Abstract
A library of novel naproxen based 1,3,4-oxadiazole derivatives (8–16 and 19–26) has been synthesized and screened for cytotoxicity as EGFR inhibitors. Among the synthesized hybrids, compound2-(4-((5-((S)-1-(2-methoxynaphthalen-6-yl)ethyl)-1,3,4-oxadiazol-2-ylthio)methyl)-1H-1,2,3-triazol-1-yl)phenol(15) was the most potent compound against MCF-7 and HepG2cancer cells with IC50 of 2.13 and 1.63 µg/mL, respectively, and was equipotent to doxorubicin (IC50 1.62 µg/mL) towards HepG2. Furthermore, compound 15 inhibited EGFR kinase with IC50 0.41 μM compared to standard drug Erlotinib (IC50 0.30 μM). The active compound induces a high percentage of necrosis towards MCF-7, HePG2 and HCT 116 cells. The docking studies, DFT and MEP also supported the biological data. These results demonstrated that these synthesized naproxen hybrids have EGFR inhibition effects and can be used as leads for cancer therapy.
1. Introduction
Cancer is a devastating disease characterized by uncontrolled cell division and proliferation [1]. It is a major health burden globally and ranked second after cardiovascular diseases [2]. Although the development of anticancer drugs has advanced in recent years, due to their side effects such as drug resistance and non-differentiation between cancerous and normal cells, there is still an urgent need to discover more effective anticancer drugs which can only target cancer cells with reduced side effects [3,4].
Epidermal Growth Factor Receptor (EGFR) is an attractive target in cancer therapy due to its involvement in cell proliferation and differentiation [5,6]. A number of EGFR inhibitors such as Lapatinib, Gefitinib, Afatinib and Erlotinib have been developed for use in cancer therapy [7] (Figure 1) as EGFR is overexpressed or constitutively active due to mutataions in various cancers. EGFR inhibitors are small molecule tyrosine kinase inhibitors which block receptor signalling by interring with ATP binding to the receptor or monoclonal antibodies which bind to the extracellular region of the receptor, thereby inhibiting its dimerisation and autophosphorylation [8]. However, due to drug resistance and toxicity related to currently available EGFR inhibitors [9] there is an urgent need to develop new EGFR inhibitors which can overcome these side effects.
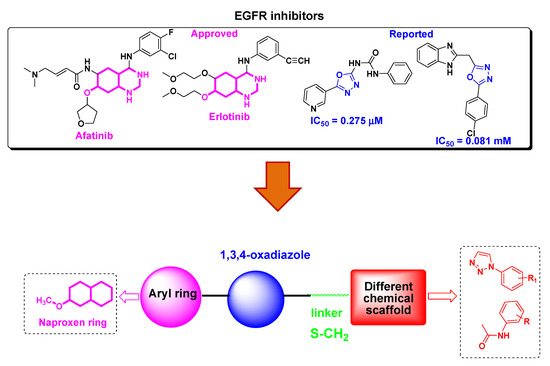
Figure 1.
Structures of EGFR inhibitors and design of naproxen-1,3,4-oxadiazole derivatives.
Naproxen is a well knownnon-steroidal antiinflammatory drug (NSAIDs) that acts as a COX inhibitor [10]. In addition to theiranti-inflammatory activity, naproxen derivatives have been explored for anticancer activity [11,12,13,14,15]. For example, Naproxen-based urea and propanamide derivatives have shown promising inhibition in colon cancer cells [16], protection in bladder cancer proliferation [17] and inhibitors of VEGFR-2 and histone deacetylase enzymes [18,19]. Heterocycles form the basic skeleton for a number of molecules of biological interests [20]. 1,3,4-oxadiazole is an important scaffold as it exhibits remarkable pharmacological activities such as anticancer, antiinflammatory, antimicrobial, antiviral, etc [21,22,23,24]. Furthermore, this moiety shows an antiproliferative effect through EGFR kinase inhibition (Figure 1) [25,26,27,28].
In continuation of our previously reported work to find new leads with potential anticancer activity [29,30,31], we thought to explore naproxen incorporated 1,3,4-oxadiazole derivatives for anticancer potential. To the best of our knowledge, the anticancer effect of naproxen derivatives as EGFR has not been reported yet. Therefore, we report the synthesis of naproxen based 1,3,4-oxadiazole hybrids, their cytotoxicity, in vitro EGFR inhibition and computational studies.
2. Materials and Methods
2.1. Chemistry
2.1.1. General
Naproxen was purchased from Sigma Aldrich (St. Louis, MO, USA). 1H NMR and 13C NMR was carried out on a Bruker spectrometer at 850 MHz in DMSO or CDCl3 solvents. IR was performedon a Thermo Scientific iS 50 using the ATR technique. The melting point was recorded on a Stuart SMP40 machine. The molecular weights of the synthesized compounds were measured on a mass spectrometer (LCQ Fleet-LCF10605) and elemental analyses were performed on a LEECO Elementar Analyzer.
2.1.2. Synthesis of Final Compounds (8–16)
The target compounds (8–16) were prepared according to our previous work [31] with fewmodifications. The key intermediate 5 (0.001 mole) was charged into a round bottom flask (100 mL) followed by the addition of ter.butanol:water (2:1, 50 mL). The reaction mixture was stirred at 40–50 °C and then cooled to room temperature. Copper sulphate pentahydrate and sodium ascorbate (0.001 mole each) wereadded to the reaction mixture followed by the addition of different aromatic azides (0.0012 mole), and stirring was continued for 4–12 h. To the reaction mixture, water (50 mL) was added and products were isolated by extracting twice with dichloromethane (DCM, 50 mL), and the DCM layer was washed with water and dried over anhydrous sodium sulphate. The MDC layer was concentrated and recrystallized using MDC and cyclohexane (Scheme 1).
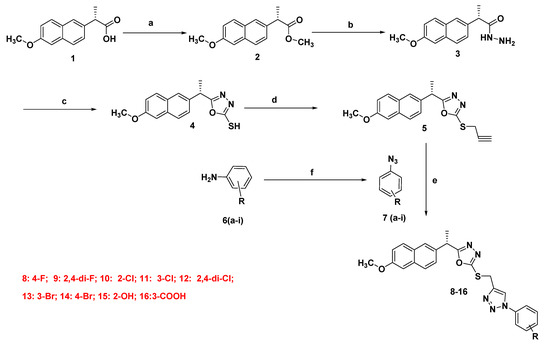
Scheme 1.
