Structured Waters Mediate Small Molecule Binding to G-Quadruplex Nucleic Acids
Abstract
:1. Introduction
2. Results
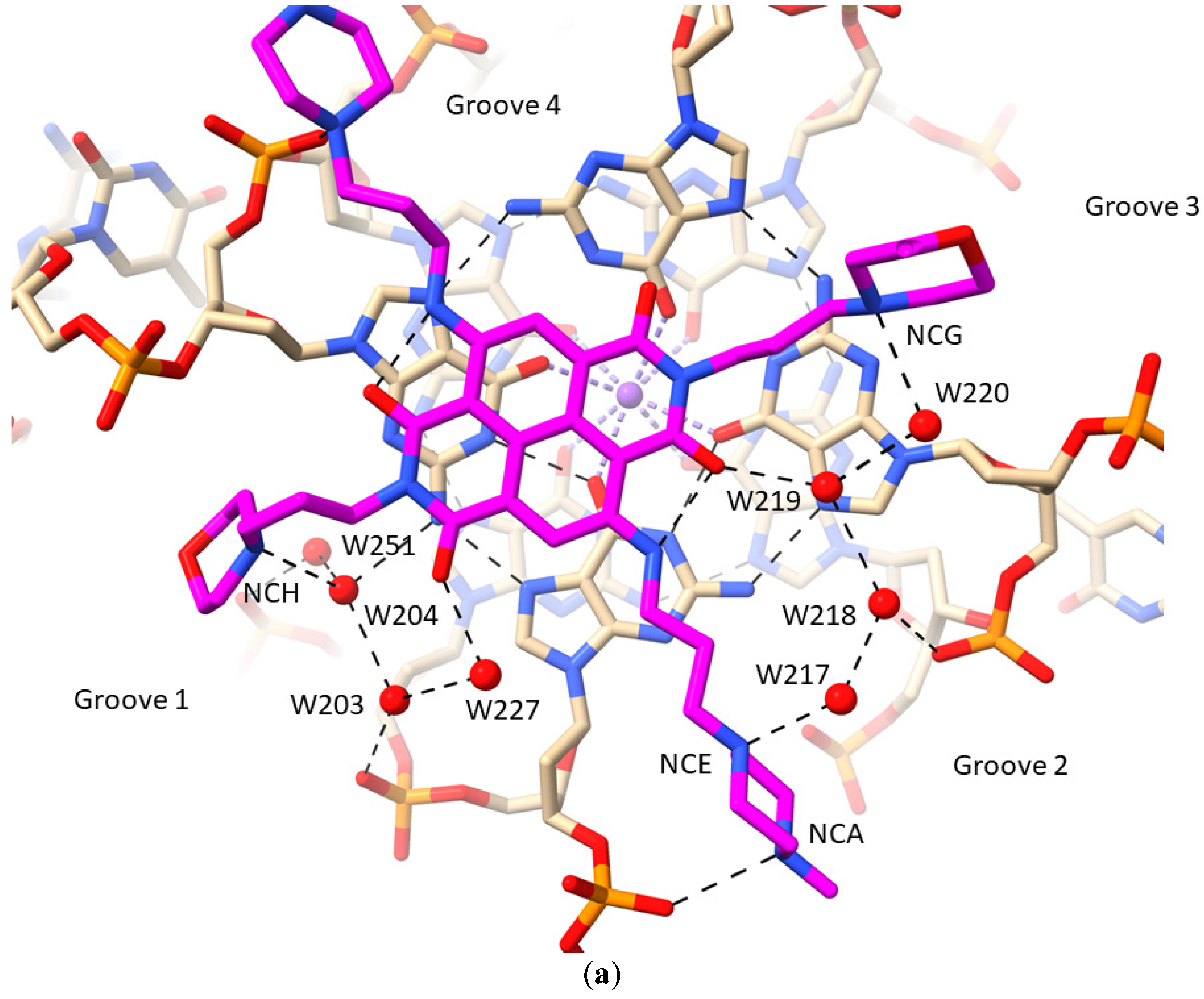
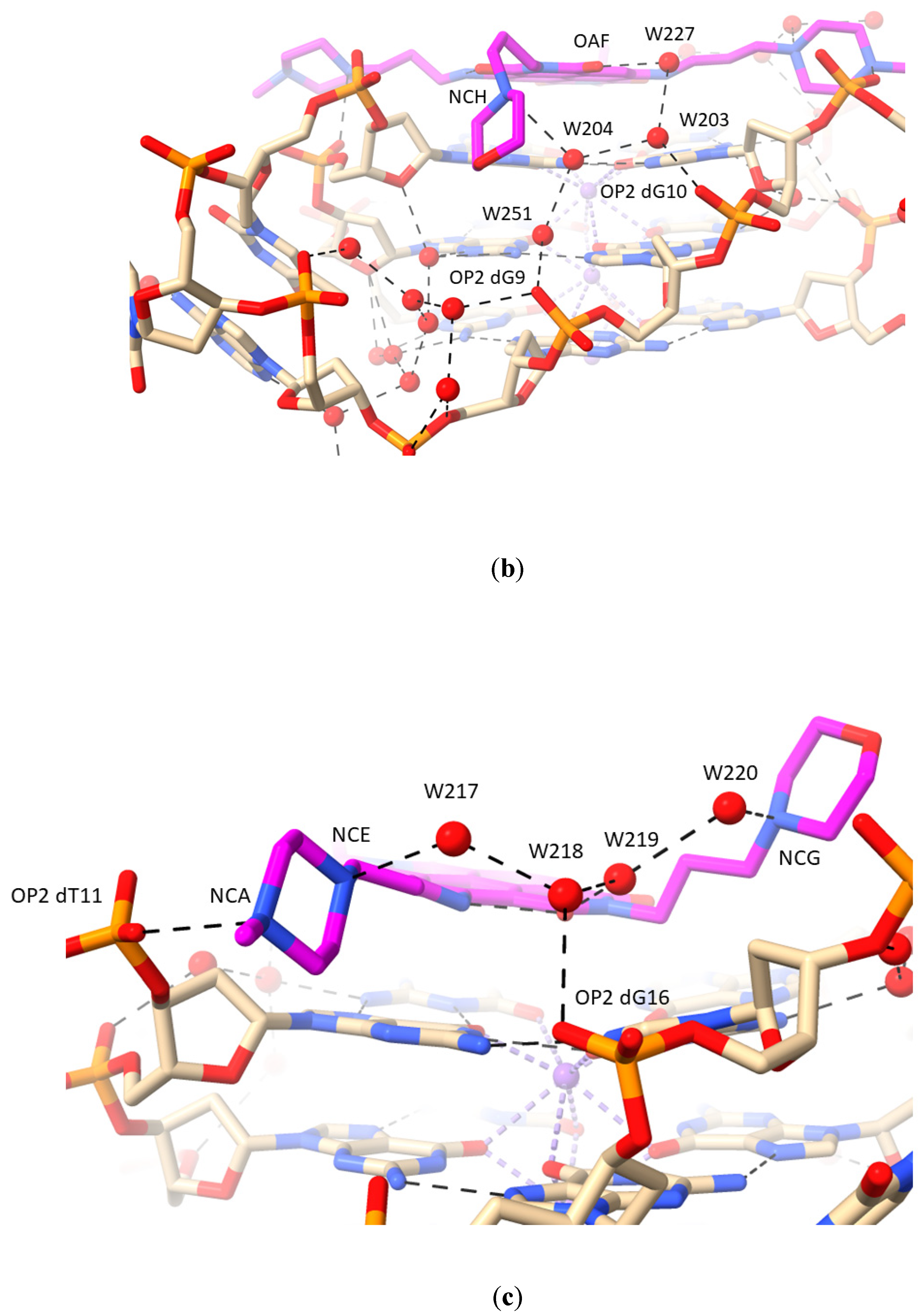
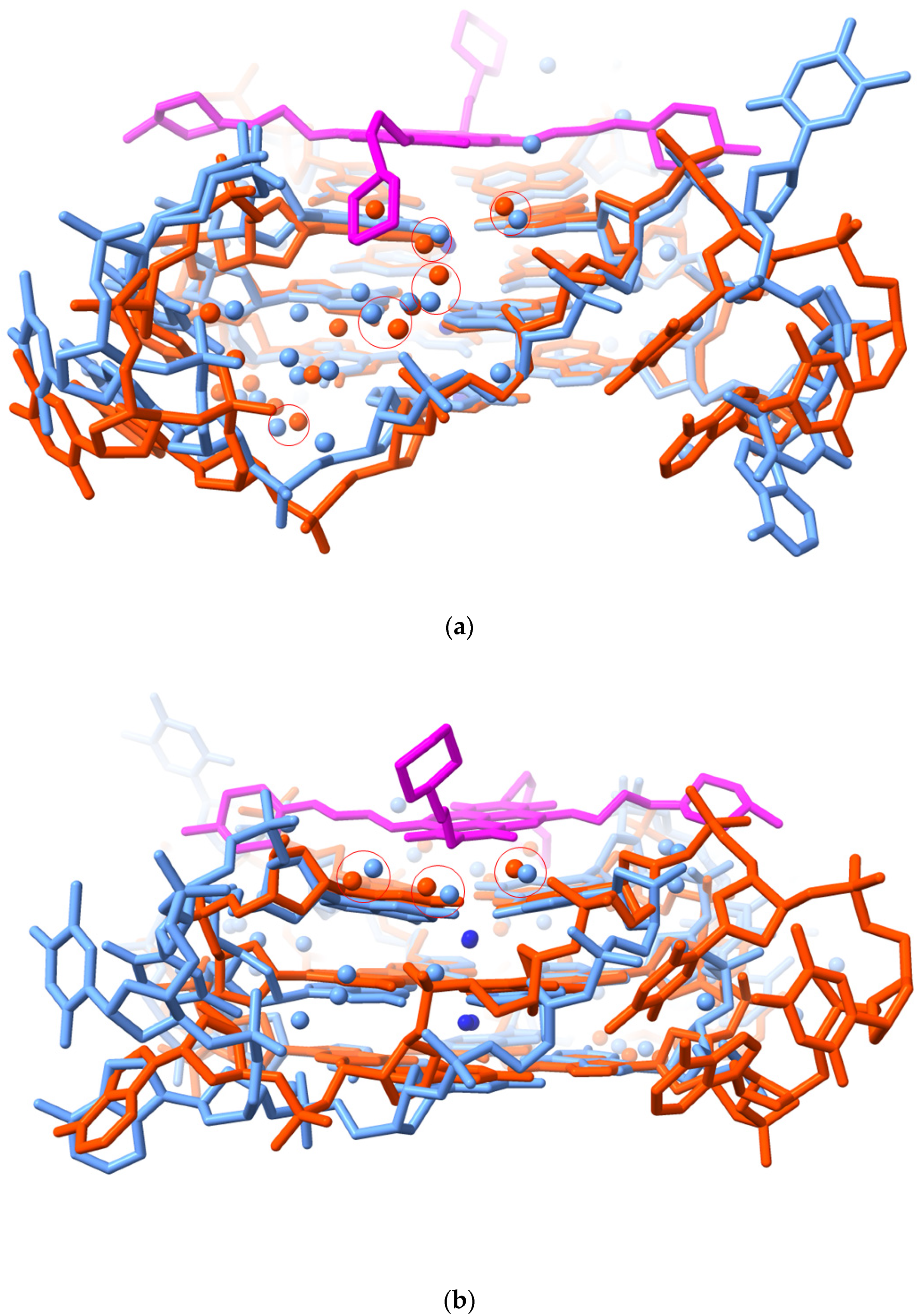
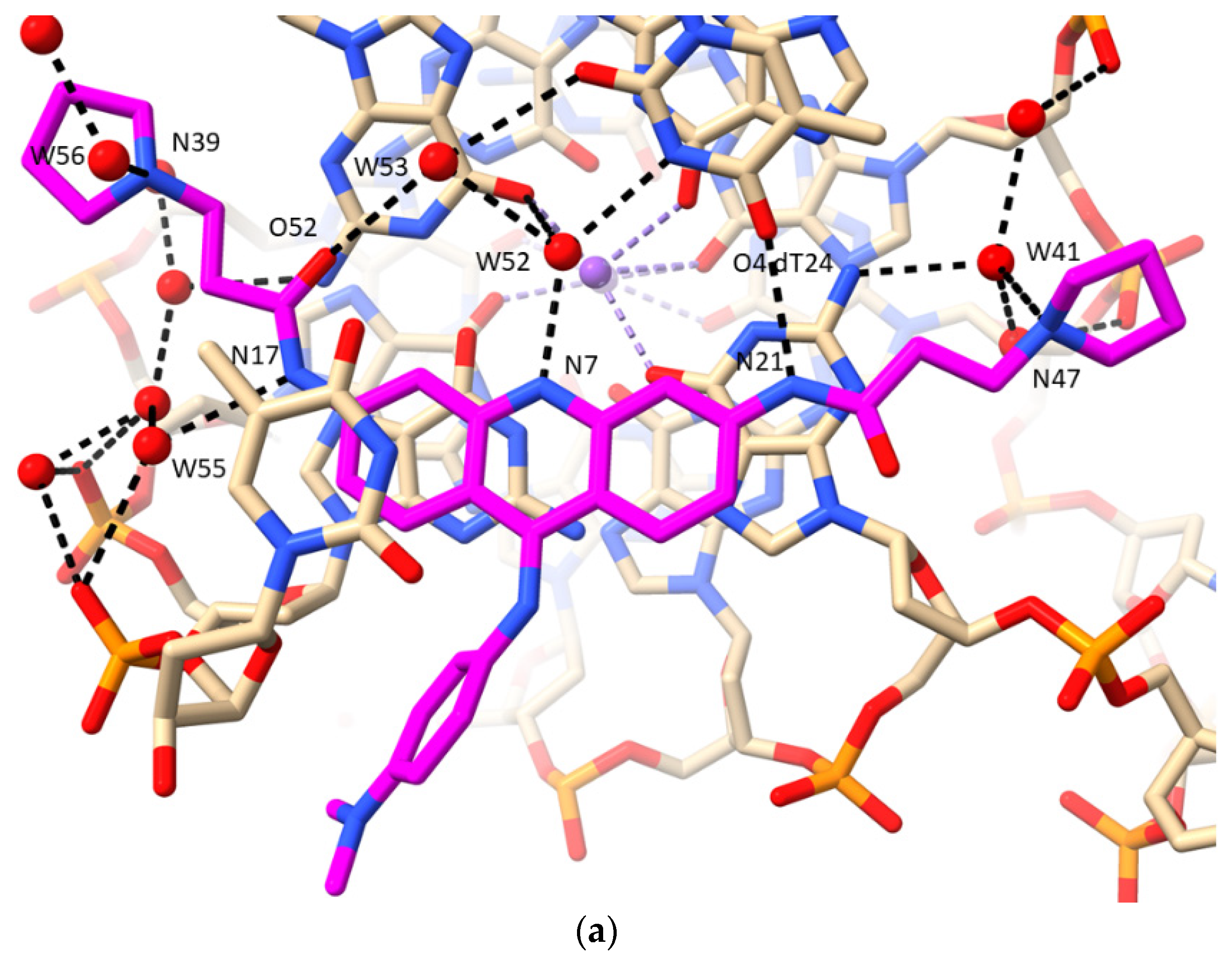
2.1. MM41 Side Chain Contacts and Water Environment
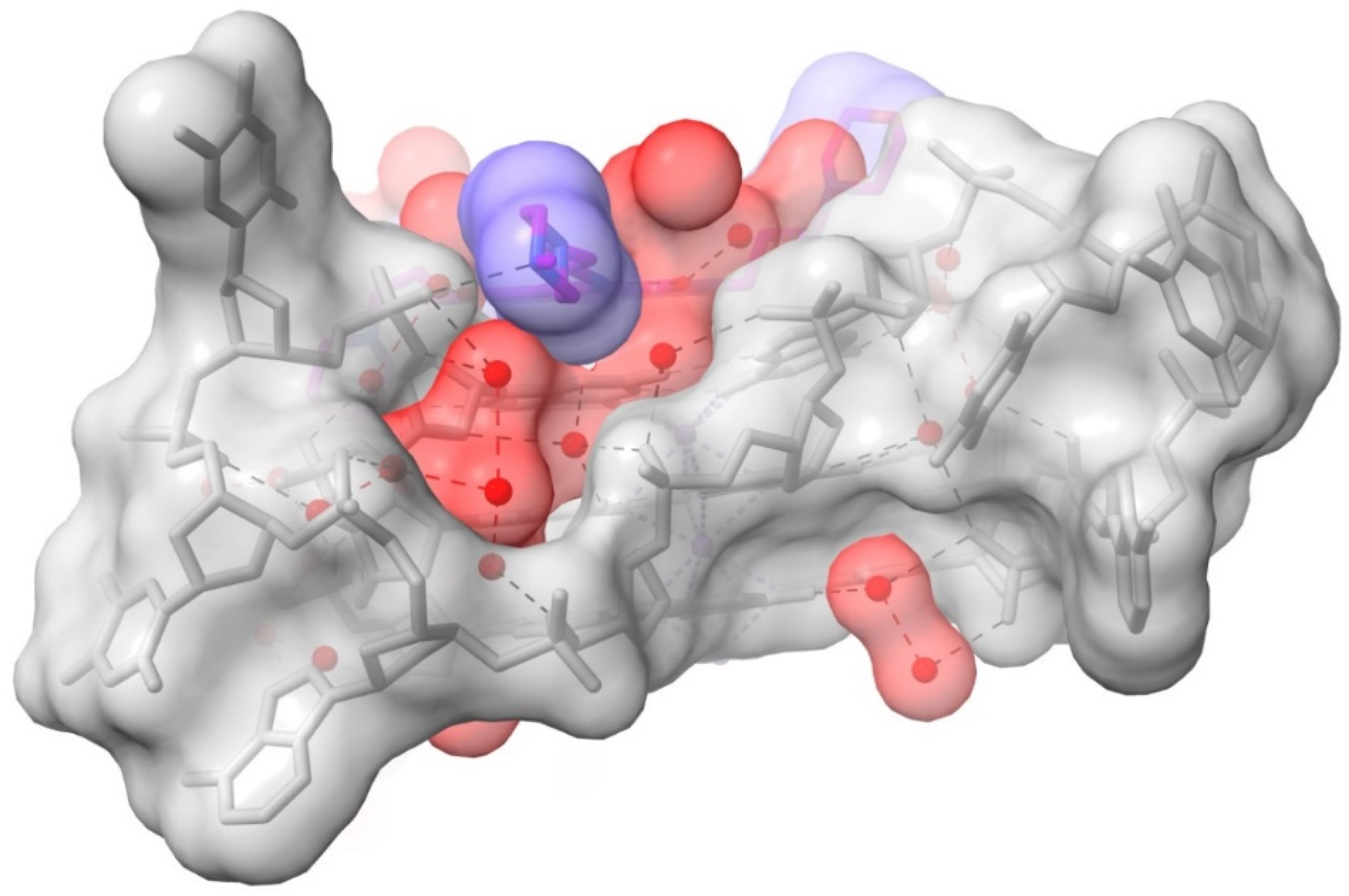
2.2. MM41 and Water Mobility
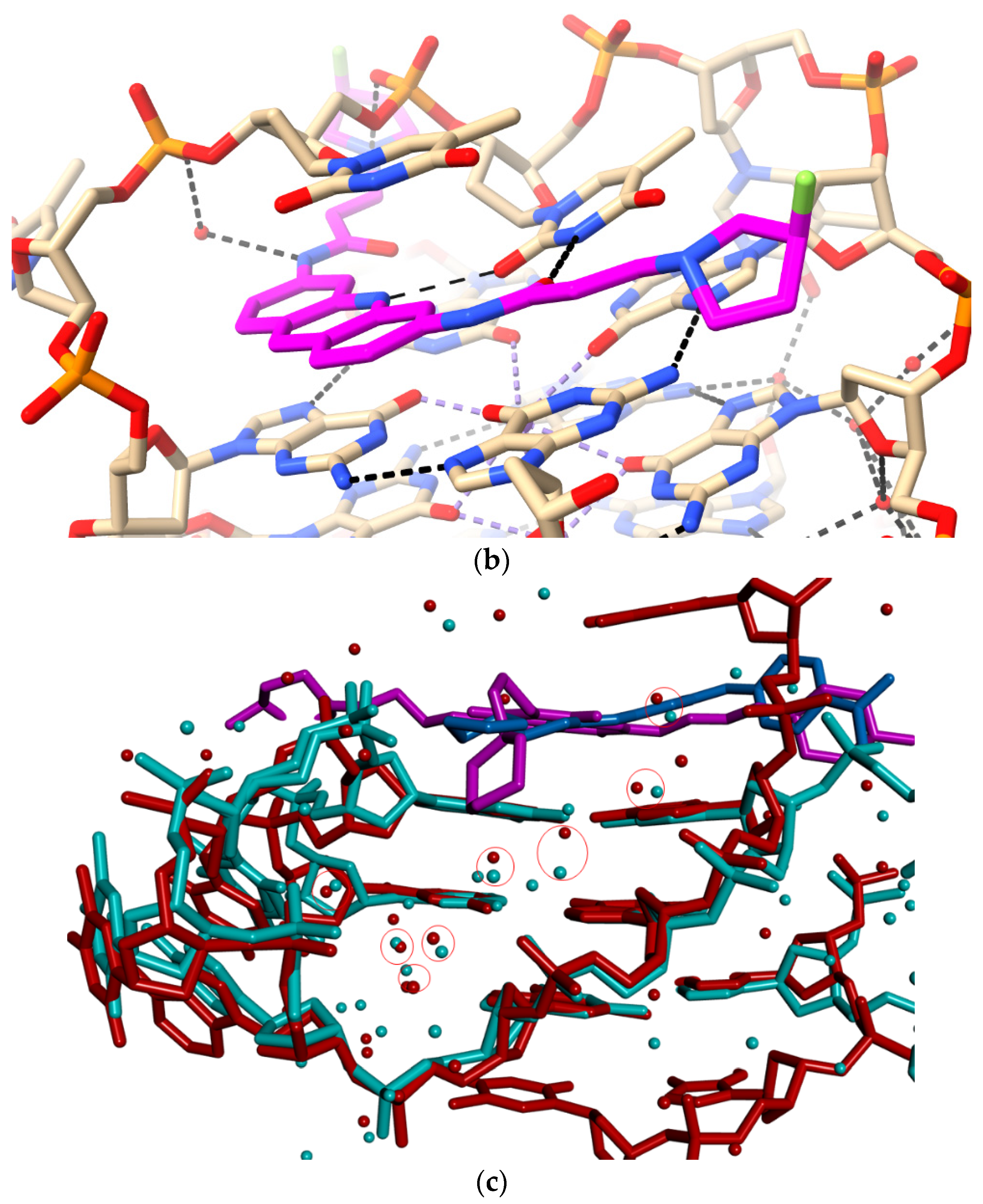
2.3. Water Mediation in Acridine-G-Quadruplex Structures
3. Discussion
- The morpholino end groups of MM41, which are assumed to be basic in the buffering conditions of the crystallization experiment and in biological solution, do not directly contact the GQ. Hydrogen bonding/electrostatic interactions with negative backbone phosphate groups were anticipated but were not observed. Instead, the basic ring nitrogen in each morpholino group hydrogen bonds to one of a group of four water molecules positioned in the mouth of the relevant grooves (1 and 3). The waters are in hydrogen bond contact with backbone phosphates. Similarly, the basic pyrrolidino side chain terminal groups of BRACO19 do not directly contact phosphate groups in its GQ complex, with water mediation being observed in the crystal environment.
- A nitrogen atom on both N-methyl-piperazine groups of MM41, by contrast, directly hydrogen bonds to a backbone phosphate oxygen atom, implying greater basicity than morpholino for this end group.
- The water clusters associated with the two morpholino groups of MM41 are highly conserved between the native and the MM41-bound GQ structures. There is also conservation of a number of the ligand-associated waters between the MM41 and BRACO19 structures, and by implication, between the native and BRACO19 structures.
4. Materials and Methods
- Resolution ≤ 2.5 Å,
- Having at least one water molecule contacting a ligand,
- Hydrogen bonds were accepted in a structure if
- donor-acceptor distances ≤3.25 Å
- donor-hydrogen…acceptor angles were ≤30° from ideality, and
- 4.
- Relevance to current drug discovery.
