Ubiquitin-Specific Proteases: Players in Cancer Cellular Processes
Abstract
1. Introduction
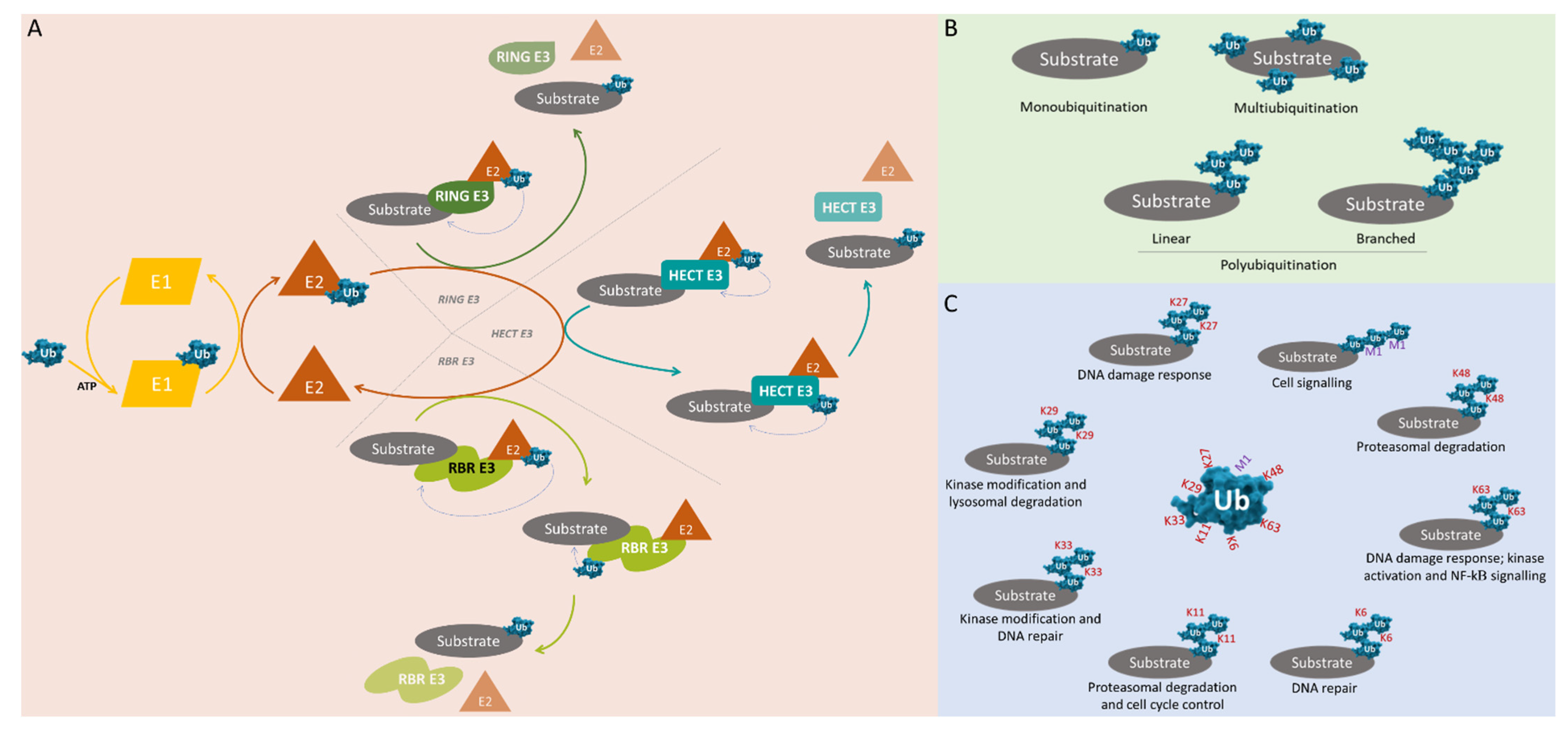
2. Ubiquitin and Ubiquitination
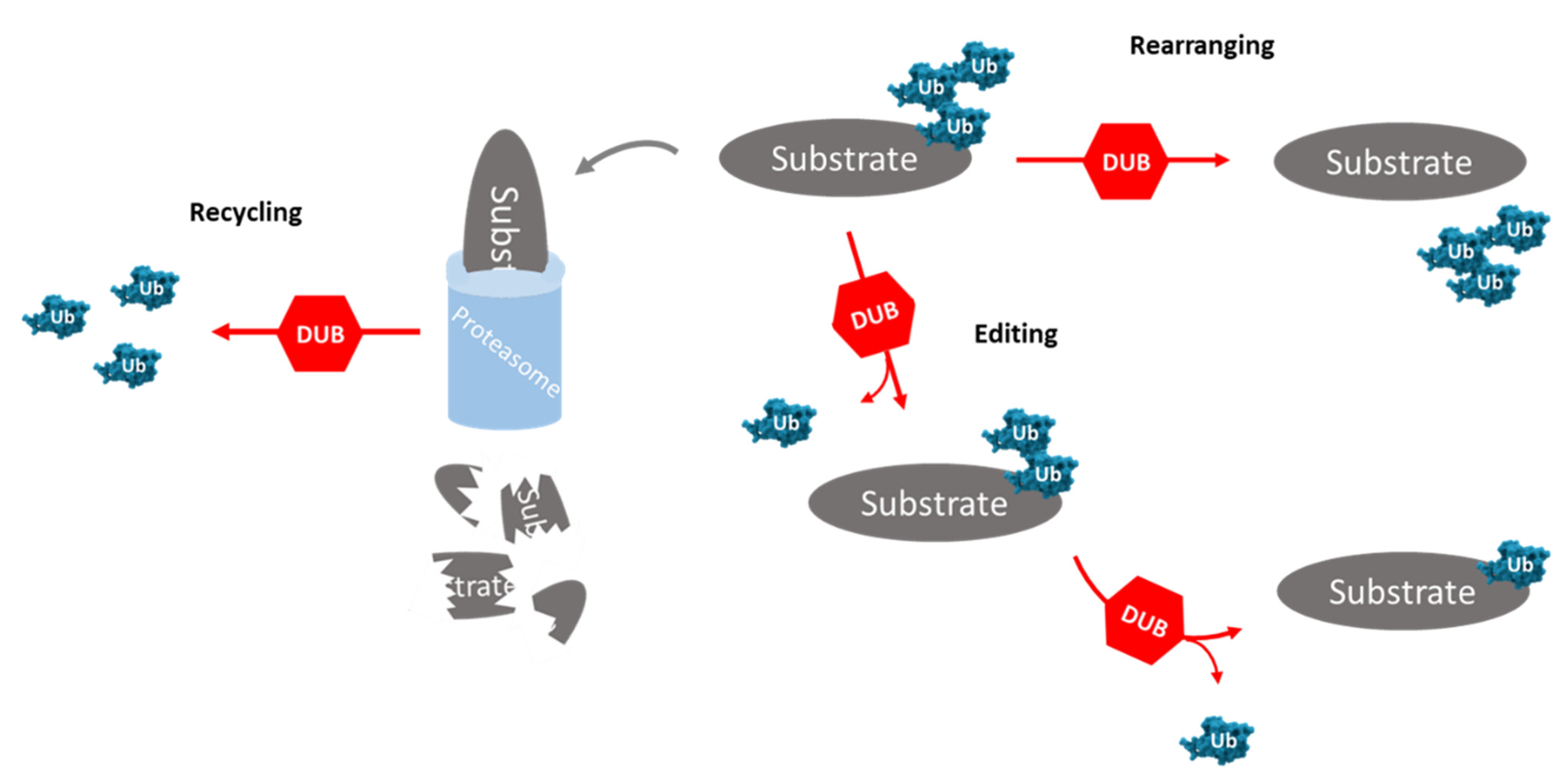
3. Deubiquitinating Enzymes
4. Association of Ubiquitin-Specific Proteases (USPs) with Cancer-Related Processes
4.1. Interactions between USPs and Target Proteins Involved in Cell Cycle Progression
| USPs | Targets | Model | Effects | Refs |
|---|---|---|---|---|
| USP2 | Cyclin A1 | T24 cells (bladder carcinoma) | USP2a overexpression induces cellular invasion and proliferation | [75] |
| Cyclin D1 | HCT116 (colorectal carcinoma) | Knockdown of USP2a inhibits cellular growth by G1/S arrest | [76] | |
| USP3 | CDC25A | HeLa cells (cervical adenocarcinoma) | USP3 knockdown provokes a delay in cell cycle progression and reduces the tumor growth in mice bearing tumor xenografts | [60] |
| Klf5 | HCC1937, HCC1806 and SUM149PT cells (breast carcinoma) | Knockdown of USP3 leads to a decrease in cellular proliferation and invasion | [63] | |
| USP7 | PHF8–cyclin A2 | MCF7 (breast carcinoma) | Knockdown of USP7 reduces cellular proliferation | [64] |
| Knockdown of USP7 reduces tumoral growth in mice bearing tumor xenografts | ||||
| PLK1 | DU145 and VCaP cells (prostate adenocarcinoma) | Knockdown of USP7 leads to a decrease in cellular proliferation and viability, and an interruption of the G2/M cell cycle | [77] | |
| USP17 | ELK1 | HEK293T cells (kidney) # | Knockdown of USP17 inhibits cellular proliferation and stops the G1/S cell cycle | [67] |
| SET8–p21 | MCF7 (breast carcinoma) | Knockdown of USP17 induces G1 phase arrest and apoptosis | [68] | |
| USP22 | Cyclin B1 | HCT116 cells (colorectal carcinoma) | USP22 knockdown prevents G2/M cell cycle progression and inhibits cellular proliferation | [73] |
| Knockdown of USP22 decreases tumoral growth in mice bearing tumor xenografts | ||||
| p15, p21 and cyclin D2 | HEPG2 cells (hepatocellular carcinoma) | Knockdown of USP22 reduces cellular viability and promotes G0/G1 cell cycle arrest and apoptosis | [78] | |
| USP39 | CDK1 and cyclin B1 | HO-8910 and SKOV3 cells (ovarian carcinoma) | USP39 knockdown induces the arrest of the G2/M cell cycle and inhibits cellular proliferation | [79] |
| TT cells (thyroid carcinoma) | Knockdown of USP39 inhibits cellular proliferation and induces G2/M arrest | [80] | ||
| Unknown | SMMC-7721 cells (hepatocellular carcinoma) | USP39 knockdown inhibits cellular proliferation, stops G2/M cell cycle transition | [81] | |
| SW1116 and HCT116 cells (colorectal carcinoma) | Knockdown of USP39 blocks cellular proliferation and G2/M phase | [82] | ||
| USP42 | Cyclin D1 and Cyclin E1 | AGS and MKN-45 cells (gastric adenocarcinoma) | Suppression of USP42 provokes G0/G1 cell cycle arrest and inhibits cellular proliferation | [83] |
| AGS cells (xenografts) | Knockdown of USP42 suppresses tumoral growth in mice bearing tumor xenografts | |||
| USP50 | Hsp90 | U2OS (bone osteosarcoma) | Knockdown of USP50 blocks cells in G2/M | [74] |
4.2. Importance of USPs in DNA Damage Repair Mechanisms
| USPs | Targets | Cell Model Association | Effects | Refs |
|---|---|---|---|---|
| USP1 | FANCD2, PCNA | MM.1S cells (multiple myeloma) | Knockdown of USP1 reduces its viability | [100] |
| HEK293T cells (kidney) # | Knockdown of USP1 leads to cellular protection towards chromosomal aberrations | [71] | ||
| USP3 | ChK1 | HCT116 (colorectal carcinoma) | Knockdown of USP3 decreases apoptosis | [101] |
| USP4 | CtIP, MRN complex | HCT116 (colorectal carcinoma) | Knockdown of USP4 sensitizes it to DNA-damage-inducing agents | [90] |
| U2OS (bone osteosarcoma) | Knockdown of USP4 inhibits DNA damage repair | |||
| USP7 | MDC1 | HeLa cells (cervical adenocarcinoma) | Knockdown of USP7 prevented cellular proliferation | [102] |
| SiHa cells (cervical carcinoma; xenografts) | Knockdown of USP7 suppressed tumoral growth in mice bearing tumor xenografts | |||
| USP9x | Claspin | U2OS (bone osteosarcoma) | USP9x loss of expression leads to accumulation of DNA damage | [92] |
| USP11 | XPC | HaCaT cells (skin) # | Knockdown of USP11 inhibits DNA damage repair | [96] |
| SPRTN | A549 (lung adenocarcinoma) U2OS (bone osteosarcoma) | USP11 cells is required for survival upon DNA–protein crosslinks | [103] | |
| USP20 | Claspin | MGC-803 cells (gastric adenocarcinoma) | Knockdown of USP20 promotes cellular proliferation | [93] |
| USP21 | BRCA2 | HuH1 cells (hepatocellular carcinoma; xenografts) | USP21 knockdown decreases tumoral growth in mice bearing tumor xenografts | [104] |
| USP34 | RNF168 | HeLa cells (cervical adenocarcinoma) | Knockdown of USP34 reduces DNA damage response and cell survival after irradiation | |
| USP51 | H2A | U2OS (bone osteosarcoma) | Knockdown of USP51 increases DNA damage | [105] |
4.3. Role of USPs in Chromatin Remodelling
| USPs | Targets | Cell Model Association | Effects | Refs |
|---|---|---|---|---|
| USP7 | EZH2–PRC2 | DUI145 and PC3 cells (prostate adenocarcinoma) | Overexpression of USP7 increases migration and invasion and inhibits apoptosis | [118] |
| PC3 cells (prostate adenocarcinoma; xenografts) | USP7 knockdown inhibits tumoral growth in mice bearing tumor xenografts | |||
| USP11 | H2A, H2B | HeLa cells (cervical adenocarcinoma) | USP11 knockdown induces apoptosis; USP11 knockdown reduces cell clonogenic survival in irradiated cells | [117] |
| USP16 | H2A | Hematopoietic stem cells (HSC) (bone marrow) # | USP16 knockdown leads to an increase in cellular quiescence; USP16 regulates haematopoiesis and HSC functions | [119] |
| USP21 | EZH2 | 5637 and T24 cells (bladder carcinoma) | USP21 overexpression promotes proliferation, invasion and migration | [120] |
4.4. Consequences of the Interactions between USPs and Proteins Associated with Several Signaling Pathways
4.4.1. TP53
| USPs | Targets | Cell Model Association | Effects | Refs |
|---|---|---|---|---|
