Inclusion Complexes of Rifampicin with Native and Derivatized Cyclodextrins: In Silico Modeling, Formulation, and Characterization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
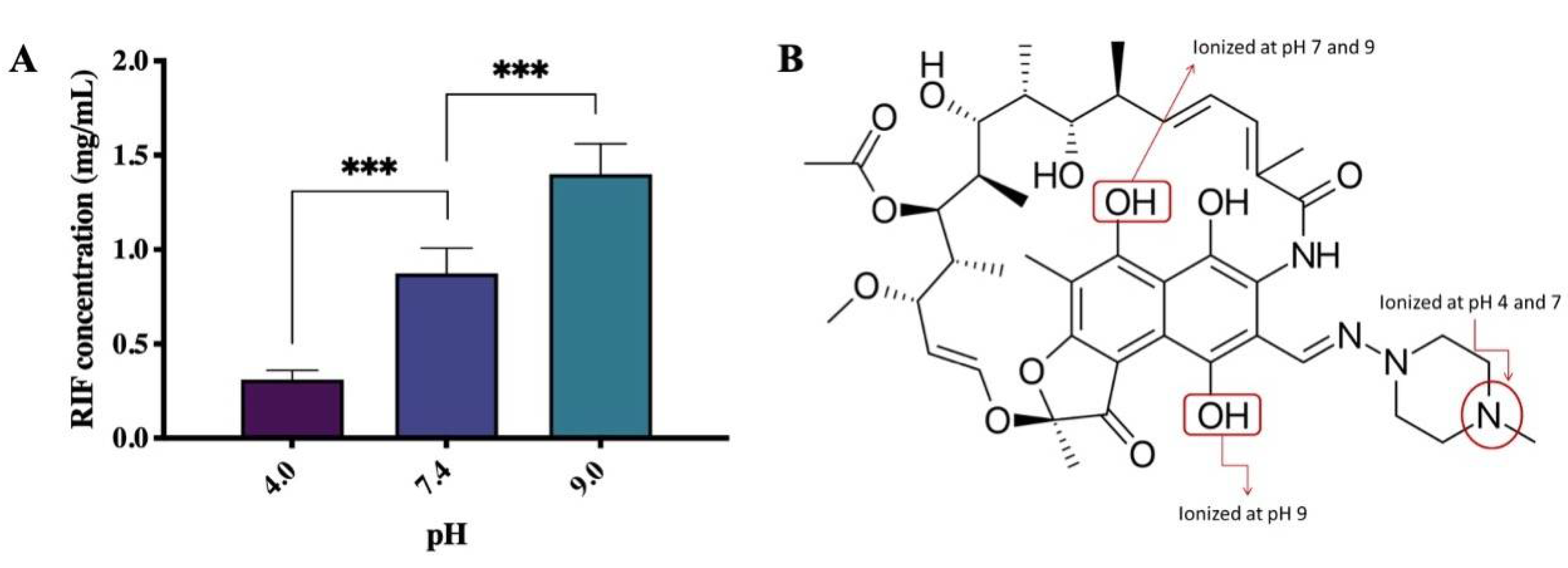
2.2. Saturation Solubility
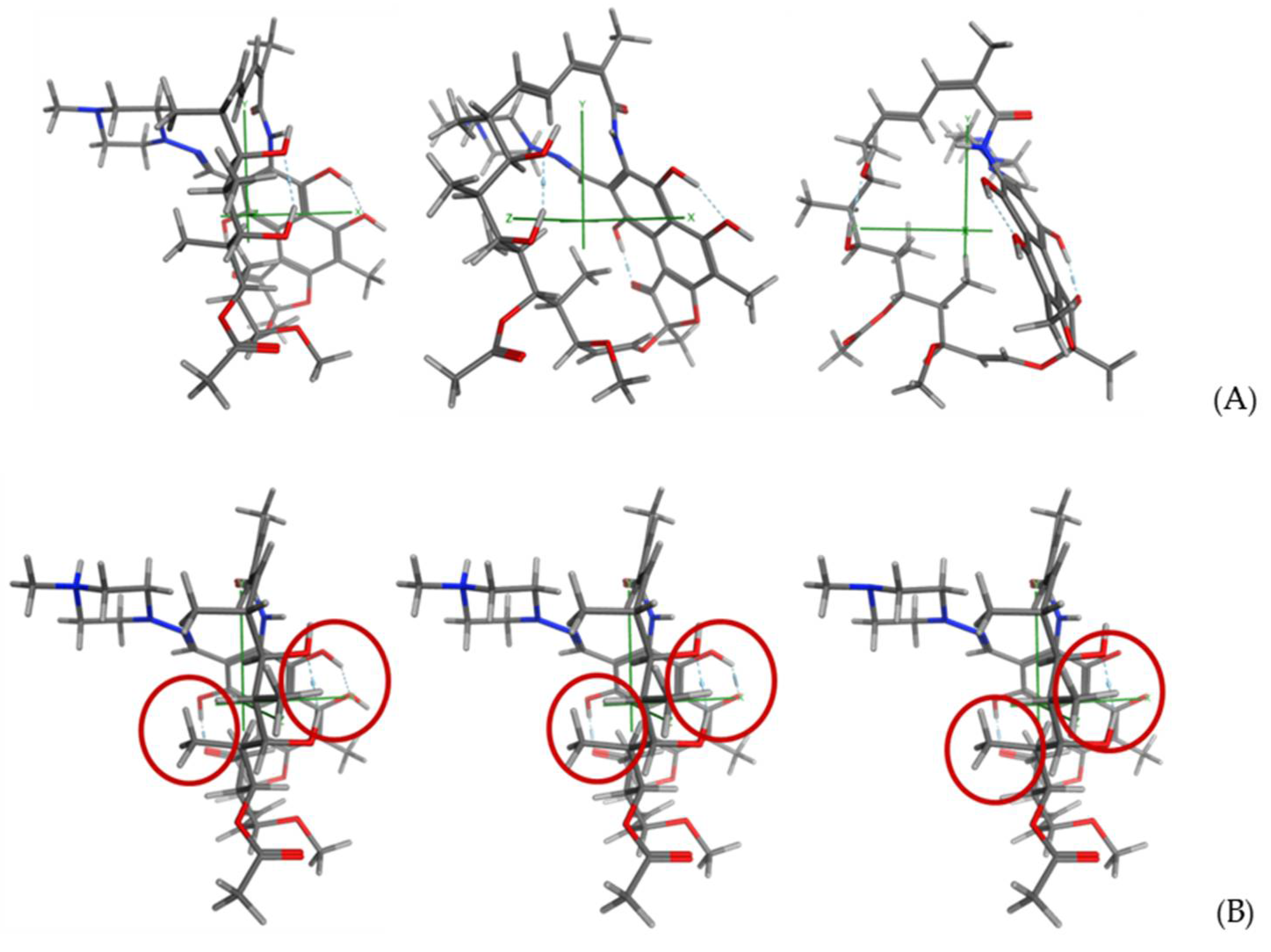
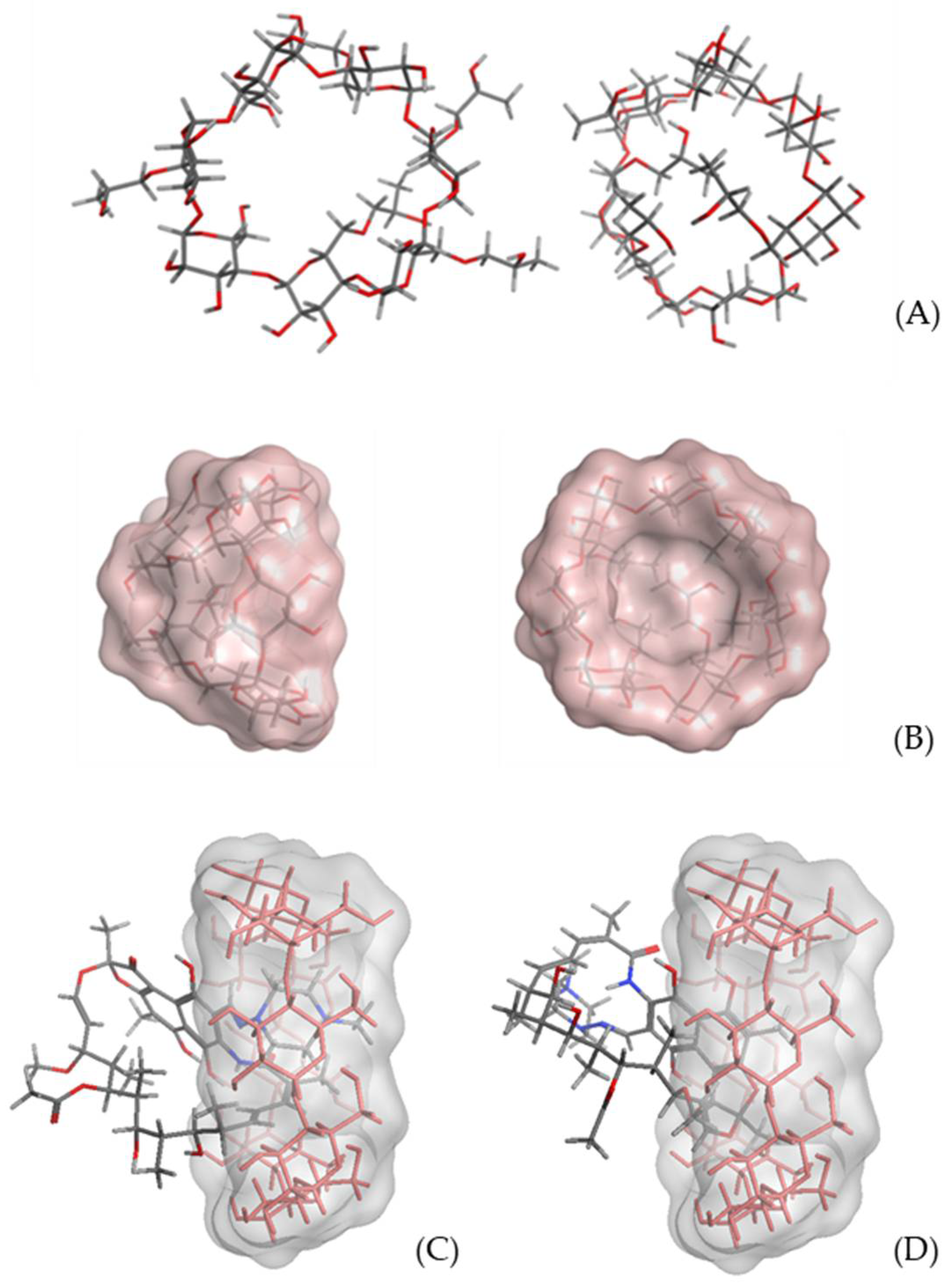
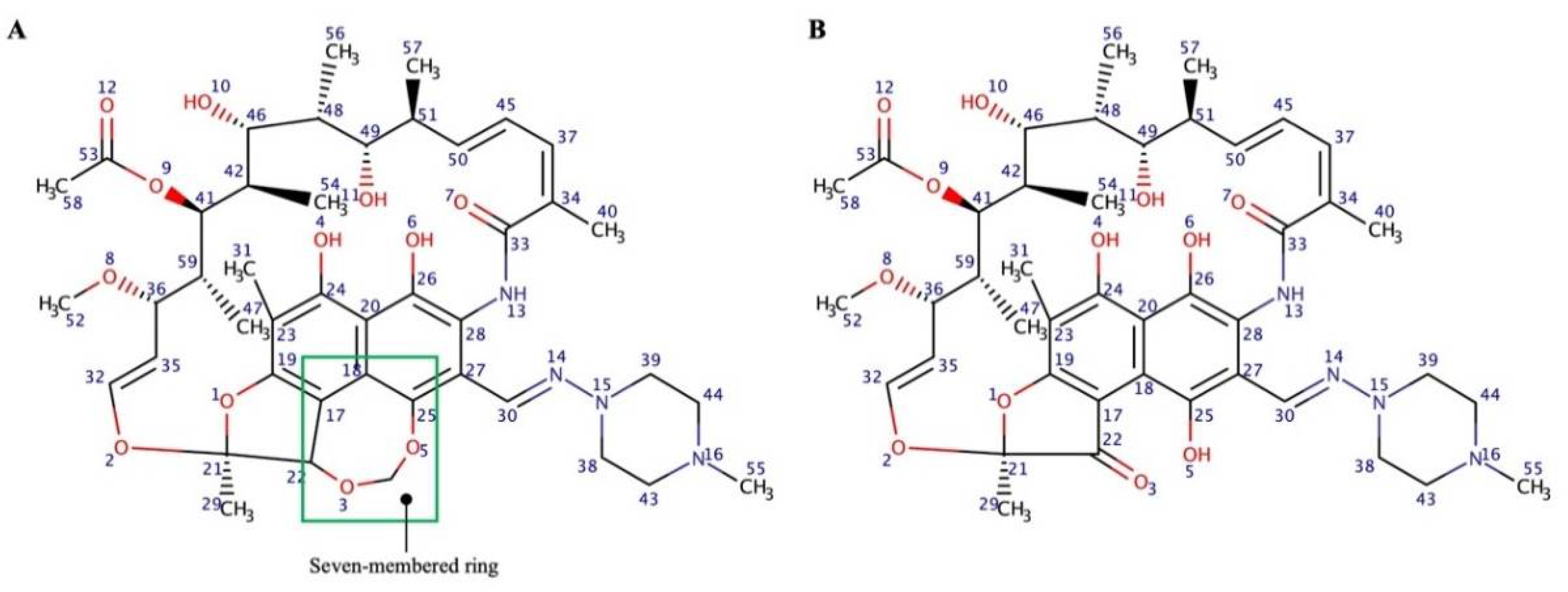
2.3. Molecular Modeling Study
2.4. Phase Solubility
2.5. Constant Calculations
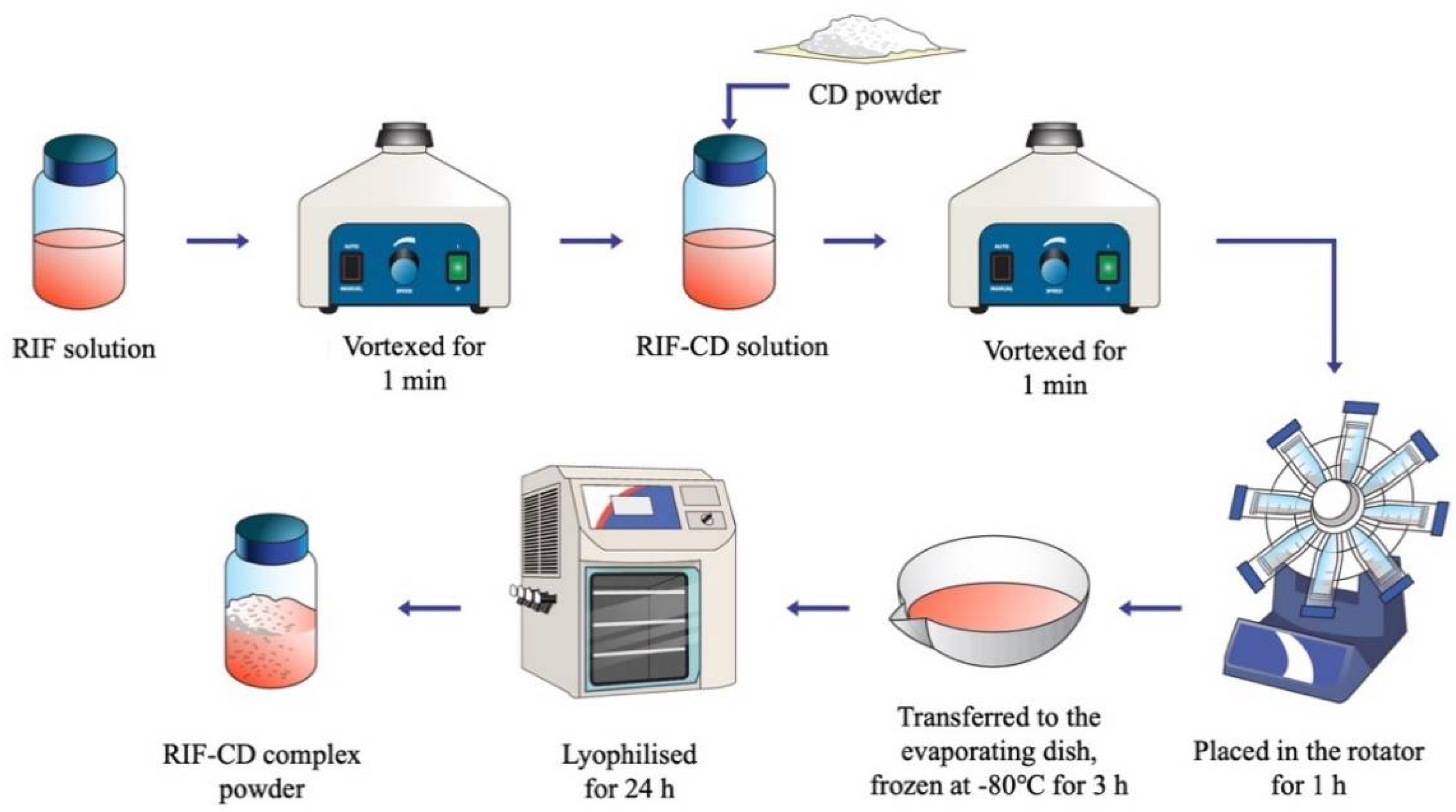
2.6. Preparation of RIF-CD Complexes
2.7. Characterization of Inclusion Complexes
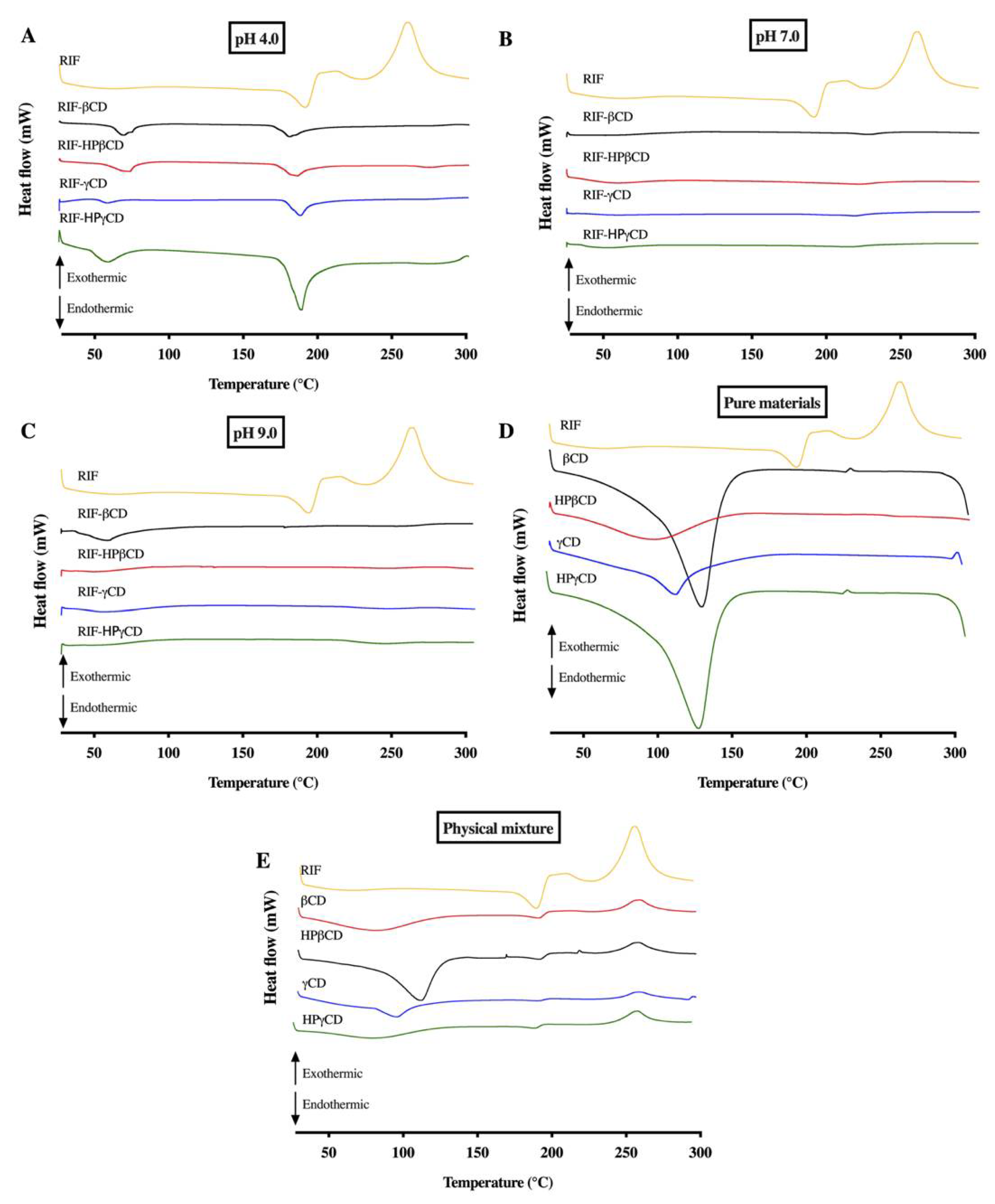
2.7.1. Differential scanning calorimetry
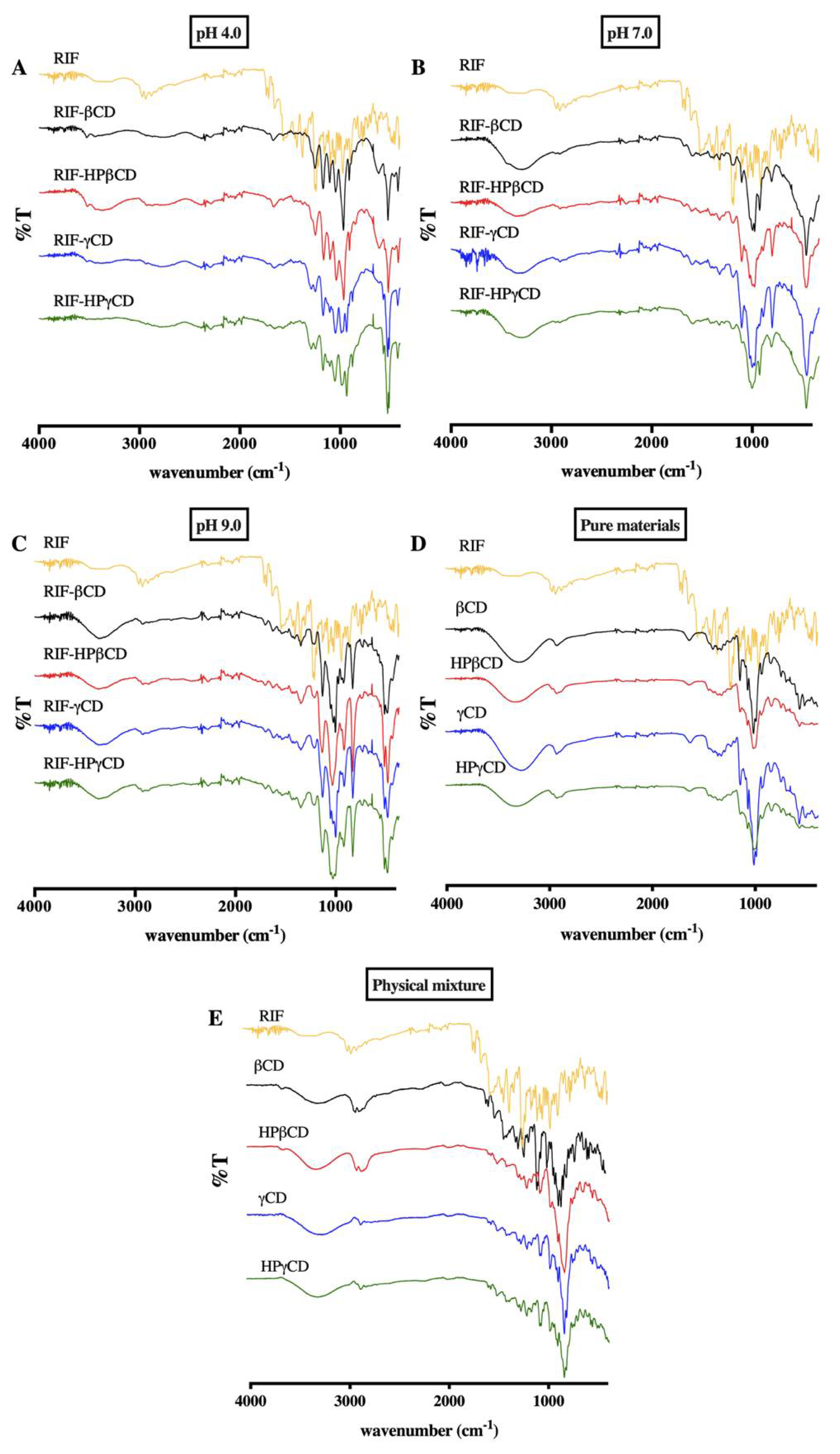
2.7.2. FT-IR Spectroscopy
2.7.3. X-ray Diffraction Analysis
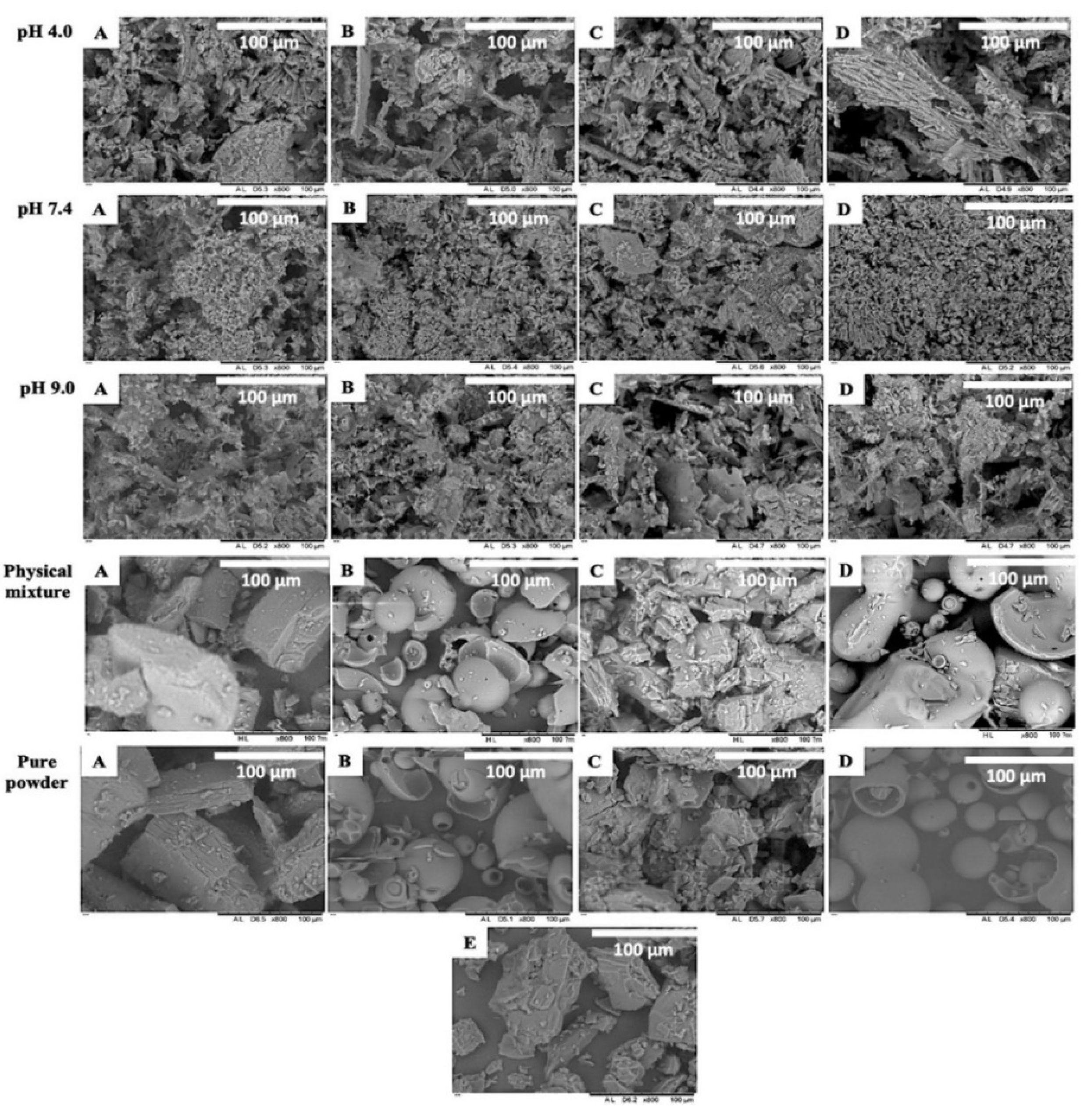
2.7.4. Morphology and Structure
2.8. In Vitro Release Study
2.9. In Vitro Antibacterial Activity
2.10. Instrumentation and Chromatographic Conditions for RIF Quantification
2.11. Statistical Analysis
3. Results and Discussion
3.1. Saturation Solubility
3.2. Molecular Modeling Studies
3.3. Phase Solubility Study
3.4. Determination of Constant Values: Stability Constant, Complexation Efficiency, and Solubility Enhancement Ratio
3.5. Characterization of Inclusion Complexes
3.5.1. Differential Scanning Calorimetry
3.5.2. FT-IR Spectroscopy
3.5.3. X-ray Diffraction Analysis
3.5.4. Morphology and Structure
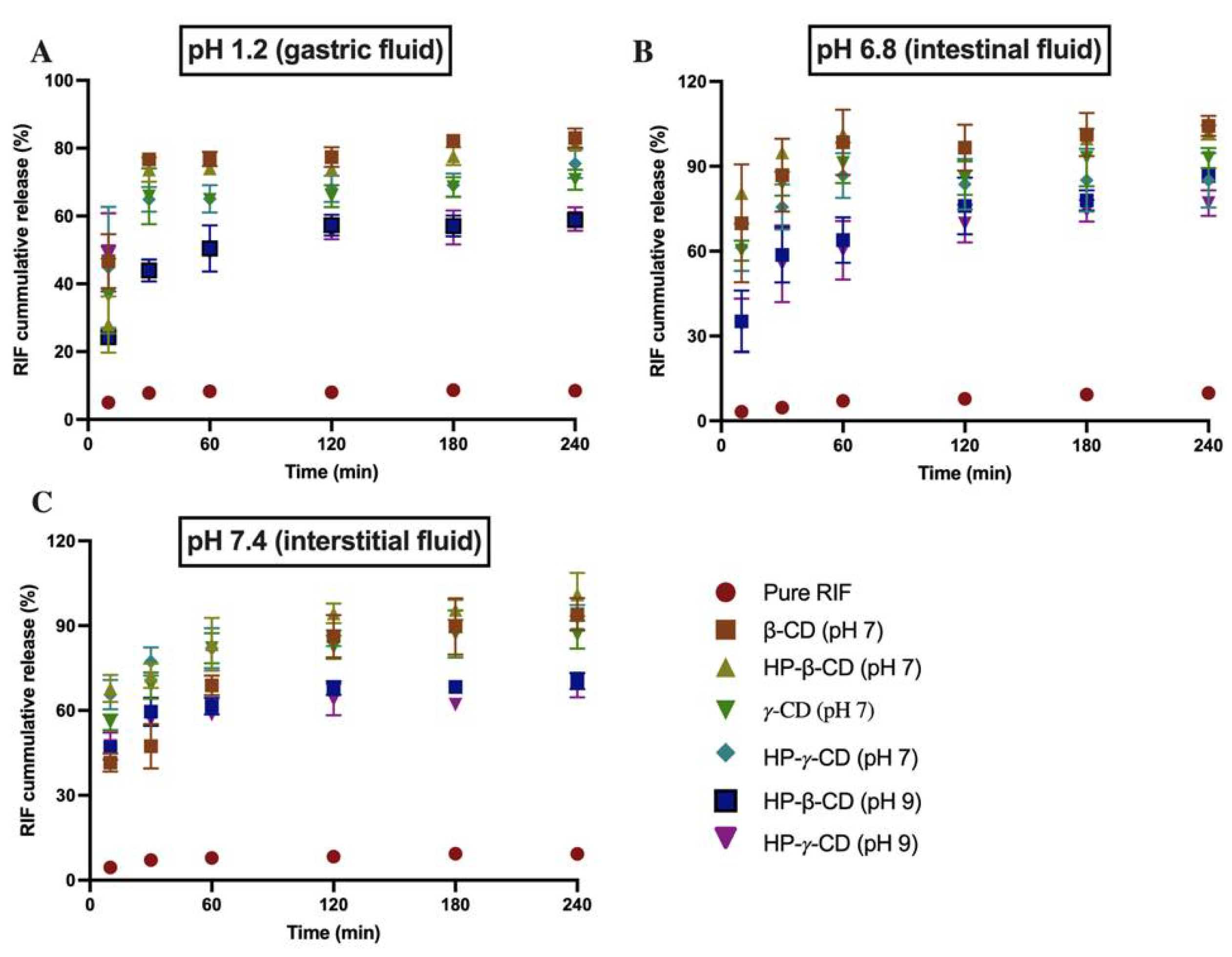
3.6. Dissolution Study
