Abstract
The transforming growth factor beta (TGFβ) pathway could modulate the Duchenne muscular dystrophy (DMD) phenotype. This meta-analysis aims to estimate the association of genetic variants involved in the TGFβ pathway, including the latent transforming growth factor beta binding protein 4 (LTBP4) and secreted phosphoprotein 1 (SPP1) genes, among others, with age of loss of ambulation (LoA) and cardiac function in patients with DMD. Meta-analyses were conducted for the hazard ratio (HR) of LoA for each genetic variant. A subgroup analysis was performed in patients treated exclusively with glucocorticoids. Eight studies were included in the systematic review and four in the meta-analyses. The systematic review suggests a protective effect of LTBP4 haplotype IAAM (recessive model) for LoA. It is also suggested that the SPP1 rs28357094 genotype G (dominant model) is associated with early LoA in glucocorticoids-treated patients. The meta-analysis of the LTBP4 haplotype IAAM showed a protective association with LoA, with an HR = 0.78 (95% CI: 0.67–0.90). No association with LoA was observed for the SPP1 rs28357094. The LTBP4 haplotype IAAM is associated with a later LoA, especially in the Caucasian population, while the SPP1 rs28357094 genotype G could be associated with a poor response to glucocorticoids. Future research is suggested for SPP1 rs11730582, LTBP4 rs710160, and THBS1 rs2725797.
Keywords:
Duchenne muscular dystrophy; polymorphism; TGFβ; SPP1; LTBP4; systematic review; meta-analysis 1. Introduction
Duchenne muscular dystrophy (DMD) is an X-linked recessive lethal disease that predominantly affects males, with an incidence of one case per 3500–9000 live births [1]. It is caused by mutations, insertions, or deletions of the dystrophin/DMD gene (location Xp21.2-p21.1) resulting in an absence of the cytoskeletal protein dystrophin. Dystrophin is an actin-binding protein that, through the α/β-dystroglycan complex, links the cytoskeleton to the extracellular matrix protein laminin, stabilizing the surface of muscle cells and preventing their rupture during contraction and relaxation cycles. Therefore, this is essential for the strength, stability, and functionality of the myofibers [2,3]. The absence of dystrophin in the muscle tissue causes a destabilization of the dystrophin-associated glycoprotein complex. This leads to, during contraction and relaxation cycles, sarcolemmal instability, and a decrease in force transmission by the sarcomere occurs. In dystrophic muscle there is an infiltration of mononuclear cells, variation in fiber size, centrally located nuclei, and degeneration and replacement of muscle tissue by fibrotic tissue. This leads to a progressive loss of muscle strength, due to the destruction of muscle tissue, along with the difficulty of muscle contraction of the remaining muscle, due to fibrotic tissue and muscle contractures. Ultimately, loss of ambulation (LoA) occurs in late childhood or early adolescence, in addition to other comorbidities such as cardiac and respiratory failure that reduce quality of life and life expectancy. Despite the optimization of glucocorticoid treatment and other pharmacological and nonpharmacological therapies, the prognosis remains poor [2,3,4,5,6,7,8,9]. However, some variability in the progression of DMD has been observed, even after considering improved treatments.
There are common genetic variants outside of the dystrophin gene and that are not pathological, which could influence disease progression through an anti-fibrotic or pro-fibrotic effect. The transforming growth factor beta (TGFβ) pathway is a complex signaling pathway that has been proposed as a candidate for modifying DMD progression, especially the age at LoA and cardiac remodeling. TGFβ is a cytokine that binds to the type II TGFβ receptor, which together with the type I TGFβ receptor forms a heterotetrameric complex, and the type I TGFβ receptor is phosphorylated at a region rich in glycine and serine residues, resulting in activation. Type I TGFβ then phosphorylates certain Smad proteins at C-terminal serine residues in a conserved C-terminal Ser-Ser-X-Ser motif. These phosphorylated Smads oligomerize with Smad4, and these complexes act on target genes, promoting homeostasis, cell differentiation, and tissue regeneration, among others [10]. At the muscle level, TGFβ1 and related cytokines, such as myostatin, a molecule belonging to the TGFβ superfamily, interfere with muscle growth and differentiation factors, such as myoblast determination protein 1 and insulin-like growth factor 2 [11]. This mechanism may also explain the cardiac remodeling that occurs with myocardial fibrosis [12,13].
In mdx models, plasma and muscle levels of TGFβ1 and the levels of TGFβ1 and connective tissue growth factor (CTGF) in the sarcoplasm of muscle cells and mesenteric interstitium are increased and correlate with the degree of fibrosis [14]. Two genes whose genetic variants might exert an effect on the phenotype are latent transforming growth factor beta binding protein 4 (LTBP4, location 19q13.2) and secreted phosphoprotein 1, also known as osteopontin (SPP1, location 4q22.1). LTBP4 binds to TGFβ, reducing its activity [15]. The homozygous IAAM haplotype (V194I, T787A, T820A, and T1140M) might create more stable binding, reducing TGFβ activity [16]. Meanwhile, SPP1 stimulates TGFβ production and increased inflammatory cell infiltration [15]. Although in mdx models, Spp1 genetic variants resulting in low basal activity are associated with an improved phenotype [17], the opposite result tends to occur in humans, showing the complexity of this pathway. Thus, for SPP1, the rs28357094 genetic variant, which is located in the SPP1 promoter, has been proposed as a possible disease modifier [16].
In 2017, a systematic review suggested the association of some of these genetic variants with LoA [18]. Since then, new studies have been published that may improve the available evidence [19,20,21,22,23,24,25,26]. Therefore, this systematic review and meta-analysis aims to estimate the association of SPP1 rs28357094, LTBP4 haplotype IAAM, and other genes directly involved in the TGFβ pathway with the age at LoA and cardiac function in patients with DMD.
2. Materials and Methods
This systematic review was conducted according to the Meta-analyses Of Observational Studies in Epidemiology (MOOSE) guidelines [27] and the Cochrane Collaboration Handbook [28], and it was registered in PROSPERO (registration number: CRD42018111191).
2.1. Search Strategy
An electronic systematic search was performed in MEDLINE (via PubMed), EMBASE, Web of Science, and Cochrane Library databases from inception until July 2021. Grey literature, including OpenGrey, Theseo, Networked Digital Library of Theses and Dissertations, and Google Scholar, was also searched. The search terms included dystrophy, Duchenne, DMD, Duchenne muscular dystrophy, dystrophinopathy, polymorphism, osteopontin, spp1, latent transforming growth factor, ltbp4, thrombospondin, thbs1, ambulation, loss of ambulation, LoA, cardiomyopathy, ventricular dysfunction, heart failure, cardiac function, and heart function. References included in previous reviews and in the included studies were screened. If necessary, the studies’ authors were contacted. The specific search is described in Appendix A.
