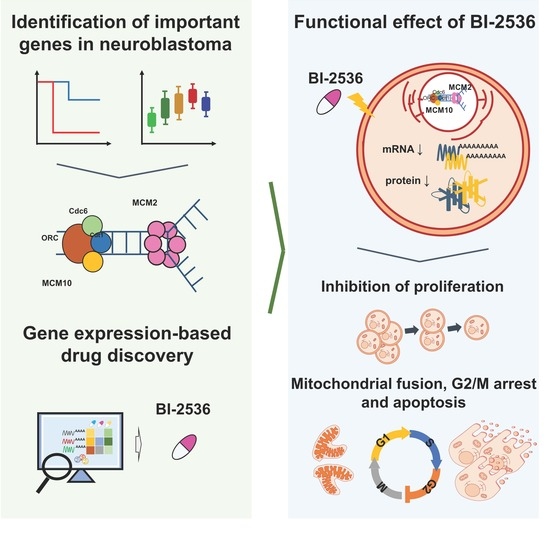
BI-2536 Promotes Neuroblastoma Cell Death via Minichromosome Maintenance Complex Components 2 and 10
Abstract
:1. Introduction
2. Results
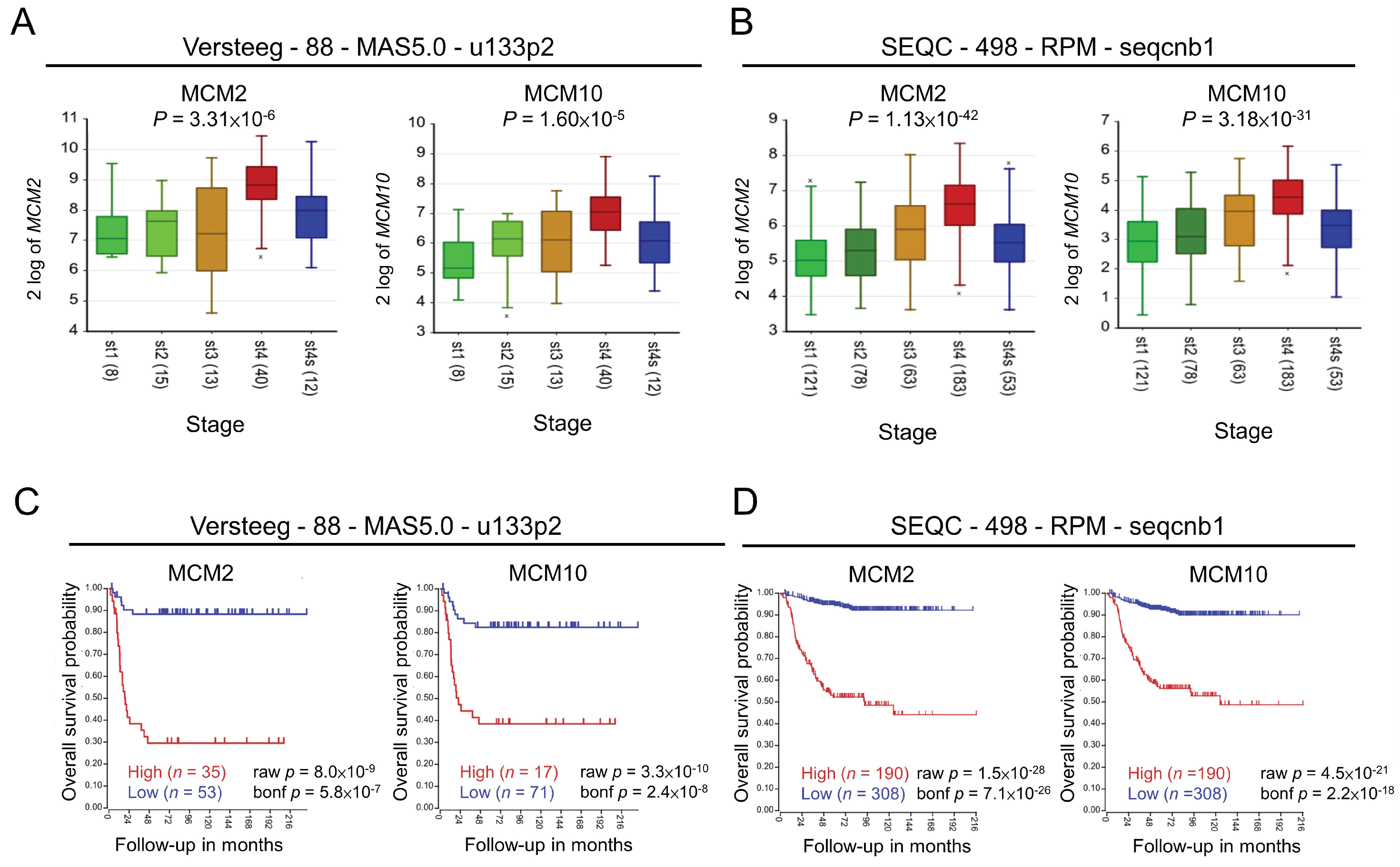
2.1. Role of MCM2 and MCM10 in Neuroblastoma
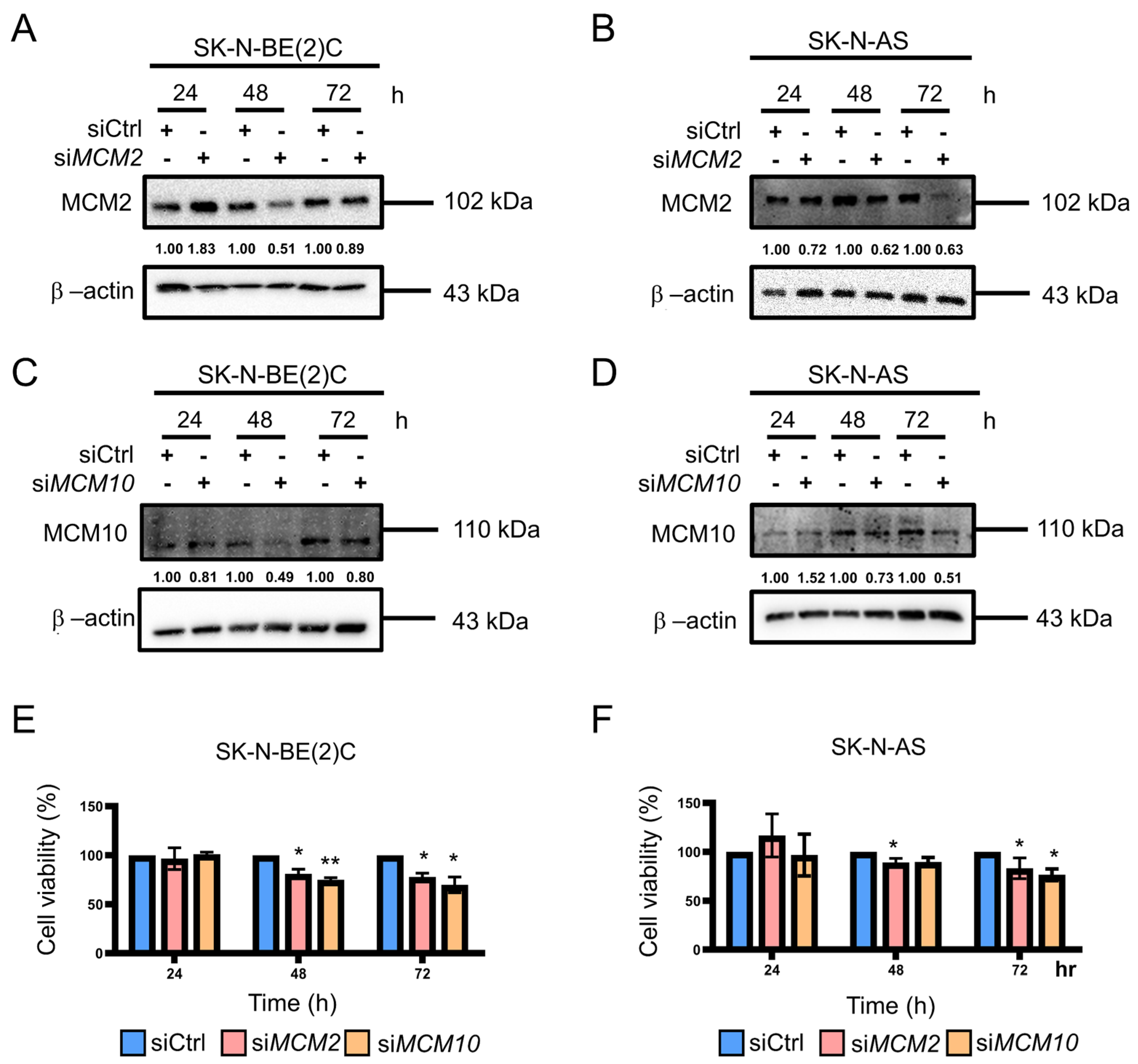
2.2. Knockdown of MCM2 and MCM10 Suppressed Neuroblastoma Cell Growth
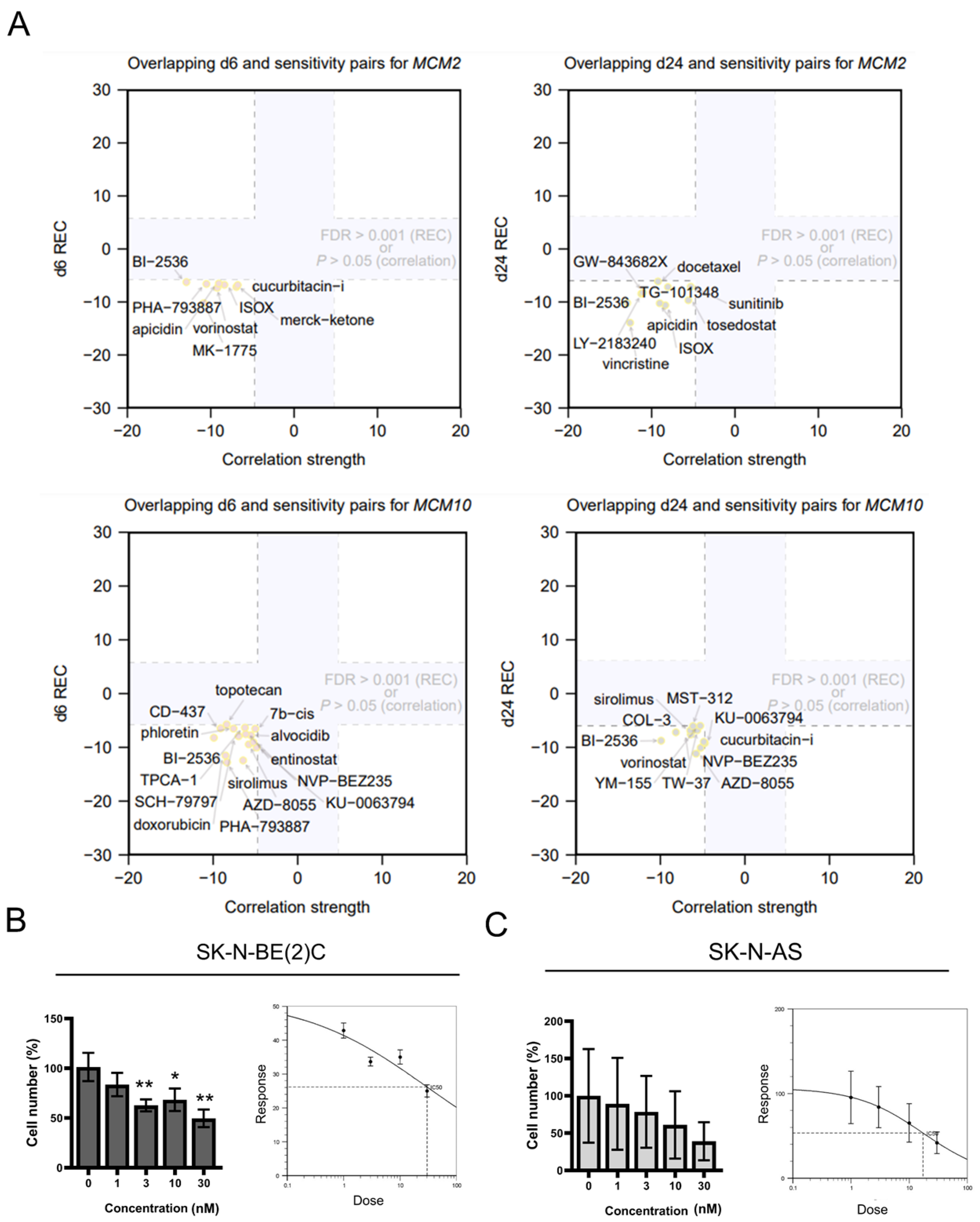
2.3. Inhibition of MCM2 and MCM10 for Neuroblastoma Therapy
2.4. Cytotoxicity of BI-2536 in Neuroblastoma Cell Lines
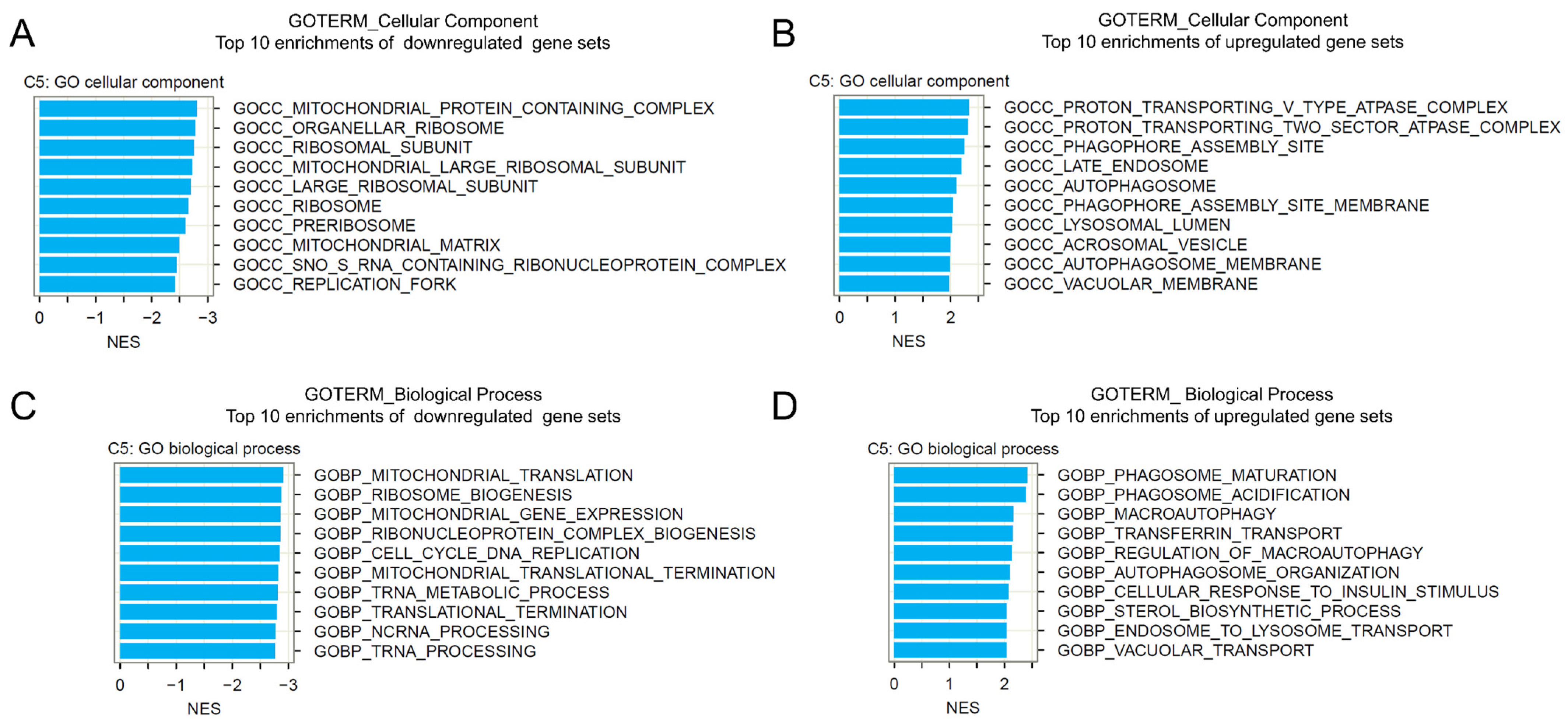
2.5. Gene Set Enrichment Analysis of BI-2536-Induced mRNA Recurrences
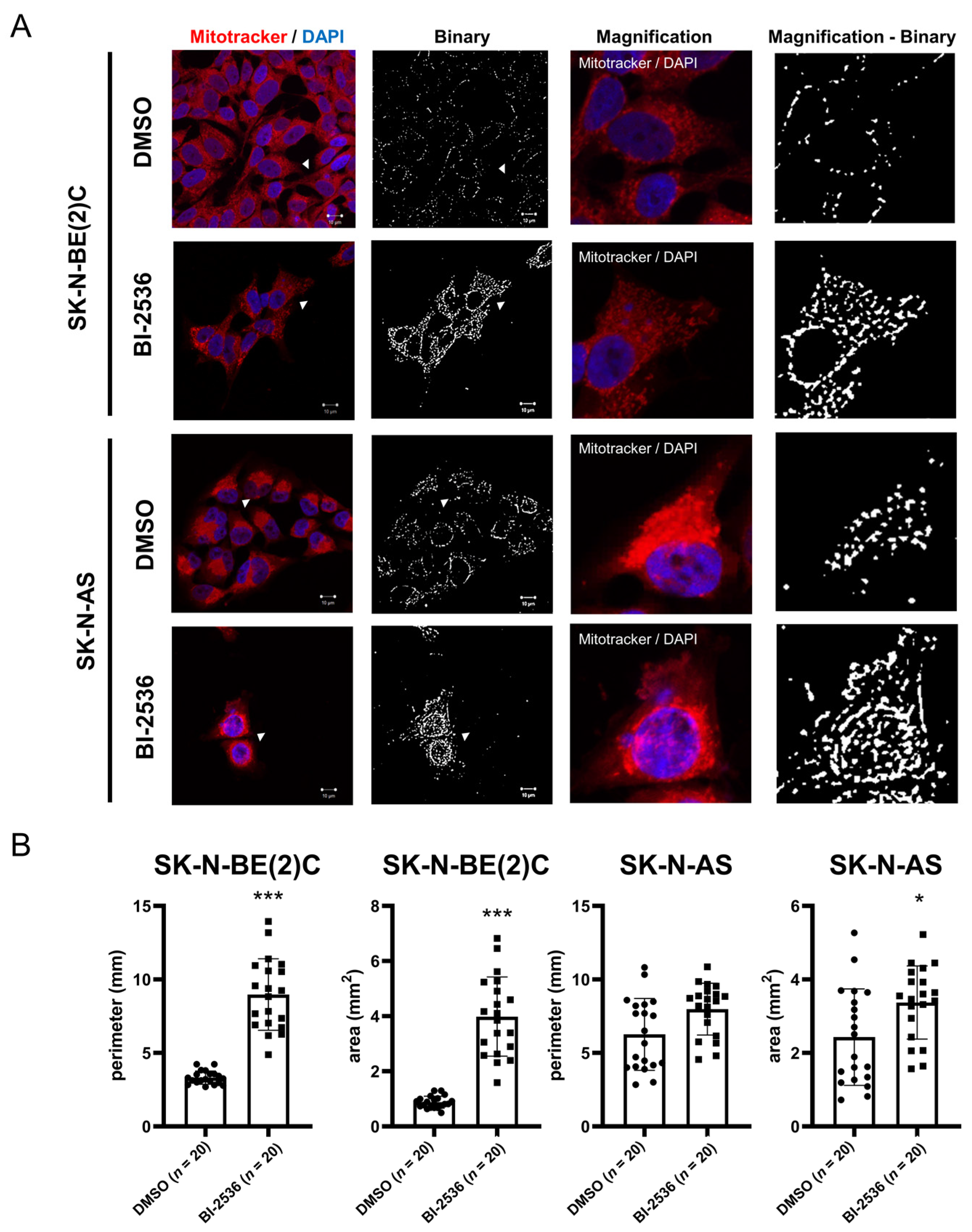
2.6. BI-2536-Induced Mitochondrial Fusion in Neuroblastoma Cells
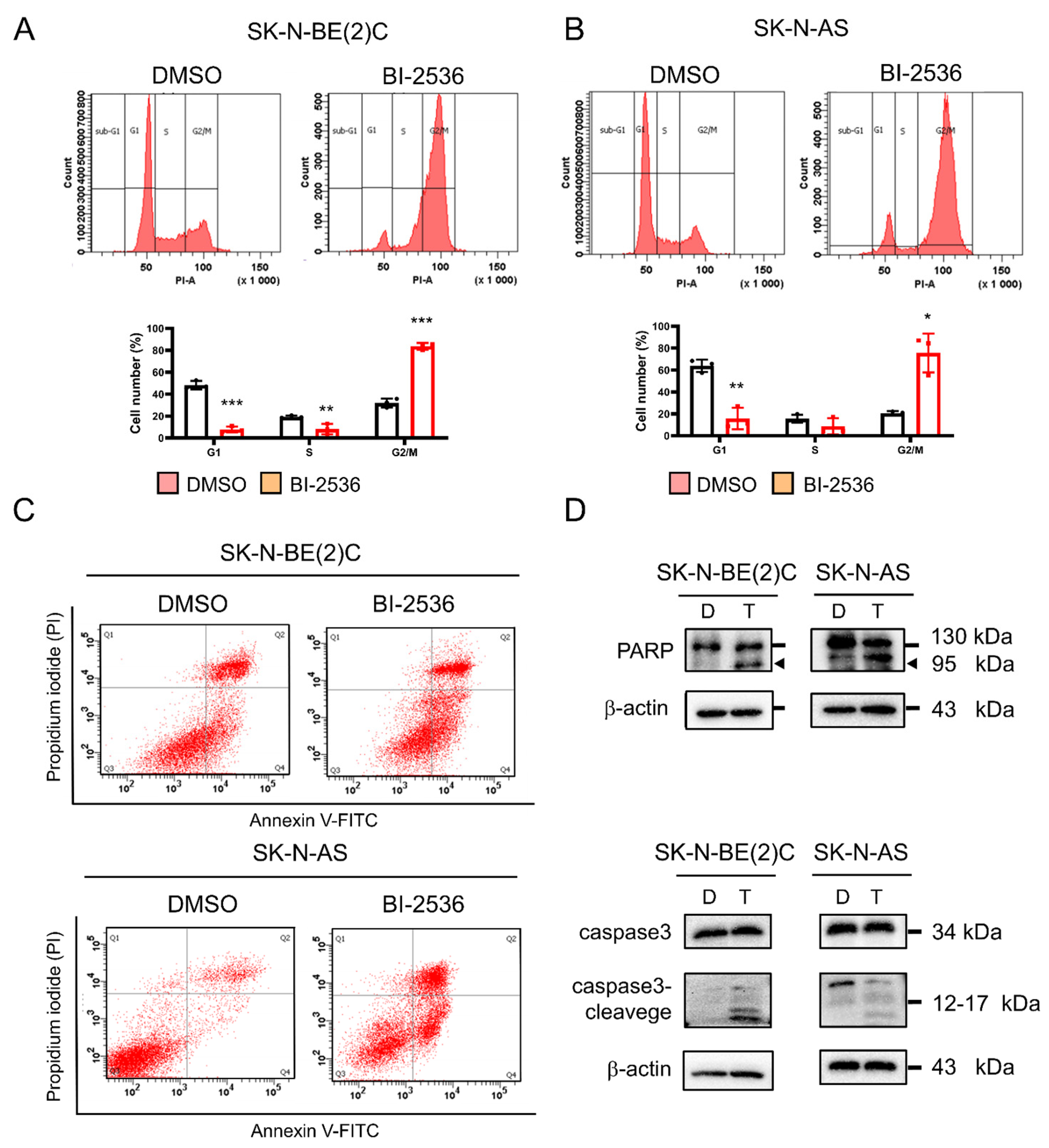
2.7. BI-2536-Induced Cell Cycle Arrest at the G2/M Phase and Apoptosis
3. Discussion and Conclusions
4. Materials and Methods
4.1. Kaplan–Meier Curves of Overall Survival of Patients with Neuroblastoma
4.2. RNA Interference
4.3. Drug Prediction
4.4. Cell Culture
4.5. Drug Treatment
4.6. RNA Extraction and Reverse Transcription
4.7. qRT-PCR
4.8. Cell Lysis and Protein Extraction
4.9. Western Blot Analysis
4.10. Colony Formation Assay
4.11. Cell Differentiation and Calculation of Neurites
4.12. Gene Set Enrichment Analysis
4.13. Immunocytochemistry and Mitochondrial Image Analysis
4.14. Cell Cycle Analysis
4.15. Apoptosis Assay