3.7. Antibacterial Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fogel, N. Tuberculosis: A disease without boundaries. Tuberculosis 2015, 95, 527–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Global Tuberculosis Report 2019; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Cruz-Knight, W.; Blake-Gumbs, L. Tuberculosis: An overview. Prim. Care Clin. Off. Pract. 2013, 40, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Araújo-Mariz, C.; Lopes, E.P.; Acioli-Santos, B.; Maruza, M.; Montarroyos, U.R.; De Ximenes, R.A.A.; Lacerda, H.R.; Miranda-Filho, D.D.B.; De Militão De Albuquerque, M.F.P. Hepatotoxicity during treatment for tuberculosis in people living with HIV/AIDS. PLoS ONE 2016, 11, e0157725. [Google Scholar] [CrossRef] [PubMed]
- Tostmann, A.; Boeree, M.J.; Aarnoutse, R.E.; De Lange, W.C.M.; Van Der Ven, A.J.A.M.; Dekhuijzen, R. Antituberculosis drug-induced hepatotoxicity: Concise up-to-date review. J. Gastroenterol. Hepatol. 2008, 23, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Saukkonen, J.J.; Cohn, D.L.; Jasmer, R.M.; Schenker, S.; Jereb, J.A.; Nolan, C.M.; Peloquin, C.A.; Gordin, F.M.; Nunes, D.; Strader, D.B.; et al. An official ATS statement: Hepatotoxicity of antituberculosis therapy. Am. J. Respir. Crit. Care Med. 2006, 174, 935–952. [Google Scholar] [CrossRef]
- Zargar, S.A.; Thapa, B.R.; Sahni, A.; Mehta, S. Rifampicin—Induced upper gastrointestinal bleeding. Postgrad. Med. J. 1990, 66, 310–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodman, L.S.; Gilman, A.; Hardman, J.G.; Gilman, A.G.; Limbird, L.E. Goodman & Gilman’s the Pharmacological Basis of Therapeutics, 12th ed.; McGraw-Hill: New York, NY, USA, 2011; pp. 1–35. ISBN 978-0-07-162442-8. [Google Scholar]
