Individual and Combined Effects of Engineered Peptides and Antibiotics on Pseudomonas aeruginosa Biofilms
Abstract
:1. Introduction
2. Materials and Methods
2.1 Strains, Media and Chemicals
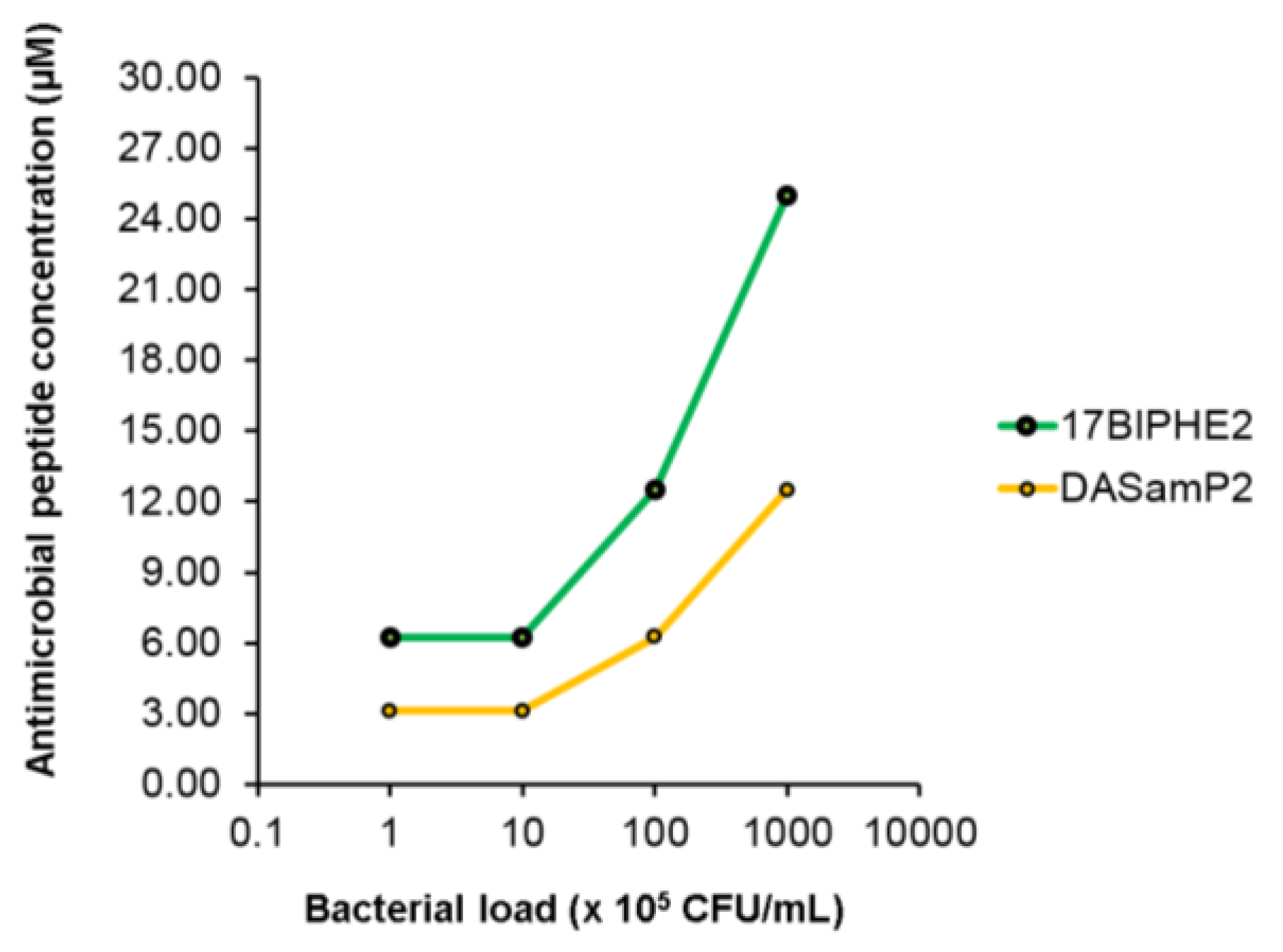
2.2. Measurement of the Minimal Inhibitory Concentration (MIC)
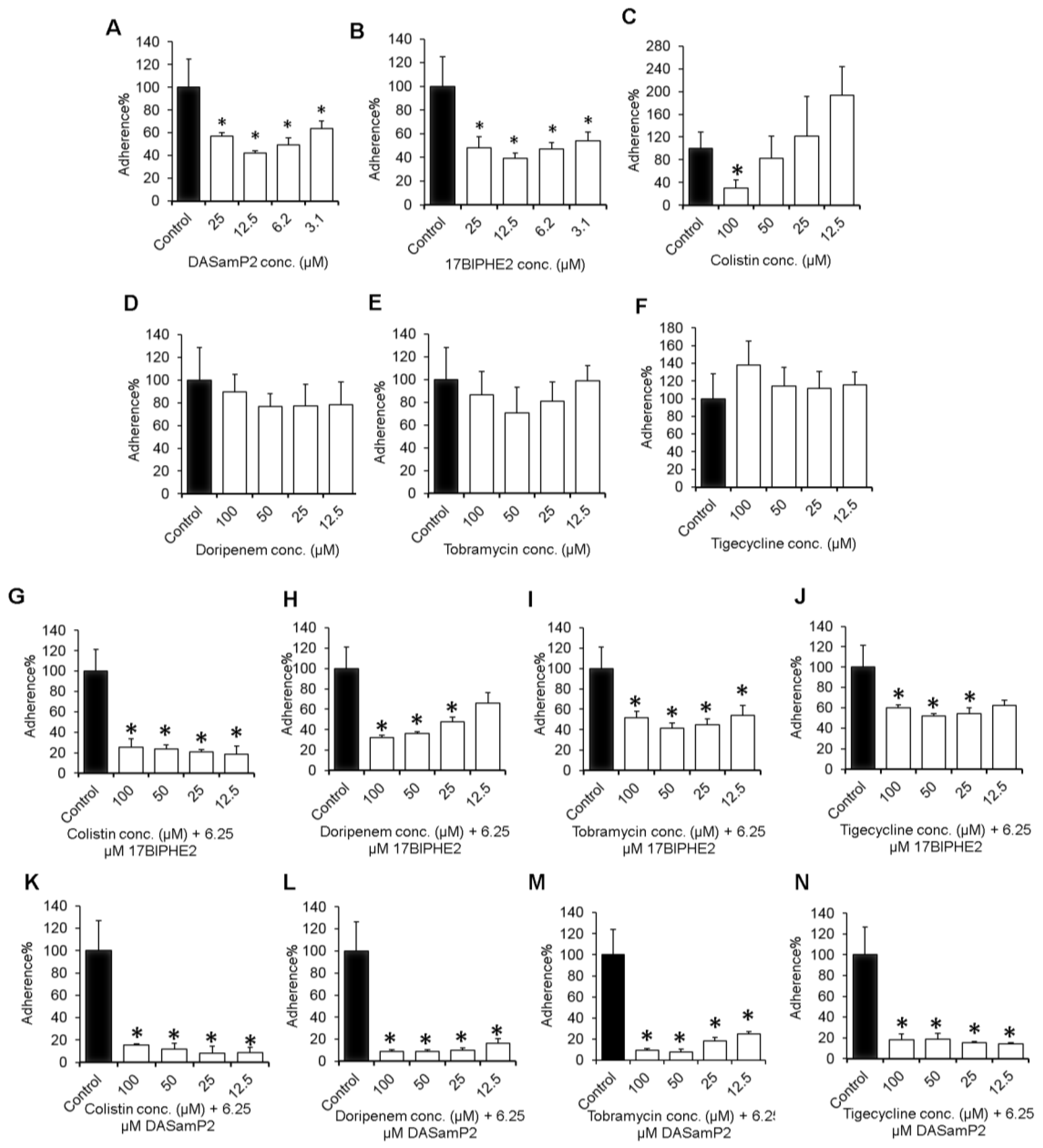
2.3. Effects on Initial Adhesion of Bacteria
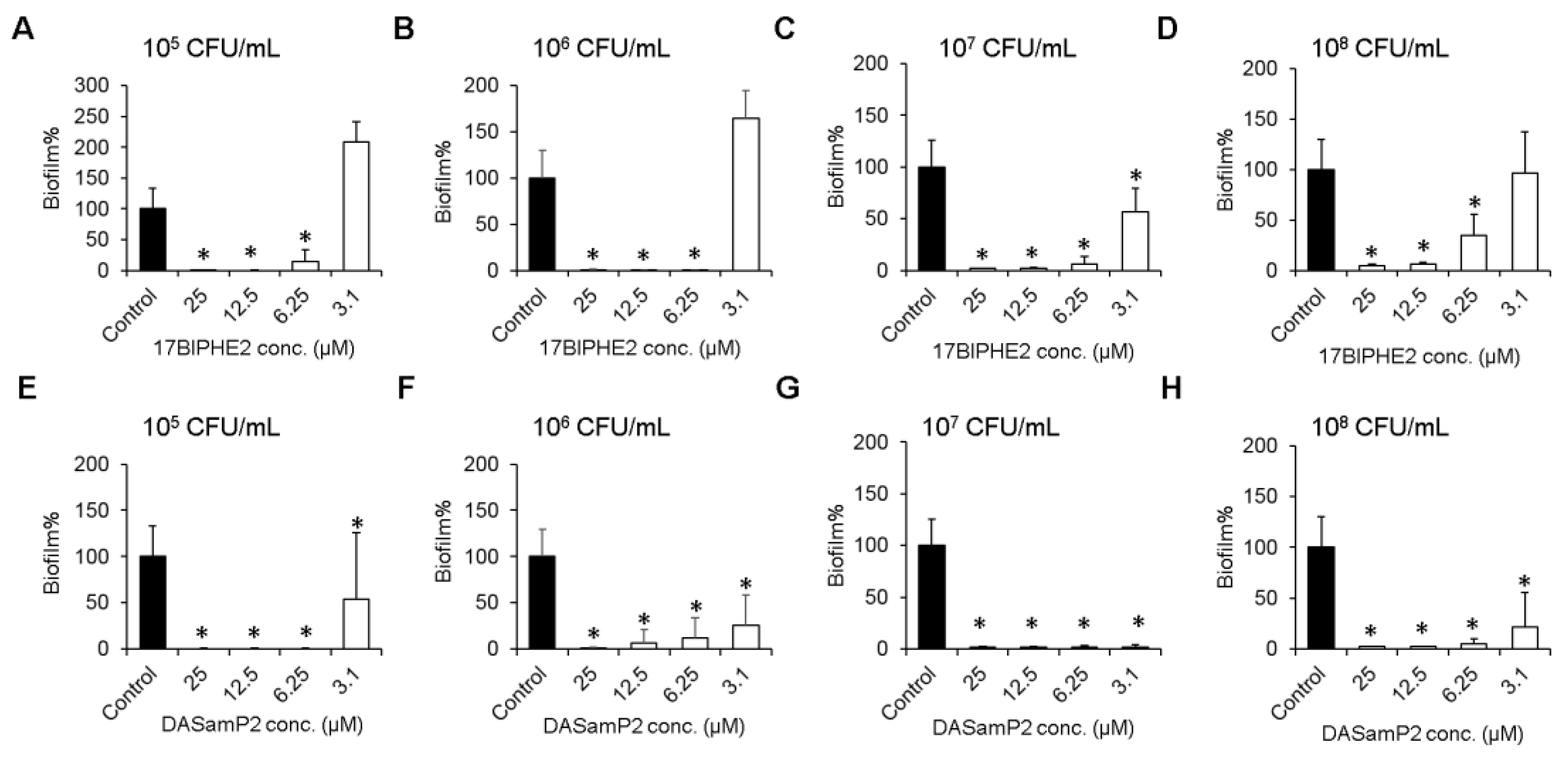
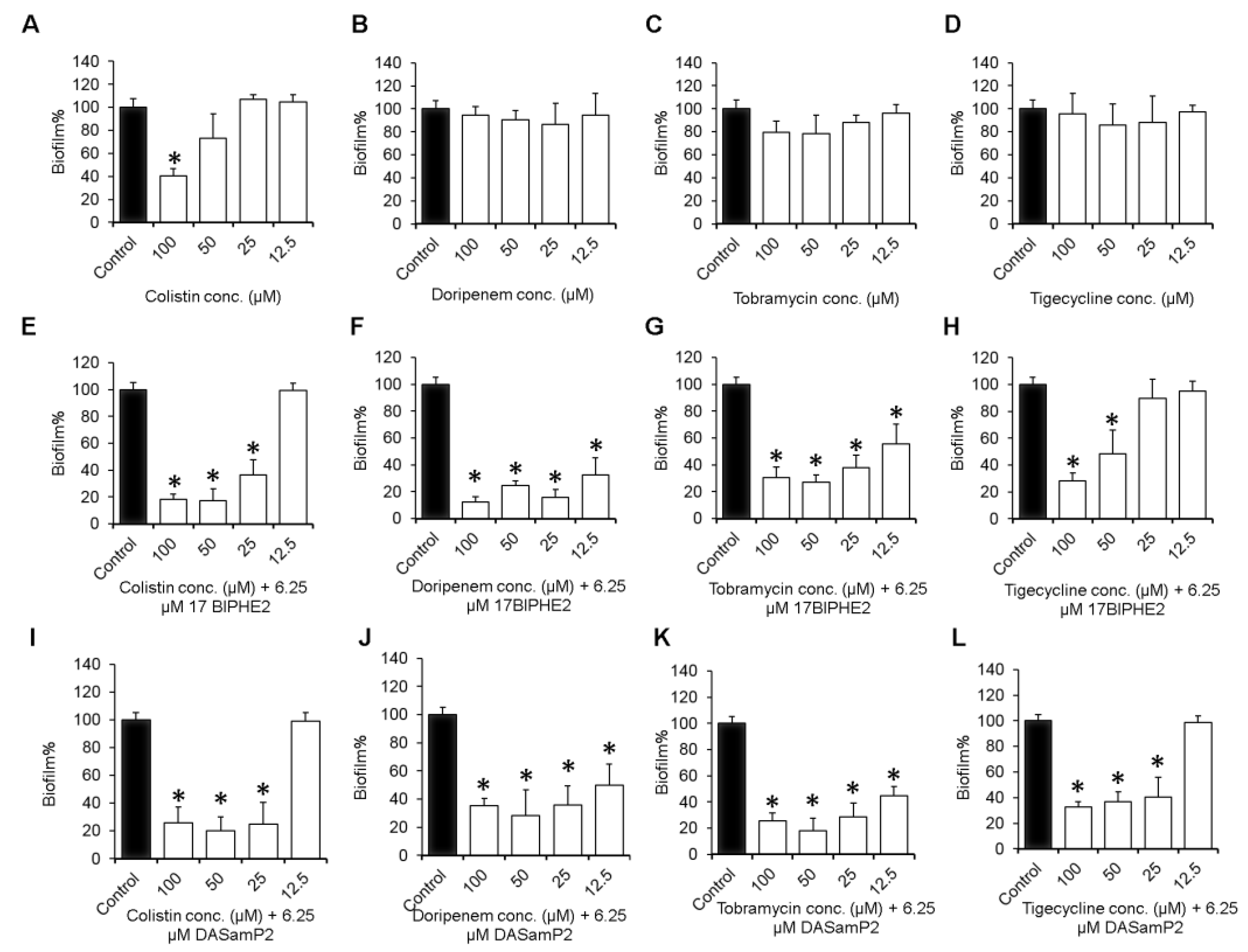
2.4. Inhibition of Biofilm Formation
2.5. Disruption of Established Biofilms
2.6. Immature and Mature Biofilm Disruption
2.7. Live and Dead Staining Assays of Established Biofilms by Confocal Laser Scanning Microscopy
3. Results
3.1. Antimicrobial Activity against Planktonic P. aeruginosa
3.2. Influence on Initial Adhesion of P. aeruginosa
3.3. Effects on Biofilm Growth of P. aeruginosa
3.4. Impact on the Established Biofilms of P. aeruginosa
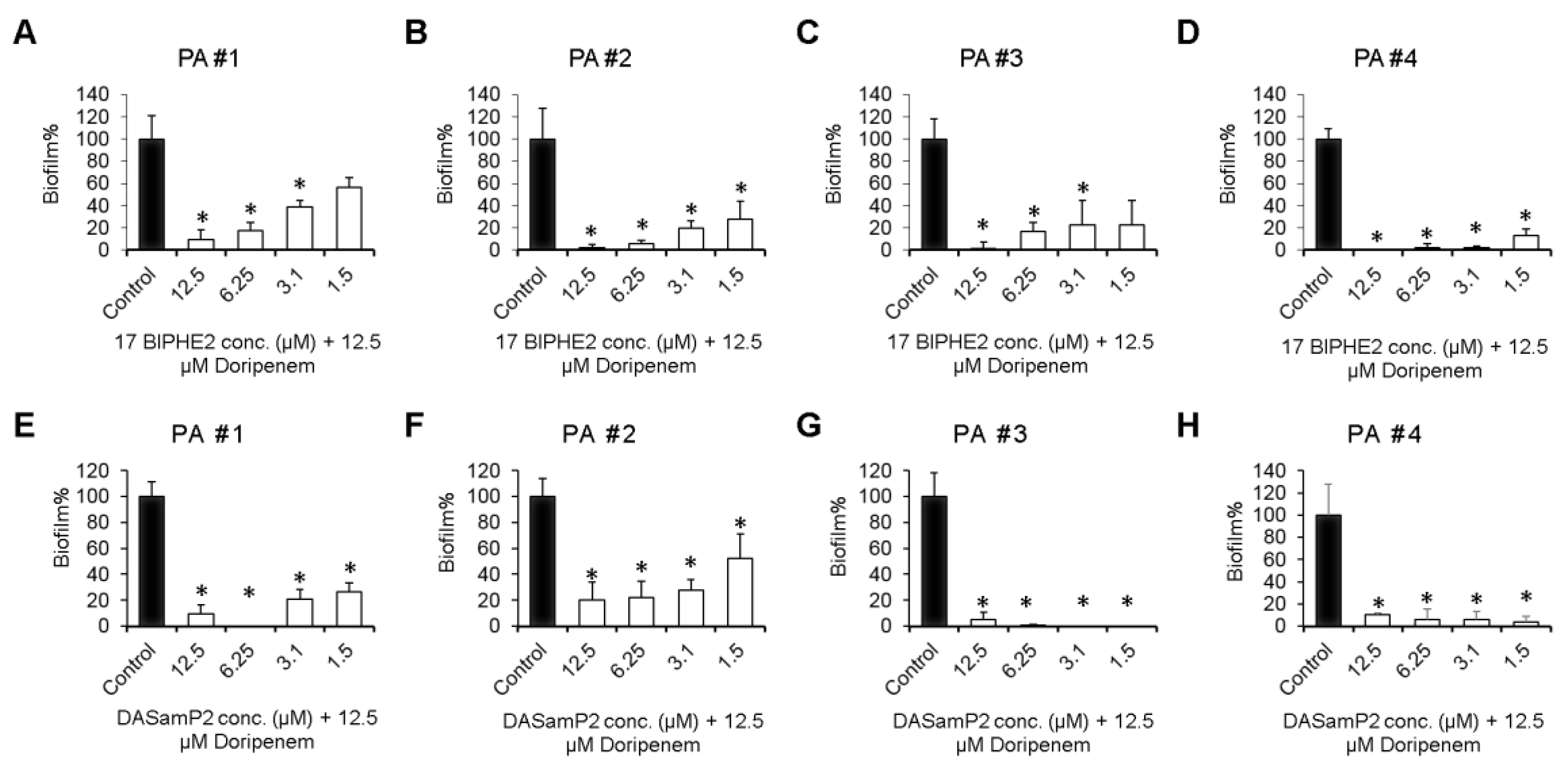
3.5. Disruption of the Preformed Biofilms of P. aeruginosa Clinical Isolates
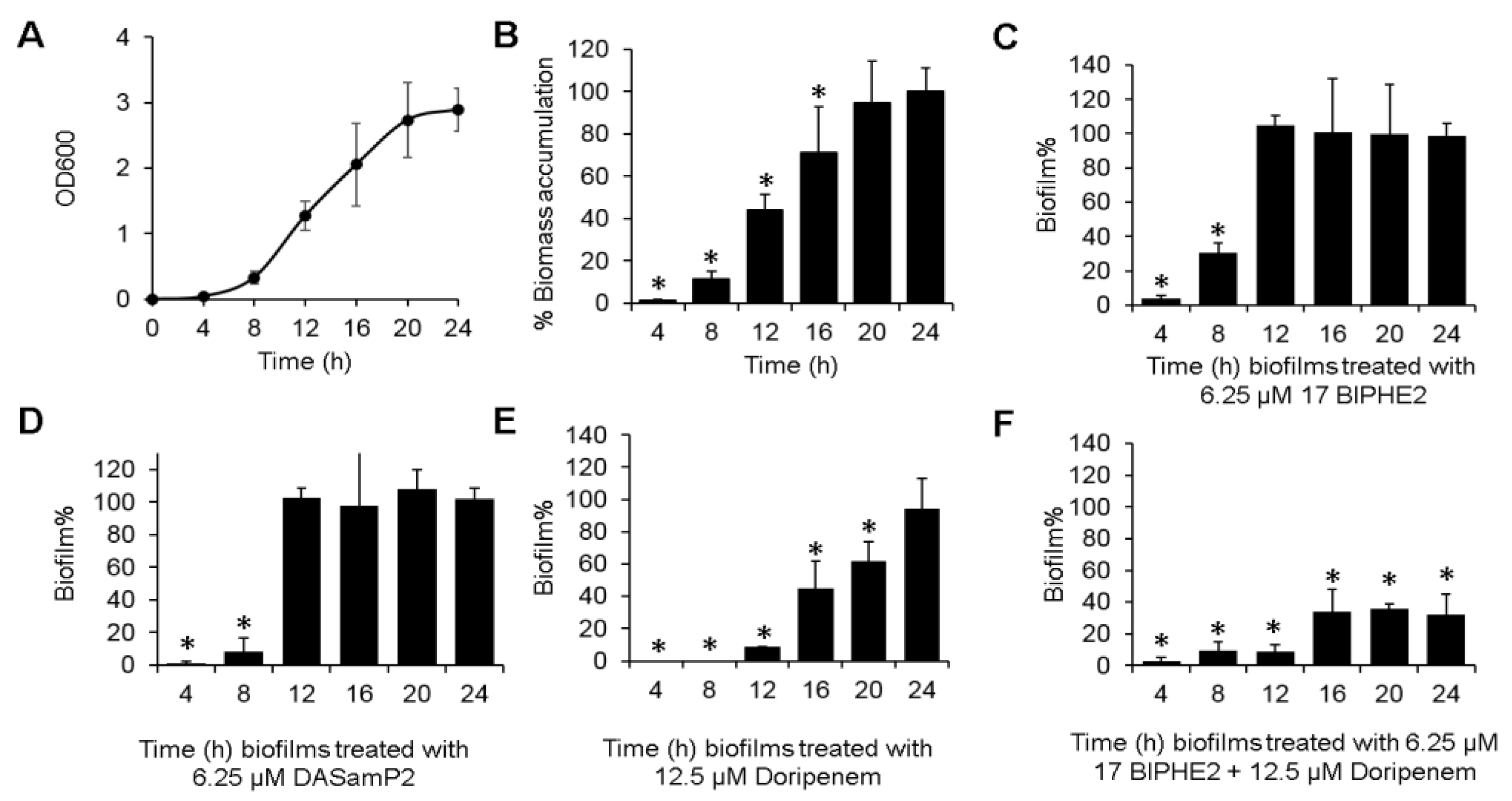
3.6. Time-dependent Biofilm Formation and Treatment Methods
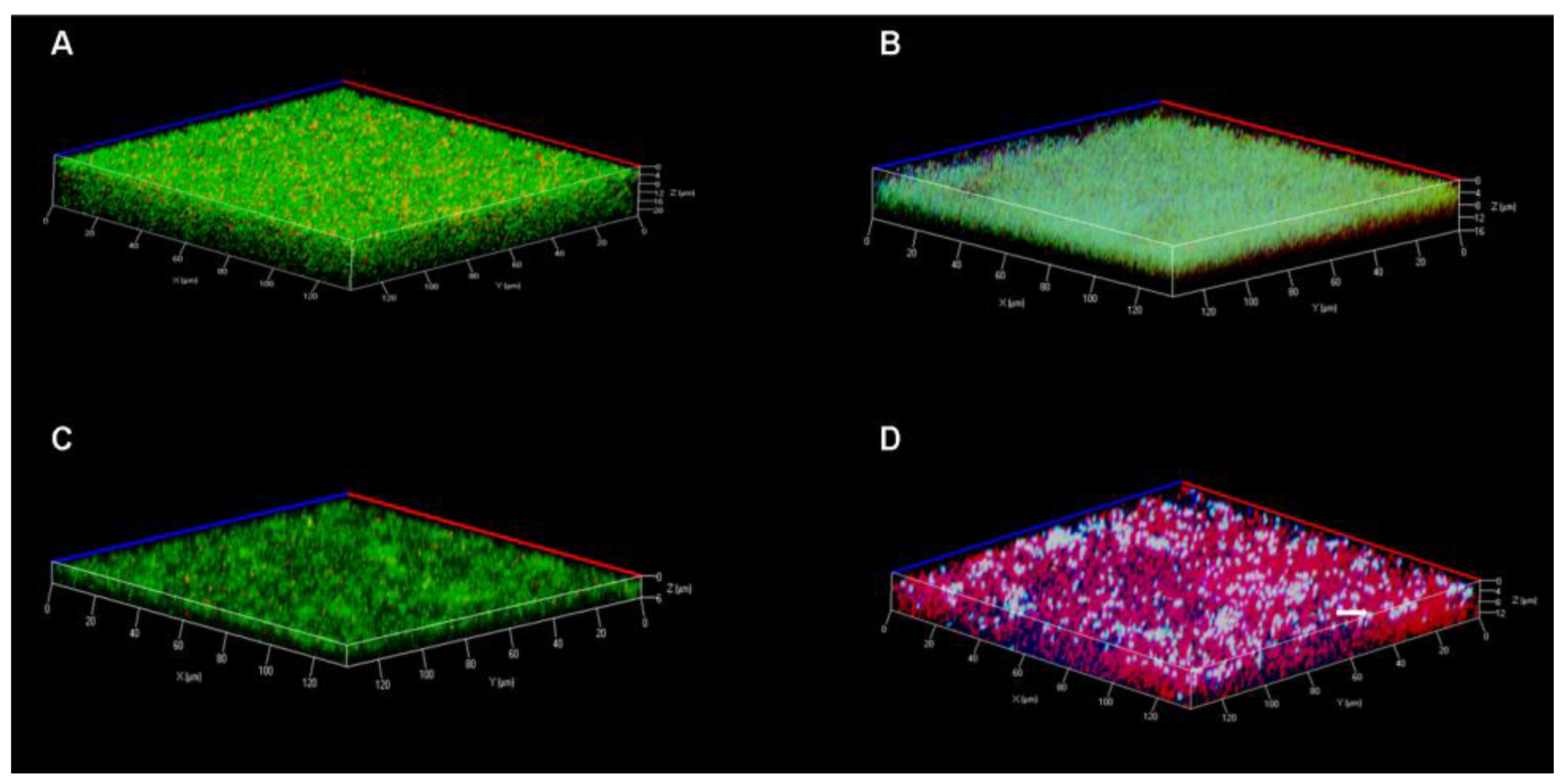
3.7. Confocal Laser Scanning Microscopy
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Park, S.C.; Park, Y.; Hahm, K.S. The role of antimicrobial peptides in preventing multidrug-resistant bacterial infections and biofilm formation. Int. J. Mol. Sci. 2011, 12, 5971–5992. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.J.; Records, A.R.; Orr, M.W.; Linden, S.B.; Lee, V.T. Catheter-associated urinary tract infection by Pseudomonas aeruginosa is mediated by exopolysaccharide-independent biofilms. Infect. Immun. 2014, 82, 2048–2058. [Google Scholar] [CrossRef] [PubMed]
- Buhl, M.; Peter, S.; Willmann, M. Prevalence and risk factors associated with colonization and infection of extensively drug-resistant Pseudomonas aeruginosa: A systematic review. Expert Rev. Anti. Infect. Ther. 2015, 13, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Hoyle, B.D.; Costerton, J.W. Bacterial resistance to antibiotics: The role of biofilms. Prog. Drug Res. 1991, 37, 91–105. [Google Scholar] [PubMed]