Synthetic route of target compounds 8–16. Reagent and conditions: (a) MeOH, H2SO4, 60–70 °C, 10 h; (b) NH2·NH2·H2O, EtOH, 80–90 °C, 8 h; (c) i- KOH, EtOH, CS2, stir, 24 h, ii- 90–100 °C, 10h; (d) K2CO3, acetone, propargyl bromide, 50–60 °C, 4 h; (e) H2O-tBuOH (1:1), CuSO4·5H2O, Na ascorbate, r.t., 4–12 h; (f) NaNO2, HCl, NaN3, stir, 0–(−5) °C.
2.1.3. Synthesis of Final Compounds Bearing Acetamide (19–26)
To a clean round bottom flask (100 mL), intermediate 4 (0.001 mole) was added followed by addition of 100 mL dry acetone and anhydrous potassium carbonate (0.0012 mole). The reaction mixture was stirred for 1 h at 50–60 °C then different chloro acetamides (0.0011 mole) were added, andstirring continued untilcompletion of the reaction. After completion of the reaction, the reaction mixture was filtered and the filtrate was concentrated and poured into water (50 mL). The compounds were isolated with ethylacetate (50 mL × 2). The combined organic layer was concentrated and finally recrystallized with petroleum ether and ethyl acetate (Scheme 2).
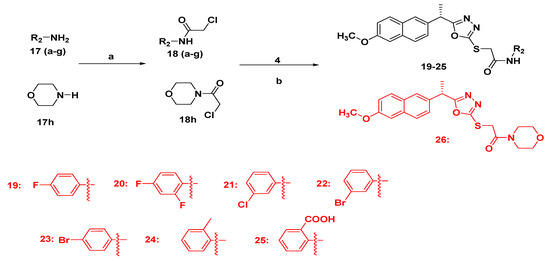
Scheme 2.
Synthetic route of target compounds 19–26. Reagents and conditions: (a) ClCH2COOCH2CH3, TEA, DCM, 0–5 °C, 10 h; (b) acetone, K2CO3, 50–60 °C, 10–12 h.
2.1.4. Analytical Data
The analytical data including physical appearance, % yield and melting point of all final compounds 8–16 and 19–26 are present in Supplementary Material S1. The spectra (1H NMR, 13C NMR and mass) are also enclosed.
2.2. Biological Activities
2.2.1. Antiproliferative Activity
The cytotoxicity of the new isolated compounds was evaluated against MCF-7 (breast cells), HepG2 (Hepatocellular) and HCT-116 (colorectal cells) human cancer cells using sulphorhodamine B assay (SRB). This assay was performed as previously published [32].
2.2.2. Apoptosis Using Acridine Orange/Ethidium Bromide Staining
AO and EtBr are DNA binding dyes. They have been used for detection of the morphological features of apoptotic and necrotic cells. The stainings were performed according the protocol as previously published [33].
2.2.3. EGFR Kinase Activity
In vitroEGFR inhibitory activity of most active compounds was evaluated by Enzyme Linked immunoabsorbent assay (ELISA) using a EGFR Erb B1 kinase kit (Sigma) with Erlotinib as a positive control according to the reported method [34]. Briefly, solutions of kinase/peptide and ATP were prepared prior to use. The solution on the plate were carefully mixed and incubated for 1 h at 25 °C. Then, 5 mL of this prepared solution was added into 96 well plates, incubated for 1 h and determined by an ELISA Reader (Perkin Elmer, Waltham, MA, USA). Each experiment was repeated three times and IC50 represented as mean ± S.E, was calculated using Graph Pad Prism 5.
2.2.4. Statistical Analysis
Data were presented as mean standard deviation unless otherwise indicated. Significance of the statistical analysis was acceptable to a level of p < 0.05.
2.3. Computational Details and Molecular Docking
The Jaguar package used for DFT calculations also applied for ligand preparation [35]. The geometrical optimizations and chemical reactivity were estimated using B3LYP/MIDIX level using 6-311++G** basis set. The optimized geometry structures were implemented in the Maestro package, followed by minimization using an OPLS force field. TM protein was obtained in 3D crystal structure (PDB ID: 1M17), then minimized using an OPLS force field. The binding site was investigated by theheCASTp package. Glide software was utilized for the generation of the Receptor grid and Docking simulation process, and the remaining procedure was conducted as reported in previous research [31].
3. Results and Discussion
3.1. Chemistry
The synthetic path of naproxen hybrids 8–16 and 19–26 are presented in Scheme 1 and Scheme 2. The key intermediates 4 and 5 were prepared according to the reported methods with little modification [30]. Firstly, Naproxen 1 was converted into methyl ester 2 by reacting with methanol in presence of concentrated sulphuric acid as catalyst. The methyl ester 2 was refluxed with hydrazine monohydrate in methanol to yield hydrazide 3, which was further reacted with carbon disulphide in presence of potassium hydroxide in absolute ethanol at room temperature overnight and then refluxed for 12 h to obtainthe key intermediate 4. This key intermediate 4 was reacted with propargyl bromide in presence of potassium carbonate using dry acetone as solvent to obtainother key intermediate 5. The target compounds 8–16 were synthesized via Click dipolar cycloaddition approach from 5 using different aromatic azides in presence of tert BuOH:H2O (1:1) as a solvent. Other target compounds 19–26 were obtained by S-alkylation of key intermediate 4 with different chloroacetamide using acetone and potassium carbonate. The structures of all final hybrids were confirmed by spectroscopic techniques and elemental analysis. The structure of final compounds 8–16 displayed characteristic peaks for 1,2,3 triazolyl protons in the range δ 8.10–8.46 ppm in 1H NMR and 1,3,4-oxadiazole ring in the range δ 159.46–165.89 ppm in 13C NMR supporting their formation. In compounds 19–26, the absence of triazolyl proton which was present in 8–16 in 1H NMR and appearance of a new singlet for NH in the range δ 8.58–9.47 ppm in 1H NMR and downshielded C=O peaks in the range δ 171.04–176.59 ppm in 13C NMR confirmed the presence of NH-C=O group in hybrids 19–26. The linker S-CH2 between heterocycles appeared in range δ 3.94–4.64 ppm as a singlet in 1H NMR and δ 25.84–37.69 ppm in 13C NMR spectra. The electron ionization mass spectra were in agreement with the structures of all final compounds.
