Probing Allosteric Hsp70 Inhibitors by Molecular Modelling Studies to Expedite the Development of Novel Combined F508del CFTR Modulators
Abstract
1. Introduction
2. Results and Discussion
2.1. Exploring In Silico the Human HSP70 Protein
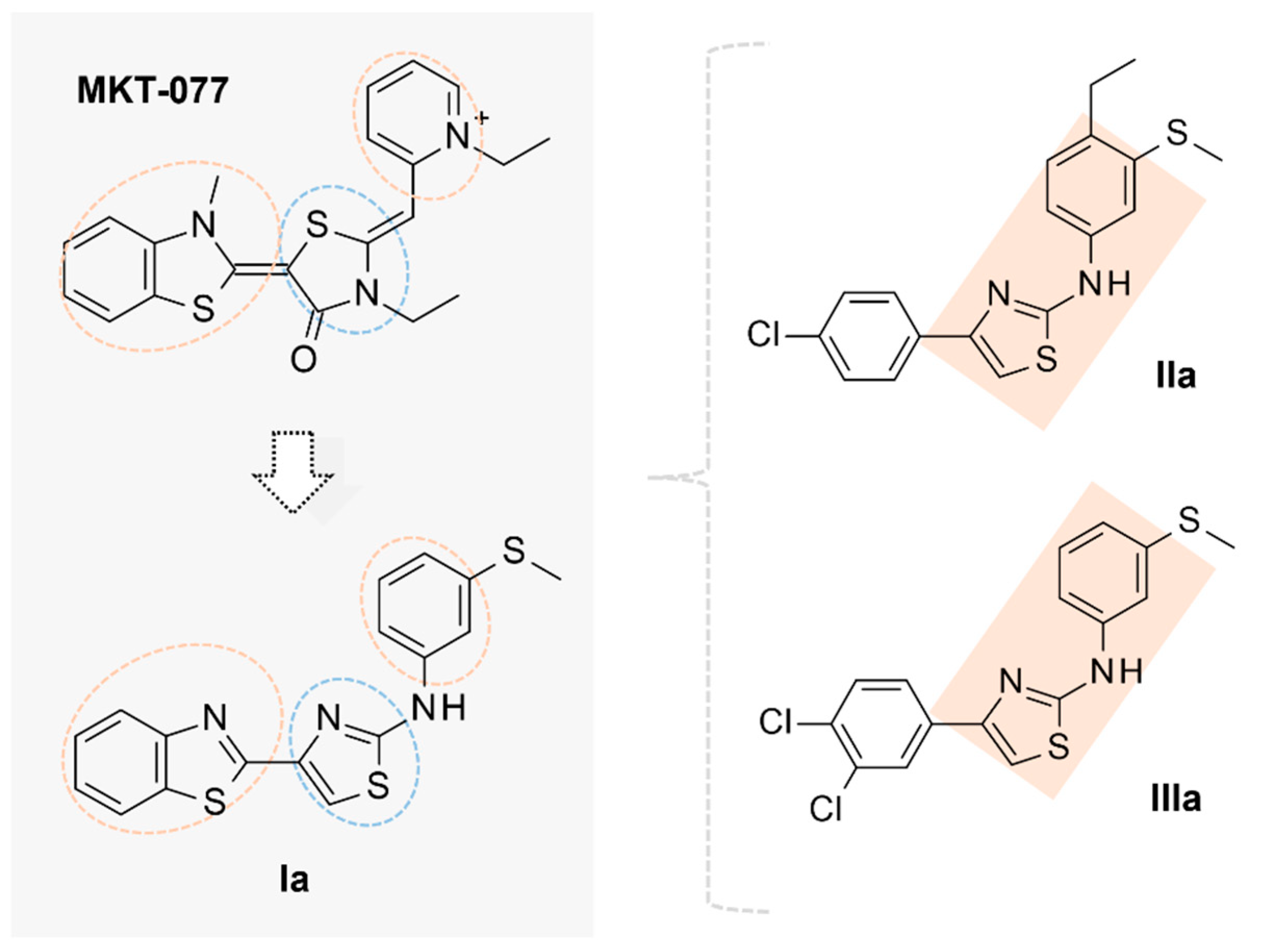
2.2. Molecular Docking Studies of MKT-077 and Related Analogues
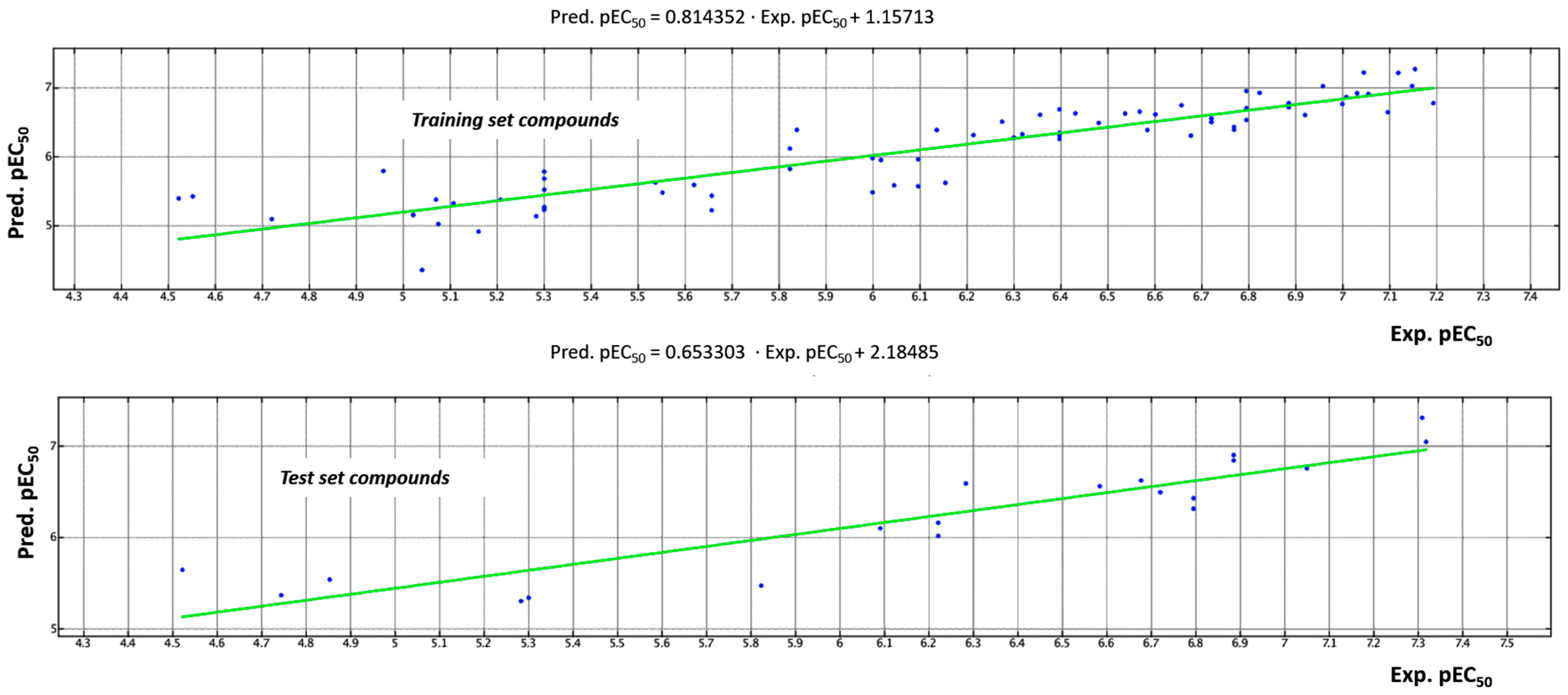
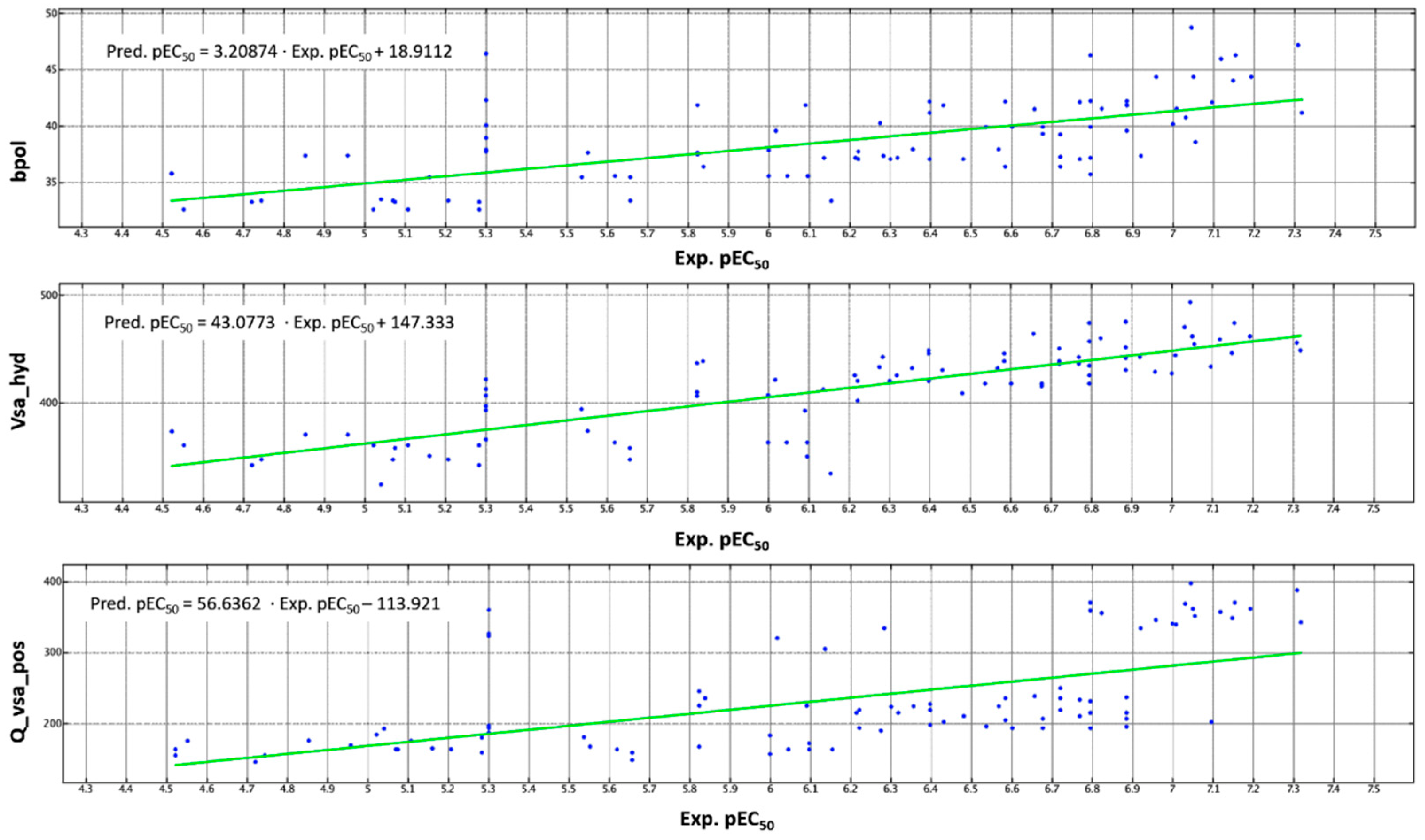
2.3. QSAR Analyses
2.4. Virtual Screening of AATs as Putative HSP70 Inhibitors
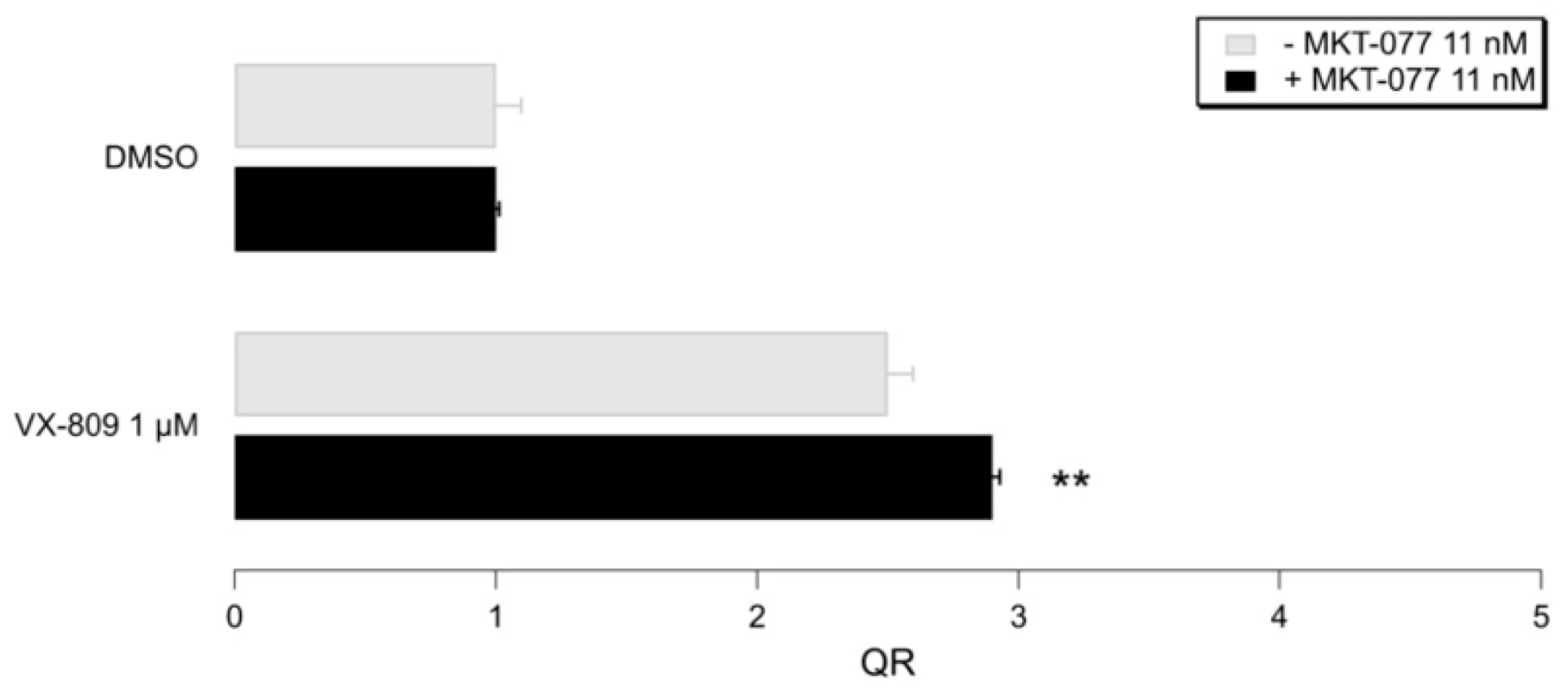
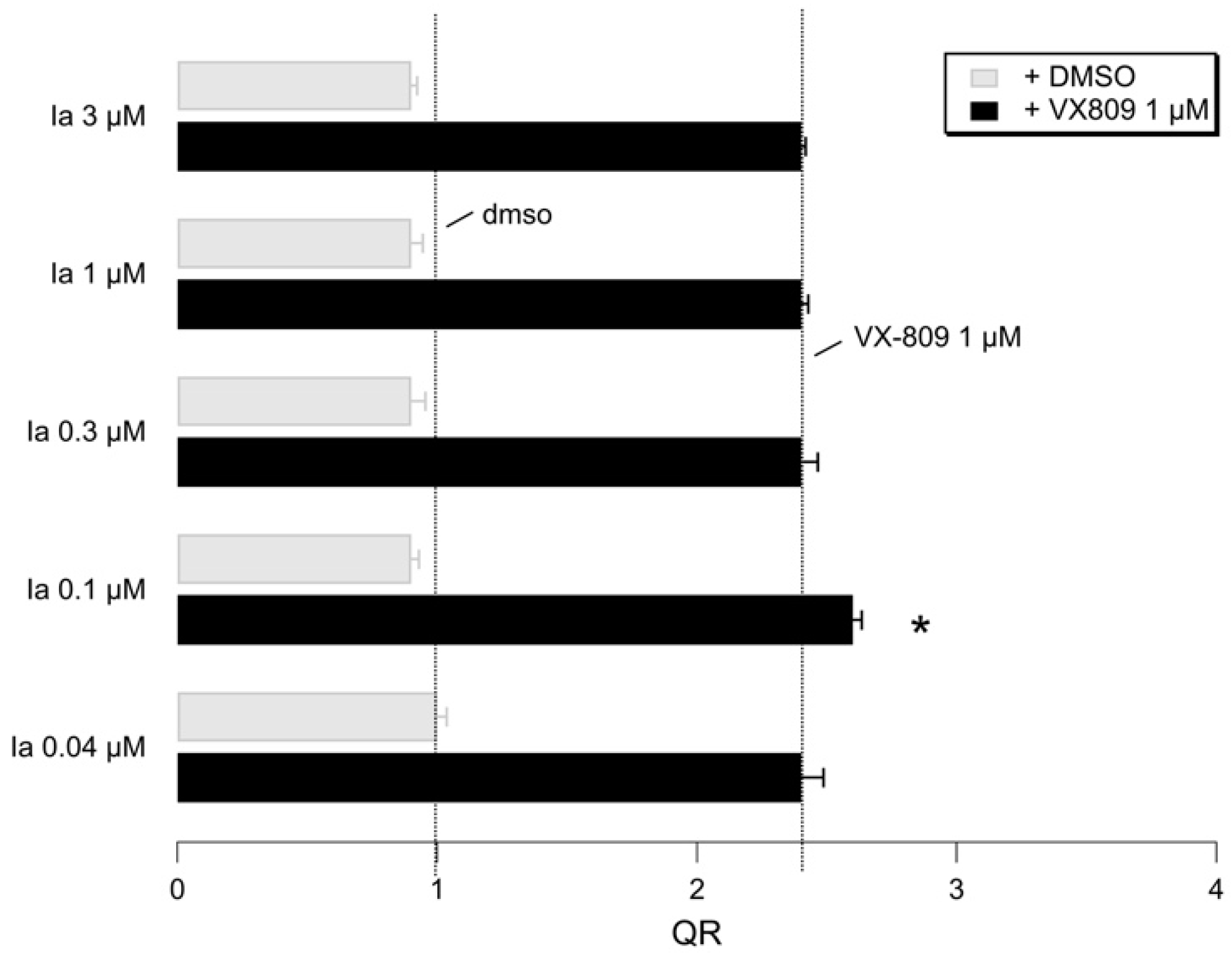
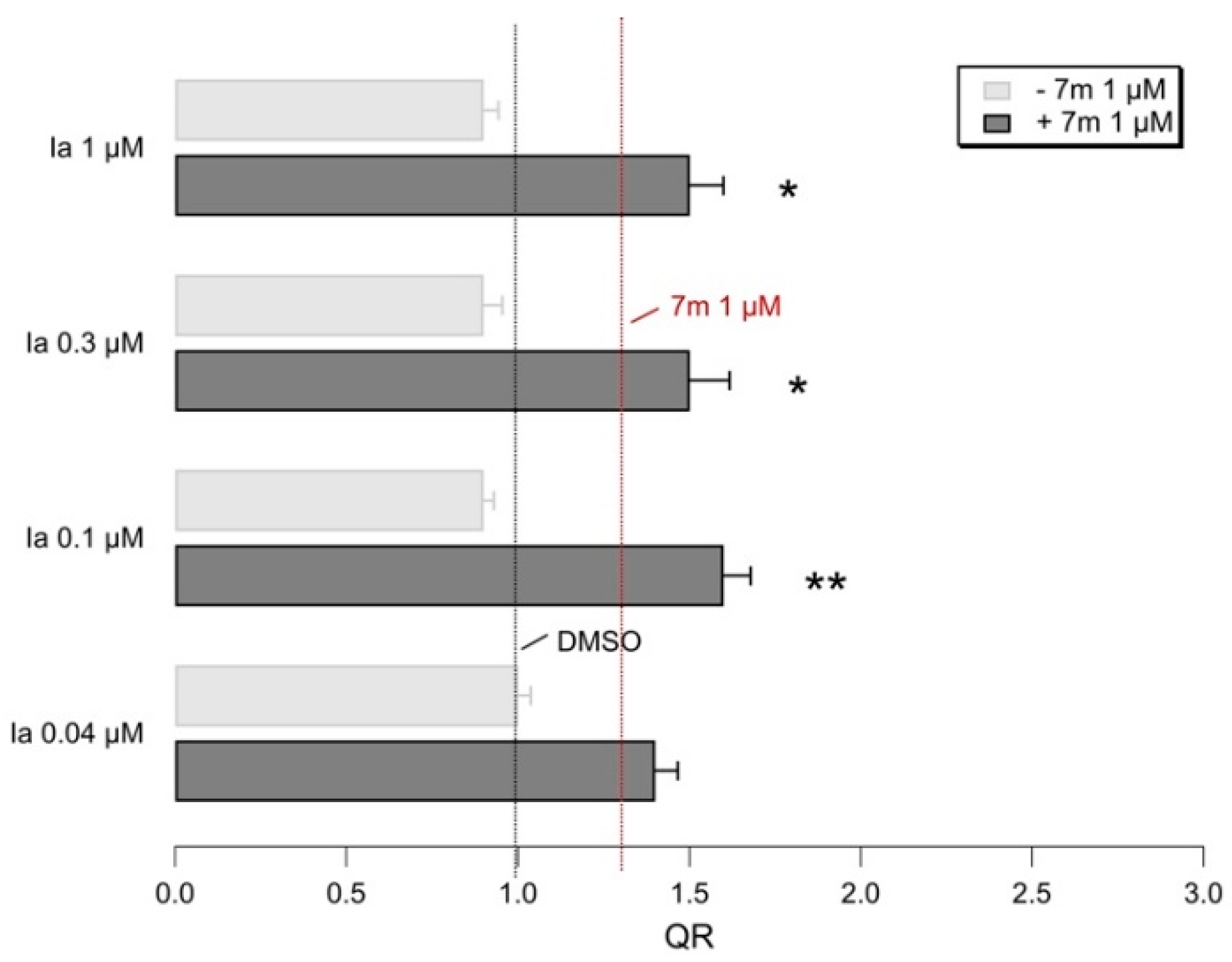
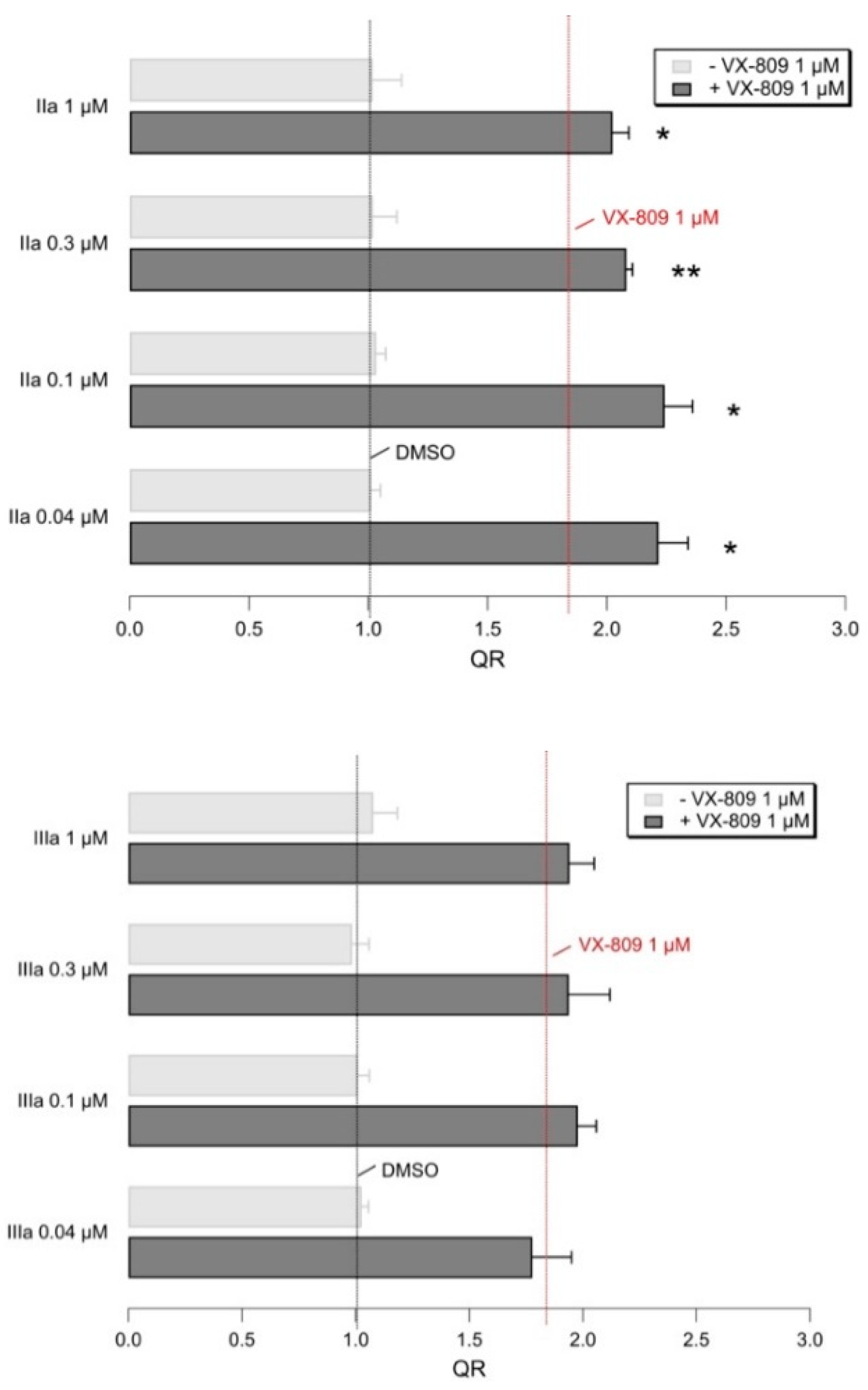
2.5. Biological Evaluation of I–IIIa in Presence of F508del CFTR Correctors
2.6. In Silico PK Profile Prediction
3. Material and Methods
3.1. Ligand Preparation
3.2. Protein Preparation and In Silico Analysis
3.3. Molecular Docking Studies
3.4. QSAR Analysis
3.5. In Silico Evaluation ADME Properties
3.6. Biological Evaluation
3.6.1. Cell Culture
3.6.2. Fluorescence Assay for CFTR Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Elborn, J.S. Cystic fibrosis. Lancet 2016, 388, 2519–2531. [Google Scholar] [CrossRef]
- Cheng, S.H.; Gregory, R.J.; Marshall, J.; Paul, S.; Souza, D.W.; White, G.A.; O’Riordan, C.R.; Smith, A.E. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell 1990, 63, 827–834. [Google Scholar] [CrossRef]
- Veit, G.; Avramescu, R.G.; Chiang, A.N.; Houck, S.A.; Cai, Z.; Peters, K.W.; Hong, J.S.; Pollard, H.B.; Guggino, W.B.; Balch, W.E.; et al. From CFTR biology toward combinatorial pharmacotherapy: Expanded classification of cystic fibrosis mutations. Mol. Biol. Cell 2016, 27, 424–433. [Google Scholar] [CrossRef]
- Dean, M.; Rzhetsky, A.; Alikmets, R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001, 11, 1156–1166. [Google Scholar] [CrossRef]
- Gadsby, D.C.; Naim, A.C. Control of CFTR channel gating by phosphorylation and nucleotide hydrolysis. Physiol. Rev. 1999, 79, S77–S107. [Google Scholar] [CrossRef]
- Anderson, M.P.; Berger, H.A.; Rich, D.P.; Gregory, R.J.; Smith, A.E.; Welsh, M.J. Nucleoside triphosphates are required to open the CFTR chloride channel. Cell 1991, 67, 775–784. [Google Scholar] [CrossRef]
- Berger, H.A.; Travis, S.M.; Welsh, M.J. Regulation of the cystic fibrosis transmembrane conductance regulator Cl- channel by specific protein kinases and protein phosphatases. J. Biol. Chem. 1993, 268, 2037–2047. [Google Scholar] [CrossRef]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 2002, 15, 194–222. [Google Scholar] [CrossRef]
- Bhatt, J.M. Treatment of pulmonary exacerbations in cystic fibrosis. Eur. Respir. Rev. 2013, 22, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Moscowitz, S.M.; Chmiel, J.F.; Sernen; Cheng, E.; Gibson, R.L.; Marshall, S.G.; Cutting, G.R. Clinical practice and genetic counseling for cystic fibrosis and CFTR-related disorders. Genet. Med. 2008, 10, 851–868. [Google Scholar] [CrossRef]
- Fajac, I.; De Boeck, K. New horizons for cystic fibrosis treatment. Pharm Thera 2017, 170, 205–211. [Google Scholar] [CrossRef]
- Cai, Z.W.; Liu, J.; Li, H.Y.; Sheppard, D.N. Targeting F508del-CFTR to develop new rational therapies for cystic fibrosis. Acta Pharm. Sin. 2011, 32, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Kym, P.R.; Wang, X.; Pizzonero, M.; Van der Plas, S. Recent Progress in the Discovery and Development of Small-Molecule Modulators of CFTR. Prog. Med. Chem. 2018, 57, 235–276. [Google Scholar] [CrossRef]
