05, CHCl3)] and the reverse CD curve (Figure 4A), 3 is clearly distinct from others [21]. Taking advantage of MMFF94 energy minimization in the Chem3D 16.0 suite [22], the dihedral angle ω16-N,C-16,C-13,C-12 of 3 should be the same no matter the lone pair on the 16-N atom pointing toward the α or β direction. However, the helical conformation of the γ-lactone ring and aprophine is poised in an M-configuration (Figure S1 in the Supporting Information). In the ECD calculation for all possible stereoisomers of 3 (Figure 4B), only is 3m-16R (the mirror isomer of 3 with 16R) in line with the experimental CD spectrum of 3 concerning the binary positive [π→π* transitions at 234 and 251 nm] and negative [n→π* transitions at 279 and 303 nm] Cotton effects. Altogether, the most plausible configuration for 3 is 16R, 7’S, ent-neo-TC, not least considering both the NOESY correlations and the absence of NOESY correlations of H-16/H-6′β/H-7′.
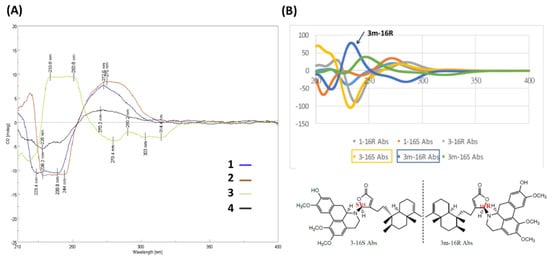
Figure 4.
Stereochemistry of 1–4 determined by CD/ECD. (A) Experimental CD spectra of 1–4. (B) Calculated ECD comparison between 1 and 3. The calculated ECD curves of different stereoisomers of 1 and 3 in B are shown as 1-16R, light blue; 1-16S, orange; 3-16R, gray; 3-16S, yellow orange; 3-16R, gray; 3-16S, yellow orange; 3m-16R (the mirror isomer of 3 with 16R), blue; 3m-16S (the mirror isomer of 3 with 16S), green.
Similarly, polyalongarin D (4) was first analyzed by HRESIMS with m/z 598.35281 [M + H]+ (calcd. for C38H49NO5, 598.35270). On the basis of 1H- and 13C NMR (Table 1), four methyls at δH 0.75 (d, J = 5.5 Hz), 0.77 (s), 1.00 (s), 1.57 (s), and two sp2 methines at δC 5.16 (br s) and 5.98 (s) were spotted; four olefinic carbons at δC 118.6, 120.1, 144.6, 169.0, and one carboxyl carbon at δC 171.4 were additionally identified, together suggesting that 4 is a cleordane–alkaloid congener with a molecular formula of C38H47NO5 and 16 DOU. Apart from the signals for a typical clerodane, a characteristic AA′XX′ proton system δH 6.90 (d, J = 10.0 Hz), 6.33 (d, J = 10.0 Hz), 6.44 (d, J = 10.0 Hz), and 7.04 (d, J = 10.0 Hz) at the down field indicates two pairs of aromatic ortho-protons, which are further ascertained by HMBC correlations [H-9′ (H-13′)/C-8′, C-10′ (C-12′), C-11′, C-13′ (C-9′), C-14′, and C-15′; and H-10′ (H-12′)/C-9′ (C-13′), C-11′, C-12′ (C-10′) and C-14′]. All together establish a unique spiro conformation. On the other hand, specific sets of cross peaks (H-3′/C-1′, C-2′, C-4′, C-5′, C-16′; H-8′/C-9′, C-13′, C-14′, C-15′, C-16′; 1′-OMe/C-1′; 2′-OMe/C-2′) identified from HMBC spectrum are elucidated, which correlates the alkaloid to a stepharine moiety [23]. Moreover, the optical rotation [–61.0 (c 0.05, CHCl3)] of 4 echoes a similar CD curve in analogy to those of 1 and 2, suggesting the stereochemistry of 4 follows 5R, 8R, 9S, 10R, 16R, and 7′R. Summing up, compound 4 is a new type of hybrid featuring an unusual chemical architecture of a clerodane–proaporphine alkaloid.
The biosynthetic pathways for these hybrids 1-4 are put forward and shown in Figure S2 in the Supporting Information.
2.2. Anti-DENV2 Activities of Polyalongarins A–D (1–4)
Concerning their biological activity, the anti-DENV2 activity of polyalongarins A–D (1–4) was examined using post-infection assay. The DENV2-infected Huh-7 cells were incubated with each of 1–4 at a concentration of 5, 10, or 20 μM for 72 hrs (0.2 μM celastrol and 0.1% DMSO served as positive control and mock control, respectively) [24]. It was defined active when a compound exhibited >50% inhibition of DENV at a given concentration specified above. All compounds 1–4 showed >50% inhibition of DENV2 RNA expression within the testing range (Figure 5), and the IC50 values for 1–4 were, respectively, determined to be 4.46, 2.80, 3.24, and 6.36 μM with sorted inhibitory effect in order as 2 > 3 > 1 > 4 in agreement with the western blotting analysis. In the meantime, the uninfected Huh-7 cells were incubated with each individual 1–4 at a range of concentrations for cytotoxicity evaluation, of which the result suggests no or very low cytotoxicity (Table 2). Based on previous reports [25], a compound that diminishes the DENV activity by 90% but presents a 50% cytotoxic concentration (CC50) > 15 μM is considered a “hit”. By this definition, compounds 1–4 are good hits without doubt.
