Rhynchosia volubilis Promotes Cell Survival via cAMP-PKA/ERK-CREB Pathway
Abstract
1. Introduction
2. Results
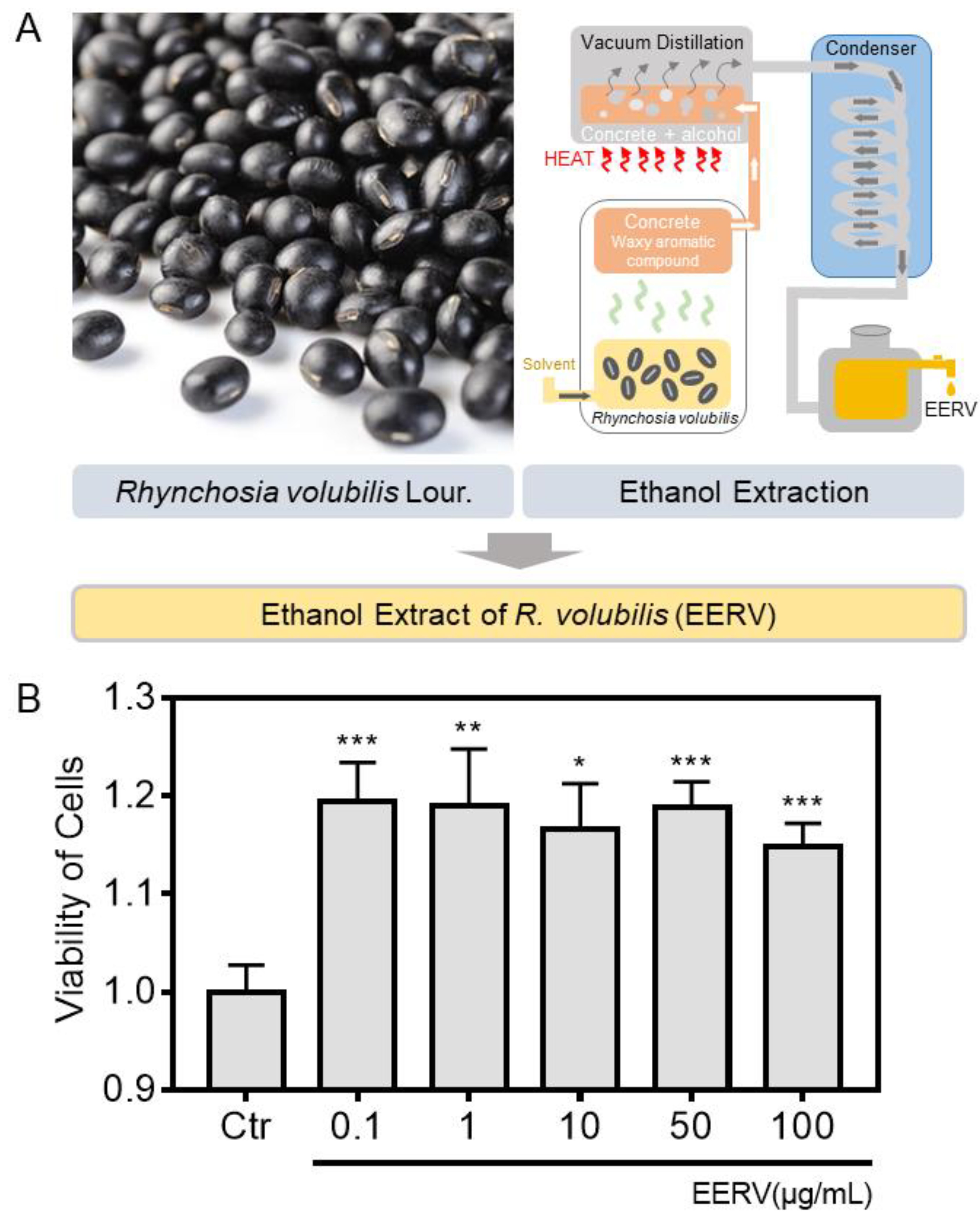
2.1. Effects of EERV on Cell Viability
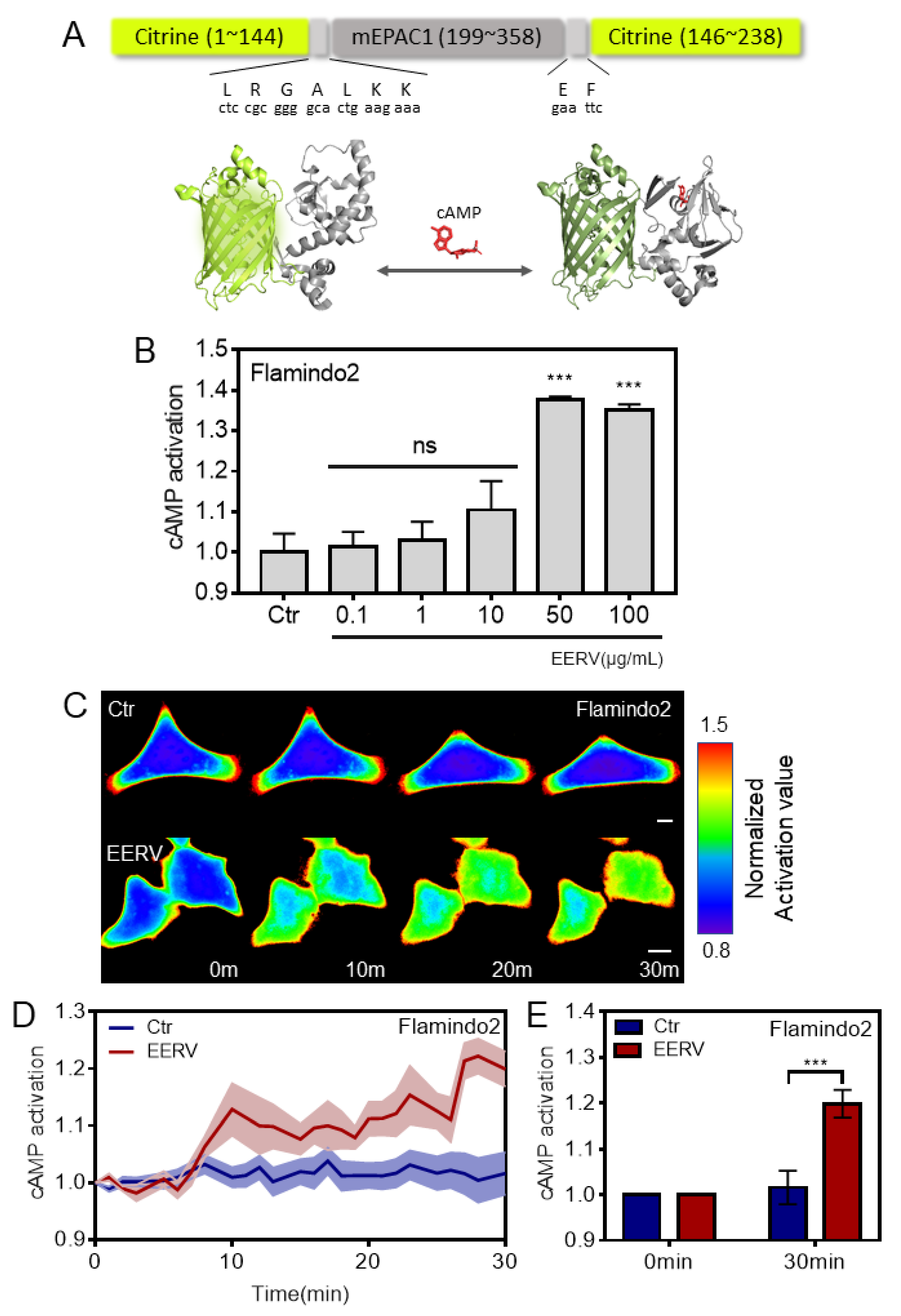
2.2. EERV Induces Intracellular cAMP Concentration Increase
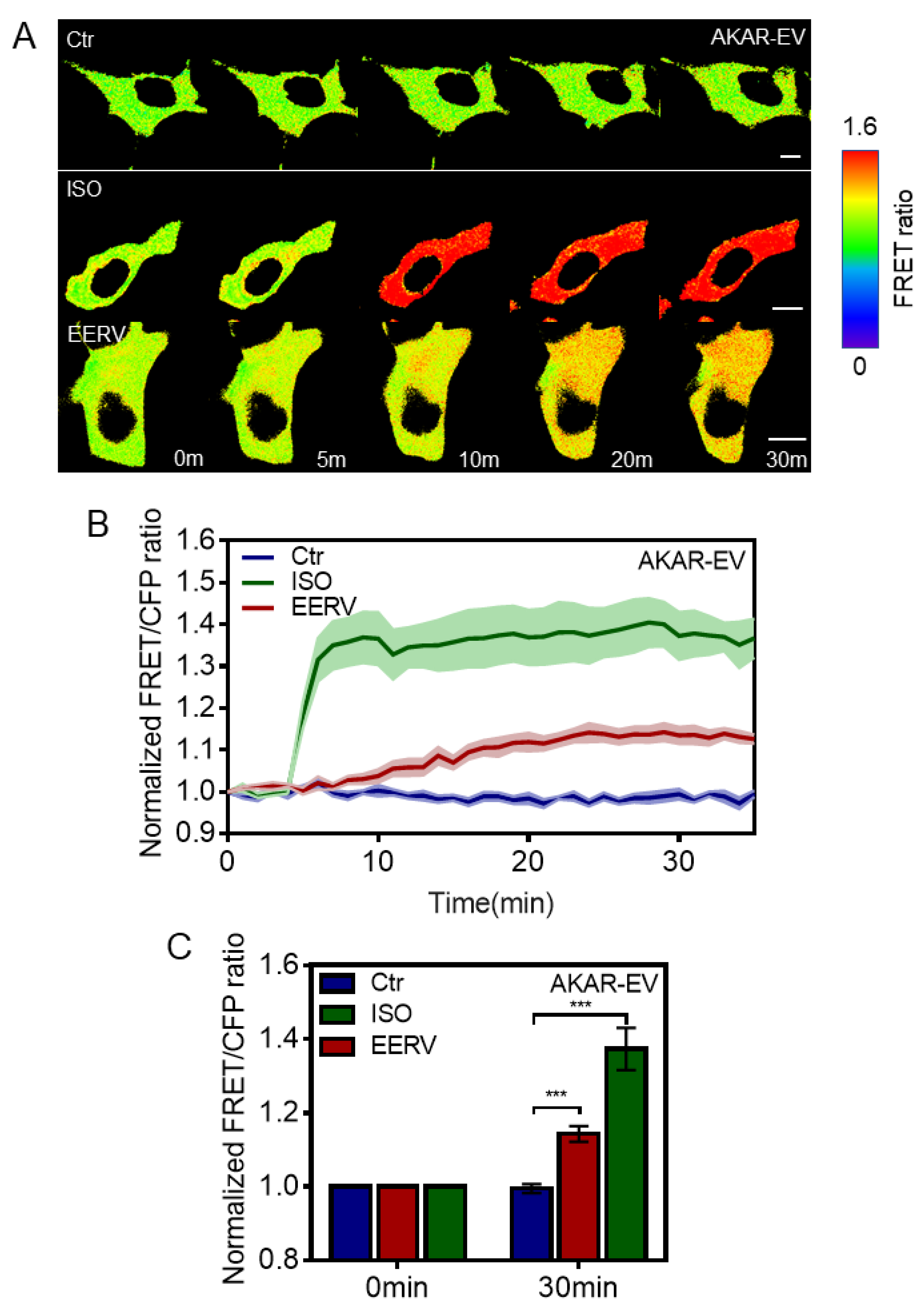
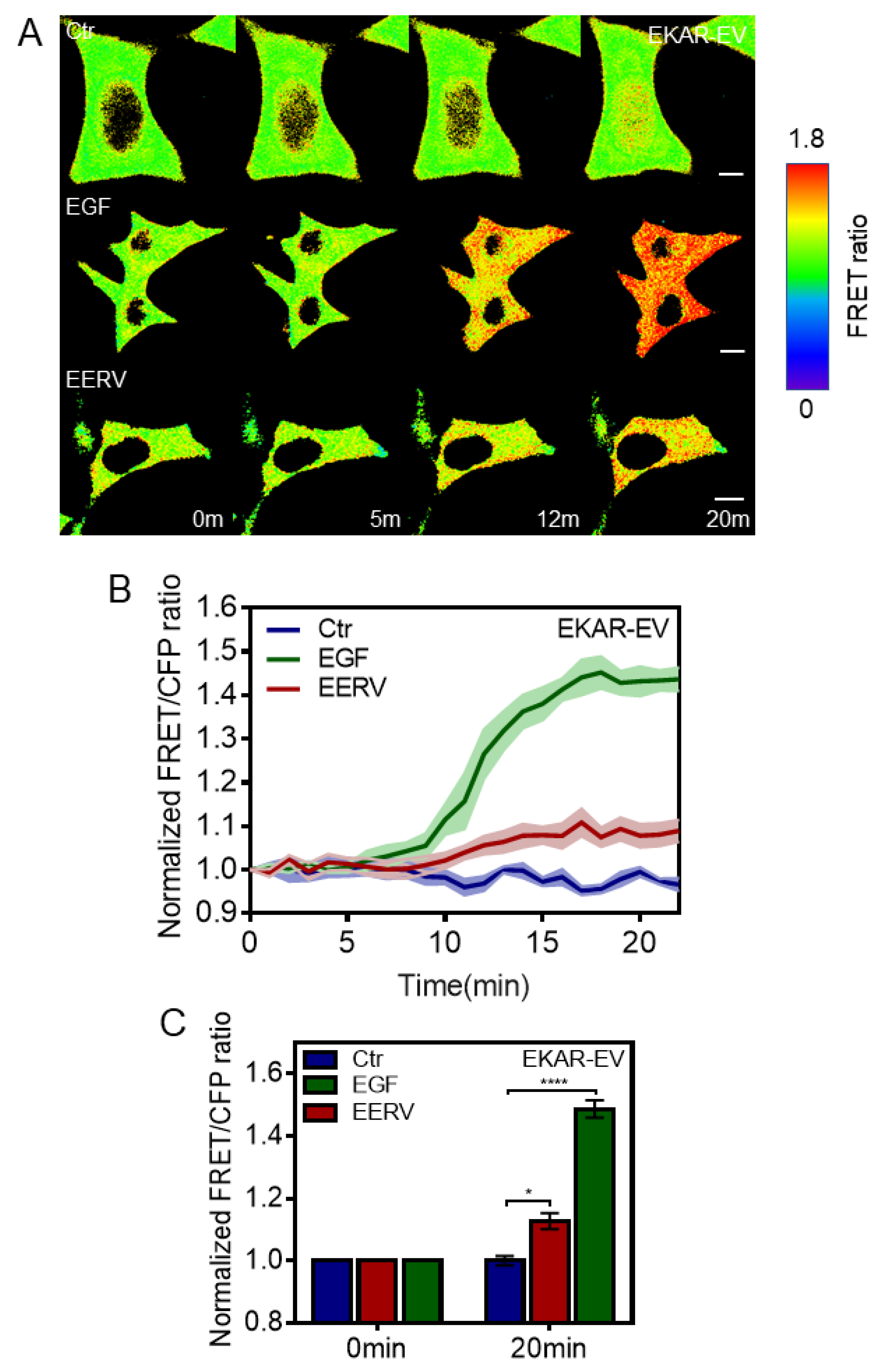
2.3. EERV-Induced cAMP Regulates PKA and ERK Activation
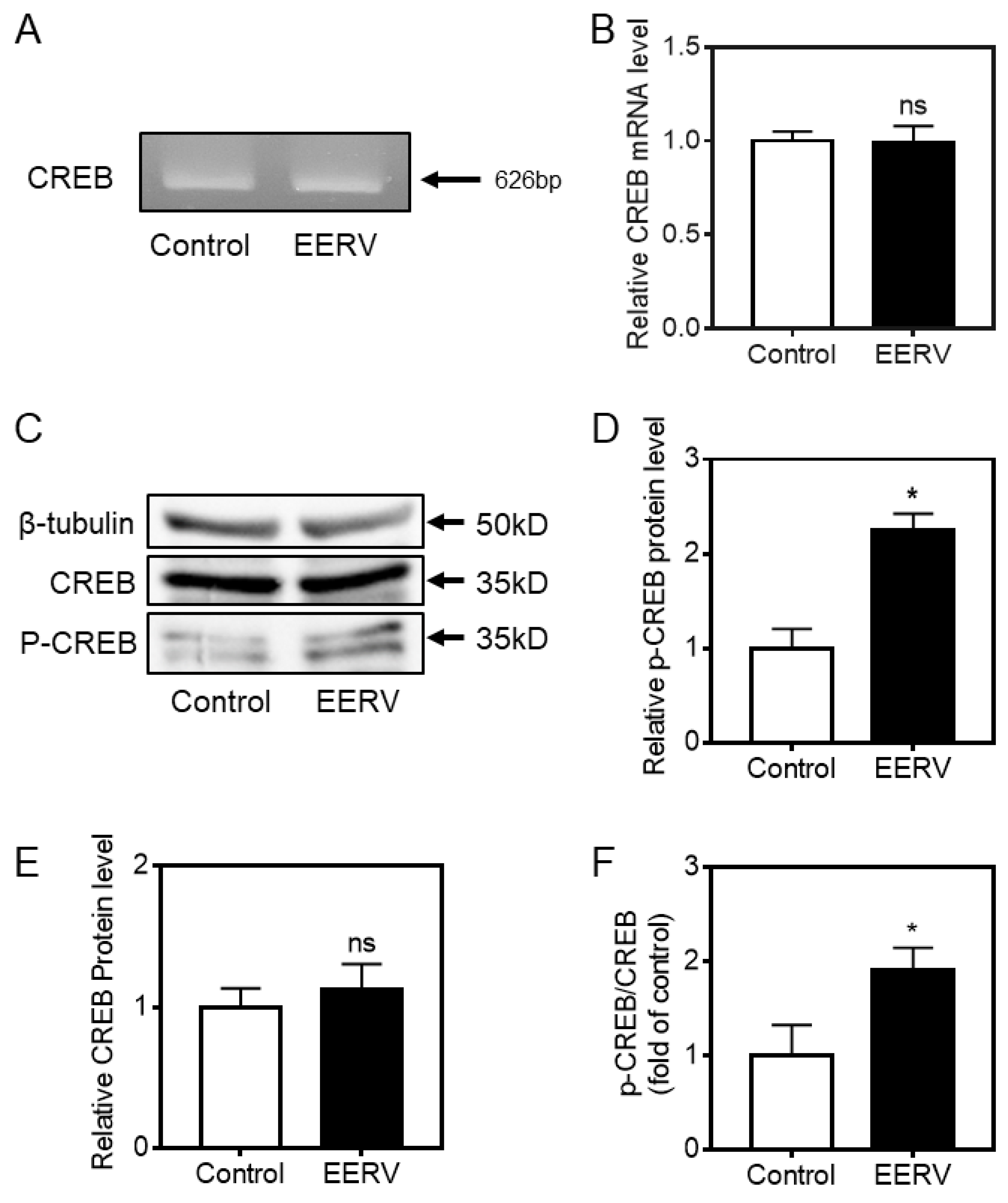
2.4. Increasing CREB Phosphorylation by EERV
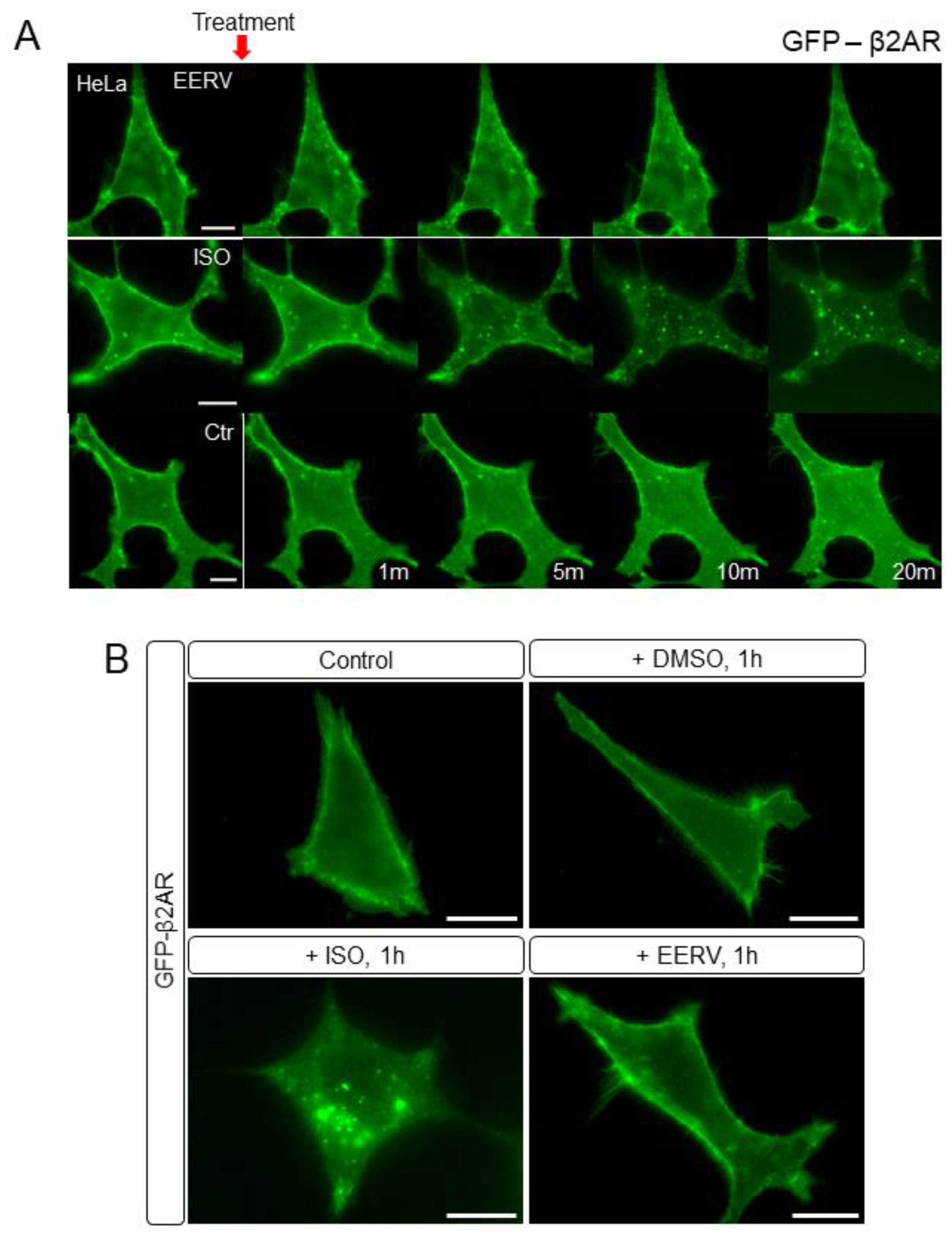
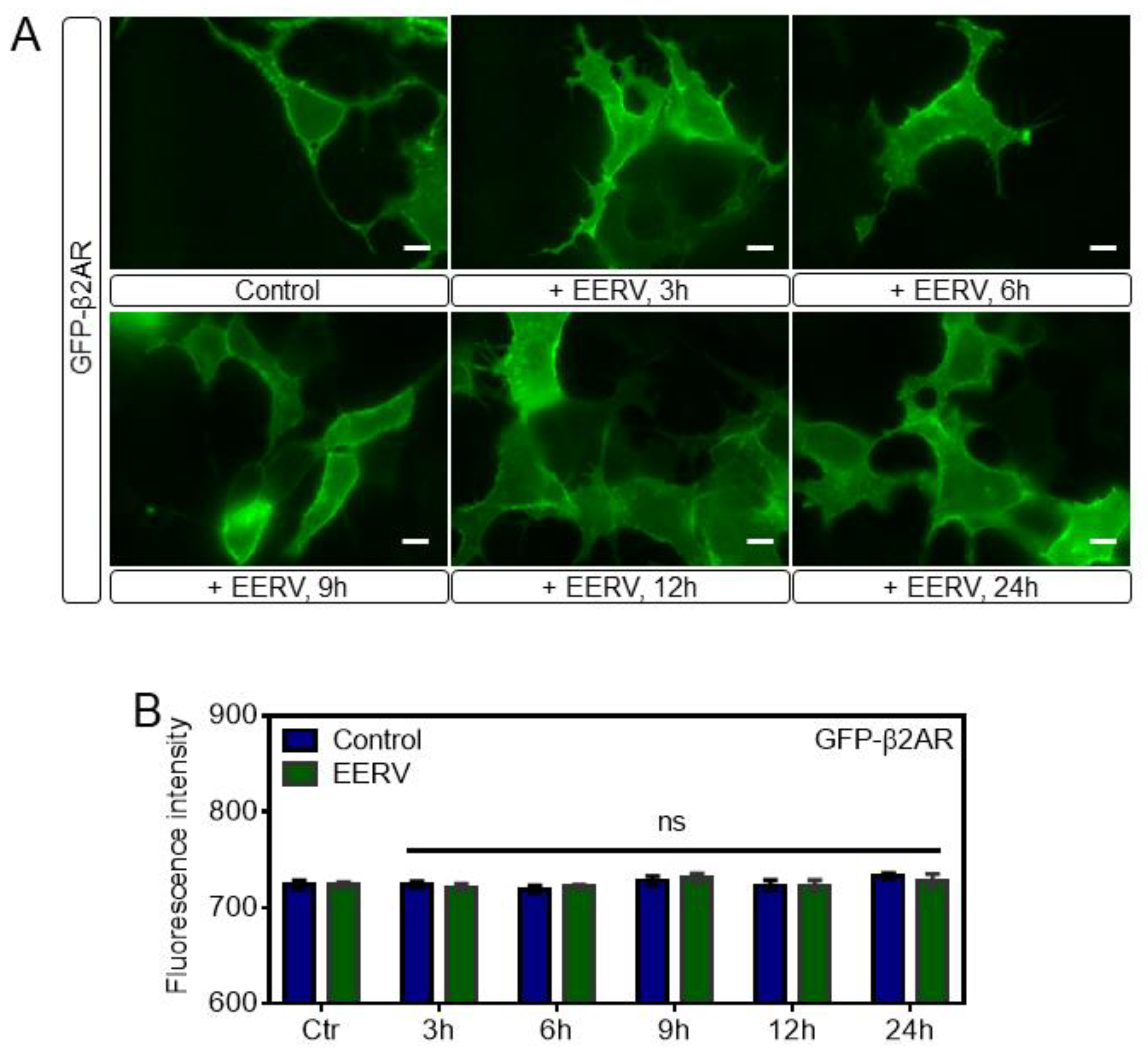
2.5. EERV Does Not Activate Adrenoceptor Beta-2
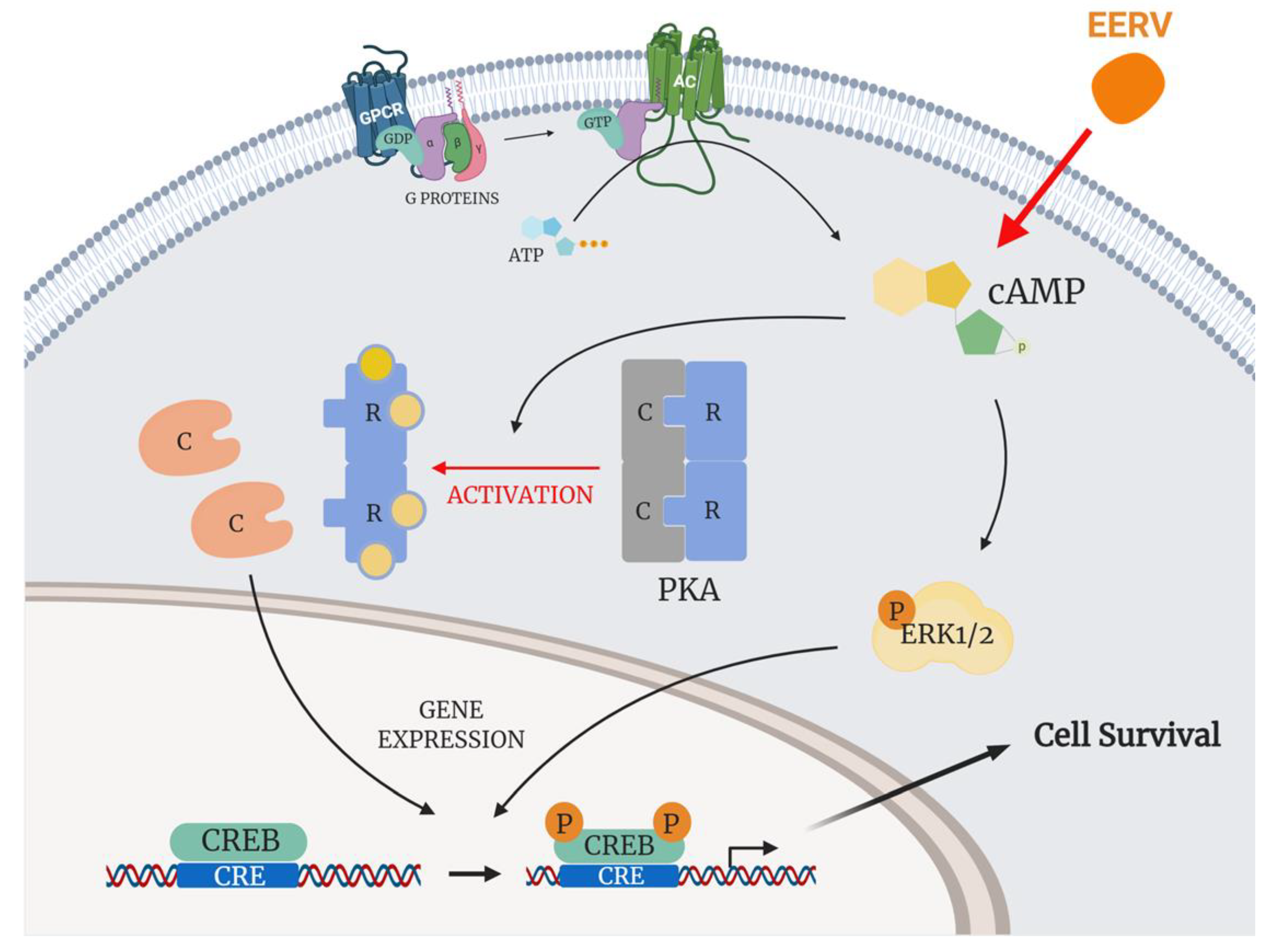
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. DNA Construction and Plasmids
4.3. Plant Material and Chemicals
4.4. Viability Assay
4.5. Fluorescence Intensity Detection
4.6. Image Acquisition and Microscopy
4.7. RT-PCR Experiment
4.8. Western Blot Experiment
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Laraia, L.; Robke, L.; Waldmann, H. Bioactive Compound Collections: From Design to Target Identification. Chem 2018, 4, 705–730. [Google Scholar] [CrossRef]
- Koehn, F.E.; Carter, G.T. The Evolving Role of Natural Products in Drug Discovery. Nat. Rev. Drug Discov. 2005, 4, 206–220. [Google Scholar] [CrossRef]
- Kingston, D.G.I. Modern Natural Products Drug Discovery and Its Relevance to Biodiversity Conservation. J. Nat. Prod. 2011, 74, 496–511. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L.; Edrada-ebel, R.; Quinn, R.J. The Re-emergence of Natural Products for Drug Discovery in the Genomics Era. Nat. Rev. Drug Discov. 2015, 14, 119–129. [Google Scholar] [CrossRef]
- Nomura, D.K.; Maimone, T.J. Target Identi Fi Cation of Bioactive Covalently Acting Natural Products. Act.-Based Protein Profiling 2018, 420, 351–374. [Google Scholar] [CrossRef]
- Kim, K.-S.; Kim, M.-J.; Lee, K.-A.; Kwon, D.-Y. Physico-Chemical Properties of Korean Traditional Soybeans. Korean J. Food Sci. Technol. 2003, 35, 335–341. [Google Scholar]
- Myung, J.; Hwang, I. Functional Components and Antioxidative Activities of Soybean Extracts. Korea Soybean Dig. 2008, 25, 23–29. [Google Scholar]
- Kim, S.J.; Shin, J.; Cho, M.; Oh, Y.; Park, N.; Lee, S.H.; Science, F.; Kim, S.J.; Shin, J.; Cho, M.; et al. Antioxidant Activity and Isoflavone Profile of Rhynchosia Nolubilis Seeds Pickled in Vinegar (Chokong). Food Sci. Biotechnol. 2007, 16, 444–450. [Google Scholar]
- Kinjo, J.; Nagao, S.; Tanaka, T.; Nonaka, G.I.; Okabe, H. Antiproliferative Constituents in the Plant 8. Seeds of Rhynchosia volubilis. Biol. Pharm. Bull. 2001, 24, 1443–1445. [Google Scholar] [CrossRef][Green Version]
