Characterization of LDD-2633 as a Novel RET Kinase Inhibitor with Anti-Tumor Effects in Thyroid Cancer
Abstract
1. Introduction
2. Results
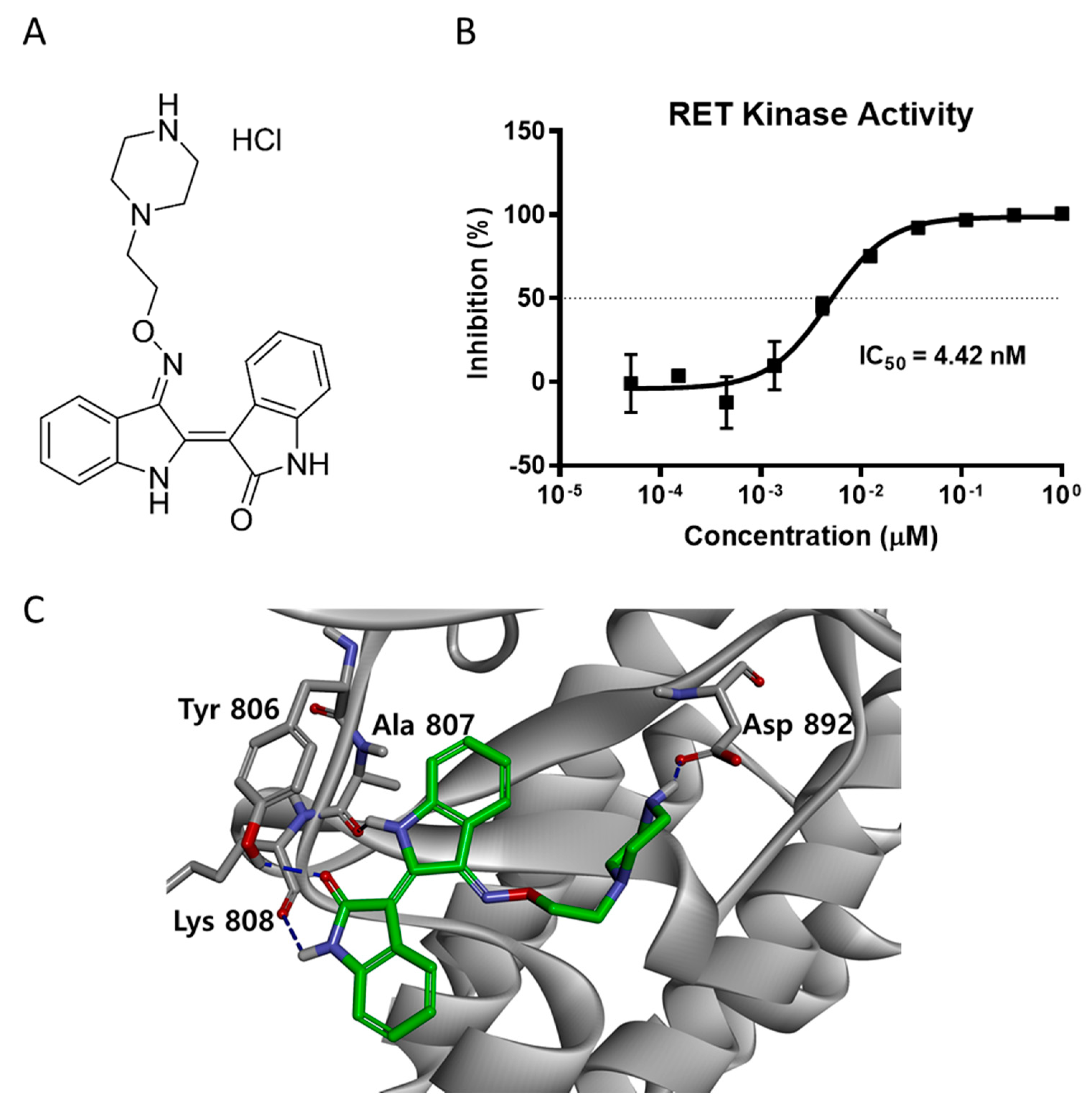
2.1. LDD-2633 Inhibits the Kinase Activity of RET In Vitro
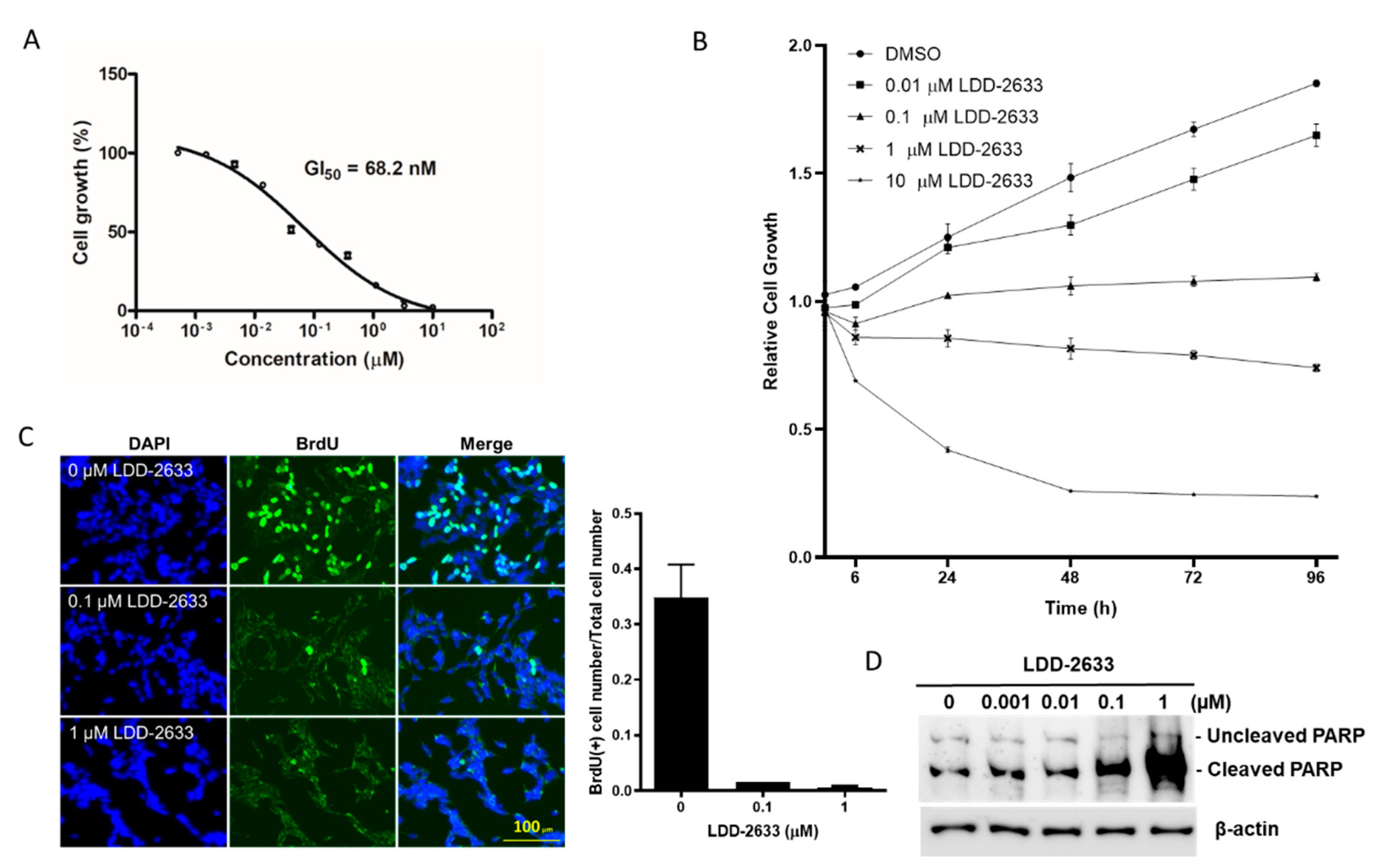
2.2. LDD-2633 Suppresses the Growth of TT MTC Cells
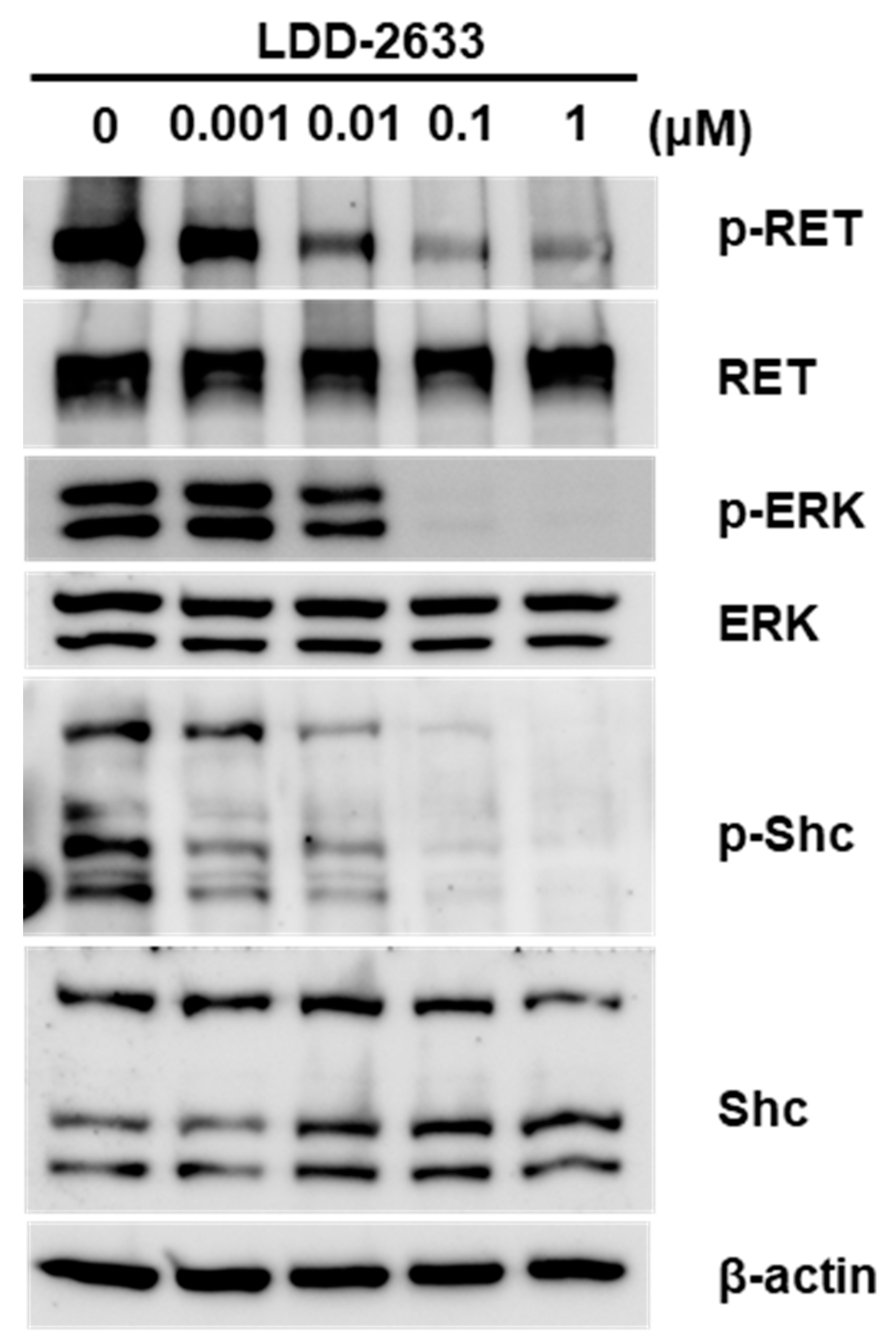
2.3. LDD-2633 Suppresses RET Signaling Pathway
2.4. LDD-2633 Exerts Anti-Tumor Effects In Vivo in TT Cell Xenografts
3. Discussion
4. Materials and Methods
4.1. Compounds
4.2. Cell Culture
4.3. Cell Viability Assay and BrdU Incorporation Assay
4.4. Molecular Modeling
4.5. In Vitro Kinase Assay
4.6. Immunoblotting Analysis
4.7. Mouse Tumor Xenograft
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tuttle, R.M.; Ball, D.W.; Byrd, D.; Dilawari, R.A.; Doherty, G.M.; Duh, Q.Y.; Ehya, H.; Farrar, W.B.; Haddad, R.I.; Kandeel, F.; et al. Wirth, and Network National Comprehensive Cancer—“Thyroid Carcinoma”. J. Natl. Compr. Cancer Netw. 2010, 8, 1228–1274. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Hu, Z.I.; Lai, G.G.Y.; Tan, D.S.W. Targeting RET-driven cancers: Lessons from evolving preclinical and clinical landscapes. Nat. Rev. Clin. Oncol. 2017, 15, 151–167. [Google Scholar] [CrossRef]
- DeVita, V.T.; Lawrence, T.S.; Rosenberg, S.A. (Eds.) Devita, Hellman, and Rosenberg’s Cancer: Principles & Practice of Oncology; Wolters Kluwer: Philadelphia, PA, USA, 2015. [Google Scholar]
