1-C Metabolism—Serine, Glycine, Folates—In Acute Myeloid Leukemia
Abstract
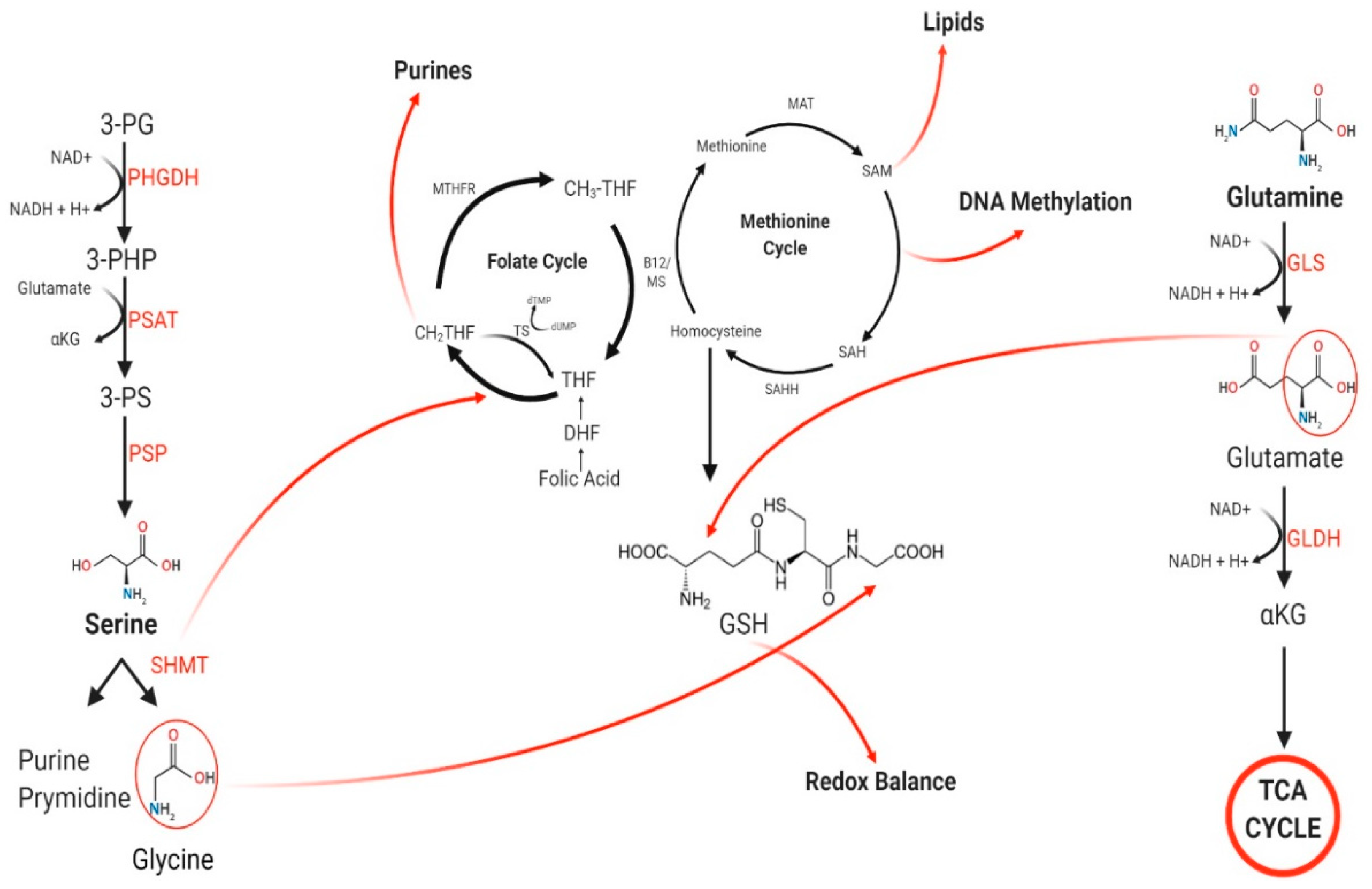
1. Introduction
2. Metabolic Dysfunction in AML
3. De Novo Serine/Glycine Biosynthesis
3.1. Serine
3.2. Glycine
4. 1-C Metabolism
4.1. Folate Cycle
4.2. Methionine Cycle
5. Glutamine Metabolism
Glutamine Metabolism and Cancer
6. Glutamine Contribution to 1-C Metabolism
6.1. Glutamine, Serine, and 1-C Interaction
6.2. TP73
7. Target-Based Therapy Using Clinically Available Agents
7.1. Glutamine Metabolism Modulators/Inhibitors
7.2. Serine Biosynthesis Modulators/Inhibitors
7.3. Anti-Folates and Anti-Metabolites
8. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Kreitz, J.; Schönfeld, C.; Seibert, M.; Stolp, V.; Alshamleh, I.; Oellerich, T.; Steffen, B.; Schwalbe, H.; Schnütgen, F.; Kurrle, N.; et al. Metabolic Plasticity of Acute Myeloid Leukemia. Cells 2019, 8, 805. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol. Life Sci. 2016, 73, 377–392. [Google Scholar] [CrossRef]
- Lukey, M.J.; Katt, W.P.; Cerione, R.A. Targeting amino acid metabolism for cancer therapy. Drug Discov. Today 2017, 22, 796–804. [Google Scholar] [CrossRef]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef]
- Ducker, G.S.; Rabinowitz, J.D. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017, 25, 27–42. [Google Scholar] [CrossRef]
- Mullarky, E.; Lairson, L.L.; Cantley, L.C.; Lyssiotis, C.A. A novel small-molecule inhibitor of 3phosphoglycerate dehydrogenase. Mol. Cell Oncol. 2016, 3, e1164280. [Google Scholar] [CrossRef]
- Reid, M.A.; Allen, A.E.; Liu, S.; Liberti, M.V.; Liu, P.; Liu, X.; Dai, Z.; Gao, X.; Wang, Q.; Liu, Y.; et al. Serine synthesis through PHGDH coordinates nucleotide levels by maintaining central carbon metabolism. Nat. Commun. 2018, 9, 5442. [Google Scholar] [CrossRef] [PubMed]
- Shuvalov, O.; Petukhov, A.; Daks, A.; Fedorova, O.; Vasileva, E.; Barlev, N.A. One-carbon metabolism and nucleotide biosynthesis as attractive targets for anticancer therapy. Oncotarget 2017, 8, 23955–23977. [Google Scholar] [CrossRef]
- Gao, X.; Lee, K.; Reid, M.A.; Sanderson, S.M.; Qiu, C.; Li, S.; Liu, J.; Locasale, J.W. Serine Availability Influences Mitochondrial Dynamics and Function through Lipid Metabolism. Cell Rep. 2018, 22, 3507–3520. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Fu, J.; Du, J.; Xu, W. The role of D-3-phosphoglycerate dehydrogenase in cancer. Int. J. Biol. Sci. 2020, 16, 1495–1506. [Google Scholar] [CrossRef]