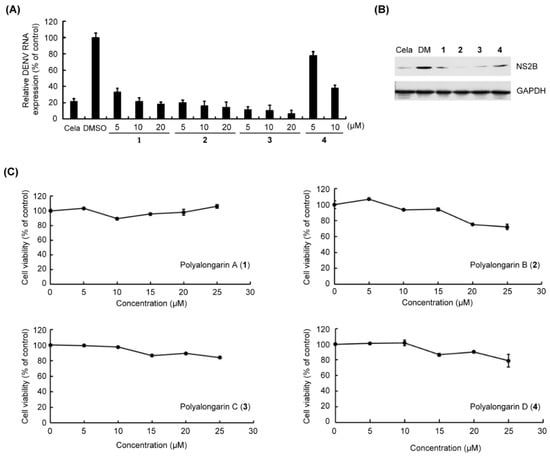
Figure 5.
In vitro anti-DENV2 assay for 1–4. (A) DENV2-infected/uninfected Huh-7 cells were incubated with celastrol (Cela, 0.2 μM, positive control) or indicated concentrations of 1–4 (5 to 25 μM) for 72 h. Total DENV2 RNA expression (%) was extracted and quantified. The DENV2 RNA level was measured by RT-qPCR. The DENV2 RNA expression was normalized by cellular GAPDH mRNA. (B) DENV2-infected Huh-7 cells were treated with 10 μM of 1–4. The NS2B protein expression was analyzed by western blotting with anti-NS2B and anti-GAPDH antibodies (a loading control). (C) The cell viability (%) was determined by the XTT method. Treatment with 0.1% DMSO served as a mock control. The results are expressed as the means ± standard errors of the means (SEM) of triplicate experiments.

Table 2.
Anti-DENV-2 activity of polyalongarins A–D (1–4).
2.3. Molecular Docking Analysis of Polyalongarins and DENV2 NS2B-NS3pro
The NS2B-NS3 protease is essential for flavivirus infections and their sequences are highly conserved (Figure S3 in the Supporting Information), highlighting the NS2B-NS3pro protease as an ideal antiviral target. To date, only a handful of apo/liganded structures of DENV NS2-NS3 proteases from the PDB database (Figure S4 in the Supporting Information) are available, indicating their difficulty in crystallization. For the docking study, compounds 1–4 were individually modeled into DENV2 NS2B-NS3pro (PDB ID: 6MO1) by using AutoDock 4.2 and AutoDock Vina under the PyRx suite for probing the most possible binding site and evaluating their interactions with NS2B-NS3pro. To our surprise, compound 2, for instance, neither stays in the anticipated NS3pro active site (H51, D75, and S135) nor interacts with NS2B (residues 48–71) as shown in Figure 6. Instead, it enters into an unexpected binding pocket constituted by residues (Met1049–Asn1167), wherein Lys1074 adapts double conformations in cooperation with the “open–active” or “closed–inactive” form of NS2B-NS3pro. Namely, the ε-NH2 group of Lys1074 is in close proximity to three O atoms of 1′,2′,3′-trimethoxyl groups of 2 through a constellation of H-bonds in the closed-inactive model, thereby revealing that 2 is an allosteric inhibitor specific to the DENV2 NS2B-NS3 protease. In addition to 2, the other clerodane–aporphine/proaporphine hybrids also display comparably as allosteric regulators of the NS2B-NS3 protease (Figure S5 in the Supporting Information) with an overall high anti-DENV2 activity but no observable cytotoxicity.
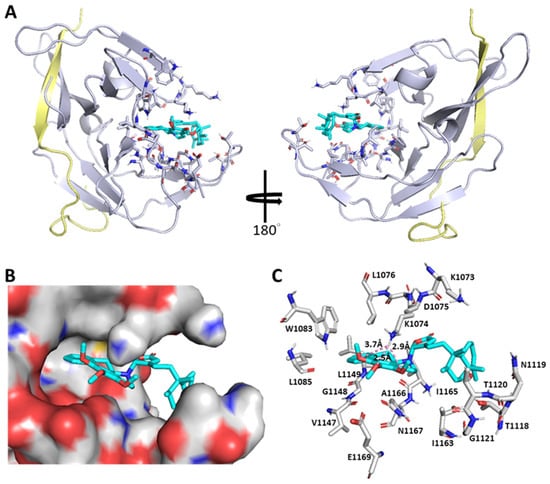
Figure 6.
Computational modeling for polyalongarin B (2) bound to the allosteric site of NS2B-NS3pro. (A) DENV NS2B-NS3pro in complex with 2. Gray, NS3pro; yellow, NS2B. (B) Compound 2 is located in the pocket of the allosteric site (shown in electrostatic surface). (C) Compound 2 interacts with the allosteric-site residues. Cyan, compound 2. The default analytical distance for the ligand within its binding site is set less than 5 Å. Figures are prepared using PyMOL 2.0 software (Warren L. DeLano, PyMOL 2.0 software; Schrödinger, Inc. 2017).