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [Green Version]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [Green Version]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA secondary structures: Stability and function of G-quadruplex structures. Nature Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The structure and function of DNA G-quadruplexes. Trends Chem. 2019, 2, 123–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winnerdy, F.R.; Phan, A.T. Quadruplex structure and diversity. Ann. Rep. Med. Chem. 2020, 54, 45–73. [Google Scholar]
- Todd, A.K.; Johnston, M.; Neidle, S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, E.; Hardin, C.C.; Walk, S.K.; Tinoco, I., Jr.; Blackburn, E.H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Biochemistry 1987, 51, 899–908. [Google Scholar]
- Williamson, J.R. G-quartet structures in telomeric DNA. Ann. Rev. Biophys. 1994, 23, 703–730. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [Green Version]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nature Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef]
- Spiegel, J.; Cuesta, S.M.; Adhikari, S.; Hänsel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol. 2021, 22, 117. [Google Scholar] [CrossRef] [PubMed]
- Hansel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nature Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef] [Green Version]
- Huppert, J.L.; Bugaut, A.; Kumari, S.; Balasubramanian, S. G-quadruplexes: The beginning and end of UTRs. Nucleic Acids Res. 2008, 36, 6260–6268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bugaut, A.; Balasubramanian, S. 5′-UTR RNA G-quadruplexes: Translation regulation and targeting. Nucleic Acids Res. 2012, 40, 4727–4741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.S.M.; Ghanem, L.R.; Barash, Y. Integrative analysis reveals RNA G-quadruplexes in UTRs are selectively constrained and enriched for functional associations. Nat. Commun. 2020, 11, 527. [Google Scholar] [CrossRef] [Green Version]
- Marchetti, C.; Zyner, K.G.; Ohnmacht, S.A.; Robson, M.; Haider, S.M.; Morton, J.P.; Marsico, G.; Vo, T.; Laughlin-Toth, S.; Ahmed, A.A.; et al. Targeting multiple effector pathways in pancreatic ductal adenocarcinoma with a G-quadruplex-binding small molecule. J. Med. Chem. 2018, 61, 2500–2517. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.A.; Angell, R.; Oxenford, S.; Worthington, J.; Williams, N.; Barton, N.; Fowler, T.G.; O’Flynn, D.E.; Sunose, M.; McConville, M.; et al. Asymmetrically substituted quadruplex-binding naphthalene diimide showing potent activity in pancreatic cancer models. ACS Med. Chem. Lett. 2020, 11, 1634–1644. [Google Scholar] [CrossRef]
- Zyner, K.G.; Mulhearn, D.S.; Adhikari, S.; Martínez Cuesta, S.; Di Antonio, M.; Erard, N.; Hannon, G.J.; Tannahill, D.; Balasubramanian, S. Genetic interactions of G-quadruplexes in humans. eLife 2019, 8, e46793. [Google Scholar] [CrossRef] [PubMed]
- Hänsel-Hertsch, R.; Simeone, A.; Shea, A.; Hui, W.W.I.; Zyner, K.G.; Marsico, G.; Rueda, O.M.; Bruna, A.; Martin, A.; Zhang, X.; et al. Landscape of G-quadruplex DNA structural regions in breast cancer. Nature Genet. 2020, 52, 878–883. [Google Scholar] [CrossRef]
- Shen, J.; Varshney, D.; Simeone, A.; Zhang, X.; Adhikari, S.; Tannahill, D.; Balasubramanian, S. Promoter G-quadruplex folding precedes transcription and is controlled by chromatin. Genome Biol. 2021, 22, 143. [Google Scholar] [CrossRef]
- Perrone, R.; Nadai, M.; Poe, J.A.; Frasson, I.; Palumbo, M.; Palù, G.; Smithgall, T.E.; Richter, S.N. Formation of a unique cluster of G-quadruplex structures in the HIV-1 Nef coding region: Implications for antiviral activity. PLoS ONE 2013, 8, e73121. [Google Scholar]
- Ruggiero, E.; Richter, S.N. Viral G-quadruplexes: New frontiers in virus pathogenesis and antiviral therapy. Ann. Rep. Med. Chem. 2020, 54, 101–131. [Google Scholar]
- Abiri, A.; Lavigne, M.; Rezaei, M.; Nikzad, S.; Zare, P.; Mergny, J.-L.; Rahimi, H.R. Unlocking G-quadruplexes as antiviral targets. Pharmacol. Rev. 2021, 73, 897–923. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Qin, G.; Niu, J.; Wang, Z.; Wang, C.; Ren, J.; Qu, X. Targeting RNA G-quadruplex in SARS-CoV-2: A promising therapeutic target for COVID-19? Angew. Chem. Int. Ed. Engl. 2021, 60, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Belmonte-Reche, E.; Serrano-Chacón, I.; Gonzalez, C.; Gallo, J.; Bañobre-López, M. Potential G-quadruplexes and i-Motifs in the SARS-CoV-2. PLoS ONE 2021, 16, e0250654. [Google Scholar] [CrossRef] [PubMed]
- Rawal, P.; Kummarasetti, V.B.; Ravindran, J.; Kumar, N.; Halder, K.; Sharma, R.; Mukerji, M.; Das, S.K.; Chowdhury, S. Genome-wide prediction of G4 DNA as regulatory motifs: Role in Escherichia coli global regulation. Genome Res. 2006, 16, 644–655. [Google Scholar] [CrossRef] [Green Version]
- Holder, I.T.; Hartig, J.S. A matter of location: Influence of G-quadruplexes on Escherichia coli gene expression. Chem. Biol. 2014, 21, 1511–1521. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Wojtowicz, D.; Bowers, A.A.; Levens, D.; Benham, C.J.; Przytycka, T.M. The genome-wide distribution of non-B DNA motifs is shaped by operon structure and suggests the transcriptional importance of non-B DNA structures in Escherichia coli. Nucleic Acids Res. 2014, 41, 5965–5977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, P.; Kim, N.; Kumari, M.; Verma, S.; Sharma, T.K.; Yadav, V.; Kumar, A. G-quadruplex structures in bacteria—Biological relevance and potential as antimicrobial target. J. Bacteriol. 2021, 203, e0057720. [Google Scholar] [CrossRef] [PubMed]
- Smargiasso, N.; Gabelica, V.; Damblon, C.; Rosu, F.; De Pauw, E.; Teulade-Fichou, M.-P.; Rowe, J.A.; Claessens, A. Putative DNA G-quadruplex formation within the promoters of Plasmodium falciparum var genes. BMC Genom. 2009, 10, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, L.M.; Monsell, K.R.; Noulin, F.; Famodimu, M.T.; Smargiasso, N.; Damblon, C.; Horrocks, P.; Merrick, C.J. G-quadruplex DNA motifs in the malaria parasite plasmodium falciparum and their potential as novel antimalarial drug targets. Antimicrob. Agents Chemother. 2018, 62, e01828-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gazanion, E.; Lacroix, L.; Alberti, P.; Gurung, P.; Wein, S.; Cheng, M.; Mergny, J.-L.; Gomes, A.R.; Lopez-Rubio, J.J. Genome wide distribution of G-quadruplexes and their impact on gene expression in malaria parasites. PLoS Genet. 2020, 16, e1008917. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Henderson, A.; Wu, Y.; Huang, Y.C.; Chavez, E.A.; Platt, J.; Johnson, F.B.; Brosh, R.M., Jr.; Sen, D.; Lansdorp, P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2013, 42, 860–869. [Google Scholar] [CrossRef] [Green Version]
- Biffi, G.; Di Antonio, M.; Tannahill, D.; Balasubramanian, S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014, 6, 75–80. [Google Scholar] [CrossRef]