- Polet, F.; Corbet, C.; Pinto, A.; Rubio, L.I.; Martherus, R.; Bol, V.; Drozak, X.; Grégoire, V.; Riant, O.; Feron, O. Reducing the serine availability complements the inhibition of the glutamine metabolism to block leukemia cell growth. Oncotarget 2016, 7, 1765–1776. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.H.; Glüxam, T.; Schlerka, A.; Bauer, K.; Grandits, A.M.; Hackl, H.; Dovey, O.; Zöchbauer-Müller, S.; Cooper, J.L.; Vassiliou, G.S.; et al. SOCS2 is part of a highly prognostic 4-gene signature in AML and promotes disease aggressiveness. Sci. Rep. 2019, 9, 9139. [Google Scholar] [CrossRef]
- Ducker, G.S.; Ghergurovich, J.M.; Mainolfi, N.; Suri, V.; Jeong, S.K.; Friedman, A.; Manfredi, M.G.; Gitai, Z.; Kim, H.; Rabinowitz, J.D.; et al. Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 2017, 114, 11404–11409. [Google Scholar] [CrossRef]
- Maddocks, O.D.; Berkers, C.R.; Mason, S.M.; Zheng, L.; Blyth, K.; Gottlieb, E.; Vousden, K.H. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 2013, 493, 542–546. [Google Scholar] [CrossRef] [PubMed]
- Amelio, I.; Cutruzzolá, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef]
- García-Cañaveras, J.C.; Lancho, O.; Ducker, G.S.; Ghergurovich, X.X.; Silva-Diz, V.; Minuzzo, S.; Indraccolo, S.; Kim, H.; Herranz, D.; Rabinowitz, J.D. SHMT inhibition is effective and synergizes with methotrexate in T-cell acute lymphoblastic leukemia. Leukemia 2020. [Google Scholar] [CrossRef]
- Yang, M.; Vousden, K.H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 2016, 16, 650–662. [Google Scholar] [CrossRef]
- Lyon, P.; Strippoli, V.; Fang, B.; Cimmino, L. B Vitamins and One-Carbon Metabolism: Implications in Human Health and Disease. Nutrients 2020, 12, 2867. [Google Scholar] [CrossRef]
- Nilsson, R.; Jain, M.; Madhusudhan, N.; Sheppard, N.G.; Strittmatter, L.; Kampf, C.; Huang, J.; Asplund, A.; Mootha, V.K. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014, 5, 3128. [Google Scholar] [CrossRef] [PubMed]
- Pikman, Y.; Puissant, A.; Alexe, G.; Furman, A.; Chen, L.M.; Frumm, S.M.; Ross, L.; Fenouille, N.; Bassil, C.F.; Lewis, C.A.; et al. Targeting MTHFD2 in acute myeloid leukemia. J. Exp. Med. 2016, 213, 1285–1306. [Google Scholar] [CrossRef] [PubMed]
- De Jonge, R.; Hooijberg, J.H.; van Zelst, B.D.; Jansen, G.; van Zantwijk, C.H.; Kaspers, G.J.; Peters, G.J.; Ravindranath, Y.; Pieters, R.; Lindemans, J. Effect of polymorphisms in folate-related genes on in vitro methotrexate sensitivity in pediatric acute lymphoblastic leukemia. Blood 2005, 106, 717–720. [Google Scholar] [CrossRef]
- Dulucq, S.; St-Onge, G.; Gagné, V.; Ansari, M.; Sinnett, D.; Labuda, D.; Moghrabi, A.; Krajinovic, M. DNA variants in the dihydrofolate reductase gene and outcome in childhood ALL. Blood 2008, 111, 3692–3700. [Google Scholar] [CrossRef]
- Kotb, M.; Geller, A.M. Methionine adenosyltransferase: Structure and function. Pharmacol. Ther. 1993, 59, 125–143. [Google Scholar] [CrossRef]