- Durante, C.; Haddy, N.; Baudin, E.; Leboulleux, S.; Hartl, D.; Travagli, J.P.; Caillou, B.; Ricard, M.; Lumbroso, J.D.; De Vathaire, F.; et al. Long-Term Outcome of 444 Patients with Distant Metastases from Papillary and Follicular Thyroid Carcinoma: Benefits and Limits of Radioiodine Therapy. J. Clin. Endocrinol. Metab. 2006, 91, 2892–2899. [Google Scholar] [CrossRef] [PubMed]
- Roman, S.; Lin, R.; Sosa, J.A. Prognosis of Medullary Thyroid Carcinoma: Demographic, Clinical, and Pathologic Predictors of Survival in 1252 Cases. Cancer 2006, 107, 2134–2142. [Google Scholar] [CrossRef]
- Romei, C.; Ciampi, R.; Elisei, C.R.R.C.R. A comprehensive overview of the role of the RET proto-oncogene in thyroid carcinoma. Nat. Rev. Endocrinol. 2016, 12, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Ichikawa, H.; Totoki, Y.; Yasuda, K.; Hiramoto, M.; Nammo, T.; Sakamoto, H.; Tsuta, K.; Furuta, K.; Shimada, Y.; et al. KIF5B-RET fusions in lung adenocarcinoma. Nat. Med. 2012, 18, 375–377. [Google Scholar] [CrossRef]
- Nikiforov, Y.E.; Rowland, J.M.; Bove, K.E.; Monforte-Munoz, H.; Fagin, J.A. Distinct pattern of ret oncogene rearrangements in morphological variants of radiation-induced and sporadic thyroid papillary carcinomas in children. Cancer Res. 1997, 57, 1690–1694. [Google Scholar]
- Nikiforov, Y.E. RET/PTC Rearrangement in Thyroid Tumors. Endocr. Pathol. 2002, 13, 03–16. [Google Scholar] [CrossRef] [PubMed]