- Du Toit, L.C.; Pillay, V.; Danckwerts, M.P. Tuberculosis chemotherapy: Current drug delivery approaches. Respir. Res. 2006, 7, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saokham, P.; Muankaew, C.; Jansook, P.; Loftsson, T. Solubility of cyclodextrins and drug/cyclodextrin complexes. Molecules 2018, 23, 1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maheriya, M.; Cyclodextrin, P. A Promising Candidate in Enhancing Oral Bioavailability of poorly Water Soluble Drugs. MOJ Bioequivalence Bioavailab. 2017, 3, 60–63. [Google Scholar] [CrossRef] [Green Version]
- Loftsson, T.; Jarho, P.; Másson, M.; Järvinen, T. Cyclodextrins in drug delivery. Expert Opin. Drug Deliv. 2005, 2, 335–351. [Google Scholar] [CrossRef]
- Larraneta, E.; Isasi, J.R. Self-Assembled supramolecular gels of reverse poloxamers and cyclodextrins. Langmuir 2012, 28, 12457–12462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larrañeta, E.; Isasi, J.R. Non-covalent hydrogels of cyclodextrins and poloxamines for the controlled release of proteins. Carbohydr. Polym. 2014, 102, 674–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loftsson, T.; Hreinsdóttir, D.; Másson, M. Evaluation of cyclodextrin solubilization of drugs. Int. J. Pharm. 2005, 302, 18–28. [Google Scholar] [CrossRef]
- Chadha, R.; Saini, A.; Gupta, S.; Arora, P.; Thakur, D.; Jain, D.V.S. Encapsulation of rifampicin by natural and modified β-cyclodextrins: Characterization and thermodynamic parameters. J. Incl. Phenom. Macrocycl. Chem. 2010, 67, 109–116. [Google Scholar] [CrossRef]