- Kim, H.R.; Kim, S.; Kim, S.J.; Jeong, S.I.; Kim, S.Y. Rhynchosia Volubilis and Beta Vulgaris Modulate UV-Induced Retinal Pigment Epithelial Cells Damage and Ocular Injury in Mouse. Korean J. Pharmacogn. 2020, 51, 131–138. [Google Scholar]
- Yim, J.H.; Lee, O.; Choi, U.; Kim, Y. Antinociceptive and Anti-Inflammatory Effects of Ethanolic Extracts of Glycine Max (L.) Merr and Rhynchosia nulubilis Seeds. Int. J. Mol. Sci. 2009, 10, 4742–4753. [Google Scholar] [CrossRef]
- Jeon, J.; Kang, S.W.; Um, B.; Kim, C.Y.; Jeon, J.; Kang, S.W.; Um, B.; Kim, C.Y. Preparative Isolation of Antioxidant Flavonoids from Small Black Soybeans by Centrifugal Partition Chromatography and Sequential Solid-Phase Extraction Preparative Isolation of Antioxidant Flavonoids from Small Black Soybeans by Centrifugal Partition Chro. Sep. Sci. Technol. 2014, 49, 2756–2764. [Google Scholar] [CrossRef]
- Jeong, H.; Young, I.; Ahn, C.; Lee, S.; Gyu, H. Purification and Identification of Adipogenesis Inhibitory Peptide from Black Soybean Protein Hydrolysate. Peptides 2007, 28, 2098–2103. [Google Scholar] [CrossRef]
- Kim, J.; Um, S.J.; Woo, J.; Kim, J.Y.; Kim, H.A.; Jang, K.H.; Kang, S.A.; Lim, B.O.; Kang, I.; Choue, R.W.; et al. Comparative Effect of Seeds of Rhynchosia Volubilis and Soybean on MG-63 Human Osteoblastic Cell Proliferation and Estrogenicity. Life Sci. 2005, 78, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Park, L.; Oh, Y.; Lee, S.H.; Youn, K.; Kim, S.J.; Science, F.; Shin, J.; Park, L.; Oh, Y.; et al. Inhibition of Lipid Accumulation in 3T3-L1 Adipocytes by Extract of Seeds Pickled in Vinegar. Food Sci. Biotechnol. 2008, 17, 425–429. [Google Scholar]
- Vitro, A.I. N-Butanol Extract of Rhynchosia Volubilis Lour: A Potent Spermicidal. J. Huazhong Univ. Sci. Technol. 2014, 34, 398–402. [Google Scholar] [CrossRef]
- Guo, Y.; Yin, W.; Liu, P.; Yan, M.; Sun, Q. Separation of Isoflavones in Deer Patchouli by Ionized Liquid Extraction. J. Agric. Food Chem. 2012, 60, 3432–3440. [Google Scholar] [CrossRef]
- Hong, S.H.; Sim, M.J.; Kim, Y.C. Melanogenesis-Promoting Effects of Rhynchosia Nulubilis and Rhynchosia Volubilis Ethanol Extracts in Melan-a Cells. Toxicol. Res. 2016, 32, 141–147. [Google Scholar] [CrossRef][Green Version]
- Legumes, G.; Yoshida, K.; Sato, Y.; Okuno, R.; Kameda, K.; Isobe, M.; Kondo, T. Structural Analysis and Measurement of Anthocyanins from Colored Seed Coats of Isolation and Structural Analysis of Anthocyanins From. Biosci. Biotechnol. Biochem. 1996, 60, 589–593. [Google Scholar]
- Bae, E.-A.; Moon, G.-S. A Study on the Antioxidative Activities of Korean Soybeans. J. Korean Soc. Food Sci. Nutr. 1997, 26, 203–208. [Google Scholar]
