Anti-Cancer Effects of an Optimised Combination of Ginsenoside Rg3 Epimers on Triple Negative Breast Cancer Models
Abstract
1. Introduction
2. Results
2.1. Response Surface Methodology Modelling
2.2. Rg3 Inhibits Migration but Not Proliferation in TNBC Cell Lines
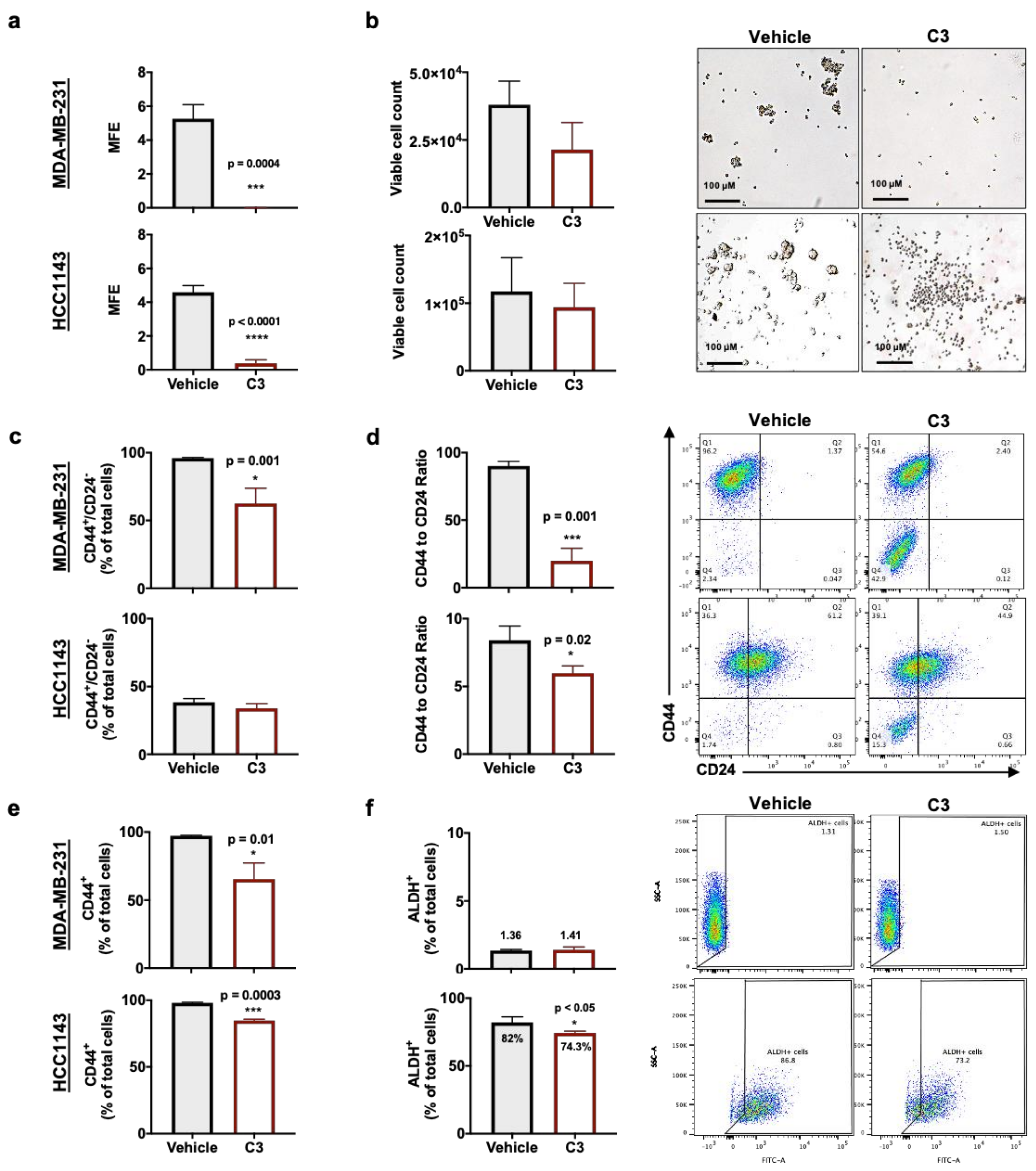
2.3. Rg3 Decreases Mammosphere Formation Efficiency (MFE) in TNBC 3D Models, via Decreasing ‘Stemness’ of the Cells
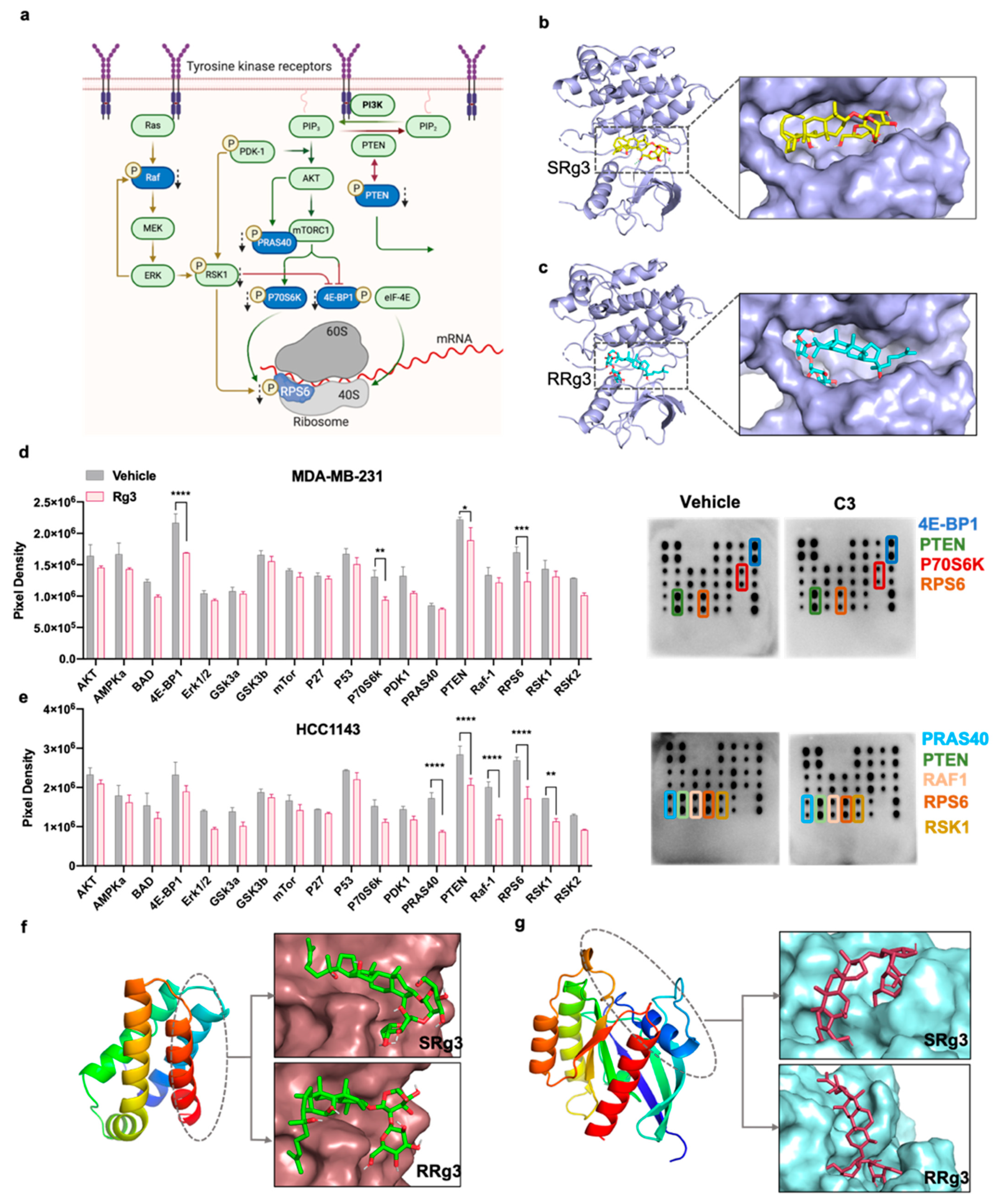
2.4. Effect of Rg3 on Akt/mTOR Signalling
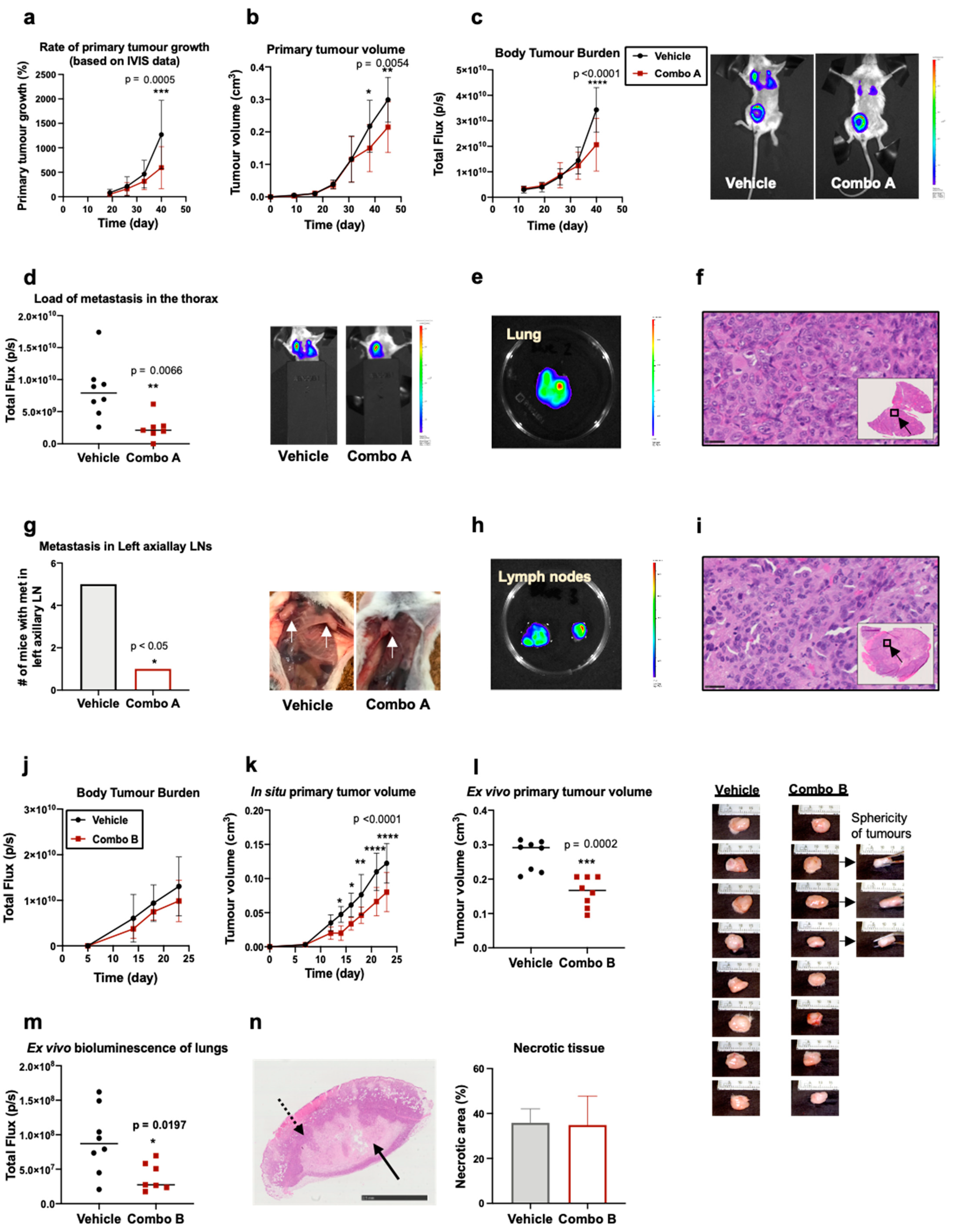
2.5. In Vivo Evaluation of the Efficacy of Rg3 Combo
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture of Adherent Cells
4.3. Circular Scratch Migration Assay
4.4. Response Surface Methodology (RSM)
4.5. Transwell Migration Assay
4.6. Proliferation Assay
4.7. Culture of Mammospheres
4.8. Mammosphere Formation Efficiency (MFE)
4.9. Cell Viability Analysis
4.10. Expression of Stem Cell Markers
4.11. AKT Pathway Phosphorylation Array
4.12. In Silico Molecular Docking
4.13. Developing the Mouse Model of mTNBC
4.14. Drug Administration and Toxicity Assessment
4.15. IVIS Imaging and Tumour Size Measurement
4.16. H&E Staining and Proliferative Area Measurement
4.17. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into molecular classifications of triple-negative breast cancer: Improving patient selection for treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef] [PubMed]
- Biswas, T.; Efird, J.T.; Prasad, S.; Jindal, C.; Walker, P.R. The survival benefit of neoadjuvant chemotherapy and pCR among patients with advanced stage triple negative breast cancer. Oncotarget 2017, 8, 112712–112719. [Google Scholar] [CrossRef]
- Groza, I.-M.; Braicu, C.; Jurj, A.; Zanoaga, O.; Lajos, R.; Chiroi, P.; Cojocneanu, R.; Paun, D.; Irimie, A.; Korban, S.S.; et al. Cancer-Associated Stemness and Epithelial-to-Mesenchymal Transition Signatures Related to Breast Invasive Carcinoma Prognostic. Cancers 2020, 12, 3053. [Google Scholar] [CrossRef]
- Li, W.; Ma, H.; Zhang, J.; Zhu, L.; Wang, C.; Yang, Y. Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Nakhjavani, M.; Hardingham, J.E.; Palethorpe, H.M.; Price, T.J.; Townsend, A.R. Druggable Molecular Targets for the Treatment of Triple Negative Breast Cancer. J. Breast Cancer 2019, 22, 341–361. [Google Scholar] [CrossRef]