- Li, G.G.; Somwar, R.; Joseph, J.; Smith, R.S.; Hayashi, T.; Martin, L.; Franovic, A.; Schairer, A.; Martin, E.; Riely, G.J.; et al. Antitumor Activity of RXDX-105 in Multiple Cancer Types with RET Rearrangements or Mutations. Clin. Cancer Res. 2017, 23, 2981–2990. [Google Scholar] [CrossRef]
- Subbiah, V.; Cote, G.J. Advances in Targeting RET-Dependent Cancers. Cancer Discov. 2020, 10, 498–505. [Google Scholar] [CrossRef]
- Leclerc, S.; Garnier, M.; Hoessel, R.; Marko, D.; Bibb, J.A.; Snyder, G.L.; Greengard, P.; Biernat, J.; Wu, Y.Z.; Mandelkow, E.M.; et al. Indirubins Inhibit Glycogen Synthase Kinase-3 Beta and Cdk5/P25, Two Protein Kinases Involved in Abnormal Tau Phosphorylation in Alzheimer’s Disease. A Property Common to Most Cyclin-Dependent Kinase Inhibitors? J. Biol. Chem. 2001, 276, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Hoessel, R.; Leclerc, S.; Endicott, J.A.; Nobel, M.E.M.; Lawrie, A.; Tunnah, P.; Leost, M.; Damiens, E.; Marie, D.; Marko, D.; et al. Indirubin, the active constituent of a Chinese antileukaemia medicine, inhibits cyclin-dependent kinases. Nat. Cell Biol. 1999, 1, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Myrianthopoulos, V.; Kritsanida, M.; Gaboriaud-Kolar, N.; Magiatis, P.; Ferandin, Y.; Durieu, E.; Lozach, O.; Cappel, D.; Soundararajan, M.; Filippakopoulos, P.; et al. Novel Inverse Binding Mode of Indirubin Derivatives Yields Improved Selectivity for DYRK Kinases. ACS Med. Chem. Lett. 2012, 4, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Ndolo, K.M.; Park, K.R.; Lee, H.J.; Bin Yoon, K.; Kim, Y.-C.; Han, S.-Y. Characterization of the Indirubin Derivative LDD970 as a Small Molecule Aurora Kinase A Inhibitor in Human Colorectal Cancer Cells. Immune Netw. 2017, 17, 110–115. [Google Scholar] [CrossRef]