3. Discussion
Given plentiful examples on opium alkaloids and clerodane-type diterpenoids in literature, a plausible biosynthetic pathway for aporphine/proaporphine alkaloids and clerodanes can be proposed, in which hybrid products 1–4 are joined with a C–N bond that may be achieved by a 1,2-dehydroreticuline reductase (DRR) homologue-mediated nucleophilic addition reaction (Figure S2 in the Supporting Information). Such a coupling reaction is reminiscent of the biosynthesis of imidazoyl hybrid conidiogenone B, where an α,β-hydrolase Con-ABH puts an imidazole-containing alkaloid (meleagrin) and a diterpenoid (conidiogenone) together via an aza-Michael addition reaction [26]. The isomerization from S- to R-configuration of the chiral center C7′ in the coclaurine alkaloid intermediate is likely achievable by recruiting two enzymes, 1,2-dehydroreticuline synthase (DRS) and DRR [27,28], whereby clerodanes may be subject to structural reconfiguration into a normal (chair–chair–“normal”), ent (chair–chair–“antipodal”), syn (chair–boat–“normal”), or syn-ent (chair–boat–“antipodal”) conformation [20,29]. In bioactive evaluation, compounds 1–4 showed strong anti-DENV2 activity (EC50 values: 2.8–6.4 μM and CC50 values: 50.4–200 μM) as a result of suppressing NS2B-NS3pro expression. In molecular docking analysis (polyalongarins vs. DENV2 NS2B-NS3pro), the simulation reveals that these four ligands do not hold the catalytic triad (H51, D75, and S135) at bay but instated dock at the cave lined by specific residues (M1049, H1051, W1063, K1073, K1074, D1075, L1076, T1120, G1148, L1149, G1151, I1165, A1166, and N1167) (Figure 6), suggesting that polyalongarins are potent allosteric regulators inhibiting the protease activity of DENV2 NS2B-NS3. Lys1074 and Asn1167 residues are the allosteric domain of pocket in NS2B-NS3pro and critical for its activity [4,30]. Also, it has also been reported that aminopyrazine derivatives could bind to the pocket of NS2B-NS3pro and affects its activity [4].
The anti-DENV2 assay and the in silico analysis are in a position to gauge the order of inhibitory strength in 2 > 3 > 1 > 4. Based on their structure–activity–relationship (NS2B-NS3pro and 1–4), Lys1074, Leu1149, and Asn1167 are highlighted as particularly critical and potent for the allosteric regulation. Three OMe groups in aprophine of 2 may make 2, getting a little closer to Lys1074. In aprophine of 3, the additional 11′-OH and the absence of 3′-OMe may together usher this ligand to lean toward Asn1167. In contrast, the dioxomethylene substitution of 1 and the bulky spiro moiety of 4 may prohibit their interactions with Lys1074, thus resulting in relatively poor NS2B inhibitory activities (Figure S5). Together, our results indicate that the inhibitory target of compounds 1–4 is the NS2B-NS3 protease. The activity of NS2B-NS3pro has been reported to be critical for the replication of DENV [31] and therefore an attractive target for the development of anti-dengue drugs [32]. Here, we provide evidence that clerodane–aporphine/proaporphine-type hybrids isolated from the barks of P. longifolia var. pendula are unique and potent DENV NS2B-NS3 protease inhibitors favorabe for developing into anti-dengue agents.
4. Materials and Methods
4.1. General
In structural elucidation, the optical rotations were determined using a JASCO P-1020 polarimeter (Jasco Co., Tokyo, Japan); the ultraviolet spectra (UV) were recorded on a JASCO V530 UV/VIS spectrophotometer; the circular dichroism spectroscopy (CD) were recorded on a JASCO J-815 spectrophotometer; the infrared spectra (IR) were recorded on a Mattson Genesis II FT-IR spectrophotometer (Thermo Nicolet, Madison, WI, USA); electrospray ionization mass (ESIMS) and high-resolution ESIMS data were measured using a VG Biotech Quattro 5022 mass spectrometer (VG Biotech, UK); nuclear magnetic resonance (NMR) spectra were collected on a Varian Unity Inova-400 MHz NMR system; the single crystal X-ray diffraction of pure compounds was determined by Bruker APEX II X-ray single crystal diffractometer. In chemical isolation and separation, column chromatography (CC) was applied including silica gel 60 (70–230 and 230–400 mesh, Merck, Darmstadt, Germany), Sephadex LH-20 (Pharmacia), and pre-coated silica gel TLC plates (aluminum Merck Silica gel 60 F254). HPLC purification was performed on a Hitachi L-2,130 Intelligent pump with a Hitachi L2420 UV-Vis detector, equipped with a 250 × 20 mm i.d. preparative Ascentis Si (5 μm) and Purospher STAR RP-18 (5 μm). In biological evaluation, cell viability/cytotoxicity assay was evaluated using CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA) according to the manufacturer’s instructions, where the color intensity was detected at 490 nm using a 550 BioRad plate-reader for determining the number of viable cells in proliferation. Computational modeling analyzed by PyMOL 2.0 software (creator is Warren L. DeLano; published by Schrödinger, Inc. 2017; and the latest vision is PyMOL 2.5 vision).
