LRP1B: A Giant Lost in Cancer Translation
Abstract
1. Low-Density Lipoprotein (LDL) Receptor (LDRL)-Related Protein 1B (LRP1B)
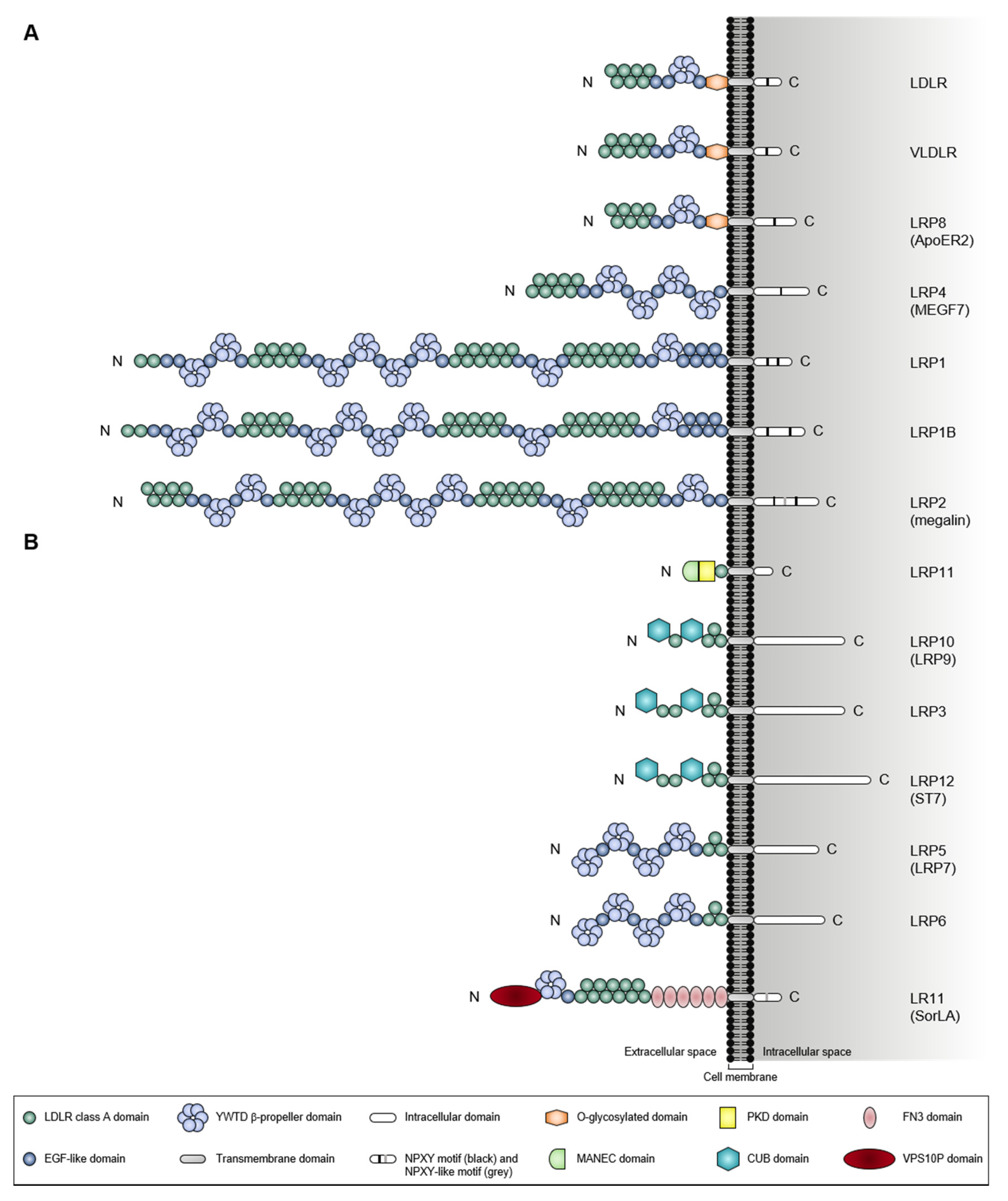
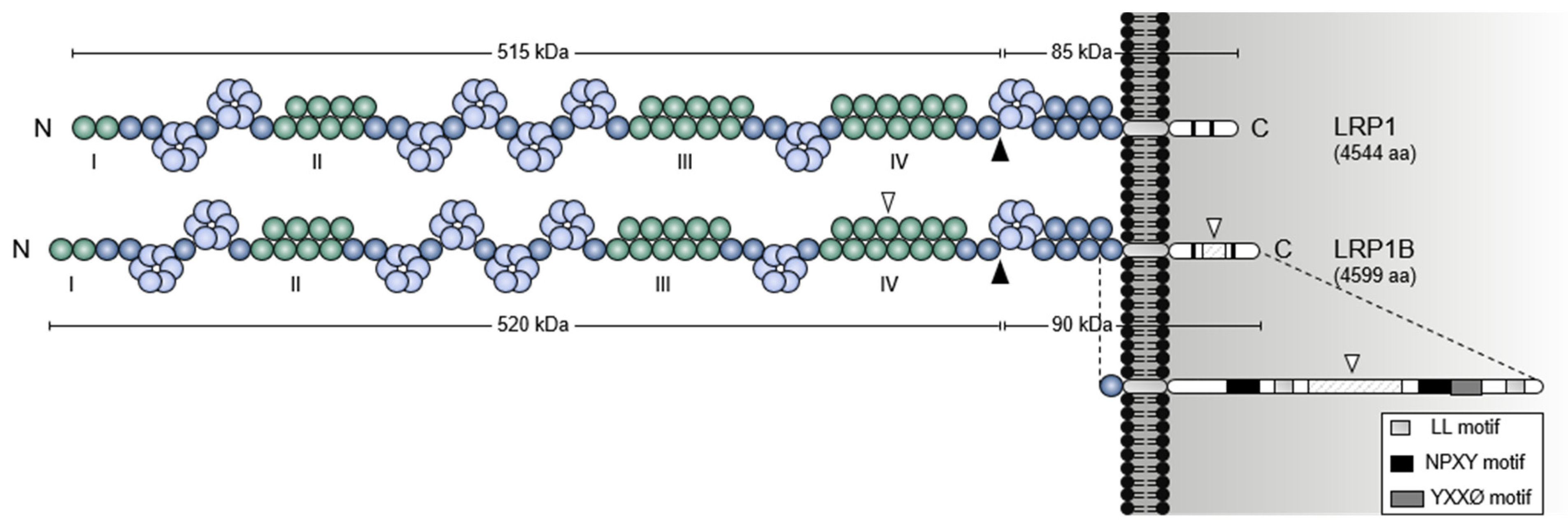
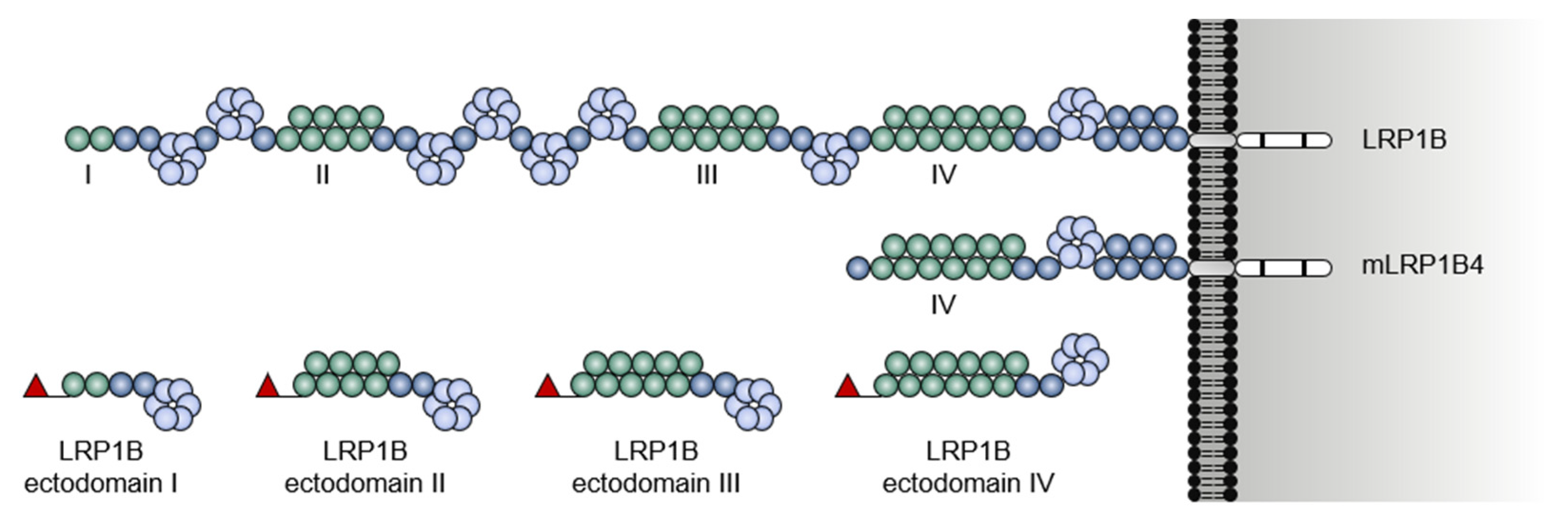
1.1. A Giant Member from a Large Receptor Family
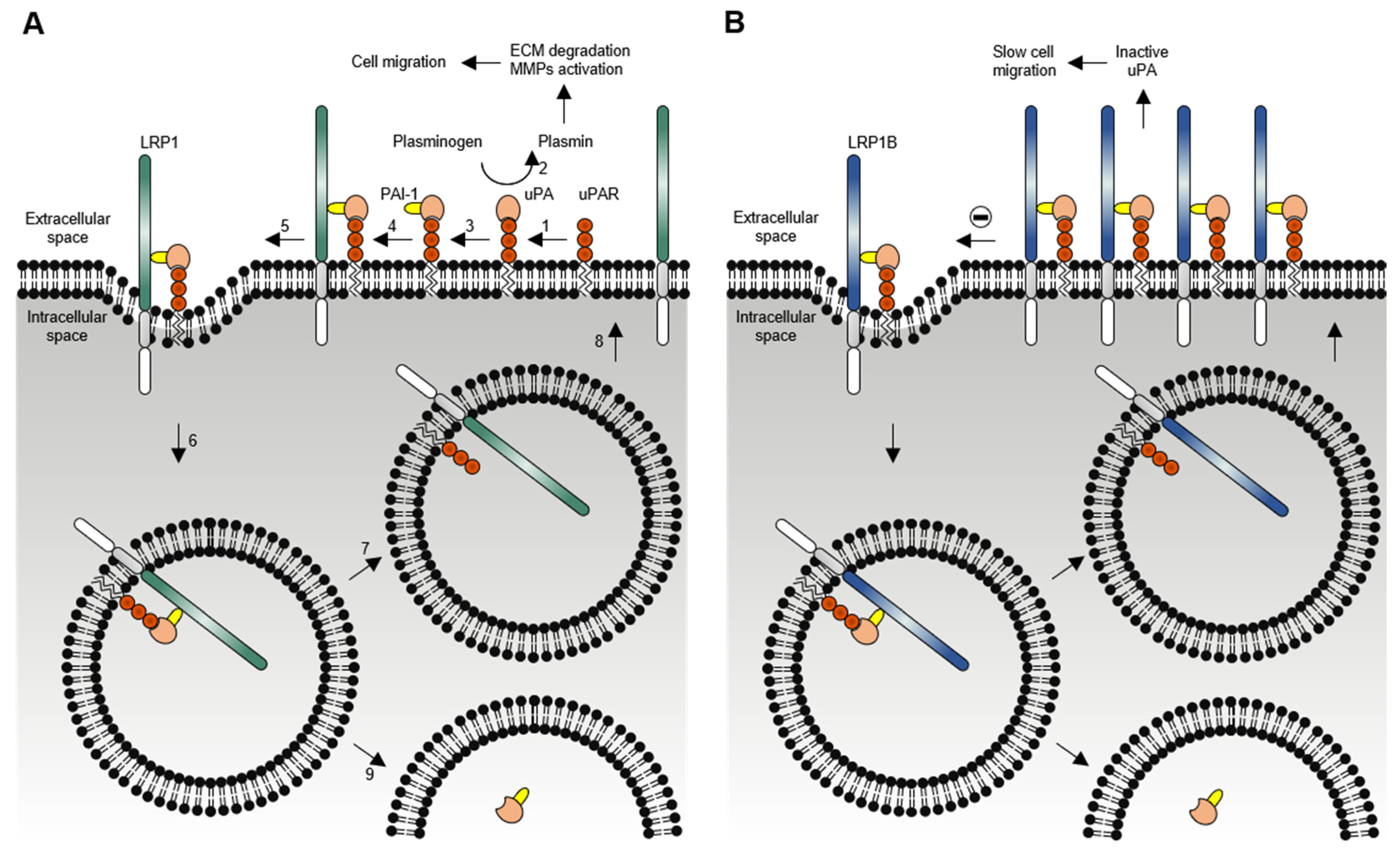
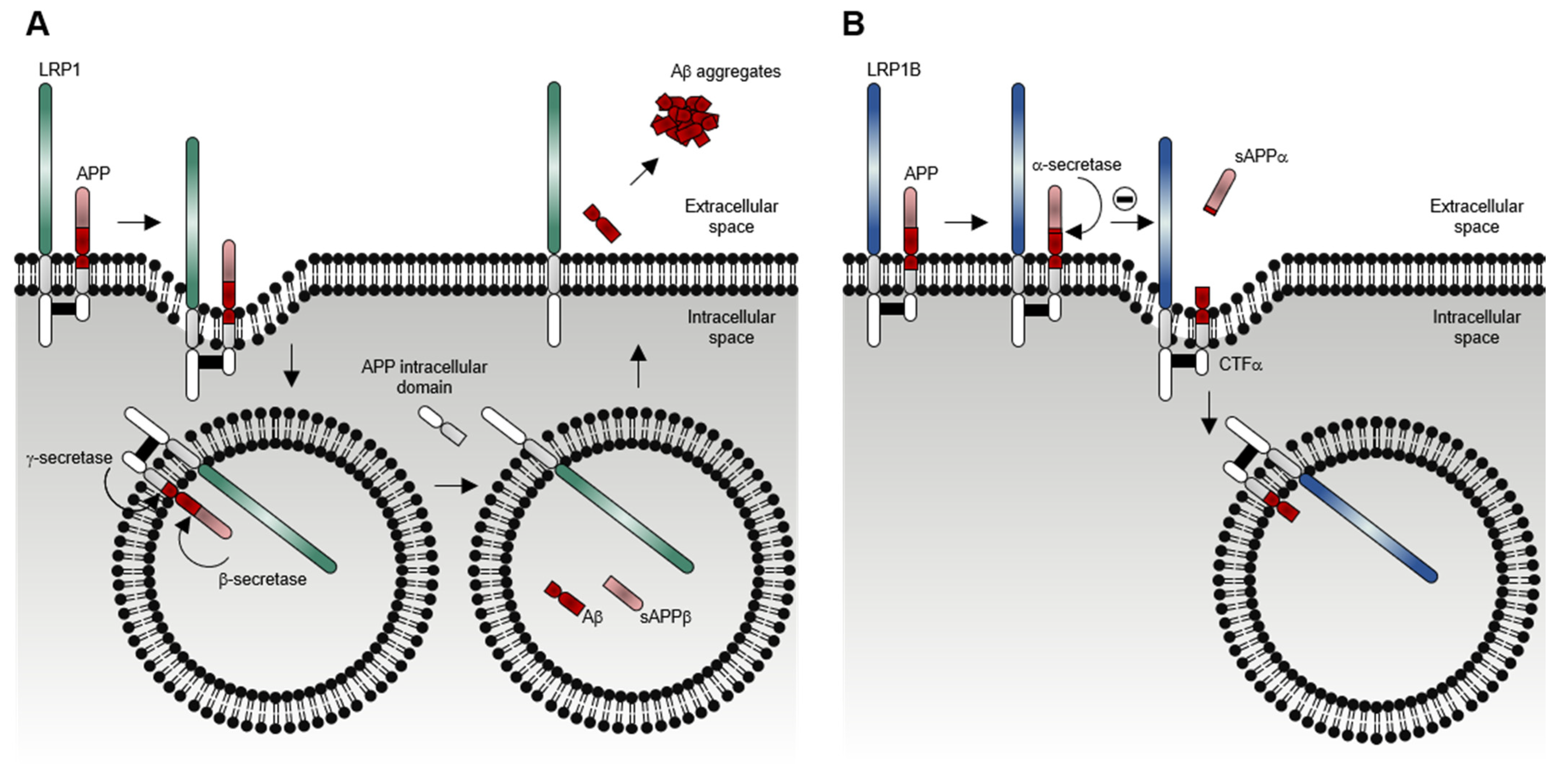
1.2. LRP1B Function
2. LRP1B Impairment in Cancer
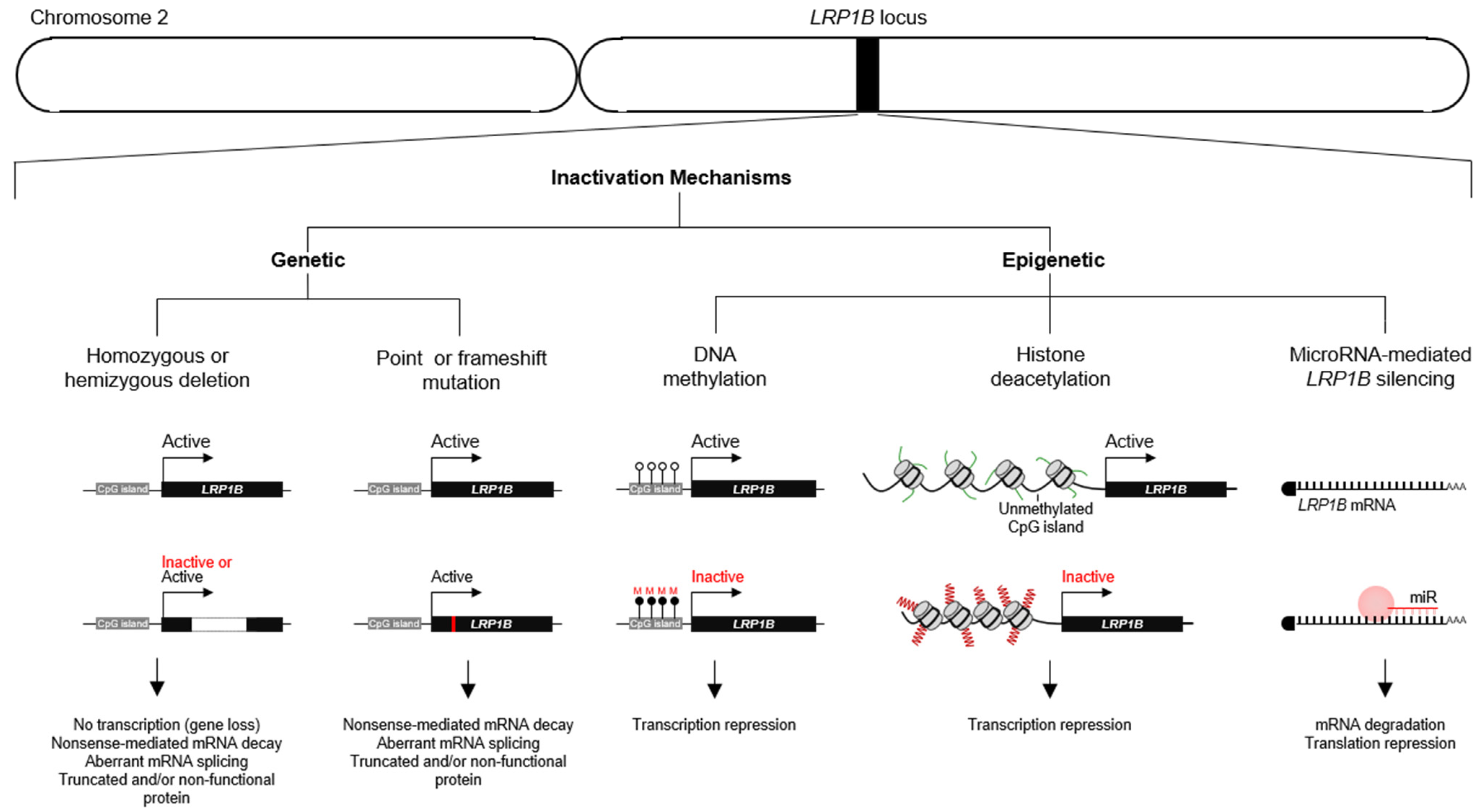
2.1. LRP1B Expression and Roles in Cancer
2.2. LRP1B as a Therapeutic Target, Prognostic Factor, and Predictor of Response to Therapy
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, C.X.; Musco, S.; Lisitsina, N.M.; Forgacs, E.; Minna, J.D.; Lisitsyn, N.A. LRP-DIT, A Putative Endocytic Receptor Gene, is Frequently Inactivated in Non-Small Cell Lung Cancer Cell Lines. Cancer Res. 2000, 60, 1961–1967. [Google Scholar]
- Liu, C.X.; Musco, S.; Lisitsina, N.M.; Yaklichkin, S.Y.; Lisitsyn, N.A. Genomic Organization of a New Candidate Tumor Suppressor Gene, LRP1B. Genomics 2000, 69, 271–274. [Google Scholar] [CrossRef]
- Prazeres, H.; Salgado, C.; Duarte, C.; Soares, P. LRP1B (low density lipoprotein receptor-related protein 1B). Atlas Genet. Cytogenet. Oncol. Haematol. 2014, 18, 93–101. [Google Scholar] [CrossRef][Green Version]
- Strickland, D.K.; Gonias, S.L.; Argraves, W.S. Diverse Roles For The LDL Receptor Family. Trends Endocrinol. Metab. 2002, 13, 66–74. [Google Scholar] [CrossRef]
- May, P.; Woldt, E.; Matz, R.L.; Boucher, P. The LDL Receptor-Related Protein (LRP) Family: An Old Family of Proteins With New Physiological Functions. Ann. Med. 2007, 39, 219–228. [Google Scholar] [CrossRef]
- Dieckmann, M.; Dietrich, M.F.; Herz, J. Lipoprotein Receptors—An Evolutionarily Ancient Multifunctional Receptor Family. Biol. Chem. 2010, 391, 1341–1363. [Google Scholar] [CrossRef] [PubMed]
- Willnow, T.E.; Hammes, A.; Eaton, S. Lipoproteins And Their Receptors in Embryonic Development: More Than Cholesterol Clearance. Development 2007, 134, 3239–3249. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Willnow, T.E.; Christ, A.; Hammes, A. Endocytic Receptor-Mediated Control of Morphogen Signaling. Development 2012, 139, 4311–4319. [Google Scholar] [CrossRef] [PubMed]
- Andersen, O.M.; Dagil, R.; Kragelund, B.B. New Horizons For Lipoprotein Receptors: Communication By β-Propellers. J. Lipid Res. 2013, 54, 2763–2774. [Google Scholar] [CrossRef] [PubMed]
- Willnow, T.E.; Nykjaer, A.; Herz, J. Lipoprotein Receptors: New Roles For Ancient Proteins. Nat. Cell Biol. 1999, 1, E157–E162. [Google Scholar] [CrossRef]
- Brown, S.D.; Twells, R.C.; Hey, P.J.; Cox, R.D.; Levy, E.R.; Soderman, A.R.; Metzker, M.L.; Caskey, C.T.; Todd, J.A.; Hess, J.F. Isolation And Characterization of LRP6, A Novel Member of The Low Density Lipoprotein Receptor Gene Family. Biochem. Biophys. Res. Commun. 1998, 248, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, L.; Madsen, P.; Moestrup, S.K.; Lund, A.H.; Tommerup, N.; Nykjaer, A.; Sottrup-Jensen, L.; Gliemann, J.; Petersen, C.M. Molecular Characterization of A Novel Human Hybrid-Type Receptor That Binds The Alpha2-Macroglobulin Receptor-Associated Protein. J. Biol. Chem. 1996, 271, 31379–31383. [Google Scholar] [CrossRef] [PubMed]
- Fjorback, A.W.; Seaman, M.; Gustafsen, C.; Mehmedbasic, A.; Gokool, S.; Wu, C.; Militz, D.; Schmidt, V.; Madsen, P.; Nyengaard, J.R.; et al. Retromer Binds The FANSHY Sorting Motif in SorLA To Regulate Amyloid Precursor Protein Sorting And Processing. J. Neurosci. 2012, 32, 1467–1480. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X.; Li, Y.; Obermoeller-McCormick, L.M.; Schwartz, A.L.; Bu, G. The putative tumor suppressor LRP1B, a novel member of the low density lipoprotein (LDL) receptor family, exhibits both overlapping and distinct properties with the LDL receptor-related protein. J. Biol. Chem. 2001, 276, 28889–28896. [Google Scholar] [CrossRef] [PubMed]
- Herz, J.; Kowal, R.C.; Goldstein, J.L.; Brown, M.S. Proteolytic Processing of The 600 Kd Low Density Lipoprotein Receptor-Related Protein (LRP) Occurs in A Trans-Golgi Compartment. EMBO J. 1990, 9, 1769–1776. [Google Scholar] [CrossRef]