- Choi, S.-J.; Lee, J.E.; Jeong, S.-Y.; Im, I.; Lee, S.-D.; Lee, E.-J.; Lee, S.K.; Kwon, S.-M.; Ahn, S.-G.; Yoon, J.-H.; et al. 5,5′-Substituted Indirubin-3′-oxime Derivatives as Potent Cyclin-Dependent Kinase Inhibitors with Anticancer Activity. J. Med. Chem. 2010, 53, 3696–3706. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, J.; Jeong, P.; Choi, J.; Baek, J.; Ahn, S.J.; Moon, Y.; Heo, J.D.; Choi, Y.H.; Chin, Y.W.; et al. Discovery of a Flt3 Inhibitor Ldd1937 as an Anti-Leukemic Agent for Acute Myeloid Leukemia. Oncotarget 2018, 9, 924–936. [Google Scholar] [CrossRef]
- Mologni, L.; Rostagno, R.; Brussolo, S.; Knowles, P.P.; Kjaer, S.; Murray-Rust, J.; Rosso, E.; Zambon, A.; Scapozza, L.; McDonald, N.Q.; et al. Synthesis, structure–activity relationship and crystallographic studies of 3-substituted indolin-2-one RET inhibitors. Bioorganic Med. Chem. 2010, 18, 1482–1496. [Google Scholar] [CrossRef]
- Knowles, P.P.; Murray-Rust, J.; Kjaer, S.; Scott, R.P.; Hanrahan, S.; Santoro, M.; Ibáñez, C.F.; McDonald, N.Q. Structure and Chemical Inhibition of the RET Tyrosine Kinase Domain. J. Biol. Chem. 2006, 281, 33577–33587. [Google Scholar] [CrossRef]
- Carlomagno, F.; Salvatore, D.; Santoro, M.; De Franciscis, V.; Quadro, L.; Panariello, L.; Colantuoni, V.; Fusco, A. Point mutation of the RET proto-oncogene in the TT human medullary thyroid carcinoma cell line. Biochem. Biophys. Res. Commun. 1995, 207, 1022–1028. [Google Scholar] [CrossRef]
