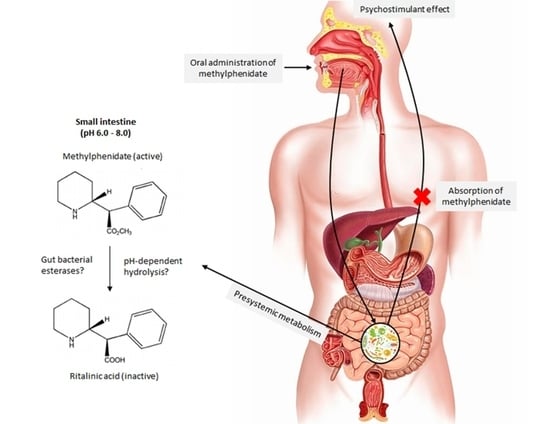
Stability of Methylphenidate under Various pH Conditions in the Presence or Absence of Gut Microbiota
Abstract
:1. Introduction
2. Results
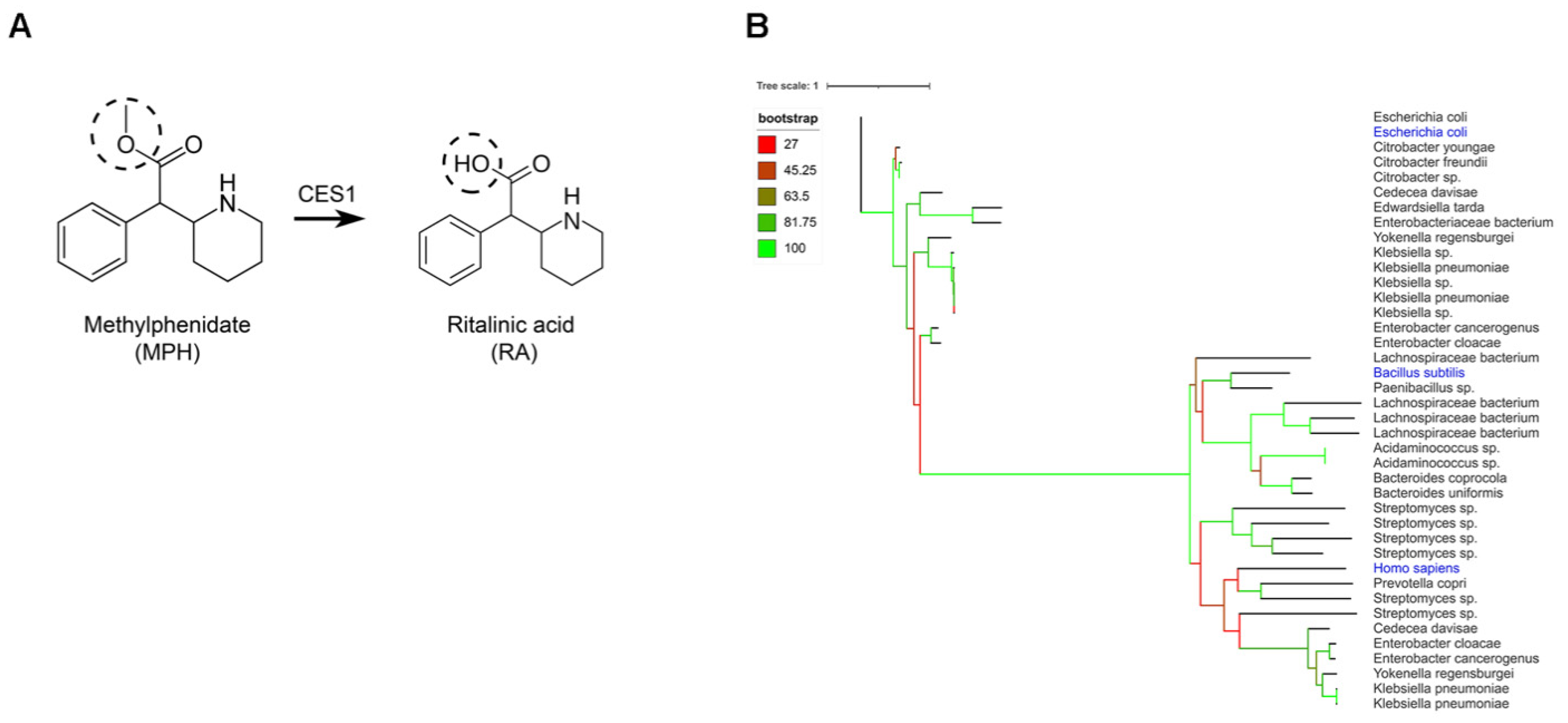
2.1. Gut Bacteria Harbor Homologues for the Human CES1 Enzyme Responsible for the Metabolization of Methylphenidate
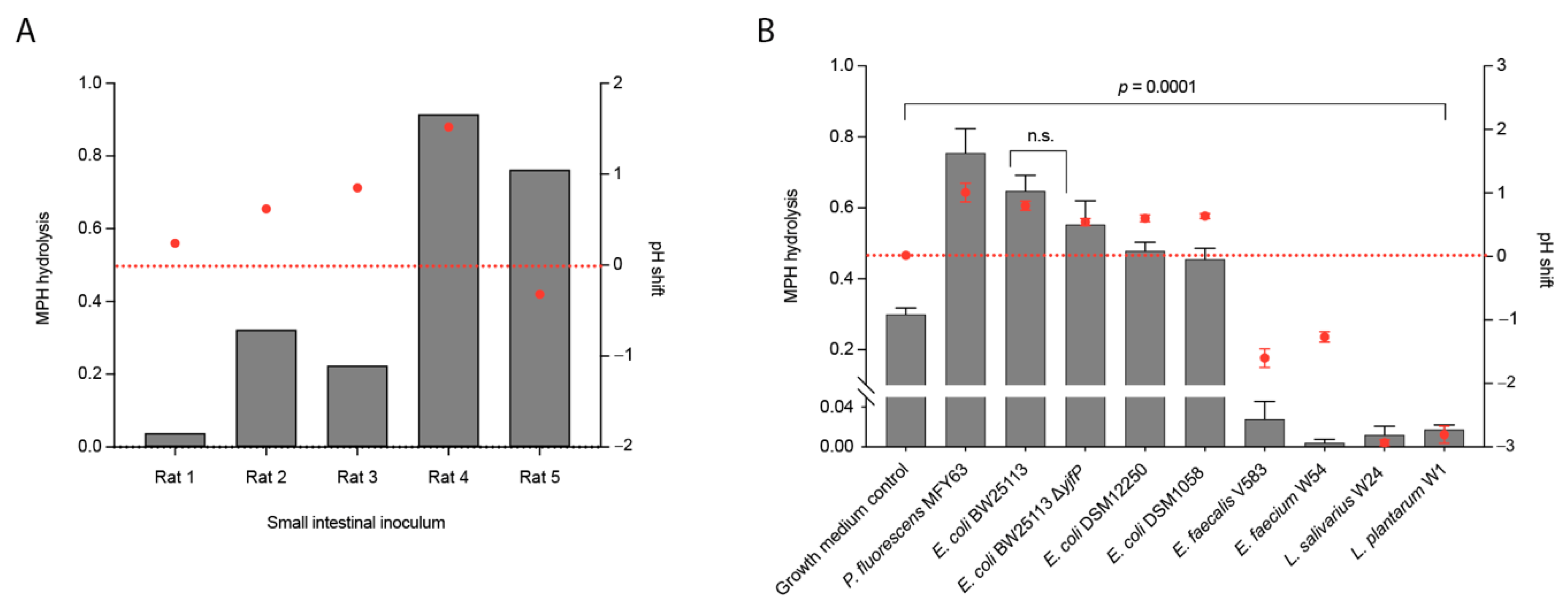
2.2. Conversion of Methylphenidate into Ritalinic Acid in Complex and Pure Bacterial Culture
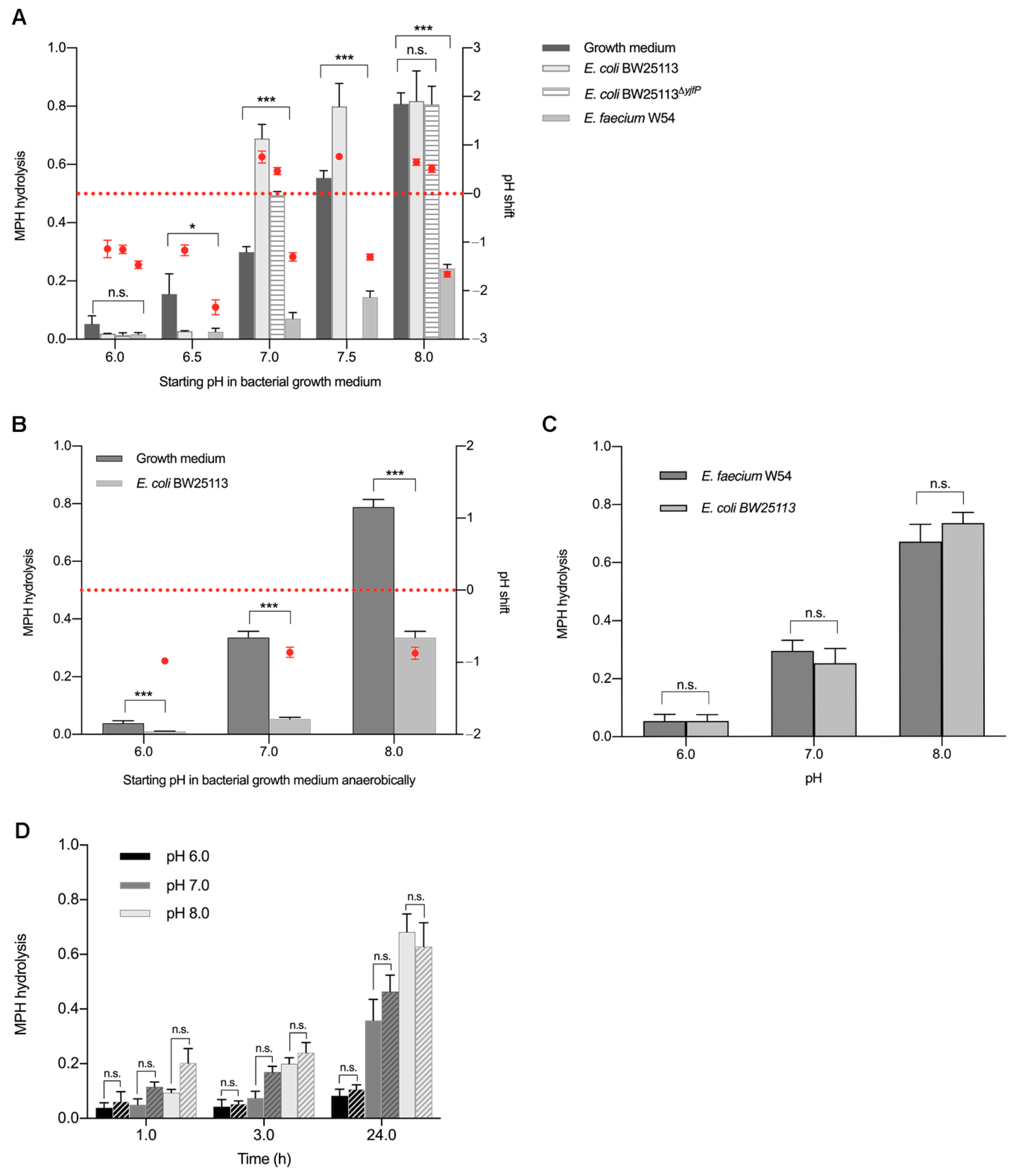
2.3. pH Shifts during Bacterial Growth Cause Hydrolysis of Methylphenidate in Bacterial Pure Cultures (An)aerobically
3. Discussion
4. Material and Methods
4.1. Bioinformatics
4.2. Rat Luminal Content
4.3. Pure Bacterial Cultures
4.4. Amplification of yjfP Esterase by PCR
4.5. HPLC-MS/MS Sample Preparation and Analysis
4.6. Calibration Standards and Biological Matrices
4.7. Cell Lysate Preparation and Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Faraone, S.V.; Sergeant, J.; Gillberg, C.; Biederman, J. The Worldwide Prevalence of ADHD: Is It an American Condition? World Psychiatry 2003, 2, 104–113. Available online: http://www.ncbi.nlm.nih.gov/pubmed/16946911 (accessed on 27 July 2021). [PubMed]
- Del Campo, N.; Chamberlain, S.R.; Sahakian, B.J.; Robbins, T.W. The roles of dopamine and nor-adrenaline in the pathophysiology and treatment of attention-deficit/hyperactivity disorder. Biol. Psychiatry 2011, 69, e145–e157. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.S.; Baek, D.J.; Baek, S.S. Effect of exercise on hyperactivity, impulsivity and dopamine D2 receptor expression in the substantia nigra and striatum of spontaneous hypertensive rats. J. Exerc. Nutr. Biochem. 2014, 18, 379–384. [Google Scholar] [CrossRef]