- Linares, J.F.; Gustafsson, I.; Baquero, F.; Martinez, J.L. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. USA. 2006, 103, 19484–19489. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, L.R.; D'Argenio, D.A.; MacCoss, M.J.; Zhang, Z.; Jones, R.A.; Miller, S.I. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 2005, 436, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Mishra, B.; Lau, K.; Lushnikova, T.; Golla, R.; Wang, X. Antimicrobial peptides in 2014. Pharmaceuticals (Basel) 2015, 8, 123–150. [Google Scholar] [CrossRef] [PubMed]
- Bergen, P.J.; Bulitta, J.B.; Forrest, A.; Tsuji, B.T.; Li, J.; Nation, R.L. Pharmacokinetic/pharmacodynamic investigation of colistin against Pseudomonas aeruginosa using an in vitro model. Antimicrob. Agents Chemother. 2010, 54, 3783–3789. [Google Scholar] [CrossRef] [PubMed]
- Beringer, P. The clinical use of colistin in patients with cystic fibrosis. Curr. Opin. Pulm. Med. 2001, 7, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Tre-Hardy, M.; Vanderbist, F.; Traore, H.; Devleeschouwer, M.J. In vitro activity of antibiotic combinations against Pseudomonas aeruginosa biofilm and planktonic cultures. Int. J. Antimicrob. Agents 2008, 31, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Dosler, S.; Karaaslan, E. Inhibition and destruction of Pseudomonas aeruginosa biofilms by antibiotics and antimicrobial peptides. Peptides 2014, 62, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Lora-Tamayo, J.; Murillo, O.; Bergen, P.J.; Nation, R.L.; Poudyal, A.; Luo, X.; Yu, H.Y.; Ariza, J.; Li, J. Activity of colistin combined with doripenem at clinically relevant concentrations against multidrug-resistant Pseudomonas aeruginosa in an in vitro dynamic biofilm model. J. Antimicrob. Chemother. 2014, 69, 2434–2442. [Google Scholar] [CrossRef] [PubMed]
- Bergen, P.J.; Tsuji, B.T.; Bulitta, J.B.; Forrest, A.; Jacob, J.; Sidjabat, H.E.; Paterson, D.L.; Nation, R.L.; Li, J. Synergistic killing of multidrug-resistant Pseudomonas aeruginosa at multiple inocula by colistin combined with doripenem in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob. Agents Chemother. 2011, 55, 5685–5695. [Google Scholar] [CrossRef] [PubMed]
- Tarquinio, K.; Confreda, K.; Shurko, J.; LaPlante, K. Activities of tobramycin and polymyxin e against Pseudomonas aeruginosa biofilm-coated medical grade endotracheal tubes. Antimicrob. Agents Chemother. 2014, 58, 1723–1729. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, G.; Yang, L.; Wu, H.; Song, Z.; Wang, H.; Hoiby, N.; Ulrich, M.; Molin, S.; Riethmuller, J.; Doring, G. Colistin-tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. J. Infect. Dis. 2010, 202, 1585–1592. [Google Scholar] [CrossRef] [PubMed]