- Patil, J.S.; Suresh, S. Physicochemical characterization, in vitro release and permeation studies of respirable rifampicin-cyclodextrin inclusion complexes. Indian J. Pharm. Sci. 2009, 71, 638–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tewes, F.; Brillault, J.; Couet, W.; Olivier, J.C. Formulation of rifampicin-cyclodextrin complexes for lung nebulization. J. Control. Release 2008, 129, 93–99. [Google Scholar] [CrossRef]
- He, D.; Deng, P.; Yang, L.; Tan, Q.; Liu, J.; Yang, M.; Zhang, J. Molecular encapsulation of rifampicin as an inclusion complex of hydroxypropyl-β-cyclodextrin: Design, characterization and in vitro dissolution. Colloids Surf. B Biointerfaces 2013, 103, 580–585. [Google Scholar] [CrossRef]
- Bruno, I.J.; Cole, J.C.; Edgington, P.R.; Kessler, M.; Macrae, C.F.; McCabe, P.; Pearson, J.; Taylor, R. New software for searching the Cambridge Structural Database and visualizing crystal structures. Acta Crystallogr. B. 2002, 58, 389–397. [Google Scholar] [CrossRef]
- Macrae, C.F.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Shields, G.P.; Taylor, R.; Towler, M.; Van De Streek, J. Mercury: Visualization and analysis of crystal structures. J. Appl. Crystallogr. 2006, 39, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [Green Version]
- Labute, P. Protonate3D: Assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins 2009, 75, 187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, P.; Ma, X.; Wu, D.; Li, S.; Xu, K.; Tang, B.; Li, H. Posaconazole/hydroxypropyl-β-cyclodextrin host-guest system: Improving dissolution while maintaining antifungal activity. Carbohydr. Polym. 