- Kang, S.-A.; Jang, K.-H.; Cho, Y.; Hong, K.; Suh, J.-H.; Choue, R. Effects of Artificial Stomach Fluid and Digestive Enzymes on the Aglycone Isoflavone Contents of Soybean and Black Bean (Rhynchosia Molubilis: Yak-Kong). Korean J. Nutr. 2003, 36, 32–39. [Google Scholar]
- Odaka, H.; Arai, S.; Inoue, T.; Kitaguchi, T. Genetically-Encoded Yellow Fluorescent CAMP Indicator with an Expanded Dynamic Range for Dual-Color Imaging. PLoS ONE 2014, 9, e100252. [Google Scholar] [CrossRef]
- Mita, M.; Ito, M.; Harada, K.; Sugawara, I.; Ueda, H.; Tsuboi, T.; Kitaguchi, T. Green Fluorescent Protein-Based Glucose Indicators Report Glucose Dynamics in Living Cells. Anal. Chem. 2019, 91, 4821–4830. [Google Scholar] [CrossRef] [PubMed]
- Jares-erijman, E.A.; Jovin, T.M. FRET Imaging. Nat. Biotechnol. 2003, 21, 1387–1395. [Google Scholar] [CrossRef]
- Miyawaki, A. Visualization of the Spatial and Temporal Dynamics of Intracellular Signaling. Dev. Cell 2003, 4, 295–305. [Google Scholar] [CrossRef]
- Komatsu, N.; Aoki, K.; Yamada, M.; Yukinaga, H.; Fujita, Y.; Kamioka, Y.; Matsuda, M. Development of an Optimized Backbone of FRET Biosensors for Kinases and GTPases. Mol. Biol. Cell 2011, 22, 4647–4656. [Google Scholar] [CrossRef]
- Woo, S.; Kim, K.; Hyun, C.; Jae, S.; Kyeom, T. A Standardized Extract of Rhynchosia Volubilis Lour. Exerts a Protective e Ff Ect on Benzalkonium Chloride-Induced Mouse Dry Eye Model. J. Ethnopharmacol. 2018, 215, 91–100. [Google Scholar] [CrossRef]
- Matsukawa, T.; Motojima, H.; Sato, Y.; Takahashi, S.; Villareal, M.O. Upregulation of Skeletal Muscle PGC-1 α through the Elevation of Cyclic AMP Levels by Cyanidin- 3-Glucoside Enhances Exercise Performance. Nat. Publ. Gr. 2017, 7, 44799. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ha, T.; Du, Q.; Zhao, Z.; Liu, Y.; Zou, J.; Hua, W. International Immunopharmacology Genistein Protects against DSS-Induced Colitis by Inhibiting NLRP3 in Fl Ammasome via TGR5-CAMP Signaling. Int. Immunopharmacol. 2019, 71, 144–154. [Google Scholar] [CrossRef]
- Bartsch, O.; Bartlick, B.; Ivell, R. Relaxin Signalling Links Tyrosine Phosphorylation to Phosphodiesterase and Adenylyl Cyclase Activity. Mol. Hum. Reprod. 2014, 7, 799–809. [Google Scholar] [CrossRef]
- Cheng, X.; Ji, Z.; Tsalkova, T.; Mei, F. Epac and PKA: A Tale of Two Intracellular CAMP Receptors Overview of the CAMP Second Messenger System Epac, a New Intracellular CAMP Receptor. Acta Biochim. Biophys. Sin. 2008, 40, 651–662. [Google Scholar] [CrossRef]
- Introduction, I.; Neurons, A.; Endocrine, B.; Phosphodiesterases, B.; Properties, T.; Neurons, A.; Neurons, B. PKA-Dependent and PKA-Independent Pathways for CAMP-Regulated Exocytosis. Physiol. Rev. 2021, 85, 1303–1342. [Google Scholar] [CrossRef]