3.2. In Silico ADME/Pharmacokinetics Studies
The pharmacological and pharmacokinetic properties of a compound are important parameters in drug development and discovery as theyreduce the time and cost in the development of a drug [23]. The molecule must satisfy Lipinski and Veber’s rules to be developed as an effective orally available drug. Any violation of Lipinski and Veber’s rules might lead to problems with bioavailability, permeability, solubility, etc [36]. The final compounds have been subjected toin silicoADME studies to predict their physicochemical and pharmacokinetics parameters.
The in silico pharmacokinetics data (Table 1) showed that all the target compounds except 9, 12, 13 and 14 indicated easy transportation as they displayed a molecular weight below 500. These compounds depicted percentage absorption in the range of 60.19–73.62%, suggesting good absorption by the human body. The physicochemical parameters such as hydrogen bond acceptor (less than 5)/donor (less than 10) required for a drug development were found to be in the range of 5–8 and 0–2, respectively, for the final hybrids. Permeability of a drug is another important parameter which decides the fate of a drug and should be less than 5. The final compounds exhibited good permeability as indicated by logP values in the range of 2.92–4.67. However, the standard drug, doxorubicin, indicated three violations of the Lipinski rules as it exhibited MW of 543.52, nROTB = 5, HBA/HBD-12/6, TPSA = 206.07, ilogP = 2.58, low GI absorption and % abs of 37.90. These results revealed that the compounds 8–16 and 19–26 possess desired pharmacokinetics properties for the development of a lead molecule.

Table 1.
Physicochemical and pharmacokinetic properties of final compounds 8–16 and 19–26.
3.3. Biological Activities
3.3.1. Antiproliferative Activity
The antiproliferative activity of the target compounds was assessed on three cancer lines, MCF-7, HepG2 and HCT-116 cells using sulforhodamine assay [32]. The cytotoxic results displayed that the compounds against tested tumor cell lines showed variable cytotoxic activity (Table 2). Compound 15 bearing hydroxyl group at the ortho position of the triazolyl ring was the most potent compound against MCF-7 and HepG2cancer cells with IC50 of 2.13 and 1.63 µg/mL, respectively, and was equipotent to doxorubicin (IC50 1.62 µg/mL) towards HepG2 (Figure 2). The halogen-substituted 1,2,3-triazolyl analogues displayed good cytotoxicity with IC50 in the range 5.05–15.84 µg/mL against MCF-7 cells and 3.53–25.49 µg/mL towards HepG2, whereas only compound 9 was active against HCT-116 with IC50 7.46 µg/mL.

Table 2.
Cytotoxicity of final compounds 8–16 and 19–26.
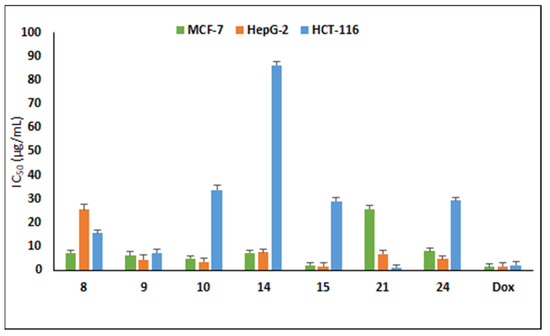
Figure 2.
Cytotoxicity of active compounds on MCF-7, HepG2 and HCT-116 cancer cells. Data represent three independent experiments in mean ± SD. Dox: doxorubicin (positive control).
Naproxen-bearing oxadiazole and acetamide moieties were less effective in exerting the anticancer effect on the tested cell lines. Compound 21 possessing 3-Cl exhibited prominent antiproliferative activity with two-fold (IC50 1.24 µg/mL) cytotoxicity to doxorubicin (IC50 2.11 µg/mL) on colorectal HCT-116 cells, whereas compound 24 was active against MCF-7 (IC50 8.15 µg/mL) and HepG2 (IC50 4.88 µg/mL). Other compounds displayed moderate activity with IC50 less than 100 µg/mL and two compounds (19 and 22) were inactive towards the tested cancer cells with IC50 above 100 µg/mL. From these data, it can be noted that these synthesized naproxen analogues have the capability to kill cancerous cells.
3.3.2. In Vitro EGFR Activity
EGFR overexpression is reported to play a significant role in the uncontrolled proliferation of different cancers.Therefore inhibition of EGFR has been a captivating target for cancer therapy. Compounds with promising cytotoxicity against the tested cell line were selected for EGFR kinase activity and werecompared with reference drug, erlotinib. From the results EGFR inhibitory activity (Table 3), all hybrids displayed remarkable inhibitory effects on EGFR with IC50 ranging between 0.41–7.31 μM. 1,2,3-triazole incorporated naproxen hybrids were more active in EGFR inhibition than acetamide containing naproxen hybrids. Two compounds, 15 and 10, were the most potent compounds, with IC50 0.41 μM and 0.67 μM, respectively, in inhibiting EGFR kinase compared to standard drug Erlotinib (IC50 0.30 μM). Naproxen hybrids-bearing acetamide groups, 21 and 24, revealed good inhibition with IC50 of 0.82 μM and 1.08 μM, respectively. It was noted that position of substituent on the phenyl ring linked to 1,2,3-triazole played significant role in EGFR inhibition. Compound 15 (2-OH) and 10 (2-Cl) bearing substituents at ortho position exhibited significant EGFR inhibition than meta substituted hybrids (16 and 13) and the activity was further diminished by para substituted hybrids (8 and 14). This trend in activity may be attributed to the close binding of ortho substituted group with the receptor. It was also observed that disubstituted hybrids (9, IC50 0.71 μM) were more potent in EGFR inhibition than monosubstituted (8, IC50 7.31 μM). Replacement of bromo group (13) with COOH group (16) enhanced the EGFR activity. These results demonstrated that these hybrids have EGFR inhibition effects and well correlates with the in vitro antiproliferative data.

Table 3.
Inhibitory activity of derivatives on EGFR.
