NHERF1/EBP50 as a Target for Modulation of MRP Function in HepG2 Cells
Abstract
1. Introduction
2. Results
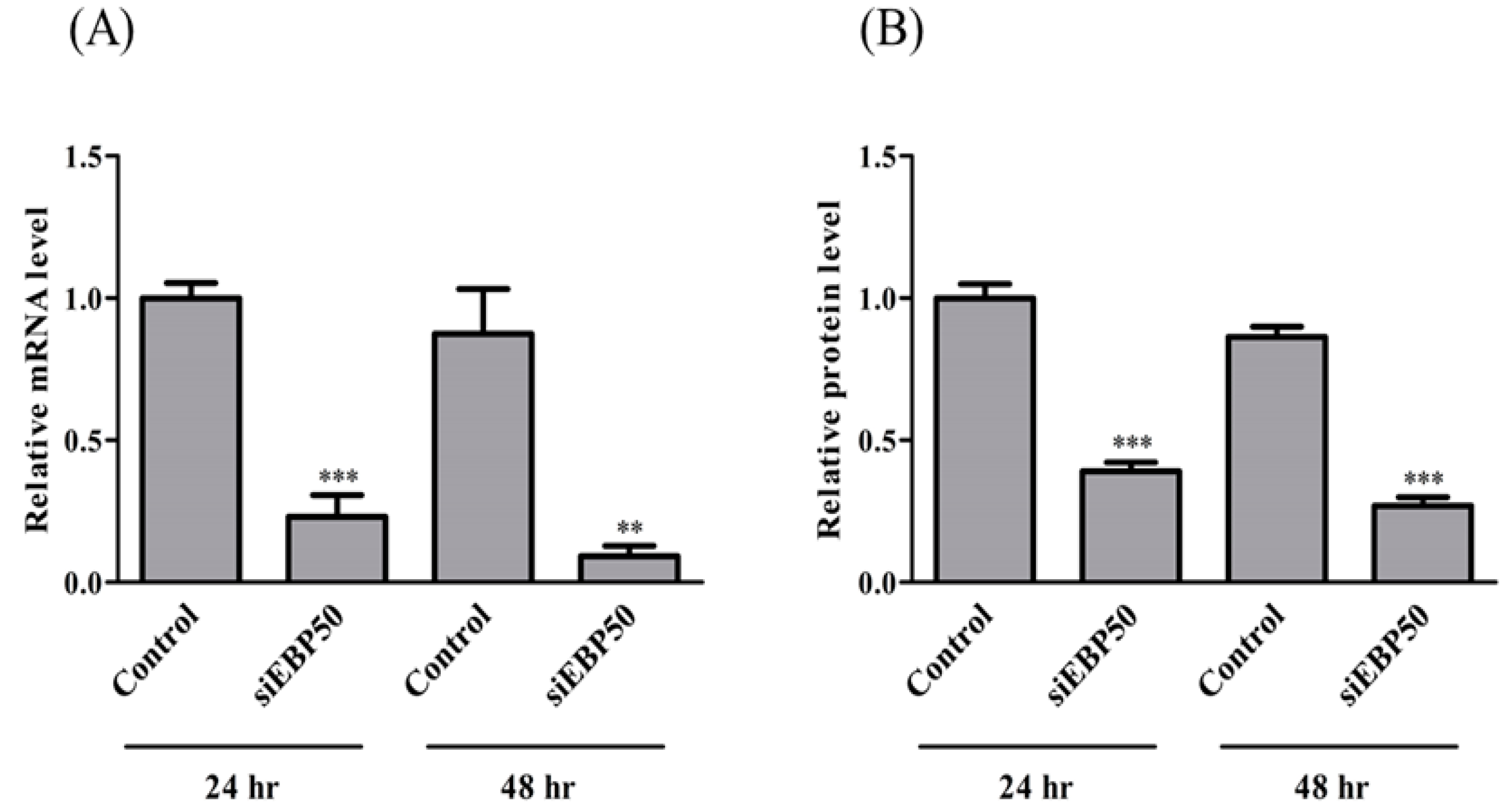
2.1. EBP50 Knockdown Affected Both mRNA and Protein Levels in HepG2 Cells with siRNA Transfection
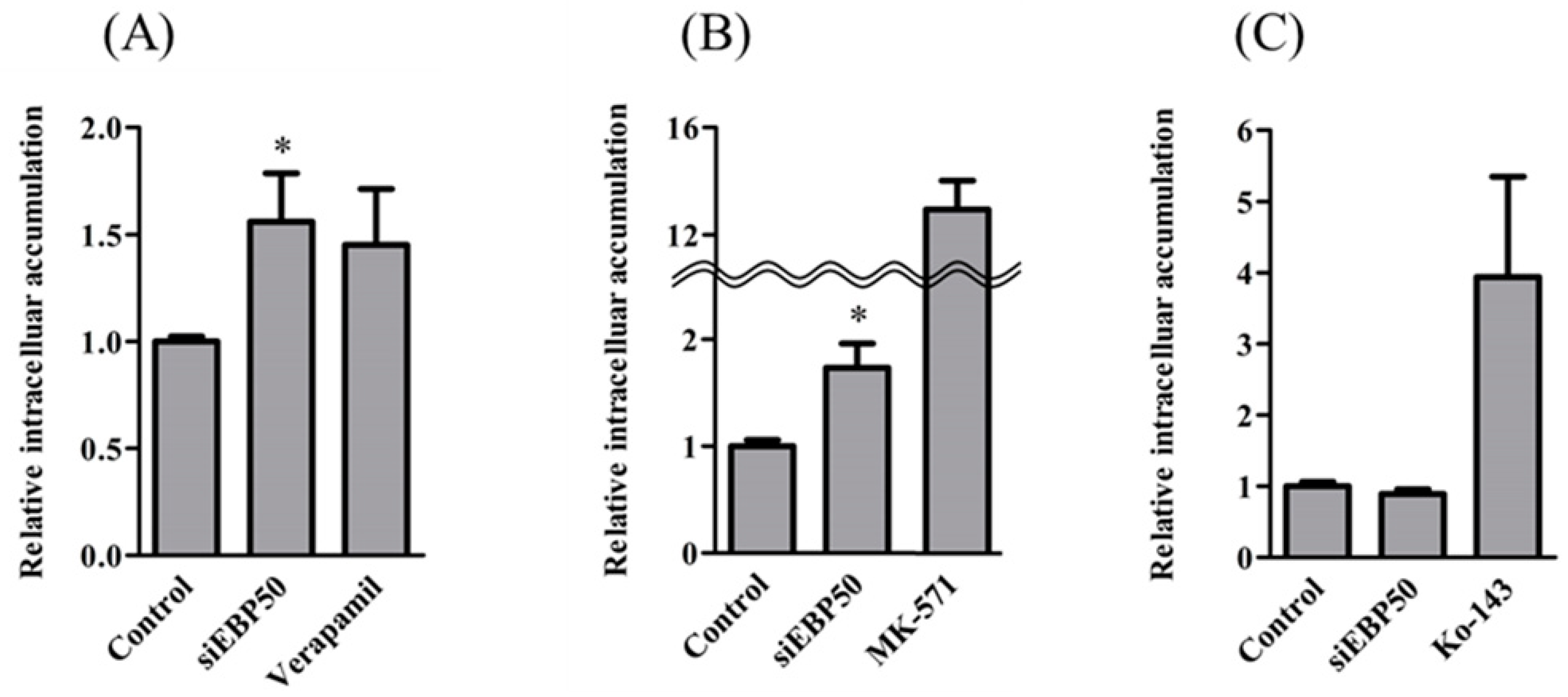
2.2. Activities of P-gp and MRP—But Not BCRP—Increased in HepG2EBP50KD Cells
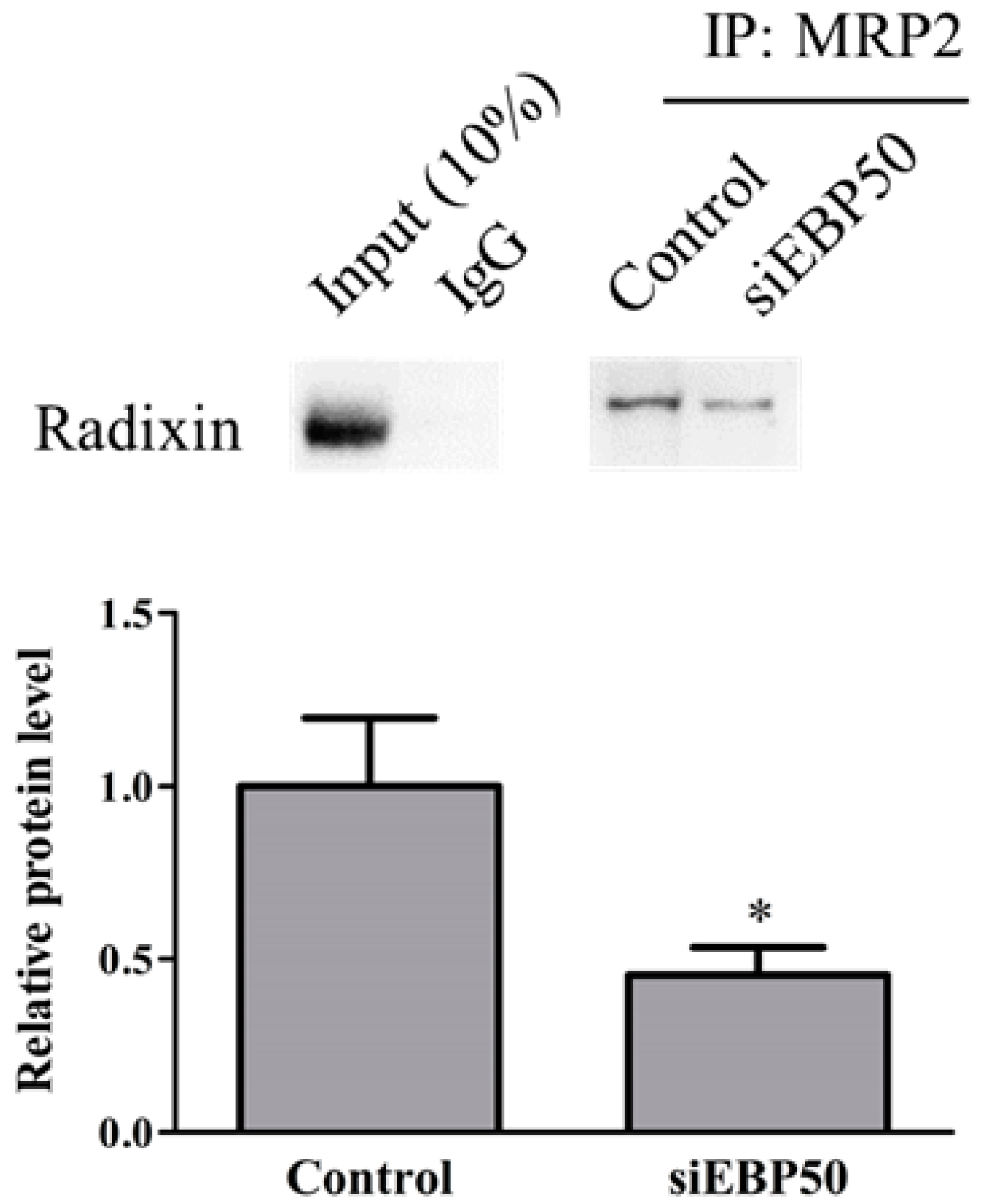
2.3. EBP50 KD Led to Decreased Interaction between MRP2 and Radixin in HepG2 Cells
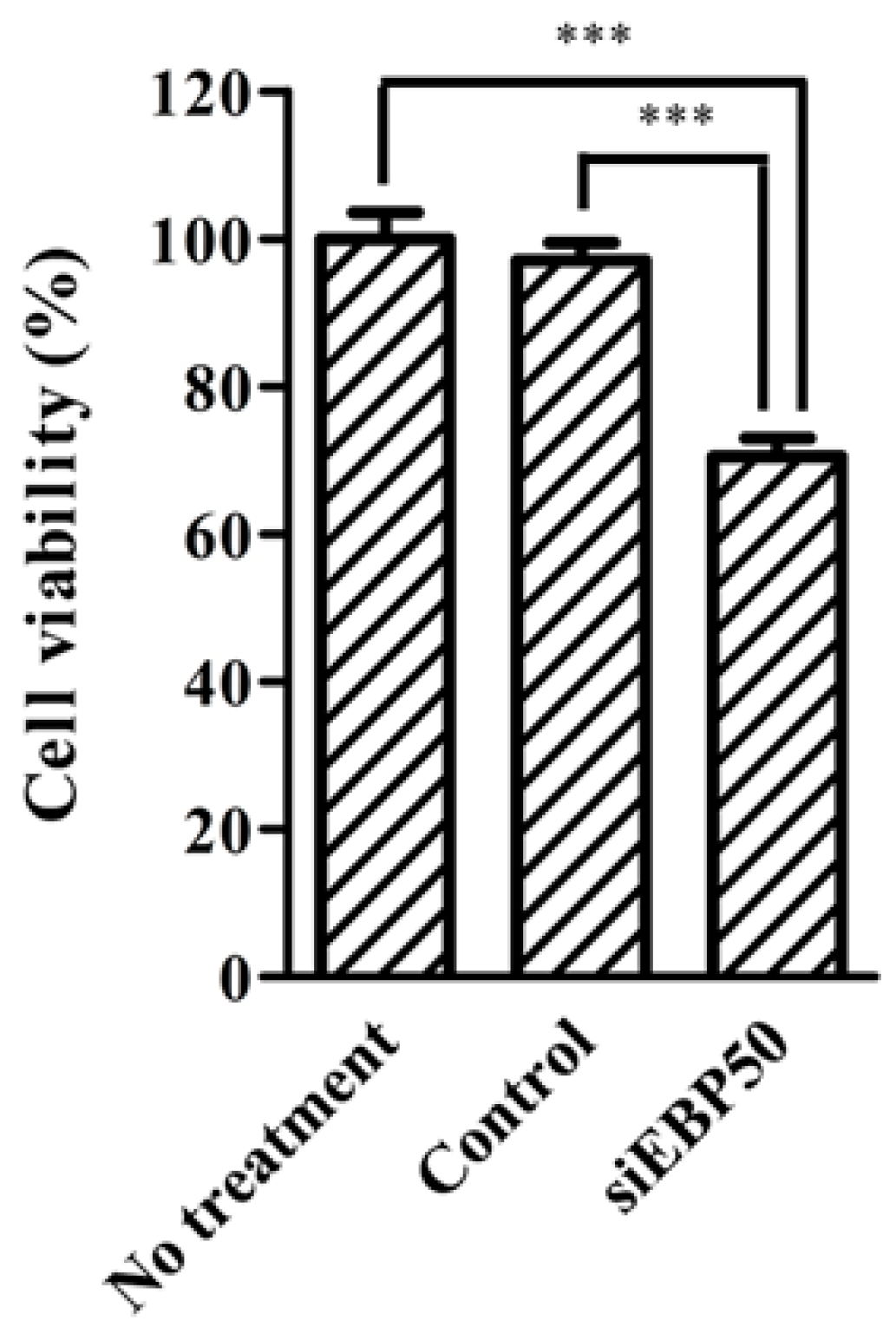
2.4. HepG2EBP50KD Cells Showed Increased MTX Efficiency
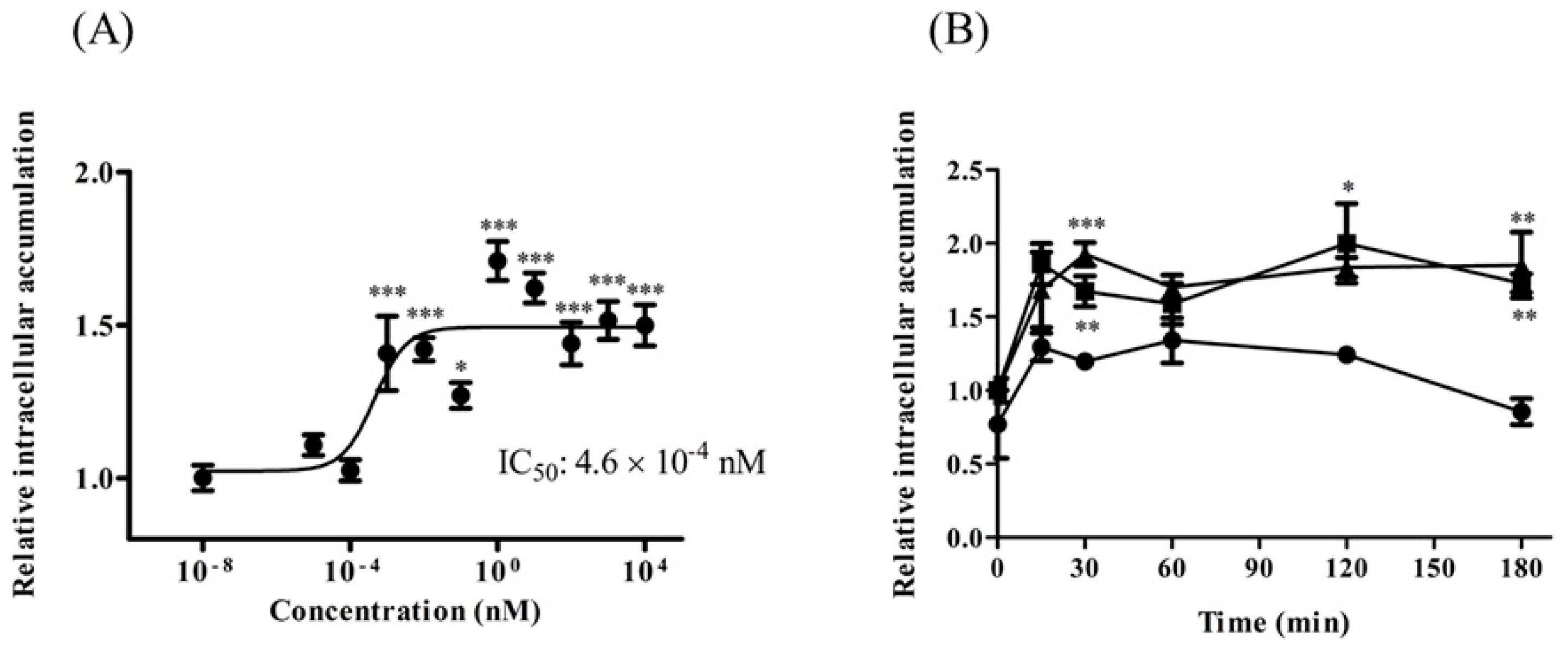
2.5. TAT-PDZ1 Peptide Inhibited MRP Activity in HepG2 Cells
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture and siRNA or Peptide Treatment
4.3. Determination of mRNA Levels by Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
4.4. Determination of Protein Levels by LC-MS/MS-Based Targeted Proteomics
4.5. Transporter Activity Assays
4.6. Immunoprecipitation Analyses
4.7. HepG2 Cell Viability after MTX Treatment