2.2. Inclusion/Exclusion Criteria
The inclusion criteria were as follows: (1) participants—males with DMD, with no age restrictions; (2) exposure—(i) LTBP4 haplotypes (i.e., IAAM/IAAM vs. others), occasionally a single SNP may be considered due to linkage disequilibrium; (ii) SPP1 genetic variants; (iii) other genetic variants in genes directly involved in the TGFβ pathway; (3) outcomes—LoA (main outcome), defined as the age at which the person requires continued use of a wheelchair because of an inability to walk independently, and cardiac function (secondary outcome), including age of onset of dilated cardiomyopathy (DCM) and age of onset of myocardial dysfunction. No language limitations were imposed.
The exclusion criteria were as follows: (1) participants—a population that was not mainly affected by DMD and the effect of genotype on participants with DMD could not be estimated; (2) exposure—genetic variants not directly involved in the TGFβ pathway.
The literature search was conducted independently by two reviewers (CP-M and IC-R), and disagreements were solved by consensus or with a third reviewer (VM-V).
2.3. Data Extraction
An ad hoc table was conducted with the following data extracted from the selected studies: (1) reference (authors and publication year), (2) exposure (gene and genetic variant or haplotype), (3) country, (4) sample size, (5) design, and (6) outcomes (LoA and cardiac function). The data extraction was conducted individually by two reviewers (CP-M and IC-R).
2.4. Risk of Bias Assessment
To assess the risk of bias, we used the Quality Assessment Tool for Observational Cohort and Cross-Sectional Studies from Study Quality Assessment Tools [29]. This tool has a 14-item checklist. Each item receives a score of good, fair, or poor and includes the following domains: research question, study population, groups recruited from the same population and uniform eligibility criteria, sample size justification, exposure assessed prior to measurement, sufficient timeframe to observe an effect, different levels of the exposure of interest, exposure measures and assessment, repeated exposure assessment, outcome measures, blinding of outcome assessors, follow-up rate, and statistical analysis.
The risk of bias assessment was conducted independently by two reviewers (CP-M and IC-R), and disagreements were solved by consensus or with a third reviewer (VM-V).
2.5. Grading the Quality of Evidence
The Grading of Recommendations, Assessment, Development and Evaluation tool (GRADE) was used to assess the strength of the evidence for the main outcome included in the meta-analyses [30]. Depending on the study design, risk of bias, inconsistency, indirect evidence, imprecision, publication bias, larger effect, possible confounding variables, and dose–response gradient, the GRADE tool rates the strength of evidence as high, moderate, low and very low for each intervention and outcome.
2.6. Data Synthesis
A narrative synthesis and an ad hoc table were generated for the main outcome. Additionally, forest plots were used to graphically depict the hazard ratio (HR) of LoA with their 95% confidence intervals (95% CIs) for each genetic variant. The forest plots differentiated between patients treated with or without glucocorticoids.
When two or more HRs for the LoA associated with a specific genetic variant and their 95% CI were available, a fixed-effects meta-analysis was performed [31]. If the confidence interval or standard error was not reported, it was calculated from the standard deviation or estimated from the p-value [32]. Heterogeneity, as measured by the I2 test [28,33], was considered not important <40%, substantial 30–60%, important 50–90%, and considerable >75%. The p-value of heterogeneity was also considered to determine whether heterogeneity was significant. The meta-analyses differentiated whether glucocorticoid-treated patients, patients who were not treated with glucocorticoids, or both types of patients were included.
STATA SE software, version 15 (StataCorp, College Station, TX, USA), was used to conduct the statistical analyses.
2.7. Infographics
An infographic of the manuscript was created using Canva software (Figure A1).
3. Results
Of the 259 studies identified, 8 met the inclusion criteria and were included in the systematic review [19,20,21,22,23,24,25,26], and 4 were included in the meta-analysis (Table 1 and Figure 1) [20,21,22,24]. Four studies were excluded for various reasons (Supplementary Table S1) [34,35,36,37].

Table 1.
Characteristics of the included studies.
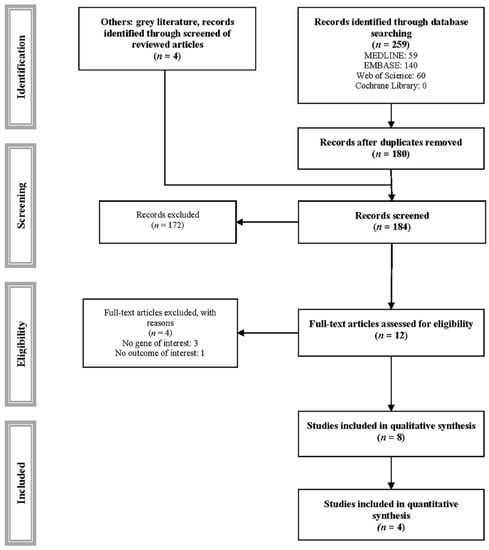
Figure 1.
PRISMA flowchart of study selection.
The main characteristics of the included studies are summarized in Table 1. Three studies used cohorts from the United States [22,25,26], one from China [21], one from the Cooperative International Neuromuscular Research Group (CINRG) [20], one from Italy [19], one from Italy and CINRG [23], and one from France, Italy, the Netherlands and the United Kingdom [24]. A total of 2050 individuals were analyzed, although the use of CINRG cohorts may overrepresent the sample. Five studies analyzed the association of LTBP4 haplotypes with LoA [19,20,21,22,24], five analyzed the association of SPP1 genotypes with LoA [19,20,21,23,24], one assessed the interaction and association of LTBP4 and THBS1 genotypes with LoA [26], two investigated the association of the LTBP4 genotype with cardiac function [19,25], and one analyzed the association of the SPP1 genotype with cardiac function [19].
3.1. Loss of Ambulation—Kaplan–Meier Analyses
Table 2 shows the Kaplan–Meier analyses of the included studies. No study showed an association of the LTBP4 haplotype IAAM (recessive model) vs. other haplotypes with LoA [19,20,21]. SPP1 rs28357094 showed an association with LoA of 1–1.2 years in favor of the genotype T (recessive model) in two of the three studies [20,23]. Finally, an interaction between LTBP4 rs710160 genotype CC and THBS1 rs2725797 genotype TT may result in a protective effect [26].

Table 2.
Studies including as outcome age of loss of ambulation (Kaplan–Meier analyses).
Regarding the glucocorticoid subgroup, only one study showed an association of LTBP4 haplotype IAAM vs. other haplotypes, with a difference of 1.8 years in favor of the IAAM haplotype [22]. For SPP1 rs28357094, one of the two studies showed an association [20], while rs28357094 showed no association in the nonglucocorticoid subgroup [21]. Finally, SPP1 rs11730582 also showed an association, with the C genotype being protective (dominant model) [21].
3.2. Loss of Ambulation—Cox Regression Analyses
Table 3 and Figure 2 show Cox regression analyses of the included studies. The LTBP4 haplotype IAAM was associated with an HR = 0.77 (p = 0.01) and HR = 0.52 (95% CI: 0.34–0.78) compared with other haplotypes [22,24], while in other studies it showed no significant effect [20,21]. SPP1 rs28357094 showed no association [20,24].

Table 3.
Studies including as outcome age of loss of ambulation (Cox regression analyses).