- Wang, X.; Liu, B.; Searle, X.; Yeung, C.; Bogdan, A.; Greszler, S.; Singh, A.; Fan, Y.; Swensen, A.M.; Vortherms, T.; et al. Discovery of 4-[(2R,4R)-4-({[1-(2,2-Difluoro-1,3-benzodioxol-5-yl)cyclopropyl]carbonyl}amino)-7-(difluoromethoxy)-3,4-dihydro-2H-chromen-2-yl]benzoic Acid (ABBV/GLPG-2222), a Potent Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Corrector for the Treatment of Cystic Fibrosis. J. Med. Chem. 2018, 61, 1436–1449. [Google Scholar] [CrossRef]
- Jih, K.Y.; Hwang, T.C. Vx-770 Potentiates CFTR Function by Promoting Decoupling between the Gating Cycle and ATP Hydrolysis Cycle. Proc. Natl. Acad. Sci. USA 2013, 110, 4404–4409. [Google Scholar] [CrossRef]
- Okiyoneda, T.; Veit, G.; Dekkers, J.F.; Bagdany, M.; Soya, N.; Xu, H.; Roldan, A.; Verkman, A.S.; Kurth, M.; Simon, A.; et al. Mechanism-based corrector combination restores DF508-CFTR folding and function. Nat. Chem. Biol. 2013, 9, 444–454. [Google Scholar] [CrossRef]
- Farinha, C.M.; King-Underwood, J.; Sousa, M.; Correia, A.R.; Henriques, B.J.; Roxo-Rosa, M.; Da Paula, A.C.; Williams, J.; Hirst, S.; Gomes, C.M.; et al. Revertants, low temperature and correctors reveal the rescue mechanism of F508del-CFTR via VX-809 and suggest multiple agents for a complete correction. Chem. Biol. 2013, 20, 943–955. [Google Scholar] [CrossRef] [PubMed]
- Amaral, M.D.; Farinha, C.M. CFTR Rescuing mutant CFTR: A multi-task approach to a better outcome in treating cystic fibrosis. Curr. Pharm. Des. 2013, 19, 3497–3508. [Google Scholar] [CrossRef] [PubMed]
- Veit, G.; Xu, H.; Dreano, E.; Avramescu, R.G.; Bagdany, M.; Beitel, L.K.; Roldan, A.; Hancock, M.A.; Lay, C.; Li, W.; et al. Structure-guided combination therapy to potently improve the function of mutant CFTRs. Nat. Med. 2018, 24, 1732–1742. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Pacheco, M. CFTR modulators: Shedding light on precision medicine for cystic fibrosis. F. Phar. 2016, 7, 275. [Google Scholar] [CrossRef]
- Van Goor, F.; Hadida, S.; Grootenhuis, P.D.; Burton, B.; Cao, D.; Neuberger, T.; Turnbull, A.; Singh, A.; Joubran, J.; Hazlewood, A.; et al. Rescue of CF airway cell function in vitro by a CFTR potentiator, VX-770. Proc. Natl. Acad. Sci. USA 2009, 106, 18825–18830. [Google Scholar] [CrossRef] [PubMed]
- Van Goor, R.; Yu, H.; Burton, B.; Hoffman, B.J. Effect of ivacaftor on CFTR forms with missense mutations associated with defects in protein processing or function. J. Cyst. Fibros. 2014, 13, 29–36. [Google Scholar] [CrossRef]
- Hoppe, J.E.; Chilvers, M.; Ratjen, F.; McNamara, J.J.; Owen, C.A.; Tian, S.; Zahigian, R.; Cornell, A.G.; McColley, S.A. Long-term safety of lumacaftor-ivacaftor in children aged 2-5 years with cystic fibrosis homozygous for the F508del-CFTR mutation: A multicentre, phase 3, open-label, extension study. Lancet Respir. Med. 2021, 9, 977–988. [Google Scholar] [CrossRef]
- Kuek, S.L.; Ranganathan, S.C.; Harrison, J.; Robinson, P.J.; Shanthikumar, S. Optimism with Caution: Elexacaftor-Tezacaftor-Ivacaftor in Patients with Advanced Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2021, 204, 371–372. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Moskowitz, S.M.; Brown, C.; Horsley, A.; Mall, M.A.; McKone, E.F.; Plant, B.J.; Prais, D.; Ramsey, B.W.; Taylor-Cousar, J.L.; et al. VX16-659-101 Study Group. VX-659-Tezacaftor-Ivacaftor in Patients with Cystic Fibrosis and One or Two Phe508del Alleles. N. Engl. J. Med. 2018, 379, 1599–1611. [Google Scholar] [CrossRef] [PubMed]