4.2. Plant Material
The barks of Polyalthia longifolia (Sonn.) Thwaites var. pendula (synonym: Monoon longifolium var. pendula) were collected from Wanluan township, Pingtung County, Taiwan, in July 2015, and identified by one of the authors (Y.-S. Lin). A voucher specimen (No. ME2015-BST01) has been deposited in the Department of Biological Science and Technology, Meiho University, Taiwan.
4.3. Extraction and Isolation
The freeze-drying barks of P. longifolia var. pendula (9.5 kg) were grinded and extracted with 20 L of methanol (MeOH) at room temperature three times, and the combined extracts were concentrated in vacuum. The combined MeOH extract (310 g) was suspended into 3% aqueous solution of tartaric acid, and then partitioned with ethyl acetate (EtOAc). After adjusting the acidic solution to pH 9 by saturated Na2CO3 solution, the mixture was further partitioned with CH2Cl2. The CH2Cl2 extract was washed with double distilled water and concentrated in reduced pressure to obtain an alkaloid-containing extract (72 g). This crude alkaloid extract was partitioned between n-hexane and 75% MeOH aqueous (1:1) to afford a n-hexane layer (39 g) and a MeOH (aq.) layer (33 g). The MeOH layer was subjected to silica gel flash chromatography with a n-hexane/EtOAc solvent system (1:0, 20:1, 10:1, 8:1, 6:1, 5:1, 4:1, 3:1, 2:1, 1:1, 0:1), and ten fractions (PL1~PL10) were obtained. The PL2 fraction (3.2 g) was further separated by an open column prepared by silica gel to give five sub-fractions (PL2-1~PL2-5). The sub-fraction PL2-2 was re-suspended for crystallization with MeOH/CH2Cl2 solution, and spindle crystals were obtained as polyalongarin A (1, 16.1 mg). The sub-fraction PL2-3 was successively subjected to normal-phase HPLC (eluting with n-hexane/EtOAc = 5:1) and preparative TLC (eluting with n-hexane/acetone = 5:2) to purify polyalongarin B (2, 11.1 mg). The sub-fraction PL2-4 (2.1 g) was chromatographed by a Sephadex LH-20 column with elution of CH2Cl2/MeOH (1:1), and seven fractional fractions (PL2-4-1~PL2-4-7) were given. Of them, polyalongarin D (4, 4.7 mg) was isolated after purification of PL2-4-3 (480 mg) via reverse-phase C18 HPLC (MeOH/H2O/acetonitrile = 55:40:5). Finally, the sub-fraction PL2-6 (4.9 g) was fractionated by using Sephadex LH-20 column chromatography, which was eluted with CH2Cl2/MeOH (1:1) and then further purified by normal-phase HPLC with n-hexane/EtOAc (2:1) to obtained polyalongarin C (3, 13.1 mg).
4.4. Spectral Measurements
Polyalongarin A (1): Colorless needle crystal; [α] −181.0 (c 0.05, CHCl3); UV (CDCl3) λmax (log ε) 273 (3.41), 225 (3.93) nm; IR (KBr) νmax 2939, 2860, 1755, 1447, 1252, 1220, 1139, 1047 cm−1; 1H-NMR (500 MHz, CDCl3) and 13C-NMR (125 MHz, CDCl3) spectroscopic data, see Table 1; ESIMS m/z 566 [M + H]+; HRESIMS m/z 566.32658 [M + H]+ (calcd. for C37H44NO4, 566.32649).
Polyalongarin B (2): White amorphous powder; [α] −36.0 (c 0.05, CHCl3); UV (CDCl3) λmax (log ε) 273 (1.042), 213 (3.41) nm; IR (KBr) νmax 3346, 2362, 2296, 1639, 1267, 1055, 1014 cm−1; 1H-NMR (500 MHz, CDCl3) and 13C-NMR (125 MHz, CDCl3) spectroscopic data, see Table 1; ESIMS m/z 612 [M + H]+; HRESIMS m/z 612.36844 [M + H]+ (calcd. for C39H50NO5, 612.36835).
Polyalongarin C (3): White amorphous powder; [α] +35.4 (c 0.05, CHCl3); UV (CDCl3) λmax (log ε) 302 (3.09), 281 (3.14), 217 (3.60) nm; IR (KBr) νmax 3349, 2959, 2362, 1740, 1644, 1251, 1135, 1068 cm−1; 1H-NMR (500 MHz, CDCl3) and 13C-NMR (125 MHz, CDCl3) spectroscopic data, see Table 1; ESIMS m/z 628 [M + H]+; HRESIMS m/z 628.36295 [M + H]+ (calcd. for C39H50NO6, 628.32326).