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fouassier, L.; Duan, C.; Feranchak, A.P.; Yun, C.H.C.; Sutherland, E.; Simon, F. Ezrin-radixin-moesin-binding phosphoprotein 50 is expressed at the apical membrane of rat liver epithelia. Hepatology 2001, 33, 166–176. [Google Scholar] [CrossRef]
- Morales, F.C.; Takahashi, Y.; Kreimann, E.L.; Georgescu, M.-M. Ezrin-radixin-moesin (ERM)-binding phosphoprotein 50 organizes ERM proteins at the apical membrane of polarized epithelia. Proc. Natl. Acad. Sci. USA 2004, 101, 17705–17710. [Google Scholar] [CrossRef]
- Kikuchi, S.; Hata, M.; Fukumoto, K.; Yamane, Y.; Matsui, T.; Tamura, A. Radixin deficiency causes conjugated hyperbilirubinemia with loss of Mrp2 from bile canalicular membranes. Nat. Genet. 2002, 31, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, W.; Soroka, C.J.; Mennone, A.; Harry, K.; Weinman, E.J. NHERF-1 binds to Mrp2 and regulates hepatic Mrp2 expression and function. J. Biol. Chem. 2010, 285, 19299–19307. [Google Scholar] [CrossRef] [PubMed]
- Kawase, A.; Sakata, M.; Yada, N.; Nakasaka, M.; Shimizu, T.; Kato, Y. Decreased Radixin Function for ATP-Binding Cassette Transporters in Liver in Adjuvant-Induced Arthritis Rats. J. Pharm. Sci. 2014, 103, 4058–4065. [Google Scholar] [CrossRef] [PubMed]