- Keating, D.; Marigowda, G.; Burr, L.; Daines, C.; Mall, M.A.; McKone, E.F.; Ramsey, B.W.; Rowe, S.M.; Sass, L.A.; Tullis, E.; et al. VX16-445-001 Study Group VX-445-Tezacaftor-Ivacaftor in patients with cystic fibrosis and one or two Phe508del Alleles. N. Engl. J. Med. 2018, 379, 1612–1620. [Google Scholar] [CrossRef]
- Ridley, K.; Condren, M. Elexacaftor-Tezacaftor-Ivacaftor: The First Triple-Combination Cystic Fibrosis Transmembrane Conductance Regulator Modulating Therapy. Pediatr. Pharmacol. Ther. 2020, 25, 192–197. [Google Scholar] [CrossRef]
- Shaughnessy, C.A.; Zeitlin, P.L.; Bratcher, P.E. Elexacaftor is a CFTR potentiator and acts synergistically with ivacaftor during acute and chronic treatment. Sci. Rep. 2021, 11, 19810. [Google Scholar] [CrossRef] [PubMed]
- Balch, W.E.; Morimoto, R.I.; Dillin, A.; Kelly, J.W. Proteostasis adaptation for disease intervention. Science 2008, 319, 916–919. [Google Scholar] [CrossRef]
- Bagdany, M.; Veit, G.; Fukuda, R.; Avramescu, R.G.; Okiyoneda, T.; Baaklini, I.; Singh, J.; Sovak, G.; Xu, H.; Apaja, P.M.; et al. Chaperones rescue the energetic landscape of mutant CFTR at single molecule and in cell. Nat. Commun. 2017, 8, 398. [Google Scholar] [CrossRef] [PubMed]
- Kim Chiaw, P.; Hantouche, C.; Wong, M.J.H.; Matthes, E.; Robert, R.; Hanrahan, J.W.; Shrier, A.; Young, J.C. Hsp70 and DNAJA2 limit CFTR levels through degradation. PLoS ONE. 2019, 14, e220984. [Google Scholar] [CrossRef] [PubMed]
- Baaklini, I.; de Campos Gonçalves, C.; Lukacs, G.L.; Young, J.C. Selective Binding of HSC70 and its Co-Chaperones to Structural Hotspots on CFTR. Sci. Rep. 2020, 10, 4176. [Google Scholar] [CrossRef] [PubMed]
- Young, J.C. The role of the cytosolic HSP70 chaperone system in diseases caused by misfolding and aberrant trafficking of ion channels. Dis. Model. Mech. 2014, 7, 319–329. [Google Scholar] [CrossRef][Green Version]
- Loo, M.A.; Jensen, T.J.; Cui, L.; Hou, Y.; Chang, X.B.; Riordan, J.R. Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 1998, 17, 6879–6887. [Google Scholar] [CrossRef]
- Propper, D.; Braybrooke, J.; Taylor, D.; Lodi, R.; Styles, P.; Cramer, J.; Collins, W.; Levitt, N.; Talbot, D.; Ganesan, T.; et al. Phase I trial of the selective mitochondrial toxin MKT 077 in chemo-resistant solid tumours. Ann. Oncol. 1999, 10, 923–927. [Google Scholar] [CrossRef]
- Li, X.; Srinivasan, S.R.; Connarn, J.; Ahmad, A.; Young, Z.T.; Kabza, A.M.; Zuiderweg, E.R.; Sun, D.; Gestwicki, J.E. Analogs of the Allosteric Heat Shock Protein 70 (Hsp70) Inhibitor, MKT-077, as Anti-Cancer Agents. ACS Med. Chem. Lett. 2013, 4, 1042–1047. [Google Scholar] [CrossRef]
- Shao, H.; Li, X.; Moses, M.A.; Gilbert, L.A.; Kalyanaraman, C.; Young, Z.T.; Chernova, M.; Journey, S.N.; Weissman, J.S.; Hann, B.; et al. Exploration of Benzothiazole Rhodacyanines as Allosteric Inhibitors of Protein-Protein Interactions with Heat Shock Protein 70 (Hsp70). J. Med. Chem. 2018, 61, 6163–6177. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The Protein Data Bank. Acta Cryst. Sect. D 2002, 58, 899–907. [Google Scholar] [CrossRef]