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ries, L.A.G.; Smith, M.A.; Gurney, J.G.; Linet, M.; Tamra, T.; Young, J.L.; Bunin, G.R. Cancer Incidence and Survival among Children and Adolescents: United States SEER Program 1975–1995, National Cancer Institute, SEER Program. Available online: https://seer.cancer.gov/archive/publications/childhood/ (accessed on 27 September 2021).
- Friedman, G.K.; Castleberry, R.P. Changing trends of research and treatment in infant neuroblastoma. Pediatr. Blood Cancer 2007, 49 (Suppl S7), 1060–1065. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.M.; Rasalkar, D.D.; Hu, Y.J.; Cheng, F.W.; Li, C.K.; Chu, W.C. Clinical presentations and imaging findings of neuroblastoma beyond abdominal mass and a review of imaging algorithm. Br. J. Radiol. 2011, 84, 81–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodeur, G.M.; Pritchard, J.; Berthold, F.; Carlsen, N.L.; Castel, V.; Castelberry, R.P.; De Bernardi, B.; Evans, A.E.; Favrot, M.; Hedborg, F.; et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J. Clin. Oncol. 1993, 11, 1466–1477. [Google Scholar] [CrossRef] [PubMed]
- Smith, V.; Foster, J. High-Risk Neuroblastoma Treatment Review. Children 2018, 5, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, T.; Sicinski, P. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer 2017, 17, 93–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, G.K.; Shah, M.A. Targeting the cell cycle: A new approach to cancer therapy. J. Clin. Oncol. 2005, 23, 9408–9421. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Cheng, E.; Wu, H.; Li, N.; Yung, P.Y.; Gao, N.; Tye, B.K. Open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer. Nat. Struct. Mol. Biol. 2017, 24, 300–308. [Google Scholar] [CrossRef]
- Lee, J.K.; Hurwitz, J. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc. Natl. Acad. Sci. USA 2001, 98, 54–59. [Google Scholar] [CrossRef] [Green Version]
- Homesley, L.; Lei, M.; Kawasaki, Y.; Sawyer, S.; Christensen, T.; Tye, B.K. Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 2000, 14, 913–926. [Google Scholar] [CrossRef]
- Simon, N.E.; Schwacha, A. The Mcm2-7 replicative helicase: A promising chemotherapeutic target. Biomed. Res. Int. 2014, 2014, 549719. [Google Scholar] [CrossRef] [Green Version]
- Giaginis, C.; Vgenopoulou, S.; Vielh, P.; Theocharis, S. MCM proteins as diagnostic and prognostic tumor markers in the clinical setting. Histol. Histopathol. 2010, 25, 351–370. [Google Scholar]
- Yu, S.; Wang, G.; Shi, Y.; Xu, H.; Zheng, Y.; Chen, Y. MCMs in Cancer: Prognostic Potential and Mechanisms. Anal. Cell. Pathol. 2020, 2020, 3750294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, M. The MCM complex: Its role in DNA replication and implications for cancer therapy. Curr. Cancer Drug Targets 2005, 5, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.T.; Hsieh, C.H.; Lee, W.C.; Liu, Y.L.; Yang, T.S.; Hsu, W.M.; Oyang, Y.J.; Huang, H.C.; Juan, H.F. Therapeutic Targeting of Non-oncogene