2016, 142, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Loftsson, T.; Brewster, M.E. Cyclodextrins as functional excipients: Methods to enhance complexation efficiency. J. Pharm. Sci. 2012, 101, 3019–3032. [Google Scholar] [CrossRef] [PubMed]
- Anjani, Q.K.; Permana, A.D.; Cárcamo-Martínez, Á.; Domínguez-Robles, J.; Tekko, I.A.; Larrañeta, E.; Vora, L.K.; Ramadon, D.; Donnelly, R.F. Versatility of hydrogel-forming microneedles in in vitro transdermal delivery of tuberculosis drugs. Eur. J. Pharm. Biopharm. 2021, 158, 294–312. [Google Scholar] [CrossRef] [PubMed]
- Migdadi, E.M.; Courtenay, A.J.; Tekko, I.A.; McCrudden, M.T.C.; Kearney, M.C.; McAlister, E.; McCarthy, H.O.; Donnelly, R.F. Hydrogel-forming microneedles enhance transdermal delivery of metformin hydrochloride. J. Control. Release 2018, 285, 142–151. [Google Scholar] [CrossRef]
- Klein, S. The use of biorelevant dissolution media to forecast the in vivo performance of a drug. AAPS J. 2010, 12, 397–406. [Google Scholar] [CrossRef] [Green Version]
- Radaelli, M.; da Silva, B.P.; Weidlich, L.; Hoehne, L.; Flach, A.; da Costa, L.A.M.A.; Ethur, E.M. Antimicrobial activities of six essential oils commonly used as condiments in Brazil against clostridium perfringens. Braz. J. Microbiol. 2016, 47, 424–430. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, S.; Panchagnula, R. Dissolution test as a surrogate for quality evaluation of rifampicin containing fixed dose combination formulations. Int. J. Pharm. 2004, 287, 97–112. [Google Scholar] [CrossRef]