- Tseng, T.-Y.; Chien, C.-H.; Chu, J.-F.; Huang, W.-C.; Lin, M.-Y.; Chang, C.-C.; Chang, T.-C. Fluorescent probe for visualizing guanine-quadruplex DNA by fluorescence lifetime imaging microscopy. J. Biomed. Opt. 2013, 18, 101309. [Google Scholar] [CrossRef] [Green Version]
- Laguerre, A.; Hukezalie, K.; Winckler, P.; Katranji, F.; Chanteloup, G.; Pirrotta, M.; Perrier-Cornet, J.-M.; Wong, J.M.Y.; Monchaud, D. Visualization of RNA-quadruplexes in live cells. J. Am. Chem. Soc. 2015, 137, 8521–8525. [Google Scholar] [CrossRef]
- Summers, P.A.; Lewis, B.W.; Gonzalez-Garcia, J.; Porreca, R.M.; Lim, A.H.M.; Cadinu, P.; Martin-Pintado, N.; Mann, D.J.; Edel, J.B.; Vannier, J.B.; et al. Visualising G-quadruplex DNA dynamics in live cells by fluorescence lifetime imaging microscopy. Nat. Commun. 2021, 12, 162. [Google Scholar] [CrossRef]
- Di Antonio, M.; Ponjavic, A.; Radzevičius, A.; Ranasinghe, R.T.; Catalano, M.; Zhang, X.; Shen, J.; Needham, L.M.; Lee, S.F.; Klenerman, D.; et al. Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat. Chem. 2020, 12, 832–837. [Google Scholar] [CrossRef]
- Long, W.; Zheng, B.-X.; Huang, X.-H.; She, M.-T.; Liu, A.-L.; Zhang, K.; Wong, W.-L.; Lu, Y.-J. Molecular recognition and imaging of human telomeric G-quadruplex DNA in live cells: A systematic advancement of thiazole orange scaffold to enhance binding specificity and inhibition of gene expression. J. Med. Chem. 2021, 64, 2125–2138. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, J.; Wild, A.T.; Wu, W.H.; Shah, R.; Danussi, C.; Riggins, G.J.; Kannan, K.; Sulman, E.P.; Chan, T.A.; et al. G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat. Commun. 2019, 10, 943. [Google Scholar] [CrossRef] [Green Version]
- Lerner, L.K.; Sale, J.E. Replication of G quadruplex DNA. Genes 2019, 10, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amato, J.; Miglietta, G.; Morigi, R.; Iaccarino, N.; Locatelli, A.; Leoni, A.; Novellino, E.; Pagano, B.; Capranico, G.; Randazzo, A. Monohydrazone based G-quadruplex selective ligands induce DNA damage and genome instability in human cancer cells. J. Med. Chem. 2020, 63, 3090–3103. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug. Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigo, R.; Palumbo, M.; Sissi, C. G-quadruplexes in human promoters: A challenge for therapeutic applications. Biochim. Biophys. Acta 2017, 1861, 1399–1413. [Google Scholar] [CrossRef]
- Song, J.H.; Kang, H.-J.; Luevano, L.A.; Gokhale, V.; Wu, K.; Pandey, R.; Sherry Chow, H.-H.; Hurley, L.H.; Kraft, A.S. Small-molecule-targeting hairpin loop of hTERT promoter G-quadruplex induces cancer cell death. Cell Chem. Biol. 2019, 26, 1110–1121. [Google Scholar] [CrossRef]
- Boddupally, P.V.; Hahn, S.; Beman, C.; De, B.; Brooks, T.A.; Gokhale, V.; Hurley, L.H. Anticancer activity and cellular repression of c-MYC by the G-quadruplex-stabilizing 11-piperazinylquindoline is not dependent on direct targeting of the G-quadruplex in the c-MYC promoter. J. Med. Chem. 2012, 55, 6076–6086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calabrese, D.R.; Chen, X.; Leon, E.C.; Gaikwad, S.M.; Phyo, Z.; Hewitt, W.M.; Alden, S.; Hilimire, T.A.; He, F.; Michalowski, A.M.; et al. Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex. Nat. Commun. 2018, 9, 4229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.B.; Elsayed, M.S.A.; Wu, G.; Deng, N.; Cushman, M.; Yang, D. Indenoisoquinoline topoisomerase inhibitors strongly bind and stabilize the MYC promoter G-quadruplex and downregulate MYC. J. Am. Chem. Soc. 2019, 141, 11059–11070. [Google Scholar] [CrossRef] [PubMed]
- Lavrado, J.; Brito, H.; Borralho, P.M.; Ohnmacht, S.A.; Kim, N.S.; Leitão, C.; Pisco, S.; Gunaratnam, M.; Rodrigues, C.M.; Moreira, R.; et al. KRAS oncogene repression in colon cancer cell lines by G-quadruplex binding indolo[3,2-c]quinolines. Sci. Rep. 2015, 5, 9696. [Google Scholar] [CrossRef] [Green Version]
- Brito, H.; Martins, A.C.; Lavrado, J.; Mendes, E.; Francisco, A.P.; Santos, S.A.; Ohnmacht, S.A.; Kim, N.S.; Rodrigues, C.M.; Moreira, R.; et al. KRAS oncogene in colon cancer cells with 7-carboxylate indolo[3,2-b]quinoline tri-alkylamine derivatives. PLoS ONE 2015, 10, e0126891. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.-D.; Ou, T.-M.; Lu, Y.-J.; Li, Z.; Xu, Z.; Xi, C.; Tan, J.-H.; Huang, S.-L.; An, L.-K.; Li, D.; et al. Turning off transcription of the bcl-2 gene by stabilizing the bcl-2 promoter quadruplex with quindoline derivatives. J. Med. Chem. 2010, 53, 4390–4398. [Google Scholar] [CrossRef] [PubMed]
- Gunaratnam, M.; Collie, G.W.; Reszka, A.P.; Todd, A.K.; Parkinson, G.N.; Neidle, S. A naphthalene diimide G-quadruplex ligand inhibits cell growth and down-regulates BCL-2 expression in an imatinib-resistant gastrointestinal cancer cell line. Bioorg. Med. Chem. 2018, 26, 2958–2964. [Google Scholar] [CrossRef] [PubMed]
- Pelliccia, S.; Amato, J.; Capasso, D.; Di Gaetano, S.; Massarotti, A.; Piccolo, M.; Irace, C.; Tron, G.C.; Pagano, B.; Randazzo, A.; et al. Bio-inspired dual-selective BCL-2/c-MYC G-quadruplex binders: Design, synthesis, and anticancer activity of drug-like imidazo[2,1-i]purine derivatives. J. Med. Chem. 2020, 63, 2035–2050. [Google Scholar] [CrossRef] [PubMed]
- Gunaratnam, M.; Swank, S.; Haider, S.M.; Galesa, K.; Reszka, A.P.; Beltran, M.; Cuenca, F.; Fletcher, J.A.; Neidle, S. Targeting human gastrointestinal stromal tumor cells with a quadruplex-binding small molecule. J. Med. Chem. 2009, 52, 3774–3783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLuckie, K.I.; Waller, Z.A.; Sanders, D.A.; Alves, D.; Rodriguez, R.; Dash, J.; McKenzie, G.J.; Venkitaraman, A.R.; Balasubramanian, S. G-quadruplex-binding benzo[a]phenoxazines down-regulate c-KIT expression in human gastric carcinoma cells. J. Am. Chem. Soc. 2011, 133, 2658–2663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosiol, N.; Juranek, S.; Brossart, P.; Heine, A.; Paeschke, K. G-quadruplexes: A promising target for cancer therapy. Mol. Cancer 2021, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Monchaud, D.; Teulade-Fichou, M.-P. A hitchhiker’s guide to G-quadruplex ligands. Org. Biomol. Chem. 2008, 6, 627–636. [Google Scholar] [CrossRef]
- Müller, S.; Rodriguez, R. G-quadruplex interacting small molecules and drugs: From bench towards bedside. Expert. Rev. Clin. Pharmacol. 2014, 7, 663–679. [Google Scholar] [CrossRef]
- Islam, M.K.; Jackson, P.J.; Rahman, K.M.; Thurston, D.E. Recent advances in targeting the telomeric G-quadruplex DNA sequence with small molecules as a strategy for anticancer therapies. Future Med. Chem. 2016, 8, 1259–1290. [Google Scholar] [CrossRef]
- Neidle, S. Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 2016, 59, 5987–6011. [Google Scholar] [CrossRef]
- Duarte, A.R.; Cadoni, E.; Ressurreição, A.S.; Moreira, R.; Paulo, A. Design of modular G-quadruplex ligands. ChemMedChem 2018, 13, 869–893. [Google Scholar] [CrossRef]