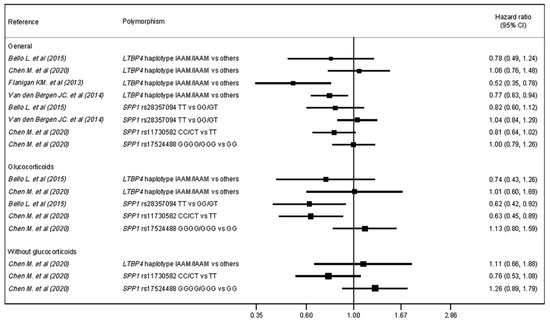
Figure 2.
Forest plot for hazard ratio of LoA by genetic variant (polymorphism) and glucocorticoid subgroup.
Regarding the glucocorticoid subgroup, the LTBP4 haplotype IAAM was not associated with HR compared with other haplotypes [20,21]. A comparison of the SPP1 rs28357094 TT genotype with the GG/GT genotypes revealed an HR = 0.62 (95% CI = 0.42–0.92) [20]. Finally, regarding SPP1 rs11730582 and rs17524488, only rs11730582 genotype C showed a protective association, with an HR = 0.63 (95% CI: 0.45, 0.89) [21].
3.3. Cardiac Function
Table 4 shows Kaplan–Meier analyses of the included studies. LTBP4 rs10880 was not significantly associated with the age at onset of DCM or myocardial dysfunction [19,25]. However, there was a trend toward a higher percentage of patients without myocardial dysfunction with the rs10880 TT genotype [25]. In the glucocorticoid subgroup, the rs10880 genotype T (recessive model) was associated with a later age of onset of DCM (log-rank p < 0.05) [19]. SPP1 rs28357094 was not associated with the age at onset of DCM or myocardial dysfunction, although the genotype T tended to be harmful [19].

Table 4.
Studies including cardiac function as an outcome.
3.4. Assessment of the Risk of Bias
The studies fulfilled between 64.3% and 71.4% of the quality criteria proposed by the Quality Assessment Tool for Observational Cohort and Cross-Sectional Studies from Study Quality Assessment Tools. The objective of the study was not correctly specified in 12.5% of the studies. No study justified the sample size used to achieve a specific statistical power, and the authors used the available sample. An evaluation of different exposure levels or their changes over time was not necessary. Finally, blinding of the analysts was not considered necessary in any study. The complete risk assessment is available in Supplementary Table S2.
3.5. Evidence Assessment
Using the GRADE tool, the LTBP4 haplotype IAAM/IAAM, SPP1 rs28357094 TT vs. GG/GT and LTBP4 haplotype IAAM/IAAM vs. other haplotypes in glucocorticoid-treated patients showed low certainty of evidence. The complete assessment is detailed in Supplementary Table S3.
3.6. Meta-Analysis
Three meta-analyses were conducted that included the following studies: four studies of LTBP4 haplotype IAAM compared with other haplotypes in glucocorticoid-treated and nonglucocorticoid-treated patients, two studies of the involvement of SPP1 rs28357094 in glucocorticoid-treated and nonglucocorticoid-treated patients, and two studies of LTBP4 haplotype IAAM compared with other haplotypes in patients who were exclusively treated with glucocorticoids. Due to the scarcity of studies, we were unable to conduct meta-analyses of the remaining genotypes or of patients who were not treated with glucocorticoids.
The meta-analysis showed a protective association of the LTBP4 haplotype IAAM, with an HR = 0.78 (95% CI: 0.67, 0.90) (Figure 3A), while SPP1 rs28357094 showed no association (Figure 3B). In the glucocorticoid subgroup, no association was observed for the LTBP4 haplotype IAAM/IAAM (Figure 3C).
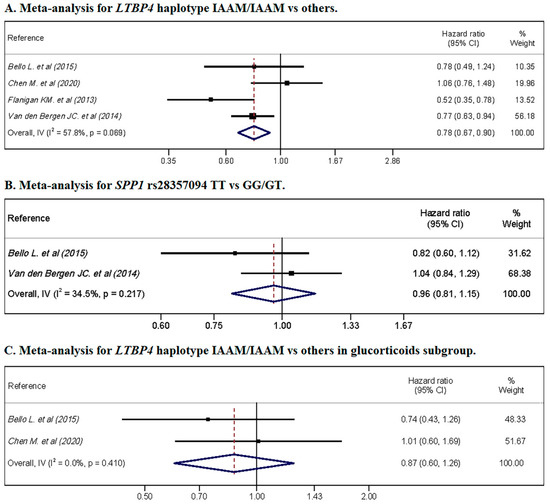
Figure 3.
Meta-analyses for hazard ratio of LoA by genetic variant. (A) shows the pooled for LTBP4 haplotype IAAM/IAAM vs. others, without glucocorticoids restrictions; (B) shows the pooled for SPP1 rs28357094 genotype TT vs. GG/GT, without glucocorticoids restrictions; (C) shows the forest plot for LTBP4 haplotype IAAM/IAAM vs. others in glucocorticoids-treated cohorts.
Heterogeneity was substantial for the LTBP4 haplotype IAAM, including glucocorticoid-treated and glucocorticoid-untreated patients, with a heterogeneity of I2 = 57.8% (p = 0.069). Furthermore, substantial heterogeneity was not observed for SPP1 rs28357094 in glucocorticoid-treated and glucocorticoid-untreated patients or LTBP4 in the glucocorticoid subgroup, with I2 = 34.5% (p = 0.217) and I2 = 0.0% (p = 0.410), respectively.
4. Discussion
4.1. Main Findings
This systematic review and meta-analysis provides an overview of the evidence supporting the associations of genetic variants with LoA and cardiac function. Our results suggest a notable effect of genetic variants involved in the TGFβ pathway, especially LTBP4 haplotype IAAM (in the recessive model), but not in patients who were exclusively treated with glucocorticoids, probably due to confounding factors. SPP1 rs28357094 did not display a significant association. However, the use of glucocorticoids by patients carrying the SPP1 rs28357094 genetic variant potentially increased the association in favor of genotype T (recessive model). Further research on THBS1 genetic variants is needed, and the limited evidence available indicates an interaction with LTBP4 that might improve the DMD phenotype. Finally, more research is needed on other proinflammatory and profibrotic pathways that might exert some effect and distort the effect observed in our study.