- Xia, P.; Xu, X.-Y. PI3K/Akt/mTOR signaling pathway in cancer stem cells: From basic research to clinical application. Am. J. Cancer Res. 2015, 5, 1602–1609. [Google Scholar]
- Li, X.; Zhou, N.; Wang, J.; Liu, Z.; Wang, X.; Zhang, Q.; Liu, Q.; Gao, L.; Wang, R. Quercetin suppresses breast cancer stem cells (CD44+/CD24−) by inhibiting the PI3K/Akt/mTOR-signaling pathway. Life Sci. 2018, 196, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Zotano, A.; Mayer, I.A.; Arteaga, C.L. PI3K/AKT/mTOR: Role in breast cancer progression, drug resistance, and treatment. Cancer Metastasis Rev. 2016, 35, 515–524. [Google Scholar] [CrossRef]
- Nakhjavani, M.; Hardingham, J.E.; Palethorpe, H.M.; Tomita, Y.; Smith, E.; Price, T.J.; Townsend, A.R. Ginsenoside Rg3: Potential molecular targets and therapeutic indication in metastatic breast cancer. Medicines 2019, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Nakhjavani, M.; Smith, E.; Townsend, A.R.; Price, T.J.; Hardingham, J.E. Anti-Angiogenic Properties of Ginsenoside Rg3. Molecules 2020, 25, 4905. [Google Scholar] [CrossRef] [PubMed]
- Nakhjavani, M.; Palethorpe, H.M.; Tomita, Y.; Smith, E.; Price, T.J.; Yool, A.J.; Pei, J.V.; Townsend, A.R.; Hardingham, J.E. Stereoselective anti-cancer activities of ginsenoside Rg3 on triple negative breast cancer cell models. Pharmaceuticals 2019, 12, 117. [Google Scholar] [CrossRef] [PubMed]
- Aboushady, D.; Parr, M.K.; Hanafi, R.S. Quality-by-Design Is a Tool for Quality Assurance in the Assessment of Enantioseparation of a Model Active Pharmaceutical Ingredient. Pharmaceuticals 2020, 13, 364. [Google Scholar] [CrossRef]
- Zhao, W.; Sachsenmeier, K.; Zhang, L.; Sult, E.; Hollingsworth, R.E.; Yang, H. A new bliss independence model to analyze drug combination data. J. Biomol. Screen. 2014, 19, 817–821. [Google Scholar] [CrossRef]
- Chui, C.H.; Wong, R.S.M.; Cheng, G.Y.M.; Lau, F.Y.; Kok, S.H.L.; Cheng, C.H.; Cheung, F.; Tang, W.K.; Teo, I.T.N.; Chan, A.S.C. Antiproliferative ability of a combination regimen of crocodile egg extract, wild radix ginseng and natural Ganoderma lucidum on acute myelogenous leukemia. Oncol. Rep. 2006, 16, 1313–1316. [Google Scholar] [CrossRef]