3.3.3. Apoptosis Studies
The MCF-7, HepG2 and HCT 116 cancer cells were categorized by AO/EtBr dual staining [33]. The different colors of the cells indicate different stages of apoptosis. Normal green nucleus indicates living cells; bright green nucleus with fragments of chromatin indicates early apoptotic stage; orange nuclei along with fragmented chromatin fibres shows late apoptosis and uniformly orange nuclei indicate necrotic cells. After 48 h of staining, the potent compounds were treated and examined under a fluorescent microscope. Regularly stained green with normal, round, intact nuclei and cytoplasm indicates the viability of the normal cell (control). As shown in Figure 3, compound 9 and 10 caused a significant rate of cell death in early apoptosis on all the tested three cell lines. However, the percentage of cell death decreased after treatment with compound 15 against HePG2 and HCT 116 cancer cells.
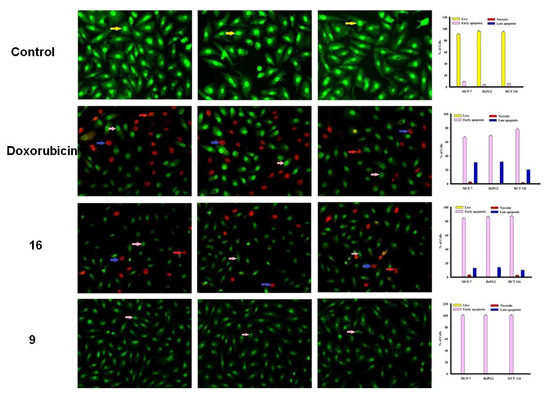
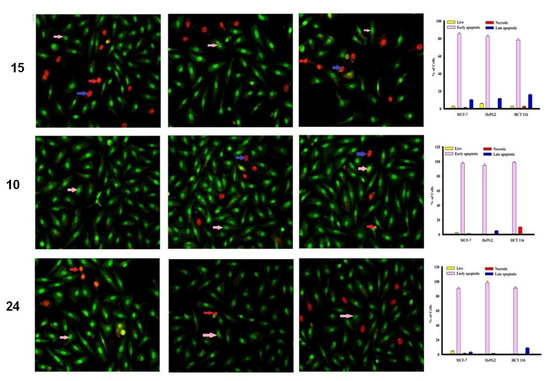
Figure 3.
Cell apoptosis observed using fluorescence microscope. Cells were treated with compound 16, 9, doxorubicin and control with acridine orange-ethyl bromide staining. Cells were treated with compound 15, 10 and 24 with acridine orange-ethyl bromide staining.
In addition, compound 16 induces a high percentage of necrosis towards MCF-7 and HePG2 cells as well as compound 15 with HCT 116 cells, while compound 9 does not cause death of the necrosis pathway in all cancer cells (Figure 3). Late apoptosis was observed at a high rate after treatment with compound 16 on MCF-7 and HePG2 cells and compound 15 induces more late apoptosis in HCT 116 cells compared to other cancer cells.
3.4. Computational Study
3.4.1. Electronic Properties
The highest occupied/lowest unoccupied molecular orbitals (HOMOs/LUMOs) of naproxen derivatives are probed at B3LYP/6–31G** level (Figure 4). HOMO area was observed over terminal naproxenyl groups whereas LUMO zone was found to caped over the opposite terminal scaffold in 8–11 and 19–26 compounds indicating intra-molecular charge transfer (ICT) was observed from HOMOs to LUMOs. The activity of compounds is also strictly associated to the spatial distribution of occupied molecular orbital enlightening the most credible locations in order that certainly attacked by reactive agents. The FMOs superimposed considerably, which is revealing the particularly reactive nature of the drugs. The energies of FMOs, for instance HOMO (EHOMO), LUMO (ELUMO), as well as HOMO-LUMO energy gaps (Egap) are noteworthy parameters to probe molecules’ electronic characteristics. The EHOMO, ELUMO, and Egap of recently synthesized derivatives along with reference drug are displayed in Table S1.
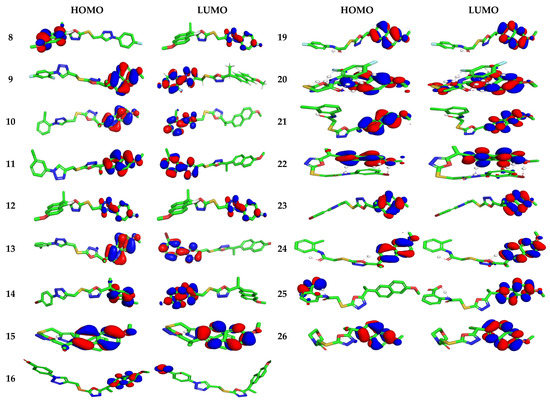
Figure 4.
Ground state charge density of FMOs of studied derivatives with contour value = 0.032.
To determine the activity, the measurement of global chemical reactivity descriptors (GCRD) is an essential consideration which is developed as a reported reference [37,38]. In these parts, we estimated the large number of GCRD parameters specified by energy of HOMO with LUMO orbitals such aselectrophilicity index (ω), softness (S), electronegativity (χ), chemical potential (µ) as well as chemical hardness (η) with the help of HOMO/LUMO energies, vertical ionization energy (IP) and vertical electron affinity (EA). The growing HOMO energy is attributed to high efficacy for biomolecules and good electrophile parameters. In addition, decreasing LUMO-energy is related to good nucleophile character for the molecule [39]. The η of phytochemicals is interconnected to aromaticity [40].
3.4.2. Molecular Electrostatic and Ionization Potential Maps
The molecular electrostatic potential (MEP) is used to explore the reactivity of synthesized hybrids and is measured experimentally by diffraction approaches [41]. MEP maps are important to visualize a region charged within molecules. In Figure 5, the MEP map of the 8–11 and 19–26 compounds is shown in color visualization. Red color defines high negative potential regions that benefit from electrophilic attacks, while blue indicates high positive potential regions that prefer nucleophilic attacks. The MEP is reduced by ordering green > blue > yellow > orange > red, the red indicates a more intense repulsion while blue defines sufficient attraction. The difference between colors provides useful information about the ability of these atomic sites to produce electrophilic and nucleophilic attacks with the receptor. The swelling of green zones may be due to the nucleophilicity power of the compounds. As expected in 8–11 and 19–26 compounds, the green and yellow regions, respectively, are shielded upon aromatic and triazole rings, respectively [42].