- Willnow, T.E.; Moehring, J.M.; Inocencio, N.M.; Moehring, T.J.; Herz, J. The Low-Density-Lipoprotein Receptor-Related Protein (LRP) Is Processed By Furin In Vivo And In Vitro. Biochem. J. 1996, 313 Pt 1, 71–76. [Google Scholar] [CrossRef]
- Cam, J.A.; Zerbinatti, C.V.; Knisely, J.M.; Hecimovic, S.; Li, Y.H.; Bu, G.J. The Low Density Lipoprotein Receptor-Related Protein 1B Retains Beta-Amyloid Precursor Protein At The Cell Surface And Reduces Amyloid-Beta Peptide Production. J. Biol. Chem. 2004, 279, 29639–29646. [Google Scholar] [CrossRef]
- Marschang, P.; Brich, J.; Weeber, E.J.; Sweatt, J.D.; Shelton, J.M.; Richardson, J.A.; Hammer, R.E.; Herz, J. Normal Development And Fertility of Knockout Mice Lacking The Tumor Suppressor Gene LRP1B Suggest Functional Compensation By LRP1. Mol. Cell. Biol. 2004, 24, 3782–3793. [Google Scholar] [CrossRef]
- Tanaga, K.; Bujo, H.; Zhu, Y.J.; Kanaki, T.; Hirayama, S.; Takahashi, K.; Inoue, M.; Mikami, K.; Schneider, W.J.; Saito, Y. LRP1B Attenuates The Migration of Smooth Muscle Cells By Reducing Membrane Localization of Urokinase And PDGF Receptors. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 1422–1428. [Google Scholar] [CrossRef]
- Li, Y.H.; Lu, W.Y.; Bu, G.J. Striking Differences of LDL Receptor-Related Protein 1B Expression in Mouse And Human. Biochem. Biophys. Res. Commun. 2005, 333, 868–873. [Google Scholar] [CrossRef]
- Beer, A.G.; Zenzmaier, C.; Schreinlechner, M.; Haas, J.; Dietrich, M.F.; Herz, J.; Marschang, P. Expression of a recombinant full-length LRP1B receptor in human non-small cell lung cancer cells confirms the postulated growth-suppressing function of this large LDL receptor family member. Oncotarget 2016, 7, 68721–68733. [Google Scholar] [CrossRef] [PubMed]
- Knisely, J.M.; Li, Y.; Grifflith, J.M.; Geuze, H.J.; Schwartz, A.L.; Bu, G.J. Slow Endocytosis of The LDL Receptor-Related Protein 1B: Implications For A Novel Cytoplasmic Tail Conformation. Exp. Cell Res. 2007, 313, 3298–3307. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of The Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- The Human Protein Atlas. Available online: http://www.proteinatlas.org (accessed on 14 June 2021).
- Li, Y.H.; Knisely, J.M.; Lu, W.Y.; McCormick, L.M.; Wang, J.Y.; Henkin, J.; Schwartz, A.L.; Bu, G.J. Low Density Lipoprotein (LDL) Receptor-Related Protein 1B Impairs Urokinase Receptor Regeneration on The Cell Surface And Inhibits Cell Migration. J. Biol. Chem. 2002, 277, 42366–42371. [Google Scholar] [CrossRef]
- Pastrana, D.V.; Hanson, A.J.; Knisely, J.; Bu, G.J.; Fitzgerald, D.J. LRP1B Functions As A Receptor For Pseudomonas Exotoxin. Biochim. Biophys. Acta Mol. Basis Dis. 2005, 1741, 234–239. [Google Scholar] [CrossRef]
- Haas, J.; Beer, A.G.; Widschwendter, P.; Oberdanner, J.; Salzmann, K.; Sarg, B.; Lindner, H.; Herz, J.; Patsch, J.R.; Marschang, P. LRP1B Shows Restricted Expression in Human Tissues And Binds To Several Extracellular Ligands, Including Fibrinogen And ApoE—Carrying Lipoproteins. Atherosclerosis 2011, 216, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Herz, J.; Strickland, D.K. LRP: A Multifunctional Scavenger And Signaling Receptor. J. Clin. Investig. 2001, 108, 779–784. [Google Scholar] [CrossRef]
- Bu, G. Receptor-Associated Protein: A Specialized Chaperone And Antagonist For Members of The LDL Receptor Gene Family. Curr. Opin. Lipidol. 1998, 9, 149–155. [Google Scholar] [CrossRef] [PubMed]
- De Haas, C.J.C. New Insights Into The Role of Serum Amyloid P Component, A Novel Lipopolysaccharide-Binding Protein. FEMS Immunol. Med. Microbiol. 1999, 26, 197–202. [Google Scholar] [CrossRef]
- Schvartz, I.; Seger, D.; Shaltiel, S. Vitronectin. Int. J. Biochem. Cell Biol. 1999, 31, 539–544. [Google Scholar] [CrossRef]
- Mosesson, M.W. Fibrinogen And Fibrin Structure And Functions. J. Thromb. Haemost. 2005, 3, 1894–1904. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Pang, Z.P.; Shin, O.; Südhof, T.C. Synaptotagmin-1 Functions As A Ca2+ Sensor For Spontaneous Release. Nat. Neurosci. 2009, 12, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Scheiman, J.; Tseng, J.C.; Zheng, Y.; Meruelo, D. Multiple Functions of The 37/67-kd Laminin Receptor Make It A Suitable Target For Novel Cancer Gene Therapy. Mol. Ther. 2010, 18, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, H.W.; Cavacini, L. Structure And Function of Immunoglobulins. J. Allergy Clin. Immunol. 2010, 125, S41–S52. [Google Scholar] [CrossRef] [PubMed]