4.2. Interpretation
In dystrophic mice there is a 36-base-pair insertion/deletion site in a proline-rich domain of Ltbp4. The 12 amino acid insertion confers resistance to proteolysis of the TGFβ–LTBP4 complex, reducing TGFβ activity and therefore muscle fibrosis. Although this indel is not present in humans, the haplotypes mentioned in the previous sections (i.e., IAAM and VTTT) represent more than 80% of the total of all possible combinations in the human population. The haplotype IAAM behaves like the 36-base-pair insertion in Ltbp4 of dystrophic mice, giving LTBP4 increased binding avidity to TGFβ, thereby reducing its activity [38]. Our meta-analysis shows that the LTBP4 haplotype IAAM (or the rs10880 genotype T) is a protective factor resulting in prolonged ambulation. However, in patients treated exclusively with glucocorticoids, the haplotype IAAM had no effect, probably due to the inclusion of only two studies, one including patients from China, in which the LTBP4 haplotype IAAM does not appear to be associated with LoA, perhaps due to the genetic/ethnic factors described below. Considering the Kaplan–Meier analyses, Barp A et al. and Chen M et al. did not observe an association, while Bello L et al. and Flanigan KM et al. reported a trend toward benefit for the haplotype IAAM, especially in the glucocorticoid-treated population. Interestingly, the cohorts in the former two studies (Barp A et al. and Chen M et al.) lost ambulation earlier than the cohorts in the latter two studies (Bello L et al. and Flanigan KM et al.), which might imply differences in the care and management or genetic/ethnicity of these patients. Thus, in the cohort analyzed by Barp A et al., the use of glucocorticoids was still not a universal standard, and a relatively low use of these drugs was reported. Chen M et al. investigated Asian cohorts, in which the effect of the LTBP4 haplotype IAAM was different than that on Caucasian cohorts. Furthermore, previous evidence [38] has identified rs710160 as a genetic variant that may modulate the association obtained for the LTBP4 haplotype IAAM with LoA. Thus, rs710160 genotype C together with the IAAM haplotype leads to less profibrotic signaling, potentially resulting in milder DMD. Additionally, rs710160 has a significant linkage disequilibrium with rs10880 in Caucasian populations but not in other populations, which might explain why the haplotype IAAM has a higher association with LoA in the Caucasian population but not in the Asian population, as described in the study by Chen M et al.
Thrombospondin-1, encoded by the THBS1 gene (location 15q14), is a potent activator of the latent TGFβ complex. To activate TGF-β1, thrombospondin-1 interacts with the N-terminal region of the latency-associated protein, which binds non-covalently to TGF-β1. Thus, a trimolecular complex is formed, a conformational change occurs, and the reactivity of TGF-β1 is altered. TGF-β1 activation by thrombospondin-1 may be essential for the development of the heart, liver, bones, testes, and hematopoietic systems, among other tissues and organs [39]. Moreover, it exerts an anti-angiogenic effect by decreasing the activities of the nitric oxide and vascular endothelial growth factor (VEGF) pathway, and blocking endothelial cell migration, [39,40], which is harmful in animal models [41]. Finally, thrombospondin-1 could be useful in maintaining low but constant levels of active TGF-β1 [39]. In our study, THBS1 rs2725797 genotype T is associated with lower thrombospondin-1 activity, with an additive effect with LTBP4 rs10880 allele C, which may improve the phenotype of the disease, as suggested by the data presented in this review.
SPP1 is a cytokine secreted by macrophages and myoblasts. It belongs to the family of small integrin-binding ligand N-linked glycoprotein secreted phosphoproteins and is expressed in numerous tissues in response to tissue damage/regeneration and inflammatory response. Alternative splicing and post-translational modifications make it a difficult cytokine to study. In mdx models without osteopontin, mice had reduced TGFβ levels, fibrosis, and increased strength, with reduced infiltration of inflammatory cells, neutrophils, and natural-killer T cells and increased numbers of regulatory T cells and M2 macrophages. Although the reduction in osteopontin could delay regeneration in acute damage, in mdx models (and probably in DMD) it decreases fibrosis caused by chronic damage [38,42]. Although in our study SPP1 rs28357094 was not associated with LoA in a population that was not stratified by glucocorticoid use, in the glucocorticoid subgroup, it appears to exert a glucocorticoid-dependent effect on LoA in regression analyses. In fact, in subgroup analyses stratified by glucocorticoid treatment, this genetic variant showed no association with LoA in the glucocorticoid-untreated subgroup or when a low percentage of glucocorticoid-treated patients was included. SPP1 rs28357094 genotype G was detrimental to patients using glucocorticoids, with earlier LoA. Allele G is associated with lower osteopontin transcriptional activity under basal conditions [43]. However, SPP1 likely contains glucocorticoid receptor elements, which modulate osteopontin expression through interactions with NF-kB, estrogens, and glucocorticoids. Thus, the SPP1 rs28357094 genotype G might increase osteopontin expression by 3-fold in the presence of glucocorticoids, increasing profibrotic signaling [44]. SPP1 rs11730582 might also use a similar mechanism. Genotype C, which results in higher initial osteopontin levels [45], is significantly associated with a later LoA in glucocorticoid-treated patients, suggesting that its effect is glucocorticoid-dependent, similar to rs28357094. Therefore, SPP1 rs28357094 and perhaps rs11730582 are postulated to function not as disease modifiers but as predictors of a good or poor response to glucocorticoid treatment.
Regarding cardiac complications, no clear association with genetic variants can be observed due to the few included studies. LTBP4 rs10880 genotype T tended to delay the age of DCM onset, with a significant association in patients treated with glucocorticoids [19]. This result is consistent with the data obtained for LoA. This association may be due to an additive effect of the LTBP4 haplotype IAAM and glucocorticoids, but this requires further research. In contrast, another study did not detect an association of LTBP4 with the development of left ventricular dysfunction. Interestingly, it was found that, among patients without left ventricular dysfunction, there was a higher proportion of patients with the rs10880 genotype T [25]. These discrepancies are, perhaps, due to the relatively small sample size, the age of the cohorts, or the categorization of patients into glucocorticoid-treated/untreated groups. Furthermore, the SPP1 rs28357094 genotype T tended to be harmful in the development of DCM, including in the glucocorticoid subgroup [19]. This fact is interesting, since it is in the opposite direction to what happens in skeletal muscle. Conversely, it is in agreement with what has been observed in animal models, in which an overexpression of Spp1 causes myocarditis and myocardial dilation [46]. Future research is needed in this regard. Cardiac complications, and especially heart failure, are one of the leading causes of death in patients with DMD. Both the SPP1 genotype and glucocorticoids use could have some negative impact on the development of dilated cardiomyopathy. However, it should be noted that this does not necessarily imply progression to heart failure, and especially glucocorticoids could delay the onset of heart failure through other mechanisms.
The importance of the TGFβ pathway in the DMD phenotype may have several clinical implications: first, drug development aimed at the downregulation of TGFβ signaling to reduce muscle fibrosis. This approach includes angiotensin 1–7, halofuginone, anti-TGFβ1 antibodies, ixazomib, and angiotensin-II type 1 receptor blockers, which antagonize or downmodulate the TGFβ pathway with promising results in animal models [47,48,49,50,51]. However, some of these drugs have produced undesirable pleiotropic effects, which might be a problem in achieving a clinical benefit [49]. Another pathway is myostatin inhibition using compounds such as follistatin, ACE-031, domagrozumab, and the GDF11 propeptide that inhibit or antagonize the effect of myostatin [52,53,54,55,56,57]. However, the results from animal models and patients with DMD have raised some concerns related to other biological functions of myostatin that affect the metabolism and oxidative capacity of muscle fibers [52,58]. Second, genotyping of patients with DMD for LTBP4 and SPP1 could be considered, especially in the Caucasian population. Although knowledge of the genotype of LTBP4 haplotype/rs10880 and SPP1 rs28357094 would not alter the medical treatment of the patient, it can potentially provide the clinician and the patient and their family with more individualized prognosis as to their possible evolution and expected response to glucocorticoids. Third, genotyping can be considered in clinical trials of new drugs, such as treatments designed to restore dystrophin expression. Perhaps, by subgroup analysis according to genotype, part of the variability found in the results could be explained. It is possible that the patient’s genotype can determine the response to small changes in dystrophin expression. Finally, the study of genetic variants in unknown DMD patients could be very interesting. Clinical DMD is well established, with diagnosis in early childhood. However, there are rare cases of dystrophinopathies, including Becker muscular dystrophy (BMD) and DMD, that remain undiagnosed until their debut in adolescence or adulthood, mainly due to cardiac involvement. [59,60,61,62,63]. Moreover, there are phenotypically intermediate forms of diagnosed dystrophinopathies, with slowly evolving DMD or rapidly evolving BMD [38]. These exceptional cases could be an opportunity to study the effect of genetic factors (known and unknown) as well as environmental factors on the progression of DMD and other dystrophinopathies.