- Kawase, A.; Inoue, Y.; Hirosoko, M.; Sugihara, Y.; Shimada, H.; Iwaki, M. Decrease in multidrug resistance-associated protein 2 activities by knockdown of phosphatidylinositol 4-phosphate 5-kinase in hepatocytes and cancer cells. J. Pharm. Pharm. Sci. 2019, 22, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Stemmer-Rachamimov, A.O.; Wiederhold, T.; Nielsen, G.P.; James, M.; Pinney-Michalowski, D.; Roy, J.E. NHE-RF, a merlin-interacting protein, is primarily expressed in luminal epithelia, proliferative endometrium, and estrogen receptor-positive breast carcinomas. Am. J. Pathol. 2001, 158, 57–62. [Google Scholar] [CrossRef]
- Shibata, T.; Chuma, M.; Kokubu, A.; Sakamoto, M.; Hirohashi, S. EBP50, a β-catenin-associating protein, enhances Wnt signaling and is over-expressed in hepatocellular carcinoma. Hepatology 2003, 38, 178–186. [Google Scholar] [CrossRef]
- Cardone, R.A.; Bellizzi, A.; Busco, G.; Weinman, E.J.; Dell’Aquila, M.E.; Casavola, V. The NHERF1 PDZ2 domain regulates PKA-RhoA-p38-mediated NHE1 activation and invasion in breast tumor cells. Mol. Biol. Cell 2007, 18, 1768–1780. [Google Scholar] [CrossRef]