Polyalongarin D (4): White amorphous powder; [α] −61.0 (c 0.05, CHCl3); UV (CDCl3) λmax (log ε) 285 (2.63), 234 (3.34), 213 (3.72); IR (KBr) νmax 3736, 3388, 2929, 2355, 2285, 1747, 1657, 1540, 1089 cm−1; 1H-NMR (500 MHz, CDCl3) and 13C-NMR (125 MHz, CDCl3) spectroscopic data, see Table 1; ESIMS m/z 598 [M + H]+; HRESIMS m/z 598.35281 [M + H]+ (calcd. for C38H49NO5, 598.35270).
4.5. X-ray Crystallographic Data of Polyalongarin A (1)
A colorless crystal of polyalongarin A (1) was obtained by simple evaporation from CH2Cl2/MeOH solution. The data was collected on a Bruker APEX II X-ray single crystal diffractometer with MoKα radiation (λ = 1.54178 Å) at 200 K in the Instrumentation Center of National Taiwan Normal University, Taiwan. Crystal data: crystal size 0.20 × 0.11 × 0.05 mm3, C37H43NO4, monoclinic, a = 12.2259(2) Å, b = 9.50360(10) Å, c = 13.0577(2) Å, V = 1494.23(4) Å3, space group P21, Z = 4, density (calculated) 1.257 Mg/m3, absorption coefficient 0.635 mm−1, F(000) = 608, and 1761 reflections were collected for crystal structural analysis by 5242 independent reflections [Rint = 0.0485] with I > 2σ (I). The data were solved by using the “direct method” (normal model), and the chemical structure was refined by the “full-matrix least-squares” procedure on F2 values. All non-hydrogen atoms were refined anisotropically within thermal parameters, and the hydrogen atom positions were idealized geometrically and attached to ride on the parent carbon atoms with C–H distances (0.98–1.00 Å). The final indices (I > 2σ(I)) were R1 = 0.0383, wR2 = 0.0942 with goodness-of-fit = 1.044. The absolute structure parameter is -002(17). Crystal data and structure refinement are shown on Table S1 in the Supporting Information. The final X-ray model is shown in Figure 3. The crystal data of polyalongarin A (1) have been deposited in the Cambridge Crystallographic Data Centre, CCDC number: 1939242.
4.6. Cytotoxic Assay
Huh-7 cells were seeded in 96-well plates at a density 5 × 104 cells/well in 120 μL of culture medium and incubated for 6 hr in a humidified CO2 (5%) atmosphere. Then, respectively, treated Huh-7 cells with 0.1% DMSO (positive control) and analytes at indicated concentrations, followed by incubation at 37 °C for an additional 72 hr. Then, the XTT method was applied to cell viability evaluation: cells were dyed by 2,3-bis(2-methyloxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide and recorded by an enzyme-linked immunosorbent assay (ELISA) plate reader (at UV 490 nm). Three independent experiments were performed at each treatment concentration, and all data are presented as mean ± standard error of the mean (SEM).
4.7. In Vitro Anti-DENV2 Assay
Huh-7 human liver carcinoma cell line, purchased from Bioresources Collection and Research Center, Taiwan, were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% non-essential amino acids (NEAA), and 1% penicillin and streptomycin at 37 °C in a humidified 5% CO2 incubator. The DENV serotype 2 (strain 16681) was directly from the Centers for Disease Control, Department of Health, Taiwan, and was amplified in Aedes albopictus C6/36 cell line (CRL-1660, ATCC, USA). C6/36 cells were cultured in RPMI 1640 medium supplemented with 2% FBS. Huh-7 cells were seeded at a density of 5 × 104 cells/mL in the 24-well plate for 16–20 h and then infected with DENV2 at a multiplicity of infection (MOI) of 0.1 PFU/cell for 2 h at 37 °C. The viral titer was determined in a plaque assay in the Huh-7 cells. Then, DENV2-infected Huh-7 cells were washed with PBS and refilled, respectively, with DMEM−2% FBS medium containing 2 μM celastrol (positive control), 0.1% DMSO (mock control), and various indicating concentration of test compounds. After incubation at 37 °C for 72 h, DENV2-infected Huh-7 cells were harvested and cell lysates were prepared for quantification of DENV RNA by RT-qPCR. Total cellular RNA was extracted using Total RNA Miniprep Purification Kit (GMbiolab, Taiwan) according to the manufacturer’s instructions.
4.8. Western Blotting Analysis
Cell lysates (20 μg) were loaded onto and assessed by SDS-PAGE, and transferred to an electroblotting PVDF membrane that had been incubated overnight with rabbit polyclonal antibodies against NS2B (1:3,000; GeneTex, Irvine, CA, USA) and anti-GAPDH antibodies (1:10,000; GeneTex, Irvine, CA, USA) serving as loading control.