Dependencies in High-risk Neuroblastoma. Clin. Cancer Res. 2019, 25, 4063–4078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.T.; Hsieh, C.H.; Chung, Y.H.; Oyang, Y.J.; Huang, H.C.; Juan, H.F. Perturbational Gene-Expression Signatures for Combinatorial Drug Discovery. iScience 2019, 15, 291–306. [Google Scholar] [CrossRef] [Green Version]
- Müller, I.; Larsson, K.; Frenzel, A.; Oliynyk, G.; Zirath, H.; Prochownik, E.V.; Westwood, N.J.; Henriksson, M.A. Targeting of the MYCN protein with small molecule c-MYC inhibitors. PLoS ONE 2014, 9, e97285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zirath, H.; Frenzel, A.; Oliynyk, G.; Segerström, L.; Westermark, U.K.; Larsson, K.; Munksgaard Persson, M.; Hultenby, K.; Lehtiö, J.; Einvik, C.; et al. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc. Natl. Acad. Sci. USA 2013, 110, 10258–10263. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Winger, A.J.; Knowlton, A.A. Mitochondrial dynamic changes in health and genetic diseases. Mol. Biol. Rep. 2014, 41, 7053–7062. [Google Scholar] [CrossRef] [Green Version]
- Deng, M.; Sun, J.; Xie, S.; Zhen, H.; Wang, Y.; Zhong, A.; Zhang, H.; Lu, R.; Guo, L. Inhibition of MCM2 enhances the sensitivity of ovarian cancer cell to carboplatin. Mol. Med. Rep. 2019, 20, 2258–2266. [Google Scholar] [CrossRef] [Green Version]