- Stanzani, M.; Tumietto, F.; Giannini, M.B.; Bianchi, G.; Nanetti, A.; Vianelli, N.; Arpinati, M.; Giovannini, M.; Bonifazi, F.; Bandini, G.; et al. Successful treatment of multi-resistant Pseudomonas aeruginosa osteomyelitis after allogeneic bone marrow transplantation with a combination of colistin and tigecycline. J. Med. Microbiol. 2007, 56, 1692–1695. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.A.; Lee, J.E.; Huh, W.; Peck, K.R.; Kim, Y.G.; Kim, D.J.; Oh, H.Y. Predictors of acute kidney injury associated with intravenous colistin treatment. Int. J. Antimicrob. Agents 2010, 35, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Minardi, D.; Ghiselli, R.; Cirioni, O.; Giacometti, A.; Kamysz, W.; Orlando, F.; Silvestri, C.; Parri, G.; Kamysz, E.; Scalise, G.; et al. The antimicrobial peptide tachyplesin III coated alone and in combination with intraperitoneal piperacillin-tazobactam prevents ureteral stent pseudomonas infection in a rat subcutaneous pouch model. Peptides 2007, 28, 2293–2298. [Google Scholar] [CrossRef] [PubMed]
- Chernish, R.N.; Aaron, S.D. Approach to resistant gram-negative bacterial pulmonary infections in patients with cystic fibrosis. Curr. Opin. Pulm. Med. 2003, 9, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Berditsch, M.; Jäger, T.; Strempel, N.; Schwartz, T.; Overhage, J.; Ulrich, A.S. Synergistic interaction between two cyclic membrane-active peptides toward 17 multidrug-resistant P. aeruginosa and biofilms Antimicrob. Agents Chemother. 2015, 59, 5288. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente-Núñez, C.; Cardoso, M.H.; de Souza, C.E.; Franco, O.L.; Hancock, R.E.W. Synthetic antibiofilm peptides. Biochim. Biophys. Acta. 2016, 1858, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Rudilla, H.; Fusté, E.; Cajal, Y.; Rabanal, F.; Vinuesa, T.; Viñas, M. Synergistic Antipseudomonal Effects of Synthetic Peptide AMP38 and Carbapenems. Molecules 2016, 21, 1223. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies; CABI: Oxfordshire, UK, 2010. [Google Scholar]