- Poon, I.K.; Patel, K.K.; Davis, D.S.; Parish, C.R.; Hulett, M.D. Histidine-Rich Glycoprotein: The Swiss Army Knife of Mammalian Plasma. Blood 2011, 117, 2093–2101. [Google Scholar] [CrossRef]
- Zheng, H.; Koo, E.H. Biology And Pathophysiology of The Amyloid Precursor Protein. Mol. Neurodegener. 2011, 6, 27. [Google Scholar] [CrossRef]
- Marzec, M.; Eletto, D.; Argon, Y. GRP94: An HSP90-Like Protein Specialized For Protein Folding And Quality Control in The Endoplasmic Reticulum. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 774–787. [Google Scholar] [CrossRef]
- Aisina, R.B.; Mukhametova, L.I. Structure And Function of Plasminogen/Plasmin System. Russ. J. Bioorganic Chem. 2014, 40, 590–605. [Google Scholar] [CrossRef]
- DiGiacomo, V.; Meruelo, D. Looking Into Laminin Receptor: Critical Discussion Regarding The Non-Integrin 37/67-kDa Laminin Receptor/RPSA Protein. Biol. Rev. Camb. Philos. Soc. 2016, 91, 288–310. [Google Scholar] [CrossRef]
- Matukumalli, S.R.; Tangirala, R.; Rao, C.M. Clusterin: Full-Length Protein And One of Its Chains Show Opposing Effects on Cellular Lipid Accumulation. Sci. Rep. 2017, 7, 41235. [Google Scholar] [CrossRef]
- Nykjaer, A.; Petersen, C.M.; Møller, B.; Jensen, P.H.; Moestrup, S.K.; Holtet, T.L.; Etzerodt, M.; Thøgersen, H.C.; Munch, M.; Andreasen, P.A.; et al. Purified Alpha 2-Macroglobulin Receptor/LDL Receptor-Related Protein Binds Urokinase/Plasminogen Activator Inhibitor Type-1 Complex. Evidence That The Alpha 2-Macroglobulin Receptor Mediates Cellular Degradation of Urokinase Receptor-Bound Complexes. J. Biol. Chem. 1992, 267, 14543–14546. [Google Scholar] [CrossRef]
- Conese, M.; Nykjaer, A.; Petersen, C.M.; Cremona, O.; Pardi, R.; Andreasen, P.A.; Gliemann, J.; Christensen, E.I.; Blasi, F. Alpha-2 Macroglobulin Receptor/LDL Receptor-Related Protein(LRP)-Dependent Internalization of The Urokinase Receptor. J. Cell Biol. 1995, 131, 1609–1622. [Google Scholar] [CrossRef]
- Nykjaer, A.; Conese, M.; Christensen, E.I.; Olson, D.; Cremona, O.; Gliemann, J.; Blasi, F. Recycling of The Urokinase Receptor Upon Internalization of The uPA:Serpin Complexes. EMBO J. 1997, 16, 2610–2620. [Google Scholar] [CrossRef]
- Czekay, R.P.; Kuemmel, T.A.; Orlando, R.A.; Farquhar, M.G. Direct Binding of Occupied Urokinase Receptor (uPAR) To LDL Receptor-Related Protein Is Required For Endocytosis of uPAR And Regulation of Cell Surface Urokinase Activity. Mol. Biol. Cell 2001, 12, 1467–1479. [Google Scholar] [CrossRef]
- Yasuda, J.; Whitmarsh, A.J.; Cavanagh, J.; Sharma, M.; Davis, R.J. The JIP Group of Mitogen-Activated Protein Kinase Scaffold Proteins. Mol. Cell. Biol. 1999, 19, 7245. [Google Scholar] [CrossRef]
- Han, D.C.; Shen, T.L.; Miao, H.; Wang, B.; Guan, J.L. EphB1 Associates With Grb7 And Regulates Cell Migration. J. Biol. Chem. 2002, 277, 45655–45661. [Google Scholar] [CrossRef]
- Terashima, A.; Pelkey, K.A.; Rah, J.; Suh, Y.H.; Roche, K.W.; Collingridge, G.L.; McBain, C.J.; Isaac, J.T.R. An Essential Role For PICK1 in NMDA Receptor-Dependent Bidirectional Synaptic Plasticity. Neuron 2008, 57, 872–882. [Google Scholar] [CrossRef]
- Chu, P.Y.; Li, T.K.; Ding, S.T.; Lai, I.R.; Shen, T.L. EGF-Induced Grb7 Recruits And Promotes Ras Activity Essential For The Tumorigenicity of Sk-Br3 Breast Cancer Cells. J. Biol. Chem. 2010, 285, 29279–29285. [Google Scholar] [CrossRef] [PubMed]
- Volk, L.; Kim, C.; Takamiya, K.; Yu, Y.; Huganir, R. Developmental Regulation of Protein Interacting With C Kinase 1 (PICK1) Function in Hippocampal Synaptic Plasticity And Learning. Proc. Natl. Acad. Sci. USA 2010, 107, 21784–21789. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, D.; Scarpa, M.; Tessari, A.; Uka, R.; Amari, F.; Lee, C.; Richmond, T.; Foray, C.; Sheetz, T.; Braddom, A.; et al. Ran Binding Protein 9 (RanBP9) Is A Novel Mediator of Cellular DNA Damage Response in Lung Cancer Cells. Oncotarget 2016, 7, 18371–18383. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.S.; Ali, R.; Khanday, F.A. Syntrophins Entangled in Cytoskeletal Meshwork: Helping To Hold It All Together. Cell Prolif. 2019, 52, e12562. [Google Scholar] [CrossRef] [PubMed]