- Volkow, N.D.; Fowler, J.S.; Wang, G.J.; Ding, Y.S.; Gatley, S.J. Mechanism of action of methylphenidate: Insights from PET imaging studies. J. Atten. Disord. 2002, 6, S31–S43. [Google Scholar] [CrossRef] [PubMed]
- Castells, X.; Ramos-Quiroga, J.A.; Rigau, D.; Bosch, R.; Nogueira, M.; Vidal, X.; Casas, M. Efficacy of Methylphenidate for Adults with Attention-Deficit Hyperactivity Disorder: A Meta-Regression Analysis. CNS Drugs 2011, 25, 157–169. Available online: http://www.ncbi.nlm.nih.gov/pubmed/21254791 (accessed on 27 July 2021). [CrossRef]
- Kimko, H.C.; Cross, J.T.; Abernethy, D.R. Pharmacokinetics and Clinical Effectiveness of Methylphenidate. Clin. Pharmacokinet. 1999, 37, 457–470. Available online: http://link.springer.com/10.2165/00003088-199937060-00002 (accessed on 27 July 2021). [CrossRef]
- Ermer, J.C.; Adeyi, B.A.; Pucci, M.L. Pharmacokinetic Variability of Long-Acting Stimulants in the Treatment of Children and Adults with Attention-Deficit Hyperactivity Disorder. CNS Drugs 2010, 24, 1009–1025. Available online: http://link.springer.com/10.2165/11539410-000000000-00000 (accessed on 27 July 2021). [CrossRef]
- Maldonado, R. Comparison of the pharmacokinetics and clinical efficacy of new extended-release formulations of methylphenidate. Expert Opin. Drug Metab. Toxicol. 2013, 9, 1001–1014. [Google Scholar] [CrossRef]
- Zhu, H.J.; Patrick, K.S.; Yuan, H.J.; Wang, J.S.; Donovan, J.L.; DeVane, C.L.; Malcolm, R.; Johnson, J.A.; Youngblood, G.L.; Sweet, D.H.; et al. Two CES1 Gene Mutations Lead to Dysfunctional Carboxylesterase 1 Activity in Man: Clinical Significance and Molecular Basis. Am. J. Hum. Genet. 2008, 82, 1241–1248. [Google Scholar] [CrossRef] [Green Version]