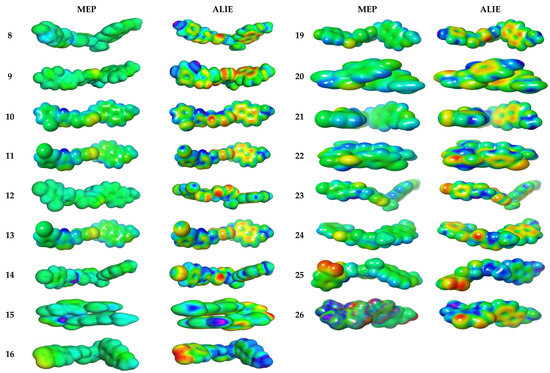
Figure 5.
Molecular electrostatic potential surfaces views of studied derivatives with contour value = 0.032.
ALIE (average local ionization energy map) is more preferable than MEP for prediction of favorable molecular sites to electrophilic attacks. The red color is bounded to both nitrogen atoms in triazole moiety in all compounds 8–11 and 19–26. The electron density in yellow color has high spreading over all molecular backbone for all hybrids 8–11 and 19–26. The blue color is more localized in terminal phenyl groups opposite to naproxenyl moiety; thereby, these groups are prone to nucleophilic attacks. The most hypersensitive action is displayed in compounds 8, 9, 10, 15, 16 and 24 towards electrophilic attackviasurrounding biological media.
3.4.3. Molecular Docking
The comparative study for protein sequences using sequence alignment offers valuable data throughout structural and functional analysis through revealing sequence of the structure–function relationships. Three-dimensional (3D) representation of the EGFR kinase domain was established. The mGen THERADER was applied togenerate a 3D loop structure of PDB 1M17 complexed with Erlotinib, which was used as framework for docking experiment [43]. The X-ray structures of the rough model for the EGFR template werecompared with the target protein using comparative SWISS MODEL [44]. The 3D model was further checked and verified using Ramachandran plot at PROCHECK [45]. The amino acids of EGFR active pocket (Lys721, Phe699, Val702, Cys773, Leu820, Asn818, Asp783, Asn784, Gln958, Gln962, Met96, Met964, Leu97722) were identified using CASTp [44] (Figure 6).
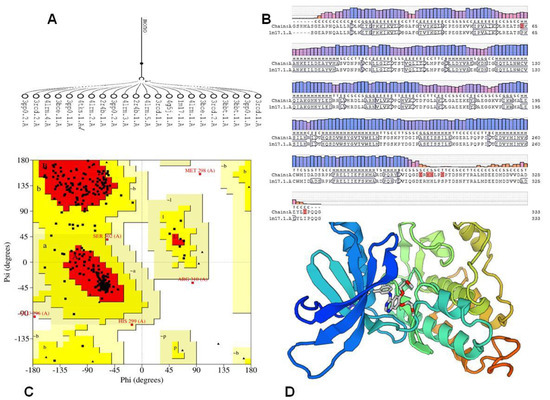
Figure 6.
(A) Alignment of EGFR sequence with rough models. (B) Alignment of Homologues model of 1M17. (C) generated model. (D) Ramachandran Plot: (A,B,L) Most favoured regions in [red] with 86.1%, (a,b,l,p) Additional allowed regions in yellow with 12.2%, generously allowed regions 0.7 [pale yellow], Disallowed regions [white].
In order to find suitable binding modes of the investigated naproxenyl oxadiazols hybrids 8–11 and 19–26 against EGFR kinase, docking studies were simulated using the Schrödinger software [46]. The reproducibility of the software was validated through redocking of hybrids. The investigated hybrids were successfully redocked with a root mean square deviation (RMSD) less than 2 Å. The fingerprint for ligand-protein-interactions was estimated based on the docking score ΔG. All calculated energies of the docking simulation were summarized in Table 4. The hybrids 8–11 and 19–26 were successfully capped into the active zone of the enzymes. The docked poses of complexes were obtained and used for energy-minimized by a molecular-mechanics (Oples3e) force field, until neutralization. The poses were selected depending on the lowest binding free energy ΔG with the lowest root means quart deviation (RMSD) between the pose before and after refinement. Finally, the highest scoring function for the tested compounds was applied to evaluate the binding affinities of the tested compounds.

Table 4.
Docking scores (kcal/mol) of Erlotinib, 8–11 and 19–26 hybrids.
All hybrids 8–11 and 19–26 showed higher binding affinity (−6.3 to −7.14 Kcal/mol.) than Erlotinib (−6.2 Kcal/mol.). The calculated RMSDs are lower than 2 degrees, which reflects the accuracy of the docking process. The most potent compounds, 9, 10, 12, 15, 16, 21 and 24, were selected to clarify their molecular interaction mechanisms for in-depth analysis. The compounds 9, 10, 15, 16 and 24 have promising binding affinities through formation higher number of H & π-π interaction (3 to 5) against EGFR active when compared to Erlotinib (two H-bond),while other compounds 15 and 21 showed the same number of interactions with template drug. It was found that most potent compounds, 9, 10, 12, 15, 16, 21 and 24, were stabilized in binding pocket similar to Erlotinib. The compounds 9, 15, 16 and 24 were arranged in parallel style with important Lys7251 to form a strong H-bond interaction, while compounds 9, 12 and 15 capped binding pocket by formation other important H-bond with Asn818 and localized by perpendicular mode with their amino acid (Figure 7 and Figure 8). The presence of the hydrophobic interactions observed in 9, 10, 21 and 24 represents the higher binding affinities through the receptor-ligands with ranged ligand efficiency (−2.95 to −6.74).
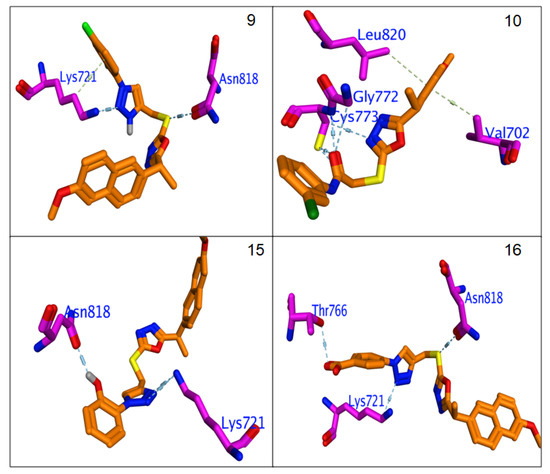
Figure 7.