- Suda, J.; Zhu, L.; Karvar, S. Phosphorylation of radixin regulates cell polarity and Mrp-2 distribution in hepatocytes. Am. J. Physiol. Cell Physiol. 2011, 300, C416–C424. [Google Scholar]
- Hegedüs, T.; Sessler, T.; Scott, R.; Thelin, W.; Bakos, É.; Váradi, A. C-terminal phosphorylation of MRP2 modulates its interaction with PDZ proteins. Biochem. Biophys. Res. Commun. 2003, 302, 454–461. [Google Scholar] [CrossRef]
- Hoque, M.T.; Cole, S.P.C. Down-regulation of Na+/H+ exchanger regulatory factor 1 increases expression and function of multidrug resistance protein 4. Cancer Res. 2008, 68, 4802–4809. [Google Scholar] [CrossRef]
- Kano, T.; Wada, S.; Morimoto, K.; Kato, Y.; Ogihara, T. Effect of knockdown of ezrin, radixin, and moesin on P-glycoprotein function in HepG2 cells. J. Pharm. Sci. 2011, 100, 5308–5314. [Google Scholar] [CrossRef]
- Pokharel, D.; Padula, M.; Lu, J.; Jaiswal, R.; Djordjevic, S.; Bebawy, M. The Role of CD44 and ERM Proteins in Expression and Functionality of P-glycoprotein in Breast Cancer Cells. Molecules 2016, 21, 290. [Google Scholar] [CrossRef]
- Cunningham, A.D.; Qvit, N.; Mochly-Rosen, D. Peptides and peptidomimetics as regulators of protein–Protein interactions. Curr. Opin. Struct. Biol. 2017, 44, 59–66. [Google Scholar] [CrossRef]