- Pesce, E.; Bellotti, M.; Liessi, N.; Guariento, S.; Damonte, G.; Cichero, E.; Galatini, A.; Salis, A.; Gianotti, A.; Pedemonte, N.; et al. Synthesis and structure-activity relationship of aminoarylthiazole derivatives as correctors of the chloride transport defect in cystic fibrosis. Eur. J. Med. Chem. 2015, 99, 14–35. [Google Scholar] [CrossRef] [PubMed]
- Liessi, N.; Cichero, E.; Pesce, E.; Arkel, M.; Salis, A.; Tomati, V.; Paccagnella, M.; Damonte, G.; Tasso, B.; Galietta, L.J.V.; et al. Synthesis and biological evaluation of novel thiazole- VX-809 hybrid derivatives as F508del correctors by QSAR-based filtering tools. Eur. J. Med. Chem. 2018, 144, 179–200. [Google Scholar] [CrossRef]
- Parodi, A.; Righetti, G.; Pesce, E.; Salis, A.; Tasso, B.; Urbinati, C.; Tomati, V.; Damonte, G.; Rusnati, M.; Pedemonte, N.; et al. Discovery of novel VX-809 hybrid derivatives as F508del-CFTR correctors by molecular modeling, chemical synthesis and biological assays. Eur. J. Med. Chem. 2020, 208, 112833. [Google Scholar] [CrossRef]
- Flaherty, K.M.; DeLuca-Flaherty, C.; McKay, D.B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 1990, 346, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Rousaki, A.; Miyata, Y.; Jinwal, U.K.; Dickey, C.A.; Gestwicki, J.E.; Zuiderweg, E.R. Allosteric drugs: The interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J. Mol. Biol. 2011, 411, 614–632. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, S.; Assimon, V.A.; Young, Z.T.; Morra, G.; Shao, H.; Taylor, I.R.; Gestwicki, J.E.; Colombo, G.A. Local Allosteric Network in Heat Shock Protein 70 (Hsp70) Links Inhibitor Binding to Enzyme Activity and Distal Protein-Protein Interactions. ACS Chem Biol. 2018, 11, 3142–3152. [Google Scholar] [CrossRef]
- Molecular Operating Environment (MOE); Chemical Computing Group ULC, 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7. 2021. Available online: http://www.chemcomp.com/ (accessed on 9 December 2021).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, S.; Min, D.; Kim, W. Comprehensive Survey of Recent Drug Discovery Using Deep Learning. Int. J. Mol. Sci. 2021, 18, 9983. [Google Scholar] [CrossRef] [PubMed]
- Shaker, B.; Ahmad, S.; Lee, J.; Jung, C.; Na, D. In silico methods and tools for drug discovery. Comput. Biol. Med. 2021, 137, 104851. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Li, J.Y.; Shi, X.X.; Wang, Z.; Hao, G.F.; Yang, G.F. Web-Based Quantitative Structure-Activity Relationship Resources Facilitate Effective Drug Discovery. Top. Curr. Chem. 2021, 6, 37. [Google Scholar] [CrossRef]
- Guariento, S.; Tonelli, M.; Espinoza, S.; Gerasimov, A.S.; Gainetdinov, R.R.; Cichero, E. Rational design, chemical synthesis and biological evaluation of novel biguanides exploring species-specificity responsiveness of TAAR1 agonists. Eur. J. Med. Chem. 2018, 146, 171–184. [Google Scholar] [CrossRef]
- Righetti, G.; Casale, M.; Liessi, N.; Tasso, B.; Salis, A.; Tonelli, M.; Millo, E.; Pedemonte, N.; Fossa, P.; Cichero, E. Molecular Docking and QSAR Studies as Computational Tools Exploring the Rescue Ability of F508del CFTR Correctors. Int. J. Mol. Sci. 2020, 21, 8084. [Google Scholar] [CrossRef] [PubMed]
- Righetti, G.; Casale, M.; Tonelli, M.; Liessi, N.; Fossa, P.; Pedemonte, N.; Millo, E.; Cichero, E. New Insights into the Binding Features of F508del CFTR Potentiators: A Molecular Docking, Pharmacophore Mapping and QSAR Analysis Approach. Pharmaceuticals 2020, 12, 445. [Google Scholar] [CrossRef]