- Stage, C.; Dalhoff, K.; Rasmussen, H.B.; Schow Guski, L.; Thomsen, R.; Bjerre, D.; Ferrero-Miliani, L.; Madsen, M.B.; Jürgens, G. The im-pact of human CES1 genetic variation on enzyme activity assessed by ritalinic acid/methylphenidate ratios. Basic Clin. Pharmacol. Toxicol. 2019, 125, 54–61. [Google Scholar] [PubMed]
- Stage, C.; Jürgens, G.; Guski, L.S.; Thomsen, R.; Bjerre, D.; Ferrero-Miliani, L.; Lyauk, Y.K.; Rasmussen, H.B.; Dalhoff, K.; INDICES Consortium. The impact of CES1 genotypes on the pharmacokinetics of methylphenidate in healthy Danish subjects. Br. J. Clin. Pharmacol. 2017, 83, 1506–1514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Duan, J.; Fisher, J. Application of physiologically based absorption modeling to characterize the pharmacokinetic profiles of oral extended release methylphenidate products in adults. PLoS ONE 2016, 11, e0164641. [Google Scholar] [CrossRef] [PubMed]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramya, S.L.; Venkatesan, T.; Srinivasa Murthy, K.; Jalali, S.K.; Verghese, A. Environmental Microbiology Detection of carboxylesterase and esterase activity in culturable gut bacterial flora isolated from diamondback moth, Plutella xylostella (Linnaeus), from India and its possible role in indoxacarb degradation. Braz. J. Microbiol. 2016, 47, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Johns, N.; Wrench, A.; Loto, F.; Valladares, R.; Lorca, G.; Gonzalez, C.F. The Escherichia coli yjfP Gene Encodes a Carboxylesterase Involved in Sugar Utilization during Diauxie. J. Mol. Micro-Biol. Biotechnol. 