- Coley, A.A.; Gao, W. PSD-95 Deficiency Disrupts PFC-Associated Function And Behavior During Neurodevelopment. Sci. Rep. 2019, 9, 9486. [Google Scholar] [CrossRef] [PubMed]
- Shiroshima, T.; Oka, C.; Kawaichi, M. Identification of LRP1B-Interacting Proteins And Inhibition of Protein Kinase C Alpha-Phosphorylation of LRP1B By Association With PICK1. FEBS Lett. 2009, 583, 43–48. [Google Scholar] [CrossRef]
- Ranganathan, S.; Liu, C.X.; Migliorini, M.M.; Von Arnim, C.A.; Peltan, I.D.; Mikhailenko, I.; Hyman, B.T.; Strickland, D.K. Serine And Threonine Phosphorylation of The Low Density Lipoprotein Receptor-Related Protein By Protein Kinase Calpha Regulates Endocytosis And Association With Adaptor Molecules. J. Biol. Chem. 2004, 279, 40536–40544. [Google Scholar] [CrossRef]
- Brown, M.S.; Ye, J.; Rawson, R.B.; Goldstein, J.L. Regulated Intramembrane Proteolysis: A Control Mechanism Conserved From Bacteria To Humans. Cell 2000, 100, 391–398. [Google Scholar] [CrossRef]
- Liu, C.X.; Ranganathan, S.; Robinson, S.; Strickland, D.K. Gamma-Secretase-Mediated Release of The Low Density Lipoprotein Receptor-Related Protein 1B Intracellular Domain Suppresses Anchorage-Independent Growth of Neuroglioma Cells. J. Biol. Chem. 2007, 282, 7504–7511. [Google Scholar] [CrossRef]
- Brown, L.C.; Tucker, M.D.; Sedhom, R.; Schwartz, E.B.; Zhu, J.; Kao, C.; Labriola, M.K.; Gupta, R.T.; Marin, D.; Wu, Y.; et al. LRP1B mutations are associated with favorable outcomes to immune checkpoint inhibitors across multiple cancer types. J. Immunother. Cancer 2021, 9, e001792. [Google Scholar] [CrossRef]
- Cowin, P.A.; George, J.; Fereday, S.; Loehrer, E.; Van Loo, P.; Cullinane, C.; Etemadmoghadam, D.; Ftouni, S.; Galletta, L.; Anglesio, M.S.; et al. LRP1B Deletion in High-Grade Serous Ovarian Cancers Is Associated With Acquired Chemotherapy Resistance To Liposomal Doxorubicin. Cancer Res. 2012, 72, 4060–4073. [Google Scholar] [CrossRef]
- Dionisio de Sousa, I.J.; Cunha, A.I.; Saraiva, I.A.; POrtugal, R.V.; Gimba, E.R.P.; Guimarães, M.; Prazeres, H.; Lopes, J.M.; Soares, P.; Lima, R.T. LRP1B expression as a putative predictor of response to pegylated liposomal doxorubicin treatment in ovarian cancer. Pathobiology. in press. [CrossRef]
- Liu, F.; Hou, W.; Liang, J.; Zhu, L.; Luo, C. LRP1B mutation: A novel independent prognostic factor and a predictive tumor mutation burden in hepatocellular carcinoma. J. Cancer 2021, 12, 4039–4048. [Google Scholar] [CrossRef]
- Prazeres, H.; Torres, J.; Rodrigues, F.; Pinto, M.; Pastoriza, M.C.; Gomes, D.; Cameselle-Teijeiro, J.; Vidal, A.; Martins, T.C.; Sobrinho-Simoes, M.; et al. Chromosomal, Epigenetic And MicroRNA-Mediated Inactivation of LRP1B, A Modulator of The Extracellular Environment of Thyroid Cancer Cells. Oncogene 2011, 36, 146. [Google Scholar] [CrossRef]
- Sousa, I.; Rodrigues, F.; Prazeres, H.; Lima, R.T.; Soares, P. Liposomal therapies in oncology: Does one size fit all? Cancer Chemother. Pharm. 2018, 82, 741–755. [Google Scholar] [CrossRef]
- Tabouret, E.; Labussiere, M.; Alentorn, A.; Schmitt, Y.; Marie, Y.; Sanson, M. LRP1B Deletion Is Associated With Poor Outcome For Glioblastoma Patients. J. Neurol. Sci. 2015, 358, 440–443. [Google Scholar] [CrossRef] [PubMed]
- Langbein, S.; Szakacs, O.; Wilhelm, M.; Sukosd, F.; Weber, S.; Jauch, A.; Beltran, A.L.; Alken, P.; Kalble, T.; Kovacs, G. Alteration of The LRP1B Gene Region Is Associated With High Grade of Urothelial Cancer. Lab. Investig. 2002, 82, 639–643. [Google Scholar] [CrossRef][Green Version]
- Pineau, P.; Marchio, A.; Nagamori, S.; Seki, S.; Tiollais, P.; Dejean, A. Homozygous Deletion Scanning in Hepatobiliary Tumor Cell Lines Reveals Alternative Pathways For Liver Carcinogenesis. Hepatology 2003, 37, 852–861. [Google Scholar] [CrossRef] [PubMed]
- Hirai, Y.; Utsugi, K.; Takeshima, N.; Kawamata, Y.; Furuta, R.; Kitagawa, T.; Kawaguchi, T.; Hasumi, K.; Noda, S.T. Putative Gene Loci Associated With Carcinogenesis And Metastasis of Endocervical Adenocarcinomas of Uterus Determined By Conventional And Array-Based CGH. Am. J. Obstet. Gynecol. 2004, 191, 1173–1182. [Google Scholar] [CrossRef] [PubMed]