Binding interactions of most potent Naproxen-1,2,3-triazole hybrids 9, 10, 15 and 16 against EGFR synthase (1M17).
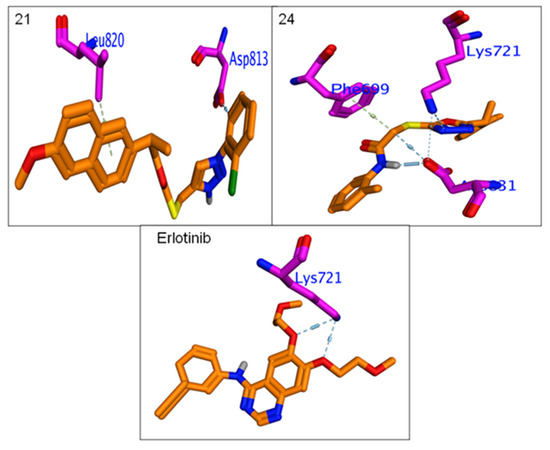
Figure 8.
Binding interactions of most potent Naproxen-acetamide hybrids 21, 24 andreference drug, Erlotinibagainst EGFR synthase (1M17).
4. Conclusions
A library of novel naproxen based 1,3,4-oxadiazole hybrids (8–16 and 19–26) has been synthesized and screened for cytotoxicity as EGFR inhibitors. Among the synthesized hybrids, compound2-(4-((5-((S)-1-(2-methoxynaphthalen-6-yl)ethyl)-1,3,4-oxadiazol-2-ylthio)methyl)-1H-1,2,3-triazol-1-yl)phenol (15) was the most potent compound against MCF-7 and HepG2 cancer cells with IC50 of 2.13 and 1.63 µg/mL, respectively, and was equipotent to doxorubicin (IC50 1.62 µg/mL) towards HepG2. Furthermore, this compound 15 inhibited EGFR kinase with IC50 0.41 μM compared to standard drug Erlotinib (IC50 0.30 μM). The active compound induces a high percentage of necrosis towards MCF-7, HePG2 and HCT 116 cells. The docking studies, DFT and MEP also supported the biological data. From these results, it can be inferred that these naproxen hybrids have the potential to inhibit EGFR kinase by binding with adenosine triphosphate (ATP) through competitive inhibition in tyrosine kinase domain.
Supplementary Materials
The analytical data, physical appearance, melting point, %yield, 1H NMR, 13C NMR and Mass spectra of final derivatives are available online at https://www.mdpi.com/article/10.3390/ph14090870/s1, Figures S1–S17: 1H NMR spectra, Figures S18–S34: 13C NMR spectra, Figures S35–S50: Mass spectra.
Author Contributions
S.N. and M.M.A. designed the experiment and supervision. S.E.I.E. and M.Y.A. performed the biological assay, M.M.A., N.I.A., S.Y.M.A. and A.S.A.A. carried out the synthesis, A.A.E. and M.A.A. carried out the computational study, A.M.M., N.I.A. and S.N. analyzed the data, characterization of compounds and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
Authors acknowledge financial support from Taif University Researchers Supporting Project (Number TURSP-2020/44). The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through General Group Research Project under grant number RGP. 2/66/42.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and supplementary material.
Acknowledgments
Authors are thankful to Albaha University for providing the required facilities for this work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- El-Gohary, N.S.; Gabra, M.T.; Shaaban, M.I. Synthesis, molecular modeling and biological evaluation of new pyrazolo[3,4-b]pyridine analogs as potential antimicrobial, antiquorum-sensing and anticancer agents. Bioorg. Chem. 2019, 89, 102976. [Google Scholar] [CrossRef]
- Alghamdi, A.A.; Nazreen, S. Synthesis, characterization and cytotoxic study of 2-hydroxy benzothiazole incorporated 1,3,4-oxadiazole derivatives. Egypt. J. Chem. 2020, 63, 471–482. [Google Scholar] [CrossRef]
- Rajanarendar, E.; Reddy, M.N.; Krishna, S.R.; Reddy, K.G.; Reddy, Y.N.; Rajam, M.V. Design, synthesis, in vitro antimicrobial and anticancer activity of novel methylenebis-isoxazolo[4,5-b]azepines derivatives. Eur. J. Med. Chem. 2012, 50, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, N.; Bloom, D.; Bloom, R.; Stein, R. Breakaway: The Global Burden of Cancer: Challenges and Opportunities. Econ. Intell. Unit. 2009, 1–73. [Google Scholar]
- Bianco, R.; Gelardi, T.; Damiano, V.; Ciardiello, F.; Tortora, G. Rational bases for the development of EGFR inhibitors for cancer treatment. Int. J. Biochem. Cell Biol. 2007, 39, 1416–1431. [Google Scholar] [CrossRef] [PubMed]
- Regad, T. Targeting RTK Signaling Pathways in Cancer. Cancers 2015, 7, 1758–1784. [Google Scholar] [CrossRef]
- Higgins, B.; Kolinsky, K.; Smith, M.; Beck, G.; Rashed, M.; Adames, V.; Linn, M.; Wheeldon, E.; Gand, L.; Birnboeck, H.; et al. Antitumor activity of erlotinib (OSI-774, Tarceva) alone or in combination in human non-small cell lung cancer tumor xenograft models. Anticancer Drugs 2014, 15, 503–512. [Google Scholar] [CrossRef]
- Celik, I.; Ayhan-Kılcıgil, G.; Guven, B.; Kara, Z.; Gurkan-Alp, S.; Karayel, A.; Onay-Besikci, A. Design, synthesis and docking studies of benzimidazole derivatives as potential EGFR inhibitors. Eur. J. Med. Chem. 2019, 173, 240–249. [Google Scholar] [CrossRef]
- Sequist, L.V.; Waltman, B.A.; Dias-Santagata, D.; Digumarthy, S.; Turke, A.B.; Fidias, P.; Bergethon, K.; Shaw, A.T.; Gettinger, S.; Cosper, A.K.; et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 2011, 3, 75ra26. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, M.R.; Lanzarini, C.M.; Ricci-Junior, E. Preparation, in vitro characterization and in vivo release of naproxen loaded in poly-caprolactone nanoparticles. Pharm. Dev. Technol. 2011, 16, 12–21. [Google Scholar] [CrossRef]