| USP2 | Mdm2–p53 | NTERA-2 cells (testicular embryonal carcinoma) | USP2a knockdown induces apoptosis | [123] |
| LNCaP and DU145 cells (prostate adenocarcinoma) | Knockdown of USP2a induces apoptosis | [133] | ||
| MyLa2000 and Hut-78 cells (T-cell lymphoma) | Knockdown of USP2a promotes apoptosis in MyLa2000 cells and decreases p53-dependent apoptosis | [135] | ||
| USP3 | p53 | U2OS cells (osteosarcoma) IMR90 cells (lung) # | Knockdown of USP3 decreases p53 levels and increases cellular proliferation | [104] |
| USP4 | p53–ARF-BP1 | Mouse embryonic fibroblasts (MEFs) # | USP4 silencing leads to early senescence, retarded growth and resistance to oncogene transformation in USP4-deficient MEF cells | [124] |
| p53 | A2058 and 451Lu cells (melanoma) | USP4 knockdown reduces invasion, migration and apoptosis | [131] | |
| Upregulation of USP4 increases invasion, migration and apoptosis | ||||
| USP7 | Mdm2–p53 | NHF-1 (human fibroblasts) # IMR90 cells (lung) # | Slight reduction in USP7 levels destabilizes p53 levels in NHF-1 and IMR90 | [65] |
| U2OS cells (bone osteosarcoma) | Severe reduction in USP7 levels stabilizes p53 levels | |||
| USP10 | Mdm2–p53 | CAKI-1 and CAKI-2 cells (renal carcinoma) | Increase in USP10 levels inhibits colony formation and cell proliferation | [128] |
| USP29 | p53 | HCT116 (colorectal carcinoma) HeLa (cervival adenocarcinoma) U2OS (bone osteossarcoma) | USP29 provokes p53 accumulation and apoptosis | [136] |
| USP39 | p53 | HO-8910 and SKOV3 cells (ovarian carcinoma) | Knockdown of USP39 increases p53 levels and stops the G2/M cell cycle phase | [79] |
| USP42 | p53 | U2OS cells (osteosarcoma) | Downregulation of USP42 reduces p53 levels during the initial stress response phases in U2OS | [137] |
4.4.2. Wnt/β-Catenin
| USPs | Targets | Cell Model Association | Effects | Refs |
|---|---|---|---|---|
| USP3 | MMP2 | HGC27 cells (gastric carcinoma and xenografts) | Knockdown of USP3 suppresses proliferation and migration, and promotes G1 cell cycle arrest | [59] |
| Knockdown of USP3 decreases tumor growth in mice bearing tumor xenografts | ||||
| SK-GT-2 cells (gastric adenocarcinoma) | Overexpression of USP3 leads to increased migration and invasion | |||
| USP4 | β-catenin | HCT116 cells (colorectal carcinoma) | USP4 knockdown decreases proliferation and invasion | [140] |
| USP5 | β-catenin | A549, H1299 and 95-D (lung adenocarcinoma and xenografts) | USP5 overexpression increases cellular proliferation in A549 cells | [145] |
| Knockdown of USP5 decreases cellular proliferation in H1299 cells | ||||
| USP5 knockdown decreases tumoral growth in mice bearing tumor xenografts from H1299 cells | ||||
| Knockdown of USP5 reduces β-catenin transcriptional activity and inhibits invasion and migration in H1299 and 95-D cells | [150] | |||
| USP7 | Axin1 | HEK293T cells (kidney) # | Knockdown of USP7 decreases Axin levels leading to an increase in β-catenin levels and Wnt signaling activation | [151] |
| USP14 | Disheveled (Dvl) | HEK293T cells # (not cancer) (kidney) | Inhibition of USP14 increases polyubiquitination of Dvl, hindering the progression of Wnt signaling | [142] |
| USP15 | APC | HeLa cells (cervical adenocarcinoma) | Knockdown of USP15 decreases APC levels and increases β-catenin levels | [152] |
| USP21 | TCF7 | hTERT-HPNE E6/E7 cells (pancreas cancer cells, xenografts) | USP21 overexpression promotes cellular proliferation and tumoral progression in mice bearing tumor xenografts | [153] |
| USP34 | Axin1 | HEK293T cells (kidney) # | Knockdown of USP34 decreases Axin1 levels and increases levels of β-catenin in HEK293T cells | [154] |
| USP39 | β-catenin | HO-8910 and SKOV3 (ovarian carcinoma) | USP39 knockdown decreases β-catenin levels and inhibits cellular migration and invasion | [79] |
| β-catenin, TCF4, MMP2 and MMP9 | HT29 and SW480 cells (colorectal adenocarcinoma) | USP39 knockdown reduces the expression of β-catenin, TCF4, MMP2 and MMP9, and prevents migration and invasion | [146] | |
| USP44 | Axin1 | HT29 and HCT116 cells (colorectal adenocarcinoma/carcinoma) | USP4 overexpression increases Axin1 levels and decreases β-catenin, c-Myc and cyclin D1 levels, and also inhibits cellular proliferation and promoting apoptosis | [155] |
4.4.3. Receptor Tyrosine Kinases (RTKs)
| USPs | Targets | Cell Model Association | Effects | Refs |
|---|---|---|---|---|
| USP8 | EGFR, ERBB3 and c-Met | Mouse embryonic fibroblasts (MEFs) # | Inhibition of cellular proliferation in USP8-deficient MEFs | [161] |
| H1975 and H1650 (non-small cell lung cancer cells resistant to gefitinib) CCD-8Lu (lung fibroblasts) # HBTEC (human bronchial/tracheal epithelial cells) # | USP8 knockdown reduces cell viability of gefitinib-resistant cells, but not in non-tumoral lung cells | [163] | ||
| LRIG1 (c-Met regulation) | EBC1 cells (lung squamous cell carcinoma) | USP8 overexpression reduces LRIG1—c-Met degradation induced by SAIT301 | [164] | |
| EGFR | PL16T cells (lung adenocarcinoma) | Overexpression of USP8 increases EGFR activity and cellular proliferation | [165] | |
| VEGFR2 | HUVEC (human umbilical vein endothelial cells) # | USP8 knockdown impaired VEGF-A signaling via proteolysis of VEGFR2 into a 120 kDa VEGFR2 fragment | [166] | |
| USP9x | Eps15 (EGFR) | HeLa cells (cervical adenocarcinoma) | USP9x indirectly deregulates EGFR signaling; USP9x increases Eps15 monoubiquitination, supporting EGFR internalization and delaying EGFR signaling | [167] |
| USP18 | miR-7 (EGFR) | T98G (glioblastoma) HeLa (cervival adenocarcinoma) | USP18 knockdown increases miR-7 activity, decreases EGFR levels as well as cellular proliferation and induces apoptosis | [159] |
5. First Steps in USP Inhibition Envisioning (Cancer) Therapeutic Applications
5.1. Inhibitors Targeting USP7
5.2. Inhibitors Targeting USP14
5.3. Inhibitors of Other USPs
6. General Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Cancer. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 25 May 2021).