2015, 25, 412–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zock, J.; Cantwell, C.; Swartling, J.; Hodges, R.; Pohl, T.; Sutton, K.; Rosteck, P., Jr.; McGilvray, D.; Queener, S. The Bacillus subtilis pnbA gene encoding p-nitrobenzyl esterase: Cloning, sequence and high-level expression in Escherichia coli. Gene 1994, 151, 37–43. [Google Scholar] [CrossRef]
- Nugent, S.G.; Kumar, D.; Rampton, D.S.; Evans, D.F. Intestinal luminal pH in inflammatory bowel disease: Possible determinants and implications for therapy with aminosalicylates and other drugs. Gut 2001, 48, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Abuhelwa, A.Y.; Williams, D.B.; Upton, R.N.; Foster, D.J.R. Food, gastrointestinal pH, and models of oral drug absorption. Eur. J. Pharm. Biopharm. 2017, 112, 234–248. [Google Scholar] [CrossRef]
- Kohl, K.D.; Stengel, A.; Samuni-Blank, M.; Dearing, M.D. Effects of Anatomy and Diet on Gastro-intestinal pH in Rodents. J. Exp. Zool. Part A Ecol. Genet. Physiol. 2013, 319, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Lyauk, Y.K.; Stage, C.; Bergmann, T.K.; Ferrero-Milliani, L.; Bjerre, D.; Thomsen, R.; Dalhoff, K.P.; Rasmussen, H.B.; Jürgens, G. Population Pharmacokinetics of Methylphenidate in Healthy Adults Emphasizing Novel and Known Effects of Several Carboxylesterase 1 (CES1) Variants. Clin. Transl. Sci. 2016, 9, 337–345. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5351003/pdf/CTS-9-337.pdf (accessed on 27 July 2021). [CrossRef] [PubMed] [Green Version]
- Xie, Y.; Hu, F.; Xiang, D.; Lu, H.; Li, W.; Zhao, A.; Huang, L.; Wang, R. The metabolic effect of gut microbiota on drugs. Drug Metab. Rev. 2020, 52, 139–156. [Google Scholar] [CrossRef]