- Fatoba, S.T.; Tognetti, S.; Berto, M.; Leo, E.; Mulvey, C.M.; Godovac-Zimmermann, J.; Pommier, Y.; Okorokov, A.L. Human SIRT1 regulates DNA binding and stability of the Mcm10 DNA replication factor via deacetylation. Nucleic Acids Res. 2013, 41, 4065–4079. [Google Scholar] [CrossRef] [Green Version]
- Fotedar, R.; Diederich, L.; Fotedar, A. Apoptosis and the cell cycle. Prog. Cell Cycle Res. 1996, 2, 147–163. [Google Scholar] [PubMed]
- Finkel, T. Signal transduction by mitochondrial oxidants. J. Biol. Chem. 2012, 287, 4434–4440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, A.R.; Band, L.R.; Murray, J.A.H. Double or Nothing? Cell Division and Cell Size Control. Trends Plant Sci. 2019, 24, 1083–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wibowo, A.; Becker, C.; Durr, J.; Price, J.; Spaepen, S.; Hilton, S.; Putra, H.; Papareddy, R.; Saintain, Q.; Harvey, S.; et al. Partial maintenance of organ-specific epigenetic marks during plant asexual reproduction leads to heritable phenotypic variation. Proc. Natl. Acad. Sci. USA 2018, 115, e9145–e9152. [Google Scholar] [CrossRef] [Green Version]
- Alhmoud, J.F.; Woolley, J.F.; Al Moustafa, A.E.; Malki, M.I. DNA Damage/Repair Management in Cancers. Cancers 2020, 12, 1050. [Google Scholar] [CrossRef]
- Bruck, I.; Kaplan, D.L. Origin single-stranded DNA releases Sld3 protein from the Mcm2-7 complex, allowing the GINS tetramer to bind the Mcm2-7 complex. J. Biol. Chem. 2011, 286, 18602–18613. [Google Scholar] [CrossRef] [Green Version]