- Kennard, R.W.; Stone, L.A. Computer aided design of experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Boggia, R.; Forina, M.; Fossa, P.; Mosti, L. Zupan’s descriptors in QSAR applied to the study of a new class of cardiotonic agents. Farmaco 1997, 52, 411–419. [Google Scholar]
- Miyata, Y.; Li, X.; Lee, H.-F.; Umesh, K.J.; Sharan, R.S.; Sandlin, P.S.; Young, T.Z.; Brodsky, J.L.; Dickey, C.A.; Sun, D.; et al. Synthesis and initial evaluation of YM-08, a blood-brain barrier permeable derivative of the heat shock protein 70 (Hsp70) inhibitor MKT-077, which reduces tau levels. ACS Chem. Neurosci. 2013, 4, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Van de Waterbeemd, H.; Gifford, E. ADMET in Silico Modelling: Towards Prediction Paradise? Nat. Rev. Drug Discov. 2003, 2, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Francesconi, V.; Cichero, E.; Kanov, E.V.; Laurini, E.; Pricl, S.; Gainetdinov, R.R.; Tonelli, M. Novel 1-Amidino-4-Phenylpiperazines as Potent Agonists at Human Taar1 Receptor: Rational Design, Synthesis, Biological Evaluation and Molecular Docking Studies. Pharmaceuticals 2020, 13, 391. [Google Scholar] [CrossRef]
- Tonelli, M.; Espinoza, S.; Gainetdinov, R.R.; Cichero, E. Novel Biguanide-Based Derivatives Scouted as TAAR1 Agonists: Synthesis, Biological Evaluation, ADME Prediction and Molecular Docking Studies. Eur. J. Med. Chem. 2017, 127, 781–792. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Needleman, S.B.; Wunsch, C.D. A General Method Applicable to the Search for Similarities in the Amino Acid Sequences of Two Proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Ryu, J.; Park, R.; Kim, D.S. Molecular surfaces on proteins via beta shapes. Comput. Aided Des. 2007, 39, 1042–1057. [Google Scholar] [CrossRef]
- Lee, B.; Richards, F.M. The Interpretation of Protein Structures: Estimation of Static Accessibility. J. Mol. Biol. 1971, 55, 379–400. [Google Scholar] [CrossRef]
- Richards, F.M. Areas, Volumes, Packing and Protein Structure. Annu. Rev. Biophys. Bioeng. 1977, 6, 151–176. [Google Scholar] [CrossRef] [PubMed]
- Connolly, M.L. Solvent-Accessible Surfaces of Proteins and Nucleic Acids. Science 1983, 221, 709–713. [Google Scholar] [CrossRef]
- Rocchia, W.; Sridharan, S.; Nicholls, A.; Alexov, E.; Chiabrera, A.; Honig, B. Rapid Grid-Based Construction of the Molecular Surface and the Use of Induced Surface Charge to Calculate Reaction Field Energies: Applications to the Molecular Systems and Geometric Objects. J. Comput. Chem. 2002, 23, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Brandas, C.; Ludovico, A.; Parodi, A.; Moran, O.; Millo, E.; Cichero, E.; Baroni, D. NBD2 Is Required for the Rescue of Mutant F508del CFTR by a Thiazole-Based Molecule: A Class II Corrector for the Multi-Drug Therapy of Cystic Fibrosis. Biomolecules 2021, 10, 1417. [Google Scholar] [CrossRef] [PubMed]
- Forina, M.; Leardi, R.; Armanino, C.; Lanteri, S. (Eds.) PARVUS: An. Extendable Package of Programs for Data Exploration, Classification and Correlation; Elsevier: Amsterdam, The Netherlands, 1988. [Google Scholar]
- ACD/Percepta Platform; Advanced Chemistry Development, Inc.: Toronto, ON, Canada, 2015. Available online: www.acdlabs.com (accessed on 8 December 2021).