4.9. In Silico Molecular Docking by PyRx
The molecular docking studies were performed by using PyRx 0.8 software [33], a suite of established open-source function including: AutoDock 4.2 and AutoDock Vina [34] are used as a docking software; AutoDockTools is used to generate input files; whereas Open Babel is used for importing SDF files, removing salts and energy minimization. AutoDock 4.2, AutoDock vina, and AutoDock Tools 1.5.7 are created by the Center for Computational Structural Biology in the Scripps Research Institute. Prior to molecular docking, the protein molecules were prepared by using a standard protocol for the structural preparation tool of AutoDock 4.2 that included addition of hydrogen atoms and assignment of charges (Compute Gasteiger–Marsili:Ligand). AutoDock 4.2 and AutoDock Vina, implementing a Lamarckian Genetic Algorithm (LGA), were employed for molecular docking studies. AutoDock Tools 1.5.7 was used to generate a grid box around the active site with other parameters set as default in the input PDBQT files of protein. All the docking conformations were clustered together ground on RMSD, and the one with the most favorable free energy or the highest percentage frequency was selected as representative. 3D coordinates of ligands, polyalongarins A–D (1–4), were obtained using Open Babel for MMFF94 energy minimization. The dataset of compounds 1–4 were subjected to structure-based virtual screening against the selected protein, DENV2 NS2B-NS3 protease, and the protein was kept rigid during docking calculation. Finally, the virtually screened best conformation poses and binding affinities of each ligands were ensured (Tables S3 and S4 in the Supporting Information), and the interactions between protein and ligands were visualized and illustrated using PyMOL 2.0 software (Figure S5 in the Supporting Information). In these computational simulations of protein–ligand interactions, the defaulted distance of ligand within the active site is less than 5Å; protein is presented in a cartoon or hydrophobicity surface model; the residues of active site and ligands are drawn in sticks.
5. Conclusions
In this study, we presented four hybrid polyalongarins A–D (1–4) isolated from the barks of Taiwanese P. longifolia (Sonn.) Thwaites var. pendula which exhibited amazing inhibitory activity against DENV2 with EC50 ranging from 2.8–6.4 μM and CC50 ranging from 50.4–200 μM. The inhibitory target of 1–4 was revealed to be NS2B by proteomic analysis. On the basis of in silico simulation, estimated binding affinities and predicted interactions between the allosteric-site residues of the DENV2 NS2B-NS3 protease (PDB ID: 6MO1) and compounds 1–4, we conclude that the clerodane–aporphine/proaporphine type hybrids are unique and potent DENV NS2B-NS3 protease inhibitors suitable for developing into anti-Dengue agents.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph15101218/s1, Figure S1: The ω16-N,C-16,C-13,C-12 torsional angles of cis-1 (A), trans-1 (B), cis-3m-16R (C), and trans-3m-16R (D). Figure S2: Proposed biosynthetic pathways of 1–4. Figure S3: Protein sequence alignments of various flavivirus NS2B-NS3 proteases including DENV serotypes 1-4, Zika virus, West Nile virus (WNV), and Murray Valley encephalitis virus (MVEV). Figure S4: Protein crystal structures of NS2B-NS3 proteases in DENV serotypes 1–4. Figure S5: AutoDock 4.2 of binding simulation between DENV2 NS2B-NS3 protease and 1–4. Table S1: Crystal data and structure refinement for 1. Table S2: Atomic coordinates and equivalent isotropic displacement parameters for 1. U(eq) is defined as one third of the trace of the orthogonalized U tensor. Table S3: The Predicted Binding Affinity and RMSD bounds of 1–4. Table S4: Python Shell Information of 1-4 in Different Models.; Spectra: Mass, IR, UV, CD, and NMR spectra of compounds 1–4. References [35,36,37] are cited in the Supplementary Materials.
Author Contributions
Conceptualization, T.-L.L., Y.-S.L. and C.-C.L.; methodology, Y.-S.L. and C.-C.L.; software, I.-W.L., Y.-C.W., G.-Y.L. and J.-C.L.; validation, I.-W.L., G.-Y.L., J.-C.L. and C.-C.L.; formal analysis, S.-P.L., H.-J.S., C.-I.L., C.-Y.K. and Z.-Y.L.; investigation, C.-I.C., Y.-Y.C., C.-I.L., C.-Y.K. and Z.-Y.L.; resources, C.-I.C., Y.-S.L. and C.-C.L.; data curation, I.-W.L., Y.-S.L. and C.-C.L.; writing—original draft preparation, I.-W.L., G.-Y.L. and J.-C.L.; writing—review and editing, T.-L.L., Y.-S.L. and C.-C.L.; visualization, I.-W.L., G.-Y.L., J.-C.L. and C.-C.L.; supervision, T.-L.L., Y.-S.L. and C.-C.L.; project administration, Y.-S.L. and C.-C.L.; funding acquisition, Y.-S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ministry of Science and Technology, Republic of China, grant number MOST 107-2320-B-276-001.