- Nakhjavani, M.; Smith, E.; Yeo, K.; Palethorpe, H.M.; Tomita, Y.; Price, T.J.; Townsend, A.R.; Hardingham, J.E. Anti-Angiogenic Properties of Ginsenoside Rg3 Epimers: In Vitro Assessment of Single and Combination Treatments. Cancers 2021, 13, 2223. [Google Scholar] [CrossRef] [PubMed]
- Grimshaw, M.J.; Cooper, L.; Papazisis, K.; Coleman, J.A.; Bohnenkamp, H.R.; Chiapero-Stanke, L.; Taylor-Papadimitriou, J.; Burchell, J.M. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008, 10, R52. [Google Scholar] [CrossRef]
- Yousefnia, S.; Ghaedi, K.; Seyed Forootan, F.; Nasr Esfahani, M.H. Characterization of the stemness potency of mammospheres isolated from the breast cancer cell lines. Tumor Biol. 2019, 41, 1010428319869101. [Google Scholar] [CrossRef]
- Zou, W.; Yang, Y.; Zheng, R.; Wang, Z.; Zeng, H.; Chen, Z.; Yang, F.; Wang, J. Association of CD44 and CD24 phenotype with lymph node metastasis and survival in triple-negative breast cancer. Int. J. Clin. Exp. Pathol. 2020, 13, 1008. [Google Scholar] [PubMed]
- Hiraga, T.; Ito, S.; Nakamura, H. Side population in MDA-MB-231 human breast cancer cells exhibits cancer stem cell-like properties without higher bone-metastatic potential. Oncol. Rep. 2011, 25, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.; Zhang, D.; Bartholomeusz, C.; Doihara, H.; Hortobagyi, G.N.; Ueno, N.T. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res. Treat. 2012, 136, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Bahhnassy, A.; Mohanad, M.; Shaarawy, S.; Ismail, M.F.; El-Bastawisy, A.; Ashmawy, A.M.; Zekri, A.R. Transforming growth factor-β, insulin-like growth factor I/insulin-like growth factor I receptor and vascular endothelial growth factor-A: Prognostic and predictive markers in triple-negative and non-triple-negative breast cancer. Mol. Med. Rep. 2015, 12, 851–864. [Google Scholar] [CrossRef] [PubMed]
- Toyama, T.; Yamashita, H.; Kondo, N.; Okuda, K.; Takahashi, S.; Sasaki, H.; Sugiura, H.; Iwase, H.; Fujii, Y. Frequently increased epidermal growth factor receptor (EGFR) copy numbers and decreased BRCA1 mRNA expression in Japanese triple-negative breast cancers. BMC Cancer 2008, 8, 309. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-W.; Storey, K.B. Regulation of the mTOR signaling network in hibernating thirteen-lined ground squirrels. J. Exp. Biol. 2012, 215, 1720–1727. [Google Scholar] [CrossRef] [PubMed]