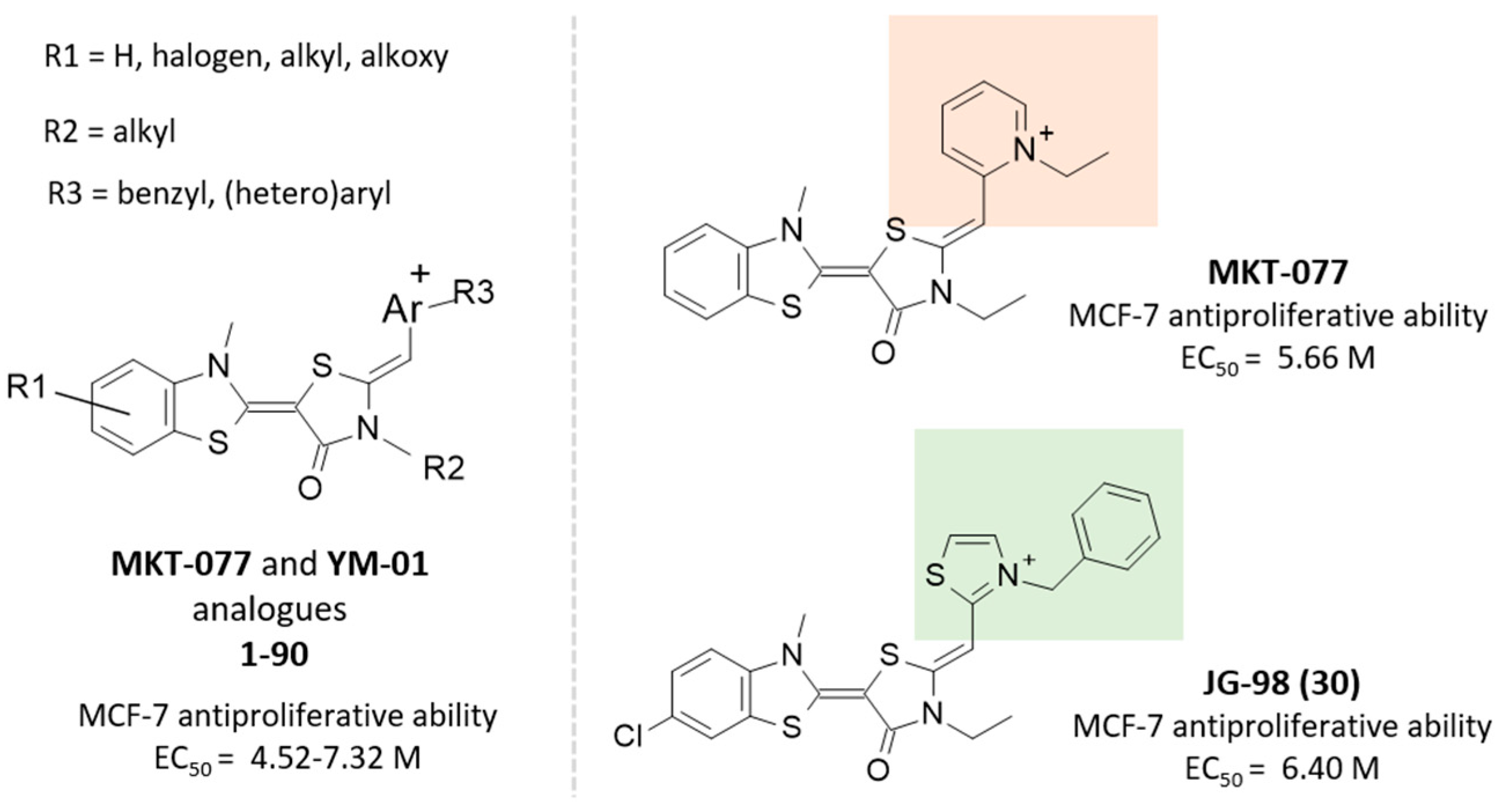

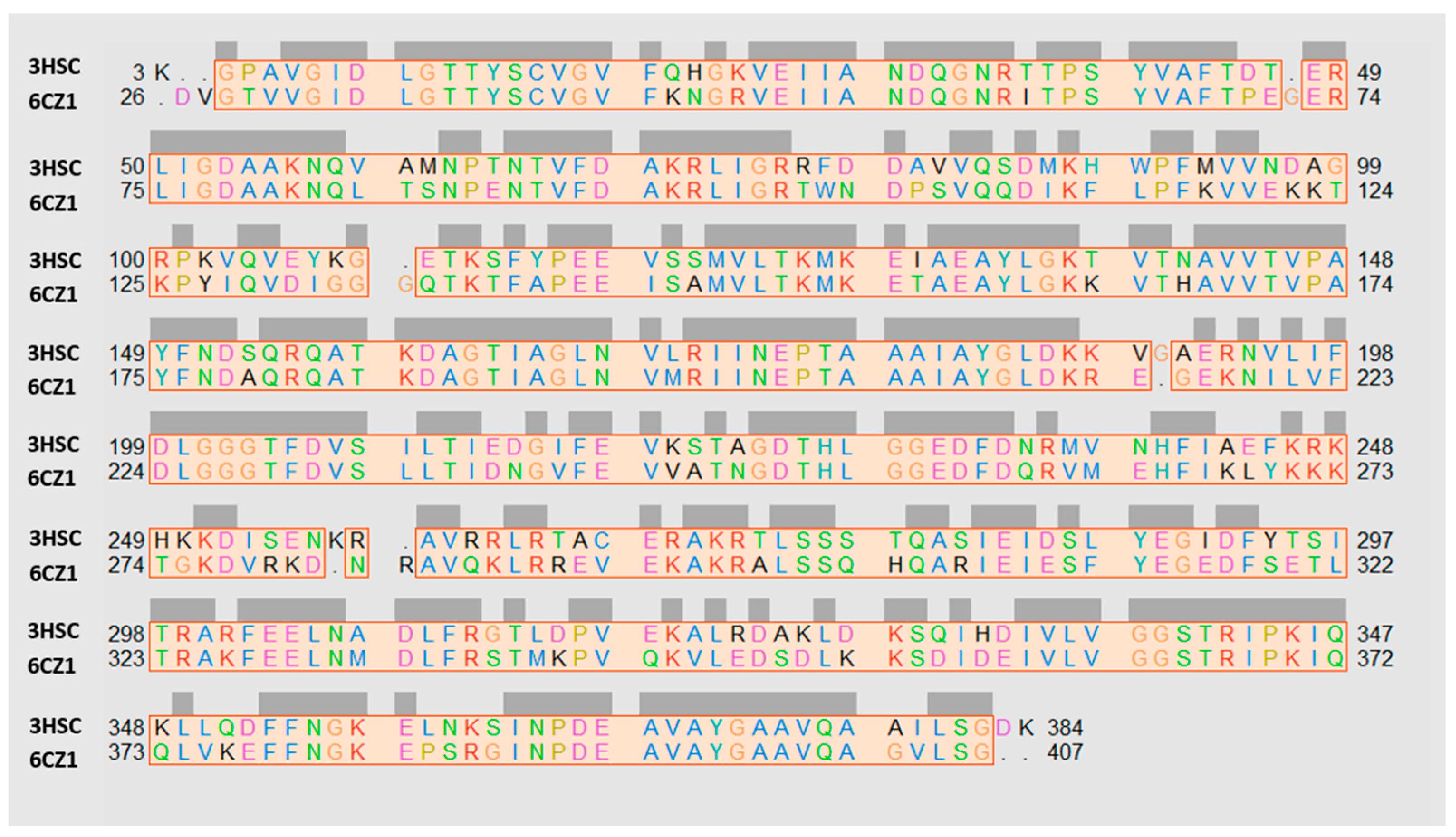
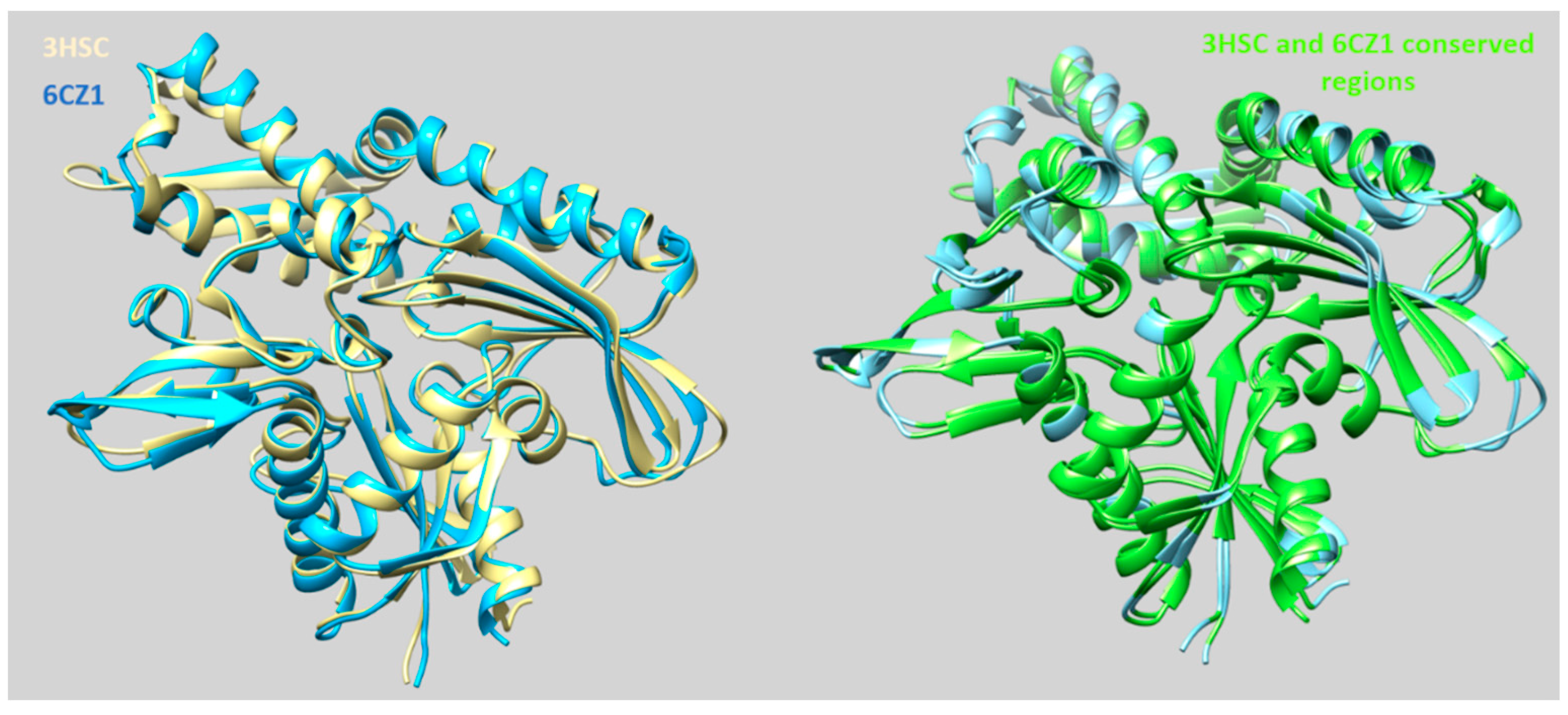

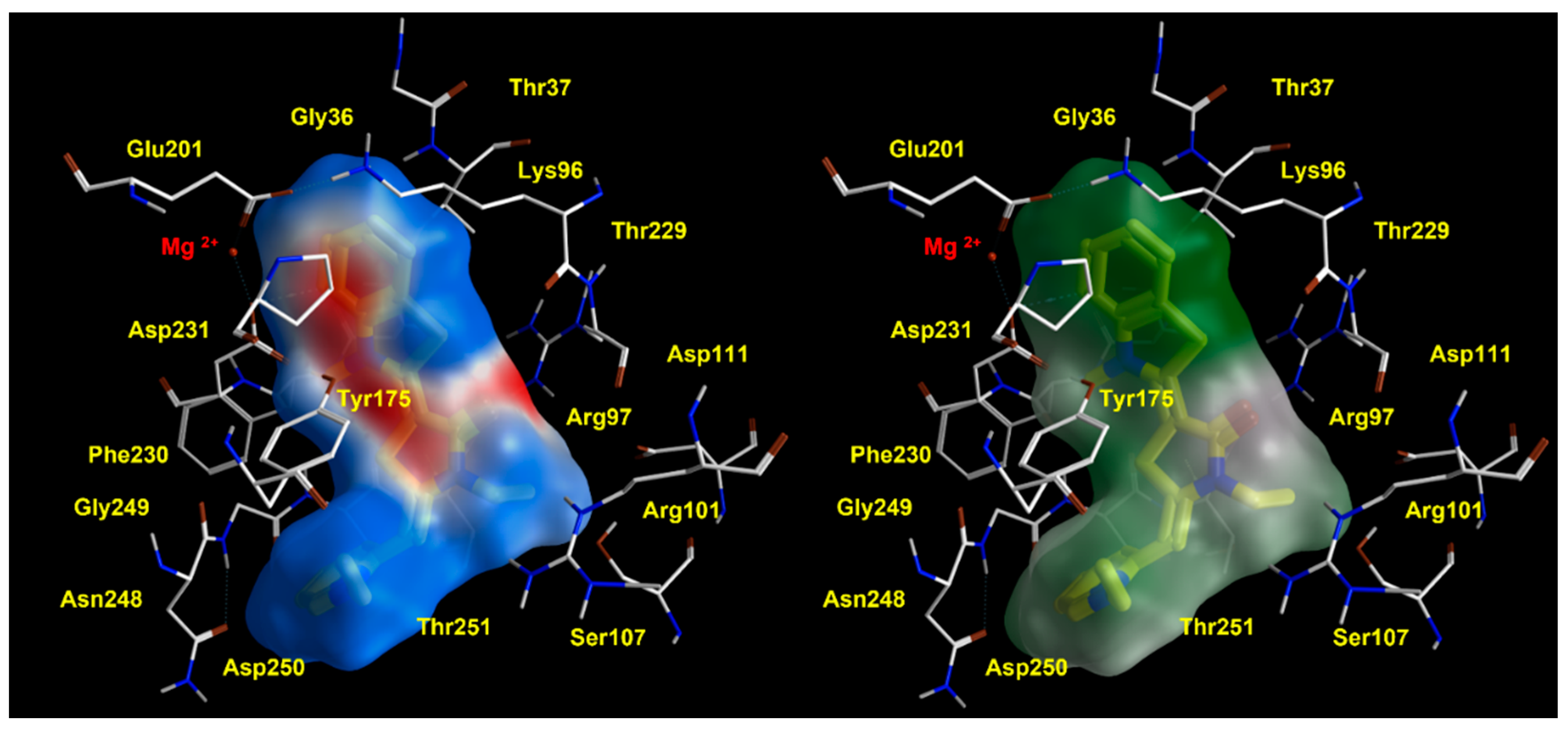
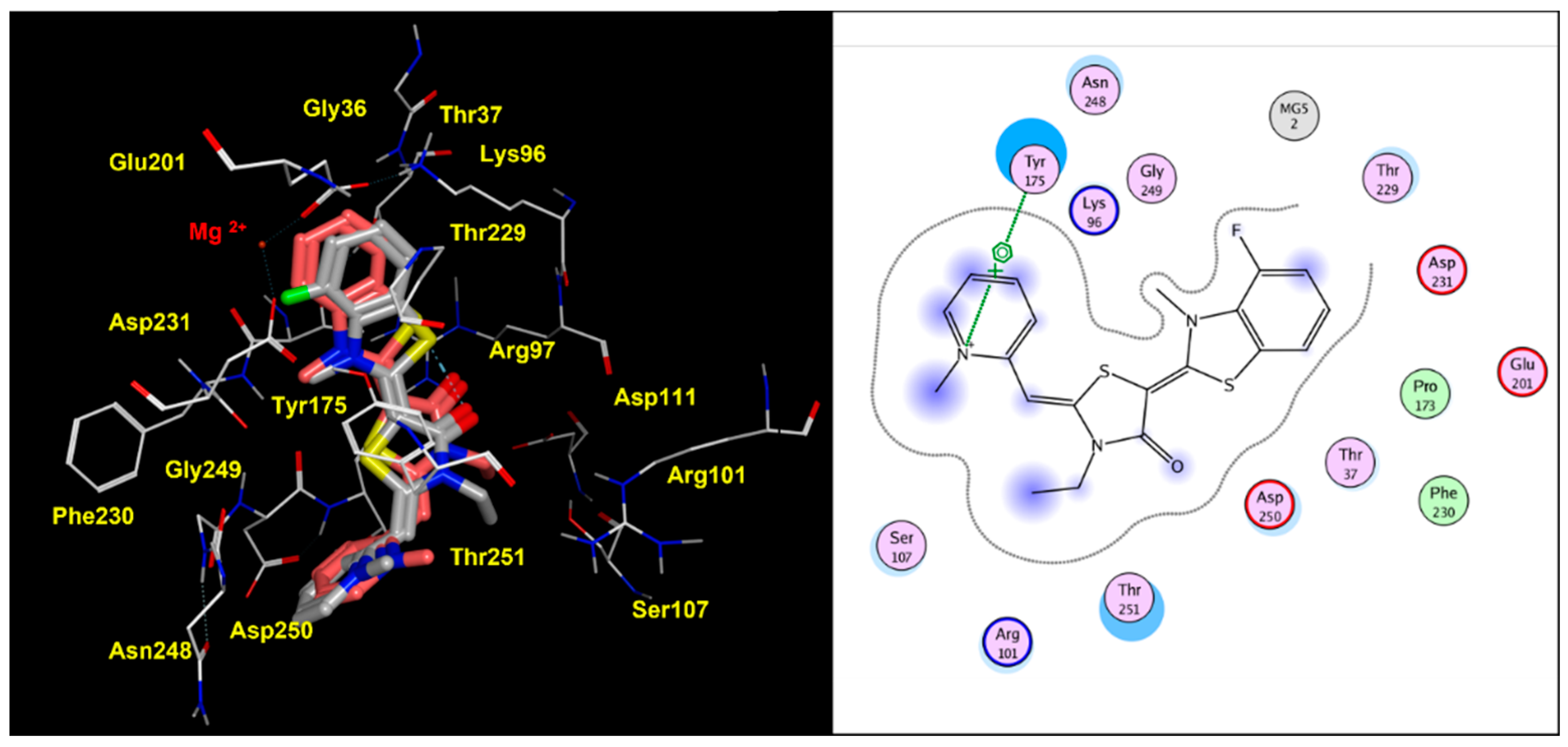
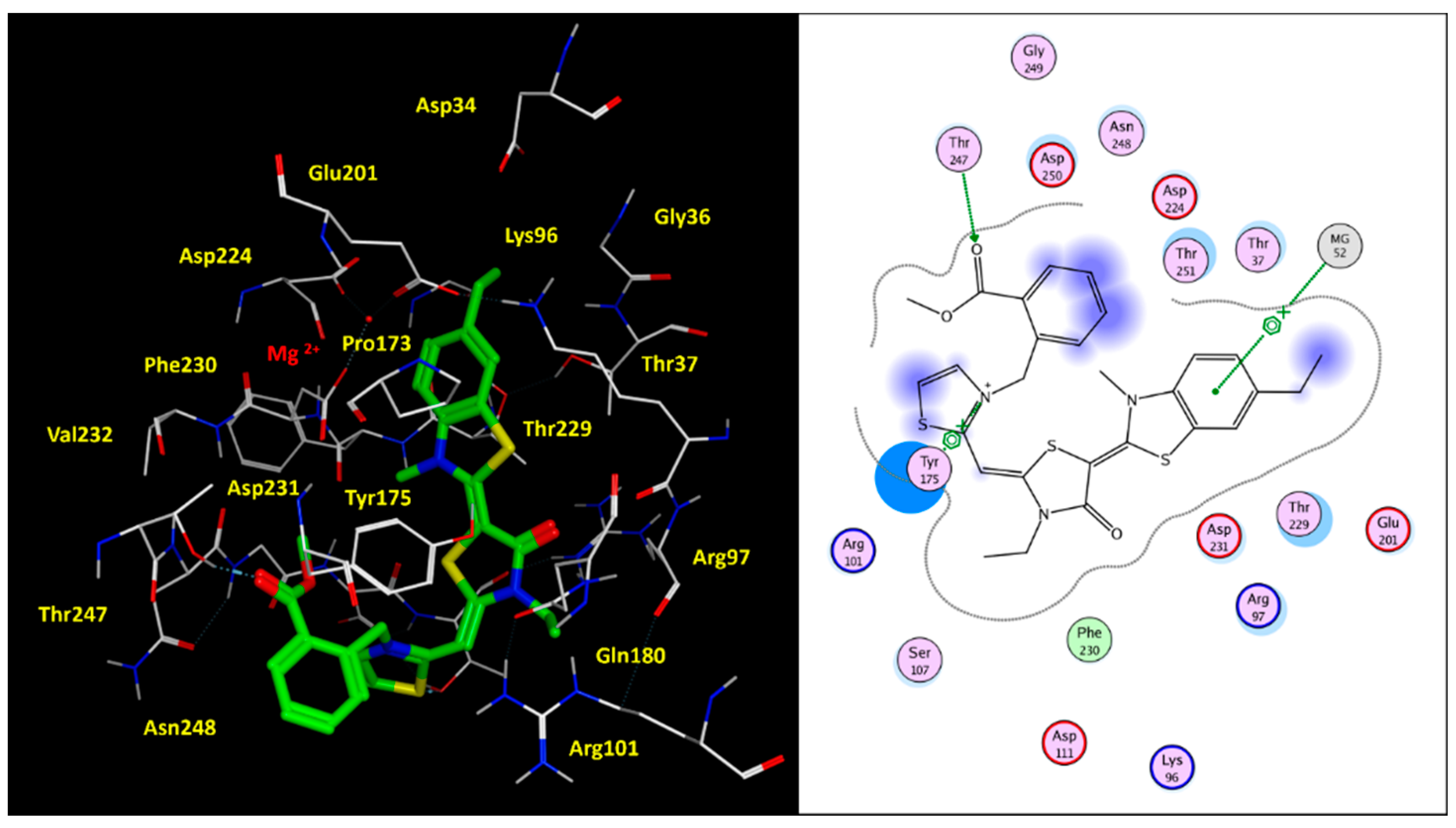
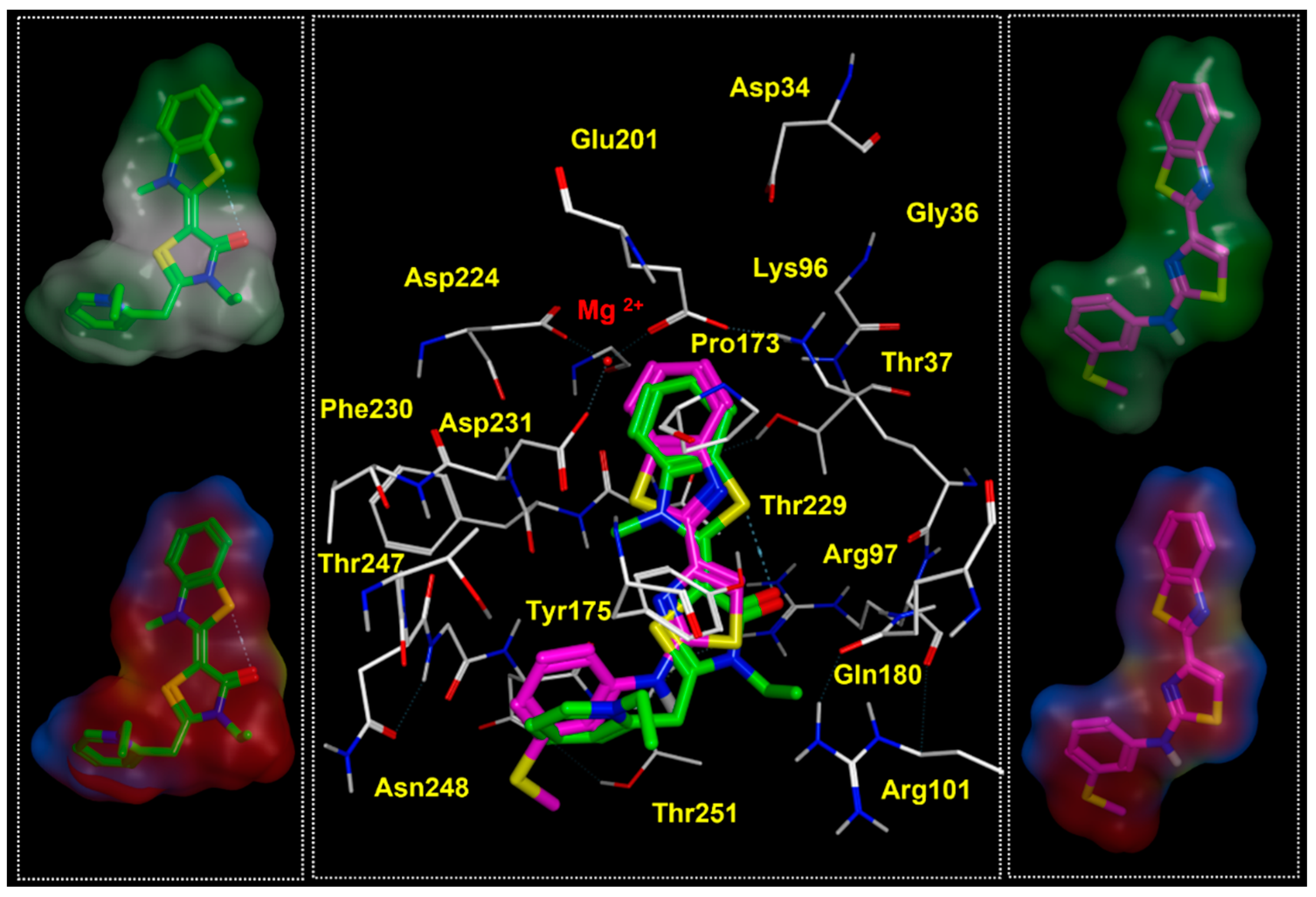
| Protein PDB Code | Binding Site: Pairwise Amino Acids | ||||||
|---|---|---|---|---|---|---|---|
| 3HSC | K71 | R72 | V82 | P147 | Y149 | F150 | T222 |
| 6CZ1 | K96 | R97 | S107 | P173 | Y175 | F176 | T247 |
| 3HSC | A223 | G224 | D225 | T226 | H227 | L228 | |
| 6CZ1 | N248 | G249 | D250 | T251 | H252 | L263 | |
| Descriptor | Type | Series | RI |
|---|---|---|---|
| bpol | Sum of the absolute value of the difference between atomic polarizabilities of all bonded atoms in the molecule (including implicit hydrogens). | 2D-I a | 0.293554 |
| SlogP_VSA1 | Sum of vi, such that Li is in −0.4, −0.2. | 2D-II b | 0.320719 |
| SlogP_VSA3 | Sum of vi, such that Li is in (0,0.1). | 2D-II b | 0.138811 |
| Q_VSA_POS | Total positive van der Waals surface area. This is the sum of the vi, such that qi is non-negative. The vi are calculated using a connection table approximation. | 2D-V | 0.112977 |
| vsa_hyd | Approximation to the sum of VDW surface areas of hydrophobic atoms (Å2). | 2D-VI | 0.289878 |
| E_str | Bond stretch potential energy. | 3D-I | 1.000000 |
| E_strain | Local strain energy: the current energy minus the value of the energy at a near local minimum. | 3D-I | 0.726455 |