- Barve, A.; Vega, A.; Shah, P.P.; Ghare, S.; Casson, L.; Wunderlich, M.; Siskind, L.J.; Beverly, L.J. Perturbation of methionine/S-adenosylmethionine metabolism as a novel vulnerability in MLL rearranged leukemia. Cells 2019, 8, 1322. [Google Scholar] [CrossRef]
- Ueland, P.M.; Hustad, S.; Schneede, J.; Refsum, H.; Vollset, S.E. Biological and clinical implications of the MTHFR C677T polymorphism. Trends Pharmacol. Sci. 2001, 22, 195–201. [Google Scholar] [CrossRef]
- Carr, D.F.; Whiteley, G.; Alfirevic, A.; Pirmohamed, M.; FolATED Study Team. Investigation of inter-individual variability of the one-carbon folate pathway: A bioinformatic and genetic review. Pharmacogenomics J. 2009, 9, 291–305. [Google Scholar] [CrossRef]
- Steluti, J.; Carvalho, A.M.; Carioca, A.A.F.; Miranda, A.; Gattás, G.J.F.; Fisberg, R.M.; Marchioni, D.M. Genetic Variants Involved in One-Carbon Metabolism: Polymorphism Frequencies and Differences in Homocysteine Concentrations in the Folic Acid Fortification Era. Nutrients 2017, 9, 539. [Google Scholar] [CrossRef]
- Willems, L.; Jacque, N.; Jacquel, A.; Neveux, N.; Maciel, T.T.; Lambert, M.; Schmitt, A.; Poulain, L.; Green, A.S.; Uzunov, M.; et al. Inhibiting glutamine uptake represents an attractive new strategy for treating acute myeloid leukemia. Blood 2013, 122, 3521–3532. [Google Scholar] [CrossRef] [PubMed]
- Emadi, A.; Zokaee, H.; Sausville, E.A. Asparaginase in the treatment of non-ALL hematologic malignancies. Cancer Chemother. Pharmacol. 2014, 73, 875–883. [Google Scholar] [CrossRef]
- Emadi, A. Exploiting AML vulnerability: Glutamine dependency. Blood 2015, 126, 1269–1270. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Pavlova, N.N.; Thompson, C.B. Cancer cell metabolism: The essential role of the nonessential amino acid, glutamine. EMBO J. 2017, 36, 1302–1315. [Google Scholar] [CrossRef]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to clinic: Glutamine metabolism to cancer. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef]
- Zhou, T.; Yang, Y.; Chen, Q.; Xie, L. Glutamine Metabolism Is Essential for Stemness of Bone Marrow Mesenchymal Stem Cells and Bone Homeostasis. Stem Cells Int. 2019, 2019, 8928934. [Google Scholar] [CrossRef]
- Farhadi, P.; Yarani, R.; Dokaneheifard, S.; Mansouri, K. The emerging role of targeting cancer metabolism for cancer therapy. Tumour Biol. 2020, 42. [Google Scholar] [CrossRef] [PubMed]
- Jewell, J.L.; Kim, Y.C.; Russell, R.C.; Yu, F.X.; Park, H.W.; Plouffe, S.W.; Tagliabracci, V.S.; Guan, K.L. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science 2015, 347, 194–198. [Google Scholar] [CrossRef]