- Ruvinsky, I.; Sharon, N.; Lerer, T.; Cohen, H.; Stolovich-Rain, M.; Nir, T.; Dor, Y.; Zisman, P.; Meyuhas, O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005, 19, 2199–2211. [Google Scholar] [CrossRef] [PubMed]
- Romeo, Y.; Zhang, X.; Roux, P.P. Regulation and function of the RSK family of protein kinases. Biochem. J. 2012, 441, 553–569. [Google Scholar] [CrossRef]
- Anjum, R.; Blenis, J. The RSK family of kinases: Emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 2008, 9, 747–758. [Google Scholar] [CrossRef]
- Hekman, M.; Fischer, A.; Wennogle, L.P.; Wang, Y.K.; Campbell, S.L.; Rapp, U.R. Novel C-Raf phosphorylation sites: Serine 296 and 301 participate in Raf regulation. FEBS Lett. 2005, 579, 464–468. [Google Scholar] [CrossRef]
- Cardillo, T.M.; Trisal, P.; Arrojo, R.; Goldenberg, D.M.; Chang, C.-H. Targeting both IGF-1R and mTOR synergistically inhibits growth of renal cell carcinoma in vitro. BMC Cancer 2013, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lamhamedi-Cherradi, S.-E.; Menegaz, B.A.; Ramamoorthy, V.; Vishwamitra, D.; Wang, Y.; Maywald, R.L.; Buford, A.S.; Fokt, I.; Skora, S.; Wang, J. IGF-1R and mTOR blockade: Novel resistance mechanisms and synergistic drug combinations for Ewing sarcoma. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef]
- Ji, P.; Zhang, Y.; Wang, S.-J.; Ge, H.-L.; Zhao, G.-P.; Xu, Y.-C.; Wang, Y. CD44hiCD24lo mammosphere-forming cells from primary breast cancer display resistance to multiple chemotherapeutic drugs. Oncol. Rep. 2016, 35, 3293–3302. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Giatromanolaki, A.; Sivridis, E.; Fiska, A.; Koukourakis, M.I. The CD44+/CD24− phenotype relates to ‘triple-negative’state and unfavorable prognosis in breast cancer patients. Med. Oncol. 2011, 28, 745–752. [Google Scholar] [CrossRef]
- Shipitsin, M.; Campbell, L.L.; Argani, P.; Weremowicz, S.; Bloushtain-Qimron, N.; Yao, J.; Nikolskaya, T.; Serebryiskaya, T.; Beroukhim, R.; Hu, M. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007, 11, 259–273. [Google Scholar] [CrossRef]