- Bos, J.L. Epac Proteins: Multi-Purpose CAMP Targets. Trends Biochem. Sci. 2006, 31, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Fourth, N.; Dale, E.; Augustine, G.J.; Fitzpatrick, D.; Hall, W.C.; Lamantia, A.; Mcnamara, J.O.; White, L.E. What Have I-Pods Gotta Do with It? J. Int. Neuropsychol. Soc. 2008, 14, 667–668. [Google Scholar]
- Silva, A.J.; Kogan, J.H.; Frankland, P.W. Creb and Memory. Annu. Rev. Neurosci. 1998, 21, 127–148. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.M.F. Regulation and Organization of Adenylyl Cyclases and CAMP. Biochem. J. 2003, 529, 517–529. [Google Scholar] [CrossRef]
- In, P.; Science, B. Cyclic-3 J, 5 J -Nucleotide Phosphodiesterase Isozymes in Cell Biology and Pathophysiology of the Kidney. Kidney Int. 1999, 55, 29–62. [Google Scholar] [CrossRef]
- Kotani, S.; Tanaka, H.; Yasuda, H.; Todokoro, K. Regulation of APC Activity by Phosphorylation and Regulatory Factors. J. Cell Biol. 1999, 146, 791–800. [Google Scholar] [CrossRef]
- Brushia, R.J.; Walsh, D.A. Phosphorylase Kinase: The Complexity of Its Regulation Is Reflected in the Complexity of Its Structure. Front. Biosci. 1999, 4, 618–641. [Google Scholar] [CrossRef]
- Mcdonald, B.J.; Amato, A.; Connolly, C.N.; Benke, D.; Moss, S.J.; Smart, T.G. Adjacent Phosphorylation Sites on GABA A Receptor β Subunits Determine Regulation by CAMP-Dependent Protein Kinase. Nat. Neurosci. 1998, 1, 23–28. [Google Scholar] [CrossRef]
- Zhou, R.; Cao, X.; Watson, C.; Miao, Y.; Guo, Z.; Forte, J.G.; Yao, X. Characterization of Protein Kinase A-Mediated Phosphorylation of Ezrin in Gastric Parietal Cell Activation. J. Biol. Chem. 2003, 278, 35651–35659. [Google Scholar] [CrossRef]
- Gerits, N.; Mikalsen, T.; Kostenko, S.; Shiryaev, A.; Johannessen, M.; Moens, U. Modulation of F-Actin Rearrangement by the Cyclic AMP/CAMP-Dependent Protein Kinase (PKA ) Pathway Is Mediated by MAPK-Activated Protein Kinase 5 and Requires PKA-Induced Nuclear Export of MK5. J. Biol. Chem. 2007, 282, 37232–37243. [Google Scholar] [CrossRef]
- Takimoto, E.; Soergel, D.G.; Janssen, P.M.L.; Stull, L.B.; Kass, D.A.; Murphy, A.M. Kinase A Phosphorylation Sites. Circ. Res. 2004, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Diviani, D. Modulation of Cardiac Function by A-Kinase Anchoring Proteins. Curr. Opin. Pharmacol. 2021, 8, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Phosphorylation, B.A.D.; Sastry, K.S.R.; Karpova, Y.; Prokopovich, S.; Smith, A.J.; Essau, B.; Gersappe, A.; Carson, J.P.; Weber, M.J.; Register, T.C.; et al. Epinephrine Protects Cancer Cells from Apoptosis via Activation of CAMP-Dependent Protein Kinase. J. Biol. Chem. 2007, 282, 14094–14100. [Google Scholar] [CrossRef]