Institutional Review Board Statement
The authors declare that their research did not include any human subjects or animal experiments.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The Instrumentation Center of National Taiwan Normal University is appreciated for X-ray measurements and database inspection.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pierson, T.C.; Diamond, M.S. The continued threat of emerging flaviviruses. Nat. Microbiol. 2020, 5, 796–812. [Google Scholar] [CrossRef] [PubMed]
- Vannice, K.S.; Wilder-Smith, A.; Barrett, A.D.T.; Carrijo, K.; Cavaleri, M.; Silva, A.; Durbin, A.P.; Endy, T.; Harris, E.; Innis, B.L.; et al. Clinical development and regulatory points for consideration for second-generation live attenuated dengue vaccines. Vaccine 2018, 36, 3411–3417. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, M.; Ghosh, S.; Bell, J.A.; Sherman, W.; Hardy, J.A. Allosteric Inhibition of the NS2B-NS3 Protease from Dengue Virus. ACS Chem. Biol. 2013, 8, 2744–2752. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Huo, T.; Lin, Y.L.; Nie, S.; Wu, F.; Hua, Y.; Wu, J.; Kneubehl, A.R.; Vogt, M.B.; Rico-Hesse, R.; et al. Discovery, X-ray crystallography and antiviral activity of allostericinhibitors of flavivirus NS2B-NS3 protease. J. Am. Chem. Soc. 2019, 141, 6832–6836. [Google Scholar] [CrossRef]
- Erbel, P.; Schiering, N.; D’Arcy, A.; Renatus, M.; Kroemer, M.; Lim, S.P.; Yin, Z.; Keller, T.H.; Vasudevan, S.G.; Hommel, U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 2006, 13, 372–373. [Google Scholar] [CrossRef]
- Tietze, L.F.; Bell, H.P.; Chandrasekhar, S. Natural product hybrids as new leads for drug discovery. Angew. Chem. Int. Ed. 2003, 42, 3996–4028. [Google Scholar] [CrossRef]
- Viegas-Junior, C.; Danuello, A.; Bolzani, V.d.S.; Barreiro, E.J.; Fraga, C.A.M. Molecular Hybridization: A useful tool in the design of new drug prototypes. Curr. Med. Chem. 2007, 14, 1829–1852. [Google Scholar] [CrossRef]
- Liu, J.; Liu, A.; Hu, Y. Enzymatic dimerization in the biosynthetic pathway of microbial natural products. Nat. Prod. Rep. 2021, 38, 1469–1505. [Google Scholar] [CrossRef]
- Sashidhara, K.V.; Singh, S.P.; Kant, R.; Maulik, P.R.; Sarkar, J.; Kanojiya, S.; Kumar, K.R. Cytotoxic cycloartane triterpene and rare isomeric bisclerodane diterpenes from the leaves of Polyalthia longifolia var. pendula. Bioorg. Med. Chem. Lett. 2010, 20, 5767–5771. [Google Scholar] [CrossRef]
- Kanokmedhakul, S.; Kanokmedhakul, K.; Yodbuddee, D.; Phonkerd, N. New antimalarial bis-dehydroaporphine alkaloids from Polyalthia debilis. J. Nat. Prod. 2003, 66, 616–619. [Google Scholar] [CrossRef]
- Huan, T.C.; Shieh, W.C.; Keng, H.; Tsai, J.L.; Hsieh, C.F. Flora of Taiwan, 2nd ed.; Department of Botany, National Taiwan University: Taipei, Taiwan, 1993. [Google Scholar]
- Misra, P.; Sashidhara, K.V.; Singh, S.P.; Kumar, A.; Gupta, R.; Chaudhaery, S.S.; Gupta, S.S.; Majumder, H.K.; Saxena, A.K.; Dube, A. 16α-Hydroxycleroda-3,13(14)Z-dien-15,16-olide from Polyalthia longifolia: A safe and orally active antileishmanial agent. Br. J. Pharmacol. 2016, 159, 1143–1150. [Google Scholar] [CrossRef]
- Pu, D.B.; Zhang, X.J.; Bi, D.W.; Gao, J.B.; Yang, Y.; Li, X.L.; Lin, J.; Li, X.N.; Zhang, R.H.; Xiao, W.L. Callicarpins, two classes of rearranged ent-clerodane diterpenoids from callicarpa plants blocking NLRP3 inflammasome-induced pyroptosis. J. Nat. Prod. 2020, 83, 2191–2199. [Google Scholar] [CrossRef]
- Chang, F.R.; Hwang, T.L.; Yang, Y.L.; Li, C.E.; Wu, C.C.; Issa, H.H.; Hsieh, W.B.; Wu, Y.C. Anti-inflammatory and cytotoxic diterpenes from formosan Polyalthia longifolia var. pendula. Planta Med. 2006, 72, 1344–1347. [Google Scholar] [CrossRef]
- Annan, K.; Ekuadzi, E.; Asare, C.; Sarpong, K.; Pistorius, D.; Oberer, L.; Gyan, B.A.; Ofori, M. Antiplasmodial constituents from the stem bark of Polyalthia longifolia var pendula. Phytochem. Lett. 2015, 11, 28–31. [Google Scholar] [CrossRef]
- Gbedema, S.Y.; Bayor, M.T.; Annan, K.; Wright, C.W. Clerodane diterpenes from Polyalthia longifolia (Sonn) Thw. var. pendula: Potential antimalarial agents for drug resistant Plasmodium falciparum infection. J. Ethnopharmacol. 2015, 169, 176–182. [Google Scholar] [CrossRef]