- Oh, Y.S.; Heo, K.; Kim, E.K.; Jang, J.H.; Bae, S.S.; Park, J.B. Dynamic relocalization of NHERF1 mediates chemotactic migration of ovarian cancer cells toward lysophosphatidic acid stimulation. Exp. Mol. Med. 2017, 49, e351. [Google Scholar] [CrossRef]
- Ma, Q.; Jiao, Y.; Hao, Y.; Yan, S.; Lyu, N.; Gao, H. Targeting of NHERF1 through RNA interference inhibits the proliferation and migration of metastatic prostate cancer cells. Oncol. Lett. 2016, 11, 1149–1154. [Google Scholar] [CrossRef]
- Saponaro, C.; Vagheggini, A.; Scarpi, E.; Centonze, M.; Catacchio, I.; Popescu, O. NHERF1 and tumor microenvironment: A new scene in invasive breast carcinoma. J. Exp. Clin. Cancer Res. 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Kawase, A.; Fujii, A.; Negoro, M.; Akai, R.; Ishikubo, M.; Komura, H. Differences in cytochrome P450 and nuclear receptor mRNA levels in liver and small intestines between SD and DA rats. Drug Metab. Pharmacokinet. 2008, 23, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Kawase, A.; Yamada, A.; Gamou, Y.; Tahara, C.; Takeshita, F.; Murata, K. Increased effects of ginsenosides on the expression of cholesterol 7α-hydroxylase but not the bile salt export pump are involved in cholesterol metabolism. J. Nat. Med. 2013, 67, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Hersman, E.M.; Bumpus, N.N. A targeted proteomics approach for profiling murine cytochrome P450 expression. J. Pharmacol. Exp. Ther. 2014, 349, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Kawase, A.; Tateishi, S.; Kazaoka, A. Profiling of hepatic metabolizing enzymes and nuclear receptors in rats with adjuvant arthritis by targeted proteomics. Biopharm Drug Dispos. 2018, 39, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Kawase, A.; Inoue, Y.; Nakazaki, S.; Koizumi, E.; Iwaki, M. Radixin knockdown improves the accumulation and efficiency of methotrexate in tumor cells. Oncol. Rep. 2019, 42, 283–290. [Google Scholar] [CrossRef]
- Lin, L.; Xu, W.; Liang, H.; He, L.; Liu, S.; Li, Y. Construction of pH-sensitive lysozyme/pectin nanogel for tumor methotrexate delivery. Colloids Surf. B Biointerfaces 2015, 126, 459–466. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primer Sequence (5′−3′) | Product Size (bp) |