- Powers, E.T.; Morimoto, R.I.; Dillin, A.; Kelly, J.W.; Balch, W.E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009, 78, 959–991. [Google Scholar] [CrossRef]
- Fulda, S.; Rajalingam, K.; Dikic, I. Ubiquitylation in immune disorders and cancer: From molecular mechanisms to therapeutic implications. EMBO Mol. Med. 2012, 4, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Vega, I.A.; Martin, Y.; Smits, V.A. USP7 controls Chk1 protein stability by direct deubiquitination. Cell Cycle 2014, 13, 3921–3926. [Google Scholar] [CrossRef] [PubMed]
- Pfoh, R.; Lacdao, I.K.; Saridakis, V. Deubiquitinases and the new therapeutic opportunities offered to cancer. Endocr. Relat. Cancer 2015, 22, T35–T54. [Google Scholar] [CrossRef] [PubMed]
- Morrow, J.K.; Lin, H.K.; Sun, S.C.; Zhang, S. Targeting ubiquitination for cancer therapies. Future Med. Chem. 2015, 7, 2333–2350. [Google Scholar] [CrossRef]
- Manasanch, E.E.; Orlowski, R.Z. Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 2017, 14, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Meng, T.; Chen, L.; Wei, W.; Wang, P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct. Target Ther. 2020, 5, 11. [Google Scholar] [CrossRef]
- Keusekotten, K.; Elliott, P.R.; Glockner, L.; Fiil, B.K.; Damgaard, R.B.; Kulathu, Y.; Wauer, T.; Hospenthal, M.K.; Gyrd-Hansen, M.; Krappmann, D.; et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell 2013, 153, 1312–1326. [Google Scholar] [CrossRef]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Wertz, I.E.; Wang, X. From Discovery to Bedside: Targeting the Ubiquitin System. Cell Chem. Biol. 2019, 26, 156–177. [Google Scholar] [CrossRef]
- Morreale, F.E.; Walden, H. Types of Ubiquitin Ligases. Cell 2016, 165, 248–248.e1. [Google Scholar] [CrossRef]
- Buetow, L.; Huang, D.T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol. Cell Biol. 2016, 17, 626–642. [Google Scholar] [CrossRef]
- Bonifacino, J.S.; Weissman, A.M. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol. 1998, 14, 19–57. [Google Scholar] [CrossRef]
- Sadowski, M.; Suryadinata, R.; Tan, A.R.; Roesley, S.N.; Sarcevic, B. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life 2012, 64, 136–142. [Google Scholar] [CrossRef]
- Bernassola, F.; Karin, M.; Ciechanover, A.; Melino, G. The HECT family of E3 ubiquitin ligases: Multiple players in cancer development. Cancer Cell 2008, 14, 10–21. [Google Scholar] [CrossRef]
- Lo, F.Y.; Tan, Y.H.; Cheng, H.C.; Salgia, R.; Wang, Y.C. An E3 ubiquitin ligase: C-Cbl: A new therapeutic target of lung cancer. Cancer 2011, 117, 5344–5350. [Google Scholar] [CrossRef]
- Tan, Y.H.C.; Krishnaswamy, S.; Nandi, S.; Kanteti, R.; Vora, S.; Onel, K.; Hasina, R.; Lo, F.Y.; El-Hashani, E.; Cervantes, G.; et al. CBL Is Frequently Altered in Lung Cancers: Its Relationship to Mutations in MET and EGFR Tyrosine Kinases. PLoS ONE 2010, 5, e8972. [Google Scholar] [CrossRef]
- Khoo, K.H.; Verma, C.S.; Lane, D.P. Drugging the p53 pathway: Understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 2014, 13, 217–236. [Google Scholar] [CrossRef] [PubMed]
- Wade, M.; Li, Y.C.; Matani, A.S.; Braun, S.M.; Milanesi, F.; Rodewald, L.W.; Wahl, G.M. Functional analysis and consequences of Mdm2 E3 ligase inhibition in human tumor cells. Oncogene 2012, 31, 4789–4797. [Google Scholar] [CrossRef]
- Kamei, T.; Machida, K.; Nimura, Y.; Senga, T.; Yamada, I.; Yoshii, S.; Matsuda, S.; Hamaguchi, M. c-Cbl protein in human cancer tissues is frequently tyrosine phosphorylated in a tumor-specific manner. Int. J. Oncol. 2000, 17, 335–339. [Google Scholar] [CrossRef]
- Lai, A.Z.; Durrant, M.; Zuo, D.; Ratcliffe, C.D.; Park, M. Met kinase-dependent loss of the E3 ligase Cbl in gastric cancer. J. Biol. Chem. 2012, 287, 8048–8059. [Google Scholar] [CrossRef]
- Cheng, Y.; Li, G. Role of the ubiquitin ligase Fbw7 in cancer progression. Cancer Metastasis Rev. 2012, 31, 75–87. [Google Scholar] [CrossRef]
- Babaei-Jadidi, R.; Li, N.; Saadeddin, A.; Spencer-Dene, B.; Jandke, A.; Muhammad, B.; Ibrahim, E.E.; Muraleedharan, R.; Abuzinadah, M.; Davis, H.; et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J. Exp. Med. 2011, 208, 295–312. [Google Scholar] [CrossRef]
- Wei, G.; Wang, Y.; Zhang, P.; Lu, J.; Mao, J.H. Evaluating the prognostic significance of FBXW7 expression level in human breast cancer by a meta-analysis of transcriptional profiles. J. Cancer Sci. Ther. 2012, 4, 299–305. [Google Scholar] [CrossRef]
- Yokobori, T.; Mimori, K.; Iwatsuki, M.; Ishii, H.; Onoyama, I.; Fukagawa, T.; Kuwano, H.; Nakayama, K.I.; Mori, M. p53-Altered FBXW7 expression determines poor prognosis in gastric cancer cases. Cancer Res. 2009, 69, 3788–3794. [Google Scholar] [CrossRef] [PubMed]
- Yokobori, T.; Yokoyama, Y.; Mogi, A.; Endoh, H.; Altan, B.; Kosaka, T.; Yamaki, E.; Yajima, T.; Tomizawa, K.; Azuma, Y.; et al. FBXW7 mediates chemotherapeutic sensitivity and prognosis in NSCLCs. Mol. Cancer Res. 2014, 12, 32–37. [Google Scholar] [CrossRef]
- Maser, R.S.; Choudhury, B.; Campbell, P.J.; Feng, B.; Wong, K.K.; Protopopov, A.; O’Neil, J.; Gutierrez, A.; Ivanova, E.; Perna, I.; et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature 2007, 447, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, S.; Oike, Y.; Onoyama, I.; Iwama, A.; Arai, F.; Takubo, K.; Mashimo, Y.; Oguro, H.; Nitta, E.; Ito, K.; et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 2008, 22, 986–991. [Google Scholar] [CrossRef]
- Meszaros, B.; Kumar, M.; Gibson, T.J.; Uyar, B.; Dosztanyi, Z. Degrons in cancer. Sci. Signal 2017, 10, eaak9982. [Google Scholar] [CrossRef]
- Gadd, M.S.; Bulatov, E.; Ciulli, A. Serendipitous SAD Solution for DMSO-Soaked SOCS2-ElonginC-ElonginB Crystals Using Covalently Incorporated Dimethylarsenic: Insights into Substrate Receptor Conformational Flexibility in Cullin RING Ligases. PLoS ONE 2015, 10, e0131218. [Google Scholar] [CrossRef]
- Clifford, S.C.; Astuti, D.; Hooper, L.; Maxwell, P.H.; Ratcliffe, P.J.; Maher, E.R. The pVHL-associated SCF ubiquitin ligase complex: Molecular genetic analysis of elongin B and C, Rbx1 and HIF-1alpha in renal cell carcinoma. Oncogene 2001, 20, 5067–5074. [Google Scholar] [CrossRef]
- Ong, K.R.; Woodward, E.R.; Killick, P.; Lim, C.; Macdonald, F.; Maher, E.R. Genotype-phenotype correlations in von Hippel-Lindau disease. Hum. Mutat. 2007, 28, 143–149. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, T.; Simon, J.; Takada, M.; Saito, R.; Fan, C.; Liu, X.D.; Jonasch, E.; Xie, L.; Chen, X.; et al. VHL substrate transcription factor ZHX2 as an oncogenic driver in clear cell renal cell carcinoma. Science 2018, 361, 290–295. [Google Scholar] [CrossRef]
- Guo, Y.C.; Zhang, S.W.; Yuan, Q. Deubiquitinating Enzymes and Bone Remodeling. Stem Cells Int. 2018, 2018, 3712083. [Google Scholar] [CrossRef]
- Chandrasekaran, A.P.; Suresh, B.; Kim, H.H.; Kim, K.S.; Ramakrishna, S. Concise Review: Fate Determination of Stem Cells by Deubiquitinating Enzymes. Stem Cells 2017, 35, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Satija, Y.K.; Bhardwaj, A.; Das, S. A portrayal of E3 ubiquitin ligases and deubiquitylases in cancer. Int. J. Cancer 2013, 133, 2759–2768. [Google Scholar] [CrossRef]
- Wijnhoven, P.; Konietzny, R.; Blackford, A.N.; Travers, J.; Kessler, B.M.; Nishi, R.; Jackson, S.P. USP4 Auto-Deubiquitylation Promotes Homologous Recombination. Mol. Cell 2015, 60, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Leznicki, P.; Kulathu, Y. Mechanisms of regulation and diversification of deubiquitylating enzyme function. J. Cell Sci. 2017, 130, 1997–2006. [Google Scholar] [CrossRef]
- Rehman, S.A.A.; Kristariyanto, Y.A.; Choi, S.Y.; Nkosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol. Cell 2016, 63, 146–155. [Google Scholar]
- Kwasna, D.; Rehman, S.A.A.; Natarajan, J.; Matthews, S.; Madden, R.; de Cesare, V.; Weidlich, S.; Virdee, S.; Ahel, I.; Gibbs-Seymour, I.; et al. Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability. Mol. Cell 2018, 70, 150–164.e6. [Google Scholar] [CrossRef] [PubMed]
- Hermanns, T.; Pichlo, C.; Woiwode, I.; Klopffleisch, K.; Witting, K.F.; Ovaa, H.; Baumann, U.; Hofmann, K. A family of unconventional deubiquitinases with modular chain specificity determinants. Nat. Commun. 2018, 9, 799. [Google Scholar] [CrossRef]
- Darling, S.; Fielding, A.B.; Sabat-Pospiech, D.; Prior, I.A.; Coulson, J.M. Regulation of the Cell Cycle and centrosome biology by deubiquitylases. Biochem. Soc. Trans. 2017, 45, 1125–1136. [Google Scholar]
- Cotto-Rios, X.M.; Bekes, M.; Chapman, J.; Ueberheide, B.; Huang, T.T. Deubiquitinases as a Signaling Target of Oxidative Stress. Cell Rep. 2012, 2, 1475–1484. [Google Scholar] [CrossRef] [PubMed]
- Haahr, P.; Borgermann, N.; Guo, X.; Typas, D.; Achuthankutty, D.; Hoffmann, S.; Shearer, R.; Sixma, T.K.; Mailand, N. ZUFSP Deubiquitylates K63-Linked Polyubiquitin Chains to Promote Genome Stability. Mol. Cell 2018, 70, 165–174.e166. [Google Scholar] [CrossRef] [PubMed]
- Haq, S.; Ramakrishna, S. Deubiquitylation of deubiquitylases. Open Biol. 2017, 7, 170016. [Google Scholar] [CrossRef]