- Sonoda, I.; Imoto, I.; Inoue, J.; Shibata, T.; Shimada, Y.; Chin, K.; Imamura, M.; Amagasa, T.; Gray, J.W.; Hirohashi, S.; et al. Frequent Silencing of Low Density Lipoprotein Receptor-Related Protein 1B (LRP1B) Expression By Genetic And Epigenetic Mechanisms in Esophageal Squamous Cell Carcinoma. Cancer Res. 2004, 64, 3741–3747. [Google Scholar] [CrossRef] [PubMed]
- Roversi, G.; Pfundt, R.; Moroni, R.F.; Magnani, I.; van Reijmersdal, S.; Pollo, B.; Straatman, H.; Larizza, L.; Schoenmakers, E.F.P.M. Identification of Novel Genomic Markers Related To Progression To Glioblastoma Through Genomic Profiling of 25 Primary Glioma Cell Lines. Oncogene 2005, 25, 1571–1583. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Pimkhaokham, A.; Suzuki, E.; Omura, K.; Inazawa, J.; Imoto, I. Genetic Or Epigenetic Silencing of Low Density Lipoprotein Receptor-Related Protein 1B Expression in Oral Squamous Cell Carcinoma. Cancer Sci. 2006, 97, 1070–1074. [Google Scholar] [CrossRef]
- Cengiz, B.; Gunduz, M.; Nagatsuka, H.; Beder, L.; Gunduz, E.; Tarnarnura, R.; Mahmut, N.; Fukushirna, K.; Ali, M.A.S.; NaoMoto, Y.; et al. Fine Deletion Mapping of Chromosome 2q21-37 Shows Three Preferentially Deleted Regions in Oral Cancer. Oral Oncol. 2007, 43, 241–247. [Google Scholar] [CrossRef]
- Choi, Y.W.; Bae, S.M.; Kim, Y.W.; Lee, H.N.; Kim, Y.W.; Park, T.C.; Ro, D.Y.; Shin, J.C.; Shin, S.J.; Seo, J.S.; et al. Gene Expression Profiles in Squamous Cell Cervical Carcinoma Using Array-Based Comparative Genomic Hybridization Analysis. Int. J. Gynecol. Cancer 2007, 17, 687–696. [Google Scholar] [CrossRef]
- Yin, D.; Ogawa, S.; Kawamata, N.; Tunici, P.; Finocchiaro, G.; Eoli, M.; Ruckert, C.; Huynh, T.; Liu, G.T.; Kato, M.; et al. High-Resolution Genomic Copy Number Profiling of Glioblastoma Multiforme by Single Nucleotide Polymorphism DNA Microarray. Mol. Cancer Res. 2009, 7, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Kadota, M.; Yang, H.H.; Gomez, B.; Sato, M.; Clifford, R.J.; Meerzaman, D.; Dunn, B.K.; Wakefield, L.M.; Lee, M.P. Delineating Genetic Alterations For Tumor Progression in The MCF10A Series of Breast Cancer Cell Lines. PLoS ONE 2010, 5, e9201. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Otsuka, A.; Girard, L.; Sato, M.; Iwakawa, R.; Ogiwara, H.; Sanchez-Cespedes, M.; Minna, J.D.; Yokota, J. A Catalog of Genes Homozygously Deleted in Human Lung Cancer and the Candidacy of PTPRD as a Tumor Suppressor Gene. Genes Chromosomes Cancer 2010, 49, 342–352. [Google Scholar] [CrossRef]
- Brown, J.; Bothma, H.; Veale, R.; Willem, P. Genomic Imbalances in Esophageal Carcinoma Cell Lines Involve Wnt Pathway Genes. World J. Gastroenterol. 2011, 17, 2909–2923. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, J.; Mengelbier, L.H.; Elfving, P.; Nord, D.G. High-Resolution Genomic Profiling of An Adult Wilms’ Tumor: Evidence For A Pathogenesis Distinct From Corresponding Pediatric Tumors. Virchows Arch. 2011, 459, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.B.; Hu, J.R.; Duan, Y.S.; Shi, S.L.; Li, R.; Wu, H.J.; Qu, Y.P.; Li, Y. Down Expression of LRP1B Promotes Cell Migration Via RhoA/Cdc42 Pathway And Actin Cytoskeleton Remodeling in Renal Cell Cancer. Cancer Sci. 2013, 104, 817–825. [Google Scholar] [CrossRef]
- Tucker, M.D.; Zhu, J.S.; Marin, D.; Gupta, R.T.; Gupta, S.; Berry, W.R.; Ramalingam, S.; Zhang, T.; Harrison, M.; Wu, Y.; et al. Pembrolizumab in Men With Heavily Treated Metastatic Castrate-Resistant Prostate Cancer. Cancer Med. 2019, 8, 4644–4655. [Google Scholar] [CrossRef]
- Ding, L.; Getz, G.; Wheeler, D.A.; Mardis, E.R.; McLellan, M.D.; Cibulskis, K.; Sougnez, C.; Greulich, H.; Muzny, D.M.; Morgan, M.B.; et al. Somatic Mutations Affect Key Pathways in Lung Adenocarcinoma. Nature 2008, 455, 1069–1075. [Google Scholar] [CrossRef]
- Nikolaev, S.I.; Rimoldi, D.; Iseli, C.; Valsesia, A.; Robyr, D.; Gehrig, C.; Harshman, K.; Guipponi, M.; Bukach, O.; Zoete, V.; et al. Exome Sequencing Identifies Recurrent Somatic MAP2K1 and MAP2K2 Mutations in Melanoma. Nat. Genet. 2011, 44, 133–139. [Google Scholar] [CrossRef]
- Lee, S.; Lee, J.; Sim, S.H.; Lee, Y.; Moon, K.C.; Lee, C.; Park, W.Y.; Kim, N.K.; Lee, S.H.; Lee, H. Comprehensive Somatic Genome Alterations of Urachal Carcinoma. J. Med. Genet. 2017, 54, 572–578. [Google Scholar] [CrossRef]