- Noguchi, Y.; Yuan, Z.; Bai, L.; Schneider, S.; Zhao, G.; Stillman, B.; Speck, C.; Li, H. Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model. Proc. Natl. Acad. Sci. USA 2017, 114, e9529–e9538. [Google Scholar] [CrossRef] [Green Version]
- Baxley, R.M.; Bielinsky, A.K. Mcm10: A Dynamic Scaffold at Eukaryotic Replication Forks. Genes 2017, 8, 73. [Google Scholar] [CrossRef]
- Wharton, S.B.; Chan, K.K.; Anderson, J.R.; Stoeber, K.; Williams, G.H. Replicative Mcm2 protein as a novel proliferation marker in oligodendrogliomas and its relationship to Ki67 labelling index, histological grade and prognosis. Neuropathol. Appl. Neurobiol. 2001, 27, 305–313. [Google Scholar] [CrossRef]
- Dudderidge, T.J.; Stoeber, K.; Loddo, M.; Atkinson, G.; Fanshawe, T.; Griffiths, D.F.; Williams, G.H. Mcm2, Geminin, and KI67 define proliferative state and are prognostic markers in renal cell carcinoma. Clin. Cancer Res. 2005, 11, 2510–2517. [Google Scholar] [CrossRef] [Green Version]
- Burger, M. MCM2 and MCM5 as prognostic markers in colon cancer: A worthwhile approach. Dig. Dis. Sci. 2009, 54, 197–198. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.D.; Wang, L. MCM10 facilitates the invaded/migrated potentials of breast cancer cells via Wnt/β-catenin signaling and is positively interlinked with poor prognosis in breast carcinoma. J. Biochem. Mol. Toxicol. 2019, 33, e22330. [Google Scholar] [CrossRef] [PubMed]
- Li, W.M.; Huang, C.N.; Ke, H.L.; Li, C.C.; Wei, Y.C.; Yeh, H.C.; Chang, L.L.; Huang, C.H.; Liang, P.I.; Yeh, B.W.; et al. MCM10 overexpression implicates adverse prognosis in urothelial carcinoma. Oncotarget 2016, 7, 77777–77792. [Google Scholar] [CrossRef] [Green Version]