4.3. Limitations
Some limitations should be acknowledged. First, the scarcity of studies included in the meta-analyses limits the statistical power and external validity of the results in larger cohorts of patients with DMD. Second, due to the limited number of studies, we were unable to perform publication bias, metaregression, or sensitivity analyses. This could question the association for LTBP4 and LoA. In systematic reviews, it is not uncommon for the first published studies to show stronger associations than subsequent ones, and they are even less likely to be published unless they question previous findings. Third, in some studies, the LTBP4 haplotype was determined using a single SNP (mainly rs10880). Despite the large linkage disequilibrium, it could slightly underestimate the observed result. Fourth, in the Kaplan–Meier survival analysis, we were not always able to consider ethnic differences, the effects of other genetic variants, or a different proportion of glucocorticoid-treated patients, which would potentially modify the observed association. Fifth, participants who are candidates for exon 8 or 44 skipping tend to have slower disease progression [64,65]. Most studies did not consider this possible confounding genotype, and although it is possible that there are no statistically significant differences in the prevalence of these genotypes in the included studies, it cannot be ruled out, considering the ethnic and geographical diversity of the participants, overestimating or underestimating the effect observed for the genetic variants studied. Sixth, other genetic variants, such as LTBP4 rs710160, and their possible differences in prevalence in different populations, might modify the association expected for the LTBP4 haplotype IAAM. Seventh, some confidence intervals of hazard ratios were estimated from the p-value, which might differ from the true confidence interval. Eighth, in general, the authors did not determine the level of fibrosis of the patients’ muscle tissue, which, in theory, should correlate negatively with LoA or cardiac function. This is probably due to the complexity of the procedures to obtain this information.
5. Conclusions
Some of the variability in the DMD phenotype can be explained by the genetic variants directly involved in the TGFβ pathway. Thus, the LTBP4 haplotype IAAM may predict a subsequent LoA, independent of glucocorticoid use, although glucocorticoids may exert an additive effect on this outcome. This haplotype might also be associated with better cardiac function, although more research is needed to confirm or reject this potential association. SPP1 rs28357094 and possibly rs11730582 exert a glucocorticoid-dependent effect, with the rs28357094 genotype G serving as a predictor of a poor response. Further research is needed to establish the possible interaction of LTBP4 rs710160 and THBS1 rs2725797. Moreover, future studies for the LTBP4 haplotype IAAM in non-Caucasian cohorts are required to confirm the findings in this review, as well as the possibility that the association of the LTBP4 haplotype IAAM and LoA is conditioned by the genetic variant rs710160, being relevant in non-Caucasian populations. Knowledge of the effect of the TGFβ pathway on the DMD phenotype provides a possibility of developing new treatments, and genotyping of the genetic variants involved could provide the clinician and the patient with more information on their possible progression or response to certain pharmacological treatments.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ph14080798/s1, Table S1: Excluded studies with reasons, Table S2: Risk of bias assessment, Table S3: Grades of Recommendation, Assessment, Development, and Evaluation of HR of LoA.
Author Contributions
Conceptualization: C.P.-M.; methodology: C.P.-M. and I.C.-R.; data curation and investigation: C.P.-M., I.C.-R.; formal analysis: C.P.-M., A.S.-L., I.S.-D., M.L.-L.-T.; validation and visualization: A.S.-L., I.S.-D. and M.L.-L.-T.; writing—original draft preparation: C.P.-M., I.C.-R., V.M.-V.; writing—review and editing: all authors; supervision: I.C.-R. and V.M.-V.; funding acquisition: V.M.-V.; project administration: V.M.-V. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the European Regional Development Fund. C.P.-M. is supported by a grant from the Universidad de Castilla-La Mancha (2018-CPUCLM-7939).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated and analyzed are available from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
Appendix A.1. Medline, EMBASE, Web of Science, Cochrane Library
(“dystrophy” OR “Duchenne” OR “DMD” OR “Duchenne muscular dystrophy” OR “dystrophinopathy”) AND (“polymorphism” OR “osteopontin” OR “spp1” OR “latent transforming growth factor” OR “ltbp4” OR “thrombospondin” or “thbs1”) AND (“ambulation” OR “loss of ambulation” OR “LoA” OR “cardiomyopathy” OR “ventricular dysfunction” OR “heart failure” OR “cardiac function” OR “heart function”).
Appendix A.2. Grey Literature
Not specified.
Appendix B
Figure A1.
Infographic summary of the manuscript.