- Mishra, B.; Wang, G. The importance of amino acid composition in natural amps: An evolutional, structural, and functional perspective. Front. Immunol. 2012, 3, 221. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Reiling, S.; Zarena, D.; Wang, G. Host Defense antimicrobial peptides as antibiotics: Design and application strategies. Curr. Opin. Chem. Biol. 2017, 38, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Lushnikova, T.; Golla, R.M.; Wang, X.; Wang, G. Design and surface immobilization of short anti-biofilm peptides. Acta. Biomater. 2017, 49, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Hanke, M.L.; Mishra, B.; Lushnikova, T.; Heim, C.E.; Chittezham Thomas, V.; Bayles, K.W.; Kielian, T. Transformation of human cathelicidin ll-37 into selective, stable, and potent antimicrobial compounds. ACS Chem. Biol. 2014, 9, 1997–2002. [Google Scholar] [CrossRef] [PubMed]
- Menousek, J.; Mishra, B.; Hanke, M.L.; Heim, C.E.; Kielian, T.; Wang, G. Database screening and in vivo efficacy of antimicrobial peptides against methicillin-resistant Staphylococcus aureus USA300. Int. J. Antimicrob. Agents 2012, 39, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Han, H.; Miller, D.W.; Wang, G. Solution structures of human LL-37 fragments and NMR-based identification of a minimal membrane-targeting antimicrobial and anticancer region. J. Am. Chem. Soc. 2006, 128, 5776–5785. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Structures of human host defense cathelicidin LL-37 and its smallest antimicrobial peptide KR-12 in lipid micelles. J. Biol. Chem. 2008, 283, 32637–32643. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Junior, J.C.B.; Mishra, B.; Lushnikova, T.; Epand, R.M.; Wang, G. Arginine-lysine positional swap of the LL-37 peptides reveals evolutional advantages of the native sequence and leads to bacterial probes. Biochim. Biophys. Acta. 2017, 1859, 1350–1361. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, G. APD: The antimicrobial peptide database. Nucleic Acids Res. 2004, 32, D590–D592. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD2: The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009, 37, D933–D937. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Improved methods for classification, prediction, and design of antimicrobial peptides. Methods Mol. Biol. 2015, 1268, 43–66. [Google Scholar] [PubMed]