- Da Silva, P.L.; Guimarães, L.; Pliego, J.R. Revisiting the mechanism of neutral hydrolysis of esters: Water autoionization mechanisms with acid or base initiation pathways. J. Phys. Chem. B 2013, 117, 6487–6497. [Google Scholar] [CrossRef] [PubMed]
- Conway, T.; Krogfelt, K.A.; Cohen, P.S. The Life of Commensal Escherichia coli in the Mammalian Intestine. EcoSal Plus 2004, 1. [Google Scholar] [CrossRef]
- Dubin, K.; Pamer, E.G. Enterococci and their interactions with the intestinal microbiome. Microbiol. Spectr. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Auchtung, J.M.; Robinson, C.D.; Britton, R.A. Cultivation of stable, reproducible microbial com-munities from different fecal donors using minibioreactor arrays (MBRAs). Microbiome 2015, 3, 42. Available online: http://www.microbiomejournal.com/content/3/1/42 (accessed on 27 July 2021). [CrossRef] [PubMed] [Green Version]
- Maier, L.; Pruteanu, M.; Kuhn, M.; Zeller, G.; Telzerow, A.; Anderson, E.E.; Brochado, A.R.; Fernandez, K.C.; Dose, H.; Mori, H.; et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018, 555, 623–628. [Google Scholar] [CrossRef]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A.L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019, 570, 462–467. Available online: http://www.nature.com/articles/s41586-019-1291-3 (accessed on 27 July 2021). [CrossRef]
- Padmanabhan, G.R. Methylphenidate Hydrochloride. Anal. Profiles Drug Subst. 1981, 10, 473–497. Available online: https://www.sciencedirect.com/science/article/pii/S0099542808606486 (accessed on 27 July 2021).