- Asamitsu, S.; Bando, T.; Sugiyama, H. Ligand design to acquire specificity to intended G-quadruplex structures. Chemistry 2019, 25, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Savva, L.; Georgiades, S.N. Recent developments in small-molecule ligands of medicinal relevance for harnessing the anticancer potential of G-quadruplexes. Molecules 2021, 26, 841. [Google Scholar] [CrossRef]
- Harrison, R.J.; Gowan, S.M.; Kelland, L.R.; Neidle, S. Human telomerase inhibition by substituted acridine derivatives. Bioorg. Med. Chem. Lett. 1999, 9, 2463–2468. [Google Scholar] [CrossRef]
- Read, M.A.; Wood, A.A.; Harrison, J.R.; Gowan, S.M.; Kelland, L.R.; Dosanjh, H.S.; Neidle, S. Molecular modelling studies on G-quadruplex complexes of telomerase inhibitors: Structure-activity relationships. J. Med. Chem. 1999, 42, 4538–4546. [Google Scholar] [CrossRef]
- Read, M.A.; Harrison, R.J.; Romagnoli, B.; Tanious, F.A.; Gowan, S.H.; Reszka, A.P.; Wilson, W.D.; Kelland, L.R.; Neidle, S. Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc. Natl. Acad. Sci. USA 2001, 98, 4844–4849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heald, R.A.; Modi, C.; Cookson, J.C.; Hutchinson, I.; Laughton, C.A.; Gowan, S.M.; Kelland, L.R.; Stevens, M.F.G. Antitumor polycyclic acridines. 8. Synthesis and telomerase-inhibitory activity of methylated pentacyclic acridinium salts. J. Med. Chem. 2002, 45, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.M.; Parkinson, G.N.; Neidle, S. Structure of a G-quadruplex–ligand complex. J. Mol. Biol. 2003, 326, 117–125. [Google Scholar] [CrossRef]
- Burger, A.M.; Dai, F.; Schultes, C.M.; Reszka, A.P.; Moore, M.J.; Double, J.A.; Neidle, S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005, 65, 1489–1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, M.J.; Schultes, C.M.; Cuesta, J.; Cuenca, F.; Gunaratnam, M.; Tanious, F.A.; Wilson, W.D.; Neidle, S. Trisubstituted acridines as G-quadruplex telomere targeting agents. Effects of extensions of the 3,6- and 9-side chains on quadruplex binding, telomerase activity, and cell proliferation. J. Med. Chem. 2006, 49, 582–599. [Google Scholar] [CrossRef] [PubMed]
- Campbell, N.H.; Parkinson, G.N.; Reszka, A.P.; Neidle, S. Structural basis of DNA quadruplex recognition by an acridine drug. J. Am. Chem. Soc. 2008, 130, 6722–6724. [Google Scholar] [CrossRef] [PubMed]
- Campbell, N.H.; Patel, M.; Tofa, A.B.; Ghosh, R.; Parkinson, G.N.; Neidle, S. Selectivity in ligand recognition of G-quadruplex loops. Biochemistry 2009, 48, 1675–1680. [Google Scholar] [CrossRef]
- Collie, G.W.; Sparapani, S.; Parkinson, G.N.; Neidle, S. Structural basis of telomeric RNA quadruplex-acridine ligand recognition. J. Am. Chem. Soc. 2011, 133, 2721–2728. [Google Scholar] [CrossRef]
- Cuenca, F.; Greciano, O.; Gunaratnam, M.; Haider, S.; Munnur, D.; Nanjunda, R.; Wilson, W.D.; Neidle, S. Tri- and tetra-substituted naphthalene diimides as potent G-quadruplex ligands. Bioorg. Med. Chem. Lett. 2008, 18, 1668–1673. [Google Scholar] [CrossRef]
- Hampel, S.M.; Sidibe, A.; Gunaratnam, M.; Riou, J.-F.; Neidle, S. Tetrasubstituted naphthalene diimide ligands with selectivity for telomeric G-quadruplexes and cancer cells. Bioorg. Med. Chem. Lett. 2010, 20, 6459–6463. [Google Scholar] [CrossRef]
- Gunaratnam, M.; de la Fuente, M.; Hampel, S.M.; Todd, A.K.; Reszka, A.P.; Schatzlein, A.; Neidle, S. Targeting pancreatic cancer with a G-quadruplex ligand. Bioorg. Med. Chem. 2011, 19, 7151–7157. [Google Scholar] [CrossRef] [PubMed]
- Collie, G.W.; Promontorio, R.; Hampel, S.M.; Micco, M.; Neidle, S.; Parkinson, G.N. Structural basis for telomeric G-quadruplex naphthalene diimide ligand targeting. J. Am. Chem. Soc. 2012, 134, 2723–2731. [Google Scholar] [CrossRef]
- Micco, M.; Collie, G.W.; Dale, A.G.; Ohnmacht, S.A.; Pazitna, I.; Gunaratnam, M.; Reszka, A.P.; Neidle, S. Structure-based design and evaluation of naphthalene diimide G-quadruplex ligands as telomere targeting agents in pancreatic cancer cells. J. Med. Chem. 2013, 56, 2959–2974. [Google Scholar] [CrossRef]
- Nadai, M.; Cimino-Reale, G.; Sattin, G.; Doria, F.; Butovskaya, E.; Zaffaroni, N.; Freccero, M.; Palumbo, M.; Richter, S.N.; Folini, M. Assessment of gene promoter Gquadruplex binding and modulation by a naphthalene diimide derivative in tumor cells. Int. J. Oncol. 2015, 46, 369–380. [Google Scholar] [CrossRef] [Green Version]
- Ohnmacht, S.A.; Marchetti, C.; Gunaratnam, M.; Besser, R.J.; Haider, S.M.; Di Vita, G.; Lowe, H.L.; Mellinas-Gomez, M.; Diocou, S.; Robson, M.; et al. A G-quadruplex-binding compound showing anti-tumour activity in an in vivo model for pancreatic cancer. Sci. Rep. 2015, 5, 11385. [Google Scholar] [CrossRef] [Green Version]
- Spinello, A.; Barone, G.; Grunenberg, J. Molecular recognition of naphthalene diimide ligands by telomeric quadruplex-DNA: The importance of the protonation state and mediated hydrogen bonds. Phys. Chem. Chem. Phys. 2016, 18, 2871–2877. [Google Scholar] [CrossRef] [Green Version]
- Lopergolo, A.; Perrone, R.; Tortoreto, M.; Doria, F.; Beretta, G.L.; Zuco, V.; Freccero, M.; Borrello, M.G.; Lanzi, C.; Richter, S.N.; et al. Targeting of RET oncogene by naphthalene diimide-mediated gene promoter G-quadruplex stabilization exerts anti-tumor activity in oncogene-addicted human medullary thyroid cancer. Oncotarget 2016, 7, 49649–49663. [Google Scholar] [CrossRef] [Green Version]
- Răsădean, D.M.; Sheng, B.; Dash, J.; Pantoş, G.D. Amino-acid-derived naphthalenediimides as versatile G-quadruplex binders. Chemistry 2017, 23, 8491–8499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Street, S.; Chin, D.; Hollingworth, G.; Berry, M.; Morales, J.C.; Galan, M.C. Divalent naphthalene diimide ligands display high selectivity for the human telomeric G-quadruplex in K+ buffer. Chemistry 2017, 23, 6953–6958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Recagni, M.; Greco, M.L.; Milelli, A.; Minarini, A.; Zaffaroni, N.; Folini, M.; Sissi, C. Distinct biological responses of metastatic castration resistant prostate cancer cells upon exposure to G-quadruplex interacting naphthalenediimide derivatives. Eur. J. Med. Chem. 2019, 177, 401–413. [Google Scholar] [CrossRef]
- Pirota, V.; Nadai, M.; Doria, F.; Richter, S.N. Naphthalene diimides as multimodal G-quadruplex-selective ligands. Molecules 2019, 24, 426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vo, T.; Oxenford, S.; Angell, R.; Marchetti, C.; Ohnmacht, S.A.; Wilson, W.D.; Neidle, S. Substituted naphthalenediimide compounds bind selectively to two human quadruplex structures with parallel topology. ACS Med. Chem. Lett. 2020, 11, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Trajkovski, M.; Doria, F.; Freccero, M.; Plavec, J.; Montesarchio, D. On the interaction of an anticancer trisubstituted naphthalene diimide with G-quadruplexes of different topologies: A structural insight. Nucleic Acids Res. 2020, 48, 12380–12393. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Wang, C.; Wang, Y.; Li, C.; Hou, J.; Zhang, F.; Kang, C.; Gao, L. Topological conversion of human telomeric G-quadruplexes from hybrid to parallel form induced by naphthalene diimide ligands. Int. J. Biol. Macromol. 2021, 167, 1048–1058. [Google Scholar] [CrossRef]