- Thomson, D.M.; Herway, S.T.; Fillmore, N.; Kim, H.; Brown, J.D.; Barrow, J.R.; Winder, W.W.; Barrow, J.R.; Winder, W.W. AMP-Activated Protein Kinase Phosphorylates Transcription Factors of the CREB Family. J. Appl. Physiol. 2021, 84602, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, N.; Terai, K.; Imanishi, A.; Kamioka, Y.; Sumiyama, K. A Platform of BRET-FRET Hybrid Biosensors for Optogenetics, Chemical Screening, and in vivo Imaging. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Lynch, M.J.; Baillie, G.S.; Mohamed, A.; Li, X.; Maisonneuve, C.; Klussmann, E.; Van Heeke, G.; Houslay, M.D. RNA Silencing Identifies PDE4D5 as the Functionally Relevant CAMP Phosphodiesterase Interacting with Βarrestin to Control the Protein Kinase A/AKAP79-Mediated Switching of the Β2-Adrenergic Receptor to Activation of ERK in HEK293B2 Cells. J. Biol. Chem. 2005, 280, 33178–33189. [Google Scholar] [CrossRef]
- Lim, J.A.; Juhnn, Y.S. Isoproterenol Increases Histone Deacetylase 6 Expression and Cell Migration by Inhibiting ERK Signaling via PKA and Epac Pathways in Human Lung Cancer Cells. Exp. Mol. Med. 2016, 48, e204–e211. [Google Scholar] [CrossRef]
- Di Benedetto, G.; Zoccarato, A.; Lissandron, V.; Terrin, A.; Li, X.; Houslay, M.D.; Baillie, G.S.; Zaccolo, M. Protein Kinase A Type i and Type II Define Distinct Intracellular Signaling Compartments. Circ. Res. 2008, 103, 836–844. [Google Scholar] [CrossRef]
- Salehi, B.; Cappellini, F.; Zorzan, D.; Imran, M.; Sener, B.; Kilic, M.; El-shazly, M.; Fahmy, N.M.; Al-sayed, E.; Martorell, M.; et al. The Therapeutic Potential of Anthocyanins: Current Approaches Based on Their Molecular Mechanism of Action. Front. Pharmacol. 2020, 11, 1300. [Google Scholar] [CrossRef]
- Zhu, W.; Jia, Q.; Wang, Y.; Zhang, Y.; Xia, M. Free Radical Biology & Medicine The Anthocyanin Cyanidin-3-O–β–Glucoside, a Fl Avonoid, Increases Hepatic Glutathione Synthesis and Protects Hepatocytes against Reactive Oxygen Species during Hyperglycemia: Involvement of a CAMP–PKA-Dependent Sig. Free Radic. Biol. Med. 2012, 52, 314–327. [Google Scholar] [CrossRef]
- Fang, J.; Luo, Y.; Jin, S.; Yuan, K.; Guo, Y. Ameliorative e Ff Ect of Anthocyanin on Depression Mice by Increasing Monoamine Neurotransmitter and Up-Regulating BDNF Expression. J. Funct. Foods 2020, 66, 103757. [Google Scholar] [CrossRef]
- Guo, Y.; Pan, W.; Liu, S.B.; Shen, Z.; Xu, Y.; Hu, L.L. ERK/MAPK Signalling Pathway and Tumorigenesis (Review). Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef] [PubMed]
- Chambard, J.; Lefloch, R.; Pouysségur, J.; Lenormand, P. ERK Implication in Cell Cycle Regulation. Biochim. Biophys. Acta 2007, 1773, 1299–1310. [Google Scholar] [CrossRef]
- Cosgaya, J.M.; Scsucova, S.; Aranda, A. Rapid Effects of Retinoic Acid on CREB and ERK Phosphorylation in Neuronal Cells. Mol. Biol. Cell 2004, 15, 5583–5592. [Google Scholar] [CrossRef]