- Maru, Y.; Tanaka, N.; Ohira, M.; Itami, M.; Hippo, Y.; Nagase, H. Identification of Novel Mutations in Japanese Ovarian Clear Cell Carcinoma Patients Using Optimized Targeted NGS For Clinical Diagnosis. Gynecol. Oncol. 2017, 144, 377–383. [Google Scholar] [CrossRef]
- Xiao, D.K.; Li, F.Q.; Pan, H.; Liang, H.; Wu, K.; He, J.X. Integrative Analysis of Genomic Sequencing Data Reveals Higher Prevalence of LRP1B Mutations in Lung Adenocarcinoma Patients With COPD. Sci. Rep. 2017, 7, 2121. [Google Scholar] [CrossRef]
- Corre, J.; Cleynen, A.; du Pont, S.R.; Buisson, L.; Bolli, N.; Attal, M.; Munshi, N.; Avet-Loiseau, H. Multiple Myeloma Clonal Evolution in Homogeneously Treated Patients. Leukemia 2018, 32, 2636–2647. [Google Scholar] [CrossRef]
- Konukiewitz, B.; Jesinghaus, M.; Steiger, K.; Schlitter, A.M.; Kasajima, A.; Sipos, B.; Zamboni, G.; Welchert, W.; Pfarr, N.; Kloppel, G. Pancreatic Neuroendocrine Carcinomas Reveal A Closer Relationship To Ductal Adenocarcinomas Than To Neuroendocrine Tumors G3. Hum. Pathol. 2018, 77, 70–79. [Google Scholar] [CrossRef]
- Leung, E.Y.; Askarian-Amiri, M.E.; Singleton, D.C.; Ferraro-Peyret, C.; Joseph, W.R.; Finlay, G.J.; Brooms, R.J.; Kakadia, P.M.; Bohlander, S.K.; Marshall, E.; et al. Derivation of Breast Cancer Cell Lines Under Physiological (5%) Oxygen Concentrations. Front. Oncol. 2018, 8, 425. [Google Scholar] [CrossRef]
- Wolff, R.K.; Hoffman, M.D.; Wolff, E.C.; Herrick, J.S.; Sakoda, L.C.; Samowitz, W.S.; Slattery, M.L. Mutation Analysis of Adenomas And Carcinomas of The Colon: Early And Late Drivers. Genes Chromosomes Cancer 2018, 57, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chong, W.; Wu, Q.; Yao, Y.; Mao, M.; Wang, X. Association of LRP1B Mutation With Tumor Mutation Burden And Outcomes in Melanoma And Non-Small Cell Lung Cancer Patients Treated With Immune Check-Point Blockades. Front. Immunol. 2019, 10, 1113. [Google Scholar] [CrossRef]
- Elgendy, M.; Fusco, J.P.; Segura, V.; Lozano, M.D.; Minucci, S.; Echeveste, J.I.; Gurpide, A.; Andueza, M.; Melero, I.; Sanmamed, M.F.; et al. Identification of Mutations Associated With Acquired Resistance To Sunitinib in Renal Cell Cancer. Int. J. Cancer 2019, 145, 1991–2001. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, Y.; Zhang, Y.; Yu, Y.F.; Chen, H.; Liu, K.; Yao, M.; Wang, K.; Gu, W.G.; Shou, T. Comprehensive Genomic Profiling of Small Cell Lung Cancer in Chinese Patients And The Implications For Therapeutic Potential. Cancer Med. 2019, 8, 4338–4347. [Google Scholar] [CrossRef] [PubMed]
- Lan, S.W.; Li, H.; Liu, Y.; Ma, L.X.; Liu, X.H.; Liu, Y.; Yan, S.; Cheng, Y. Somatic Mutation of LRP1B Is Associated With Tumor Mutational Burden in Patients With Lung Cancer. Lung Cancer 2019, 132, 154–156. [Google Scholar] [CrossRef]
- Li, B.J.; Liu, C.X.; Cheng, G.X.; Peng, M.L.; Qin, X.S.; Liu, Y.; Li, Y.Z.; Qin, D.C. LRP1B Polymorphisms Are Associated with Multiple Myeloma Risk in a Chinese Han Population. J. Cancer 2019, 10, 577–582. [Google Scholar] [CrossRef]
- Zhao, X.; Lei, Y.; Li, G.; Cheng, Y.; Yang, H.; Xie, L.; Long, H.; Jiang, R. Integrative Analysis of Cancer Driver Genes in Prostate Adenocarcinoma. Mol. Med. Rep. 2019, 19, 2707–2715. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.T.; Hu, H.G.; Cai, W.; Xu, J.H.; Hu, W.X.; Weng, X.Y.; Qin, X.; Huang, Y.Q.; Han, W.D.; Hu, Y.T.; et al. High-Risk Stage III Colon Cancer Patients Identified By A Novel Five-Gene Mutational Signature Are Characterized By Upregulation of IL-23A And Gut Bacterial Translocation of The Tumor Microenvironment. Int. J. Cancer 2020, 146, 2027–2035. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Yang, B.; Liu, J.; Wu, W.; Ling, Y. Case Report of Acute Myeloid Leukemia With “WT1, ATRX, CEBPA, CSMD1, IKZF1, And LRP1B Mutation And Translocation Between Chromosome 1 And 19” Developing from Philadelphia-Negative Chronic Myeloid Leukemia after TKI Therapy. Medicine 2020, 99, e18888. [Google Scholar] [CrossRef]
- Rahmatpanah, F.B.; Carstens, S.; Guo, J.; Sjahputera, O.; Taylor, K.H.; Duff, D.; Shi, H.; Davis, J.W.; Hooshmand, S.I.; Chitma-Matsiga, R.; et al. Differential DNA Methylation Patterns of Small B-Cell Lymphoma Subclasses With Different Clinical Behavior. Leukemia 2006, 20, 1855–1862. [Google Scholar] [CrossRef]