| vsurf_DD23 | Contact distances of vsurf_DDmin (three descriptors). | 3D-IV c | 0.148377 |
| MCF-7 Inhibitors | E_Strmean | SlogP_VSA1mean | bpolmean | Q_VSA_POSmean | Vsa_hydmean | pEC50 |
|---|---|---|---|---|---|---|
| Pyridine-based MKT-077 analogues | 18.4938 | 9.0545 | 34.7501 | 167.2119 | 361.5962 | 4.72–6.10 |
| Thiazole-based MKT-077 analogues | 3.5747 | 11.0400 | 42.7463 | 322.0256 | 453.5605 | >6.80 |
| MCF-7 Inhibitors | bpolmean | Q_VSA_POSmean | Vsa_hydmean | pEC50 |
|---|---|---|---|---|
| Five-membered ring-based MKT-077 analogues | 40.9062 | 341.1889 | 436.2678 | 6.92–7.00 |
| Benzyl-containing MKT-077 analogues | 43.2063 | 317.2347 | 457.8837 | 6.82–7.32 |
| Compound | MW a | HBA b | HBD c | nRot bond d | TPSA e | cLogP f |
|---|---|---|---|---|---|---|
| MKT-077 | 396.56 | 1 | 0 | 3 | 27.43 | −0.88 |
| YM-01 | 382.53 | 1 | 0 | 2 | 27.43 | −1.31 |
| JG98 | 499.10 | 1 | 0 | 4 | 27.43 | 2.59 |
| JG231 | 584.03 | 1 | 0 | 4 | 27.43 | 3.04 |
| JG345 | 550.75 | 2 | 0 | 7 | 53.73 | 2.55 |
| Ia | 355.51 | 2 | 1 | 4 | 37.81 | 4.92 |
| IIa | 360.93 | 1 | 1 | 5 | 24.92 | 5.97 |
| IIIa | 367.32 | 1 | 1 | 4 | 24.92 | 5.70 |
| Compound | HIA (%) a | Vd (l/kg) b | %PPB c | LogKa HAS d | %F (Oral) e |
|---|---|---|---|---|---|
| MKT-077 | 2% | 2.8 | 76.62 | 2.79 | 0.0 |
| YM-01 | 3% | 2.4 | 73.28 | 2.75 | 0.0 |
| JG98 | 51 | 4.7 | 97.96 | 3.75 | 0.0 |
| JG231 | 74 | 5.1 | 98.10 | 3.61 | 0.0 |
| JG345 | 49 | 4.1 | 96.86 | 3.44 | 0.0 |
| Ia | 100 | 4.3 | 99.59 | 4.91 | 83.9 |