- Abraham, B.K.; Fritz, P.; McClellan, M.; Hauptvogel, P.; Athelogou, M.; Brauch, H. Prevalence of CD44+/CD24−/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin. Cancer Res. 2005, 11, 1154–1159. [Google Scholar] [PubMed]
- Sheridan, C.; Kishimoto, H.; Fuchs, R.K.; Mehrotra, S.; Bhat-Nakshatri, P.; Turner, C.H.; Goulet, R.; Badve, S.; Nakshatri, H. CD44+/CD24-breast cancer cells exhibit enhanced invasive properties: An early step necessary for metastasis. Breast Cancer Res. 2006, 8, R59. [Google Scholar] [CrossRef] [PubMed]
- Van Phuc, P.; Nhan, P.L.C.; Nhung, T.H.; Tam, N.T.; Hoang, N.M.; Tue, V.G.; Thuy, D.T.; Ngoc, P.K. Downregulation of CD44 reduces doxorubicin resistance of CD44+ CD24− breast cancer cells. OncoTargets Ther. 2011, 4, 71. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oh, J.; Yoon, H.-J.; Jang, J.-H.; Kim, D.-H.; Surh, Y.-J. The standardized Korean Red Ginseng extract and its ingredient ginsenoside Rg3 inhibit manifestation of breast cancer stem cell–like properties through modulation of self-renewal signaling. J. Ginseng Res. 2019, 43, 421–430. [Google Scholar] [CrossRef]
- Xu, H.; Tian, Y.; Yuan, X.; Wu, H.; Liu, Q.; Pestell, R.G.; Wu, K. The role of CD44 in epithelial–mesenchymal transition and cancer development. Onco Targets Ther. 2015, 8, 3783. [Google Scholar] [CrossRef]
- Chen, J.; Shao, R.; Li, F.; Monteiro, M.; Liu, J.P.; Xu, Z.P.; Gu, W. PI 3K/Akt/mTOR pathway dual inhibitor BEZ 235 suppresses the stemness of colon cancer stem cells. Clin. Exp. Pharmacol. Physiol. 2015, 42, 1317–1326. [Google Scholar] [CrossRef]
- Lu, C.; Makala, L.; Wu, D.; Cai, Y. Targeting translation: eIF4E as an emerging anticancer drug target. Expert Rev. Mol. Med. 2016, 18. [Google Scholar] [CrossRef]
- Rutkovsky, A.C.; Yeh, E.S.; Guest, S.T.; Findlay, V.J.; Muise-Helmericks, R.C.; Armeson, K.; Ethier, S.P. Eukaryotic initiation factor 4E-binding protein as an oncogene in breast cancer. BMC Cancer 2019, 19, 1–15. [Google Scholar] [CrossRef]
- Pons, B.; Peg, V.; Vázquez-Sánchez, M.Á.; López-Vicente, L.; Argelaguet, E.; Coch, L.; Martínez, A.; Hernández-Losa, J.; Armengol, G.; Ramon y Cajal, S. The effect of p-4E-BP1 and p-eIF4E on cell proliferation in a breast cancer model. Int. J. Oncol. 2011, 39, 1337–1345. [Google Scholar] [CrossRef] [PubMed]