- Suresh, B.; Lee, J.; Kim, K.S.; Ramakrishna, S. The Importance of Ubiquitination and Deubiquitination in Cellular Reprogramming. Stem Cells Int. 2016, 2016, 6705927. [Google Scholar] [CrossRef]
- Goto, Y.; Zeng, L.; Yeom, C.J.; Zhu, Y.; Morinibu, A.; Shinomiya, K.; Kobayashi, M.; Hirota, K.; Itasaka, S.; Yoshimura, M.; et al. UCHL1 provides diagnostic and antimetastatic strategies due to its deubiquitinating effect on HIF-1alpha. Nat. Commun. 2015, 6, 6153. [Google Scholar] [CrossRef]
- Hou, J.; Liu, G.; Yuan, Y.; Wang, D.; Jiao, P.; Xing, L.; Pan, Y. Increased Jab1/COPS5 is associated with therapeutic response and adverse outcome in lung cancer and breast cancer patients. Oncotarget 2017, 8, 97504–97515. [Google Scholar] [CrossRef][Green Version]
- Sun, J.; Shi, X.; Mamun, M.A.A.; Gao, Y. The role of deubiquitinating enzymes in gastric cancer. Oncol. Lett. 2020, 19, 30–44. [Google Scholar] [CrossRef]
- Ye, Y.; Scheel, H.; Hofmann, K.; Komander, D. Dissection of USP catalytic domains reveals five common insertion points. Mol. Biosyst. 2009, 5, 1797–1808. [Google Scholar] [CrossRef]
- Nijman, S.M.; Luna-Vargas, M.P.; Velds, A.; Brummelkamp, T.R.; Dirac, A.M.; Sixma, T.K.; Bernards, R. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005, 123, 773–786. [Google Scholar] [CrossRef]
- Clague, M.J.; Barsukov, I.; Coulson, J.M.; Liu, H.; Rigden, D.J.; Urbe, S. Deubiquitylases from genes to organism. Physiol. Rev. 2013, 93, 1289–1315. [Google Scholar] [CrossRef]
- Lambies, G.; de Herreros, A.G.; Diaz, V.M. The role of DUBs in the post-translational control of cell migration. Essays Biochem. 2019, 63, 579–594. [Google Scholar]
- Pal, A.; Young, M.A.; Donato, N.J. Emerging potential of therapeutic targeting of ubiquitin-specific proteases in the treatment of cancer. Cancer Res. 2014, 74, 4955–4966. [Google Scholar] [CrossRef]
- Brinton, L.T.; Sloane, H.S.; Kester, M.; Kelly, K.A. Formation and role of exosomes in cancer. Cell Mol. Life Sci. 2015, 72, 659–671. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Nicassio, F.; Corrado, N.; Vissers, J.H.A.; Areces, L.B.; Bergink, S.; Marteijn, J.A.; Geverts, B.; Houtsmuller, A.B.; Vermeulen, W.; di Fiore, P.P.; et al. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 2007, 17, 1972–1977. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.L.; Lin, C.C.; Chen, H.K.; Hseu, Y.C.; Hung, S.T.; Sun, D.P.; Uen, Y.H.; Lin, K.Y. Ubiquitin-specific protease 3 overexpression promotes gastric carcinogenesis and is predictive of poor patient prognosis. Cancer Sci. 2018, 109, 3438–3449. [Google Scholar] [CrossRef]
- Das, S.; Chandrasekaran, A.P.; Suresh, B.; Haq, S.; Kang, J.H.; Lee, S.J.; Kim, J.; Kim, J.; Lee, S.; Kim, H.H.; et al. Genome-scale screening of deubiquitinase subfamily identifies USP3 as a stabilizer of Cdc25A regulating Cell Cycle in cancer. Cell Death Differ. 2020, 27, 3004–3020. [Google Scholar] [CrossRef]
- Nilsson, I.; Hoffmann, I. Cell Cycle regulation by the Cdc25 phosphatase family. Prog. Cell Cycle Res. 2000, 4, 107–114. [Google Scholar]
- Qin, J.; Zhou, Z.; Chen, W.; Wang, C.; Zhang, H.; Ge, G.; Shao, M.; You, D.; Fan, Z.; Xia, H.; et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat. Commun. 2015, 6, 8471. [Google Scholar] [CrossRef]
- Wu, Y.; Qin, J.; Li, F.; Yang, C.; Li, Z.; Zhou, Z.; Zhang, H.; Li, Y.; Wang, X.; Liu, R.; et al. USP3 promotes breast cancer cell proliferation by deubiquitinating KLF5. J. Biol. Chem. 2019, 294, 17837–17847. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, S.; Song, N.; Li, X.; Liu, L.; Yang, S.; Ding, X.; Shan, L.; Zhou, X.; Su, D.; et al. Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. J. Clin. Investig. 2016, 126, 2205–2220. [Google Scholar] [CrossRef]
- Li, M.; Brooks, C.L.; Kon, N.; Gu, W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell 2004, 13, 879–886. [Google Scholar] [CrossRef]
- Shin, J.M.; Yoo, K.J.; Kim, M.S.; Kim, D.; Baek, K.H. Hyaluronan- and RNA-binding deubiquitinating enzymes of USP17 family members associated with cell viability. BMC Genom. 2006, 7, 292. [Google Scholar] [CrossRef][Green Version]
- Ducker, C.; Chow, L.K.Y.; Saxton, J.; Handwerger, J.; McGregor, A.; Strahl, T.; Layfield, R.; Shaw, P.E. De-ubiquitination of ELK-1 by USP17 potentiates mitogenic gene expression and cell proliferation. Nucleic Acids Res 2019, 47, 4495–4508. [Google Scholar] [CrossRef]
- Fukuura, K.; Inoue, Y.; Miyajima, C.; Watanabe, S.; Tokugawa, M.; Morishita, D.; Ohoka, N.; Komada, M.; Hayashi, H. The ubiquitin-specific protease USP17 prevents cellular senescence by stabilizing the methyltransferase SET8 and transcriptionally repressing p21. J. Biol. Chem. 2019, 294, 16429–16439. [Google Scholar] [CrossRef]
- McFarlane, C.; Kelvin, A.A.; de la Vega, M.; Govender, U.; Scott, C.J.; Burrows, J.F.; Johnston, J.A. The deubiquitinating enzyme USP17 is highly expressed in tumor biopsies, is Cell Cycle regulated, and is required for G1-S progression. Cancer Res. 2010, 70, 3329–3339. [Google Scholar] [CrossRef]
- Zheng, N.; Dai, X.; Wang, Z.; Wei, W. A new layer of degradation mechanism for PR-Set7/Set8 during Cell Cycle. Cell Cycle 2016, 15, 3042–3047. [Google Scholar] [CrossRef]
- Nijman, S.M.; Huang, T.T.; Dirac, A.M.; Brummelkamp, T.R.; Kerkhoven, R.M.; D’Andrea, A.D.; Bernards, R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell 2005, 17, 331–339. [Google Scholar] [CrossRef]
- Bonacci, T.; Emanuele, M.J. Dissenting degradation: Deubiquitinases in Cell Cycle and cancer. Semin. Cancer Biol. 2020, 67, 145–158. [Google Scholar] [CrossRef]
- Lin, Z.H.; Tan, C.; Qiu, Q.; Kong, S.Y.; Yang, H.; Zhao, F.; Liu, Z.J.; Li, J.P.; Kong, Q.F.; Gao, B.X.; et al. Ubiquitin-specific protease 22 is a deubiquitinase of CCNB1. Cell Discov. 2015, 1, 1–16. [Google Scholar] [CrossRef]
- Aressy, B.; Jullien, D.; Cazales, M.; Marcellin, M.; Bugler, B.; Burlet-Schiltz, O.; Ducommun, B. A screen for deubiquitinating enzymes involved in the G(2)/M checkpoint identifies USP50 as a regulator of HSP90-dependent Wee1 stability. Cell Cycle 2010, 9, 3815–3822. [Google Scholar] [CrossRef]
- Kim, J.; Kim, W.J.; Liu, Z.; Loda, M.; Freeman, M.R. The ubiquitin-specific protease USP2a enhances tumor progression by targeting cyclin A1 in bladder cancer. Cell Cycle 2012, 11, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Zhao, W.; Gu, W. Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol. Cell 2009, 36, 469–476. [Google Scholar] [CrossRef]
- Peng, Y.; Liu, Y.; Gao, Y.; Yuan, B.; Qi, X.; Fu, Y.; Zhu, Q.; Cao, T.; Zhang, S.; Yin, L.; et al. USP7 is a novel Deubiquitinase sustaining PLK1 protein stability and regulating chromosome alignment in mitosis. J. Exp. Clin. Cancer Res. 2019, 38, 468. [Google Scholar] [CrossRef]
- Ling, S.B.; Sun, D.G.; Tang, B.; Guo, C.; Zhang, Y.; Liang, R.; Wang, L.M. Knock-down of USP22 by small interfering RNA interference inhibits HepG2 cell proliferation and induces Cell Cycle arrest. Cell Mol. Biol. 2012, 58, 1803–1808. [Google Scholar]
- Yan, C.; Yuan, J.; Xu, J.; Zhang, G.; Li, X.; Zhang, B.; Hu, T.; Huang, X.; Mao, Y.; Song, G. Ubiquitin-specific peptidase 39 regulates the process of proliferation and migration of human ovarian cancer via p53/p21 pathway and EMT. Med. Oncol. 2019, 36, 95. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Yang, S.; Guo, K.; Ma, B.; Wang, Y. Reduced USP39 expression inhibits malignant proliferation of medullary thyroid carcinoma in vitro. World J. Surg. Oncol. 2015, 13, 255. [Google Scholar] [CrossRef]
- Pan, Z.; Pan, H.; Zhang, J.; Yang, Y.; Liu, H.; Yang, Y.; Huang, G.; Ni, J.; Huang, J.; Zhou, W. Lentivirus mediated silencing of ubiquitin specific peptidase 39 inhibits cell proliferation of human hepatocellular carcinoma cells in vitro. Biol. Res. 2015, 48, 18. [Google Scholar] [CrossRef]
- Xing, Z.; Sun, F.; He, W.; Wang, Z.; Song, X.; Zhang, F. Downregulation of ubiquitin-specific peptidase 39 suppresses the proliferation and induces the apoptosis of human colorectal cancer cells. Oncol. Lett. 2018, 15, 5443–5450. [Google Scholar] [PubMed]
- Hou, K.; Zhu, Z.; Wang, Y.; Zhang, C.; Yu, S.; Zhu, Q.; Yan, B. Overexpression and Biological Function of Ubiquitin-Specific Protease 42 in Gastric Cancer. PLoS ONE 2016, 11, e0152997. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Grant, S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Cimprich, K.A.; Cortez, D. ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008, 9, 616–627. [Google Scholar] [CrossRef]
- Gerelchuluun, A. DNA Damage, Repair Mechanisms, and Chromosomal Aberrations. In Proton Beam Radiotherapy; Springer: Singapore, 2020; pp. 183–208. [Google Scholar]
- Kee, Y.; Huang, T.T. Role of Deubiquitinating Enzymes in DNA Repair. Mol. Cell. Biol. 2016, 36, 524–544. [Google Scholar] [CrossRef] [PubMed]
- Carter, R.J.; Parsons, J.L. Base Excision Repair, a Pathway Regulated by Posttranslational Modifications. Mol. Cell. Biol. 2016, 36, 1426–1437. [Google Scholar] [CrossRef] [PubMed]