- Bhutia, Y.D.; Ganapathy, V. Glutamine transporters in mammalian cells and their functions in physiology and cancer. Biochim. Biophys. Acta 2016, 1863, 2531–2539. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J.; Duffy, L.; Stratton, R.; Ford, D.; O’Reilly, S. Metabolic reprogramming of glycolysis and glutamine metabolism are key events in myofibroblast transition in systemic sclerosis pathogenesis. J. Cell Mol. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Le, A.; Lane, A.N.; Hamaker, M.; Bose, S.; Gouw, A.; Barbi, J.; Tsukamoto, T.; Rojas, C.J.; Slusher, B.S.; Zhang, H.; et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012, 15, 110–121. [Google Scholar] [CrossRef]
- Jacque, N.; Ronchetti, A.M.; Larrue, C.; Meunier, G.; Birsen, R.; Willems, L.; Saland, E.; Decroocq, J.; Maciel, T.T.; Lambert, M.; et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood 2015, 126, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Egler, R.A.; Ahuja, S.P.; Matloub, Y. L-asparaginase in the treatment of patients with acute lymphoblastic leukemia. J. Pharmacol. Pharmacother. 2016, 7, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Gross, M.I.; Demo, S.D.; Dennison, J.B.; Chen, L.; Chernov-Rogan, T.; Goyal, B.; Janes, J.R.; Laidig, G.J.; Lewis, E.R.; Li, J.; et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 2014, 13, 890–901. [Google Scholar] [CrossRef]
- Gregory, M.A.; Nemkov, T.; Park, H.J.; Zaberezhnyy, V.; Gehrke, S.; Adane, B.; Jordan, C.T.; Hansen, K.C.; D’Alessandro, A.; DeGregori, J. Targeting Glutamine Metabolism and Redox State for Leukemia Therapy. Clin. Cancer Res. 2019, 25, 4079–4090. [Google Scholar] [CrossRef]
- Schulte, M.L.; Fu, A.; Zhao, P.; Li, J.; Geng, L.; Smith, S.T.; Kondo, J.; Coffey, R.J.; Johnson, M.O.; Rathmell, J.C.; et al. Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat Med. 2018, 24, 194–202. [Google Scholar] [CrossRef]
- Van Geldermalsen, M.; Wang, Q.; Nagarajah, R.; Marshall, A.D.; Thoeng, A.; Gao, D.; Ritchie, W.; Feng, Y.; Bailey, C.G.; Deng, N.; et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene 2016, 35, 3201–3208. [Google Scholar] [CrossRef]
- Hanaford, A.R.; Alt, J.; Rais, R.; Wang, S.Z.; Kaur, H.; Thorek, D.L.J.; Eberhart, C.G.; Slusher, B.S.; Martin, A.M.; Raabe, E.H. Orally bioavailable glutamine antagonist prodrug JHU-083 penetrates mouse brain and suppresses the growth of MYC-driven medulloblastoma. Transl. Oncol. 2019, 12, 1314–1322. [Google Scholar] [CrossRef]
- Napoli, M.; Flores, E.R. Another case for diet restriction: TAp73-expressing medulloblastomas are stunted by glutamine withdrawal. Genes Dev. 2017, 31, 1715–1716. [Google Scholar] [CrossRef] [PubMed]
- Amelio, I.; Markert, E.K.; Rufini, A.; Antonov, A.V.; Sayan, B.S.; Tucci, P.; Agostini, M.; Mineo, T.C.; Levine, A.J.; Melino, G. p73 regulates serine biosynthesis in cancer. Oncogene 2014, 33, 5039–5046. [Google Scholar] [CrossRef] [PubMed]