- Hartman, N.W.; Lin, T.V.; Zhang, L.; Paquelet, G.E.; Feliciano, D.M.; Bordey, A. mTORC1 Targets the Translational Repressor 4E-BP2, but Not S6 Kinase 1/2, to Regulate Neural Stem Cell Self-Renewal In Vivo. Cell Rep. 2013, 5, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Abraham, J.; Flynn, D.; Castranova, V.; Shi, X.; Qian, Y. Individualized survival and treatment response predictions for breast cancers using phospho-EGFR, phospho-ER, phospho-HER2/neu, phospho-IGF-IR/In, phospho-MAPK, and phospho-p70S6K proteins. Int. J. Biol. Marker. 2007, 22, 1–11. [Google Scholar] [CrossRef]
- Xiao, L.; Wang, Y.C.; Li, W.S.; Du, Y. The role of mTOR and phospho-p70S6K in pathogenesis and progression of gastric carcinomas: An immunohistochemical study on tissue microarray. J. Exp. Clin. Cancer Res. 2009, 28, 152. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Tan, Z.; Gao, J.; Wu, W.; Liu, L.; Jin, W.; Cao, Y.; Zhao, S.; Zhang, W.; Qiu, Z. Hyperphosphorylation of ribosomal protein S6 predicts unfavorable clinical survival in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2015, 34, 126. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.-Z.; Deng, A.-M.; Li, L.-H.; Liu, G.-Y.; Wu, G.-Y. Prognostic role of phospho-PRAS40 (Thr246) expression in gastric cancer. Arch. Med. Res. 2014, 10, 149. [Google Scholar] [CrossRef]
- Shipitsin, M.; Small, C.; Giladi, E.; Siddiqui, S.; Choudhury, S.; Hussain, S.; Huang, Y.E.; Chang, H.; Rimm, D.L.; Berman, D.M. Automated quantitative multiplex immunofluorescence in situ imaging identifies phospho-S6 and phospho-PRAS40 as predictive protein biomarkers for prostate cancer lethality. Proteome Sci. 2014, 12, 40. [Google Scholar] [CrossRef]
- Zhao, H.; Martin, T.A.; Davies, E.L.; Ruge, F.; Yu, H.; Zhang, Y.; Teng, X.; Jiang, W.G. The Clinical Implications of RSK1-3 in Human Breast Cancer. Anticancer Res. 2016, 36, 1267–1274. [Google Scholar]
- Ludwik, K.A.; Campbell, J.P.; Li, M.; Li, Y.; Sandusky, Z.M.; Pasic, L.; Sowder, M.E.; Brenin, D.R.; Pietenpol, J.A.; O’Doherty, G.A. Development of a RSK inhibitor as a novel therapy for triple-negative breast cancer. Mol. Cancer Ther. 2016, 15, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Herbert, S.P.; Costa, G. Sending messages in moving cells: mRNA localization and the regulation of cell migration. Essays. Biochem. 2019, 63, 595–606. [Google Scholar] [CrossRef]