- Patil, M.; Pabla, N.; Dong, Z. Checkpoint kinase 1 in DNA damage response and Cell Cycle regulation. Cell Mol. Life Sci. 2013, 70, 4009–4021. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, H.; Wang, X.; Tian, Q.; Hu, Z.; Peng, C.; Jiang, P.; Wang, T.; Guo, W.; Chen, Y.; et al. The Deubiquitylating Enzyme USP4 Cooperates with CtIP in DNA Double-Strand Break End Resection. Cell Rep. 2015, 13, 93–107. [Google Scholar] [CrossRef]
- Bianco, J.N.; Bergoglio, V.; Lin, Y.L.; Pillaire, M.J.; Schmitz, A.L.; Gilhodes, J.; Lusque, A.; Mazieres, J.; Lacroix-Triki, M.; Roumeliotis, T.I.; et al. Overexpression of Claspin and Timeless protects cancer cells from replication stress in a checkpoint-independent manner. Nat. Commun. 2019, 10, 910. [Google Scholar] [CrossRef]
- McGarry, E.; Gaboriau, D.; Rainey, M.D.; Restuccia, U.; Bachi, A.; Santocanale, C. The Deubiquitinase USP9X Maintains DNA Replication Fork Stability and DNA Damage Checkpoint Responses by Regulating CLASPIN during S-Phase. Cancer Res. 2016, 76, 2384–2393. [Google Scholar] [CrossRef]
- Wang, C.; Yang, C.; Ji, J.; Jiang, J.; Shi, M.; Cai, Q.; Yu, Y.; Zhu, Z.; Zhang, J. Deubiquitinating enzyme USP20 is a positive regulator of Claspin and suppresses the malignant characteristics of gastric cancer cells. Int. J. Oncol. 2017, 50, 1136–1146. [Google Scholar] [CrossRef]
- Paull, T.T.; Rogakou, E.P.; Yamazaki, V.; Kirchgessner, C.U.; Gellert, M.; Bonner, W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000, 10, 886–895. [Google Scholar] [CrossRef]
- Stucki, M.; Clapperton, J.A.; Mohammad, D.; Yaffe, M.B.; Smerdon, S.J.; Jackson, S.P. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 2005, 123, 1213–1226. [Google Scholar] [CrossRef]
- Shah, P.; Qiang, L.; Yang, S.; Soltani, K.; He, Y.Y. Regulation of XPC deubiquitination by USP11 in repair of UV-induced DNA damage. Oncotarget 2017, 8, 96522–96535. [Google Scholar] [CrossRef]
- Yu, M.; Liu, K.; Mao, Z.; Luo, J.; Gu, W.; Zhao, W. USP11 is a negative regulator to γH2AX ubiquitylation by RNF8/RNF168. J. Biol. Chem. 2016, 291, 959–967. [Google Scholar] [CrossRef]
- He, J.; Zhu, Q.; Wani, G.; Sharma, N.; Han, C.; Qian, J.; Pentz, K.; Wang, Q.E.; Wani, A.A. Ubiquitin-specific protease 7 regulates nucleotide excision repair through deubiquitinating XPC protein and preventing XPC protein from undergoing ultraviolet light-induced and VCP/p97 protein-regulated proteolysis. J. Biol. Chem. 2014, 289, 27278–27289. [Google Scholar] [CrossRef]
- Sy, S.M.; Jiang, J.; Sun, O.W.; Deng, Y.; Huen, M.S. The ubiquitin specific protease USP34 promotes ubiquitin signaling at DNA double-strand breaks. Nucleic Acids Res. 2013, 41, 8572–8580. [Google Scholar] [CrossRef] [PubMed]
- Das, D.S.; Das, A.; Ray, A.; Song, Y.; Samur, M.K.; Munshi, N.C.; Chauhan, D.; Anderson, K.C. Blockade of Deubiquitylating Enzyme USP1 Inhibits DNA Repair and Triggers Apoptosis in Multiple Myeloma Cells. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 4280–4289. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.C.; Shieh, S.Y. Deubiquitinating enzyme USP3 controls CHK1 chromatin association and activation. Proc. Natl. Acad. Sci. USA 2018, 115, 5546–5551. [Google Scholar] [CrossRef]
- Su, D.; Ma, S.; Shan, L.; Wang, Y.; Wang, Y.; Cao, C.; Liu, B.; Yang, C.; Wang, L.; Tian, S.; et al. Ubiquitin-specific protease 7 sustains DNA damage response and promotes cervical carcinogenesis. J. Clin. Investig. 2018, 128, 4280–4296. [Google Scholar] [CrossRef] [PubMed]
- Perry, M.; Kollala, S.S.; Biegert, M.; Su, G.; Kodavati, M.; Mallard, H.; Kreiling, N.; Holbrook, A.; Ghosal, G. USP11 deubiquitinates monoubiquitinated SPRTN to repair DNA-protein crosslinks. bioRxiv 2020. [Google Scholar] [CrossRef]
- Fu, S.; Shao, S.; Wang, L.; Liu, H.; Hou, H.; Wang, Y.; Wang, H.; Huang, X.; Lv, R. USP3 stabilizes p53 protein through its deubiquitinase activity. Biochem. Biophys. Res. Commun. 2017, 492, 178–183. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, H.; Liu, J.; Cheruiyot, A.; Lee, J.H.; Ordog, T.; Lou, Z.; You, Z.; Zhang, Z. USP51 deubiquitylates H2AK13,15ub and regulates DNA damage response. Genes Dev. 2016, 30, 946–959. [Google Scholar] [CrossRef]
- Lee, J.S.; Shukla, A.; Schneider, J.; Swanson, S.K.; Washburn, M.P.; Florens, L.; Bhaumik, S.R.; Shilatifard, A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 2007, 131, 1084–1096. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef]
- Machida, S.; Sekine, S.; Nishiyama, Y.; Horikoshi, N.; Kurumizaka, H. Structural and biochemical analyses of monoubiquitinated human histones H2B and H4. Open Biol. 2016, 6, 160090. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.A.; Kingston, R.E. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.M.; Francis, N.J. Inhibition of Chromatin Remodeling by Polycomb Group Protein Posterior Sex Combs Is Mechanistically Distinct from Nucleosome Binding. Biochemistry 2010, 49, 9438–9448. [Google Scholar] [CrossRef][Green Version]
- Davarinejad, H. Characterization of E2E Ubiquitin-Conjugating Enzymes and Ubiquitin-Specific Protease 7 (USP7) in Histone H2A Ubiquitination; YORKspace Institutional Repository; York University: Toronto, ON, Canada, 2017; Available online: https://yorkspace.library.yorku.ca/xmlui/handle/10315/34543 (accessed on 6 December 2020).
- Gagarina, V.; Bojagora, A.; Lacdao, I.K.; Luthra, N.; Pfoh, R.; Mohseni, S.; Chaharlangi, D.; Tan, N.; Saridakis, V. Structural Basis of the Interaction Between Ubiquitin Specific Protease 7 and Enhancer of Zeste Homolog 2. J. Mol. Biol. 2020, 432, 897–912. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Varthi, M.; Sykes, S.M.; Phillips, C.; Warzecha, C.; Zhu, W.; Wyce, A.; Thorne, A.W.; Berger, S.L.; McMahon, S.B. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell 2008, 29, 102–111. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Pfeiffer, H.K.; Thorne, A.W.; McMahon, S.B. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle 2008, 7, 1522–1524. [Google Scholar] [CrossRef]
- Johnsen, S.A. The enigmatic role of H2Bub1 in cancer. FEBS Lett. 2012, 586, 1592–1601. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.D.; Cui, B.B.; Sun, L.Y.; Zheng, H.Q.; Huang, Q.; Tong, J.X.; Zhang, Q.F. The co-expression of USP22 and BMI-1 may promote cancer progression and predict therapy failure in gastric carcinoma. Cell Biochem. Biophys. 2011, 61, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Ting, X.; Xia, L.; Yang, J.G.; He, L.; Si, W.Z.; Shang, Y.F.; Sun, L.Y. USP11 acts as a histone deubiquitinase functioning in chromatin reorganization during DNA repair. Nucleic Acids Res. 2019, 47, 9721–9740. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Chu, M.; Lin, M.; He, Y.; Wang, Z. USP7 stabilizes EZH2 and enhances cancer malignant progression. Am. J. Cancer Res. 2020, 10, 299–313. [Google Scholar]
- Gu, Y.; Jones, A.E.; Yang, W.; Liu, S.; Dai, Q.; Liu, Y.; Swindle, C.S.; Zhou, D.; Zhang, Z.; Ryan, T.M.; et al. The histone H2A deubiquitinase Usp16 regulates hematopoiesis and hematopoietic stem cell function. Proc. Natl. Acad. Sci. USA 2016, 113, E51–E60. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, B.; Chen, D. USP21 promotes cell proliferation and metastasis through suppressing EZH2 ubiquitination in bladder carcinoma. OncoTargets Ther. 2017, 10, 681–689. [Google Scholar] [CrossRef]
- Mijit, M.; Caracciolo, V.; Melillo, A.; Amicarelli, F.; Giordano, A. Role of p53 in the Regulation of Cellular Senescence. Biomolecules 2020, 10, 420. [Google Scholar] [CrossRef]
- Li, X.; Dou, Q.P. Deubiquitylating Enzymes. Encycl. Mol. Pharmacol. 2020, 1–8. [Google Scholar] [CrossRef]
- Stevenson, L.F.; Sparks, A.; Allende-Vega, N.; Xirodimas, D.P.; Lane, D.P.; Saville, M.K. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007, 26, 976–986. [Google Scholar] [CrossRef]
- Zhang, X.; Berger, F.G.; Yang, J.; Lu, X. USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO J. 2011, 30, 2177–2189. [Google Scholar] [CrossRef] [PubMed]
- Mungamuri, S.K.; Qiao, R.F.; Yao, S.; Manfredi, J.J.; Gu, W.; Aaronson, S.A. USP7 Enforces Heterochromatinization of p53 Target Promoters by Protecting SUV39H1 from MDM2-Mediated Degradation. Cell Rep. 2016, 14, 2528–2537. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Saridakis, V.; Sarkari, F.; Duan, S.; Wu, T.; Arrowsmith, C.H.; Frappier, L. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat. Struct. Mol. Biol. 2006, 13, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Ghosh, M.K. Cell death and deubiquitinases: Perspectives in cancer. Biomed. Res. Int. 2014, 2014, 435197. [Google Scholar] [CrossRef]
- Yuan, J.; Luo, K.; Zhang, L.; Cheville, J.C.; Lou, Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 2010, 140, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Hock, A.K.; Vigneron, A.M.; Vousden, K.H. Ubiquitin-specific peptidase 42 (USP42) functions to deubiquitylate histones and regulate transcriptional activity. J. Biol. Chem. 2014, 289, 34862–34870. [Google Scholar] [CrossRef] [PubMed]
- Schober, A. Establishment and Characterisation of USP29 as a Novel Regulator of Hypoxia Inducible Factor α; The University of Liverpool Repository, University of Liverpool: Liverpool, UK, 2016; Available online: https://livrepository.liverpool.ac.uk/3007229/ (accessed on 30 November 2020).