- Dean, S.N.; Bishop, B.M.; van Hoek, M.L. Natural and synthetic cathelicidin peptides with anti-microbial and anti-biofilm activity against Staphylococcus aureus. BMC Microbiol. 2011, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Kielhofner, M.; Atmar, R.L.; Hamill, R.J.; Musher, D.M. Life-threatening Pseudomonas aeruginosa infections in patients with human immunodeficiency virus infection. Clin. Infect. Dis. 1992, 14, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Saye, D.E. Recurring and antimicrobial-resistant infections: Considering the potential role of biofilms in clinical practice. Ostomy Wound. Manage. 2007, 53, 46–48. [Google Scholar] [PubMed]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013, 136, 1–58. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Kapaskelis, A.M.; Kouranos, V.D.; Kakisi, O.K.; Athanassa, Z.; Karageorgopoulos, D.E. Outcome of antimicrobial therapy in documented biofilm-associated infections: A review of the available clinical evidence. Drugs 2009, 69, 1351–1361. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007, 6, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Potron, A.; Poirel, L.; Nordmann, P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents 2015, 45, 568–585. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, L.; Gooderham, W.J.; Bains, M.; McPhee, J.B.; Wiegand, I.; Hancock, R.E. Adaptive resistance to the “last hope” antibiotics polymyxin b and colistin in pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob. Agents Chemother. 2010, 54, 3372–3382. [Google Scholar] [CrossRef] [PubMed]
- Bergen, P.J.; Li, J.; Nation, R.L.; Turnidge, J.D.; Coulthard, K.; Milne, R.W. Comparison of once-, twice- and thrice-daily dosing of colistin on antibacterial effect and emergence of resistance: Studies with Pseudomonas aeruginosa in an in vitro pharmacodynamic model. J. Antimicrob. Chemother. 2008, 61, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Bulitta, J.B.; Yang, J.C.; Yohonn, L.; Ly, N.S.; Brown, S.V.; D'Hondt, R.E.; Jusko, W.J.; Forrest, A.; Tsuji, B.T. Attenuation of colistin bactericidal activity by high inoculum of Pseudomonas aeruginosa characterized by a new mechanism-based population pharmacodynamic model. Antimicrob. Agents Chemother. 2010, 54, 2051–2062. [Google Scholar] [CrossRef] [PubMed]