- Willett, M.; Brocard, M.; Davide, A.; Morley, S.J. Translation initiation factors and active sites of protein synthesis co-localize at the leading edge of migrating fibroblasts. Biochem. J. 2011, 438, 217–227. [Google Scholar] [CrossRef][Green Version]
- Mollard, S.; Fanciullino, R.; Giacometti, S.; Serdjebi, C.; Benzekry, S.; Ciccolini, J. In vivo bioluminescence tomography for monitoring breast tumor growth and metastatic spreading: Comparative study and mathematical modeling. Sci. Rep. 2016, 6, 36173. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Li, G.; Zhang, P.; Zhuang, X.; Hu, G. A CD44v+ subpopulation of breast cancer stem-like cells with enhanced lung metastasis capacity. Cell Death Dis. 2017, 8, e2679. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Kim, T.W.; Singh, S.V. Ginsenoside Rh2-mediated G 1 phase cell cycle arrest in human breast cancer cells is caused by p15 Ink4B and p27 Kip1-dependent inhibition of cyclin-dependent kinases. Pharm. Res. 2009, 26, 2280–2288. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.H.; Park, J.Y.; Lee, D.; Kwak, J.Y.; Park, E.H.; Kim, K.H.; Park, H.-J.; Kim, H.Y.; Jang, H.J.; Ham, J. Inhibitory effects of ginseng sapogenins on the proliferation of triple negative breast cancer MDA-MB-231 cells. Bioorganic Med. Chem. Lett. 2014, 24, 5409–5412. [Google Scholar] [CrossRef]
- Peng, B.; He, R.; Xu, Q.; Yang, Y.; Hu, Q.; Hou, H.; Liu, X.; Li, J. Ginsenoside 20(S)-protopanaxadiol inhibits triple-negative breast cancer metastasis in vivo by targeting EGFR-mediated MAPK pathway. Pharmacol. Res. 2019, 142, 1–13. [Google Scholar] [CrossRef]
- Smith, E.; Palethorpe, H.M.; Tomita, Y.; Pei, J.V.; Townsend, A.R.; Price, T.J.; Young, J.P.; Yool, A.J.; Hardingham, J.E. The Purified Extract from the Medicinal Plant Bacopa monnieri, Bacopaside II, Inhibits Growth of Colon Cancer Cells In Vitro by Inducing Cell Cycle Arrest and Apoptosis. Cells 2018, 7, 81. [Google Scholar] [CrossRef]