- Guo, W.; Ma, J.; Pei, T.; Zhao, T.; Guo, S.; Yi, X.; Liu, Y.; Wang, S.; Zhu, G.; Jian, Z. Up-regulated deubiquitinase USP 4 plays an oncogenic role in melanoma. J. Cell Mol. Med. 2018, 22, 2944–2954. [Google Scholar] [CrossRef]
- Sun, X.X.; Dai, M.S. Deubiquitinating enzyme regulation of the p53 pathway: A lesson from Otub1. World J. Biol. Chem. 2014, 5, 75–84. [Google Scholar]
- Priolo, C.; Tang, D.; Brahamandan, M.; Benassi, B.; Sicinska, E.; Ogino, S.; Farsetti, A.; Porrello, A.; Finn, S.; Zimmermann, J.; et al. The isopeptidase USP2a protects human prostate cancer from apoptosis. Cancer Res. 2006, 66, 8625–8632. [Google Scholar] [CrossRef]
- Taylor, W.R.; Stark, G.R. Regulation of the G2/M transition by p53. Oncogene 2001, 20, 1803–1815. [Google Scholar] [CrossRef]
- Wei, T.; Biskup, E.; Gjerdrum, L.M.; Niazi, O.; Odum, N.; Gniadecki, R. Ubiquitin-specific protease 2 decreases p53-dependent apoptosis in cutaneous T-cell lymphoma. Oncotarget 2016, 7, 48391–48400. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chung, H.J.; Vogt, M.; Jin, Y.; Malide, D.; He, L.; Dundr, M.; Levens, D. JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J. 2011, 30, 846–858. [Google Scholar] [CrossRef] [PubMed]
- Hock, A.K.; Vigneron, A.M.; Carter, S.; Ludwig, R.L.; Vousden, K.H. Regulation of p53 stability and function by the deubiquitinating enzyme USP42. EMBO J. 2011, 30, 4921–4930. [Google Scholar] [CrossRef] [PubMed]
- Murillo-Garzon, V.; Kypta, R. WNT signalling in prostate cancer. Nat. Rev. Urol. 2017, 14, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Driehuis, E.; Clevers, H. WNT signalling events near the cell membrane and their pharmacological targeting for the treatment of cancer. Br. J. Pharmacol. 2017, 174, 4547–4563. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.-I.; Kim, H.H.; Yoon, J.H.; Park, W.S.; Hahn, M.-J.; Kim, H.C.; Chung, C.H.; Kim, K.K. Ubiquitin specific protease 4 positively regulates the WNT/β-catenin signaling in colorectal cancer. Mol. Oncol. 2015, 9, 1834–1851. [Google Scholar] [CrossRef]
- Zhao, B.; Schlesiger, C.; Masucci, M.G.; Lindsten, K. The ubiquitin specific protease 4 (USP4) is a new player in the Wnt signalling pathway. J. Cell Mol. Med. 2009, 13, 1886–1895. [Google Scholar] [CrossRef]
- Jung, H.; Kim, B.G.; Han, W.H.; Lee, J.H.; Cho, J.Y.; Park, W.S.; Maurice, M.M.; Han, J.K.; Lee, M.J.; Finley, D.; et al. Deubiquitination of Dishevelled by Usp14 is required for Wnt signaling. Oncogenesis 2013, 2, e64. [Google Scholar] [CrossRef]
- Chou, C.K.; Chang, Y.T.; Korinek, M.; Chen, Y.T.; Yang, Y.T.; Leu, S.; Lin, I.L.; Tang, C.J.; Chiu, C.C. The Regulations of Deubiquitinase USP15 and Its Pathophysiological Mechanisms in Diseases. Int. J. Mol. Sci. 2017, 18, 483. [Google Scholar] [CrossRef]
- Vlasschaert, C.; Xia, X.; Gray, D.A. Selection preserves Ubiquitin Specific Protease 4 alternative exon skipping in therian mammals. Sci. Rep. 2016, 6, 20039. [Google Scholar] [CrossRef]
- Ma, X.; Qi, W.; Pan, H.; Yang, F.; Deng, J. Overexpression of USP5 contributes to tumorigenesis in non-small cell lung cancer via the stabilization of beta-catenin protein. Am. J. Cancer Res. 2018, 8, 2284–2295. [Google Scholar] [PubMed]
- Yuan, X.; Sun, X.; Shi, X.; Wang, H.; Wu, G.; Jiang, C.; Yu, D.; Zhang, W.; Xue, B.; Ding, Y. USP39 promotes colorectal cancer growth and metastasis through the Wnt/beta-catenin pathway. Oncol. Rep. 2017, 37, 2398–2404. [Google Scholar] [CrossRef]
- Jung, O.; Lee, J.; Lee, Y.J.; Yun, J.M.; Son, Y.J.; Cho, J.Y.; Ryou, C.; Lee, S.Y. Timosaponin AIII inhibits migration and invasion of A549 human non-small-cell lung cancer cells via attenuations of MMP-2 and MMP-9 by inhibitions of ERK1/2, Src/FAK and beta-catenin signaling pathways. Bioorg. Med. Chem. Lett. 2016, 26, 3963–3967. [Google Scholar] [CrossRef]
- Wu, B.; Crampton, S.P.; Hughes, C.C. Wnt signaling induces matrix metalloproteinase expression and regulates T cell transmigration. Immunity 2007, 26, 227–239. [Google Scholar] [CrossRef]
- Schmalhofer, O.; Brabletz, S.; Brabletz, T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009, 28, 151–166. [Google Scholar] [CrossRef]
- Xue, S.; Wu, W.; Wang, Z.; Lu, G.; Sun, J.; Jin, X.; Xie, L.; Wang, X.; Tan, C.; Wang, Z.; et al. USP5 Promotes Metastasis in Non-Small Cell Lung Cancer by Inducing Epithelial-Mesenchymal Transition via Wnt/beta-Catenin Pathway. Front. Pharmacol. 2020, 11, 668. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Lu, B.; Zamponi, R.; Charlat, O.; Aversa, R.; Yang, Z.; Sigoillot, F.; Zhu, X.; Hu, T.; Reece-Hoyes, J.S.; et al. USP7 inhibits Wnt/beta-catenin signaling through promoting stabilization of Axin. Nat. Commun. 2019, 10, 4184. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Langelotz, C.; Hetfeld-Pechoc, B.K.; Schwenk, W.; Dubiel, W. The COP9 signalosome mediates beta-catenin degradation by deneddylation and blocks adenomatous polyposis coli destruction via USP15. J. Mol. Biol. 2009, 391, 691–702. [Google Scholar] [CrossRef]
- Hou, P.; Ma, X.; Zhang, Q.; Wu, C.J.; Liao, W.; Li, J.; Wang, H.; Zhao, J.; Zhou, X.; Guan, C.; et al. USP21 deubiquitinase promotes pancreas cancer cell stemness via Wnt pathway activation. Genes Dev. 2019, 33, 1361–1366. [Google Scholar] [CrossRef]
- Lui, T.T.; Lacroix, C.; Ahmed, S.M.; Goldenberg, S.J.; Leach, C.A.; Daulat, A.M.; Angers, S. The ubiquitin-specific protease USP34 regulates axin stability and Wnt/beta-catenin signaling. Mol. Cell. Biol. 2011, 31, 2053–2065. [Google Scholar] [CrossRef]
- Huang, T.; Zhang, Q.; Ren, W.; Yan, B.; Yi, L.; Tang, T.; Lin, H.; Zhang, Y. USP44 suppresses proliferation and enhances apoptosis in colorectal cancer cells by inactivating the Wnt/beta-catenin pathway via Axin1 deubiquitination. Cell Biol. Int. 2020, 44, 1651–1659. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Ito, F. Receptor tyrosine kinases and targeted cancer therapeutics. Biol. Pharm. Bull. 2011, 34, 1774–1780. [Google Scholar] [CrossRef]
- Critchley, W.R.; Pellet-Many, C.; Ringham-Terry, B.; Harrison, M.A.; Zachary, I.C.; Ponnambalam, S. Receptor Tyrosine Kinase Ubiquitination and De-Ubiquitination in Signal Transduction and Receptor Trafficking. Cells 2018, 7, 22. [Google Scholar] [CrossRef]
- Yan, M.; Zhao, C.H.; Wei, N.; Wu, X.Q.; Cui, J.L.; Xing, Y.L. High Expression of Ubiquitin-Specific Protease 8 (USP8) Is Associated with Poor Prognosis in Patients with Cervical Squamous Cell Carcinoma. Med. Sci. Monit. 2018, 24, 4934–4943. [Google Scholar] [CrossRef] [PubMed]
- Duex, J.E.; Comeau, L.; Sorkin, A.; Purow, B.; Kefas, B. USP18 regulates epidermal growth factor (EGF) receptor expression and cancer cell survival via microRNA-7. J. Biol. Chem. 2011, 286, 25377–25386. [Google Scholar] [CrossRef]
- Zheng, L.; Mustachio, L.; Chen, Y.; Liu, X.; Roszik, J.; Kurie, J.; Dmitrovsky, E. Loss of USP18 represses invasion and metastasis of lung cancer. In Proceedings of the American Association for Cancer Research Annual Meeting, Washington, DC, USA, 1–5 April 2017. [Google Scholar]
- Zhang, Y.Z.; Xia, M.F.; Jin, K.; Wang, S.F.; Wei, H.; Fan, C.M.; Wu, Y.F.; Li, X.L.; Li, X.Y.; Li, G.Y.; et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol. Cancer 2018, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Niendorf, S.; Oksche, A.; Kisser, A.; Lohler, J.; Prinz, M.; Schorle, H.; Feller, S.; Lewitzky, M.; Horak, I.; Knobeloch, K.P. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol. Cell. Biol. 2007, 27, 5029–5039. [Google Scholar] [CrossRef]
- Byun, S.; Lee, S.Y.; Lee, J.; Jeong, C.H.; Farrand, L.; Lim, S.; Reddy, K.; Kim, J.Y.; Lee, M.H.; Lee, H.J.; et al. USP8 Is a Novel Target for Overcoming Gefitinib Resistance in Lung Cancer. Clin. Cancer Res. 2013, 19, 3894–3904. [Google Scholar] [CrossRef]
- Oh, Y.M.; Lee, S.B.; Choi, J.; Suh, H.Y.; Shim, S.; Song, Y.J.; Kim, B.; Lee, J.M.; Oh, S.J.; Jeong, Y.; et al. USP8 modulates ubiquitination of LRIG1 for Met degradation. Sci. Rep. 2014, 4, 1–8. [Google Scholar] [CrossRef]
- Kim, Y.; Shiba-Ishii, A.; Nakagawa, T.; Husni, R.E.; Sakashita, S.; Takeuchi, T.; Noguchi, M. Ubiquitin-specific protease 8 is a novel prognostic marker in early-stage lung adenocarcinoma. Pathol. Int. 2017, 67, 292–301. [Google Scholar] [CrossRef]
- Smith, G.A.; Fearnley, G.W.; Abdul-Zani, I.; Wheatcroft, S.B.; Tomlinson, D.C.; Harrison, M.A.; Ponnambalam, S. VEGFR2 Trafficking, Signaling and Proteolysis is Regulated by the Ubiquitin Isopeptidase USP8. Traffic 2016, 17, 53–65. [Google Scholar] [CrossRef]
- Savio, M.G.; Wollscheid, N.; Cavallaro, E.; Algisi, V.; di Fiore, P.P.; Sigismund, S.; Maspero, E.; Polo, S. USP9X Controls EGFR Fate by Deubiquitinating the Endocytic Adaptor Eps15. Curr. Biol. 2016, 26, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Frezza, M.; Schmitt, S.; Kanwar, J.; Dou, Q.P. Bortezomib as the first proteasome inhibitor anticancer drug: Current status and future perspectives. Curr. Cancer Drug Targets 2011, 11, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Dou, Q.P.; Zonder, J.A. Overview of proteasome inhibitor-based anti-cancer therapies: Perspective on bortezomib and second generation proteasome inhibitors versus future generation inhibitors of ubiquitin-proteasome system. Curr. Cancer Drug Targets 2014, 14, 517–536. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, S.; Cai, C.Y.; Assaraf, Y.G.; Guo, H.Q.; Cui, Q.; Wei, L.; Huang, J.J.; Ashby, C.R., Jr.; Chen, Z.S. Targeting the ubiquitin-proteasome pathway to overcome anti-cancer drug resistance. Drug Resist. Updat. 2020, 48, 100663. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, M.; Cesselli, D.; di Loreto, C.; la Marra, F.; Schneider, C.; Demarchi, F. USP1 (ubiquitin specific peptidase 1) targets ULK1 and regulates its cellular compartmentalization and autophagy. Autophagy 2019, 15, 613–630. [Google Scholar] [CrossRef]
- Dexheimer, T.S.; Rosenthal, A.S.; Liang, Q.; Chen, J.; Villamil, M.A.; Kerns, E.H.; Simeonov, A.; Jadhav, A.; Zhuang, Z.; Maloney, D.J. Discovery of ML323 as a Novel Inhibitor of the USP1/UAF1 Deubiquitinase Complex. In Probe Reports from the NIH Molecular Libraries Program; National Center for Biotechnology Information: Bethesda, MD, USA, 2010. [Google Scholar]
- Liang, Q.; Dexheimer, T.S.; Zhang, P.; Rosenthal, A.S.; Villamil, M.A.; You, C.; Zhang, Q.; Chen, J.; Ott, C.A.; Sun, H.; et al. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat. Chem. Biol. 2014, 10, 298–304. [Google Scholar] [CrossRef]
- Ma, A.; Tang, M.; Zhang, L.; Wang, B.; Yang, Z.; Liu, Y.; Xu, G.; Wu, L.; Jing, T.; Xu, X.; et al. USP1 inhibition destabilizes KPNA2 and suppresses breast cancer metastasis. Oncogene 2019, 38, 2405–2419. [Google Scholar] [CrossRef]
- Mistry, H.; Hsieh, G.; Buhrlage, S.J.; Huang, M.; Park, E.; Cuny, G.D.; Galinsky, I.; Stone, R.M.; Gray, N.S.; D’Andrea, A.D.; et al. Small-molecule inhibitors of USP1 target ID1 degradation in leukemic cells. Mol. Cancer Ther. 2013, 12, 2651–2662. [Google Scholar] [CrossRef]