- Nishioka, C.; Ikezoe, T.; Takeshita, A.; Yang, J.; Tasaka, T.; Yang, Y.; Kuwayama, Y.; Komatsu, N.; Togitani, K.; Koeffler, H.P.; et al. ZD6474 induces growth arrest and apoptosis of human leukemia cells, which is enhanced by concomitant use of a novel MEK inhibitor, AZD6244. Leukemia 2007, 21, 1308–1310. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.-W.; Wang, A.-N.; Liao, H.-A.; Chen, C.-Y.; Hou, H.-A.; Hu, C.-Y.; Tien, H.-F.; Ou, D.-L.; Lin, L.-I. Cabozantinib is selectively cytotoxic in acute myeloid leukemia cells with FLT3-internal tandem duplication (FLT3-ITD). Cancer Lett. 2016, 376, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ivaturi, V.; Sabato, P.; Gobburu, J.V.S.; Greer, J.M.; Wright, J.J.; Smith, B.D.; Pratz, K.W.; Rudek, M.A.; on behalf of the ETCTN-6745 study team. Sorafenib Dose Recommendation in Acute Myeloid Leukemia Based on Exposure-FLT3 Relationship. Clin. Transl. Sci. 2018, 11, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Jeong, P.; Moon, Y.; Lee, J.-H.; Lee, S.-D.; Park, J.; Lee, J.; Kim, J.; Lee, H.J.; Kim, N.Y.; Choi, J.; et al. Discovery of orally active indirubin-3′-oxime derivatives as potent type 1 FLT3 inhibitors for acute myeloid leukemia. Eur. J. Med. Chem. 2020, 195, 112205. [Google Scholar] [CrossRef] [PubMed]
- Markham, A. Pralsetinib: First Approval. Drugs 2020, 80, 1865–1870. [Google Scholar] [CrossRef] [PubMed]