References
- Mah, J.K.; Korngut, L.; Dykeman, J.; Day, L.; Pringsheim, T.; Jette, N. A Systematic Review and Meta-Analysis on the Epidemiology of Duchenne and Becker Muscular Dystrophy. Neuromuscul. Disord. 2014, 24, 482–491. [Google Scholar] [CrossRef]
- Holland, A.; Carberry, S.; Ohlendieck, K. Proteomics of the Dystrophin-Glycoprotein Complex and Dystrophinopathy. Curr. Protein Pept. Sci. 2014, 14, 680–697. [Google Scholar] [CrossRef]
- Warner, L.E.; DelloRusso, C.T.; Crawford, R.W.; Rybakova, I.N.; Patel, J.R.; Ervasti, J.M.; Chamberlain, J.S. Expression of Dp260 in Muscle Tethers the Actin Cytoskeleton to the Dystrophin-Glycoprotein Complex and Partially Prevents Dystrophy. Hum. Mol. Genet. 2002, 11, 1095–1105. [Google Scholar] [CrossRef]
- Birnkrant, D.J.; Bushby, K.; Bann, C.M.; Apkon, S.D.; Blackwell, A.; Brumbaugh, D.; Case, L.E.; Clemens, P.R.; Hadjiyannakis, S.; Pandya, S.; et al. Diagnosis and Management of Duchenne Muscular Dystrophy, Part 1: Diagnosis, and Neuromuscular, Rehabilitation, Endocrine, and Gastrointestinal and Nutritional Management. Lancet Neurol. 2018, 17, 251–267. [Google Scholar] [CrossRef] [Green Version]
- Goemans, N. How Glucocorticoids Change Life in Duchenne Muscular Dystrophy. Lancet 2018, 391, 406–407. [Google Scholar] [CrossRef]
- D’Amario, D.; Amodeo, A.; Adorisio, R.; Tiziano, F.D.; Leone, A.M.; Perri, G.; Bruno, P.; Massetti, M.; Ferlini, A.; Pane, M.; et al. A Current Approach to Heart Failure in Duchenne Muscular Dystrophy. Heart 2017, 103, 1770–1779. [Google Scholar] [CrossRef] [PubMed]
- Garg, S. Management of Scoliosis in Patients with Duchenne Muscular Dystrophy and Spinal Muscular Atrophy: A Literature Review. J. Pediatr. Rehabil. Med. 2016, 9, 23–29. [Google Scholar] [CrossRef]
- Bushby, K.; Finkel, R.; Birnkrant, D.J.; Case, L.E.; Clemens, P.R.; Cripe, L.; Kaul, A.; Kinnett, K.; McDonald, C.; Pandya, S.; et al. Diagnosis and Management of Duchenne Muscular Dystrophy, Part 2: Implementation of Multidisciplinary Care. Lancet Neurol. 2010, 9, 177–189. [Google Scholar] [CrossRef]
- Pascual-Morena, C.; Cavero-Redondo, I.; Álvarez-Bueno, C.; Mesas, A.E.; Pozuelo-Carrascosa, D.; Martínez-Vizcaíno, V. Restorative Treatments of Dystrophin Expression in Duchenne Muscular Dystrophy: A Systematic Review. Ann. Clin. Transl. Neurol. 2020, 7, acn3.51149. [Google Scholar] [CrossRef]
- Souchelnytskyi, S.; Rönnstrand, L.; Heldin, C.H.; ten Dijke, P. Phosphorylation of Smad Signaling Proteins by Receptor Serine/Threonine Kinases. Methods Mol. Biol. 2001, 124, 107–120. [Google Scholar] [CrossRef]
- Gardner, S.; Alzhanov, D.; Knollman, P.; Kuninger, D.; Rotwein, P. TGF-β Inhibits Muscle Differentiation by Blocking Autocrine Signaling Pathways Initiated by IGF-II. Mol. Endocrinol. 2011, 25, 128–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobaczewski, M.; Chen, W.; Frangogiannis, N.G. Transforming Growth Factor (TGF)-β Signaling in Cardiac Remodeling. J. Mol. Cell. Cardiol. 2011, 51, 600–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, Y.; Meng, K.; Pu, Y.; Zhang, X. Transforming Growth Factor Beta (TGF-β) Mediates Cardiac Fibrosis and Induces Diabetic Cardiomyopathy. Diabetes Res. Clin. Pract. 2017, 133, 124–130. [Google Scholar] [CrossRef]
- Ismaeel, A.; Kim, J.S.; Kirk, J.S.; Smith, R.S.; Bohannon, W.T.; Koutakis, P. Role of Transforming Growth Factor-β in Skeletal Muscle Fibrosis: A Review. Int. J. Mol. Sci. 2019, 20, 2446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quattrocelli, M.; Capote, J.; Ohiri, J.C.; Warner, J.L.; Vo, A.H.; Earley, J.U.; Hadhazy, M.; Demonbreun, A.R.; Spencer, M.J.; McNally, E.M. Genetic Modifiers of Muscular Dystrophy Act on Sarcolemmal Resealing and Recovery from Injury. PLoS Genet. 2017, 13, e1007070. [Google Scholar] [CrossRef]
- Vo, A.H.; McNally, E.M. Modifier Genes and Their Effect on Duchenne Muscular Dystrophy. Curr. Opin. Neurol. 2015, 28, 528–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vetrone, S.A.; Montecino-Rodriguez, E.; Kudryashova, E.; Kramerova, I.; Hoffman, E.P.; Liu, S.D.; Miceli, M.C.; Spencer, M.J. Osteopontin Promotes Fibrosis in Dystrophic Mouse Muscle by Modulating Immune Cell Subsets and Intramuscular TGF-β. J. Clin. Investig. 2009, 119, 1583–1594. [Google Scholar] [CrossRef] [Green Version]
- Barakat-Haddad, C.; Shin, S.; Candundo, H.; Lieshout, P.; Van Martino, R. A Systematic Review of Risk Factors Associated with Muscular Dystrophies. Neurotoxicology 2017, 61, 55–62. [Google Scholar] [CrossRef]
- Barp, A.; Bello, L.; Politano, L.; Melacini, P.; Calore, C.; Polo, A.; Vianello, S.; Soraru, G.; Semplicini, C.; Pantic, B.; et al. Genetic Modifiers of Duchenne Muscular Dystrophy and Dilated Cardiomyopathy. PLoS ONE 2015, 10, e0141240. [Google Scholar] [CrossRef]
- Bello, L.; Kesari, A.; Gordish-Dressman, H.; Cnaan, A.; Morgenroth, L.P.; Punetha, J.; Duong, T.; Henricson, E.K.; Pegoraro, E.; McDonald, C.M.; et al. Genetic Modifiers of Ambulation in the Cooperative International Neuromuscular Research Group Duchenne Natural History Study. Ann. Neurol. 2015, 77, 684–696. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Wang, L.; Li, Y.; Chen, Y.; Zhang, H.; Zhu, Y.; He, R.; Li, H.; Lin, J.; Zhang, Y.; et al. Genetic Modifiers of Duchenne Muscular Dystrophy in Chinese Patients. Front. Neurol. 2020, 11, 721. [Google Scholar] [CrossRef]
- Flanigan, K.M.; Ceco, E.; Lamar, K.-M.; Kaminoh, Y.; Dunn, D.M.; Mendell, J.R.; King, W.M.; Pestronk, A.; Florence, J.M.; Mathews, K.D.; et al. LTBP4 Genotype Predicts Age of Ambulatory Loss in Duchenne Muscular Dystrophy. Ann. Neurol. 2013, 73, 481–488. [Google Scholar] [CrossRef] [Green Version]
- Pegoraro, E.; Hoffman, E.P.; Piva, L.; Gavassini, B.F.; Cagnin, S.; Ermani, M.; Bello, L.; Soraru, G.; Pacchioni, B.; Bonifati, M.D.; et al. SPP1 Genotype is a Determinant of Disease Severity in Duchenne Muscular Dystrophy. Neurology 2011, 76, 219–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van den Bergen, J.C.; Hiller, M.; Bohringer, S.; Vijfhuizen, L.; Ginjaar, H.B.; Chaouch, A.; Bushby, K.; Straub, V.; Scoto, M.; Cirak, S.; et al. Validation of Genetic Modifiers for Duchenne Muscular Dystrophy: A Multicentre Study Assessing SPP1 and LTBP4 Variants. J. Neurol. Neurosurg. Psychiatry 2015, 86, 1060–1065. [Google Scholar] [CrossRef] [Green Version]
- Van Dorn, C.S.; Puchalski, M.D.; Weng, H.-Y.; Bleyl, S.B.; Butterfield, R.J.; Williams, R.V. DMD Mutation and LTBP4 Haplotype do not Predict Onset of Left Ventricular Dysfunction in Duchenne Muscular Dystrophy. Cardiol. Young 2018, 28, 910–915. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.B.; Vieland, V.J.; Dunn, D.M.; Kaminoh, Y.; Flanigan, K.M. Long-Range Genomic Regulators of THBS1 and LTBP4 Modify Disease Severity in Duchenne Muscular Dystrophy. Ann. Neurol. 2018, 84, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Stroup, D.F.; Berlin, J.A.; Morton, S.C.; Olkin, I.; Williamson, G.D.; Rennie, D.; Moher, D.; Becker, B.J.; Sipe, T.A.; Thacker, S.B. Meta-Analysis of Observational Studies in Epidemiology: A Proposal for Reporting. J. Am. Med. Assoc. 2000, 283, 2008–2012. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.; Green, S. Cochrane Handbook for Systematic Reviews of Interventions: Cochrane Book Series; John Wiley and Sons: Hoboken, NJ, USA, 2008; ISBN 9780470699515. [Google Scholar]
- Study Quality Assessment Tools|NHLBI, NIH. Available online: https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools (accessed on 15 April 2021).