- Elhenawy, A.A.; Al-Harbi, M.; Moustafa, G.O.; El-Gazzar, M.A.; Abdel-Rahman, R.F.; Salim, A.E. Synthesis, comparative docking, and pharmacological activity of naproxen amino acid derivatives as possible anti-inflammatory and analgesic agents. Drug Des. Dev. Ther. 2019, 13, 1773–1790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cıkla, P.; Ozsavcı, D.; Ozakpınar, O.B.; Sener, A.; Cevik, O.; Ozbas-Turan, S.; Akbuga, J.; Sahin, F.; Kucukguzel, S.G. Synthesis, Cytotoxicity, and Pro-Apoptosis Activity of Etodolac Hydrazide Derivatives as Anticancer Agents. Arch. Pharm. Chem. Life Sci. 2013, 346, 367–379. [Google Scholar] [CrossRef]
- Kucukguzel, S.G.; Koc, D.; Suzgun, P.C.; Ozsavci, D.; Ozakpinar, O.B.; Tiber, P.M.; Orun, O.; Erzincan, P.; Erdem, S.S.; Sahin, F. Synthesis of Tolmetin Hydrazide–Hydrazones and Discovery of a Potent Apoptosis Inducer in Colon Cancer Cells. Arch. Pharm. Chem. Life Sci. 2015, 348, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Han, M.I.; Bekci, H.; Uba, A.I.; Yildrim, Y.; Karasulu, E.; Cumaoglu, A.; Karasulu, H.Y.; Yelecksi, K.; Yilmaz, O.; Kucukguzel, S.G. Synthesis, molecular modeling, in vivo study, and anticancer activity of 1,2,4-triazole containing hydrazide-hydrazones derived from(S)-naproxen. Arch. Pharm. 2019, 352, 1800365. [Google Scholar] [CrossRef] [PubMed]
- Birgul, K.; Yıldırım, Y.; Karasulu, H.Y.; Karasulu, E.; Uba, A.I.; Yelekci, K.; Bekci, H.; Cumaoglu, A.; Kabasakal, L.; Yilmaz, O.; et al. Synthesis, molecular modeling, in vivo study and anticancer activity against prostate cancer of (+) (S)-naproxen derivatives. Eur. J. Med. Chem. 2020, 208, 112841. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, M.M.; Ismail, M.M.; Eissa, S.; Ammar, Y. Design and synthesis of some novel 6-methoxynaphthalene derivatives with Potential anticancer activity. Der Pharma. Chem. 2012, 4, 1552–1566. [Google Scholar]
- Lubet, R.A.; Steele, V.E.; Juliana, M.M.; Grubbs, C.J. Screening agents for preventive efficacy in a bladder cancer model: Study design, end points, and gefitinib and naproxen efficacy. J. Urol. 2010, 183, 1598. [Google Scholar] [CrossRef]
- Han, M.I.; Atalay, P.; Tunc, C.U.; Unal, G.; Dayan, S.; Aydın, O.; Kuçukguzel, S.G. Design and synthesis of novel (S)-Naproxen hydrazide-hydrazones as potent VEGFR-2 inhibitors and their evaluation in vitro/in vivo breast cancer models. Bioorg. Med. Chem. 2021, 37, 116097. [Google Scholar] [CrossRef]
- Chen, P.C.; Patil, V.; Guerrant, W.; Green, P.; Oyelere, A.K. Synthesis and structure-activity relationship of histone deacetylase (HDAC) inhibitors with triazole-linked cap group. Bioorg. Med. Chem. 2008, 16, 4839–4853. [Google Scholar] [CrossRef]
- Dua, R.; Shrivastava, S.; Sonwane, S.K.; Srivastava, S.K. Pharmacological Significance of Synthetic Heterocycles Scaffold: A Review. Adv. Biol. Res. 2011, 5, 120–144. [Google Scholar]
- Nazreen, S. Design, synthesis, and molecular docking studies of thiazolidinediones as PPAR-γ agonists and thymidylate synthase inhibitors. Arch. Der Pharm. 2021, e2100021. [Google Scholar] [CrossRef]
- Chawla, G.; Naaz, B.; Siddiqui, A.A. Exploring 1,3,4-Oxadiazole Scaffold for Anti-inflammatory and Analgesic Activities: A Review of Literature From 2005–2016. Mini. Rev. Med. Chem. 2018, 18, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Alghmadi, A.A.; Alam, M.M.; Nazreen, S. In silico ADME predictions and in vitro antibacterial evaluation of 2-hydroxy benzothiazole-based 1,3,4-oxadiazole derivatives. Turk. J. Chem. 2020, 44, 1068–1084. [Google Scholar] [CrossRef] [PubMed]
- Albratty, M.; El-Sharkawy, K.A.; Alhazmi, H.A. Synthesis and evaluation of some new 1,3,4-oxadiazoles bearing thiophene, thiazole, coumarin, pyridine and pyridazine derivatives as antiviral agents. Acta. Pharm. 2019, 69, 261–276. [Google Scholar] [CrossRef] [Green Version]
- Abou-Seri, S.M. Synthesis and biological evaluation of novel 2,4′-bis substituted diphenylamines as anticancer agents and potential epidermal growth factor receptor tyrosine kinase inhibitors. Eur. J. Med. Chem. 2010, 45, 4113–4121. [Google Scholar] [CrossRef]
- Akhtar, M.J.; Siddiqui, A.A.; Khan, A.A.; Ali, Z.; Dewangan, R.P.; Pasha, S.; Yar, M.S. Design, synthesis, docking and QSAR study of substituted benzimidazole linked oxadiazole as cytotoxic agents, EGFR and erbB2 receptor inhibitors. Eur. J. Med. Chem. 2017, 126, 853–869. [Google Scholar] [CrossRef]
- El-Sayed, N.A.; Nour, M.S.; Alaraby Salem, M.; Arafa, R.K. New oxadiazoles with selective-COX-2 and EGFR dual inhibitory activity: Design, synthesis, cytotoxicity evaluation and in silico studies. Eur. J. Med. Chem. 2019, 183, 111693. [Google Scholar] [CrossRef] [PubMed]