- Dean, S.N.; Walsh, C.; Goodman, H.; van Hoek, M.L. Analysis of mixed biofilm (Staphylococcus aureus and Pseudomonas aeruginosa) by laser ablation electrospray ionization mass spectrometry. Biofouling 2015, 31, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Dean, S.N.; Bishop, B.M.; van Hoek, M.L. Susceptibility of Pseudomonas aeruginosa biofilm to alpha-helical peptides: d-enantiomer of LL-37. Front. Microbiol. 2011, 2, 128. [Google Scholar] [CrossRef] [PubMed]
- Overhage, J.; Campisano, A.; Bains, M.; Torfs, E.C.; Rehm, B.H.; Hancock, R.E. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 2008, 76, 4176–4182. [Google Scholar] [CrossRef] [PubMed]
- Wnorowska, U.; Niemirowicz, K.; Myint, M.; Diamond, S.L.; Wroblewska, M.; Savage, P.B.; Janmey, P.A.; Bucki, R. Bactericidal activities of cathelicidin LL-37 and select cationic lipids against the hypervirulent Pseudomonas aeruginosa strain LESB58. Antimicrob. Agents Chemother. 2015, 59, 3808–3815. [Google Scholar] [CrossRef] [PubMed]
- Souza, B.M.; Mendes, M.A.; Santos, L.D.; Marques, M.R.; Cesar, L.M.; Almeida, R.N.; Pagnocca, F.C.; Konno, K.; Palma, M.S. Structural and functional characterization of two novel peptide toxins isolated from the venom of the social wasp Polybia. paulista. Peptides 2005, 26, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Wang, G. Ab initio design of potent anti-MRSA peptides based on database filtering technology. J. Am. Chem. Soc. 2012, 134, 12426–12429. [Google Scholar] [CrossRef] [PubMed]
- Di Luca, M.; Maccari, G.; Maisetta, G.; Batoni, G. BaAMPs: The database of biofilm-active antimicrobial peptides. Biofouling 2015, 31, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Bowler, L.L.; Zhanel, G.G.; Ball, T.B.; Saward, L.L. Mature Pseudomonas aeruginosa biofilms prevail compared to young biofilms in the presence of ceftazidime. Antimicrob. Agents Chemother. 2012, 56, 4976–4979. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, R.; Wadman, M.W.; Dohm, M.T.; Czyzewski, A.M.; Spormann, A.M.; Barron, A.E. Antimicrobial peptoids are effective against Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2011, 55, 3054–3057. [Google Scholar] [CrossRef] [PubMed]
- Nagant, C.; Pitts, B.; Nazmi, K.; Vandenbranden, M.; Bolscher, J.G.; Stewart, P.S.; Dehaye, J.P. Identification of peptides derived from the human antimicrobial peptide LL-37 active against biofilms formed by Pseudomonas aeruginosa using a library of truncated fragments. Antimicrob. Agents Chemother. 2012, 56, 5698–5708. [Google Scholar] [CrossRef] [PubMed]