- Looney, A.-M.; Nawaz, K.; Webster, R.M. Tumour-agnostic therapies. Nat. Rev. Drug Discov. 2020, 19, 383–384. [Google Scholar] [CrossRef] [PubMed]
- Ndolo, K.M.; An, S.J.; Park, K.R.; Lee, H.J.; Bin Yoon, K.; Kim, Y.-C.; Han, S.-Y. Discovery of an Indirubin Derivative as a Novel c-Met Kinase Inhibitor with In Vitro Anti-Tumor Effects. Biomol. Ther. 2019, 27, 216–221. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.J.; Jeong, P.; Moon, Y.; Choi, J.; Heo, J.D.; Kim, Y.-C.; Han, S.-Y. Characterization of LDD-2633 as a Novel RET Kinase Inhibitor with Anti-Tumor Effects in Thyroid Cancer. Pharmaceuticals 2021, 14, 38. https://doi.org/10.3390/ph14010038
Lee HJ, Jeong P, Moon Y, Choi J, Heo JD, Kim Y-C, Han S-Y. Characterization of LDD-2633 as a Novel RET Kinase Inhibitor with Anti-Tumor Effects in Thyroid Cancer. Pharmaceuticals. 2021; 14(1):38. https://doi.org/10.3390/ph14010038
Chicago/Turabian StyleLee, Hyo Jeong, Pyeonghwa Jeong, Yeongyu Moon, Jungil Choi, Jeong Doo Heo, Yong-Chul Kim, and Sun-Young Han. 2021. "Characterization of LDD-2633 as a Novel RET Kinase Inhibitor with Anti-Tumor Effects in Thyroid Cancer" Pharmaceuticals 14, no. 1: 38. https://doi.org/10.3390/ph14010038
APA StyleLee, H. J., Jeong, P., Moon, Y., Choi, J., Heo, J. D., Kim, Y.-C., & Han, S.-Y. (2021). Characterization of LDD-2633 as a Novel RET Kinase Inhibitor with Anti-Tumor Effects in Thyroid Cancer. Pharmaceuticals, 14(1), 38. https://doi.org/10.3390/ph14010038