- Shaywitz, A.J.; Greenberg, M.E. Creb: A Stimulus -Induced Transcription Factor Activated by Adiverse Array of Extracellular. Annu. Rev. Biochem. 1999, 68, 821–861. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.L.; Cha, Y.Y.; Lee, E.S. Effect of Germinated Seed of Rhynchosia Volubilis on Cathepsin-K in Ovariectomized Rats. Korean J. Oreintal Physiol. Pathlogy 2005, 19, 184–190. [Google Scholar]
- Memarzadeh, E.; Luther, T.; Heidari-soureshjani, S. Effect and Mechanisms of Medicinal Plants on Dry Eye Disease: A Systematic Review. J. Clin. Diagn. Res. 2018, 12, 10–13. [Google Scholar] [CrossRef]
- Tareq, M.; Khan, H.; Ather, A. Molecules from Nature: Modulating the Expression of Estrogen Receptor Genes in Breast Cancer Cells. Brain Behav. Immun. 2006, 143–150. [Google Scholar]
- Kolmus, K.; Tavernier, J.; Gerlo, S. Β2-Adrenergic Receptors in Immunity and Inflammation: Stressing NF-ΚB. Brain Behav. Immun. 2015, 45, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Spadari, R.C.; Cavadas, C.; de Carvalho, A.E.T.S.; Ortolani, D.; de Moura, A.L.; Vassalo, P.F. Role of Beta-Adrenergic Receptors and Sirtuin Signaling in the Heart during Aging, Heart Failure, and Adaptation to Stress. Cell. Mol. Neurobiol. 2018, 38, 109–120. [Google Scholar] [CrossRef] [PubMed]
- He, J.J.; Zhang, W.H.; Liu, S.L.; Chen, Y.F.; Liao, C.X.; Shen, Q.Q.; Hu, P. Activation of β-Adrenergic Receptor Promotes Cellular Proliferation in Human Glioblastoma. Oncol. Lett. 2017, 14, 3846–3852. [Google Scholar] [CrossRef][Green Version]
- Johnson, M. Molecular Mechanisms of Β2-Adrenergic Receptor Function, Response, and Regulation. J. Allergy Clin. Immunol. 2006, 117, 18–24. [Google Scholar] [CrossRef]
- Irannejad, R.; Tomshine, J.C.; Tomshine, J.R.; Chevalier, M.; Mahoney, J.P.; Steyaert, J.; Rasmussen, S.G.F.; Sunahara, R.K.; El-Samad, H.; Huang, B.; et al. Conformational Biosensors Reveal GPCR Signalling from Endosomes. Nature 2013, 495, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Di Certo, M.G.; Batassa, E.M.; Casella, I.; Serafino, A.; Floridi, A.; Passananti, C.; Molinari, P.; Mattei, E. Delayed Internalization and Lack of Recycling in a Beta2- Adrenergic Receptor Fused to the G Protein Alpha-Subunit. BMC Cell Biol. 2008, 9, 56. [Google Scholar] [CrossRef]
- Lohse, M.J.; Calebiro, D. Cell Biology: Receptor Signals Come in Waves. Nature 2013, 495, 457–458. [Google Scholar] [CrossRef] [PubMed]
- Ordo, R.M.; Ordo, A.A.L. Design and Quality Control of a Pharmaceutical Formulation Containing Natural Products with Antibacterial, Antifungal and Antioxidant Properties. Int. J. Pharm. 2009, 378, 51–58. [Google Scholar] [CrossRef]