- Thibault, A.; Samid, D.; Tompkins, A.C.; Figg, W.D.; Cooper, M.R.; Hohl, R.J.; Trepel, J.; Liang, B.; Patronas, N.; Venzon, D.J.; et al. Phase I study of lovastatin, an inhibitor of the mevalonate pathway, in patients with cancer. Clin. Cancer Res. 1996, 2, 483–491. [Google Scholar] [PubMed]
- Paulson, C.N.; John, K.; Baxley, R.M.; Kurniawan, F.; Orellana, K.; Francis, R.; Sobeck, A.; Eichman, B.F.; Chazin, W.J.; Aihara, H.; et al. The anti-parasitic agent suramin and several of its analogues are inhibitors of the DNA binding protein Mcm10. Open Biol. 2019, 9, 190117. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Teng, Y.; Yang, F.; Wang, M.; Hong, X.; Ye, L.G.; Gao, Y.N.; Chen, G.Y. MCM2 is a therapeutic target of lovastatin in human non-small cell lung carcinomas. Oncol. Rep. 2015, 33, 2599–2605. [Google Scholar] [CrossRef] [Green Version]
- Xiao, D.; Yue, M.; Su, H.; Ren, P.; Jiang, J.; Li, F.; Hu, Y.; Du, H.; Liu, H.; Qing, G. Polo-like Kinase-1 Regulates Myc Stabilization and Activates a Feedforward Circuit Promoting Tumor Cell Survival. Mol. Cell 2016, 64, 493–506. [Google Scholar] [CrossRef] [Green Version]
- Ashrafi, G.; Schwarz, T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013, 20, 31–42. [Google Scholar] [CrossRef] [Green Version]