- Issaenko, O.A.; Amerik, A.Y. Chalcone-based small-molecule inhibitors attenuate malignant phenotype via targeting deubiquitinating enzymes. Cell Cycle 2012, 11, 1804–1817. [Google Scholar] [CrossRef]
- Davis, M.I.; Pragani, R.; Fox, J.T.; Shen, M.; Parmar, K.; Gaudiano, E.F.; Liu, L.; Tanega, C.; McGee, L.; Hall, M.D.; et al. Small Molecule Inhibition of the Ubiquitin-specific Protease USP2 Accelerates cyclin D1 Degradation and Leads to Cell Cycle Arrest in Colorectal Cancer and Mantle Cell Lymphoma Models. J. Biol. Chem. 2016, 291, 24628–24640. [Google Scholar] [CrossRef]
- Okada, K.; Ye, Y.Q.; Taniguchi, K.; Yoshida, A.; Akiyama, T.; Yoshioka, Y.; Onose, J.; Koshino, H.; Takahashi, S.; Yajima, A.; et al. Vialinin A is a ubiquitin-specific peptidase inhibitor. Bioorg. Med. Chem. Lett. 2013, 23, 4328–4331. [Google Scholar] [CrossRef]
- Kapuria, V.; Peterson, L.F.; Fang, D.; Bornmann, W.G.; Talpaz, M.; Donato, N.J. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010, 70, 9265–9276. [Google Scholar] [CrossRef] [PubMed]
- Potu, H.; Peterson, L.F.; Pal, A.; Verhaegen, M.; Cao, J.; Talpaz, M.; Donato, N.J. USP5 links suppression of p53 and FAS levels in melanoma to the BRAF pathway. Oncotarget 2014, 5, 5559–5569. [Google Scholar] [CrossRef]
- Reverdy, C.; Conrath, S.; Lopez, R.; Planquette, C.; Atmanene, C.; Collura, V.; Harpon, J.; Battaglia, V.; Vivat, V.; Sippl, W.; et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem. Biol. 2012, 19, 467–477. [Google Scholar] [CrossRef]
- Colland, F.; Formstecher, E.; Jacq, X.; Reverdy, C.; Planquette, C.; Conrath, S.; Trouplin, V.; Bianchi, J.; Aushev, V.N.; Camonis, J.; et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol. Cancer Ther. 2009, 8, 2286–2295. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, D.; Tian, Z.; Nicholson, B.; Kumar, K.G.; Zhou, B.; Carrasco, R.; McDermott, J.L.; Leach, C.A.; Fulcinniti, M.; Kodrasov, M.P.; et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012, 22, 345–358. [Google Scholar] [CrossRef]
- Tian, X.; Isamiddinova, N.S.; Peroutka, R.J.; Goldenberg, S.J.; Mattern, M.R.; Nicholson, B.; Leach, C. Characterization of selective ubiquitin and ubiquitin-like protease inhibitors using a fluorescence-based multiplex assay format. Assay Drug Dev. Technol. 2011, 9, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Altun, M.; Kramer, H.B.; Willems, L.I.; McDermott, J.L.; Leach, C.A.; Goldenberg, S.J.; Kumar, K.G.; Konietzny, R.; Fischer, R.; Kogan, E.; et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 2011, 18, 1401–1412. [Google Scholar] [CrossRef]
- Fan, Y.H.; Cheng, J.; Vasudevan, S.A.; Dou, J.; Zhang, H.; Patel, R.H.; Ma, I.T.; Rojas, Y.; Zhao, Y.; Yu, Y.; et al. USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis. 2013, 4, e867. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, A.P.; Ioannidis, S.; Krajewski, W.W.; Pinto-Fernandez, A.; Heride, C.; Martin, A.C.L.; Tonkin, L.M.; Townsend, E.C.; Buker, S.M.; Lancia, D.R.; et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 2017, 550, 481–486. [Google Scholar] [CrossRef]
- Lamberto, I.; Liu, X.; Seo, H.S.; Schauer, N.J.; Iacob, R.E.; Hu, W.; Das, D.; Mikhailova, T.; Weisberg, E.L.; Engen, J.R.; et al. Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7. Cell Chem. Biol. 2017, 24, 1490–1500.e11. [Google Scholar] [CrossRef] [PubMed]
- Coughlin, K.; Anchoori, R.; Iizuka, Y.; Meints, J.; MacNeill, L.; Vogel, R.I.; Orlowski, R.Z.; Lee, M.K.; Roden, R.B.; Bazzaro, M. Small-molecule RA-9 inhibits proteasome-associated DUBs and ovarian cancer in vitro and in vivo via exacerbating unfolded protein responses. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 3174–3186. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Umezawa, Y.; Ishida, S.; Okada, K.; Nogami, A.; Miura, O. Inhibition of USP9X induces apoptosis in FLT3-ITD-positive AML cells cooperatively by inhibiting the mutant kinase through aggresomal translocation and inducing oxidative stress. Cancer Lett. 2019, 453, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.F.; Sun, H.; Liu, Y.; Potu, H.; Kandarpa, M.; Ermann, M.; Courtney, S.M.; Young, M.; Showalter, H.D.; Sun, D.; et al. Targeting deubiquitinase activity with a novel small-molecule inhibitor as therapy for B-cell malignancies. Blood 2015, 125, 3588–3597. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, E.L.; Schauer, N.J.; Yang, J.; Lamberto, I.; Doherty, L.; Bhatt, S.; Nonami, A.; Meng, C.; Letai, A.; Wright, R.; et al. Inhibition of USP10 induces degradation of oncogenic FLT3. Nat. Chem. Biol. 2017, 13, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xia, H.; Kim, M.; Xu, L.; Li, Y.; Zhang, L.; Cai, Y.; Norberg, H.V.; Zhang, T.; Furuya, T.; et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 2011, 147, 223–234. [Google Scholar] [CrossRef]
- Burkhart, R.A.; Peng, Y.; Norris, Z.A.; Tholey, R.M.; Talbott, V.A.; Liang, Q.; Ai, Y.; Miller, K.; Lal, S.; Cozzitorto, J.A.; et al. Mitoxantrone targets human ubiquitin-specific peptidase 11 (USP11) and is a potent inhibitor of pancreatic cancer cell survival. Mol. Cancer Res. 2013, 11, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; D’Arcy, P.; Wang, X.; Ray, A.; Tai, Y.T.; Hu, Y.; Carrasco, R.D.; Richardson, P.; Linder, S.; Chauhan, D.; et al. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood 2014, 123, 706–716. [Google Scholar] [CrossRef]
- D’Arcy, P.; Brnjic, S.; Olofsson, M.H.; Fryknas, M.; Lindsten, K.; de Cesare, M.; Perego, P.; Sadeghi, B.; Hassan, M.; Larsson, R.; et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 2011, 17, 1636–1640. [Google Scholar] [CrossRef]
- Wang, X.; D’Arcy, P.; Caulfield, T.R.; Paulus, A.; Chitta, K.; Mohanty, C.; Gullbo, J.; Chanan-Khan, A.; Linder, S. Synthesis and evaluation of derivatives of the proteasome deubiquitinase inhibitor b-AP15. Chem. Biol. Drug Des. 2015, 86, 1036–1048. [Google Scholar] [CrossRef]
- Chitta, K.; Paulus, A.; Akhtar, S.; Blake, M.K.; Caulfield, T.R.; Novak, A.J.; Ansell, S.M.; Advani, P.; Ailawadhi, S.; Sher, T.; et al. Targeted inhibition of the deubiquitinating enzymes, USP14 and UCHL5, induces proteotoxic stress and apoptosis in Waldenstrom macroglobulinaemia tumour cells. Br. J. Haematol. 2015, 169, 377–390. [Google Scholar] [CrossRef]
- Wang, X.; Mazurkiewicz, M.; Hillert, E.K.; Olofsson, M.H.; Pierrou, S.; Hillertz, P.; Gullbo, J.; Selvaraju, K.; Paulus, A.; Akhtar, S.; et al. The proteasome deubiquitinase inhibitor VLX1570 shows selectivity for ubiquitin-specific protease-14 and induces apoptosis of multiple myeloma cells. Sci. Rep. 2016, 6, 26979. [Google Scholar] [CrossRef]
- Liao, Y.; Xia, X.; Liu, N.; Cai, J.; Guo, Z.; Li, Y.; Jiang, L.; Dou, Q.P.; Tang, D.; Huang, H.; et al. Growth arrest and apoptosis induction in androgen receptor-positive human breast cancer cells by inhibition of USP14-mediated androgen receptor deubiquitination. Oncogene 2018, 37, 1896–1910. [Google Scholar] [CrossRef]
- Liao, Y.; Liu, N.; Hua, X.; Cai, J.; Xia, X.; Wang, X.; Huang, H.; Liu, J. Proteasome-associated deubiquitinase ubiquitin-specific protease 14 regulates prostate cancer proliferation by deubiquitinating and stabilizing androgen receptor. Cell Death Dis. 2017, 8, e2585. [Google Scholar] [CrossRef]
- Lee, B.H.; Lee, M.J.; Park, S.; Oh, D.C.; Elsasser, S.; Chen, P.C.; Gartner, C.; Dimova, N.; Hanna, J.; Gygi, S.P.; et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 2010, 467, 179–184. [Google Scholar] [CrossRef]
- Novellasdemunt, L.; Foglizzo, V.; Cuadrado, L.; Antas, P.; Kucharska, A.; Encheva, V.; Snijders, A.P.; Li, V.S. USP7 is a tumor-specific WNT activator for APC-mutated colorectal cancer by mediating β-catenin deubiquitination. Cell Rep. 2017, 21, 612–627. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, Y.; Wang, T.; Zhang, J.; Zhou, Z.; Sun, Y.; Wang, S.; Shi, Y.; Luan, X.; Zhang, Y.; et al. The USP7 Inhibitor P5091 Induces Cell Death in Ovarian Cancers with Different P53 Status. Cell. Physiol. Biochem. 2017, 43, 1755–1766. [Google Scholar] [CrossRef]
- An, T.; Gong, Y.; Li, X.; Kong, L.; Ma, P.; Gong, L.; Zhu, H.; Yu, C.; Liu, J.; Zhou, H.; et al. USP7 inhibitor P5091 inhibits Wnt signaling and colorectal tumor growth. Biochem. Pharmacol. 2017, 131, 29–39. [Google Scholar] [CrossRef]
- Chen, P.C.; Qin, L.N.; Li, X.M.; Walters, B.J.; Wilson, J.A.; Mei, L.; Wilson, S.M. The proteasome-associated deubiquitinating enzyme Usp14 is essential for the maintenance of synaptic ubiquitin levels and the development of neuromuscular junctions. J. Neurosci. 2009, 29, 10909–10919. [Google Scholar] [CrossRef]
- Shinji, S.; Naito, Z.; Ishiwata, S.; Ishiwata, T.; Tanaka, N.; Furukawa, K.; Suzuki, H.; Seya, T.; Matsuda, A.; Katsuta, M.; et al. Ubiquitin-specific protease 14 expression in colorectal cancer is associated with liver and lymph node metastases. Oncol. Rep. 2006, 15, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, J.; Zhong, J.; Deng, Y.; Xi, Q.; He, S.; Yang, S.; Jiang, L.; Huang, M.; Tang, C.; et al. Ubiquitin-specific protease 14 (USP14) regulates cellular proliferation and apoptosis in epithelial ovarian cancer. Med. Oncol. 2015, 32, 379. [Google Scholar] [CrossRef]
- Wu, N.; Liu, C.; Bai, C.; Han, Y.P.; Cho, W.C.; Li, Q. Over-expression of deubiquitinating enzyme USP14 in lung adenocarcinoma promotes proliferation through the accumulation of beta-catenin. Int. J. Mol. Sci. 2013, 14, 10749–10760. [Google Scholar] [CrossRef]
- Bartholomeusz, G.A.; Talpaz, M.; Kapuria, V.; Kong, L.Y.; Wang, S.; Estrov, Z.; Priebe, W.; Wu, J.; Donato, N.J. Activation of a novel Bcr/Abl destruction pathway by WP1130 induces apoptosis of chronic myelogenous leukemia cells. Blood 2007, 109, 3470–3478. [Google Scholar] [CrossRef]
- Bartholomeusz, G.; Talpaz, M.; Bornmann, W.; Kong, L.Y.; Donato, N.J. Degrasyn activates proteasomal-dependent degradation of c-Myc. Cancer Res. 2007, 67, 3912–3918. [Google Scholar] [CrossRef] [PubMed]
- Iwamaru, A.; Szymanski, S.; Iwado, E.; Aoki, H.; Yokoyama, T.; Fokt, I.; Hess, K.; Conrad, C.; Madden, T.; Sawaya, R.; et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene 2007, 26, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Gansler, T.S.; Hardman, W., 3rd; Hunt, D.A.; Schaffel, S.; Hennigar, R.A. Increased expression of fatty acid synthase (OA-519) in ovarian neoplasms predicts shorter survival. Hum. Pathol. 1997, 28, 686–692. [Google Scholar] [CrossRef]
- Hunt, D.A.; Lane, H.M.; Zygmont, M.E.; Dervan, P.A.; Hennigar, R.A. MRNA stability and overexpression of fatty acid synthase in human breast cancer cell lines. Anticancer Res. 2007, 27, 27–34. [Google Scholar]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef]
- Schoenfeld, A.R.; Apgar, S.; Dolios, G.; Wang, R.; Aaronson, S.A. BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol. Cell. Biol. 2004, 24, 7444–7455. [Google Scholar] [CrossRef]
- NCBI. National Library of Medicine (US), PubChem Compound Summary for CID 4212, Mitoxantrone. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Mitoxantrone (accessed on 11 August 2021).