- Sanchez-Martin, V.; Schneider, D.A.; Ortiz-Gonzalez, M.; Soriano-Lerma, A.; Linde-Rodriguez, A.; Perez-Carrasco, V.; Gutierrez-Fernandez, J.; Cuadros, M.; González, C.; Soriano, M.; et al. Targeting ribosomal G-quadruplexes with naphthalene-diimides as RNA polymerase I inhibitors for colorectal cancer treatment. Cell Chem. Biol. 2021, 28, 1590–1601. [Google Scholar] [CrossRef]
- Platella, C.; Napolitano, E.; Riccardi, C.; Musumeci, D.; Montesarchio, D. Disentangling the structure−activity relationships of naphthalenediimides as anticancer G-quadruplex-targeting drugs. J. Med. Chem. 2021, 64, 3578–3603. [Google Scholar] [CrossRef]
- Collie, G.W.; Campbell, N.H.; Neidle, S. Loop flexibility in human telomeric quadruplex small-molecule complexes. Nucleic Acids Res. 2015, 43, 4785–4799. [Google Scholar] [CrossRef]
- Chung, W.J.; Heddi, B.; Hamon, F.; Teulade-Fichou, M.-P.; Phan, A.T. Solution structure of a G-quadruplex bound to the bisquinolinium compound Phen-DC(3). Angew. Chem. Int. Ed. Engl. 2014, 53, 999–1002. [Google Scholar] [CrossRef]
- Dai, J.; Carver, M.; Hurley, L.H.; Yang, D. Solution structure of a 2:1 quindoline-c-MYC G-quadruplex: Insights into G-quadruplex-interactive small molecule drug design. J. Am. Chem. Soc. 2011, 133, 17673–17680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papi, F.; Bazzicalupi, C.; Ferraroni, M.; Ciolli, G.; Lombardi, P.; Khan, A.Y.; Kumar, G.S.; Gratteri, P. Pyridine derivative of the natural alkaloid berberine as human telomeric G4-DNA Binder: A solution and solid-state study. ACS Med. Chem. Lett. 2020, 11, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Dickerhoff, J.; Dai, J.; Yang, D. Structural recognition of the MYC promoter G-quadruplex by a quinoline derivative: Insights into molecular targeting of parallel G-quadruplexes. Nucleic Acids Res. 2021, 49, 5905–5915. [Google Scholar] [CrossRef]
- Campbell, N.H.; Smith, D.L.; Reszka, A.P.; Neidle, S.; O’Hagan, D. Fluorine in medicinal chemistry: β-fluorination of peripheral pyrrolidines attached to acridine ligands affects their interactions with G-quadruplex DNA. Org. Biomol. Chem. 2011, 9, 1328–1331. [Google Scholar] [CrossRef]
- Ma, D.L.; Ma, V.P.; Chan, D.S.; Leung, K.H.; Zhong, H.J.; Leung, C.H. In silico screening of quadruplex-binding ligands. Methods 2012, 57, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Cosconati, S.; Marinelli, L.; Trotta, R.; Virno, A.; Mayol, L.; Novellino, E.; Olson, A.J.; Randazzo, A. Tandem application of virtual screening and NMR experiments in the discovery of brand-new DNA quadruplex groove binders. J. Am. Chem. Soc. 2009, 131, 16336–16337. [Google Scholar] [CrossRef] [PubMed]
- Cosconati, S.; Marinelli, L.; Trotta, R.; Virno, A.; De Tito, S.; Romagnoli, R.; Pagano, B.; Limongelli, V.; Giancola, C.; Baraldi, P.G.; et al. Structural and conformational requisites in DNA quadruplex groove binding: Another piece to the puzzle. J. Am. Chem. Soc. 2010, 132, 6425–6433. [Google Scholar] [CrossRef] [PubMed]
- Alcaro, S.; Musetti, C.; Distinto, S.; Casatti, M.; Zagotto, G.; Artese, A.; Parrotta, L.; Moraca, F.; Costa, G.; Ortuso, F.; et al. Identification and characterization of new DNA G-quadruplex binders selected by a combination of ligand and structure-based virtual screening approaches. J. Med. Chem. 2013, 56, 843–855. [Google Scholar] [CrossRef]
- Di Leva, F.S.; Zizza, P.; Cingolani, C.; D’Angelo, C.; Pagano, B.; Amato, J.; Salvati, E.; Sissi, C.; Pinato, O.; Marinelli, L.; et al. Exploring the chemical space of G-quadruplex binders: Discovery of a novel chemotype targeting the human telomeric sequence. J. Med. Chem. 2013, 56, 9646–9654. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.-J.; Park, H.-J. In silico identification of novel ligands for G-quadruplex in the c-MYC promoter. J. Comp. Aided Mol. Des. 2015, 29, 339–348. [Google Scholar] [CrossRef]
- Hou, J.-Q.; Chen, S.-B.; Zan, L.-P.; Ou, T.-M.; Tan, J.H.; Luyt, L.G.; Huang, Z.-S. Identification of a selective G-quadruplex DNA binder using a multistep virtual screening approach. Chem. Commun. 2015, 51, 198–201. [Google Scholar] [CrossRef]
- Monsen, R.C.; Trent, J.O. G-quadruplex virtual drug screening: A review. Biochimie 2018, 152, 134–148. [Google Scholar] [CrossRef]
- Luo, J.; Wei, W.; Waldispühl, J.; Moitessier, N. Challenges and current status of computational methods for docking small molecules to nucleic acids. Eur. J. Med. Chem. 2019, 168, 414–425. [Google Scholar] [CrossRef]
- Ortiz de Luzuriaga, I.; Lopez, X.; Gil, A. Learning to model G-quadruplexes: Current methods and perspectives. Annu. Rev. Biophys. 2021, 50, 209–243. [Google Scholar] [CrossRef]
- Roy, S.; Ali, A.; Bhattacharya, S. Theoretical insight into the library screening approach for binding of intermolecular G-quadruplex RNA and small molecules through docking and molecular dynamics simulation studies. J. Phys. Chem. B 2021, 125, 5489–5501. [Google Scholar] [CrossRef]
- Wei, D.; Parkinson, G.N.; Reszka, A.P.; Neidle, S. Crystal structure of a c-kit promoter quadruplex reveals the structural role of metal ions and water molecules in maintaining loop conformation. Nucleic Acids Res. 2012, 40, 4691–4700. [Google Scholar] [CrossRef] [Green Version]
- Stump, S.; Mou, T.C.; Sprang, S.R.; Natale, N.R.; Beall, H.D. Crystal structure of the major quadruplex formed in the promoter region of the human c-MYC oncogene. PLoS ONE 2018, 13, e0205584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, A.; Schmidberger, J.W.; Wilson, K.A.; Evans, C.W.; Hargreaves, J.A.; Grigg, M.; O’Mara, M.L.; Iyer, K.S.; Bond, C.S.; Smith, N.M. High resolution crystal structure of a KRAS promoter G-quadruplex reveals a dimer with extensive poly-A pi-stacking interactions for small-molecule recognition. Nucleic Acids Res. 2020, 48, 5766–5776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.; Yatsunyk, L.; Neidle, S. Water spines and networks in G-quadruplex structures. Nucleic Acids Res. 2021, 49, 519–528. [Google Scholar] [CrossRef]
- Clark, G.R.; Pytel, P.D.; Squire, C.J. The high-resolution crystal structure of a parallel intermolecular DNA G-4 quadruplex/drug complex employing syn glycosyl linkages. Nucleic Acids Res. 2012, 40, 5731–5738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, G.R.; Pytel, P.D.; Squire, C.J.; Neidle, S. Structure of the first parallel DNA quadruplex-drug complex. J. Am. Chem. Soc. 2003, 125, 4066–4067. [Google Scholar] [CrossRef]
- Wei, W.; Luo, J.; Waldispühl, J.; Moitessier, N. Predicting positions of bridging water molecules in nucleic acid−ligand complexes. J. Chem. Inf. Model. 2019, 59, 2941–2951. [Google Scholar] [CrossRef] [PubMed]
- de Beer, S.B.; Vermeulen, N.P.; Oostenbrink, C. The role of water molecules in computational drug design. Curr. Top. Med. Chem. 2010, 10, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Samways, M.L.; Taylor, R.D.; Bruce Macdonald, H.E.; Essex, J.W. Water molecules at protein-drug interfaces: Computational prediction and analysis methods. Chem. Soc. Rev. 2021, 50, 9104–9120. [Google Scholar] [CrossRef] [PubMed]