- Chennupati, S.K.; Chiu, A.G.; Tamashiro, E.; Banks, C.A.; Cohen, M.B.; Bleier, B.S.; Kofonow, J.M.; Tam, E.; Cohen, N.A. Effects of an LL-37-derived antimicrobial peptide in an animal model of biofilm Pseudomonas sinusitis. Am. J. Rhinol. Allergy 2009, 23, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Epand, R.F.; Mishra, B.; Lushnikova, T.; Thomas, V.C.; Bayles, K.W.; Epand, R.M. Decoding the functional roles of cationic side chains of the major antimicrobial region of human cathelicidin LL-37. Antimicrob. Agents Chemother. 2012, 56, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Zavascki, A.P.; Bulitta, J.B.; Landersdorfer, C.B. Combination therapy for carbapenem-resistant Gram-negative bacteria. Expert Rev. Anti. Infect. Ther. 2013, 11, 1333–1353. [Google Scholar] [CrossRef] [PubMed]
- Zusman, O.; Avni, T.; Leibovici, L.; Adler, A.; Friberg, L.; Stergiopoulou, T.; Carmeli, Y.; Paul, M. Systematic review and meta-analysis of in vitro synergy of polymyxins and carbapenems. Antimicrob. Agents Chemother. 2013, 57, 5104–5111. [Google Scholar] [CrossRef] [PubMed]
- Berlana, D.; Llop, J.M.; Manresa, F.; Jodar, R. Outpatient treatment of Pseudomonas aeruginosa bronchial colonization with long-term inhaled colistin, tobramycin, or both in adults without cystic fibrosis. Pharmacotherapy 2011, 31, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Spapen, H.; Jacobs, R.; Van Gorp, V.; Troubleyn, J.; Honore, P.M. Renal and neurological side effects of colistin in critically ill patients. Ann. Intensive Care. 2011, 1, 14. [Google Scholar] [CrossRef] [PubMed]
- Pompilio, A.; Scocchi, M.; Pomponio, S.; Guida, F.; Di Primio, A.; Fiscarelli, E.; Gennaro, R.; Di Bonaventura, G. Antibacterial and anti-biofilm effects of cathelicidin peptides against pathogens isolated from cystic fibrosis patients. Peptides 2011, 32, 1807–1814. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, S.; Goto, K.; Hagiwara, S.; Iwasaka, H.; Noguchi, T. Doripenem pharmacokinetics in critically ill patients receiving continuous hemodiafiltration (CHDF). Yakugaku Zasshi. 2010, 130, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Mogayzel, P.J., Jr.; Pierce, E.; Mills, J.; McNeil, A.; Loehr, K.; Joplin, R.; McMahan, S.; Carson, K.A. Accuracy of tobramycin levels obtained from central venous access devices in patients with cystic fibrosis is technique dependent. Pediatr. Nurs. 2008, 34, 464–468. [Google Scholar] [PubMed]