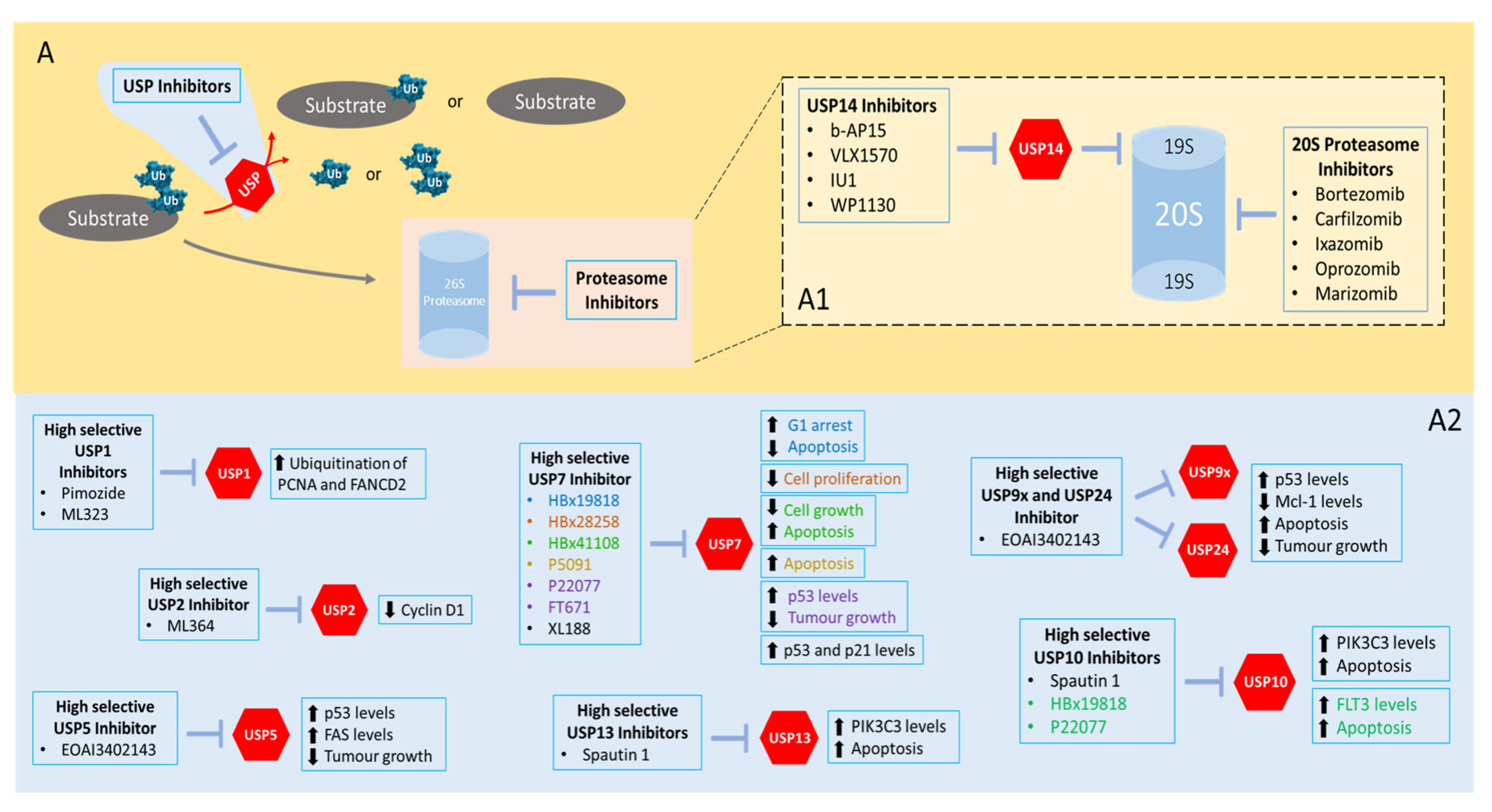
| USP | Inhibitor | Effects | Refs. |
|---|---|---|---|
| USP1 | Pimozide | Increases ubiquitination levels of PCNA and FANCD2 and inhibits growth and viability in U2OS and MCF10A cells | [171] |
| ML323 | High specificity for USP1/UAF1; increases ubiquitination levels of PCNA and FANCD2 in HEK293T and H596 cells | [172] | |
| Inhibits (synergistic effect with cisplatin) proliferation in H596 and U2OS cells | [173] | ||
| Inhibits migration/invasion ability of MCF7, MDA-MB231 and 4T1 cells; suppresses lung metastasis in mice harboring 4T1 tumors | [174] | ||
| SJB2-043 | Induces apoptosis of K562 cells | [175] | |
| SJB3-019A | A potent USP1 inhibitor, five times more potent than SJB2-043 in inducing apoptosis in K562 cells | ||
| C527 | Degrades ID1 in human U2OS cells and increases the levels of Ub-FANCD2 and Ub-FANCI in HeLa cells | ||
| USP2 | AM146 | Provokes cell cycle arrest and apoptosis in MDA-MB231 and MDA-MB468 cells | [176] |
| RA-9 | |||
| RA-14 | |||
| ML364 | Increases cyclin D1 degradation, blocks G1/S transition and inhibits cellular proliferation in colorectal cancer HCT116 and Mino (mantle cell lymphoma) cells | [177] | |
| USP4 | Vialinin A | Inhibits the enzymatic activity of USP4, USP5 and UCHL1 | [178] |
| USP5 | WP1130 | Induces ubiquitination of p53 and Mcl-1 in Z138 cells | [179] |
| Vialinin A | Inhibits the enzymatic activity of USP4, USP5 and UCHL1 | [178] | |
| EOAI3402143 (G9) | Suppresses p53 and FAS levels in A375 cells; suppresses melanoma growth in mice harboring A375 tumors | [180] | |
| RA-9 | Provokes cell cycle arrest and apoptosis in MDA-MB231 and MDA-MB468 cells | [176] | |
| RA-14 | |||
| USP7 | HBx19818 | Stabilizes p53 and promotes G1 arrest and apoptosis in HCT116 cells | [181] |
| HBx28258 | Reduces HCT116 cell proliferation, induces caspase activity and PARP cleavage and arrests HCT116 cancer cells in G1 phase | ||
| HBx41108 | Stabilizes and activates p53, inhibits HTC116 cell growth and induces p53-dependent apoptosis | [182] | |
| P5091 | Induces apoptosis in MM.1S cells | [183] | |
| P22077 | Predominantly inhibits USP7, thus regulating the apoptotic pathway of p53; suppresses neuroblastoma growth in mice harboring IMR32 tumors | [184,185,186] | |
| FT671 | Increases p53 protein levels in HCT116 and U2OS cells and stabilizes p53 in MM.1S cells; suppresses multiple myeloma growth in mice harboring MM.1S tumors | [187] | |
| XL188 | Promotes the accumulation of p53 and p21 in MCF7 and MM.1S cells | [188] | |
| USP8 | RA-9 | Decreases viability of HeLa, SiHa, CaSki, TOV21G1, SKOV3, ES-2, MDA-MB231, MDA-MB435A and MDA-MB468 cells | [176] |
| Induces apoptosis in ES-2 cells; suppresses ovarian cancer growth; and increases overall survival in mice harboring ES-2 tumors | [189] | ||
| RA-14 | Decreases viability of HeLa, SiHa, CaSki, TOV21G1, SKOV3, ES-2, MDA-MB231, MDA-MB435A and MDA-MB468 cells | [176] | |
| AM146 | |||
| USP9x | WP1130 | Reduces levels of Mcl-1 protein and stimulates apoptosis in K562 cells | [190] |
| EOAI3402143 (G9) | Induces apoptosis in MV4 11 and K562 cells | ||
| Reduces Mcl-1 levels and increases p53 levels and apoptosis in MM.1S cells; decreases tumoral growth in mice bearing tumor xenografts from MM.1S cells | [191] | ||
| USP10 | P22077 | Degrades FLT3, leading to death of HEK293T cells | [192] |
| HBx19818 | |||
| Spautin1 | Induces degradation of the PI3K3C3 complex and leads to apoptosis of K562 cells | [193] | |
| USP11 | Mitoxantrone | Increases apoptosis in PL5 cells | [194] |
| USP13 | Spautin1 | Induces degradation of the PI3K3C3 complex and leads to apoptosis of K562 cells | [193] |
| USP14 | b-AP15 | Inhibits cell growth and overcomes bortezomib resistance in MM.1S cells; anti-cancerous effect against solid tumors and multiple myeloma in vivo | [195,196] |
| VLX1570 | Analogue of b-AP15 that induces toxicity and apoptosis in OPM2, KMS11, BCWM3 and RPCI-WM1 cells | [197,198,199] | |
| WP1130 | Inhibits the activity of several DUBs such as USP5, UCH-L1, USP9x, USP14, and UCH37 | [179] | |
| IU1 | Induced cell cycle arrest and apoptosis in MDA-MB153 and MDA-MB231 cells and inhibited cell proliferation in LNCaP cells; decreases (synergistic effect with enzalutamide) tumoral growth in mice bearing tumor xenografts from MCF7 cells | [200,201,202] | |
| USP24 | EOAI3402143 (G9) | Reduces Mcl-1 levels and increases p53 levels and apoptosis in MM.1S cells; decreases tumoral growth in mice bearing tumor xenografts from MM.1S cells | [191] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz, L.; Soares, P.; Correia, M. Ubiquitin-Specific Proteases: Players in Cancer Cellular Processes. Pharmaceuticals 2021, 14, 848. https://doi.org/10.3390/ph14090848
Cruz L, Soares P, Correia M. Ubiquitin-Specific Proteases: Players in Cancer Cellular Processes. Pharmaceuticals. 2021; 14(9):848. https://doi.org/10.3390/ph14090848
Chicago/Turabian StyleCruz, Lucas, Paula Soares, and Marcelo Correia. 2021. "Ubiquitin-Specific Proteases: Players in Cancer Cellular Processes" Pharmaceuticals 14, no. 9: 848. https://doi.org/10.3390/ph14090848
APA StyleCruz, L., Soares, P., & Correia, M. (2021). Ubiquitin-Specific Proteases: Players in Cancer Cellular Processes. Pharmaceuticals, 14(9), 848. https://doi.org/10.3390/ph14090848