- B.V. Novartis Pharma. Public Assessment Report Ritalin LA Modified-Release Capsules; B.V. Novartis Pharma: Basel, Switzerland, 2016. [Google Scholar]
- Roychowdhury, K.K.; Subramanian, S. Drug-Excipient Interaction of Methylphenidate with Glycerin in Methylphenidate Oral Solution and Identification of its Transesterification Products by UPLC-MS/MS. Am. J. Anal. Chem. 2012, 7, 151–164. [Google Scholar] [CrossRef] [Green Version]
- Naveen, K.C.; Kannappan, N. A stability indicating method development and validation for determination of Methylphenidate Hydrochloride and its impurities in solid pharmaceutical oral dosage form by RP-HPLC as per ICH guidelines. J. Chem. Pharm. Res. 2015, 7, 606–629. Available online: www.jocpr.com (accessed on 27 July 2021).
- McCallum, E.S.; Lindberg, R.H.; Andersson, P.L.; Brodin, T. Stability and uptake of methylphenidate and ritalinic acid in nine-spine stickleback (Pungitius pungitius) and water louse (Asellus aquaticus). Environ. Sci. Pollut. Res. 2019, 26, 9371–9378. [Google Scholar] [CrossRef] [Green Version]
- Srinivas, N.R.; Hubbard, J.W.; McKay, G.; Hawes, E.M.; Midha, K.K. In vitro hydrolysis of RR,SS-threo-methylphenidate by blood esterases; differential and enantioselective interspecies variability. Chirality 1991, 3, 99–103. [Google Scholar] [CrossRef]
- Koziolek, M.; Grimm, M.; Becker, D.; Iordanov, V.; Zou, H.; Shimizu, J.; Wanke, C.; Garbacz, G.; Weitschies, W. Investigation of pH and Temperature Profiles in the GI Tract of Fasted Human Subjects Using the Intellicap® System. J. Pharm. Sci. 2015, 104, 2855–2863. [Google Scholar] [CrossRef] [PubMed]
- Midha, K.K.; McKay, G.; Rawson, M.J.; Korchinski, E.D.; Hubbard, J.W. Effects of food on the pharmacokinetics of methylphenidate. Pharm. Res. 2001, 18, 1185–1189. Available online: http://www.ncbi.nlm.nih.gov/pubmed/11587491 (accessed on 27 July 2021). [CrossRef] [PubMed]
- Weisler, R.H.; Stark, J.G.; Sikes, C. Fed and Fasted Administration of a Novel Extended-Release Methylphenidate Orally Disintegrating Tablet Formulation for the Treatment of ADHD. Clin. Pharmacol. Drug Dev. 2018, 7, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Zoetendal, E.G.; Raes, J.; van den Bogert, B.; Arumugam, M.; Booijink, C.C.G.M.; Troost, F.J.; Bork, P.; Wels, M.; de Vos, W.M.; Kleerebezem, M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbo-hydrates. ISME J. 2012, 6, 1415–1426. [Google Scholar] [CrossRef]
- Oliphant, K.; Allen-Vercoe, E. Macronutrient metabolism by the human gut microbiome: Major fermentation by-products and their impact on host health. Microbiome 2019, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Rios-Morales, M.; van Trijp, M.P.H.; Rösch, C.; An, R.; Boer, T.; Gerding, A.; de Ruiter, N.; Koehorst, M.; Heiner-Fokkema, M.R.; Schols, H.A.; et al. A toolbox for the comprehensive analysis of small volume human intestinal samples that can be used with gastrointestinal sampling capsules. Sci. Rep. 2021, 11, 8133. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [Green Version]