- Ivanova, E.V.; Figueroa, R.A.; Gatsinzi, T.; Hallberg, E.; Iverfeldt, K. Anchoring of FRET Sensors-A Requirement for Spatiotemporal Resolution. Sensors 2016, 16, 703. [Google Scholar] [CrossRef]
- Hochreiter, B.; Garcia, A.P.; Schmid, J.A. Fluorescent Proteins as Genetically Encoded FRET Biosensors in Life Sciences. Sensors 2015, 15, 26281–26314. [Google Scholar] [CrossRef] [PubMed]
- Hyun, K.; Kim, K.; Woo, S.; Tasnim, S.; Beom, J. Small Black Bean (Rhynchosia Volubilis) Extract Ameliorates Gut Microbial and Metabolic Perturbation in Ovariectomized Mice. J. Funct. Foods 2019, 60, 103415. [Google Scholar] [CrossRef]
- Xie, L.; Su, H.; Sun, C.; Zheng, X.; Chen, W. Trends in Food Science & Technology Recent Advances in Understanding the Anti-Obesity Activity of Anthocyanins and Their Biosynthesis in Microorganisms. Trends Food Sci. Technol. 2018, 72, 13–24. [Google Scholar] [CrossRef]
- Xie, W.; Meng, X.; Zhai, Y.; Zhou, P.; Ye, T.; Wang, Z.; Sun, G.; Sun, X. Panax Notoginseng Saponins: A Review of Its Mechanisms of Antidepressant or Anxiolytic Effects and Network Analysis on Phytochemistry. Molecules 2018, 23, 940. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, J.; Liu, F.; Tong, L.; Chen, Z.; Chen, J.; He, H. Neuroprotective e Ff Ects of Anthocyanins and Its Major Component Cyanidin- 3-O-Glucoside ( C3G ) in the Central Nervous System: An Outlined Review. Eur. J. Pharmacol. 2019, 858, 172500. [Google Scholar] [CrossRef]
- Edirisinghe, I.; Banaszewski, K.; Cappozzo, J.; Sandhya, K.; Ellis, C.L.; Tadapaneni, R.; Kappagoda, C.T.; Burton-freeman, B.M. Strawberry Anthocyanin and Its Association with Postprandial Inflammation and Insulin British Journal of Nutrition. Br. J. Nutr. 2011, 106, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Jamar, G.; Boveto, A.; Vales, L.; De Cássia, H.; Missae, L.; Vera, V.; Rosso, D.; Pellegrini, L. Bi Fi Dobacterium Spp. Reshaping in the Gut Microbiota by Low Dose of Juçara Supplementation and Hypothalamic Insulin Resistance in Wistar Rats. J. Funct. Foods 2018, 46, 212–219. [Google Scholar] [CrossRef]
- Fabroni, S.; Ballistreri, G.; Amenta, M.; Romeo, F.V.; Rapisarda, P. Screening of the Anthocyanin Profile and in vitro Pancreatic Lipase Inhibition by Anthocyanin-Containing Extracts of Fruits, Vegetables, Legumes and Cereals. J. Sci. Food Agric. 2016, 96, 4713–4723. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.W.; Kennedy, D.O.; Haskell, C.F.; Scheepens, A. A Double Blind Placebo Controlled Study Measuring the Effect of Two Berry Fruit Extracts on Mood, Cognition and Monoamine Oxidase B Inhibition in Healthy Young Adults. Appetite 2012, 59, 2012. [Google Scholar] [CrossRef]
- Walton, M.R.; Dragunow, M. Is CREB a Key to Neuronal Survival? Trends Neurosci. 2000, 23, 48–53. [Google Scholar] [CrossRef]