- Chanda, S.; Nair, R. Antimicrobial activity of Polyalthia longifolia (Sonn.) Thw. var. pendula leaf extracts against 91 clinically important pathogenic microbial strains. Chin. Med. 2010, 1, 31–38. [Google Scholar] [CrossRef]
- Khan, A.K.; Ahmed, A.; Hussain, M.; Khan, I.A.; Ali, S.A.; Farooq, A.D.; Faizi, S. Antibiofilm potential of 16-oxo-cleroda-3,13(14)E-diene-15 oic acid and its five new γ-amino γ-lactone derivatives against methicillin resistant Staphylococcus aureus and Streptococcus mutans. Eur. J. Med. Chem. 2017, 138, 480–490. [Google Scholar] [CrossRef]
- Li, R.; Morris-Natschke, S.L.; Lee, K.H. Clerodane diterpenes: Sources, structures, and biological activities. Nat. Prod. Rep. 2016, 33, 1166–1226. [Google Scholar] [CrossRef]
- Bhakuni, D.S.; Tewari, S.; Dhar, M.M. Aporphine alkaloids of annona squamosa. Phytochemistry 1972, 11, 1819–1822. [Google Scholar] [CrossRef]
- Lightner, D.A.; Gurst, J.E. Organic Conformational Analysis and Stereochemistry from Circular Dichroism Spectroscopy; John Wiley and Sons: Hoboken, NJ, USA, 2000. [Google Scholar]
- Kerwin, S.M. ChemBioOffice Ultra 2010 Suite. J. Am. Chem. Soc. 2010, 132, 2466–2467. [Google Scholar] [CrossRef]
- Flor, S.C.; Doorenbos, N.J.; Svoboda, G.H.; Knapp, J.E.; Schiff, P.L. Chemical constituents of Legnephora moorei Miers (Menispermaceae). J. Pharm. Sci. 1974, 63, 618–619. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.S.; Tseng, C.K.; Lin, C.K.; Hsu, Y.C.; Wu, Y.H.; Hsieh, C.L.; Lee, J.C. Celastrol inhibits dengue virus replication via up-regulating type I interferon and downstream interferon-stimulated responses. Antivir. Res. 2017, 137, 49–57. [Google Scholar] [CrossRef]
- Yang, C.C.; Hu, H.S.; Wu, R.H.; Wu, S.H.; Lee, S.J.; Jiaang, W.T.; Chern, J.H.; Huang, Z.S.; Wu, H.N.; Chan, C.M.; et al. A novel Dengue virus inhibitor, BP13944, discovered by high-throughput screening with dengue virus replicon cells selects for resistance in the viral NS2B/NS3 protease. Antimicrob. Agents Chemother. 2014, 58, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Hewage, R.T.; Huang, R.J.; Lai, S.J.; Lien, Y.C.; Weng, S.H.; Li, D.H.; Chen, Y.J.; Wu, S.H.; Chein, R.J.; Lin, H.C. An enzyme-mediated aza-michael addition is involved in the biosynthesis of an imidazoyl hybrid product of conidiogenone B. Org. Lett. 2021, 23, 1904–1909. [Google Scholar] [CrossRef]
- Hirata, K.; Poeaknapo, C.; Schmidt JZenk, M.H. 1,2-Dehydroreticuline synthase, the branch point enzyme opening the morphinan biosynthetic pathway. Phytochemistry 2004, 65, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- De-Eknamkul, W.; Zenk, M.H. Purification and properties of 1,2-dehydroreticuline reductase from Papaver somniferum seedlings. Phytochemistry 1992, 31, 813–821. [Google Scholar] [CrossRef]
- Peters, R.J. Two rings in them all: The labdane-related diterpenoids. Nat. Prod. Rep. 2010, 27, 1521–1530. [Google Scholar] [CrossRef]
- Timiri, A.K.; Sinha, B.N.; Jayaprakash, V. Progress and prospects on DENV protease inhibitors. Eur. J. Med. Chem. 2016, 117, 125–143. [Google Scholar] [CrossRef]
- Falgout, B.; Pethel, M.; Zhang, Y.M.; Lai, C.J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J. Virol. 1991, 65, 2467–2475. [Google Scholar] [CrossRef]
- Takagi, Y.; Matsui, K.; Nobori, H.; Maeda, H.; Sato, A.; Kurosu, T.; Orba, Y.; Sawa, H.; Hattori, K.; Higashino, K.; et al. Discovery of novel cyclic peptide inhibitors of dengue virus NS2B-NS3 protease with antiviral activity. Bioorg. Med. Chem. Lett. 2017, 27, 3586–3590. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Potter, K.C.; Zi, J.C.; Hong, Y.J.; Schulte, S.; Malchow, B.; Tantillo, D.J.; Peters, R.J. Blocking Deprotonation with Retention of Aromaticity in a Plant ent-Copalyl Diphosphate Synthase Leads to Product Rearrangement. Angew. Chem. Int. Ed. 2016, 55, 634–638. [Google Scholar] [CrossRef]
- Yang, M.; Zhu, L.P.; Li, L.; Li, J.J.; Xu, L.M.; Feng, J.; Liu, Y.L. Digital Gene Expression Analysis Provides Insight into the Transcript Profile of the Genes Involved in Aporphine Alkaloid Biosynthesis in Lotus (Nelumbo nucifera). Front. Plant Sci. 2017, 8, 80. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, Version 2.0; Schrödinger, LLC: New York, NY, USA, 2015; Available online: https://pymol.org/2/ (accessed on 15 August 2022).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).