| IIa | 100 | 4.8 | 99.68 | 5.01 | 62.2 |
| IIIa | 100 | 4.2 | 99.74 | 5.23 | 38.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabbadini, R.; Pesce, E.; Parodi, A.; Mustorgi, E.; Bruzzone, S.; Pedemonte, N.; Casale, M.; Millo, E.; Cichero, E. Probing Allosteric Hsp70 Inhibitors by Molecular Modelling Studies to Expedite the Development of Novel Combined F508del CFTR Modulators. Pharmaceuticals 2021, 14, 1296. https://doi.org/10.3390/ph14121296
Sabbadini R, Pesce E, Parodi A, Mustorgi E, Bruzzone S, Pedemonte N, Casale M, Millo E, Cichero E. Probing Allosteric Hsp70 Inhibitors by Molecular Modelling Studies to Expedite the Development of Novel Combined F508del CFTR Modulators. Pharmaceuticals. 2021; 14(12):1296. https://doi.org/10.3390/ph14121296
Chicago/Turabian StyleSabbadini, Roberto, Emanuela Pesce, Alice Parodi, Eleonora Mustorgi, Santina Bruzzone, Nicoletta Pedemonte, Monica Casale, Enrico Millo, and Elena Cichero. 2021. "Probing Allosteric Hsp70 Inhibitors by Molecular Modelling Studies to Expedite the Development of Novel Combined F508del CFTR Modulators" Pharmaceuticals 14, no. 12: 1296. https://doi.org/10.3390/ph14121296
APA StyleSabbadini, R., Pesce, E., Parodi, A., Mustorgi, E., Bruzzone, S., Pedemonte, N., Casale, M., Millo, E., & Cichero, E. (2021). Probing Allosteric Hsp70 Inhibitors by Molecular Modelling Studies to Expedite the Development of Novel Combined F508del CFTR Modulators. Pharmaceuticals, 14(12), 1296. https://doi.org/10.3390/ph14121296