- Taylor, K.H.; Kramer, R.S.; Davis, J.W.; Guo, J.; Duff, D.J.; Xu, D.; Caldwell, C.W.; Shi, H. Ultradeep Bisulfite Sequencing Analysis of DNA Methylation Patterns in Multiple Gene Promoters By 454 Sequencing. Cancer Res. 2007, 67, 8511–8518. [Google Scholar] [CrossRef]
- Taylor, K.H.; Pena-Hernandez, K.E.; Davis, J.W.; Arthur, G.L.; Duff, D.J.; Shi, H.D.; Rahmatpanah, F.B.; Sjahputera, O.; Caldwell, C.W. Large-Scale CpG Methylation Analysis Identifies Novel Candidate Genes And Reveals Methylation Hotspots in Acute Lymphoblastic Leukemia. Cancer Res. 2007, 67, 2617–2625. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.J.; Wu, C.S.; Li, H.P.; Liu, H.P.; Lu, C.Y.; Leu, Y.W.; Wang, C.S.; Chen, L.C.; Lin, K.H.; Chang, Y.S. Aberrant Methylation Impairs Low Density Lipoprotein Receptor-Related Protein 1B Tumor Suppressor Function in Gastric Cancer. Genes Chromosomes Cancer 2010, 49, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Cui, R.; Li, H.; Li, J.L. miR-500 Promotes Cell Proliferation By Directly Targetting LRP1B in Prostate Cancer. Biosci. Rep. 2019, 39, BSR20181854. [Google Scholar] [CrossRef]
- Zheng, H.Y.; Bai, L.G. Hypoxia Induced microRNA-301b-3p Overexpression Promotes Proliferation, Migration And Invasion of Prostate Cancer Cells By Targeting LRP1B. Exp. Mol. Pathol. 2019, 111, 104301. [Google Scholar] [CrossRef]
- Ding, D.; Lou, X.; Hua, D.; Yu, W.; Li, L.; Wang, J.; Gao, F.; Zhao, N.; Ren, G.; Li, L.; et al. Recurrent Targeted Genes of Hepatitis B Virus in The Liver Cancer Genomes Identified By A Next-Generation Sequencing-Based Approach. PLoS Genet. 2012, 8, e1003065. [Google Scholar] [CrossRef]
- Jiang, Y.H.; Zhu, C.Y.; He, D.; Gao, Q.L.; Tian, X.; Ma, X.; Wu, J.; Das, B.C.; Severinov, K.; Hitzeroth, I.I.; et al. Cytological Immunostaining of HMGA2, LRP1B, And TP63 As Potential Biomarkers For Triaging Human Papillomavirus-Positive Women. Transl. Oncol. 2019, 12, 959–967. [Google Scholar] [CrossRef] [PubMed]
- AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017, 7, 818–831. [CrossRef]
- Wang, Z.; Sun, P.; Gao, C.; Chen, J.; Li, J.; Chen, Z.; Xu, M.; Shao, J.; Zhang, Y.; Xie, J. Down-Regulation of LRP1B in Colon Cancer Promoted The Growth And Migration of Cancer Cells. Exp. Cell Res. 2017, 357, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hu, J.; Jin, R.; Cheng, H.; Chen, H.; Li, L.; Guo, K. Effects of LRP1B Regulated by HSF1 on Lipid Metabolism in Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2020, 7, 361–376. [Google Scholar] [CrossRef]
- Asano, Y.; Takeuchi, T.; Okubo, H.; Saigo, C.; Kito, Y.; Iwata, Y.; Futamura, M.; Yoshida, K. Nuclear localization of LDL receptor-related protein 1B in mammary gland carcinogenesis. J. Mol. Med. (Berlin, Germany) 2019, 97, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Yasufuku, I.; Saigo, C.; Kito, Y.; Yoshida, K.; Takeuchi, T. Prognostic significance of LDL receptor-related protein 1B in patients with gastric cancer. J. Mol. Histol. 2021, 52, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Sabour, L.; Sabour, M.; Ghorbian, S. Clinical Applications of Next-Generation Sequencing in Cancer Diagnosis. Pathol. Oncol. Res. 2017, 23, 225–234. [Google Scholar] [CrossRef]
- Johnson, D.B.; Frampton, G.M.; Rioth, M.J.; Yusko, E.; Xu, Y.; Guo, X.; Ennis, R.C.; Fabrizio, D.; Chalmers, Z.R.; Greenbowe, J.; et al. Targeted Next Generation Sequencing Identifies Markers of Response to PD-1 Blockade. Cancer Immunol. Res. 2016, 4, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Shen, X.; Zhang, B.; Su, R.; Cui, M.; Yan, L.; Cao, Y. Development and validation of a LRP1B mutation-associated prognostic model for hepatocellular carcinoma. Biosci. Rep. 2021, 41, BSR20211053. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Duan, Q.; Zhang, Q. Analysis of LRP1B as a potential biomarker for colorectal cancer immunotherapy. J. Clin. Oncol. 2021, 39, e15521. [Google Scholar] [CrossRef]
- Jacob, J.B.; Jacob, M.K.; Parajuli, P. Review of immune checkpoint inhibitors in immuno-oncology. Adv. Pharmacol. 2021, 91, 111–139. [Google Scholar] [CrossRef] [PubMed]
- Seidel, J.A.; Otsuka, A.; Kabashima, K. Anti-PD-1 and Anti-CTLA-4 Therapies in Cancer: Mechanisms of Action, Efficacy, and Limitations. Front. Oncol. 2018, 8, 86. [Google Scholar] [CrossRef]
- Cao, D.; Xu, H.; Xu, X.; Guo, T.; Ge, W. High tumor mutation burden predicts better efficacy of immunotherapy: A pooled analysis of 103078 cancer patients. OncoImmunology 2019, 8, e1629258. [Google Scholar] [CrossRef]