- Yamada, M.; Tanabe, K.; Wada, K.; Shimoke, K.; Ishikawa, Y.; Ikeuchi, T.; Koizumi, S.; Hatanaka, H. Differences in Survival-Promoting Effects and Intracellular Signaling Properties of BDNF and IGF-1 in Cultured Cerebral Cortical Neurons. J. Neurochem. 2001, 78, 940–951. [Google Scholar] [CrossRef] [PubMed]
- Bonni, A.; Brunet, A.; West, A.E.; Datta, S.R.; Takasu, M.A.; Greenberg, M.E. Cell Survival Promoted by the Ras-MAPK Signaling Pathway by Transcription-Dependent and -Independent Mechanisms. Science 1999, 286, 1358–1363. [Google Scholar] [CrossRef]
- Du, K.; Montminy, M. Communication CREB Is a Regulatory Target for the Protein Kinase Akt/PKB. J. Biol. Chem. 1998, 273, 32377–32379. [Google Scholar] [CrossRef]
- Mayr, B.; Montminy, M. Transcriptional Regulation by the Phosphorylation- Dependent Factor Creb. Nat. Rev. Mol. Cell Biol. 2001, 2, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Hillman, G.G.; Singh-gupta, V. Free Radical Biology & Medicine Soy Iso Fl Avones Sensitize Cancer Cells to Radiotherapy. Free Radic. Biol. Med. 2011, 51, 289–298. [Google Scholar] [CrossRef]
- Liu, D.; Jiang, H.; Grange, R. Monophosphate Signaling Pathway in Vascular Endothelial Cells and Protects Endothelial. Endocrinology 2005, 146, 1312–1320. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.A.; Yu, J.Z.; Donati, R.J.; Rasenick, M.M. β-Adrenergic Receptor Stimulation Promotes Gαs Internalization through Lipid Rafts: A Study in Living Cells. Mol. Pharmacol. 2005, 67, 1493–1504. [Google Scholar] [CrossRef]
- Tao, J.; Wang, H.Y.; Malbon, C.C. Protein Kinase A Regulates AKAP250 (Gravin) Scaffold Binding to the Β2-Adrenergic Receptor. EMBO J. 2003, 22, 6419–6429. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahn, S.-H.; Suh, J.-S.; Jang, Y.-K.; Kim, H.-S.; Choi, G.-H.; Kim, E.; Kim, T.-J. Rhynchosia volubilis Promotes Cell Survival via cAMP-PKA/ERK-CREB Pathway. Pharmaceuticals 2022, 15, 73. https://doi.org/10.3390/ph15010073
Ahn S-H, Suh J-S, Jang Y-K, Kim H-S, Choi G-H, Kim E, Kim T-J. Rhynchosia volubilis Promotes Cell Survival via cAMP-PKA/ERK-CREB Pathway. Pharmaceuticals. 2022; 15(1):73. https://doi.org/10.3390/ph15010073
Chicago/Turabian StyleAhn, Sang-Hyun, Jung-Soo Suh, Yoon-Kwan Jang, Heon-Su Kim, Gyu-Ho Choi, Eunhye Kim, and Tae-Jin Kim. 2022. "Rhynchosia volubilis Promotes Cell Survival via cAMP-PKA/ERK-CREB Pathway" Pharmaceuticals 15, no. 1: 73. https://doi.org/10.3390/ph15010073
APA StyleAhn, S.-H., Suh, J.-S., Jang, Y.-K., Kim, H.-S., Choi, G.-H., Kim, E., & Kim, T.-J. (2022). Rhynchosia volubilis Promotes Cell Survival via cAMP-PKA/ERK-CREB Pathway. Pharmaceuticals, 15(1), 73. https://doi.org/10.3390/ph15010073