- Nittinger, E.; Schneider, N.; Lamge, G.; Rarey, M. Evidence of water molecules—A statistical evaluation of water molecules based on electron density. J. Chem. Inf. Model. 2015, 55, 771–783. [Google Scholar] [CrossRef]
- Ladbury, J.E. Just add water! The effect of water on the specificity of protein-ligand binding sites and its potential application to drug design. Chem. Biol. 1996, 3, 973–980. [Google Scholar] [CrossRef] [Green Version]
- Dunitz, J.D. The entropic cost of bound water in crystals and biomolecules. Science 1994, 264, 670. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Cuenca, F.; Neidle, S. Topology conservation and loop flexibility in quadruplex-drug recognition: Crystal structures of inter- and intramolecular telomeric DNA quadruplex-drug complexes. J. Mol. Biol. 2008, 381, 1145–1156. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis: UCSF chimera visualization system. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.N.; Lee, M.P.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef]
- Neidle, S. Challenges in developing small-molecule quadruplex therapeutics. Ann. Rep. Med. Chem. 2020, 54, 517–546. [Google Scholar]
- Molnár, O.R.; Végh, A.; Somkuti, J.; Smeller, L. Characterization of a G-quadruplex from hepatitis B virus and its stabilization by binding TMPyP4, BRACO19 and PhenDC3. Sci. Rep. 2021, 11, 23243. [Google Scholar] [CrossRef] [PubMed]
- Frasson, I.; Nadai, M.; Richter, S.N. Conserved G-quadruplexes regulate the immediate early promoters of human alphaherpesviruses. Molecules 2019, 24, 2375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butovskaya, E.; Soldà, P.; Scalabrin, M.; Nadai, M.; Richter, S.N. HIV-1 nucleocapsid protein unfolds stable RNA G-quadruplexes in the viral genome and is inhibited by G-quadruplex ligands. ACS Infect. Dis. 2019, 5, 2127–2135. [Google Scholar] [CrossRef] [Green Version]
- Rudling, A.; Orro, A.; Carlsson, J. Prediction of ordered water molecules in protein binding sites from molecular dynamics simulations: The impact of ligand binding on hydration networks. J. Chem. Inf. Model. 2018, 58, 350–361. [Google Scholar] [CrossRef] [Green Version]
- Giambaşu, G.M.; Case, D.A.; York, D.M. Predicting site-binding modes of ions and water to nucleic acids using molecular solvation theory. J. Am. Chem. Soc. 2019, 141, 2435–2445. [Google Scholar] [CrossRef] [PubMed]
- Rappe, A.K.; Casewit, C.J.; Colwell, K.S.; Goddard, W.A., III; Skiff, W.M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 1992, 114, 10024–10039. [Google Scholar] [CrossRef]
- Wang, Y.H.; Yang, Q.F.; Lin, X.; Chen, D.; Wang, Z.Y.; Chen, B.; Han, H.Y.; Chen, H.D.; Cai, K.C.; Li, Q.; et al. G4LDB 2.2: A database for discovering and studying G-quadruplex and i-motif ligands. Nucleic Acids Res. 2021, gkab952. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. Software news and update Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [PubMed] [Green Version]
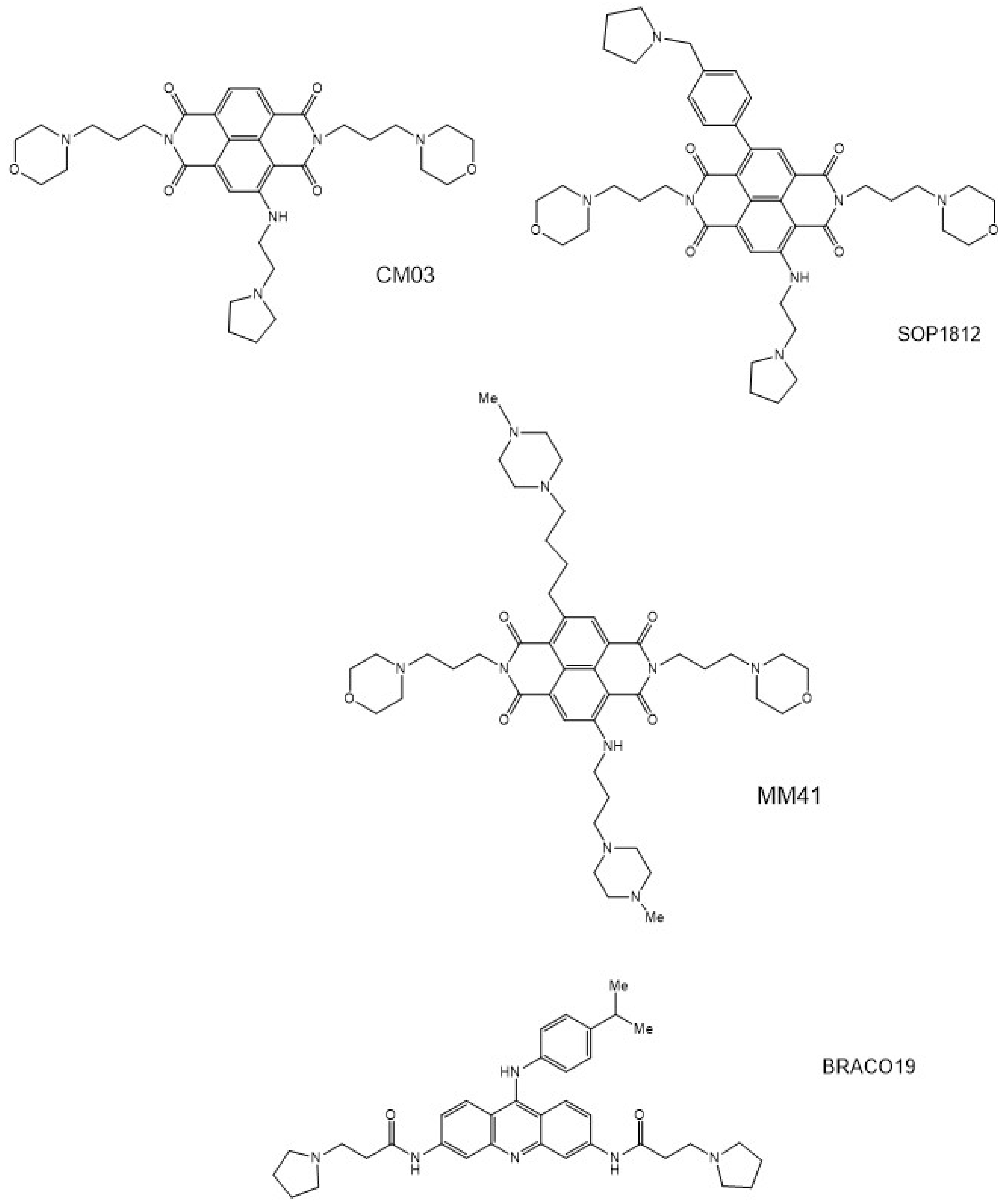
| PDB Id | G-Quadruplex Type | Compound | Resoln (Å) | No. of Waters/AU | Ref. |
|---|---|---|---|---|---|
| 3CE5 | 12-mer bimolecular human telomeric | 3,6,9- trisubstituted acridine BRACO19 | 2.5 | 54 | 74 |
| 3NZ7 | 12-mer bimolecular Oxytricha nova telomeric | 3,6- disubstituted acridine, F substituents | 1.10 | 187 | 100 |
| 3NYP | 12-mer bimolecular Oxytricha nova telomeric | 3,6- disubstituted acridine, F substituents | 1.18 | 176 | 100 |
| 3EM2 | 12-mer bimolecular Oxytricha nova telomeric | 3,6- disubstituted acridine | 2.3 | 64 | 75 |
| 3EQW | 12-mer bimolecular Oxytricha nova telomeric | 3,6- disubstituted acridine | 2.2 | 66 | 75 |
| 3EUI | 12-mer bimolecular Oxytricha nova telomeric | 3,6- disubstituted acridine | 2.2 | 159 | 75 |
| 3ERU | 12-mer bimolecular Oxytricha nova telomeric | 3,6- disubstituted acridine | 2.0 | 71 | 75 |
| 3ES0 | 12-mer bimolecular Oxytricha nova telomeric | 3,6- disubstituted acridine | 2.2 | 56 | 75 |
| 3ET8 | 12-mer bimolecular Oxytricha nova telomeric | 3,6- disubstituted acridine | 2.45 | 51 | 75 |
| 3EUM | 12-mer bimolecular Oxytricha nova telomeric | 3,6- disubstituted acridine | 1.78 | 52 | 75 |
| 1L1H | 12-mer bimolecular Oxytricha nova telomeric | 3,6- disubstituted acridine | 1.75 | 146 | 75 |
| 3UYH | 22-mer human telomeric | Tetrasubstituted naphthalene diimide MM41 | 1.95 | 51 | 81 |
| 3T5E | 22-mer human telomeric | Tetrasubstituted naphthalene diimide BMSG-SH-4 | 2.10 | 38 | 80 |
| 3CCO | 11-mer biomolecular human telomeric | Tetrasubstituted naphthalene diimide | 2.20 | 28 | 124 |
| 3CDM | 22-mer human telomeric | Tetrasubstituted naphthalene diimide | 2.10 | 158 | 124 |
| 4DA3 | 21-mer human telomeric | Tetrasubstituted naphthalene diimide MM41 | 2.40 | 25 | 81 |
| 6S15 | 12-mer bimolecular human telomeric | Pyridine derivative of berberine | 1.70 | 23 | 98 |
| (a) | |||||
|---|---|---|---|---|---|
| Atom1 | Atom2 | d1-2 | B Factor Atom1 | B Factor Atom2 | |
| Groove 1 | |||||
| NCH | W204 | 3.2 | 64 | 32 | |
| W204 | W203 | 3.0 | 32 | 32 | |
| W203 | OP2 dG10 | 2.9 | 32 | 32 | |
| W203 | W227 | 3.4 | 32 | 45 | |
| W227 | OAF | 2.5 | 45 | 29 | |
| W251 | OP2 dG9 | 2.9 | 27 | 30 | |
| W204 | N2 dG4 | 2.9 | 32 | 20 | |
| W204 | W251 | 2.8 | 32 | 27 | |
| Groove 2 | |||||
| NCA | OP2 dT11 | 3.1 | 46 | 35 | |
| NCE | W217 | 2.7 | 40 | 46 | |
| W217 | W218 | 2.6 | 46 | 48 | |
| W218 | W219 | 3.0 | 48 | 43 | |
| W219 | ODX | 2.7 | 43 | 31 | |
| W218 | OP2 dG16 | 3.4 | 48 | 45 | |
| Groove 3 | |||||
| NCG | W220 | 2.9 | 48 | 50 | |
| W219 | W220 | 2.9 | 43 | 50 | |
| Groove 4 | |||||
| NCF | OP2 dG4 | 2.9 | 58 | 39 | |
| (b) | |||||
| Atom1 | Atom2 | d1-2 | B Factor Atom1 | B Factor Atom2 | |
| Groove 1 | |||||
| N7 | W52 | 2.7 | 11 | 21 | |
| W52 | N3 dT24 | 2.9 | 21 | 12 | |
| W52 | O6 dG5 | 3.0 | 21 | 18 | |
| W52 | W53 | 3.1 | 21 | 33 | |
| W53 | O2 dT24 | 3.3 | 33 | 18 | |
| W53 | O52 | 3.1 | 33 | 14 | |
| Groove 2 | |||||
| W56 | N39 | 3.4 | 23 | 27 | |
| N17 | W55 | 3.0 | 14 | 38 | |
| W55 | O2 dT12 | 3.3 | 38 | 17 | |
| Groove 4 | |||||
| N21 | O4 dT24 | 3.0 | 13 | 17 | |
| N47 | W41 | 3.0 | 21 | 28 | |
| W41 | W44 | 2.9 | 28 | 29 | |
| W44 | OP2 dG23 | 2.5 | 29 | 21 | |
| W41 | N2 dG17 | 2.9 | 28 | 18 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neidle, S. Structured Waters Mediate Small Molecule Binding to G-Quadruplex Nucleic Acids. Pharmaceuticals 2022, 15, 7. https://doi.org/10.3390/ph15010007
Neidle S. Structured Waters Mediate Small Molecule Binding to G-Quadruplex Nucleic Acids. Pharmaceuticals. 2022; 15(1):7. https://doi.org/10.3390/ph15010007
Chicago/Turabian StyleNeidle, Stephen. 2022. "Structured Waters Mediate Small Molecule Binding to G-Quadruplex Nucleic Acids" Pharmaceuticals 15, no. 1: 7. https://doi.org/10.3390/ph15010007
APA StyleNeidle, S. (2022). Structured Waters Mediate Small Molecule Binding to G-Quadruplex Nucleic Acids. Pharmaceuticals, 15(1), 7. https://doi.org/10.3390/ph15010007