- Melino, G.; De Laurenzi, V.; Vousden, K.H. p73: Friend or foe in tumorigenesis. Nat. Rev. Cancer 2002, 2, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Pluta, A.; Nyman, U.; Joseph, B.; Robak, T.; Zhivotovsky, B.; Smolewski, P. The role of p73 in hematological malignancies. Leukemia 2006, 20, 757–766. [Google Scholar] [CrossRef]
- Lucena-Araujo, A.R.; Panepucci, R.A.; dos Santos, G.A.; Jácomo, R.H.; Santana-Lemos, B.A.; Lima, A.S.; Garcia, A.B.; Araújo, A.G.; Falcão, R.P.; Rego, E.M. The expression of DeltaΔNTP73, TATP73 and TP53 genes in acute myeloid leukaemia is associated with recurrent cytogenetic abnormalities and in vitro susceptibility to cytarabine cytotoxicity. Br. J. Haematol. 2008, 142, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Lucena-Araujo, A.R.; Kim, H.T.; Thomé, C.; Jacomo, R.H.; Melo, R.A.; Bittencourt, R.; Pasquini, R.; Pagnano, K.; Glória, A.B.; Chauffaille Mde, L.; et al. High ΔNp73/TAp73 ratio is associated with poor prognosis in acute promyelocytic leukemia. Blood 2015, 126, 2302–2306. [Google Scholar] [CrossRef]
- Emadi, A.; Law, J.Y.; Strovel, E.T.; Lapidus, R.G.; Jeng, L.J.B.; Lee, M.; Blitzer, M.G.; Carter-Cooper, B.A.; Sewell, D.; Van Der Merwe, I.; et al. Asparaginase Erwinia chrysanthemi effectively depletes plasma glutamine in adult patients with relapsed/refractory acute myeloid leukemia. Cancer Chemother. Pharmacol. 2018, 81, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Kaspers, G.J.L. Acute myeloid leukaemia niche regulates response to L-asparaginase. Br. J. Haematol. 2019, 186, 397–399. [Google Scholar] [CrossRef]
- Emadi, A.; Kapadia, B.; Bollino, D.; Bhandary, B.; Baer, M.R.; Niyongere, S.; Strovel, E.T.; Kaizer, H.; Chang, E.; Choi, E.Y.; et al. Venetoclax and pegcrisantaspase for complex karyotype acute myeloid leukemia. Leukemia 2020. [Google Scholar] [CrossRef] [PubMed]
- Locasale, J.W.; Grassian, A.R.; Melman, T.; Lyssiotis, C.A.; Mattaini, K.R.; Bass, A.J.; Heffron, G.; Metallo, C.M.; Muranen, T.; Sharfi, H.; et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 2011, 43, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Elsaadi, S.; Steiro, I.; Abdollahi, P.; Vandsemb, E.N.; Yang, R.; Slørdahl, T.S.; Rø, T.B.; Menu, E.; Sponaas, A.M.; Børset, M. Targeting phosphoglycerate dehydrogenase in multiple myeloma. Exp. Hematol. Oncol. 2021, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wan, X.; Yu, T.; Huang, Z.; Shen, C.; Qi, Q.; Xiang, S.; Chen, X.; Arbely, E.; Ling, Z.Q.; et al. Acetylation Stabilizes Phosphoglycerate Dehydrogenase by Disrupting the Interaction of E3 Ligase RNF5 to Promote Breast Tumorigenesis. Cell Rep. 2020, 32, 108021. [Google Scholar] [CrossRef]