- Neumann, I.; Pantoja, T.; Peñaloza, B.; Cifuentes, L.; Rada, G. El sistema GRADE: Un Cambio en la Forma de Evaluar la Calidad de la Evidencia y la Fuerza de Recomendaciones. Rev. Med. Chil. 2014, 142, 630–635. [Google Scholar] [CrossRef] [Green Version]
- Tufanaru, C.; Munn, Z.; Stephenson, M.; Aromataris, E. Fixed or Random Effects Meta-Analysis? Common Methodological Issues in Systematic Reviews of Effectiveness. Int. J. Evid. Based Healthc. 2015, 13, 196–207. [Google Scholar] [CrossRef] [Green Version]
- Altman, D.G.; Bland, J.M. How to Obtain the Confidence Interval from a P Value. BMJ 2011, 343, d2090. [Google Scholar] [CrossRef] [Green Version]
- Higgins, J.P.T.; Thompson, S.G. Quantifying Heterogeneity in a Meta-Analysis. Stat. Med. 2002, 21, 1539–1558. [Google Scholar] [CrossRef]
- Bello, L.; Piva, L.; Barp, A.; Taglia, A.; Picillo, E.; Vasco, G.; Pane, M.; Previtali, S.C.; Torrente, Y.; Gazzerro, E.; et al. Importance of SPP1 Genotype as a Covariate in Clinical Trials in Duchenne Muscular Dystrophy. Neurology 2012, 79, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Bello, L.; Flanigan, K.M.; Weiss, R.B.; Spitali, P.; Aartsma-Rus, A.; Muntoni, F.; Zaharieva, I.; Ferlini, A.; Mercuri, E.; Tuffery-Giraud, S.; et al. Association Study of Exon Variants in the NF-kappaB and TGFbeta Pathways Identifies CD40 as a Modifier of Duchenne Muscular Dystrophy. Am. J. Hum. Genet. 2016, 99, 1163–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagai, M.; Awano, H.; Yamamoto, T.; Bo, R.; Matsuo, M.; Iijima, K. The ACTN3 577XX Null Genotype Is Associated with Low Left Ventricular Dilation-Free Survival Rate in Patients with Duchenne Muscular Dystrophy. J. Card. Fail. 2020, 26, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Bonifati, D.M.; Witchel, S.F.; Ermani, M.; Hoffman, E.P.; Angelini, C.; Pegoraro, E. The Glucocorticoid Receptor N363S Polymorphism and Steroid Response in Duchenne Dystrophy. J. Neurol. Neurosurg. Psychiatry 2006, 77, 1177–1179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bello, L.; Pegoraro, E. The “Usual Suspects”: Genes for Inflammation, Fibrosis, Regeneration, and Muscle Strength Modify Duchenne Muscular Dystrophy. J. Clin. Med. 2019, 8, 649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, S.E.; Stellmach, V.; Murphy-Ullrich, J.E.; Ribeiro, S.M.F.; Lawler, J.; Hynes, R.O.; Boivin, G.P.; Bouck, N. Thrombospondin-1 is a Major Activator of TGF-β1 In Vivo. Cell 1998, 93, 1159–1170. [Google Scholar] [CrossRef] [Green Version]
- Lawler, P.R.; Lawler, J. Molecular Basis for the Regulation of Angiogenesis by Thrombospondin-1 and -2. Cold Spring Harb. Perspect. Med. 2012, 2, a006627. [Google Scholar] [CrossRef]
- Miyazaki, D.; Nakamura, A.; Fukushima, K.; Yoshida, K.; Takeda, S.; Ikeda, S.I. Matrix Metalloproteinase-2 Ablation in Dystrophin-Deficient mdx Muscles Reduces Angiogenesis Resulting in Impaired Growth of Regenerated Muscle Fibers. Hum. Mol. Genet. 2011, 20, 1787–1799. [Google Scholar] [CrossRef] [Green Version]
- Capote, J.; Kramerova, I.; Martinez, L.; Vetrone, S.; Barton, E.R.; Sweeney, H.L.; Miceli, M.C.; Spencer, M.J. Osteopontin Ablation Ameliorates Muscular Dystrophy by Shifting Macrophages to a Proregenerative Phenotype. J. Cell Biol. 2016, 213, 275–288. [Google Scholar] [CrossRef] [Green Version]
- Giacopelli, F.; Marciano, R.; Pistorio, A.; Catarsi, P.; Canini, S.; Karsenty, G.; Ravazzolo, R. Polymorphisms in the Osteopontin Promoter Affect Its Transcriptional Activity. Physiol. Genom. 2005, 20, 87–96. [Google Scholar] [CrossRef] [Green Version]
- Barfield, W.L.; Uaesoontrachoon, K.; Wu, C.S.; Lin, S.; Chen, Y.; Wang, P.C.; Kanaan, Y.; Bond, V.; Hoffman, E.P. Eccentric Muscle Challenge Shows Osteopontin Polymorphism Modulation of Muscle Damage. Hum. Mol. Genet. 2014, 23, 4043–4050. [Google Scholar] [CrossRef] [Green Version]
- Schultz, J.; Lorenz, P.; Ibrahim, S.M.; Kundt, G.; Gross, G.; Kunz, M. The Functional -443T/C Osteopontin Promoter Polymorphism Influences Osteopontin Gene Expression in Melanoma Cells via Binding of c-Myb Transcription Factor. Mol. Carcinog. 2009, 48, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Renault, M.A.; Robbesyn, F.; Réant, P.; Douin, V.; Daret, D.; Allières, C.; Belloc, I.; Couffinhal, T.; Arnal, J.F.; Klingel, K.; et al. Osteopontin Expression in Cardiomyocytes Induces Dilated Cardiomyopathy. Circ. Heart Fail. 2010, 3, 431–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acuña, M.J.; Pessina, P.; Olguin, H.; Cabrera, D.; Vio, C.P.; Bader, M.; Muñoz-canoves, P.; Santos, R.A.; Cabello-verrugio, C.; Brandan, E. Restoration of Muscle Strength in Dystrophic Muscle by Angiotensin-1-7 through Inhibition of TGF-β Signalling. Hum. Mol. Genet. 2014, 23, 1237–1249. [Google Scholar] [CrossRef] [Green Version]
- Pines, M.; Halevy, O. Halofuginone and Muscular Dystrophy. Histol. Histopathol. 2011, 26, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Andreetta, F.; Bernasconi, P.; Baggi, F.; Ferro, P.; Oliva, L.; Arnoldi, E.; Cornelio, F.; Mantegazza, R.; Confalonieri, P. Immunomodulation of TGF-Beta1 in mdx Mouse Inhibits Connective Tissue Proliferation in Diaphragm but Increases Inflammatory Response: Implications for Antifibrotic Therapy. J. Neuroimmunol. 2006, 175, 77–86. [Google Scholar] [CrossRef]