- Ferreira, D.A.; Ferreira, A.G.; Vizzotto, L.; Federman Neto, A.; De Oliveira, A.G. Analysis of the molecular association of rifampicin with hydroxypropyl-β-cyclodextrin. Rev. Bras. Ciências Farm. 2004, 40, 43–51. [Google Scholar] [CrossRef]
- Samuelsen, L.; Holm, R.; Lathuile, A.; Schönbeck, C. Correlation between the stability constant and pH for β-cyclodextrin complexes. Int. J. Pharm. 2019, 568, 118523. [Google Scholar] [CrossRef]
- Pereva, S.; Nikolova, V.; Angelova, S.; Spassov, T.; Dudev, T. Water inside β-cyclodextrin cavity: Amount, stability and mechanism of binding. Beilstein J. Org. Chem. 2019, 15, 1592–1600. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ye, D.; Gao, X.; Liu, W.; Li, F. Study of α-, β-and γ-Cyclodextrin/C60 Supramolecular Complexes: Influence of the Cavity Dimension toward the Host-Guest Complexes. Fuller. Nanotub. Carbon Nonstructures 2007, 15, 317–330. [Google Scholar] [CrossRef]
- Panchagnula, R.; Bedi, S.; Yasvanth, A. Polymorphism of rifampicin in FDCs: Correlation with in vitro dissolution test. Clin. Res. Regul. Aff. 2006, 23, 53–70. [Google Scholar] [CrossRef]
- Alves, R.; Reis, T.V.D.S.; Da Silva, L.C.C.; Storpírtis, S.; Mercuri, L.P.; Matos, J.D.R. Thermal behavior and decomposition kinetics of rifampicin polymorphs under isothermal and non-isothermal conditions. Brazilian J. Pharm. Sci. 2010, 46, 343–351. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Tan, W.; Zhou, F.; Li, Z.; Duan, L. The effect of phosphate buffer solutions on uniconazole complexation with hydroxypropyl-β-cyclodextrin and methyl-β-cyclodextrin. J. Incl. Phenom. Macrocycl. Chem. 2012, 73, 193–198. [Google Scholar] [CrossRef]