- Addeo, A.; Friedlaender, A.; Banna, G.L.; Weiss, G.J. TMB or not TMB as a biomarker: That is the question. Crit. Rev. Oncol./Hematol. 2021, 163, 103374. [Google Scholar] [CrossRef]
- Garutti, M.; Bonin, S.; Buriolla, S.; Bertoli, E.; Pizzichetta, M.A.; Zalaudek, I.; Puglisi, F. Find the Flame: Predictive Biomarkers for Immunotherapy in Melanoma. Cancers 2021, 13, 1819. [Google Scholar] [CrossRef]
- Li, J.; Wei, Q.; Wu, X.; Sima, J.; Xu, Q.; Wu, M.; Wang, F.; Mou, H.; Hu, H.; Zhao, J. Integrative clinical and molecular analysis of advanced biliary tract cancers on immune checkpoint blockade reveals potential markers of response. Clin. Transl. Med. 2020, 10, e118. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Ji, Q.; Cai, H.; Liang, X.; Xie, J.; Li, H.; Wang, J.; Zhu, G.; Tian, E.; et al. Clinicopathological and molecular characteristics of patients with hypermutant lung cancer: A retrospective cohort study. Oncol. Lett. 2021, 21, 329. [Google Scholar] [CrossRef]
- Wang, L.; Yan, K.; He, X.; Zhu, H.; Song, J.; Chen, S.; Cai, S.; Zhao, Y.; Wang, L. LRP1B or TP53 mutations are associated with higher tumor mutational burden and worse survival in hepatocellular carcinoma. J. Cancer 2021, 12, 217–223. [Google Scholar] [CrossRef]
- Bao, R.; Stapor, D.; Luke, J.J. Molecular correlates and therapeutic targets in T cell-inflamed versus non-T cell-inflamed tumors across cancer types. Genome Med. 2020, 12, 90. [Google Scholar] [CrossRef]


| Ligand | Description | Ligand-Binding Domain (I to IV) | Reference | |
|---|---|---|---|---|
| RAP | Chaperone | II, IV | [14,18] | |
| uPA | Serine protease | IV | [14] | |
| tPA | Serine protease | IV | [14] | |
| PAI-1 | Serine protease inhibitor | IV | [14] | |
| uPAR | Cell-surface receptor | IV | [25] | |
| APP | Cell-surface receptor | IV | [17] | |
| Sacsin | Co-chaperone | IV | [18] | |
| Endoplasmin | Chaperone | II, III | [18] | |
| Synaptotagmin-1 | Calcium ion sensor | IV | [18] | |
| DnaJ homolog subfamily A member 1 | Co-chaperone | II | [18] | |
| Glutathione S-transferase LANCL1 | Transferase | IV | [18] | |
| 40S ribosomal protein SA | Host cell receptor for virus entry, cell-surface receptor for laminin, ribonucleoprotein | II, IV | [18] | |
| Pseudomonas aeruginosa exotoxin A | P. aeruginosa toxin | IV | [26] | |
| Fibrinogen | α-chain | Substrate for thrombin, plasmin, and fibrin stabilizing factor | II | [27] |
| β-chain | IV | [27] | ||
| γ-chain | IV | [27] | ||
| HRG | Plasma glycoprotein | II | [27] | |
| Clusterin | Chaperone | II | [27] | |
| Vitronectin | Glycoprotein found in blood and the extracellular matrix | II | [27] | |
| SAP | Plasma protein | II | [27] | |
| IGKV 1-5 | Variable domain of immunoglobulin light chains | II | [27] | |
| IGHA1 | Constant region of immunoglobulin heavy chains | II | [27] | |
| HDL | apoE-containing lipoprotein | II | [27] | |
| VLDL | apoE-containing lipoprotein | II, IV | [27] | |
| Intracellular Interacting Partner | Description | Reference |
|---|---|---|
| PSD-95 | Scaffold protein | [18] |
| AIP | Co-chaperone | [18] |
| JIP-1b | Scaffold protein | [54] |
| JIP-2 | Scaffold protein | [54] |
| PICK1 | Scaffold protein | [54] |
| RanBP9 | Scaffold protein and adaptor protein | [54] |
| GRB7 | Adaptor protein | [54] |
| G2SNT | Adaptor protein | [54] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Príncipe, C.; Dionísio de Sousa, I.J.; Prazeres, H.; Soares, P.; Lima, R.T. LRP1B: A Giant Lost in Cancer Translation. Pharmaceuticals 2021, 14, 836. https://doi.org/10.3390/ph14090836
Príncipe C, Dionísio de Sousa IJ, Prazeres H, Soares P, Lima RT. LRP1B: A Giant Lost in Cancer Translation. Pharmaceuticals. 2021; 14(9):836. https://doi.org/10.3390/ph14090836
Chicago/Turabian StylePríncipe, Catarina, Isabel J. Dionísio de Sousa, Hugo Prazeres, Paula Soares, and Raquel T. Lima. 2021. "LRP1B: A Giant Lost in Cancer Translation" Pharmaceuticals 14, no. 9: 836. https://doi.org/10.3390/ph14090836
APA StylePríncipe, C., Dionísio de Sousa, I. J., Prazeres, H., Soares, P., & Lima, R. T. (2021). LRP1B: A Giant Lost in Cancer Translation. Pharmaceuticals, 14(9), 836. https://doi.org/10.3390/ph14090836