- Lee, E.M.; Kim, D.Y.; Kim, A.Y.; Lee, E.J.; Kim, S.H.; Lee, M.M.; Sung, S.E.; Park, J.K.; Jeong, K.S. Chronic Effects of Losartan on the Muscles and the Serologic Profiles of mdx Mice. Life Sci. 2015, 143, 35–42. [Google Scholar] [CrossRef]
- Micheletto, M.L.J.; Hermes, T.A.; Bertassoli, B.M.; Petri, G.; Perez, M.M.; Fonseca, F.L.A.; Carvalho, A.A.S.; Feder, D. Ixazomib an Oral Proteasome Inhibitor, Exhibits Potential Effect in Dystrophin-Deficient mdx Mice. Int. J. Exp. Pathol. 2021, 102, 11–21. [Google Scholar] [CrossRef]
- Kramerova, I.; Marinov, M.; Owens, J.; Lee, S.J.; Becerra, D.; Spencer, M.J. Myostatin Inhibition Promotes Fast Fibre Hypertrophy but Causes Loss of AMP-Activated Protein Kinase Signalling and Poor Exercise Tolerance in a Model of Limb-Girdle Muscular Dystrophy R1/2A. J. Physiol. 2020, 598, 3927–3939. [Google Scholar] [CrossRef]
- St. Andre, M.; Johnson, M.; Bansal, P.N.; Wellen, J.; Robertson, A.; Opsahl, A.; Burch, P.M.; Bialek, P.; Morris, C.; Owens, J. A Mouse Anti-Myostatin Antibody Increases Muscle Mass and Improves Muscle Strength and Contractility in the mdx Mouse Model of Duchenne Muscular Dystrophy and its Humanized Equivalent, Domagrozumab (PF-06252616), Increases Muscle Volume in Cynomolgus Monkeys. Skelet. Muscle 2017, 7, 25. [Google Scholar] [CrossRef]
- Campbell, C.; McMillan, H.J.; Mah, J.K.; Tarnopolsky, M.; Selby, K.; McClure, T.; Wilson, D.M.; Sherman, M.L.; Escolar, D.; Attie, K.M. Myostatin Inhibitor ACE-031 Treatment of Ambulatory Boys with Duchenne Muscular Dystrophy: Results of a Randomized, Placebo-Controlled Clinical Trial. Muscle Nerve 2017, 55, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.; Asari, T.; Saitoh, M.; Nirasawa, K.; Sasaki, E.; Roppongi, Y.; Nakamura, A.; Saga, Y.; Shimada, T.; Ikeyama, H.; et al. Chain-Shortened Myostatin Inhibitory Peptides Improve Grip Strength in Mice. ACS Med. Chem. Lett. 2019, 10, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Iskenderian, A.; Liu, N.; Deng, Q.; Huang, Y.; Shen, C.; Palmieri, K.; Crooker, R.; Lundberg, D.; Kastrapeli, N.; Pescatore, B.; et al. Myostatin and Activin Blockade by Engineered Follistatin Results in Hypertrophy and Improves Dystrophic Pathology in mdx Mouse more than Myostatin Blockade Alone. Skelet. Muscle 2018, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Qiao, C.; Li, J.; Xiao, B.; Li, J.; Xiao, X. A GDF11/Myostatin Inhibitor, GDF11 Propeptide-Fc, Increases Skeletal Muscle Mass and Improves Muscle Strength in Dystrophic mdx Mice. Skelet. Muscle 2019, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Rybalka, E.; Timpani, C.A.; Debruin, D.A.; Bagaric, R.M.; Campelj, D.G.; Hayes, A. The Failed Clinical Story of Myostatin Inhibitors against Duchenne Muscular Dystrophy: Exploring the Biology behind the Battle. Cells 2020, 9, 2657. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhao, R.; Yu, T.; Li, J.; Zhang, M.; Jiang, S.; Wang, L.; Zhang, G.; Li, R.; Zhu, B.; et al. Sudden Cardiac Death of Duchenne Muscular Dystrophy with NT-proBNP in Pericardial Fluid as a Useful Biomarker for Diagnosis of the Cause of Death: A Case Report. Forensic Sci. Res. 2020, 5, 165. [Google Scholar] [CrossRef]
- Nassoro, D.D.; Torres, L.; Marando, R.; Mboma, L.; Mushi, S.; Mwakyula, I.H. A Child with Duchenne Muscular Dystrophy: A Case Report of a Rare Diagnosis among Africans. Clin. Case Rep. 2020, 8, 2654–2660. [Google Scholar] [CrossRef]
- Wakefield, S.E.; Dimberg, E.L.; Moore, S.A.; Tseng, B.S. Dystrophinopathy Presenting with Arrhythmia in an Asymptomatic 34-Year-Old Man: A Case Report. J. Med. Case Rep. 2009, 3, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Cheang, I.F.; Li Li, X. Cardiac Injury from Asymptomatic Duchenne Muscular Dystrophy. J. Am. Coll. Cardiol. 2019, 73, 2190. [Google Scholar] [CrossRef]
- Navarro, G.C.; Poutvinski, V.; Alvarado, K.R.; Alvarado, F.; Padilla, C.J.I.; Rafael Calderón Guardia, H.A.; José, S.; Rica, C. Compromiso Cardiaco en Distrofias Musculares: A Propósito de un Caso. Rev. Costarric. Cardiol. 2020, 22, 35–40. Available online: http://www.scielo.sa.cr/scielo.php?script=sci_arttext&pid=S1409-41422020000100035&lng=en&nrm=iso> (accessed on 6 August 2021).
- Brogna, C.; Coratti, G.; Pane, M.; Ricotti, V.; Messina, S.; D’Amico, A.; Bruno, C.; Vita, G.; Berardinelli, A.; Mazzone, E.; et al. Long-Term Natural History Data in Duchenne Muscular Dystrophy Ambulant Patients with Mutations Amenable to Skip Exons 44, 45, 51 and 53. PLoS ONE 2019, 14, e218683. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.T.; Barthelemy, F.; Martin, A.S.; Douine, E.D.; Eskin, A.; Lucas, A.; Lavigne, J.; Peay, H.; Khanlou, N.; Sweeney, L.; et al. DMD Genotype Correlations from the Duchenne Registry: Endogenous Exon Skipping is a Factor in Prolonged Ambulation for Individuals with a Defined Mutation Subtype. Hum. Mutat. 2018, 39, 1193–1202. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).