- Paltoglou, S.; Das, R.; Townley, S.L.; Hickey, T.E.; Tarulli, G.A.; Coutinho, I.; Fernandes, R.; Hanson, A.R.; Denis, I.; Carroll, J.S.; et al. Novel Androgen Receptor Coregulator GRHL2 Exerts Both Oncogenic and Antimetastatic Functions in Prostate Cancer. Cancer Res. 2017, 77, 3417–3430. [Google Scholar] [CrossRef]
- Lombardo, Y.; de Giorgio, A.; Coombes, C.R.; Stebbing, J.; Castellano, L. Mammosphere formation assay from human breast cancer tissues and cell lines. J. Vis. Exp. 2015, e52671. [Google Scholar] [CrossRef]


| Binding Score (kJ/mol) (Number of H-Bonds) | ||||||
|---|---|---|---|---|---|---|
| Molecule | EGFR | HER-2 | IGF-1R | PDGFR | FRB | Rheb |
| SRg3 | −6.9 (2) | 2.7 (0) | −8.0 (2) | −2.8 (1) | −7.0 (1) | −8.0 (3) |
| RRg3 | −6.9 (2) | 2.7 (1) | −7.5 (0) | −2.8 (1) | −7.2 (1) | −8.0 (4) |
| Sorafenib | −9.6 (0) | −10.8 (1) | −8.9 (1) | −11.2 (1) | n.d. 1 | n.d. |
| Lenvatinib | −10.4 (1) | −9.6 (1) | −7.9 (1) | −10.1 (1) | n.d. | n.d. |
| Rapamycin | n.d. | n.d. | n.d. | n.d. | −7.6 (0) | n.d. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakhjavani, M.; Smith, E.; Palethorpe, H.M.; Tomita, Y.; Yeo, K.; Price, T.J.; Townsend, A.R.; Hardingham, J.E. Anti-Cancer Effects of an Optimised Combination of Ginsenoside Rg3 Epimers on Triple Negative Breast Cancer Models. Pharmaceuticals 2021, 14, 633. https://doi.org/10.3390/ph14070633
Nakhjavani M, Smith E, Palethorpe HM, Tomita Y, Yeo K, Price TJ, Townsend AR, Hardingham JE. Anti-Cancer Effects of an Optimised Combination of Ginsenoside Rg3 Epimers on Triple Negative Breast Cancer Models. Pharmaceuticals. 2021; 14(7):633. https://doi.org/10.3390/ph14070633
Chicago/Turabian StyleNakhjavani, Maryam, Eric Smith, Helen M. Palethorpe, Yoko Tomita, Kenny Yeo, Tim J. Price, Amanda R. Townsend, and Jennifer E. Hardingham. 2021. "Anti-Cancer Effects of an Optimised Combination of Ginsenoside Rg3 Epimers on Triple Negative Breast Cancer Models" Pharmaceuticals 14, no. 7: 633. https://doi.org/10.3390/ph14070633
APA StyleNakhjavani, M., Smith, E., Palethorpe, H. M., Tomita, Y., Yeo, K., Price, T. J., Townsend, A. R., & Hardingham, J. E. (2021). Anti-Cancer Effects of an Optimised Combination of Ginsenoside Rg3 Epimers on Triple Negative Breast Cancer Models. Pharmaceuticals, 14(7), 633. https://doi.org/10.3390/ph14070633