- Fathi, M.A.A.; Abd-El-Hafeez, A.A.; Abdelhamid, D.; Abbas, S.H.; Montano, M.M.; Abdel-Aziz, M. 1,3,4-oxadiazole/chalcone hybrids: Design, synthesis, and inhibition of leukemia cell growth and EGFR, Src, IL-6 and STAT3 activities. Bioorg. Chem. 2019, 84, 150–163. [Google Scholar] [CrossRef]
- Alzhrani, Z.M.M.; Alam, M.M.; Neamatallah, T.; Nazreen, S. Design, synthesis and in vitro antiproliferative activity of new thiazolidinedione-1,3,4-oxadiazole hybrids as thymidylate synthase inhibitors. J. Enzyme Inhib. Med. Chem. 2020, 35, 1116–1123. [Google Scholar] [CrossRef]
- Alam, M.M.; Almalki, A.S.A.; Neamatallah, T.; Ali, N.M.; Malebari, A.M.; Nazreen, S. Synthesis of New 1,3,4-Oxadiazole-Incorporated 1, 2, 3-Triazole Moieties as Potential Anticancer Agents Targeting Thymidylate Synthase and Their Docking Studies. Pharmaceuticals 2020, 13, 390. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.M.; Malebari, A.M.; Nazreen, S.; Neamatallah, T.; Almalki, A.S.A.; Elhenawy, A.A.; Obaid, R.J.; Alsherif, M.A. Design, synthesis and molecular docking studies of thymol based 1,2,3-triazole hybrids as thymidylate synthase inhibitors and apoptosis inducers against breast cancer cells. Bioorg. Med. Chem. 2021, 38, 116136. [Google Scholar] [CrossRef]
- Alarif, W.M.; Ghandourah, M.A.; Abdel-Lateff, A.; Bawakid, N.O.; Alotaibi, A.A.; Alfaifi, M.Y.; Elbehairi, S.E.I. Two new xeniolide diterpenes from the soft coral Xenia umbellata; displayed anti proliferative effects. Pharmacogn. Mag. 2020, 16, 774–779. [Google Scholar] [CrossRef]
- Althagbi, H.I.; Budiyanto, F.; Abdel-Lateff, A.; Al-Footy, K.O.; Bawakid, N.O.; Ghandourah, M.A.; Alfaifi, M.Y.; Elbehairi, S.E.I.; Alarif, W.M. Antiproliferative Isoprenoid Derivatives from the Red Sea Alcyonacean Xenia umbellata. Molecules 2021, 26, 1311. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Chen, E.; Shu, K.; Chen, W.; Zhang, G.; Yu, Y. 6-Oxooxazolidine–quinazolines as noncovalent inhibitors with the potential to target mutant forms of EGFR. Bioorg. Med. Chem. 2016, 24, 3359–3370. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Freener, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug. Del. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Elgendy, A.; Nady, H.; El-Rabiei, M.M.; Elhenawy, A.A. Understanding the adsorption performance of two glycine derivatives as novel and environmentally safe anti-corrosion agents for copper in chloride solutions: Experimental, DFT, and MC studies. RSC Adv. 2019, 9, 42120–42131. [Google Scholar] [CrossRef] [Green Version]
- El Gaafary, M.; Syrovets, T.; Mohamed, H.M.; Elhenawy, A.A.; El-Agrody, A.M.; Amr, A.E.; Ghabbour, H.A.; Almehizia, A.A. Synthesis, Cytotoxic Activity, Crystal Structure, DFT Studies and Molecular Docking of 3-Amino-1-(2,5-dichlorophenyl)-8-methoxy-1H-benzo[f]chromene-2-carbonitrile. Crystals 2021, 11, 184. [Google Scholar] [CrossRef]
- Elhenawy, A.A.; Al-Harbi, L.M.; El-Gazzar, M.A.; Khowdiary, M.M.; Ouidate, A.; Alosaimi, A.M.; Salim, A.E. Naproxenylamino acid derivatives: Design, synthesis, docking, QSAR and anti-inflammatory and analgesic activity. Biomed. Pharmacother. 2019, 116, 109024. [Google Scholar] [CrossRef] [PubMed]
- Elhenawy, A.A.; Al-Harbi, L.M.; El-Gazzar, M.A.; Khowdiary, M.M.; Moustfa, A. Synthesis, molecular properties and comparative docking and QSAR of new 2-(7-hydroxy-2-oxo-2H-chromen-4-yl) acetic acid derivatives as possible anticancer agents. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 218, 248–262. [Google Scholar] [CrossRef]
- Abusaif, M.S.; Fathy, M.; Abu-Saied, M.A.; Elhenawy, A.A.; Kashyout, A.B.; Selim, M.R.; Ammar, Y.A. New carbazole-based organic dyes with different acceptors for dye-sensitized solar cells: Synthesis, characterization, dssc fabrications and density functional theory studies. J. Mol. Struct. 2021, 1225, 129297. [Google Scholar] [CrossRef]
- Naglah, A.M.; Moustafa, G.O.; Elhenawy, A.A.; Mounier, M.M.; El-Sayed, H.; Al-Omar, M.A.; Almehizia, A.A.; Bhat, M.A. Nα-1, 3-Benzenedicarbonyl-Bis-(Amino Acid) and Dipeptide Candidates: Synthesis, Cytotoxic, Antimicrobial and Molecular Docking Investigation. Drug Des. Devel. Ther. 2021, 15, 1315–1332. [Google Scholar] [CrossRef] [PubMed]
- Stamos, J.; Sliwkowski, M.X.; Eigenbrot, C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 2002, 277, 46265–46272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laskowski, R.A.; MacArthur, M.W.; Thornton, J.M. Chapter 21.4 PROCHECK: Validation of protein-structure coordinates. Crystallogr. Biol. Macromol. 2012, F, 722–725. [Google Scholar]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrödinger Release 2018-1; 2018; Available online: https://www.mdpi.com/1420-3049/23/6/1313/htm (accessed on 27 August 2021).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).