- Zarena, D.; Mishra, B.; Lushnikova, T.; Wang, F.; Wang, G. The π Configuration of the WWW Motif of a Short Trp-rich Peptide Is Critical for Targeting Bacterial Membranes, Disrupting Preformed Biofilms and Killing Methicillin-resistant Staphylococcus aureus. Biochemistry 2017. in revision. [Google Scholar]
| Peptide | DASamP2 a | 17BIPHE2 b |
|---|---|---|
| Amino acid sequence | IKWKKLLRAAKRIL-NH2 | GXKRIVQRIKDXLRNLV-NH2 |
| Peptide template | Polybia-MPI | GF-17 |
| Changes made | D2K, D8R and Q12R | F17X and F27X; X = biphenylalanine |
| MIC (P. aeruginosa) c | 6.25 μM | 6.25 μM |
| HL50 | 75 μM | 225 μM |
| Cell selectivity | 12 | 36 |
| Compound | Minimal Inhibitory Concentration (µM) | ||||
|---|---|---|---|---|---|
| PAO1 | PA # 1 | PA # 2 | PA # 3 | PA # 4 | |
| DASamP2 | 3.1–6.25 | 6.25 | ≤ 3.1 | ≤ 3.1 | ≤ 3.1 |
| 17BIPHE2 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 |
| Colistin | 1.56 b | 12.5 | 3.1 | 1.56 | 3.1 |
| Doripenem | 0.78–1.56 | 0.78 | ≤ 0.78 | 1.56 | 0.78 |
| Tobramycin | 3.1 | 3.1 | ≤ 3.1 | ≤ 3.1 | ≤ 3.1 |
| Tigecycline | 12.5 | 25 | 25 | 25 | 12.5 |
| Biofilm Treatment Stages | Peptide Only | Antibiotic Only | Combination |
|---|---|---|---|
| Initial adhesion | √ | X | √ |
| Biofilm formation | √ | √ | ND |
| Established biofilms | √ | X | √ |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishra, B.; Wang, G. Individual and Combined Effects of Engineered Peptides and Antibiotics on Pseudomonas aeruginosa Biofilms. Pharmaceuticals 2017, 10, 58. https://doi.org/10.3390/ph10030058
Mishra B, Wang G. Individual and Combined Effects of Engineered Peptides and Antibiotics on Pseudomonas aeruginosa Biofilms. Pharmaceuticals. 2017; 10(3):58. https://doi.org/10.3390/ph10030058
Chicago/Turabian StyleMishra, Biswajit, and Guangshun Wang. 2017. "Individual and Combined Effects of Engineered Peptides and Antibiotics on Pseudomonas aeruginosa Biofilms" Pharmaceuticals 10, no. 3: 58. https://doi.org/10.3390/ph10030058
APA StyleMishra, B., & Wang, G. (2017). Individual and Combined Effects of Engineered Peptides and Antibiotics on Pseudomonas aeruginosa Biofilms. Pharmaceuticals, 10(3), 58. https://doi.org/10.3390/ph10030058