- Lucio, D.; Irache, J.M.; Font, M.; Martínez-Ohárriz, M.C. Supramolecular structure of glibenclamide and β-cyclodextrins complexes. Int. J. Pharm. 2017, 530, 377–386. [Google Scholar] [CrossRef]


| pH | CD Type | Type of Diagram | R2 | Intercept | Slope | Ks (M−1) | CE | RIF:CD Molar Ratio |
|---|---|---|---|---|---|---|---|---|
| 4.0 | βCD | AL | 0.978 | 0.400 | 0.079 | 215 | 0.09 | 1:12 |
| HP-βCD | AL | 0.983 | 0.363 | 0.164 | 541 | 0.20 | 1:6 | |
| γCD | AL | 0.983 | 0.494 | 0.485 | 2510 | 0.80 | 1:2 | |
| HP-γCD | AL | 0.991 | 0.440 | 0.245 | 738 | 0.32 | 1:4 | |
| 7.0 | βCD | AL | 0.978 | 1.171 | 0.227 | 250 | 0.29 | 1:4 |
| HP-βCD | AL | 0.961 | 1.096 | 0.480 | 848 | 0.92 | 1:2 | |
| γCD | AL | 0.965 | 0.950 | 0.549 | 1284 | 1.22 | 1:2 | |
| HP-γCD | AL | 0.959 | 1.145 | 0.435 | 674 | 0.77 | 1:2 | |
| 9.0 | βCD | - | - | - | - | - | - | |
| HP-βCD | AL | 0.942 | 1.724 | 0.528 | 649 | 1.12 | 1:2 | |
| γCD | - | - | - | - | - | - | ||
| HP-γCD | AL | 0.9366 | 1.370 | 0.497 | 722 | 0.99 | 1:2 |
| Sample | MIC (µg/mL) | |
|---|---|---|
| Staphylococcus aureus (NCTC® 10788) | Methicillin-Resistant Staphylococcus aureus (ATCC® 33593™) | |
| Pure RIF (solution) | 0.78 | 0.78 |
| Pure RIF (suspension) | 1.56 | 1.56 |
| RIF-β-CD (pH 7.0) | 0.78 | 0.78 |
| RIF-HP-β-CD (pH 7.0) | 1.56 | 1.56 |
| RIF-γCD (pH 7.0) | 1.56 | 1.56 |
| RIF-HP-γCD (pH 7.0) | 0.78 | 0.78 |