- Rider, B.J. Methotrexate. The Comprehensive Pharmacology; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–5. [Google Scholar] [CrossRef]
- Adjei, A.A. Pharmacology and mechanism of action of pemetrexed. Clin. Lung Cancer 2004, 5 (Suppl. 2), S51–S55. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Gerson, S.L. Chapter 26—Clinical Trials Using LV-P140K-MGMT for Gliomas, Gene Therapy of Cancer, 3rd ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 379–391. [Google Scholar] [CrossRef]
- Abraham, D.J. 4.04-Structure-Based Drug Design—A Historical Perspective and the Future. Comprehensive Medicina Chemistry II; Elsevier: Amsterdam, The Netherlands, 2007; pp. 65–86. [Google Scholar] [CrossRef]
| Classification | Mechanism of Action | Examples | Pre-Clinial/Clinial Usage |
|---|---|---|---|
| Glutamine Metabolism | |||
| Asparaginase | Depletes glutamine in the plasma by converting glutamine/asparagine to glutamate/aspartate and ammonia |
|
|
| Glutaminase Inhibitor | Inhibits the hydrolase enzyme that converts glutamine to glutamate |
| |
| Glutamine Transporter Inhibitor | Inhibits transport of glutamine in the cell (PM and mitochondria) |
|
|
| Glutamine Antagonist/Analog | Direct blockage of glutamine metabolism |
|
|
| Serine Biosynthesis | |||
| Phosphoglycerate dehydrogenase (PHGDH) Inhibitor | Inhibits the rate limiting step of serine biosynthesis (PHGDH) |
| |
| Serine hydroxymethyltransferase (SHMT) Inhibitor | Inhibits the enzyme (SHMT) that catalyzes the conversion of serine to glycine and initiates the folate cycle |
|
|
| 1-C Metabolism | |||
| Anti-Folates | Antagonize the actions of folic acid and inhibit key enzymes in the folate cycle including: Thymidylate synthase (TS), Dihydrofolate reductase (DHFR), Glycinamide ribonucleotide formyltransferase (GARFT) |
|
|
| Anti-Metabolites | Analogues of a normal metabolites that inhibit key enzymes in the folate cycle including: Thymidylate synthase (TS) |
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmood, K.; Emadi, A. 1-C Metabolism—Serine, Glycine, Folates—In Acute Myeloid Leukemia. Pharmaceuticals 2021, 14, 190. https://doi.org/10.3390/ph14030190
Mahmood K, Emadi A. 1-C Metabolism—Serine, Glycine, Folates—In Acute Myeloid Leukemia. Pharmaceuticals. 2021; 14(3):190. https://doi.org/10.3390/ph14030190
Chicago/Turabian StyleMahmood, Kanwal, and Ashkan Emadi. 2021. "1-C Metabolism—Serine, Glycine, Folates—In Acute Myeloid Leukemia" Pharmaceuticals 14, no. 3: 190. https://doi.org/10.3390/ph14030190
APA StyleMahmood, K., & Emadi, A. (2021). 1-C Metabolism—Serine, Glycine, Folates—In Acute Myeloid Leukemia. Pharmaceuticals, 14(3), 190. https://doi.org/10.3390/ph14030190