- Sahm, D.F.; Kissinger, J.; Gilmore, M.S.; Murray, P.R.; Mulder, R.; Solliday, J.; Clarke, B. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 1989, 33, 1588–1591. [Google Scholar] [CrossRef] [Green Version]
| Pure Cultures | Starting pH | pH after 24 h | S.D. | Complex Cultures | Starting pH | pH after 24 h | S.D. |
|---|---|---|---|---|---|---|---|
| Growth medium | 7.00 | 7.02 | 0.01 | Rat 1 | 7.00 | 7.24 | N.A. |
| P. fluorescens MFY63 | 7.00 | 8.01 | 0.15 | Rat 2 | 7.00 | 7.62 | N.A. |
| E. coli BW25113 | 7.00 | 7.80 | 0.07 | Rat 3 | 7.00 | 7.85 | N.A. |
| E. coli BW25113ΔyjfP | 7.00 | 7.54 | 0.05 | Rat 4 | 7.00 | 8.52 | N.A. |
| E. coli DSM12250 | 7.00 | 7.60 | 0.05 | Rat 5 | 7.00 | 6.68 | N.A. |
| E. coli DSM1058 | 7.00 | 7.64 | 0.04 | ||||
| E. faecalis V583 | 7.00 | 5.40 | 0.14 | ||||
| E. faecium W54 | 7.00 | 5.73 | 0.08 | ||||
| L. salivarius W24 | 7.00 | 4.07 | 0.05 | ||||
| L. plantarum W1 | 7.00 | 4.19 | 0.13 |
| Starting pH | E. coli BW25113 | E. faecium W54 | E. coli BW25113ΔyjfP | E. coli BW25113 (anaerobic) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH 24 h | S.D. | p | pH 24 h | S.D. | p | pH 24 h | S.D. | p | pH 24 h | S.D. | p | |
| 6.0 | 4.86 | 0.18 | 0.1098 | 4.20 | 0.43 | 0.1013 | 4.85 | 0.08 | 0.1797 | 5.02 | 0.12 | 0.0066 |
| 6.5 | 5.33 | 0.11 | 0.0339 | 4.30 | 0.26 | 0.0339 | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. |
| 7.0 | 7.63 | 0.07 | 0.0002 | 5.70 | 0.08 | 0.0001 | 7.46 | 0.07 | 0.0001 | 6.14 | 0.07 | 0.0003 |
| 7.5 | 8.26 | 0.06 | 0.0066 | 6.22 | 0.11 | 0.00002 | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. |
| 8.0 | 8.64 | 0.06 | 0.8851 | 6.40 | 0.16 | 0.00001 | 8.51 | 0.07 | 0.9774 | 7.13 | 0.08 | 0.0002 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aresti-Sanz, J.; Schwalbe, M.; Pereira, R.R.; Permentier, H.; El Aidy, S. Stability of Methylphenidate under Various pH Conditions in the Presence or Absence of Gut Microbiota. Pharmaceuticals 2021, 14, 733. https://doi.org/10.3390/ph14080733
Aresti-Sanz J, Schwalbe M, Pereira RR, Permentier H, El Aidy S. Stability of Methylphenidate under Various pH Conditions in the Presence or Absence of Gut Microbiota. Pharmaceuticals. 2021; 14(8):733. https://doi.org/10.3390/ph14080733
Chicago/Turabian StyleAresti-Sanz, Julia, Markus Schwalbe, Rob Rodrigues Pereira, Hjalmar Permentier, and Sahar El Aidy. 2021. "Stability of Methylphenidate under Various pH Conditions in the Presence or Absence of Gut Microbiota" Pharmaceuticals 14, no. 8: 733. https://doi.org/10.3390/ph14080733
APA StyleAresti-Sanz, J., Schwalbe, M., Pereira, R. R., Permentier, H., & El Aidy, S. (2021). Stability of Methylphenidate under Various pH Conditions in the Presence or Absence of Gut Microbiota. Pharmaceuticals, 14(8), 733. https://doi.org/10.3390/ph14080733