|---|---|---|
| EBP50 | For CCAGGATCGCATTGTGGAG | 201 |
| Rev CCATTGGTGAAGGGCACAG | ||
| GAPDH | For AGGGCTGCTTTTAACTCTGGT | 206 |
| Rev CCCCACTTGATTTTGGAGGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawase, A.; Hirosoko, M.; Sugihara, Y.; Koyama, Y.; Fukae, A.; Shimada, H.; Iwaki, M. NHERF1/EBP50 as a Target for Modulation of MRP Function in HepG2 Cells. Pharmaceuticals 2021, 14, 239. https://doi.org/10.3390/ph14030239
Kawase A, Hirosoko M, Sugihara Y, Koyama Y, Fukae A, Shimada H, Iwaki M. NHERF1/EBP50 as a Target for Modulation of MRP Function in HepG2 Cells. Pharmaceuticals. 2021; 14(3):239. https://doi.org/10.3390/ph14030239
Chicago/Turabian StyleKawase, Atsushi, Miho Hirosoko, Yuka Sugihara, Yunosuke Koyama, Ayaka Fukae, Hiroaki Shimada, and Masahiro Iwaki. 2021. "NHERF1/EBP50 as a Target for Modulation of MRP Function in HepG2 Cells" Pharmaceuticals 14, no. 3: 239. https://doi.org/10.3390/ph14030239
APA StyleKawase, A., Hirosoko, M., Sugihara, Y., Koyama, Y., Fukae, A., Shimada, H., & Iwaki, M. (2021). NHERF1/EBP50 as a Target for Modulation of MRP Function in HepG2 Cells. Pharmaceuticals, 14(3), 239. https://doi.org/10.3390/ph14030239