| RIF-HP-β-CD (pH 9.0) | 1.56 | 1.56 |
| RIF-HP-γCD (pH 9.0) | 1.56 | 1.56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anjani, Q.K.; Domínguez-Robles, J.; Utomo, E.; Font, M.; Martínez-Ohárriz, M.C.; Permana, A.D.; Cárcamo-Martínez, Á.; Larrañeta, E.; Donnelly, R.F. Inclusion Complexes of Rifampicin with Native and Derivatized Cyclodextrins: In Silico Modeling, Formulation, and Characterization. Pharmaceuticals 2022, 15, 20. https://doi.org/10.3390/ph15010020
Anjani QK, Domínguez-Robles J, Utomo E, Font M, Martínez-Ohárriz MC, Permana AD, Cárcamo-Martínez Á, Larrañeta E, Donnelly RF. Inclusion Complexes of Rifampicin with Native and Derivatized Cyclodextrins: In Silico Modeling, Formulation, and Characterization. Pharmaceuticals. 2022; 15(1):20. https://doi.org/10.3390/ph15010020
Chicago/Turabian StyleAnjani, Qonita Kurnia, Juan Domínguez-Robles, Emilia Utomo, María Font, María Cristina Martínez-Ohárriz, Andi Dian Permana, Álvaro Cárcamo-Martínez, Eneko Larrañeta, and Ryan F. Donnelly. 2022. "Inclusion Complexes of Rifampicin with Native and Derivatized Cyclodextrins: In Silico Modeling, Formulation, and Characterization" Pharmaceuticals 15, no. 1: 20. https://doi.org/10.3390/ph15010020
APA StyleAnjani, Q. K., Domínguez-Robles, J., Utomo, E., Font, M., Martínez-Ohárriz, M. C., Permana, A. D., Cárcamo-Martínez, Á., Larrañeta, E., & Donnelly, R. F. (2022). Inclusion Complexes of Rifampicin with Native and Derivatized Cyclodextrins: In Silico Modeling, Formulation, and Characterization. Pharmaceuticals, 15(1), 20. https://doi.org/10.3390/ph15010020