- Bloemberg, D.; Quadrilatero, J. Autophagy, apoptosis, and mitochondria: Molecular integration and physiological relevance in skeletal muscle. Am. J. Physiol. Cell Physiol. 2019, 317, C111–C130. [Google Scholar] [CrossRef]
- Huang, C.T.; Hsieh, C.H.; Oyang, Y.J.; Huang, H.C.; Juan, H.F. A Large-Scale Gene Expression Intensity-Based Similarity Metric for Drug Repositioning. iScience 2018, 7, 40–52. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsieh, C.-H.; Yeh, H.-N.; Huang, C.-T.; Wang, W.-H.; Hsu, W.-M.; Huang, H.-C.; Juan, H.-F. BI-2536 Promotes Neuroblastoma Cell Death via Minichromosome Maintenance Complex Components 2 and 10. Pharmaceuticals 2022, 15, 37. https://doi.org/10.3390/ph15010037
Hsieh C-H, Yeh H-N, Huang C-T, Wang W-H, Hsu W-M, Huang H-C, Juan H-F. BI-2536 Promotes Neuroblastoma Cell Death via Minichromosome Maintenance Complex Components 2 and 10. Pharmaceuticals. 2022; 15(1):37. https://doi.org/10.3390/ph15010037
Chicago/Turabian StyleHsieh, Chiao-Hui, Hsiang-Ning Yeh, Chen-Tsung Huang, Wei-Hsuan Wang, Wen-Ming Hsu, Hsuan-Cheng Huang, and Hsueh-Fen Juan. 2022. "BI-2536 Promotes Neuroblastoma Cell Death via Minichromosome Maintenance Complex Components 2 and 10" Pharmaceuticals 15, no. 1: 37. https://doi.org/10.3390/ph15010037
APA StyleHsieh, C.-H., Yeh, H.-N., Huang, C.-T., Wang, W.-H., Hsu, W.-M., Huang, H.-C., & Juan, H.-F. (2022). BI-2536 Promotes Neuroblastoma Cell Death via Minichromosome Maintenance Complex Components 2 and 10. Pharmaceuticals, 15(1), 37. https://doi.org/10.3390/ph15010037